Abstract
Rice (Oryza sativa), a monocotyledonous plant that does not cold acclimate, has evolved differently from Arabidopsis (Arabidopsis thaliana), which cold acclimates. To understand the stress response of rice in comparison with that of Arabidopsis, we developed transgenic rice plants that constitutively expressed CBF3/DREB1A (CBF3) and ABF3, Arabidopsis genes that function in abscisic acid-independent and abscisic acid-dependent stress-response pathways, respectively. CBF3 in transgenic rice elevated tolerance to drought and high salinity, and produced relatively low levels of tolerance to low-temperature exposure. These data were in direct contrast to CBF3 in Arabidopsis, which is known to function primarily to enhance freezing tolerance. ABF3 in transgenic rice increased tolerance to drought stress alone. By using the 60 K Rice Whole Genome Microarray and RNA gel-blot analyses, we identified 12 and 7 target genes that were activated in transgenic rice plants by CBF3 and ABF3, respectively, which appear to render the corresponding plants acclimated for stress conditions. The target genes together with 13 and 27 additional genes are induced further upon exposure to drought stress, consequently making the transgenic plants more tolerant to stress conditions. Interestingly, our transgenic plants exhibited neither growth inhibition nor visible phenotypic alterations despite constitutive expression of the CBF3 or ABF3, unlike the results previously obtained from Arabidopsis where transgenic plants were stunted.
Drought, high salinity, and low temperature are three important abiotic stresses that are commonly encountered by plants growing in their native environments. Upon exposure to the stresses, many genes are induced and their products are thought to function as cellular protectants of stress-induced damage (Bray, 1997; Thomashow, 1999; Shinozaki and Yamaguchi-Shinozaki, 2000). Studies on the expression of stress-regulated genes in Arabidopsis (Arabidopsis thaliana) have demonstrated the presence of several stress-response pathways involving genes that act either in an abscisic acid (ABA)-dependent or an ABA-independent manner (Shinozaki and Yamaguchi-Shinozaki, 1997). Many transcription factors were characterized and found to function in one of the pathways in Arabidopsis. CBF3/DREB1A (CBF3; Gilmour et al., 1998; Shinwari et al., 1998) and ABF3 (Kang et al., 2002) represent 2 of the characterized transcription factors that are related to the ABA-independent and ABA-dependent pathways, respectively. Promoter regions of many stress-regulated genes in Arabidopsis contain two cis-acting elements, C-repeat (CRT; Baker et al., 1994)/dehydration-responsive element (DRE; Yamaguchi Shinozaki and Shinozaki, 1994) and ABA-responsive element (ABRE; Giraudat et al., 1994) that interact with CBFs and ABFs (Jaglo-Ottosen et al., 1998; Liu et al., 1998; Kasuga et al., 1999; Kang et al., 2002), respectively. When grown under normal conditions, CBF3 in transgenic Arabidopsis enhances expression of its target genes including cor15a, rd29A, kin1, cor6.6, and cor47/rd17 (Liu et al., 1998; Kasuga et al., 1999; Maruyama et al., 2004); on the other hand, ABF3 induces ABA-related genes that encode Lea (rd29B and rab18) and protein phosphatase 2C (ABI1 and ABI2; Kang et al., 2002). As a result, it has been concluded that CBF3 enhances tolerance to freezing, drought, and high salinity, whereas ABF3 increases tolerance to drought stress alone.
The discovery of CBF3- and ABF3-related pathways in Arabidopsis provides us with strategies to improve the stress tolerance of crop plants. Transcripts of CBF-like genes were found to accumulate in response to low temperature in canola (Brassica napus), wheat (Triticum aestivum), rye (Secale cereale), and tomato (Lycopersicon esculentum) (Jaglo et al., 2001). In addition, overexpression of the Arabidopsis CBF3 in transgenic canola (Jaglo et al., 2001) and tobacco (Nicotiana tabacum; Kasuga et al., 2004) increases tolerance to freezing and drought/low-temperature exposure, respectively. Despite the fact that CBF genes and/or related responses are conserved in some plants, various plant species vary greatly in their abilities to survive adverse effects from exposure to these environmental constraints. Obviously, plants that are capable of cold acclimation have evolved differently from those that are unable to undergo cold acclimation (Jaglo et al., 2001). A typical plant from the latter is rice (Oryza sativa). In comparison to Arabidopsis and other cereals like wheat and barley (Hordeum vulgare) that cold acclimate (Wen et al., 2002), rice is much more sensitive to low temperature exposure. Interestingly, expression of OsDREB1A, a rice ortholog of CBF3, is different from that of Arabidopsis CBF3 in that it is induced not only by low temperature but also by exposure to high salinity and wounding (Dubouzet et al., 2003). In Arabidopsis, CBF3 is induced by low temperature, but it is not affected by drought or high-salinity stress (Shinwari et al., 1998; Gilmour et al., 2000). Microarray analysis of rice stress-regulated genes demonstrated the existence of 22 genes and cis-acting elements that have not been previously reported in Arabidopsis (Rabbani et al., 2003). In addition, 2 signaling pathways for low temperature stress were proposed to exist in rice, one that is responsive to a 12°C stimulus and the other that is induced by exposure to 4°C (Wen et al., 2002). These observations prompted us to examine the stress response of rice in comparison to that of Arabidopsis. We generated transgenic rice plants constitutively expressing CBF3 and ABF3 and their ectopic overexpression enhanced stress tolerance in unique ways by activating specific groups of stress-regulated genes. We also discussed the similarities and differences between rice and Arabidopsis in functional aspects of CBF3 and ABF3.
RESULTS
Production of Transgenic Rice Plants That Express Arabidopsis CBF3 and ABF3
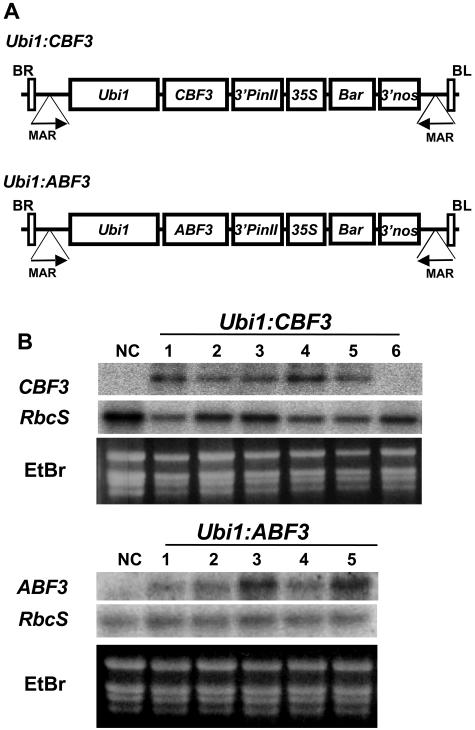
To examine the role of CBF3/DREB1A (CBF3) and ABF3 in transgenic rice plants, we constructed plasmids for rice transformation, Ubi1:CBF3 and Ubi1:ABF3 (Fig. 1A), in which the genes are under the control of the maize (Zea mays) ubiquitin1 promoter including its first intron (Ubi1; Christensen and Quail, 1996). Fifteen and 20 independent transgenic lines for Ubi1:CBF3 and Ubi1:ABF3, respectively, were obtained using the Agrobacterium-mediated transformation method (Hiei et al., 1994). Over 80% of the plants were fertile and T1, T2, T3, and T4 seeds were collected. Copy numbers and integration events of the transgene in Ubi1:CBF3 and Ubi1:ABF3 plants were determined by genomic Southern blots (Supplemental Fig. 1), which revealed that all the lines are independent and that the copy numbers of corresponding transgenes are either one or two. Expression levels of the transgenes in Ubi1:CBF3 and Ubi1:ABF3 plants were examined by RNA-blot analysis using total RNAs from leaf tissues. As shown in Figure 1B, CBF3 or ABF3 transcripts were readily detectable in all the lines tested. An exception occurred for line 6 of Ubi1:CBF3 whose hybridization signal was weak but still detectable with longer exposure times. Three or four homozygous T4 lines that contained single copy insertions of the transgene for the Ubi1:CBF3 and Ubi1:ABF3 plants, respectively, were chosen for further analysis.
Figure 1.
Production of Ubi1:CBF3 and Ubi1:ABF3 plants. A, Ubi1:CBF3 and Ubi1:ABF3 consist of the maize ubiquitin promoter (Ubi1) linked to the CBF3 and ABF3 coding regions, respectively, and the 3′-region of the potato proteinase inhibitor II gene (3′pinII), and a gene expression cassette that contains the 35S promoter, the bar coding region, and the 3′-region of the nopaline synthase gene (3′nos). The entire expression cassette is flanked by the 5′-matrix attachment region (MAR) of the chicken lysozyme gene (Phi-Van and Strätling, 1996). B, RNA gel-blot analysis was performed using total RNAs from young leaves of 6 Ubi1:CBF3 (top) and 5 Ubi1:ABF3 (bottom) lines and from NC plants. The blots were hybridized with the CBF3 and ABF3 probes (described in Supplemental Fig. 1) and reprobed with the rice RbcS gene (Kyozuka et al., 1993). Ethidium bromide (EtBr) staining of total RNA was for equal loading of RNAs.
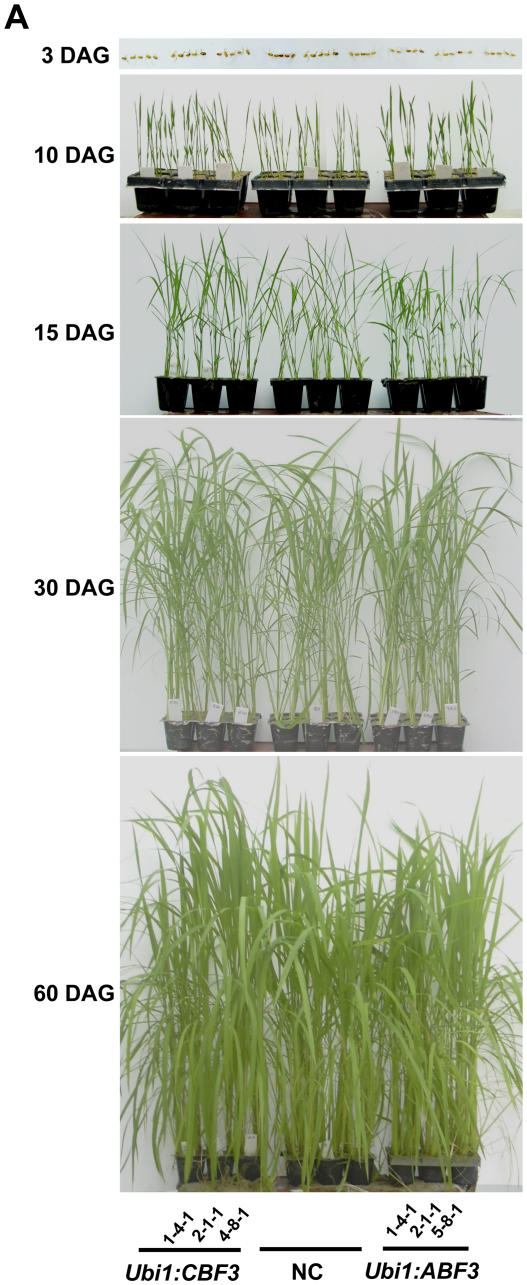
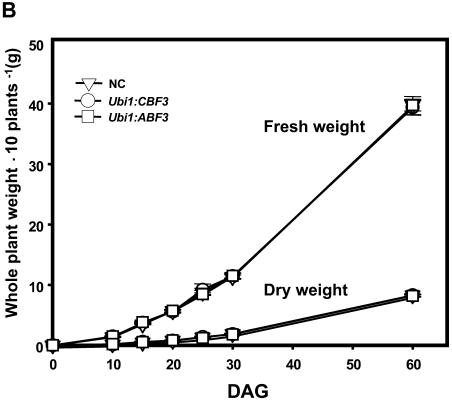
Unlike the severe stunting observed in Arabidopsis plants that overexpressed CBF3 or ABF3 (Liu et al., 1998; Kasuga et al., 1999; Gilmour et al., 2000; Kang et al., 2002), our Ubi1:CBF3 and Ubi1:ABF3 plants displayed normal growth and seed setting. Transgenic and nontransgenic seeds were germinated and their fresh and dry weights determined in the time course after germination. The transgenic plants showed normal vegetative phenotype and fertility as compared to nontransgenic control plants (Fig. 2A), without notable difference in fresh and dry weights (Fig. 2B).
Figure 2.
Growth characteristics of Ubi1:CBF3 and Ubi1:ABF3 plants. A, Growth phenotypes of 3 independent T4 homozygous lines for Ubi1:CBF3, Ubi1:ABF3, and NC plants at indicated days after germination (DAG). B, Dry weight and fresh weight accumulation of Ubi1:CBF3, Ubi1:ABF3, and NC plants. Plants grown in the greenhouse during the same time course shown in A were harvested and fresh and dry weight/10 plants measured. Each data point represents the mean ± sd of triplicate experiments with three different transgenic lines.
Stress Tolerance in Ubi1:CBF3 and Ubi1:ABF3 Plants
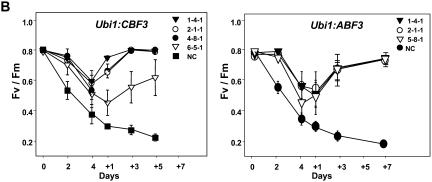
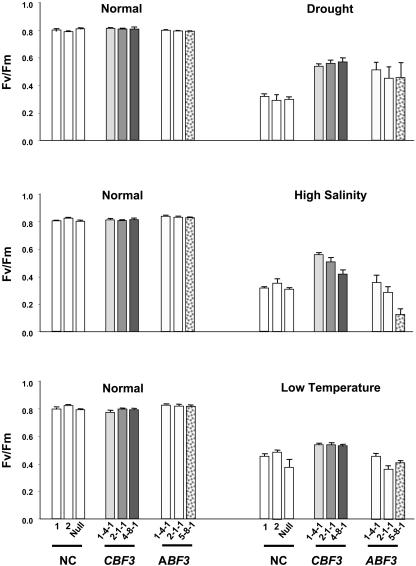
To investigate whether expression of CBF3 or ABF3 was correlated with stress tolerance in transgenic plants, 4-week-old nontransgenic control (NC) and T4 transgenic seedlings were subjected to 4 d of drought stress. After the drought treatments, plants of each line showed wilting and drought-induced rolling of young leaves with a concomitant loss of chlorophyll (Fig. 3A). In contrast to transgenic lines, NC plants exhibited leaf rolling within 2 d of the stress and exhibited considerably more visual symptoms of drought stress. After 4 d of drought stress and subsequent watering for 5 d, the growth of transgenic lines was almost identical to nonstressed control plants. In contrast, the growth of drought-stressed NC plants was severely inhibited, and these plants never recovered and finally died. With the exception of line 6-5-1, after 4 d of drought stress followed by 5 d of watering, almost all of the Ubi1:CBF3 survived, whereas 50% to 67% of the Ubi1:ABF3 plants survived (Table I). Expression level of CBF3 in the line 6-5-1 was very low (Fig. 1B). These results indicate that overexpression of Arabidopsis CBF3 or ABF3 in rice confers increased tolerance to drought stress and that the effect is greater in plants overexpressing CBF3. The enhanced drought tolerance of the transgenic plants was further verified by measuring changes in chlorophyll fluorescence. Reductions in the maximum photochemical efficiency of PSII in the dark-adapted state (Fv/Fm) were considerably larger in NC plants than in either the Ubi1:CBF3 or the Ubi1:ABF3 plants throughout the time course (Fig. 3B), thereby validating the increased tolerance to drought stress. Similarly, Fv/Fm was measured in 14-d-old transgenic and NC seedlings including one nullizygous plant after exposure to high salinity and low temperature in addition to drought stress. Levels of Fv/Fm were approximately 30% higher in Ubi1:CBF3 plants than in NC plants under drought and high salinity and 10% higher under low-temperature stress (Fig. 4). Fv/Fm levels in Ubi1:ABF3 plants were higher by 27% as compared to NC plants under drought, but were similar or even lower than NC plants under high-salinity and low-temperature treatments. In summary, CBF3 increased tolerance to drought, high salinity, and low temperature, while ABF3 increased tolerance only to drought in transgenic rice plants.
Figure 3.
Appearance of plants and changes in chlorophyll fluorescence during drought stress. A, Four and three independent T4 homozygous lines for Ubi1:CBF3 and Ubi1:ABF3, respectively, and NC seedlings were grown in the greenhouse for 4 weeks and then subjected to 4 d of drought stress followed by 5 to 7 d of watering. Eighteen plants per each line were tested. Photos were taken at 1- or 2-d intervals; + followed by number denotes days of watering. B, Fv/Fm of the transgenic and NC plants in the same time course of drought stress shown in A was measured using a pulse modulation fluorometer (mini-PAM). Fv/Fm is a measure of accumulated photooxidative damage to PSII. Each data point represents the mean ± se of triplicate experiments (n = 6).
Table I.
Survival of Ubi1:CBF3 and Ubi1:ABF3 plants under drought stress
| Plantsa | Totalb | Survivalc | Survival Rated |
|---|---|---|---|
| Ubi1:CBF3 | |||
| NC | 36 | 0 | 0% |
| 1-4-1 | 36 | 34 | 94% |
| 2-1-1 | 36 | 36 | 100% |
| 4-8-1 | 36 | 36 | 100% |
| 6-5-1 | 36 | 14 | 39% |
| Ubi1:ABF3 | |||
| NC | 36 | 3 | 8% |
| 1-4-1 | 36 | 21 | 58% |
| 2-1-1 | 36 | 24 | 67% |
| 5-8-1 | 36 | 18 | 50% |
Four-week-old soil-grown plants withheld water for 4 d followed by watering for 7 d and results scored. Plants were considered dead if there was no regrowth 7 d of rewatering. Water loss during the drought periods was similar for all pots.
Total number of plants used in each assay.
Number of survival plants.
Percent of survived plants (survival/total × 100).
Figure 4.
Changes in chlorophyll fluorescence during drought, high-salinity, and low-temperature stresses. Three independent T4 homozygous lines for Ubi1:CBF3, Ubi1:ABF3, and NC seedlings grown in the greenhouse for 14 d were subjected to various stress conditions as described: for drought stress, the seedlings were air-dried for 2 h at 28°C and for high-salinity stress seedlings were exposed to 400 mm NaCl for 2 h at 28°C. For low-temperature stress, they were exposed to 4°C for 6 h. All of the experiments were carried out under continuous light of 150 μmol m−2 s−1. Each data point represents the mean ± se of triplicate experiments (n = 6).
CBF3 and ABF3 Activate Different Groups of Stress-Related Genes in Rice
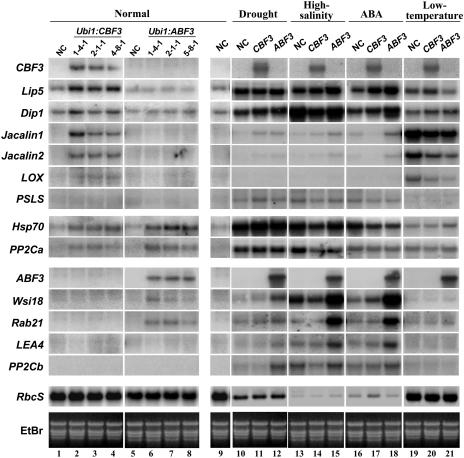
Stress-responsive genes are activated by CBFs and ABFs in Arabidopsis. To identify genes that are up-regulated by CBF3 or ABF3 in rice, global expression profiling was performed on the Ubi1:CBF3 or Ubi1:ABF3 plants in comparison with untransformed plants that were grown under normal growth conditions. The underlying assumption of this approach is that the constitutive expression of CBF3 or ABF3 in transgenic plants activates target genes whose expression levels would remain relatively low in nontransgenic plants under normal growth conditions. Profiling was conducted with the 60 K Rice Whole Genome Microarray (GreenGene Biotech, Yongin, Korea). This microarray contains 70-mer oligonucleotide probes with sequences corresponding to 58,417 known or predicted open reading frames that cover the entire rice genome. RNA samples from leaf tissues of 14-d-old transgenic and nontransgenic seedlings were used to generate Cy5- and Cy3-labeled cDNA probes, respectively, which were then hybridized to the microarray. Expression analyses with 3 replicates identified 16 different genes with 1.6-fold greater induction in transgenic plants than in nontransgenic control plants (Table II; Supplemental Table I). CBF3- or ABF3-induced expression of the candidate genes was subsequently confirmed by RNA gel-blot analysis using the same RNAs for microarray (Fig. 5, left). Our results revealed that CBF3 activates 12 genes including Lip5, Dip1, Jacalin1 and 2, and LOX, whereas ABF3 activates 7 genes including Wsi18 and Rab21. Three genes, Hsp70, PP2Ca, and a receptor kinase gene, are activated by both CBF3 and ABF3. These genes were induced at different levels in nontransgenic rice within 2 h of exposure to stress conditions (Fig. 5, right). The RbcS gene whose transcript levels rapidly decrease in response to stress treatments except for low temperature (DeRocher and Bohnert, 1993; Weatherwax et al., 1996) was used to monitor when stress-induced damage initiates. In addition to those genes that are activated by CBF3 or ABF3 under normal growth conditions, we also searched for genes that are further induced by the transcription factors under stress conditions. Expression profiling was performed on the Ubi1:CBF3 or Ubi1:ABF3 plants in comparison with nontransgenic plants that were exposed to drought stress for 2 h. As a result, we identified 15 and 29 genes that are induced further 1.6-fold or more in Ubi1:CBF3 or Ubi1:ABF3 plants, respectively, upon treatment with drought stress (Table II; Supplemental Table I). Some of the genes, Jacalin1, Jacalin2, PSLS, Wsi18, Rab21, LEA4, and PP2Cb, were confirmed by RNA gel-blot analysis using RNAs from leaf tissues of 14-d-old transgenic and nontransgenic seedlings that were exposed to drought, high salinity, ABA, and low temperature for 2 h (Fig. 5, right). Interestingly, the genes that are activated under stress conditions in the transgenic plants are different from those that are activated under normal growth conditions, except for Jacalin1, Jacalin2, Wsi18, and Rab21 that are common to both conditions. The difference may be due either to the activation of rice CBF3 and/or ABF3 homologs under stress conditions or to the increased stress tolerance of transgenic plants that was set by the activated genes under normal growth conditions. Expression of 3 CBF homologs, OsDREB1A, OsDREB1B, and OsDREB2A (Dubouzet et al., 2003), and 2 ABF homologs, OsTRAB1 (Hobo et al., 1999) and AK065873, were examined by the microarray analysis described above, which revealed that none of them was increased in expression levels by 2 h of exposure to drought stress (Supplemental Table I). These observations led us to suggest that those genes that are activated under stress conditions are not the direct targets of CBF3 and ABF3 in the transgenic plants. Instead, their increased expression under stress conditions resulted from secondary effects of enhanced stress tolerance of the transgenic plants that protected those genes from being down-regulated by stress treatments. Overall, Arabidopsis transcription factors, CBF3 and ABF3, activate 12 and 7 target genes in transgenic rice plants, respectively, which appears to render the corresponding plants acclimated for stress conditions. The target genes together with 13 and 27 additional genes are induced further upon stress treatments (Table II; Fig. 5), consequently making the transgenic plants more tolerant to stress conditions.
Table II.
List of genes that are up-regulated in Ubi1:CBF3 and Ubi1:ABF3 plants under normal and drought stress
Numbers appearing in bold are the ones that are induced 1.6-fold or more in Ubi1:CBF3 or Ubi1:ABF3 plants.
| Gene Name
|
Accession No.a
|
Normalb
|
Droughtd
|
||
|---|---|---|---|---|---|
| Ubi1:CBF3c | Ubi1:ABF3c | Ubi1:CBF3c | Ubi1:ABF3c | ||
| Jacalin1 | AK066682 | 3.56 | −1.11 | 3.04 | 2.92 |
| Jacalin2 | AK101991 | 3.32 | −1.17 | 2.42 | 2.10 |
| Dip1 | AY587109 | 1.66 | 1.13 | 1.10 | 1.39 |
| Lip5 | AB011368 | 2.32 | 1.29 | 1.20 | −1.10 |
| Lipoxygenase (LOX) | AJ270938 | 3.10 | 1.43 | −1.59 | 1.11 |
| Glutelin | AK107238 | 2.18 | −1.10 | −1.22 | 1.13 |
| Bowman Birk trypsin inhibitor1 | AK065846 | 1.83 | 1.08 | −1.19 | 1.27 |
| Bowman Birk trypsin inhibitor2 | AK105455 | 1.65 | −1.22 | −1.19 | 1.10 |
| Receptor kinase containing LRR repeats | AK119823 | 1.67 | 1.64 | 1.37 | 1.99 |
| Unknown protein | AK059202 | 1.62 | −2.07 | 1.12 | 1.26 |
| Cyt P450 | AK069394 | 1.34 | 1.11 | 5.21 | 3.30 |
| Seed imbibition protein (Sip1) | AK065100 | 1.03 | −1.03 | 2.20 | 1.12 |
| Sicrotubule-associated protein MAP65-1a | AK102553 | −1.08 | 1.06 | 1.91 | 1.22 |
| Unknown protein | NM_188534 | −1.07 | 1.09 | 1.93 | 1.06 |
| Unknown protein | AK107624 | 1.10 | −1.07 | 1.86 | 1.17 |
| Unknown protein | AK061456 | −1.03 | −1.00 | 1.68 | 1.25 |
| Polygalacturonase (PG2) | AK108477 | 1.02 | 1.09 | 1.70 | 1.47 |
| FtsJ cell division protein | AK070075 | 1.10 | 1.10 | 1.78 | −1.02 |
| mRNA cleavage factor I subunit | AK061260 | 1.03 | 1.13 | 1.79 | 1.04 |
| Beclin | AK101033 | 1.19 | 1.02 | 1.73 | 1.05 |
| Cyclophilin (CyP) | AK111654 | 1.04 | −1.01 | 1.70 | 1.21 |
| Phospho sulfolactate synthase (PSLS) | AK072958 | −1.01 | −1.04 | 1.61 | 1.02 |
| Gag-pol polyprotein | AK063408 | −1.08 | −1.18 | 1.82 | −1.01 |
| α-Expansin OsEXPA5 | AF394546 | 1.16 | 1.14 | 1.41 | 1.94 |
| Hsp70 | AK072830 | 1.99 | 1.94 | 1.39 | 1.28 |
| Protein phosphatase 2Ca (PP2Ca) | XM_463364 | 1.65 | 1.67 | 1.21 | 1.48 |
| Wsi18 | D26536 | 1.00 | 2.11 | 1.06 | 2.27 |
| RAB21 | Y00842 | −1.03 | 2.60 | 1.22 | 1.54 |
| Phosphate-induced protein 1(phi1) | AK070419 | −1.11 | 1.67 | 1.10 | −1.39 |
| Antioxidant protein | AK066452 | 1.17 | 1.68 | 1.25 | 1.15 |
| LEA4 | AK107930 | −1.01 | 1.02 | 1.08 | 4.33 |
| Protein phosphatase 2Cb (PP2Cb) | AK069274 | 1.12 | 1.05 | 1.05 | 3.45 |
| PHD-type zinc finger protein | AK059311 | −1.17 | 1.09 | 1.07 | 3.11 |
| Unknown protein | AK063747 | −1.16 | 1.09 | 1.17 | 2.83 |
| Little protein (LP1) | AK063634 | 1.09 | 1.09 | −1.22 | 3.01 |
| Unknown protein | AK072034 | 1.08 | 1.06 | −1.11 | 2.66 |
| Unknown protein | AK063680 | −1.00 | −1.05 | −1.08 | 2.63 |
| CCCH-type zinc finger protein | AK106392 | 1.37 | 1.05 | 1.40 | 2.56 |
| 26S proteasome AAA-ATPase subunit | AK103936 | 1.05 | 1.22 | −1.00 | 2.26 |
| Unknown protein | NM_197832 | −1.34 | 1.18 | −1.00 | 2.24 |
| Unknown protein | XM_480395 | 1.01 | 1.09 | 1.05 | 2.34 |
| Unknown protein | AK058851 | 1.03 | −1.00 | −1.12 | 2.38 |
| Aquaporin (TIP4) | AK121671 | 1.08 | −1.06 | 1.07 | 2.62 |
| Disease resistance protein (RPH8A) | AK072531 | 1.13 | 1.18 | −1.07 | 2.10 |
| Unknown protein | AK104155 | 1.05 | 1.00 | −1.01 | 2.20 |
| 1,4-β-d xylan xylanohydrolase | NM_184104 | 1.09 | 1.04 | 1.03 | 1.97 |
| Spore coat protein | AK108917 | 1.02 | −1.02 | 1.05 | 1.93 |
| Cellulase | XM_469540 | −1.06 | 1.07 | 1.21 | 1.98 |
| LTI6B-like | AK104060 | −1.12 | 1.09 | −1.07 | 1.88 |
| Fusarium resistance protein | AK058343 | 1.05 | 1.11 | −1.03 | 2.20 |
| ABC transporter | AK105712 | −1.08 | 1.17 | −1.19 | 1.91 |
| LEA | AK067556 | 1.13 | 1.10 | 1.22 | 1.87 |
| LEA3 | AK102039 | −1.02 | 1.17 | −1.17 | 2.14 |
GenBank accession numbers for full-length cDNA sequences of corresponding genes.
Microarrays were hybridized with Cy3- and Cy5-labeled probe pairs of either Ubi1:CBF3 and nontransgenic plants or Ubi1:ABF3 and nontransgenic plants grown in normal growth conditions.
The microarray-data sets can be found at the www.http://www.ncbi.nlm.nih.gov/geo/ (Gene Expression Omnibus, GEO). GEO accession number of microarray-data set is GSE2211.
Microarrays were hybridized with Cy3- and Cy5-labeled probe pairs of either Ubi1:CBF3 and nontransgenic plants or Ubi1:ABF3 and nontransgenic plants grown in drought-stress conditions.
Figure 5.
Induction of stress-related genes in Ubi1:CBF3 and Ubi1:ABF3 plants. Three independent T4 homozygous lines for Ubi1:CBF3, Ubi1:ABF3, and NC seedlings were grown in the greenhouse for 14 d. Transgenic and NC plants were then treated for 2 h with drought (the seedlings excised before being air-dried for 2 h), high salinity (400 mm NaCl) at the greenhouse, and with low-temperature stress (4°C) at the cold chamber under continuous light of 150 μmol m−2 s−1. For ABA treatments, 100 μm ABA was applied to each 14-d-old seedling for 2 h. RNA gel blots of total RNAs from transgenic and NC plants grown either under normal growth conditions (left) and under stress conditions (right) are indicated. RNA gel blots of NC plants grown under normal growth conditions were included on the left-hand side of each section for clarity of comparison. The blots were hybridized with probes for CBF3, ABF3, Lip5 (AB011368), Dip1 (AY587109), Jacalin1 (AK066682), Jacalin2 (AK101991), LOX (AJ270938), PSLS (AK072958), Hsp70 (CF280418), PP2Ca (CF304401), Wsi18 (D26536), Rab21 (Y00842), LEA4 (AK107930), PP2Cb (AK069274), and RbcS (Kyozuka et al., 1993). EtBr staining of total RNA was used to ensure equal RNA loading.
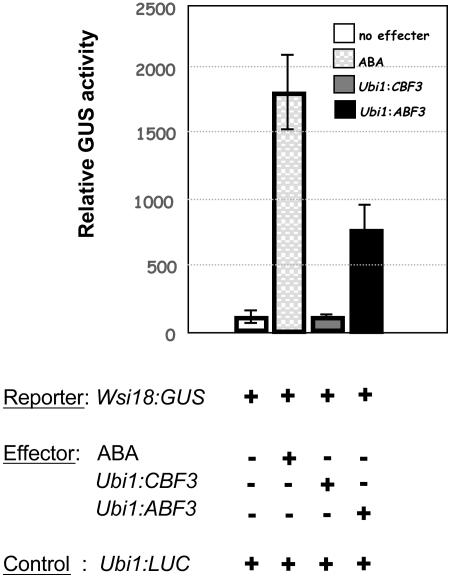
To determine the functional significance of interaction between CBF3/ABF3 and the promoters of the target genes that they induce, we chose the Wsi18 promoter as an example. Constructs containing the promoter linked to the β-glucuronidase (GUS) reporter gene and effector constructs containing CBF3 or ABF3 under the control of the Ubi1 promoter were used for transient transformation by microprojectile bombardment of 7-d-old rice seedlings. The LUC gene was used as an internal control to evaluate transformation efficiency. After particle bombardment, the samples were incubated in one-half-strength Murashige and Skoog (MS) medium either with or without ABA for 3 d. As seen in Figure 6, ABF3 activated the Wsi18 promoter and resulted in an 8-fold increase in GUS activity when compared to the activity of the promoter without the effector. Activity of the Wsi18 promoter was highly induced when ABA was applied and resulted in an 18-fold increase in relative GUS activity. CBF3, however, did not elevate expression of the Wsi18 promoter at all under the tested experimental conditions. These results are in agreement with the increased expression of Wsi18 in Ubi1:ABF3 plants, but not in Ubi1:CBF3 plants (Fig. 5), thus demonstrating that CBF3 and ABF3 in transgenic rice both enhance tolerance to stress by activating different groups of stress-regulated genes.
Figure 6.
Transactivation of the Wsi18:GUS fusion by ABF3. The Wsi18:GUS construct was cotransformed with the effector constructs, either Ubi1:CBF3 or Ubi1:ABF3, or with the expression vector alone; 4 μg of each construct with 2 μg of Ubi1:LUC as an internal standard was used in all cases. Each bar represents the mean value of the relative GUS/LUC activities from four independent experiments.
DISCUSSION
We developed transgenic rice plants constitutively expressing CBF3/DREB1A (CBF3) and ABF3. CBF3 overexpression in transgenic rice (Ubi1:CBF3) substantially elevated tolerance to drought and high salinity but had only limited effect on low-temperature stress tolerance. The minimal enhancement of low temperature tolerance was rather unexpected because CBF3 in Arabidopsis is a key regulatory factor that functions primarily in freezing tolerance by activating a battery of downstream genes (Jaglo-Ottosen et al., 1998; Kasuga et al., 1999; Fowler and Thomashow, 2002; Maruyama et al., 2004). One explanation includes differences in the composition of CBF3-regulated genes in rice. Using microarray and RNA gel-blot analyses, we identified 12 stress-regulated genes in rice that are up-regulated by CBF3. In contrast, a total of 38 stress-regulated genes were identified in Arabidopsis as the target genes of CBF3 by a similar approach (Seki et al., 2001; Fowler and Thomashow, 2002; Maruyama et al., 2004). OsDREB1A, a rice ortholog of CBF3, in transgenic Arabidopsis activates 7 CBF3-target genes (Dubouzet et al., 2003). Thus, CBF3 activates a smaller number of target genes in rice than in Arabidopsis, causing relatively lower levels of tolerance to low temperature in Ubi1:CBF3 plants. This may be due to the possibility that the Arabidopsis CBF3 cannot recognize the sequence in the promoter region of certain stress-related rice genes.
ABF3 in transgenic (Ubi1:ABF3) rice also exhibited increased tolerance to drought, but did not have enhanced tolerance to high salinity or to low-temperature stress (Fig. 4). These data were in agreement to those obtained in Arabidopsis overexpressing 35S::ABF3 that possessed enhanced drought tolerance (Kang et al., 2002). We identified 7 ABF3-target genes that are stress regulated in rice. Three of those, Hsp70, PP2Ca, and a receptor kinase gene, were also activated by CBF3 and these data allow us to suggest that ABA-dependent and ABA-independent pathways converge, at least in part, into these common target genes. Four ABF3-target genes that are stress regulated were identified in Arabidopsis (Kang et al., 2002) and none of them are activated by CBF3.
The core conserved sequences of CRT/DRE and ABRE, G/ACCGAC and ACGTG, were found in multiple copies in promoter regions of the rice target genes within 1 kb upstream of the ATG start codon (Supplemental Table II). Interestingly, CBF3-target genes carry more DREs than ABREs, while ABF3-target genes contain more ABREs than DREs. For example, Lip5, Dip1, Jacalin2, and LOX contain 2 or 3 DREs and 1 ABRE or none, whereas Rab21, Wsi18, and PP2Cb contain 1 DRE and 2 to 5 ABREs, respectively. The presence of multiple CRT/DRE elements is a common characteristic of Arabidopsis genes that are induced by CBF3 (Gilmour et al., 2000; Dubouzet et al., 2003; Maruyama et al., 2004). CBF3 binds equally well to both ACCGAC and GCCGAC, whereas OsDREB1A has higher affinity binding for GCCGAC than for ACCGAC (Sakuma et al., 2002). Similarly, HvCBF1 binds to GCCGAC more efficiently than to ACCGAC in barley (Xue, 2002). Thus, in monocots, GCCGAC appears to be the preferred binding site for CBF3 or its orthologs. CBF3-target genes contain much more GCCGAC than ACCGAC in their promoter regions (Supplemental Table II). Whether CBF3 directly binds to the promoters of the target genes in rice remains to be determined. However, our observation that CBF3 overexpression increased their transcript levels reflects that the interactions are likely to be involved. ABF3-target genes have 2 to 5 copies of ABRE in their promoters (Supplemental Table II), consistent with the observation that more than 2 copies of ABREs are required to confer ABA responsiveness (Skriver et al., 1991). The Arabidopsis rd29B promoter that lacks a DRE, but contains 2 ABREs, is induced by ABA (Uno et al., 2000) and also by ABF3 (Kang et al., 2002). Overexpression of ABF3 in our Ubi1:ABF3 plants resulted in increased expression of the target genes including Wsi18. Moreover, activity of the Wsi18 promoter was enhanced in rice seedlings by ABF3, but not by CBF3, in our transactivation assays (Fig. 6). Taken together, these results suggest that at least 2 copies of DRE or ABRE are required for CBF3 or ABF3 to activate corresponding target genes in rice.
Overexpression of Arabidopsis genes, 35S:CBF3 and the 35S:ABF3, in transgenic Arabidopsis resulted in various levels of growth inhibition under normal growth conditions (Kasuga et al., 1999; Kang et al., 2002). Similarly, overexpression of a rice gene, 35S:OsDREB1A, in transgenic Arabidopsis also caused stunted growth (Dubouzet et al., 2003). Interestingly, our Ubi1:CBF3 and Ubi1:ABF3 plants exhibited neither growth inhibition nor visible phenotypic alterations in rice, despite constitutive expression of the transgenes (Fig. 2A). This may have occurred because lower levels and/or fewer numbers of target genes are activated by CBF3 or ABF3 in rice than in Arabidopsis, and hence, the effects on plant growth might be minimized in rice. In fact, transcript levels of the target genes in our transgenic plants under normal growth conditions were lower than levels in nontransgenic plants that were treated with stress (Fig. 5). Alternatively, rice is evolutionarily more tolerant to the expression of stress-regulated genes than dicots, including Arabidopsis. This is supported by our previous observation that transgenic rice plants producing trehalose were stress tolerant and exhibited normal growth, unlike the results obtained for transgenic dicots, such as potato (Solanum tuberosum) and tobacco that were severely stunted (Jang et al., 2003). Overall, our results demonstrated that Arabidopsis genes CBF3 and ABF3 function in stress-response pathways in rice, an important agronomic crop, without causing undesirable growth phenotype.
MATERIALS AND METHODS
Plasmid Construction and Transformation of Rice
Expression plasmids, Ubi1:CBF3 and Ubi1:ABF3, contain the bar gene under the control of the cauliflower mosaic virus 35S promoter for herbicide-based selection and a pair of the matrix attachment region sequence from the chicken lysozyme gene for stable expression of transgene (Phi-Van and Strätling, 1996). The ubiquitin1 promoter, together with its intron (Ubi1), was used to drive constitutive expression (Christensen and Quail, 1996). The coding region of CBF3 was PCR-amplified from Arabidopsis (Arabidopsis thaliana) genomic DNA using a pair of primers, 5′-GATGAACTCATTTTCAGCTTTTC-3′ and 5′-ACAGTGCTCTCTTGTGGGAC-3′, and ABF3 cDNA was kindly provided by Dr. S.Y. Kim (Choi et al., 2000). The plasmids were introduced into Agrobacterium tumefaciens LBA4404 by triparental mating and embryogenic calli from mature rice (Oryza sativa) cv Nakdong seeds were transformed as previously described by Jang et al. (1999). To make Wsi18:GUS for transactivation assays, the Wsi18 promoter region was PCR-amplified from rice genomic DNA using the primers 5′-AAGCTTGAGTCATAGGGAGA-3′ and 5′-AGTGATTCCAGCCAAGTTTGGATCC-3′ (Joshee et al., 1998). The control plasmid, Ubi1:LUC, containing the firefly luciferase gene driven by the Ubi1 promoter, was obtained from Dr. Peter Quail at University of California at Berkeley.
Growth Measurements
Transgenic and nontransgenic rice (cv Nakdong) seeds were germinated in a one-half-strength MS solid medium in a growth chamber in the dark at 28°C for 3 d, transplanted into soil pots, and grown in the greenhouse (16-h-light/8-h-dark cycles) at 28°C to 30°C. Each pot (5 × 5 × 6 cm) was filled with nursery soils (Bio-media, Kyeongju, Korea) and planted with 6 seedlings. The fresh weight of plants was determined in the time course by harvesting and weighing the whole plant parts including roots of 10 plants/line. The dry weights were determined after drying the plants at 80°C for 48 h. Each experiment was repeated three times with three independent transgenic lines.
Stress Tolerance of Plants Grown in Soil
Transgenic and nontransgenic rice seeds were germinated in a one-half-strength MS solid medium in a growth chamber in the dark at 28°C for 4 d, transplanted into soil, and grown in the greenhouse (16-h-light/8-h-dark cycles) at 28°C to 30°C. Eighteen seedlings from each transgenic and nontransgenic line were grown in pots (5 × 5 × 6 cm; 6 plants/pot) for 4 weeks before performing the drought-stress experiments. For drought stress, 4-week-old NC and transgenic seedlings were subjected to 4 d of drought followed by 5 to 7 d of watering. Fv/Fm values of transgenic and NC plants were measured in the time course with a pulse modulated fluorometer (mini-PAM, Walz, Germany) as previously described (Jang et al., 2003). The numbers of the plants that survived or continued to grow were scored.
Chlorophyll Fluorescence under Conditions of Drought, High Salinity, and Low Temperature
Transgenic and nontransgenic rice seeds were germinated and grown in a one-half-strength MS solid medium for 14 d in a growth chamber (16-h-light of 150 μmol m−2 s−1/8-h-dark cycles at 28°C). Green parts of approximately 10 seedlings were cut by scissors before stress treatments in vitro. For low-temperature stress, the seedlings were incubated at 4°C water for up to 6 h under continuous light of 150 μmol m−2 s−1. For high-salinity stress treatments, they were incubated in 400 mm NaCl for 2 h at 28°C under continuous light of 150 μmol m−2 s−1 and for drought stress they were air-dried for 2 h at 28°C under continuous light of 150 μmol m−2 s−1. Fv/Fm value was measured as previously described (Artus et al., 1996; Jang et al., 2003).
60 K Rice Whole Genome DNA Chip Analysis
Expression profiling was conducted with the 60 K Rice Whole Genome Microarray. Information of the Microarray can be found at www.ggbio.com/rice60kchip.html (GreenGene Biotech). The 60 K Microarray was designed to represent all the genes in rice. In total, 60,727 oligomers were designed from gene-specific regions of both japonica and indica subsp. These include 58,417 from known and predicted genes and 66 randomized DNA. Among these, 2,310 genes were also designed as antisense oligomers. Oligomer sequences were extracted by Qiagen-Operon (Cologne, Germany) based on rice genome information from Beijing Genomics Institute (Yu et al., 2002). Oligomers were synthesized and purified by Qiagen-Operon and spotted on SuperAmine slide using facilities of the Dr. David Galbraith lab at the University of Arizona (http://ag.arizona.edu/microarray/deconvolution.html). A set of Rice 60 K Microarray is composed of 2 slides and has a total of 64,896 spot addresses. Each slide is formatted with 48 (12 × 4) blocks with addresses composed of 676 (26 × 26) spots. Blank spots (4,099) were also included for easy alignment of scanning format. Each oligomer, 70 nucleotides long and average melting temperature of 78°C, was printed in each spot address with a diameter of 100 μm.
Total RNA (100 μg) was prepared from leaf tissues of 14-d-old transgenic and nontransgenic seedlings (5–10 plants each) as reported previously (Jang et al., 2002) and the mRNA was purified from total RNAs using a Qiagen oligotex column (Qiagen, Valencia, CA). For drought stress, 14-d-old seedlings were excised from the seedlings before being air-dried for 2 h in the greenhouse under continuous light of approximately 900 to 1,000 μmol m−2 s−1. Preparation of fluorescence labeled probes and microarray hybridization was performed as procedures provided by Genisphere 3DNA Array Detection Array 50 kit (v. 2, Genisphere, Hatfield, PA). The microarray was scanned with Genepix 4000B (Axon Instruments, Union City, CA) and the quality of the chip data was analyzed with statistical R language and sma package in Bioconductor project (http://www.bioconductor.org/) implemented on Linux platform. Noncorrelation of signal and background intensities were confirmed by plotting base 2 log background intensity in x axis and base 2 log intensity subtracted with background intensity in y axis. Prior to normalization, normal distribution of Cy3 and Cy5 intensities were tested by qqplot function in R statistical language. The spatial effects on the chip during the hybridization process were checked with spatial func in sma package. The variance difference between Cy3 and Cy5 intensities within microarray was tested by the Student's t test under the assumptions firstly uniform and then nonuniform variances. The ANOVA difference of signal intensities between microarrays was performed by one-way ANOVA. Block-by-block Lowess normalization (Yang et al., 2002) and multivariate statistics such as clustering, principal component analysis, multidimensional scaling, etc. were analyzed with Acuity 3.1 (Axon Instruments). Spots with flag of 0 and a diameter greater than 51 pixel size were used for the analysis. Cy3 significant spots were determined when its log ratio is less than −0.67 [2**(−0.67) = 1.6-fold decrease] and its background subtracted intensity is higher than 500. On the contrary, Cy5 significant spots was determined when its log ratio is greater than 0.67 [2**0.67 = 1.6-fold increase] and its intensity is higher than 500. A preliminary microarray experiment using nontreated wild-type RNA labeled Cy3 and Cy5, respectively, gave less than 0.5% false positive signals in the analysis. Hybridization of different microarrays with the same mRNA samples indicated a good correlation. To assess the reproducibility of the microarray analysis, we repeated the experiment three times with independently prepared total RNA (Supplemental Table I).
RNA Gel-Blot Analysis
Transgenic and nontransgenic rice seeds were germinated on soil and grown in the greenhouse (16-h-light/8-h-dark cycles). For low-temperature stress treatment, 14-d-old seedlings were exposed to 4°C at a cold chamber for 2 h under continuous light of 150 μmol m−2 s−1. For high salinity and ABA treatment, 14-d-old seedlings were grown in a nutrient solution, 0.1% (v/v) Hyponex (Busan, Korea), for 2 d and then transferred to fresh nutrient solution containing 400 mm NaCl or 100 μm ABA for 2 h in the greenhouse under continuous light of approximately 900 to 1,000 μmol m−2 s−1. For drought stress, 14-d-old seedlings were excised from the seedlings before being air-dried for 2 h in the greenhouse under continuous light of approximately 900 to 1,000 μmol m−2 s−1. Preparation of total RNA and RNA gel-blot analyses were previously reported (Jang et al., 2002). Hybridization signals were captured by using a phosphor imager analyzer (FLA 3000, Fuji, Tokyo).
Transactivation Assay
Rice seeds were germinated and grown in one-half-strength MS solid medium for 7 to 9 d in a growth chamber with cycles of 16-h light/8-h darkness at 28°C. Green parts of approximately 3 to 4 seedlings, including leaves and sheaths, were cut by scissors and spread on a one-half-strength MS solid medium before particle bombardment. The tissues were transformed by particle bombardment with 4 μg of the Wsi18:GUS plasmid, 2 μg of Ubi1:LUC as an internal standard, and 4 μg of either Ubi1:CBF3 or Ubi1:ABF3 plasmid DNA, as previously described (Klein et al., 1987). The samples were then incubated in a MS medium, or a MS medium containing 100 μm ABA if necessary, for 72 h in the dark at 25°C before freezing in liquid nitrogen. Luciferase and GUS assays were carried out as previously described (Busk et al., 1997). Relative GUS activity was calculated as the reading of the GUS assay divided by that of the luciferase assay.
Supplementary Material
Acknowledgments
The authors thank Dr. Takuji Sasaki at the National Institute of Agrobiological Resources for providing the ESTs clones of Lip5 and Dip1 and Dr. Peter Quail at University of California at Berkley for Ubi1:LUC and the Ubi1 promoter.
This work was supported by the Ministry of Science and Technology through the Crop Functional Genomics Center (grants to J.-K.K., S.I.S., and B.H.N.), by the Biogreen21 Program (grant to J.-K.K.), and by the Korea Science and Engineering Foundation through the Plant Metabolism Research Center at Kyung-Hee University (grant to J.-K.K.).
The online version of this article contains Web-only data.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.104.059147.
References
- Artus NN, Uemura M, Steponkus PL, Gilmour SJ, Lin C, Thomashow MF (1996) Constitutive expression of the cold-regulated Arabidopsis thaliana COR15a gene affects both chloroplast and protoplast freezing tolerance. Proc Natl Acad Sci USA 93: 13404–13409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker SS, Wilhelm KS, Thomashow MF (1994) The 5′-region of Arabidopsis thaliana cor15a has cis-acting elements that confer cold-, drought- and ABA-regulated gene expression. Plant Mol Biol 24: 1–13 [DOI] [PubMed] [Google Scholar]
- Bray EA (1997) Plant responses to water deficit. Trends Plant Sci 2: 48–54 [Google Scholar]
- Busk PK, Jensen AB, Pages M (1997) Regulatory elements in vivo in the promoter of the abscisic acid responsive gene rab17 from maize. Plant J 11: 1285–1295 [DOI] [PubMed] [Google Scholar]
- Choi HI, Hong JH, Ha JO, Kang JY, Kim SY (2000) ABFs, a family of ABA-responsive element binding factors. J Biol Chem 275: 1723–1730 [DOI] [PubMed] [Google Scholar]
- Christensen AH, Quail PH (1996) Ubiquitin promoter-based vectors for high-level expression of selectable and/or screenable marker genes in monocotyledonous plants. Transgenic Res 5: 213–218 [DOI] [PubMed] [Google Scholar]
- DeRocher EJ, Bohnert HJ (1993) Development and environmental stress employ different mechanisms in the expression of a plant gene family. Plant Cell 5: 1611–1625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubouzet JG, Sakuma Y, Ito Y, Kasuga M, Dubouzet EG, Miura S, Seki M, Shinozaki K, Yamaguchi-Shinozaki K (2003) DREB genes in rice, Oryza sativa L., encode transcription activators that function in drought-, high-salt- and cold-responsive gene expression. Plant J 33: 751–763 [DOI] [PubMed] [Google Scholar]
- Fowler S, Thomashow MF (2002) Arabidopsis transcriptome profiling indicates that multiple regulatory pathways are activated during cold acclimation in addition to the CBF cold response pathway. Plant Cell 14: 1675–1690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilmour SJ, Sebolt AM, Salazar MP, Everard JD, Thomashow MF (2000) Overexpression of the Arabidopsis CBF3 transcriptional activator mimics multiple biochemical changes associated with cold acclimation. Plant Physiol 24: 1854–1865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilmour SJ, Zarka DG, Stockinger EJ, Salazar MP, Houghton JM, Thomashow MF (1998) Low temperature regulation of the Arabidopsis CBF family of AP2 transcription activators as an early step in cold-inducible COR gene expression. Plant J 16: 433–442 [DOI] [PubMed] [Google Scholar]
- Giraudat J, Parcy F, Bertauche N, Gosti F, Leung J (1994) Current advances in abscisic acid action and signaling. Plant Mol Biol 26: 1557–1562 [DOI] [PubMed] [Google Scholar]
- Hiei Y, Ohta S, Komari T, Kumashiro T (1994) Efficient transformation of rice (Oryza sativa L.) mediated by Agrobacterium and sequence analysis of the boundaries of the T-DNA. Plant J 6: 271–282 [DOI] [PubMed] [Google Scholar]
- Hobo T, Kowyama Y, Hattori T (1999) A bZIP factor, TRAB1, interacts with VP1 and mediates abscisic acid-induced transcription. Proc Natl Acad Sci USA 96: 15348–15353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaglo KR, Kleff S, Amundsen KL, Zhang X, Kaake V, Xhan JZ, Deits T, Thomashow MF (2001) Components of the Arabidopsis C-repeat/dehydration-responsive element binding factor cold-response pathway are conserved in Brassica napus and other plant species. Plant Physiol 127: 910–917 [PMC free article] [PubMed] [Google Scholar]
- Jaglo-Ottosen KR, Gilmour SJ, Zarka DG, Schabenberger O, Thomashow MF (1998) Arabidopsis CBF1 overexpression induces COR genes and enhances freezing tolerance. Science 280: 104–106 [DOI] [PubMed] [Google Scholar]
- Jang I-C, Choi W-B, Lee K-H, Song SI, Nahm BH, Kim J-K (2002) High-level and ubiquitous expression of the rice Cytochrome c gene OsCc1 and its promoter activity in transgenic plants provides a useful promoter for transgenesis of monocots. Plant Physiol 29: 1473–1481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang I-C, Nahm BH, Kim J-K (1999) Subcellular targeting of green fluorescent protein to plastids in transgenic rice plants provides a high-level expression system. Mol Breed 5: 453–461 [Google Scholar]
- Jang I-C, Oh S-J, Seo J-S, Choi W-B, Song SI, Kim CH, Kim YS, Seo H-S, Choi YD, Nahm BH, et al (2003) Expression of a bifunctional fusion of the Escherichia coli genes for trehalose-6-phosphate synthase and trehalose-6-phosphate phosphatase in transgenic rice plants increases trehalose accumulation and abiotic stress-tolerance without stunting growth. Plant Physiol 131: 516–524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joshee N, Kisaka H, Kitagawa Y (1998) Isolation and characterization of a water stress-specific genomic gene, pwsi 18, from rice. Plant Cell Physiol 39: 64–72 [DOI] [PubMed] [Google Scholar]
- Kang J-K, Choi H-I, Im M-Y, Kim SY (2002) Arabidopsis basic leucine zipper proteins that mediate stress-responsive abscisic acid signaling. Plant Cell 14: 343–357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasuga M, Liu Q, Miura S, Yamaguchi-Shinozaki K, Shinozaki K (1999) Improving plant drought, salt, and freezing tolerance by gene transfer of a single stress-inducible transcription factor. Nat Biotechnol 17: 287–291 [DOI] [PubMed] [Google Scholar]
- Kasuga M, Miura S, Shinozaki K, Yamaguchi-Shinozaki K (2004) A combination of the Arabidopsis DREB1A gene and stress-inducible rd29A promoter improved drought- and low-temperature stress tolerance in tobacco by gene transfer. Plant Cell Physiol 45: 346–350 [DOI] [PubMed] [Google Scholar]
- Klein TM, Wolf ED, Wu R, Sanford JC (1987) High velocity microprojectiles for delivering nucleic acids into living cells. Nature 327: 70–73 [PubMed] [Google Scholar]
- Kyozuka J, McElroy D, Hayakawa T, Xie Y, Wu R, Shimamoto K (1993) Light-regulated and cell-specific expression of tomato rbcS-gusA and rice rbcS-gusA fusion gene in transgenic rice. Plant Physiol 102: 991–1000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q, Kasuga M, Sakuma Y, Abe H, Miura S, Yamaguchi-Shinozaki K, Shinozaki K (1998) Two transcription factors, DREB1 and DREB2, with an EREBP/AP2 DNA binding domain separate two cellular signal transduction pathways in drought- and low-temperature-responsive gene expression, respectively, in Arabidopsis. Plant Cell 10: 1391–1406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maruyama K, Sakuma Y, Kasuga M, Ito Y, Seki M, Goda H, Shimada Y, Yoshida S, Shinozaki K, Yamaguchi-Shinozaki K (2004) Identification of cold-inducible downstream genes of the Arabidopsis DREB1A/CBF3 transcriptional factor using two microarray systems. Plant J 38: 982–993 [DOI] [PubMed] [Google Scholar]
- Phi-Van L, Strätling WH (1996) Dissection of the ability of the chicken lysozyme gene 5′ matrix attachment region to stimulate transgene expression and to dampen position effects. Biochemistry 35: 10735–10742 [DOI] [PubMed] [Google Scholar]
- Rabbani MS, Maruyama K, Abe H, Khan MA, Katsura K, Ito Y, Yoshiwara K, Seki M, Shinozaki K, Yamaguchi-Shinozaki K (2003) Monitoring expression profiles of rice genes under cold, drought, and high-salinity stresses and abscisic acid application using cDNA microarray and RNA gel-blot analyses. Plant Physiol 133: 1755–1767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakuma Y, Liu Q, Dubouzet JG, Abe H, Shinozaki K, Yamaguchi-Shinozaki K (2002) DNA-binding specificity of the ERF/AP2 domain of Arabidopsis DREBs, transcription factors involved in dehydration- and cold-inducible gene expression. Biochem Biophys Res Commun 290: 998–1009 [DOI] [PubMed] [Google Scholar]
- Seki M, Narusaka M, Abe H, Kasuga M, Yamaguchi-Shinozaki K, Carninci P, Hayashizaki Y, Shinozaki K (2001) Monitoring the expression pattern of 1300 Arabidopsis genes under drought and cold stresses by using a full-length cDNA microarray. Plant Cell 13: 61–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinozaki K, Yamaguchi-Shinozaki K (1997) Gene expression and signal transduction in water-stress response. Plant Physiol 115: 327–334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinozaki K, Yamaguchi-Shinozaki K (2000) Gene expression and signal transduction in water-stress response. Curr Opin Plant Biol 3: 217–223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinwari ZK, Nakashima K, Miura S, Kasuga M, Seki M, Yamaguchi-Shinozaki K, Shinozaki K (1998) An Arabidopsis gene family encoding DRE/CRT binding proteins involved in low-temperature-responsive gene expression. Biochem Biophys Res Commun 250: 161–170 [DOI] [PubMed] [Google Scholar]
- Skriver K, Olsen FL, Rogers JC, Mundy J (1991) Cis-acting DNA elements responsive to gibberellin and its antagonist abscisic acid. Proc Natl Acad Sci USA 88: 7266–7270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomashow MF (1999) Plant cold acclimation: freezing tolerance genes and regulatory mechanisms. Annu Rev Plant Physiol Plant Mol Biol 50: 571–599 [DOI] [PubMed] [Google Scholar]
- Uno Y, Furihata T, Abe H, Yoshida R, Shinozaki K, Yamaguchi-Shinozaki K (2000) Arabidopsis basic leucine zipper transcription factors involved in an abscisic acid-dependent signal transduction pathway under drought and high-salinity conditions. Proc Natl Acad Sci USA 97: 11632–11637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weatherwax SC, Ong MS, Degenhardt J, Bray EA, Tobin EM (1996) The interaction of light and abscisic acid in the regulation of plant gene expression. Plant Physiol 111: 363–370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen JQ, Oono K, Imai R (2002) Two novel mitogen-activated protein signaling components, OsMEK1 and OsMAP1, are involved in a moderate low-temperature signaling pathway in rice. Plant Physiol 129: 1880–1891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue GP (2002) An AP2 domain transcription factor HvCBF1 activates expression of cold-responsive genes in barley through interaction with a (G/a)(C/t)CGAC motif. Biochim Biophys Acta 1577: 63–72 [DOI] [PubMed] [Google Scholar]
- Yamaguchi-Shinozaki K, Shinozaki K (1994) A novel cis-acting element in an Arabidopsis gene is involved in responsiveness to drought, low-temperature, or high-salt stress. Plant Cell 6: 251–264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang YH, Dudoit S, Luu P, Lin DM, Peng V, Ngai J, Speed TP (2002) Normalization for cDNA microarray data: a robust composite method addressing single and multiple slide systematic variation. Nucleic Acids Res 30: e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J, Hu S, Wang J, Wong GK, Li S, Liu B, Deng Y, Dai L, Zhou Y, Zhang X, et al (2002) A draft sequence of the rice genome (Oryza sativa L. ssp. indica). Science 296: 79–92 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.