Abstract
The nuclear genome of the moss Physcomitrella patens contains two genes encoding phage-type RNA polymerases (PpRPOT1 and PpRPOT2). Each of the PpRPOT1 and PpRPOT2 transcripts possesses two in-frame AUG codons at the 5′ terminus that could act as a translational initiation site. Observation of transient and stable Physcomitrella transformants expressing the 5′ terminus of each PpRPOT cDNA fused with the green fluorescent protein gene suggested that both PpRPOT1 and PpRPOT2 are not translated from the first (upstream) AUG codon in the natural context but translated from the second (downstream) one, and that these enzymes are targeted only to mitochondria, although they are potentially targeted to plastids when translation is forced to start from the first AUG codon. The influence of the 5′-upstream sequence on the translation efficiency of the two AUG codons in PpRPOT1 and PpRPOT2 was quantitatively assessed using a β-glucuronidase reporter. The results further supported that the second AUG codon is the sole translation initiation site in Physcomitrella cells. An Arabidopsis (Arabidopsis thaliana) RPOT homolog AtRpoT;2 that possesses two initiation AUG codons in its transcripts, as do the RPOTs of P. patens, has been regarded as a dually targeted protein. When the localization of AtRpoT;2 was tested using green fluorescent protein in a similar way, AtRpoT;2 was also observed only in mitochondria in many Arabidopsis tissues. These results suggest that, despite the presence of two in-frame AUGs at the 5′ termini of RPOTs in Physcomitrella and Arabidopsis, the second AUG is specifically recognized as the initiation site in these organisms, resulting in expression of a protein that is targeted to mitochondria. This finding may change the current framework of thinking about the transcription machinery of plastids in land plants.
Plant cells contain two organelles having semiautonomous genetic systems, namely, mitochondria and plastids. They arose from eubacteria-like endosymbionts, closely related to extant α-proteobacteria and cyanobacteria, respectively (Gray, 1992, 1993; Howe et al., 1992). During the course of evolution, they lost many of their genes, while a number of genes were transferred to the cell nucleus. Therefore, the majority of the proteins involved in the biogenesis of mitochondria and plastids are encoded in the nucleus, and must be translated in the cytoplasm and then imported into the respective organelles (Schatz and Dobberstein, 1996; Neupert, 1997; Soll and Tien, 1998; Keegstra and Cline, 1999). Most of these proteins have a transit peptide at the N terminus that is necessary for their import into the target organelles (von Heijne, 1986; van Loon et al., 1988; Hand et al., 1989; Ko and Cashmore, 1989; Sidorov et al., 1999). The localization of these organellar proteins has been analyzed by in vitro import experiments using isolated organelles and in vivo experiments using a reporter, such as green fluorescent protein (GFP).
Nuclear-encoded RNA polymerases (RPOTs) consisting of a single polypeptide, which are similar to the RNA polymerase of bacteriophages T3 and T7, are widely distributed among eukaryotes and act as mitochondrial RNA polymerases (Cermakian et al., 1996, 1997). In higher plants, there is an additional plastid-targeted RPOT (Hedtke et al., 1997; Chang et al., 1999; Kobayashi et al., 2002), which is also called NEP (for nuclear-encoded polymerase). Plant RPOTs constitute a small gene family with different targeting properties. For example, Arabidopsis (Arabidopsis thaliana) RpoT;1 (AtRpoT;1) is targeted to mitochondria (Hedtke et al., 1997, 1999), while AtRpoT;3 is targeted to plastids (Hedtke et al., 1997, 1999). In addition, AtRpoT;2 was postulated to be targeted to both organelles (Hedtke et al., 2000). AtRpoT;2 possesses the property of dual targeting by the use of two probable initiation codons. Hedtke et al. (2000) reported that the polypeptide translated from the first AUG codon was targeted to the plastids and mitochondria, whereas the polypeptide translated from the second AUG was targeted only to mitochondria. Dually targeted RPOT was also identified in Nicotiana sylvestris (NsRpoT-B; Kobayashi et al., 2001a), which also contains two putative translation initiation codons. The dual targeting is thought to be effected by alternative translation initiation within a single transcript (Kobayashi et al., 2001a). In the moss Physcomitrella patens, two RPOT genes (named PpRPOT1 and PpRPOT2, respectively) have been identified (Kabeya et al., 2002; Richter et al., 2002). The translated sequence for each of these contains two putative translation initiation codons at the N terminus, just as do AtRpoT;2 and NsRpoT-B (Fig. 1A). In our previous study, the two RPOT proteins were targeted only to mitochondria (Kabeya et al., 2002). However, Richter et al. (2002) reported that the two RPOT proteins were targeted to both mitochondria and chloroplasts.
Figure 1.
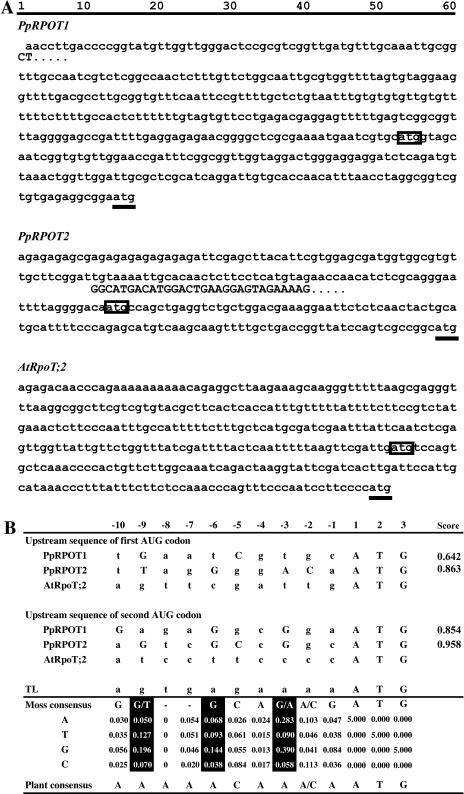
Sequence context for the putative initiation codons of PpRPOT1, PpRPOT2, and AtRpoT;2. A, The 5′ sequences upstream of the first and the second ATGs of PpRPOT1, PpRPOT2, and AtRpoT;2. Nucleotide sequences of PpRPOT1 (top), PpRPOT2 (middle), and AtRpoT;2 (bottom).The first ATG codon is surrounded by a rectangle in each sequence. The second ATG codon is marked with underline. The 5′ end of the genomic sequences of PpRPOT1 and PpRPOT2 that diverge from the cDNA sequences are shown in uppercase letters. The extensive difference in the 5′ sequence in PpRPOT2 could be due to an intron in the genomic sequence. B, Upstream context for the putative initiation codons. Top half shows actual sequences upstream of the respective ATG. The numbers above indicate nucleotide position with respect to the A of the putative initiation codon. Nucleotides corresponding to moss consensus sequence are indicated by uppercase letters. Bottom half shows moss and plant consensus sequences as well as scoring matrix for moss based on information content analysis. The score at the right side of the top half was calculated by adding respective values corresponding to the nucleotide sequence. The plant consensus was taken from Joshi et al. (1997). GenBank accession numbers are as follows: PpRPOT1 cDNA, AB055214; PpRPOT1 genomic, AJ416854; PpRPOT2 cDNA, AB055215; PpRPOT2 genomic, AJ416855; and AtRpoT;2 genomic, AJ001037.
In general, mRNA structure can influence translation initiation, e.g. the m7G cap, the length of the 5′-untranslated region (UTR), upstream open reading frame (uORF), the secondary structure of RNA, and the sequence context surrounding the initiation codon (Kozak, 1991), as well as interaction of the 5′- and the 3′-UTR (Bailey-Serres, 1999). Subcellular localization of several RPOTs was often examined with GFP-fusion protein, but the native 5′-UTR was not used in most of these targeting experiments, including the experiments by Richter et al. (2002). In our previous experiments, the native 5′-UTR was used (Kabeya et al., 2002). Effect of 5′-UTR on the translational start sites is a hypothesis that explains experimental discrepancy, and the resolution of this discrepancy will lead to physiologically important consequences on the framework of thinking about the plastid transcription machinery.
In this study, we examined the localization of the two PpRPOTs by immunoblot and enzymatic analyses, as well as targeting experiments using several GFP-fusion proteins. The results strongly indicated that the localization of these proteins is determined by the use of a unique initiation site, namely, both PpRPOT1 and PpRPOT2 proteins are translated from the second AUG codon and localized to mitochondria in Physcomitrella tissues. In addition, AtRpoT;2, so far regarded as dual-targeting RPOT in Arabidopsis, was also suggested to be localized only to mitochondria in wide plant tissues.
RESULTS
Presence of Two In-Frame AUG Codons in the 5′ Region of RPOT Sequences
The cDNA sequence for the two RPOTs in P. patens that we published previously (Kabeya et al., 2002) showed a long 5′ sequence that preceded the conserved RPOT-coding sequence (Fig. 1A). Some cDNA sequences were also available in the expressed sequence tag database (Nishiyama et al., 2003), but all of them were mapped within the cDNA sequences shown in Figure 1. Genomic sequences were published by Richter et al. (2002), and the comparison of cDNA and genomic sequences indicated that the long 5′ sequence of PpRPOT1 was shared by the two except two terminal residues (this could be due to cloning artifact or vector sequence), but that a short sequence fragment at the 5′ end of the genomic sequence of PpRPOT2 might be an intron sequence, which ends by the consensus AG. In other words, the transcription starts from an exon further upstream whose genomic sequence is still not available. Both of the cDNAs are likely full-length ones, but the 5′ end was not mapped due to unavailability of upstream genomic sequences and the low level of transcripts in the total mRNA pool.
In both RPOT sequences, the long 5′ sequence contains two in-frame ATG triplets. No further in-frame ATG is present upstream of the conserved polymerase sequence. Available sequence data indicate the transcript contains both AUG codons. No splicing variant containing only one of the AUGs is known. Therefore, the mRNA contains a long 5′-UTR (291 nucleotides) upstream of the first ATG in PpRPOT1. The length of the 5′-UTR of PpRPOT2 mRNA is 132 nucleotides. In addition, the interval between the first ATG and the second ATG was 141 and 105 nucleotides, respectively, for PpRPOT1 and PpRPOT2. In the AtRpoT;2 sequence, the 5′-UTR is 231 nucleotides long, and the interval between the two ATG codons is 117 nucleotides.
Immunoblot Analysis and Effect of Tagetitoxin on the Organellar Transcription in P. patens
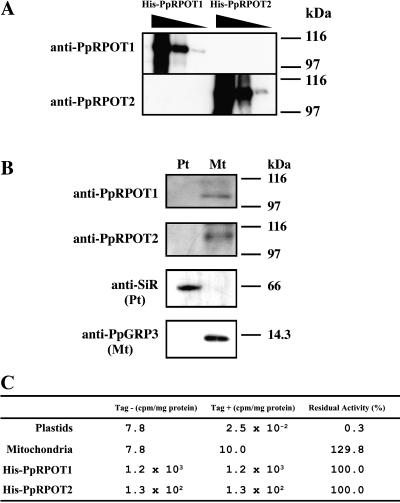
We first tested if the two PpRPOTs are present in the plastids by immunoblot analysis and by measuring the sensitivity to tagetitoxin of the transcription in isolated mitochondria and plastids of P. patens protonemata (Fig. 2, B and C, respectively). Tagetitoxin is known to inhibit transcription by plastid-encoded RNA polymerase (PEP) and bacterial RNA polymerase (Mathews and Durbin, 1990), but not the activity of RPOT. In P. patens, the core of PEP consists of plastid-encoded subunits (β, β′, and β″) and a nuclear-encoded α-subunit (Sugiura et al., 2003), but is expected to be sensitive to tagetitoxin because its structure is analogous to that of bacterial RNA polymerases. Protonemata were used in these experiments because we already showed that the two PpRPOT genes are transcribed in the protonemata (Kabeya et al., 2002). First, polyclonal antisera were raised against the glutathione S-transferase (GST)-PpRPOT1 and GST-PpRPOT2 recombinant proteins. (Primers for fusion constructs are summarized in Table I.) In these proteins, the N-terminal half of the protein sequence was used to avoid cross-reaction due to the conserved C-terminal active center domain. The cross-reaction of these antibodies with the full-length polymerase enzymes (expressed as His-tagged proteins that were used in the experiment in Fig. 2C) was indeed undetectable and estimated to be less than 1% (Fig. 2A). The two organelles were purified over Percoll gradients and then tested by immunoblot analysis with anti-PpRPOT1 or anti-PpRPOT2 (Fig. 2B). An immunoreactive protein with a molecular mass identical to that expected for PpRPOT1 (approximately 110 kD) or PpRPOT2 (approximately 108 kD) was detected in the mitochondrial fraction but not in the chloroplast fraction (Fig. 1B). Control antibodies directed against known plastid (SiR or sulfite reductase; Sato et al., 2001) and mitochondrial (PpGRP3, an RNA-binding protein; Nomata et al., 2004) proteins were also tested to confirm the purity of the two fractions. No significant cross-contamination was found (Fig. 2B). These results suggest that both PpPROT1 and PpRPOT2 proteins are localized in the mitochondria but not in the chloroplasts in the protonema.
Figure 2.
Mitochondrial localization of RPOTs in P. patens. A, Evaluation of cross-reaction of PpRPOT1 and PpRPOT2 with anti-PpRPOT1 and anti-PpRPOT2. Two His-tagged recombinant proteins (His-PpRPOT1 and His-PpRPOT2) were electrophoresed in SDS-PAGE, using 7.5% polyacrylamide gel, and then blotted for further immunoreaction. The antibodies were raised against GST-fusion polypeptides corresponding to the N-terminal half of the respective proteins that showed lower homology. Lane 1, 250 ng of His-PpRPOT1; lane 2, 25 ng of His-PpRPOT1; lane 3, 2.5 ng of His-PpRPOT1; lane 4, 250 ng of His-PpRPOT2; lane 5, 25 ng of His-PpRPOT2; and lane 6, 2.5 ng of His-PpRPOT2. B, Immunoblot analysis of isolated plastids and mitochondria from P. patens with anti-PpRPOT1, anti-PpRPOT2, and control antibodies. Proteins of plastids (Pt) and mitochondria (Mt; 20 μg of protein per lane) were analyzed by SDS-PAGE, using a 7.5% or 20% (for PpGRP3) polyacrylamide gel. The controls were as follows: SiR, sulfite reductase detected by antibodies raised against pea SiR; and PpGRP3, a Gly-rich RNA-binding protein of moss, detected by homologous antibodies. C, Effects of tagetitoxin on the transcription activity of isolated plastids, mitochondria, and recombinant PpRPOTs. Measurement of the transcription activity was carried out in the presence or absence of tagetitoxin, and [3H]UTP incorporation after 30 min of incubation was determined by liquid scintillation counting. The rightmost values indicate relative residual activities after addition of 10 μm tagetitoxin.
Table I.
Oligonucleotides used in this study
Underlines indicate restriction sites.
| No. | Oligonucleotide Name | Sequence (5′-3′) |
|---|---|---|
| For construction of GST-PpRPOT1 fusion | ||
| 1 | 1GST.F | atggatccagatcgtcgtctgattctgtg |
| 2 | 1GST.R | atgtcgacaatgcactcatgacagcagg |
| For construction of GST-PpRPOT2 fusion | ||
| 3 | 2GST.F | atagatctttgacaccattagattccgca |
| 4 | 2GST.R | atgtcgacggcattaccatatgcttgac |
| For construction of PpRPOT1-GFP fusions | ||
| 5 | 1-GFP.R | atccatggagaatccaactttagtgt |
| 6 | 1M2.F | atgtcgacgtagcaatcggtgtgttgga |
| 7 | RBCS-RPOT1M1.F | atgtcgacttcatacagaagtgagaaaaatggtagcaatcggtgtgttggaac |
| 8 | RBCS-RPOT1M2.F | atgtcgacttcatacagaagtgagaaaaatgtggagggcggcagtaagg |
| 9 | 1M48I.F | gtgtgagaggcggaatctggagggcggc |
| 10 | 1M48I.R | gccgccctccagattccgcctctcacac |
| 11 | 1M1+1.F | atgtcgacatggtagcaatcggtgtgtt |
| 12 | 1M1-10.F | atgtcgactgaatcgtgcatggtagcaa |
| For construction of PpRPOT2-GFP fusions | ||
| 13 | 2-GFP.R | taccatggtcaaggaggaagggga |
| 14 | 2M2.F | atgtcgacccagctgaggtctgctggac |
| 15 | RBCS-RPOT2M1.F | atgtcgacttcatacagaagtgagaaaaatgccagctgaggtctgctggacga |
| 16 | RBCS-RPOT2M2.F | atgtcgacttcatacagaagtgagaaaaatgtggaggtcggcagcacag |
| 17 | 2M36I.F | cagtcgccggcatctggaggtcggcag |
| 18 | 2M36I.R | ctgccgacctccagatgccggcgactg |
| For construction of AtRpoT;2-GFP fusions | ||
| 19 | At2GFP.F | atgtcgacgacatgtgagaaacagagacaaccc |
| 20 | At2+1GFP.F | atgtcgacatgtccagtgctcaaacccc |
| 21 | At2GFP.R | atccatggcctcttcggctacactcgtgtac |
| For construction of GUS fusions | ||
| 22 | 1M1GUS.F | ctcgcgaaaatgaatcgtgcatgttacgtcctgtagaaac |
| 23 | 1M1.R | gcacgattcattttcgcgag |
| 24 | 1M2GUS.F | gcggtcgtgtgagaggcggaatgttacgtcctgtagaaac |
| 25 | 1M2.R | tccgcctctcacacgaccgc |
| 26 | 2M1-GUS.F | gcagggaattttaggggacaatgttacgtcctgtagaaac |
| 27 | 2M1.R | tgtcccctaaaattccctgc |
| 28 | 2M2-GUS.F | ccggttatccagtcgccggcatgttacgtcctgtagaaac |
| 29 | 2M2.R | gccggcgactggataaccgg |
| 30 | RBCS-GUS.F | atgtcgacttcatacagaagtgagaaaaatgttacgtcctgtagaaac |
| 31 | GUS.R | atgcggccgctcattgtttgcctccctgctgcgg |
| 32 | BcaBEST Sequencing Primer M13-20 | cgacgttgtaaaacgacggccagt |
The two PpRPOTs were demonstrated to be functional RNA polymerases in a previous study (Kabeya et al., 2002). The transcription activity of the two recombinant PpRPOT enzymes was not inhibited by tagetitoxin (Fig. 2C). In mitochondria, the transcription was not inhibited by tagetitoxin or even stimulated to some extent (Fig. 2C). This result was just as expected because the two PpRPOTs are present in the mitochondria as described above, and no other type of RNA polymerase is known to function in mitochondria of higher eukaryotes. In the plastids, the transcription was almost completely (to less than 1%) inhibited by the addition of tagetitoxin. In the in vitro transcription system using the proplastid nucleoids of tobacco BY-2 cells, which contain an appreciable level of NEP, the residual transcription activity in the presence of tagetitoxin (about 50% of total activity) was ascribed to the activity of NEP (Sakai et al., 1998). In the plastids of P. patens, however, there was no measurable level of tagetitoxin-insensitive transcription activity. This is consistent with the results of immunoblot analysis that indicated the absence of RPOT enzymes in the chloroplasts. If the RPOTs are massively targeted to chloroplasts as reported by Richter et al. (2002), all these results are hard to explain. Rather, this is evidence that RPOT enzymes or NEP is absent in the plastids of P. patens protonemata.
Targeting of PpRPOT1 and PpRPOT2
Next, we reexamined targeting of GFP-fusion proteins. The discrepancy in the targeting experiments, as described in the introduction, might be due to the constructs used in the two reports. In the experiments of Richter et al. (2002), translation was forced to start from the AUG codon just downstream the translation leader sequence, which was taken from tobacco etch virus (using the vector pOL S65C; Peeters et al., 2000). The native 5′-upstream sequence was not used in their targeting experiments. By contrast, we used GFP-fused constructs containing the native 5′-upstream sequences to investigate the situation in the natural translation context. Thus, the sequence context of the translational initiation site is a likely candidate for the discrepancy. Since the nucleotide sequence context of the AUG plays a role in the efficiency of translation initiation, the 5′-upstream sequences of the first and second AUGs were compared (Fig. 1B). Although the consensus sequence of the plant context is AAAAACAA(A/C)AAUG (Joshi et al., 1997), the sequence context of both the first and second AUG were scarcely analogous to the consensus sequence, and it was difficult to estimate functional initiation codon just by such sequence comparison.
We therefore tried to obtain as much information as possible from the sequence database of P. patens. First, all available protein-encoding sequences (266 in total) were retrieved from the GenBank database. Then, the 20 nucleotides upstream of the initiation site were extracted from each entry. The information content of each site or SequenceLogo was calculated for these 214 UTR sequences and is presented in Figure 1B. Based on this statistic, we deduced moss consensus, as shown in the figure. The information content values were used as a weight matrix to calculate a score for each possible initiation site, which we hoped would represent the probability of the initiation site. The result (Fig. 1B, right column) indicated that the score was higher for the second AUG than for the first AUG in both PpRPOT1 and PpRPOT2. This is the first positive computational data suggesting that the second AUG may be used preferentially.
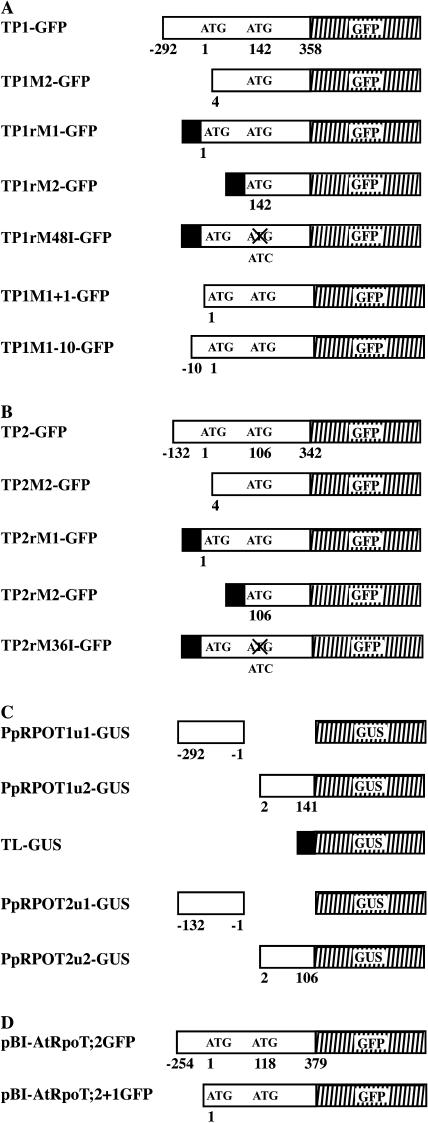
To reexamine the targeting of PpPROT1 and PpRPOT2 by GFP, we prepared several new GFP-fusion constructs using both the natural 5′-upstream sequence and the translation leader sequence (TL) taken from the RBCS 3A gene of pea (Pisum sativum; Fig. 3, A and B). In addition, in TP1rM48I-GFP and TP2rM36I-GFP, the second AUG codon was mutated, while the translation was forced to start from the first one. These constructs were introduced into moss protoplasts by polyethylene glycol-mediated transformation.
Figure 3.
Schematic representation of DNA constructs. A, PpRPOT1-GFP fusion plasmids used in P. patens protoplast transformation. B, PpRPOT2-GFP fusion plasmids. C, Plasmids for the measurement of translation efficiency. D, Plasmids for Arabidopsis transformation. Open rectangles represent the 5′ part of PpRPOT1, PpRPOT2, or AtRpoT;2 cDNAs. Vertically hatched rectangles represent reporter part, encoding either GFP (in A, B, and D) or GUS (in C). Black rectangles represent TL from the pea RBCS 3A gene. Maps are not drawn to scale for readability. In C, u1 and u2 are upstream sequences of the first and the second AUG codons, respectively. TP1 and TP2 in the construct names are used here to designate putative transit sequence (with or without upstream sequence) of PpRPOT1 and PpRPOT2, respectively. “r” indicates RBCS TL. M1 and M2 indicate the first and the second Met, respectively. Crosses over ATG indicate that the ATG was mutated to ATC (M48I and M36I mutations, respectively). “+1” indicates that the insert begins from the A of the first ATG, while “−10” indicates that the insert begins from the −10 position.
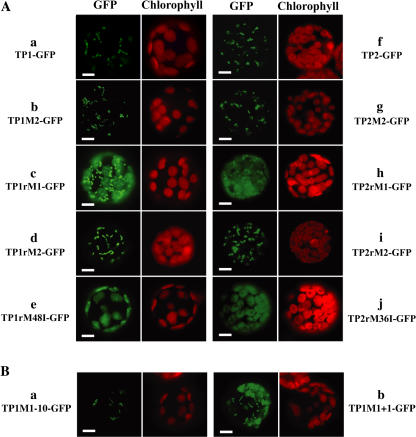
In the experiments with TP1-GFP, the construct having the entire 5′-UTR plus both Met codons (wild-type construct having the original upstream sequence), and constructs having only the second Met codon, TP1M2-GFP and TP1rM2-GFP, the fluorescence of GFP was localized to mitochondria (Fig. 4A, a, b, and d), whereas the fluorescence of GFP was localized to plastids with the construct having a mutated second Met codon, TP1rM48I-GFP (Fig. 4A, e). In analogous constructs with the PpRPOT2, namely, the construct having the entire 5′-UTR, TP2-GFP, and the constructs having the second Met codon, TP2M2-GFP and TP2rM2-GFP, the GFP fluorescence was localized to mitochondria (Fig. 4A, f, g, and i), while the fluorescence of GFP was localized to plastids with TP2rM36I-GFP (Fig. 4A, j). The fluorescence of GFP with the constructs having a translation leader that replaced the 5′-UTR, TP1rM1-GFP and TP2rM1-GFP, was localized to both mitochondria and plastids (Fig. 4A, c and h). These results indicate that forced translation from the first AUG codon using the TL of pea RBCS 3A resulted in the localization to plastids, just as in the reported results with pOL S65C vector (Richter et al., 2002). However, TP1-GFP and TP2-GFP with the natural 5′-upstream sequence were localized only to mitochondria. These results are explained if the first AUG codon is not used as the translation initiation site in vivo, although the amino acid sequence beginning from it has a property of plastid targeting sequence.
Figure 4.
Localization of PpRPOT-GFP fusion proteins in transiently transformed protoplasts. A, P. patens protoplasts were transformed with PpRPOT-GFP fusion constructs TP1-GFP (a), TP1M2-GFP (b), TP1rM1-GFP (c), TP1rM2-GFP (d), TP1rM48I-GFP (e), TP2-GFP (f), TP2M2-GFP (g), TP2rM1-GFP (h), TP2rM2-GFP (i), and TP2rM36I-GFP (j). B, P. patens protoplasts were transformed with PpRPOT1-GFP fusion constructs TP1M1−10-GFP (a) and TP1M1+1-GFP (b). Fluorescence of GFP (green) and chlorophyll (red) was observed using a fluorescence microscope BX-60 (Olympus, Tokyo) equipped with cubes U-MNIBA and U-MWU, respectively. Bar = 5 μm.
No secondary structure such as stem-loop is predicted in the 5′-upstream sequence of the two PpRPOTs with the software RNAstructure version 3.71 (Mathews et al., 1999). However, some uORFs were detected in the 5′-UTR. We examined the influence of the 5′-upstream sequence on the translation efficiency with GFP-fusion constructs (Fig. 4B, TP1M1−10-GFP; containing the 10 bp sequence upstream of the first AUG codon but no uORF, TP1M1+1-GFP; the 35S promoter directly joined to the first AUG). The results suggest that the GFP fluorescence of TP1M1−10-GFP was localized to mitochondria (Fig. 4B, a). The fluorescence of TP1M1+1-GFP was localized to both mitochondria and plastids (Fig. 4B, b). In the experiment with TP1M1−10-GFP, GFP fluorescence was observed in mitochondria as in the case of TP1-GFP containing the full-length 5′-upstream sequence of the first AUG codon. These results suggest that the uORF has no effect on the translation efficiency of the first AUG codon, and that only the 10-nucleotide sequence upstream of the first AUG is necessary to suppress translation from this site. However, this may not be suppression because not all AUGs within the mRNA act as initiation codons. What can be concluded from this experiment is that the proximal 10-nucleotide sequence but not the long 5′-UTR sequence upstream is important to determine whether translation is started from this site of RPOT mRNA.
Quantitative Estimation of the Effect of the 5′-Upstream Sequence on the Translation of PpRPOT1 and PpRPOT2
To quantitate the effects of the 5′-upstream sequence on the translation from the first and the second AUGs of PpRPOT1 and PpRPOT2, five additional plasmids with β-glucuronidase (GUS) reporter (Fig. 3C) were constructed and tested in transient expression. These constructs contain either the 5′-upstream sequence of the first (PpRPOT1u1-GUS and PpRPOT2u1-GUS) or the second AUG codon (PpRPOT1u2-GUS and PpRPOT2u2-GUS), or the TL from the pea RBCS 3A gene (TL-GUS), which were fused to the uidA gene and driven by the cauliflower mosaic virus (CaMV) 35S promoter. They were introduced into moss protoplasts by polyethylene glycol-mediated transformation. One day after transformation, GUS activity was measured. Two constructs, PpRPOT1u2-GUS and TL-GUS, gave high levels of GUS activity, whereas the GUS activity with PpRPOT1u1-GUS was at a level of pUC18 control (Table II). In contrast with PpRPOT1 constructs, PpRPOT2u2-GUS gave lower GUS activity, but this activity was significantly higher than the activity with PpRPOT2u1-GUS and pUC18 control. Therefore, it seems that translation is initiated only at the second AUG codon in both PpRPOT1 and PpRPOT2. In other words, the first AUG codon is unlikely to be recognized as a translation initiation site.
Table II.
Effects of the 5′-upstream sequences on translation efficiency
GUS activity in P. patens protoplasts that were transformed with respective plasmid is normalized with the coexpressed GFP. The GUS values are expressed in arbitrary unit, with setting the average values of expression driven by the TL-GUS as 100. Each value (±se) represents the average of three independent assays. N/A, Not applicable.
| Construct | Relative GUS Activity | P Value |
|---|---|---|
| Experiment 1 | ||
| PpRPOT1u1-GUS | 0.85 ± 0.34 | 0.51 |
| PpRPOT1u2-GUS | 93.03 ± 21.85 | 0.02 |
| TL-GUS | 100 ± 24.77 | 0.02 |
| pUC18 | 0.67 ± 0.21 | N/A |
| Experiment 2 | ||
| PpRPOT2u1-GUS | 1.10 ± 0.04 | 0.76 |
| PpRPOT2u2-GUS | 12.40 ± 0.44 | <0.01 |
| TL-GUS | 100 ± 9.75 | <0.01 |
| pUC18 | 1.18 ± 0.09 | N/A |
Subcellular Localization of PpRPOT1 and PpRPOT2 in Stably Transformed P. patens
In our transient expression experiments, both PpRPOT1 and PpRPOT2 were translated from the second AUG codon and were only localized to mitochondria in the protoplasts. However, the first AUG codon could be used as an initiation site in some particular types of cells or tissues. To address this question, we examined in detail the localization of PpRPOT1 and PpRPOT2 in various cell types and tissues in stably transformed moss (stable is used here to mean not transient or stably integrated in the chromosome). The pPpMADS-TP1GFP and pPpMADS-TP2GFP plasmids containing the natural 5′-upstream sequence of the first AUG codon and N-terminal sequence were used for the transformation. In all cell types and tissues of stably transformed P. patens, including protonemata and gametophores, GFP fluorescence was only observed in mitochondria but never in chloroplasts (Fig. 5). When immunoblot analysis was performed with plastids and mitochondria isolated from stably transformed P. patens protonemata, the GFP-fusion protein was detected in the mitochondrial fraction but not in the plastid fraction (data not shown). Thus, subcellular localization of the GFP-fusion protein in stably transformed protonemata and other tissues was the same as that in transiently transformed protoplasts, confirming that no detectable level of translation occurs from the first AUG codon of PpRPOT1 and PpRPOT2. It was concluded that the N-terminal extension in each of the two PpRPOTs (Fig. 6A) beginning from the first AUG codon is not translated in vivo.
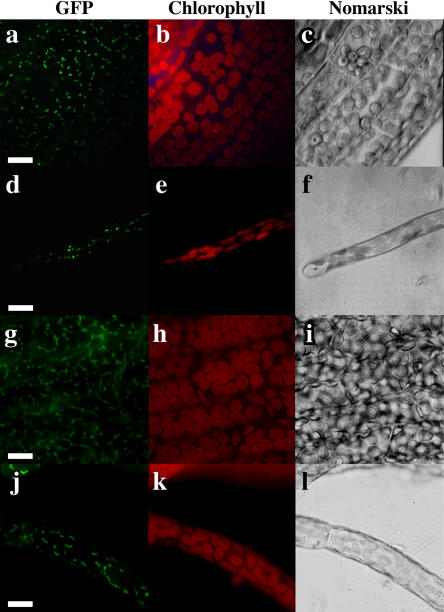
Figure 5.
Localization of PpRPOT-GFP fusion proteins in stably transformed P. patens. Localization of PpRPOT1-GFP fusion protein in a part of a gametophore (a–c) and a protonema (d–f), and of PpRPOT2-GFP fusion protein in a part of a gametophore (g–i) and a protonema (j–l). a, d, g, and j, Fluorescence of GFP; b, e, h, and k, fluorescence of chlorophyll; c, f, i, and l, Nomarski differential interference image. Fluorescence of GFP (green) and chlorophyll (red) was observed using a fluorescence microscope BX-60 (Olympus) equipped with cubes U-MNIBA and U-MWU, respectively. Bar = 5 μm.
Figure 6.
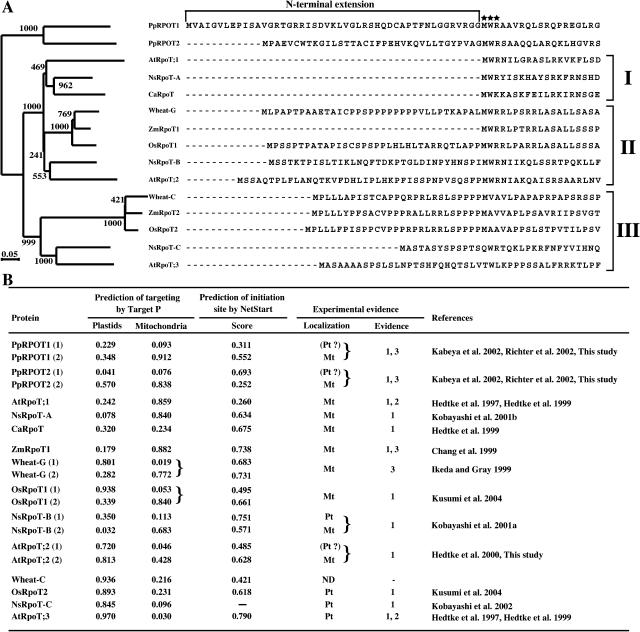
Comparison of various plastid and mitochondrial RPOTs in plants. A, Phylogenetic analysis and alignment of the N-terminal sequences of RPOT homologs. The following sequences were obtained from the GenBank database (in parentheses): PpRPOT1 and PpRPOT2 (P. patens; AB055214 and AB055215); OsRpoT2 (Oryza sativa; AB096015); AtRpoT;3 (Arabidopsis; Y08463); NsRpoT-C (N. sylvestris; AJ302020); ZmRpoT2 (Zea mays; AF127022); Wheat-C (Triticum aestivum; U34402); AtRpoT;1 (Arabidopsis; Y08137); NsRpoT-A (N. sylvestris; AJ416568); CaRpoT (Chenopodium album; Y08067); ZmRpoT1 (Z. mays; AF127021); OsRpoT1 (O. sativa; AB096014); Wheat-G (T. aestivum; AF091838); AtRpoT;2 (Arabidopsis; AJ001037); and NsRpoT-B (N. sylvestris; AJ302019). Left, Phylogenetic tree constructed by the neighbor-joining method. Amino acid sequences were used in this analysis (Kabeya et al., 2002; ALIGN_000281 in the EMBL-Align database). The numbers on the branches show bootstrap confidence levels obtained with 1,000 bootstraps. Right, Alignment of the N-terminal sequences of RPOT homologs. Asterisks indicate the conserved sequence block. B, Summary of localization of various RPOTs. If there are two putative initiation codons, (1) indicates the polypeptide translated from the first Met and (2) indicates the polypeptide translated from the second one. Results of computer prediction on the targeting by TargetP and on the probability of initiation site by NetStart are listed along with experimental results. Abbreviations for the localizations: Pt and Mt indicate plastids and mitochondria, respectively. (Pt?), Potentially targeted to plastids but no evidence for translation of such polypeptide. The following numbers are used to show experimental evidence: 1, GFP fusion; 2, in vitro import; and 3, immunoblot. ND, No available data. References cited in the figure image are as follows: Hedtke et al. (1997, 1999, 2000); Chang et al. (1999); Ikeda and Gray (1999); Kobayashi et al. (2001a, 2001b, 2002); Kabeya et al. (2002); Richter et al. (2002); Kusumi et al. (2004); and this study.
Reexamination of the Subcellular Localization of AtRpoT;2
Phylogenetic analysis of the RPOT proteins (Fig. 6A, left) suggested three major clusters, with a relationship (moss, ((Cluster I, Cluster II), Cluster III)). This result is essentially consistent with the structure of N-terminal sequences: Cluster I is characterized by the conserved sequence block (MWR) and mitochondrial localization. The conserved sequence block is not found in Cluster III, which contained plastid proteins. N-terminal extension as well as the conserved sequence block are found in the sequences in Cluster II (Fig. 6A). However, ZmRpoT1 belongs to Cluster II but lacks N-terminal extension. AtRpoT;2 and NsRpoT-B have been shown to be dually targeted to both plastids and mitochondria in experiments with GFP-fusion proteins (Hedtke et al., 2000; Kobayashi et al., 2001a). Immunological evidence suggested that Wheat-G is a mitochondrial RNA polymerase (Ikeda and Gray, 1999), but the plastid localization was not tested with GFP. The localization of Wheat-C was not reported (Fig. 6B). We chose AtRpoT;2 to test its targeting since the 5′-upstream sequence was not included in the GFP-fusion constructs in the previous report. The sequence context of two AUGs was compared to the consensus sequence of plants AAAAACAA(A/C)AAUG. However, both of the upstream sequences were not analogous to the consensus sequence (Fig. 1B). However, a computer prediction of translation initiation site with the NetStart program (Pedersen and Nielsen, 1997), which is said to be specialized for Arabidopsis, suggested that AtRpoT;2 is not translated from the first AUG codon but is translated from the second one (Fig. 6B). Analogous prediction data are presented for other plants, but they might not be correctly predicted because of limitation of neural network prediction using the Arabidopsis training data set. Two plasmids were constructed (Fig. 3D). pBI-AtRpoT;2GFP contained the 5′-upstream sequence of the first AUG codon and coding sequence (126 amino acids), and pBI-AtRpoT;2+1GFP contained the coding region and no native 5′-upstream sequence. These constructs were used to transform Arabidopsis (Fig. 7), and then subcellular localization of the GFP-fusion proteins was observed in cotyledon, leaf, and root. In the experiment with pBI-AtRpoT;2GFP, the fluorescence of GFP was localized to mitochondria (Fig. 7A, a, d, and g). By contrast, the fluorescence of AtRpoT;2+1GFP was observed in both mitochondria and plastids (Fig. 7B, j, m, and p). Additionally, we investigated into the localization of AtRpoT;2 during the early stage of seedling development and during the deetiolation process because Baba et al. (2004) reported that the mutation of AtRpoT;2 affected the light-induced accumulation of several plastid gene transcripts during early seedling development. However, the fluorescence of AtRpoT;2GFP was not detected in plastids under any conditions tested (data not shown). These results are essentially equivalent to those of the in vivo targeting experiments of PpRPOTs described above, and suggest that AtRpoT;2 may also contain a formal (or, more precisely, unused) plastid targeting sequence in its N terminus, as in the case of the two PpRPOTs.
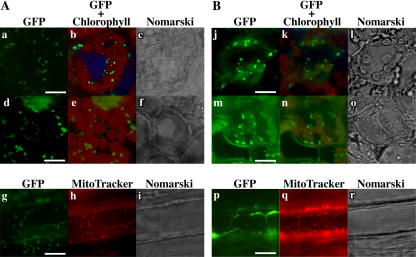
Figure 7.
Localization of AtRpoT;2-GFP fusion proteins in stably transformed Arabidopsis. A, Arabidopsis was stably transformed with pBI-AtRpoT;2GFP. Localization of AtRpoT;2-GFP fusion protein in cotyledon (a–c), leaf (d–f), and root (g–i). B, Arabidopsis was stably transformed with pBI-AtRpoT;2+1GFP. Localization of AtRpoT;2-GFP fusion protein in cotyledon (j–l), leaf (m–o), and root (p–r). In the root, no fluorescence of chlorophyll was observed. Fluorescence of MitoTracker is shown to locate mitochondria, although cell wall was also densely stained. a, d, g, j, m, and p, Fluorescence of GFP; b, e, k, and n, fluorescence of GFP and chlorophyll; h and q, fluorescence of MitoTracker; c, f, i, l, o, and r, Nomarski differential interference image. Fluorescence of GFP, chlorophyll, and MitoTracker was observed using a fluorescence microscope BX-60 (Olympus) equipped with cube U-MNIBA, U-MWU, and U-MWIG, respectively. Bar = 10 μm.
DISCUSSION
In this study, we showed that the PpRPOT1 and PpRPOT2 proteins in the moss P. patens are immunologically detected in the mitochondrial fraction but not in the plastid fraction, and that the transcription activity in the plastids is nearly completely inhibited by tagetitoxin, an inhibitor of PEP. We then confirmed the mitochondrial localization of PpRPOTs-GFP fusion proteins in transiently and stably transformed P. patens. We further investigated the translation efficiency of the 5′-upstream sequences of the first or the second AUG codon with GUS fusions. Our data indicated that PpRPOTs are translated from the second AUG codon, and such protein is targeted only to mitochondria in vivo, although the proteins are capable of targeting to plastids when translation is forced to start from the first AUG. The exclusive mitochondrial localization was confirmed in various tissues of stably transformed P. patens. Therefore, all available evidence indicates that the two PpRPOTs are targeted to mitochondria but not to plastids. Accordingly, a nuclear-encoded plastid RNA polymerase similar to mitochondrial RPOT in flowering plants, called NEP, does not exist in the plastids in P. patens. Plastid-encoded enzyme is likely the only RNA polymerase in P. patens plastids. Although we examined various cells and tissues of the transgenic moss expressing PpRPOT1-GFP and PpRPOT2-GFP, we did not detect a cell in which GFP fluorescence is localized to the plastids. The reproductive organs were not examined because they arose sporadically and rarely. They are to be examined in the future. Nevertheless, as far as the dual targeting of PpRPOTs, as originally proposed in the protonemal cells, is concerned, we can clearly say that PpRPOTs are not present in the chloroplasts of protonemal cells. Unfortunately, we have no clear answer to the question of why two AUG codons are present in these genes and why the first AUG is not used in vivo. The sequence upstream of the first AUG might not inhibit translation initiation, but simply the AUG is inactive as the initiation codon as all other AUGs within the transcript. An evolutionary view on this point is described below.
We should emphasize that the effects of the 5′-upstream sequence or translational context should be considered in experiments to examine the localization of polypeptide using GFP-fusion proteins. In a number of targeting experiments with GFP-fusion proteins, translation was forced to start from the first AUG codon without the 5′-upstream sequence. The presence or absence of the 5′-upstream sequence strongly influences the translation efficiency. In the case of PpRPOTs, the 5′-upstream sequence of the first AUG does not promote translation initiation at this site. It is generally difficult to predict the selection of the translation initiation site merely on the basis of the nucleotide sequence. Different consensus sequences are known in different organisms, e.g. UAAAAUGANAU in protozoa (Yamauchi, 1991), C(A/G)CCAUGG in vertebrates (Kozak, 1987), (A/C)(A/G)(A/C)CAUGGC in monocots, and AA(A/C)AAUGGC in dicots (Joshi et al., 1997). However, some software that predicts functional initiation codon was developed, such as NetStart (Pedersen and Nielsen, 1997), for specific species of plants. The prediction for AtRpoT;2 is in agreement with our experimental data. In addition, we developed a matrix based on information content calculated for available P. patens data obtained from GenBank entries. This approach seemed partly successful in that the score for the second AUG was higher than the score for the first AUG in both PpRPOT1 and PpRPOT2. However, the score for the first AUG of PpRPOT2 is higher than the score for the second AUG of PpRPOT1. This suggests that this method should be refined with more data. In vitro translation experiments often have been used to examine the translation initiation site, but this also is difficult since Lütcke et al. (1987) reported that the selection of translation initiation codons differs in wheat germ and reticulocyte. Thus, a heterologous system does not provide conclusive evidence. In fact, Richter et al. (2002) observed products that were translated from the first AUG and the second AUG in their in vitro translation experiments using reticulocytes, whereas we showed that the translation initiation from the first AUG is negligible in vivo. After all, in vivo experiments with constructs containing the native 5′-upstream sequences appear to be essential for identifying the translation initiation site and subcellular localization, although other experiments that have been done conventionally are also valuable.
Localization of AtRpoT;2 is an important issue in analyzing the transcription in both mitochondria and plastids in Arabidopsis. This protein possesses the N-terminal extension, as do PpRPOTs, and has so far been regarded as a dually targeted protein, while AtRpoT;1 and AtRpoT;3 are targeted to mitochondria and plastids, respectively (Hedtke et al., 2000). However, our results with GFP-fusion proteins suggested that the AtRpoT;2 transcript is translated from the second AUG but not from the first AUG in the natural context and its product uniquely localized to mitochondria in many Arabidopsis tissues (Fig. 7). Tagetitoxin test cannot be applied in Arabidopsis because the isoforms 1 and 3 are localized in mitochondria and plastids, respectively, and the effect of additional localization of isoform 2 is difficult to assess. Our results lack critical data on immunological analyses of the three RpoT isoforms in Arabidopsis organelles; however, the present situation necessarily raises questions about the regulation of translation and localization of AtRpoT;2.
Recently, Baba et al. (2004) reported that both plastid and mitochondrial transcription was affected in an AtRpoT;2 mutant, which was isolated from a population of activation-tagged T-DNA insertion lines. This mutant exhibited short roots, reduced hypocotyl length, delay in greening, and defect in light-induced accumulation of several plastid mRNAs as well as atp1 of mitochondrial mRNA. Their finding seemed consistent with the traditional idea that AtRpoT;2 is localized to both plastids and mitochondria. They analyzed organellar gene expression mostly in leaves, and, therefore, we still need some intricate explanation that compromises our data (AtRpoT;2 is localized to mitochondria in leaves, stems, and roots) and the data of the overexpressing line. It is quite probable that AtRpoT;2 in the activation line is involved in plastid transcription in some way. The AtRpoT;2 protein could be imported to plastids as well due to side effect of high level expression. Another possibility might be an indirect effect resulting from complex mitochondrion-chloroplast interactions. A mutant in the mitochondrial genome is known to cause variegation (Sakamoto et al., 1996), although no detailed mechanism is known. However, a more probable explanation for the solution of these apparent discrepancies might be that AtRpoT;2 targets to plastids as well in developing leaf cells or leaf primordia. There, this enzyme could trigger a cascade of reactions leading to normal gene expression in mature leaves. The overexpression of AtRpoT;2 could change the development of leaf cells and affects the level of gene expression in chloroplasts in mature leaves, although this enzyme is not localized in chloroplasts in mature leaves. This possibility may be solved by a strategy that develops a system that can sensitively detect changes in targeting of a given protein (see below).
In a previous study, we proposed that the creation of the NEP occurred in angiosperms after their separation from gymnosperms (Kabeya et al., 2002). The creation of the NEP in angiosperms occurred by gene duplication. The results of this study suggest that the plastid targeting sequence of the NEP might have been acquired before this gene duplication, even though it is not really used. There are two possible hypotheses on the origin of the angiosperm NEP. In one hypothesis, the plastid targeting sequence was present already before the separation of vascular plants and mosses but had remained nonfunctional (or formal) until the plastid targeting sequence was really used by activating the translation from the first AUG in angiosperms. In angiosperm NEPs such as AtRpoT;3, the second Met has been changed and no longer acts as an initiation site. The mitochondrial RPOTs, such as AtRpoT;1, lost the N-terminal extension either by mutation of the first AUG or deletion of entire extension. In another hypothesis, a formal plastid targeting sequence was added independently at various stages of evolution. In this case, the formal plastid targeting sequences of the moss are an example of an unsuccessful attempt to create a plastid protein. The NEP is a successful example, while the dually targeted RPOTs, such as AtRpoT;2, are an example of ongoing evolution by the addition of plastid targeting sequence.
A biologist's intuition favors that retention of the N-terminal extension that could serve as a plastid targeting sequence in many RpoT proteins is meaningful. This is an opinion shared by many of our colleagues. There are several solutions to this philosophical question. (1) The presence of the N-terminal extension is found in many RpoT proteins, but the sequence and length are not highly conserved. There is no selection pressure in this respect. In particular, the extension sequences in the moss are very different from those in flowering plants. (2) We do not have enough data on the presence of such N-terminal extension in plant proteins or eukaryotic proteins in general. If the presence of N-terminal extension is specific to RpoT proteins, then we will have to consider a specific role of the N-terminal extension. However, the exact N terminus of most proteins has not been determined experimentally, even though we sometimes encounter two or three Met residues in the N-terminal segment of an ORF, which is the longest reading frame that can be estimated for a given genomic sequence. (3) We will be able to challenge this hypothesis based on biologist's intuition by experimental approach. If we can detect sensitively a change in intracellular localization of a protein, we will be able to answer such a question. A possible method is to use an enzyme that functions normally in mitochondria but that causes serious damage when targeted to plastids. Various versions of such a system can be imagined, and we should try to demonstrate whether targeting to plastids of AtRpoT;2 occurs in some special types of cells during the development of plant.
In conclusion, available data suggest that the two PpRPOTs are targeted only to mitochondria due to exclusive translation from the second AUG codon and that the same is apparently true for AtRpoT;2.
MATERIALS AND METHODS
Plant Material
Gransden strain of Physcomitrella patens (Hedw.) Bruch & Schimp subsp. patens Tan was grown in the minimal medium supplemented with 5 mm diammonium (+)-tartrate as described previously (Hashimoto and Sato, 2001). Agar (0.8%) plates were used for maintaining the stock culture at 25°C. Light was provided by a bank of fluorescent lamps at a fluence rate of about 50 μmol m−2 s−1.
Seeds of Arabidopsis (Arabidopsis thaliana L.) Heynh, ecotype Columbia, were germinated in rock fiber (Nitto Bouseki, Chiba, Japan) and grown at 22°C under continuous illumination at a fluence rate of about 35 μmol m−2 s−1.
Isolation of Plastids
Seven-day-old protonemal cells were digested in Solution 1 (2.0% Driserase [Kyowa Hakko, Tokyo] and 8% mannitol) at 25°C for 30 min. Protoplasts were recovered by filtration through a 70-μm nylon mesh and centrifugation at 200g for 2 min at 4°C, followed by three washes with 8% mannitol. Isolation of chloroplasts was performed essentially according to the established method (Sato et al., 1993, 1997). The pellet was gently suspended in Grinding buffer (0.33 m sorbitol, 30 mm HEPES-KOH, pH 7.5, 2 mm EDTA, 0.1% bovine serum albumin). The protoplasts were broken by a passage through two layers of 20-μm nylon mesh. The suspension of broken protoplasts was centrifuged at 10,000g for 10 min at 4°C. The pellet was resuspended in Grinding buffer. Percoll was added to the suspension to a final concentration of 20%. The supernatant was used to prepare mitochondria (see below). Plastids were separated from nuclei and mitochondria by Percoll density gradient centrifugation (20%/40%/80%, v/v) at 10,000g for 30 min at 4°C. A green band that formed at the 40%/80% interface was collected, washed three times with Grinding buffer at 10,000g for 10 min at 4°C, and then diluted three times with TAN buffer (20 mm Tris-HCl, pH 7.5, 0.5 mm EDTA, 7 mm 2-mercaptoethanol, 0.4 mm phenylmethylsulfonyl fluoride, 1.2 mm spermidine, 500 mm Suc). The plastids were suspended in a small volume of TAN buffer containing 33% glycerol, flash frozen in liquid nitrogen, and stored at −80°C. Purity of the organelles was checked by examination under fluorescence microscope after staining with 4′,6-diamino-phenylindole as described previously (Sato et al., 1997).
Isolation of Mitochondria
Mitochondria were isolated from the broken protoplasts (see above). The supernatant after the centrifugation at 10,000g for 10 min was then centrifuged at 18,000g for 10 min at 4°C. The pellet was suspended in Grinding buffer. Percoll was added to the suspension to a final concentration of 20%. Mitochondria were purified by Percoll density gradient centrifugation (20%/33%/80%, v/v) at 18,000g for 60 min at 4°C. A yellowish turbid band that formed at the 33%/80% interface was collected, washed three times with the Grinding buffer at 18,000g for 10 min at 4°C, and then diluted three times with TAN buffer. The mitochondria were suspended in a small volume of TAN buffer containing 33% glycerol, flash frozen in liquid nitrogen, and stored at −80°C. Purity of the organelles was checked by examination under fluorescence microscope as described above.
Effects of Tagetitoxin on Organellar Transcription
Transcription activity of plastids and mitochondria was measured as incorporation of [3H]UTP. In organellar assay, a 60-mL reaction contained transcription buffer [40 mm Tris-HCl, pH 8.3, 25 mm MgCl2, 9 mm MgSO4, 30 mm (NH4)2SO4], 5 mm dithiothreitol, 0.01% Nonidet P-40, 180 μm ATP, 180 μm GTP, 180 μm CTP, 5 μm [3H]UTP (at a specific radioactivity 0.16 Gbq μmol−1 for plastids and 0.51 Gbq μmol−1 for mitochondria), 5.7 unit μL−1 RNAguard (Amersham Bioscience, Piscataway, NJ), and 60 μg of protein plastids or 24 μg of protein mitochondria. Transcription activity of recombinant PpRPOTs was measured as incorporation of [3H]UTP. For in vitro assay of purified enzymes, a 60-mL reaction contained transcription buffer (see above), 5 mm dithiothreitol, 0.01% Nonidet P-40, 180 μm ATP, 180 μm GTP, 180 μm CTP, 5 μm [3H]UTP (at a specific radioactivity 0.16 Gbq μmol−1), 5.7 unit μL−1 RNAguard, 3 μg of calf thymus DNA as the template, and 1.5 μg of recombinant PpRPOT1 or PpRPOT2. The reaction mixtures were incubated for 30 min at 25°C. Tagetitoxin (Epicentre, Madison, WI) was added to a final concentration of 10 μm in the inhibition experiments. After the reaction, 5-μL aliquots were spotted onto DEAE paper (DE-81; Whatman, Clifton, NJ). After successive washing with 5% Na2HPO4, water, and ethanol, radioactivity was determined by liquid scintillation counting.
Antibody Preparation
The DNA fragments corresponding to the amino acids 121 to 500 of PpRPOT1 and 114 to 489 of PpRPOT2 were amplified from pZL-1 and pZL-2 using the primers 1 and 2, or 3 and 4 (Table I). These PCR products were digested with SalI and BamHI or BglII, and inserted into BamHI and SalI sites of the expression vector pGEX-4T-2 (Amersham Bioscience), respectively. The resulting plasmids were named pGEX-1p and pGEX-2p, and transformed into Escherichia coli XL-1 Blue cells. The overexpression and purification with glutathione-Sepharose 4B (Amersham Bioscience) of the GST-PpRPOT1 and GST-PpRPOT2 fusion proteins were performed according to the manufacturer's directions. The fusion protein eluted from the column was further purified by gel filtration with the Superdex 75 column (Amersham Bioscience) that had been equilibrated with PBS buffer. Purified proteins were used to immunize guinea pigs. Polyclonal antisera were obtained, and the IgG fraction (after ammonium sulfate fractionation) was used in the immunoblot analysis.
For immunoblot analysis, intact plastids and mitochondria were isolated from P. patens as described above. SDS-PAGE and immunoblotting were performed using a 7.5% or 20% polyacrylamide gel as described in a previous paper (Sato et al., 1998).
GFP-Fusion Plasmids
Either of the plasmids pZL-1 or pZL-2 that contained cloned cDNA encoding PpRPOT1 and PpRPOT2 (Kabeya et al., 2002), respectively, were used as templates for PCR amplification. TP1-GFP and TP2-GFP containing the 5′-upstream sequence of the first AUG codon were amplified, respectively, using primers 5 and 32, or 13 and 32. TP1M2-GFP and TP2M2-GFP containing the 5′-upstream sequence of the second AUG codon (but downstream the first AUG codon) were amplified using primers 5 and 6, or 13 and 14. TP1rM1-GFP and TP2rM1-GFP, having TL from the pea (Pisum sativum) RBCS 3A gene in place of the 5′-upstream sequence of the first AUG codon, were generated using primers 5 and 7, or 13 and 15. TP1rM2-GFP and TP2rM2-GFP, having TL in place of the 5′-upstream sequence of the second AUG codon, were generated using primers 5 and 8, or 13 and 16. TP1rM48I-GFP, containing TL and Met-48-to-Ile mutation, was prepared by combining by PCR (with primers 5 and 7), the two partial fragments obtained by amplification using either primers 7 and 10, or primers 5 and 9. TP2rM36I-GFP, containing TL and Met-36-to-Ile mutation, was generated likewise by two successive PCR reactions, using primers 13, 15, 17, and 18. TP1+1-GFP, containing no 5′-upstream sequence of the first AUG codon, was amplified using primers 5 and 11. TP1−10-GFP, containing 10-nucleotide sequence in the 5′-upstream region of the first AUG codon, was amplified using primers 5 and 12. These PCR products were digested with NcoI and SalI and inserted into the SalI-NcoI sites of sGFPS65T (Chiu et al., 1996), which contains a synthetic GFP gene with S65T mutation and optimized codon usage for plants under the control of the CaMV 35S promoter. The plasmids pPpMADS-TP1GFP and pPpMADS-TP2GFP were constructed by inserting the 5′-upstream sequence of the first AUG codon plus the full-length transit peptide of PpRPOT1 or PpRPOT2 and a DNA fragment containing the GFP coding sequence into an expression vector pPpMADS2-7133 with E7133 promoter (Mitsuhara et al., 1996).
The DNA fragments related to AtRpoT;2 were amplified from Arabidopsis (cv Columbia) genomic DNA (laboratory stock). To construct AtRpoT;2-GFP, the 5′-upstream sequence of the first AUG codon plus the full-length transit peptide sequence was amplified using primers 19 and 21. To construct AtRpoT;2+1-GFP, the sequence coding for the full-length transit peptide was amplified using primers 20 and 21. These PCR products were digested with NcoI and SalI and inserted into the SalI-NcoI sites of sGFPS65T. These constructs were used in transient expression experiments (data not shown). The DNA fragments beginning from the CaMV 35S promoter and ending at the NOS terminator were obtained from the plasmids AtRpoT;2-GFP or AtRpoT;2+1-GFP, and were inserted into the HindIII-EcoRI sites of pBI101. These plasmids were named pBI-AtRpoT;2GFP and pBI-AtRpoT;2+1GFP, respectively.
Transient Expression of GFP-Fusion Constructs in the Moss
Thirty micrograms of the GFP-fusion plasmids were introduced into the protoplasts of P. patens by polyethylene glycol-mediated transformation (Schaefer, 1994; Nishiyama et al., 2000).
GUS-Fusion Plasmids and Measurement of Translation Activity with GUS
To generate the DNA fragment PpRPOT1u1-GUS and PpRPOT2u1-GUS containing the 5′-upstream sequence of the first AUG codon, the fragment containing the 5′-upstream sequence of the first AUG codon was amplified with primers 23 and 32 using the pZL-1 plasmid as a template, or with primers 27 and 32 using the pZL-2 plasmid as a template. The fragment containing uidA gene was amplified with primers 22 and 31, or 26 and 31, using the pBI101 plasmid as a template. Then the two fragments in each combination were combined by a second PCR with primers 31 and 32. To generate the DNA fragment PpRPOT1u2-GUS and PpRPOT2u2-GUS containing the 5′-upstream sequence of the second AUG codon (but downstream the first AUG codon), the fragment containing the 5′-upstream sequence of the second AUG codon was amplified with primers 6 and 25 using the pZL-1 as a template or with primers 14 and 29 using the pZL-2 as a template, while the fragment containing the uidA gene was amplified with primers 24 and 31, or 28 and 31, using the pBI101 plasmid as a template. Then, the two fragments in each combination were connected by amplification with primers 6 and 31 or primers 14 and 31. The DNA fragment TL-GUS containing the TL from the pea RBCS 3A gene and the GUS coding sequence was amplified with the primers 30 and 31 using the pBI101 plasmid as a template. These PCR products were digested with SalI and NotI and inserted in place of the GFP coding sequence of sGFPS65T, keeping the CaMV 35S promoter and the NOS terminator unchanged.
One of these plasmids (15 μg) and sGFPS65T as an internal standard were introduced into the moss protoplasts as described above. One day after transformation, GUS activity of the protoplasts was determined according to Jefferson et al. (1987), while the amount of GFP protein was determined by immunoblot with anti-GFP antibody (Invitrogen, Carlsbad, CA).
A two-sample t test comparing GUS activity of pUC18 and various GUS constructs was used to calculate P value. A P value of less than 0.05 was considered significant.
Stable Transformation
P. patens was transformed according to Schaefer (1994). To obtain stable transformants, pPpMADS-TP1GFP digested with NotI was introduced into the protoplasts. Transformed protoplasts were incubated for 4 d on BCDAT medium and then transferred to BCDAT medium containing 50 mg L−1 G418 (Sigma, St. Louis) for 3 weeks. The selected plants were transferred onto a medium without G418 and allowed to grow for 7 d. Then, they were transferred again onto the selection medium. After the second selection, stably transformed P. patens was confirmed by PCR analysis.
Arabidopsis was transformed according to Bechtold et al. (1993) using Agrobacterium tumefaciens strain EHA105 containing pBI-AtRpoT;2GFP or pBI-AtRpoT;2+1GFP. Transformed Arabidopsis were selected on the Murashige and Skoog medium (Murashige and Skoog, 1962) containing 1.5% Suc, 50 mg L−1 kanamycin, and 100 mg L−1 carbenicillin.
Computational and Phylogenetic Analysis
Database sequences and alignment files were manipulated using the SISEQ package version 1.30 (Sato, 2000). A total of 266 database entries for moss sequences were retrieved directly from the gbpln*.seq files in the GenBank (version 141) release using the getent command. The UTR sequence (20 bases) was extracted for each entry by the cdsnuc command. We finally used 214 UTR sequences for further calculation. SequenceLogo (Schneider and Stephens, 1990) was prepared using the alpro and makelogo programs, which were downloaded from Tom Schneider's web site (http://www.lecb.ncifcrf.gov/∼toms/) and compiled locally for Power PC G5 running under MacOS X 10.3. The information content values in the resulting logo file in ASCII postscript format were used as a scoring matrix (the values were in fact multiplied by 2.5 for drawing as SequenceLogo with a height of 5.0 cm). The score was calculated by adding the value for the corresponding nucleotide at each position from −10 to −1 with respect to A of initiation codon.
For phylogenetic analysis, alignments of amino acid sequences were constructed by ClustalX program version 1.81 (Thompson et al., 1994) with final manual adjustment. The N-terminal part was excluded from the alignment by the getclu command of SISEQ because it was highly variable. The programs used for constructing phylogenetic trees by the neighbor-joining method were PROTDIST, NEIGHBOR, SEQBOOT, and CONSENSE of the PHYLIP package (Felsenstein, 1988). Graphical representation of phylogenetic tree was made by the njplot program (Perrière and Gouy, 1996).
Acknowledgments
We thank Drs. Y. Kobayashi and Y. Niwa, University of Shizuoka, for kindly providing us with sGFPS65T plasmid, and Drs. R. Kofuji (currently at Kanazawa University) and M. Hasebe, National Institute for Basic Biology, for providing us with pPpMADS2-7133 plasmid as well as for invaluable advice in moss transformation. We are also grateful to Dr. M. Fujiwara for critical reading of the manuscript.
This work was supported in part by Grants-in-Aid from the Japan Society for the Promotion of Science (grant nos. 13440234 and 15370017 to N.S. and grant no. 16–8650 to Y.K.).
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.105.059501.
References
- Baba K, Schmidt L, Espinosa-Ruiz A, Villarejo A, Shiina T, Gardestrom P, Sane AP, Bhalerao RP (2004) Organellar gene transcription and early seedling development are affected in the rpoT;2 mutant of Arabidopsis. Plant J 38: 38–48 [DOI] [PubMed] [Google Scholar]
- Bailey-Serres J (1999) Selective translation of cytoplasmic mRNAs in plants. Trends Plant Sci 4: 142–148 [DOI] [PubMed] [Google Scholar]
- Bechtold N, Ellis J, Pelletier G (1993) In planta Agrobacterium mediated gene transfer by infiltration of adult Arabidopsis thaliana plants. C R Acad Sci Paris 316: 1194–1199 [Google Scholar]
- Cermakian N, Ikeda TM, Cedergren R, Gray MW (1996) Sequences homologous to yeast mitochondrial and bacteriophage T3 and T7 RNA polymerases are widespread throughout the eukaryotic lineage. Nucleic Acids Res 24: 648–654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cermakian N, Ikeda TM, Miramontes P, Lang BF, Gray MW, Cedergren R (1997) On the evolution of the single-subunit RNA polymerases. J Mol Evol 45: 671–681 [DOI] [PubMed] [Google Scholar]
- Chang CC, Sheen J, Bligny M, Niwa Y, Lerbs-Mache S, Stern DB (1999) Functional analysis of two maize cDNAs encoding T7-like RNA polymerases. Plant Cell 11: 911–926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu W-I, Niwa Y, Zeng W, Hirose T, Kobayashi H, Sheen J (1996) Engineered GFP as vital reporter in plants. Curr Biol 6: 325–330 [DOI] [PubMed] [Google Scholar]
- Felsenstein J (1988) Phylogenies from molecular sequences: inference and reliability. Annu Rev Genet 22: 521–565 [DOI] [PubMed] [Google Scholar]
- Gray MW (1992) The endosymbiont hypothesis revisited. Int Rev Cytol 141: 233–357 [DOI] [PubMed] [Google Scholar]
- Gray MW (1993) Origin and evolution of organellar genomes. Curr Opin Genet Dev 3: 884–890 [DOI] [PubMed] [Google Scholar]
- Hand JM, Szabo LJ, Vasconcelos AC, Cashmore AR (1989) The transit peptide of a chloroplast thylakoid membrane protein is functionally equivalent to a stromal-targeting sequence. EMBO J 8: 3195–3206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto K, Sato N (2001) Characterization of the mitochondria nad7 gene in Physcomitrella patens: similarity with angiosperm nad7 genes. Plant Sci 160: 807–815 [DOI] [PubMed] [Google Scholar]
- Hedtke B, Börner T, Weihe A (1997) Mitochondrial and chloroplast phage-type RNA polymerases in Arabidopsis. Science 277: 809–811 [DOI] [PubMed] [Google Scholar]
- Hedtke B, Börner T, Weihe A (2000) One RNA polymerase serving two genomes. EMBO Rep 1: 435–440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedtke B, Meixner M, Gillandt S, Richter E, Böerner T, Weihe A (1999) Green fluorescent protein as a marker to investigate targeting of organellar RNA polymerases of higher plants in vivo. Plant J 17: 557–561 [DOI] [PubMed] [Google Scholar]
- Howe CJ, Beanland TJ, Larkum AWD, Lockhart PJ (1992) Plastid origins. Trends Ecol Evol 7: 378–383 [DOI] [PubMed] [Google Scholar]
- Ikeda TM, Gray MW (1999) Identification and characterization of T3/T7 bacteriophage-like RNA polymerase sequences in wheat. Plant Mol Biol 40: 567–578 [DOI] [PubMed] [Google Scholar]
- Jefferson RA, Kavanagh TA, Bevan MW (1987) GUS fusions: beta-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J 6: 3901–3907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joshi CP, Zhou H, Huang X, Chiang VL (1997) Context sequences of translation initiation codon in plants. Plant Mol Biol 35: 993–1001 [DOI] [PubMed] [Google Scholar]
- Kabeya Y, Hashimoto K, Sato N (2002) Identification and characterization of two phage-type RNA polymerase cDNAs in the moss Physcomitrella patens: implication of recent evolution of nuclear-encoded RNA polymerase of plastids in plants. Plant Cell Physiol 43: 245–255 [DOI] [PubMed] [Google Scholar]
- Keegstra K, Cline K (1999) Protein import and routing systems of chloroplasts. Plant Cell 11: 557–570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ko K, Cashmore AR (1989) Targeting of proteins to the thylakoid lumen by the bipartite transit peptide of the 33 kd oxygen-evolving protein. EMBO J 8: 3187–3194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi Y, Dokiya Y, Sugita M (2001. a) Dual targeting of phage-type RNA polymerase to both mitochondria and plastids is due to alternative translation initiation in single transcripts. Biochem Biophys Res Commun 289: 1106–1113 [DOI] [PubMed] [Google Scholar]
- Kobayashi Y, Dokiya Y, Sugiura M, Niwa Y, Sugita M (2001. b) Genomic organization and organ-specific expression of a nuclear gene encoding phage-type RNA polymerase in Nicotiana sylvestris. Gene 279: 33–40 [DOI] [PubMed] [Google Scholar]
- Kobayashi Y, Dokiya Y, Kumazawa Y, Sugita M (2002) Non-AUG translation initiation of mRNA encoding plastid-targeting phage-type RNA polymerase in Nicotiana sylvestris. Biochem Biophys Res Commun 299: 57–61 [DOI] [PubMed] [Google Scholar]
- Kozak M (1987) An analysis of 5′-noncoding sequences from 699 vertebrate mRNAs. Nucleic Acids Res 15: 8125–8148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozak M (1991) An analysis of vertebrate mRNA sequences: intimations of translational control. J Cell Biol 115: 887–903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kusumi K, Yara A, Mitsui N, Tozawa Y, Iba K (2004) Characterization of a rice nuclear-encoded plastid RNA polymerase gene OsRpoTp. Plant Cell Physiol 45: 1194–1201 [DOI] [PubMed] [Google Scholar]
- Lütcke HA, Chow KC, Mickel FS, Moss KA, Kern HF, Scheele GA (1987) Selection of AUG initiation codons differs in plants and animals. EMBO J 6: 43–48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathews DE, Durbin RD (1990) Tagetitoxin inhibits RNA synthesis directed by RNA polymerases from chloroplasts and Escherichia coli. J Biol Chem 265: 493–498 [PubMed] [Google Scholar]
- Mathews DH, Sabina J, Zuker M, Turner DH (1999) Expanded sequence dependence of thermodynamic parameters improves prediction of RNA secondary structure. J Mol Biol 288: 911–940 [DOI] [PubMed] [Google Scholar]
- Mitsuhara I, Ugaki M, Hirochika H, Ohshima M, Murakami T, Gotoh Y, Katayose Y, Nakamura S, Honkura R, Nishimiya S, et al (1996) Efficient promoter cassettes for enhancer expression of foreign genes in dicotyledonous and monocotyledonous plants. Plant Cell Physiol 37: 49–59 [DOI] [PubMed] [Google Scholar]
- Murashige T, Skoog F (1962) A revised medium for rapid growth and bioassays with tobacco tissue cultures. Physiol Plant 15: 473–497 [Google Scholar]
- Neupert W (1997) Protein import into mitochondria. Annu Rev Biochem 66: 863–917 [DOI] [PubMed] [Google Scholar]
- Nishiyama T, Fujita T, Shin-I T, Seki M, Nishide H, Uchiyama I, Kamiya A, Carninci P, Hayashizaki Y, Shinozaki K, et al (2003) Comparative genomics of Physcomitrella patens gametophytic transcriptome and Arabidopsis thaliana: implication for land plant evolution. Proc Natl Acad Sci USA 100: 8007–8012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishiyama T, Hiwatashi Y, Sakakibara I, Kato M, Hasebe M (2000) Tagged mutagenesis and gene-trap in the moss, Physcomitrella patens by shuttle mutagenesis. DNA Res 7: 9–17 [DOI] [PubMed] [Google Scholar]
- Nomata T, Kabeya Y, Sato N (2004) Cloning and characterization of glycine-rich RNA-binding protein cDNAs in the moss Physcomitrella patens. Plant Cell Physiol 45: 48–56 [DOI] [PubMed] [Google Scholar]
- Peeters NM, Chapron A, Giritch A, Grandjiean O, Lancelin D, Lhomme T, Vivrel A, Small I (2000) Duplication and quadruplication of Arabidopsis thaliana cysteinyl- and asparaginyl-tRNA synthetase genes of organellar origin. J Mol Evol 50: 413–423 [DOI] [PubMed] [Google Scholar]
- Pedersen AG, Nielsen H (1997) Neural network prediction of translation initiation sites in eukaryotes: perspectives for EST and genome analysis. Proc Int Conf Intell Syst Mol Biol 5: 226–233 [PubMed] [Google Scholar]
- Perrière G, Gouy M (1996) WWW-query: an on-line retrieval system for biological sequence banks. Biochimie 78: 364–369 [DOI] [PubMed] [Google Scholar]
- Richter U, Kiessling J, Hedtke B, Decker E, Reski R, Börner T, Weihe A (2002) Two RpoT genes of Physcomitrella patens encode phage-type RNA polymerases with dual targeting to mitochondria and plastids. Gene 290: 95–105 [DOI] [PubMed] [Google Scholar]
- Sakai A, Saito C, Inada N, Kuroiwa T (1998) Transcriptional activities of the chloroplast-nuclei and proplastid-nuclei isolated from tobacco exhibit different sensitivities to tagetitoxin: implication of the presence of distinct RNA polymerase. Plant Cell Physiol 39: 928–934 [DOI] [PubMed] [Google Scholar]
- Sakamoto W, Kondo H, Murata M, Motoyoshi F (1996) Altered mitochondrial gene expression in a maternal distorted leaf mutant of Arabidopsis induced by chloroplast mutator. Plant Cell 8: 1377–1390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato N (2000) SISEQ: manipulation of multiple sequence and large database file for common platforms. Bioinformatics 16: 180–181 [DOI] [PubMed] [Google Scholar]
- Sato N, Albrieux C, Joyard J, Douce R, Kuroiwa T (1993) Detection and characterization of a plastid envelope DNA-binding protein which may anchor plastid nucleoids. EMBO J 12: 555–561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato N, Misumi O, Shinada Y, Sasaki M, Yoine M (1997) Dynamics of localization and protein composition of plastid nucleoid in light-grown pea seedlings. Protoplasma 200: 163–173 [Google Scholar]
- Sato N, Nakayama M, Hase T (2001) The 70-kDa major DNA-compacting protein of the chloroplast nucleoid is sulfite reductase. FEBS Lett 487: 347–350 [DOI] [PubMed] [Google Scholar]
- Sato N, Ohshima K, Watanabe A, Ohta N, Nishiyama Y, Joyard J, Douce R (1998) Molecular characterization of the PEND protein, a novel bZIP protein present in the envelope membrane that is the site of nucleoid replication in developing plastids. Plant Cell 10: 859–872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaefer D (1994) Molecular genetic approaches to the biology of the moss Physcomitrella patens. PhD thesis. University of Lausanne, Switzerland, http://www.unil.ch/lpc/docs/DSThesis.htm
- Schatz G, Dobberstein B (1996) Common principles of protein translocation across membranes. Science 271: 1519–1526 [DOI] [PubMed] [Google Scholar]
- Schneider TD, Stephens RM (1990) Sequence logos: a new way to display consensus sequences. Nucleic Acids Res 18: 6097–6100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sidorov VA, Kasten D, Pang SZ, Hajdukiewicz PT, Staub JM, Nehra NS (1999) Technical Advance: Stable chloroplast transformation in potato: use of green fluorescent protein as a plastid marker. Plant J 19: 209–216 [DOI] [PubMed] [Google Scholar]
- Soll J, Tien R (1998) Protein translocation into and across the chloroplastic envelope membranes. Plant Mol Biol 38: 191–207 [PubMed] [Google Scholar]
- Sugiura C, Kobayashi Y, Aoki S, Sugita C, Sugita M (2003) Complete chloroplast DNA sequence of the moss Physcomitrella patens: evidence for the loss and relocation of rpoA from the chloroplast to the nucleus. Nucleic Acids Res 15: 5324–5331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson JT, Higgins DG, Gibson TJ (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 22: 4673–4680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Heijne G (1986) Mitochondrial targeting sequences may form amphiphilic helices. EMBO J 5: 1335–1342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Loon AP, Eilers M, Baker A, Verner K (1988) Transport of proteins into yeast mitochondria. J Cell Biochem 36: 59–71 [DOI] [PubMed] [Google Scholar]
- Yamauchi K (1991) The sequence flanking translational initiation site in protozoa. Nucleic Acids Res 19: 2715–2720 [DOI] [PMC free article] [PubMed] [Google Scholar]