Abstract
Selenium (Se) plays an indispensable role in human nutrition and has been implicated to have important health benefits, including being a cancer preventative agent. While different forms of Se vary in their anticarcinogenic efficacy, Se-methylselenocysteine (SeMSC) has been demonstrated to be one of the most effective chemopreventative compounds. Broccoli (Brassica oleracea var. italica) is known for its ability to accumulate high levels of Se with the majority of the selenoamino acids in the form of Se-methylselenocysteine. Therefore, it serves as a good model to study the regulation of SeMSC accumulation in plants. A cDNA encoding selenocysteine Se-methyltransferase, the key enzyme responsible for SeMSC formation, was cloned from broccoli using a homocysteine S-methyltransferase gene probe from Arabidopsis (Arabidopsis thaliana). This clone, designated as BoSMT, was functionally expressed in Escherichia coli, and its identity was confirmed by its substrate specificity in the methylation of selenocysteine. The BoSMT gene represents a single copy sequence in the broccoli genome. Examination of BoSMT gene expression and SeMSC accumulation in response to selenate, selenite, and sulfate treatments showed that the BoSMT transcript and SeMSC synthesis were significantly up-regulated in plants exposed to selenate but were low in plants supplied with selenite. Simultaneous treatment of selenate with selenite significantly reduced SeMSC production. In addition, high levels of sulfate suppressed selenate uptake, resulting in a dramatic reduction of BoSMT mRNA level and SeMSC accumulation. Our results reveal that SeMSC accumulation closely correlated with the BoSMT gene expression and the total Se status in tissues and provide important information for maximizing the SeMSC production in this beneficial vegetable plant.
Selenium (Se) is an essential micronutrient for animals and humans, although it was once known only for its toxicity (Draize and Beath, 1935; Schwarz and Foltz, 1957). Se is a component of many enzymes and proteins in mammals (Kryukov et al., 2003). The nutritional function of Se is fulfilled by selenoenzymes/selenoproteins such as glutathione peroxidases involved in antioxidant protection (Rotruck et al., 1973), thioredoxin reductases that mediate redox regulation (Tamura and Stadtman, 1996), and iodothyronine 5′-deiodinase involved in hormonal regulation of metabolism (Larsen and Berry, 1995). Se constitutes the active sites of these selenoenzymes as the noncanonical amino acid, selenocysteine, and is crucial for their biological functions (Stadtman, 1996; Driscoll and Copeland, 2003).
In addition to its nutritional essentiality, Se has been implicated to have important health benefits. These include roles in reducing the incidence of some debilitating disorders, such as in improving male fertility and immune function (McKenzie et al., 2001; Foresta et al., 2002); in reducing viral infection (Beck et al., 2003); and in slowing the aging process (Soriano-Garcia, 2004). More recently, a large body of convincing evidence has indicated that Se acts as a cancer preventative agent when given in pharmacological amounts (Combs and Gray, 1998; Ip, 1998; Fleming et al., 2001; Whanger, 2004). A clinical trial with 1,312 Americans showed that Se supplementation reduced the incidence of cancer risks by 63% for prostate cancer, 58% for colon cancer, and 46% for lung cancer (Clark et al., 1996).
While various forms of Se offer different degrees of protection against carcinogenesis, some monomethylated forms of Se, such as Se-methylselenocysteine (SeMSC), have been shown to provide superior chemoprotective effects against cancer (Ip et al., 1991; Ganther, 1999; Ip et al., 2000; Whanger, 2002). SeMSC is easily converted to methyl selenol, an active anticancer agent, and has much reduced toxicity and low body accumulation (Medina et al., 2001; Finley et al., 2004). Such properties make this compound especially beneficial as a chemopreventative agent since increased body accumulation of Se at only 5- to 10-fold above supranutrational levels is toxic. Among various plant species, a few of them, such as a Se hyperaccumulator, Astragalus bisulcatus, and a secondary Se accumulator, broccoli (Brassica oleracea var. italica), have been found to accumulate high levels of Se with SeMSC accounting for the majority of selenoamino acids (Cai et al., 1995; Pickering et al., 2003). Thus, these plants serve as good models to investigate the molecular and biochemical basis of accumulation of this functional form of Se in plants.
In higher plants, Se is mainly taken up from soil in the form of selenate (SeO42−) or selenite (SeO32−). Se is believed to be metabolized via the sulfur (S) assimilation pathway since plants are unable to discriminate between Se and S due to their similar chemical properties (Terry et al., 2000; Ellis and Salt, 2003). While selenite is passively taken up into plants, selenate enters plant cells through a process of active transport mediated by sulfate transporters and directly competes with uptake of sulfate (Arvy, 1993; Hopper and Parker, 1999). The reduction of selenate and selenite to selenide and subsequent coupling with O-acetylserine result in the formation of selenocysteine and selenomethionine. Both of these selenoamino acids can be nonspecifically incorporated into proteins in place of Cys and Met, which contributes to Se toxicity in Se nonaccumulators (Brown and Shrift, 1982). In Se hyperaccumulators and some secondary accumulators, selenocysteine is primarily metabolized and transformed into various nonproteinogenic selenoamino acids, such as SeMSC, γ-glutamyl SeMSC, and Se-cystathionine (Terry et al., 2000; Whanger, 2002). The accumulation of these nontoxic selenoamino acids has been suggested to be the basis of Se tolerance in Se accumulators (Brown and Shrift, 1981; Neuhierl et al., 1999). Further methylation of the methylated selenocysteine or selenomethionine produces volatile Se compounds, an area that has been studied extensively for phytoremediation of Se-contaminated soils (Terry et al., 2000; Tagmount et al., 2002).
SeMSC is synthesized from selenocysteine and S-methylmethionine by the enzyme, selenocysteine Se-methyltransferase (SMT). A gene encoding SMT from A. bisulcatus (AbSMT) was successfully cloned (Neuhierl et al., 1999). This SMT enzyme belongs to a class of methyltransferases involved in metabolism of S-methylmethionine. It shares significant primary sequence homology with homocysteine S-methyltransferases (HMT) from Arabidopsis (Arabidopsis thaliana; Ranocha et al., 2000). Although both SMT and HMT catalyze methyl transfer using S-methylmethionine as the methyl donor, they exhibit remarkable Se-containing (for SMT) and S-containing (for HMT) substrate preference as a methyl acceptor in vitro (Neuhierl and Böck, 1996; Ranocha et al., 2000).
SeMSC constitutes the major peak of selenoamino acids in Se-enriched broccoli (Cai et al., 1995) and is the primary form of Se found in Se-enriched broccoli sprouts (Finley et al., 2001; Sugihara et al., 2004). Studies by Finley and coworkers (Finley et al., 2000, 2001; Finley and Davis, 2001; Davis et al., 2002) provide convincing evidence for the role of high Se broccoli in reducing cancer risk. Thus, development of approaches to increase the accumulation of SeMSC in broccoli may greatly enhance its health-promoting properties (Finley, 2003). In this study, we describe the cloning and characterization of the gene encoding SMT from broccoli. The SMT gene expression and SeMSC accumulation in response to different forms and concentrations of Se and sulfate treatments, as well as changes in plant Se status, were examined.
RESULTS
Isolation and Characterization of a cDNA Encoding SMT from Broccoli
To clone the SMT gene from broccoli, a cDNA library was constructed starting with mRNA from selenate-treated florets. Although a SMT gene from A. bisulcatus (AbSMT) has been cloned (Neuhierl et al., 1999), this gene was not suitable as a heterologous probe to isolate SMT from broccoli since the AbSMT gene specific probe did not hybridize well to the genomic DNA of broccoli (data not shown). BLAST searches revealed that AbSMT shared high sequence similarity with several methyltransferase genes. The most closely related sequence was the Arabidopsis AtHMT2 gene (Ranocha et al., 2000), which shared 68% sequence identity with AbSMT. Southern hybridization showed that AtHMT2 hybridized to the broccoli genomic DNA digested with various enzymes as a single band. Thus, AtHMT2 was used as a probe to isolate the cDNA encoding SMT from broccoli.
Screening of the broccoli cDNA library resulted in isolation of 15 positive clones. Sequence analysis of all these clones identified 3 different full-length cDNAs showing 78.2%, 84.6%, and 52.6% nucleotide sequence identity to AtHMT2. To examine whether any of these cDNA clones encoded an SMT, the function of the encoding proteins was analyzed by heterologous expression in Escherichia coli. The coding regions of these full-length cDNAs were subcloned in pTriplEx2 vector (CLONTECH, Palo Alto, CA) and transformed into the E. coli strain MTD123 (ΔyagD ΔmetE ΔmetH; Thanbichler et al., 1999), which lacks Met synthase and HMT activity and thus exhibits a very low background for SMT enzyme activity assay. In addition, the coding regions of AbSMT as well as AtHMT1 and AtHMT2 were also inserted in pTriplEx2 vector and transformed along with the empty vector into MTD123 cells as positive and negative controls. Both AtHMT1 and AtHMT2 constructs were able to complement MTD123 cells and grew well in M9 medium supplied with S-methylmethionine, which indicates that they were functional in our analysis system. Interestingly, AbSMT and 2 of the broccoli clones also grew in M9 medium supplied with S-methylmethionine. The vector alone was not able to complement the MTD123 cells.
Enzyme extracts prepared from MTD123 cells containing these plasmids were assayed for SMT activity using selenocysteine and S-methylmethionine as substrates in a semiquantitative assay. As expected, AbSMT, the positive control for the enzyme activity assay, catalyzed the methyl transfer to form SeMSC. Similarly, protein extract of a broccoli cDNA clone exhibiting 84.6% sequence identity with AtHMT2 was found to have the SMT enzyme activity in methylation of selenocysteine to produce SeMSC. This cDNA clone was designated as BoSMT. No SMT enzyme activity was detected in protein extracts from the other 2 broccoli full-length cDNA clones or from AtHMT1, AtHMT2, and the empty vector controls (data not shown).
The BoSMT cDNA (GenBank accession no. AY817737) contains an open reading frame of 1,041 bp that encodes 347 amino acid residues with a calculated molecular mass of approximately 37.9 kD. The protein sequence of BoSMT shows 65% identity with AbSMT (Fig. 1). It shares 53% and 86% identity, respectively, with AtHMT1 and AtHMT2, and 38% to E. coli HMT (YagD). BoSMT contains a consensus sequence of GGCC for a possible zinc-binding motif near the C-terminal and a conserved Cys residue upstream of the zinc-binding motif as other related methyltransferases (Ranocha et al., 2000). BoSMT has no obvious chloroplast or mitochondrial targeting sequence. Southern-blot analysis showed that the BoSMT gene specific probe hybridized to broccoli genomic DNA digested with various restriction enzymes mainly as a single band (Fig. 2), which indicates that BoSMT most probably represents a single copy gene in the broccoli genome.
Figure 1.
Sequence alignment of the deduced amino acids of BoSMT (AY817737) with those of related proteins. Conserved residues are shaded in black. Dashes depict gaps that were added for optimum alignment. The arrowhead indicates the third conserved Cys residue. The bar shows the possible zinc-binding motif. AbSMT, A. bisulcatus SMT (CAA10368); AtHMT1, AtHMT2, and AtHMT3, Arabidopsis HMT (AAF23821, AAF23822, and AAG10301); YagD, E. coli HMT (Q47690).
Figure 2.
Southern-blot analysis of BoSMT. Genomic DNA (10 μg) from broccoli leaves was digested with restriction enzymes as indicated, separated on a 0.8% agarose gel, and hybridized with the BoSMT gene specific probe. DNA size markers are noted in the left.
In Vitro Biochemical Characterization of BoSMT Expressed in E. coli
BoSMT enzyme extract from MTD123 cells catalyzed methyl transfer from S-methylmethionine to selenocysteine. The optimal pH was found to be 7.0 (data not shown). To examine the substrate preference of BoSMT, a quantitative enzyme activity assay was carried out. Since no methyl-14C-derivative of S-methylmethionine is commercially available, radiolabeled substrate of [methyl-14C]adenosylmethionine was used as a methyl donor (Neuhierl et al., 1999). The rates of methylation of a number of potential methyl acceptor substrates by BoSMT were compared (Table I). BoSMT showed the highest preference for dl-selenocysteine as a methyl acceptor. The BoSMT exhibited some activity with dl-Cys and l-Cys for the production of methylcysteine when the substrate concentration was high (10 mm). It also utilized homocysteine. These combined results suggest that the isolated gene encodes a SMT but also exhibits other methyltransferase activities in vitro.
Table I.
Specific activities of BoSMT enzyme with different methyl acceptors
Protein extracts prepared from the E. coli strain MTD123 (ΔyagD ΔmetE ΔmetH) expressing BoSMT were assayed for the enzyme activity using [methyl-14C]adenosylmethionine as the methyl donor and various Se- and S-containing substrates as methyl acceptors at the concentrations indicated in parenthesis. Data are means of five replicates ± sd.
| Methyl Acceptor (Concentration) | Methyltransferase Activity |
|---|---|
| nmol min−1 mg−1 protein | |
| dl-Selenocysteine (0.5 mm) | 5.1 ± 1.7a |
| l-Selenocysteine (0.5 mm) | 2.4 ± 0.6a |
| dl-Cystein (10 mm) | 1.8 ± 1.1b |
| l-Cysteine (10 mm) | 1.1 ± 0.6b |
| dl-Homocysteine (10 mm) | 1.0 ± 0.7c |
Product of SeMSC.
Product of methylcysteine.
Product of Met.
Increased Se Tolerance in E. coli Expressing BoSMT
To examine whether BoSMT provided protection against the toxic effect of Se in E. coli, the growth of MTD123 cells expressing BoSMT, AbSMT, or the empty vector was compared in the presence or absence of 100 μm of selenate or selenite. After 16 h of incubation with 100 μm of selenate, the growth of vector control cells was significantly reduced (94% reduction of growth). In contrast, the cells containing BoSMT or AbSMT showed Se tolerance with only a 52% or 46% reduction in growth (Fig. 3A). Similarly, enhanced Se tolerance was also observed when the cells expressing BoSMT and AbSMT were exposed to 100 μm of selenite. Selenite appears more toxic than selenate to the bacterial cell growth (Fig. 3A; pTriplEx2 control). The E. coli expressing SMT showed better protection against selenite than selenate as the SMT transformed cells had higher ratio of growth on selenite versus selenate than the empty vector control.
Figure 3.
Effects of BoSMT expression on (A) bacterial tolerance to selenate and selenite and (B) on bacterial total Se accumulation. Bacterial strain MTD123 (ΔyagD ΔmetE ΔmetH) transformed with pTriplEx2 empty vector, BoSMT, and AbSMT were grown overnight and the cell density in the absence (C) and presence of 100 μm Na2SeO4 (SeO4) or 100 μm Na2SeO3 (SeO3), and total Se accumulation in the presence of 100 μm Na2SeO4 were measured as detailed in “Materials and Methods.” The experiments were repeated at least three times with four replications. Vertical lines indicate sds.
To examine if the bacteria expressing BoSMT had increased levels of Se accumulation, the total Se concentrations were determined after exposing the cells to 100 μm of selenate for 16 h. The cell lines expressing SMT accumulated significantly higher levels of total Se than the empty vector control cells (Fig. 3B). These results established that expression of BoSMT significantly enhanced Se tolerance with an increased level of total Se accumulation in E. coli.
BoSMT Gene Expression Was Dramatically Up-Regulated by Selenate Treatment
SMT is the key enzyme for SeMSC synthesis. BoSMT gene expression in response to different forms and concentrations of Se and sulfate was examined by northern-blot analysis (Fig. 4). In leaf tissue of broccoli, the level of BoSMT mRNA was extremely low or undetectable in plants that were not exposed to Se. Treating the plants with 40 μm of selenate dramatically increased BoSMT gene expression (Fig. 4A). In contrast, only a slight accumulation of the transcript was observed for plants treated with the same concentration of selenite (Fig. 4A).
Figure 4.
Northern-blot analysis of BoSMT gene expression in plants treated with Se and sulfate. The mRNA samples (1 μg) from leaves (A–C) and florets (D) of control and treated plants were gel-fractionated and blotted. The filters were hybridized with 32P-labeled BoSMT and actin-8 control probe. A, BoSMT transcript accumulation in leaves of plants supplied with Na2SeO4 and Na2SeO3 at concentrations as indicated. B, BoSMT gene expression with increasing concentrations of selenate supply. C, BoSMT mRNA level in 40 μm selenate-treated plants with increasing concentrations of MgSO4 supply. D, BoSMT mRNA levels in florets of plants supplied with Na2SeO4 and Na2SeO3 at concentrations as indicated.
When the plants were exposed to increasing concentrations of selenate, the accumulation of BoSMT mRNA increased and reached a maximum expression at selenate levels between 20 and 40 μm. Higher concentrations (>40 μm) of selenate treatment showed a negative effect on BoSMT mRNA accumulation (Fig. 4B). At very high selenate concentrations (i.e. 100 μm), the expression of BoSMT mRNA was substantially reduced. The reduction of BoSMT gene expression in response to high concentrations (>40 μm) of selenate treatment correlated well with the general plant growth status, which showed growth inhibition progressively from slight inhibition to a more severe retardation in growth from 40 μm to 100 μm of selenate treatments.
Examination of the effect of sulfate on BoSMT gene expression showed that the addition of high levels of sulfate in growth medium significantly reduced the accumulation of BoSMT transcripts in leaves of plants. BoSMT transcript abundance became undetectable in plants exposed to 10 mm of sulfate in the presence of 40 μm of selenate (Fig. 4C). Interestingly, an increase in gene expression was observed in plants that were exposed to 0.1 mm sulfate when compared to those exposed to selenate alone.
Expression of BoSMT in florets in response to the same selenate, selenite, and sulfate treatments was also examined. In general, the patterns of BoSMT transcript accumulation were remarkably similar to those seen in leaves. The accumulation of BoSMT mRNA in florets of plants treated with 20 μm of selenate or selenite is shown in Figure 4D. In addition, accumulation of BoSMT transcripts was also observed in root tissues exposed to selenate (data not shown).
High Levels of Total Se Were Accumulated in Broccoli Exposed to Selenate
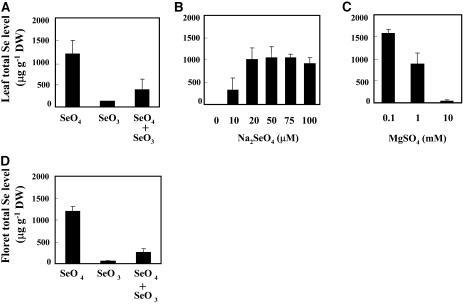
To examine whether the BoSMT gene expression corresponded with the plant Se status, the total Se concentrations in leaf and floret tissues of broccoli exposed to different forms and concentrations of Se and sulfate were determined. When plants were treated with 40 μm of selenate, the leaf tissue of broccoli accumulated high levels of Se. In contrast, significantly lower levels of Se were found in plants treated with selenite compared with selenate (130 versus 1,200 μg g−1 dry weight; Fig. 5A). Plants exposed to a combination of 20 μm of selenite and 20 μm of selenate suppressed Se accumulation to approximately 40% of that found in plants exposed to selenate alone (Fig. 5, A and B).
Figure 5.
Total Se accumulation in leaves (A–C) and florets (D) of broccoli in response to Se and sulfate treatments. A, Se concentrations in leaves of plants treated with 40 μm Na2SeO4 (SeO4), 40 μm Na2SeO3 (SeO3), and 20 μm of each SeO4 and SeO3. B, Influence of increasing concentrations of selenate supply on Se accumulation. C, Se levels in 40 μm selenate-treated plants supplied with increasing concentrations of MgSO4. D, Total Se accumulation in florets of plants exposed to 20 μm Na2SeO4 (SeO4), 20 μm Na2SeO3 (SeO3), and 10 μm of each SeO4 and SeO3. The experiments were repeated twice with at least three replications. Error bars indicate sds.
As the concentrations of selenate supply were increased in the nutrient solution, the leaf Se levels increased and reached a plateau of approximately 1,200 μg g−1 dry weight at 20 to 40 μm of selenate supply (Fig. 5B).
Sulfur in the form of sulfate competes with selenate uptake and dramatically reduces Se accumulation (Zayed et al., 1998). The addition of high levels of sulfate (i.e. 1 mm or 10 mm) showed a strong inhibitory effect on Se accumulation. At 10 mm, the Se level was less than 3% of that found in plants exposed only to the same concentration of selenate. Such a reduction is most probably due to competition at the sulfate transporters for sulfate versus selenate. Interestingly, when sulfate was supplied at low concentration (i.e. 0.1 mm) to the selenate-treated plants, Se accumulation was stimulated in leaves (Fig. 5C).
The same experiments were repeated to examine the total Se accumulation in floret tissue of broccoli. Similar overall patterns and levels of Se accumulation in response to the different treatments were found in florets compared with leaf tissue. Figure 5D shows the total Se accumulation in florets in response to 20 μm of selenate or selenate, and a combination of 10 μm of selenate and selenite.
Selenate-Treated Plants Synthesized Significantly More SeMSC
To investigate the relationship between the SeMSC accumulation and BoSMT gene expression, SeMSC from leaf and floret tissues of broccoli treated with different forms and concentrations of Se and sulfate was extracted and analyzed by HPLC. A number of extraction methods including the use of 50 mm HCl, hot 80% methanol, 100% methanol, water, and 0.43 mm phosphate buffer, pH 7.0, were compared. Extraction with 50 mm HCl was found to give consistently better results in our SeMSC quantification assay. Figure 6 depicts a typical HPLC elution profile. The SeMSC peak was very well separated from other amino acids and it was coeluted with the SeMSC standard. Inductively coupled, argon-plasma (ICP) analysis of the HPLC fraction confirmed that it contained Se.
Figure 6.
A typical HPLC elution profile of AccQTag derivatives of amino acids at an emission wavelength of 395 nm. SeMSC peak and methylcysteine (MeCys) peak are as indicated. S, Commercial SeMSC or MeCys standard; N, nontreated plants; T, selenate-treated plants.
Plants that were not treated with Se contained undetectable amounts of SeMSC (Figs. 6 and 7B). Treating the plants with 40 μm of selenate resulted in high levels of SeMSC accumulation in leaf tissue, reaching a level close to 1 μmol g−1 fresh weight. Plants treated with the same concentration of selenite, however, produced less than 8% of SeMSC found in the selenate-treated plants. Exposure of plants to 20 μm of selenate with 20 μm of selenite reduced SeMSC production to approximately 45% of that found in plants treated with only 20 μm of selenate. Such a reduction indicates that there is a negative effect of selenite supply on SeMSC production in broccoli.
Figure 7.
SeMSC accumulation in tissues of broccoli in response to Se and sulfate treatments. SeMSC was separated and quantified by HPLC. A, SeMSC accumulation in leaf tissue of broccoli plants treated with 40 μm Na2SeO4 (SeO4), 40 μm Na2SeO3 (SeO3), and 20 μm of each SeO4 and SeO3. B, Influence of increasing concentrations of selenate supply on SeMSC accumulation in leaves. C, SeMSC levels in leaves of 40 μm selenate-treated plants supplied with increasing concentration of MgSO4. D, SeMSC accumulation in florets of broccoli plants treated with 20 μm Na2SeO4 (SeO4), 20 μm Na2SeO3 (SeO3), and 10 μm of each SeO4 and SeO3. E, Influence of increasing concentrations of selenate supply on SeMSC accumulation in florets. The experiments were repeated at least three times with four replications. Error bars indicate sds.
The effect of increasing selenate supply (from 0–100 μm) on SeMSC accumulation in leaves is shown in Figure 7B. Although BoSMT gene expression started to decrease with selenate exposures above 40 μm, the accumulation of SeMSC continued to increase to a selenate concentration of 75 μm. The leaf tissue accumulated as much as 1.1 μmol g−1 fresh weight of SeMSC, which was equivalent to approximately 7.3 μmol g−1 dry weight.
When 0.1 mm of sulfate was supplied to the selenate-treated plants, the leaves of the plants accumulated higher levels of SeMSC in comparison with plants exposed to selenate alone (Fig. 7C). This finding correlated well with the effect of low sulfate addition on enhanced BoSMT gene expression and total Se accumulation. Since the presence of high concentrations of sulfate (i.e. 1 or 10 mm) significantly reduced Se accumulation and BoSMT gene expression, as expected, addition of high levels of sulfate resulted in dramatically decreased accumulation of SeMSC in leaf tissue (Fig. 7C). At 10 mm of sulfate supply (+40 μm of selenate), the SeMSC level was only about 0.5% of that seen in plants treated with selenate alone.
SeMSC accumulation in floret tissue of broccoli in response to Se and sulfate treatments showed similar patterns of changes as those for leaves. Exposure of plants to 20 μm of selenate also resulted in a higher level of SeMSC production in florets than plants treated with 20 μm of selenite (Fig. 7D). In comparison with leaf tissues, florets accumulated higher levels of SeMSC when plants were exposed to elevated levels of selenate (≥40 μm) for 1 week (Fig. 7E). Although the SeMSC levels were low in florets of broccoli exposed to low concentrations (≤20 μm) of selenate, after an extended period of selenate treatment for more than 2 weeks, the florets accumulated as much SeMSC as those exposed to high levels of selenate at 1-week time period and the plants showed no obvious abnormal growth (data not shown).
DISCUSSION
SeMSC is one of the most effective anticarcinogenic Se compounds (Ip et al., 1991; Ganther, 1999; Ip et al., 2000; Whanger, 2002). It is the major form of selenoamino acids found in Se-enriched broccoli and broccoli sprouts (Cai et al., 1995; Finley et al., 2001; Sugihara et al., 2004). Thus, identification of a broccoli cDNA encoding SMT, the key enzyme involved in the formation of SeMSC, permits a comprehensive study of gene expression in relation to SeMSC production in plants.
A full-length cDNA clone coding BoSMT was successfully identified from a broccoli cDNA library by using a heterologous HMT probe. This BoSMT clone was functionally expressed in E. coli and its identity was confirmed by its positive SMT enzyme activity in catalyzing SeMSC production. The deduced amino acid sequence of BoSMT shares high sequence identity with both the AbSMT from A. bisulcatus (Neuhierl et al., 1999) and the AtHMTs from Arabidopsis (Ranocha et al., 2000). BoSMT lacks obvious chloroplast or mitochondrial targeting sequences and appears to be a cytosolic enzyme. Although the cytosolic location of BoSMT has yet to be confirmed in the plant, such location is consistent with other published data showing that methylation of selenocysteine or selenomethionine as well as metabolism involving the synthesis of S-methylmethionine most likely takes place in the cytosol (Bourgis et al., 1999; Ranocha et al., 2000). In addition, the BoSMT protein resembles the related methyltransferases in that it contains a possible zinc-binding motif (GGCC) near the C-terminal region and an upstream conserved Cys residue. This suggests that BoSMT may have a zinc cofactor for binding and/or activating the selenol group of selenocysteine. We noticed that zinc appeared to stimulate BoSMT activity in the in vitro enzyme activity assay (data not shown). Further, it seems that the GGCC binding motif may be important for methyltransferase activity as another full-length cDNA from broccoli that shares more than 87% sequence identity with BoSMT but contains no GGCC sequence showed no SMT activity and no complementation in MTD123 cells.
The recombinant BoSMT enzyme extracts prepared from E. coli contained significantly higher enzyme activity than that of crude extracts from the plant tissues (data not shown). Thus, expression of it in E. coli provided a means to explore substrate specificity of the enzyme in vitro. Among the potential methyl acceptors tested, BoSMT catalyzed methyl transfer at significantly higher rates with the Se form of substrates compared with their S analogs. dl-Selenocysteine was the best methyl acceptor for BoSMT. Methylation of dl-Cys and l-Cys were at least 2 times less effective even when the substrate concentrations used in the assay were 20 times higher than that of selenocysteine (10 mm versus 0.5 mm, respectively). This result can be interpreted as at least a 40-fold higher substrate specificity of BoSMT for dl-selenocysteine than the methylation activities for S analog methyl acceptors. Thus, the data demonstrated that the isolated BoSMT encodes an SMT. In addition, it also has Cys methyltransferase activity in vitro despite the fact that its methylation rates on S substrates were lower than that on Se substrates.
Previously, Neuhierl and Böck (1996) reported that the AbSMT exhibited approximately 1,000-fold higher substrate specificity for the Se substrates than the S analogs. The higher affinity in discrimination of Se against S analogs for AbSMT than BoSMT provides a molecular evidence for Se accumulation difference between a Se hyperaccumulator and a Se secondary accumulator.
SMT enzyme has been proposed to play a crucial role in Se detoxification (Brown and Shrift, 1981; Neuhierl et al., 1999). Its high efficiency in methylation of selenocysteine prevents the nonspecific incorporation of selenoamino acids into proteins and other S-containing compounds, which in turn would interfere with the normal S metabolism and function. Overexpression of the AbSMT in Arabidopsis and Indian mustard (Brassica juncea) dramatically increased Se tolerance in these plants (Ellis et al., 2004; LeDuc et al., 2004), which confirms its role in Se detoxification. Here, we demonstrated that E. coli expressing the BoSMT protein also exhibited enhanced Se tolerance with increased levels of total Se accumulation in the cells. While the processes involved in transport and reduction of Se may influence Se tolerance in E. coli, it is conceivable that SMT functions in Se detoxification by diverting the cellular selenocysteine away from metabolism of S-containing proteins.
Unlike AbSMT that expressed constitutively regardless of Se treatment or tissue age (Pickering et al., 2003), BoSMT expression was extremely low or undetectable in plants not exposed to Se and dramatically up-regulated by selenate treatment in broccoli. The expression in leaf tissues was under developmental control with highest mRNA transcripts found in very young leaves (data not shown). The different expression patterns between AbSMT and BoSMT may be due to the fact that A. bisulcatus is a native plant grown on seleniferous soils, while broccoli is not restricted to Se containing soils.
In our studies, we found that BoSMT mRNA levels reached a maximum between 20 and 40 μm of selenate treatment and then decreased at higher concentrations of selenate supply (>40 μm). The reduced level of BoSMT transcript in plants exposed to high concentrations of selenate could be due to a general reduction of protein synthesis in Se-inhibited plant (Banuelos et al., 1997). While the BoSMT transcript level decreased in plants exposed to selenate concentrations greater than 40 μm, SeMSC accumulation continued to increase until plants were exposed to selenate levels of 75 μm. One possible reason for this response could be that the BoSMT protein was quite stable and subject to low levels of turnover at high selenate exposures. Thus, although the BoSMT gene expression was reduced, there would be plenty of the enzyme to interact with the substrate to form SeMSC. An additional possible reason could be that the immediate metabolic steps involved in the formation of volatile Se compounds were less active when the plant growth was inhibited by high selenate exposure, but further investigation is necessary to confirm this hypothesis. At 75 μm of selenate treatment, the plants could accumulate about 570 μg g−1 dry weight of SeMSC (7.3 μmol g−1 dry weight × 79 Se molecular weight) and 1,100 μg g−1 dry weight of total Se in leaves of broccoli. Based on the data, an estimation of up to 50% of the total Se accumulated in young leaf tissue of broccoli was converted into SeMSC by BoSMT within 1 week of selenate exposure. Previously, a study using the x-ray absorption spectroscopy showed that the majority of Se found in leaf tissue of selenate-treated broccoli remained as selenate (Zayed et al., 1998). This might have been the result of broccoli plants treated with low concentrations of selenate for a short time as we noticed here in our studies. At 10 μm of selenate treatment, SeMSC only accounted for less than 2% of total Se (Fig. 5B and Fig. 7B). We have found that SeMSC production was significantly stimulated in plants either by exposure to high selenate levels for short time periods (1 week) or exposure to lower selenate levels (10 or 20 μm) for longer time periods (3 weeks). In addition, similar as is the case with A. bisulcatus (Pickering et al., 2003), the young leaves of broccoli accumulated much higher levels of SeMSC than older leaves (data not shown). Previous reports on analysis of Se speciation of some closely related secondary Se accumulators showed that different Brassica spp. appeared to selectively accumulate different forms of Se. SeMSC was found to be the major form of Se accumulated in leaves of Chinese cabbage (Brassica oleracea capitata), composing 26.7% of the total Se species (Hamilton, 1975), while it accounted for only 3% of total Se in selenate-treated Indian mustard (Kahakachchi et al., 2004).
The lesser effect of selenite on BoSMT gene expression and SeMSC accumulation in leaf and floret tissues may be due to low rates of selenite translocation to the shoot and florets. Unlike selenate, selenite is believed to be inefficiently transported from the root to the shoot (De Souza et al., 1998; Zayed et al., 1998; Terry et al., 2000). As a result, significantly lower levels of Se accumulation were found in selenite-treated broccoli. Thus, less Se substrate would be available to BoSMT for SeMSC synthesis. Similarly, exposure of plants to a mixture of selenite and selenate suppressed Se accumulation and also resulted in low SeMSC accumulation. The availability of Se in cells appears to play a major role in determining the amount of SeMSC production in broccoli treated with selenite. In addition, as selenite has been shown to be phytotoxic to plant growth (Hopper and Parker, 1999), it is possible that selenite may inhibit BoSMT synthesis and activity through a general toxicity response. Ellis et al. (2004) demonstrated that overexpression of AbSMT in Arabidopsis resulted in a remarkable increase of total Se and SeMSC accumulation in plants exposed to selenite but not in plants treated with selenate. This was suggested to be due to the inefficient reduction of selenate to selenite in Arabidopsis that forms a rate-limiting step for Se assimilation. Broccoli appears to have relatively high efficiency in reduction of selenate to selenite. As a result, exposure of plants to selenate induced BoSMT expression and led to an increased accumulation of SeMSC in young leaf and floret tissues of broccoli.
The dramatic reduction of BoSMT message level and SeMSC accumulation in response to exposure to high levels of sulfate is most likely also due to decreased levels of available Se in plant tissues. The uptake of selenate into plant cells is mediated by active sulfate transporters; thus, sulfate and selenate directly compete for transport (Terry et al., 2000; Shibagaki et al., 2002; White et al., 2004). The selective uptake of Se and sulfate in broccoli depends on the concentrations of Se and sulfate present in the solution bathing the roots. High sulfate supply conferred a strong inhibitory effect on Se accumulation in selenate-treated plants. In line with the observed dramatic decrease in Se accumulation, here we showed that the BoSMT gene expression and SeMSC production were also significantly reduced. Such a reduction is most likely to be correlated with limited amounts of Se available in the cells. Interestingly, when plants were exposed to an additional 0.1 mm sulfate (in addition to the 0.2 mm sulfate normally found in the nutrient solution), Se accumulation and SeMSC production were increased in broccoli leaf tissue. Quite speculatively, it is possible that although S deficiency has been shown to induce expression of sulfate assimilation genes, appropriate levels of plant S status may activate sulfate assimilation at the protein level, which would stimulate more incorporation of Se into selenocysteine for SeMSC production. Another possible explanation would be the relative ratio of Se to sulfate that favors Se accumulation. It is possible that the sulfate concentration used in the nutrient solution could be suboptimal for broccoli despite the fact that the broccoli plants used in the studies showed no sulfate deficient symptoms and genes involved in sulfate metabolisms such as a high-affinity sulfate transporter, ATP sulfurylase, and Ser acetyltransferase (Leustek, 2002) exhibited no observed expression difference among the plants treated or untreated with Se and sulfate (data not shown). Slightly more sulfate fertilization could improve growth of Se-treated plants, thus promoting more efficient Se uptake, translocation, and assimilation.
The Arabidopsis plants overexpressing AbSMT were found to produce approximately equal concentration methylcysteine and SeMSC, and the AbSMT was suggested to have significant methyltransferase activity using both Cys and selenocysteine as substrates in vivo (Ellis et al., 2004). In our study, we also noticed that the accumulation of methylcysteine was closely correlated with SeMSC production in broccoli and was at significant comparable levels with SeMSC (Fig. 6). Further, the BoSMT also catalyzed the methylation of Cys in vitro to produce methylcysteine, although the specific activity of it in methylation of Cys was much lower than that with the Se analogs. These results support the previous hypothesis of Ellis et al. (2004) that the SMT may be involved in the synthesis of both SeMSC and methylcysteine. Since broccoli is considered to be a S accumulator and is not restricted to high Se soils in their natural environment, it is likely that physiologically the BoSMT catalyzes the methylation of Cys when the plants grow in no or low Se soils, but its activity on Cys may not be maximized as we did not observe methylcysteine accumulation in the control plants (Fig. 6). The BoSMT activity on S substrate might be significantly stimulated following the activation of BoSMT by appropriate levels of Se in vivo.
In summary, we have cloned a BoSMT gene from broccoli that is responsible for the formation of SeMSC, the bioactive form of Se against carcinogenesis. We have demonstrated that selenate is more effective in inducing BoSMT gene expression and SeMSC production and that the addition of selenite to selenate supply significantly reduces SeMSC accumulation. The effectiveness of Se in broccoli in reducing cancer risk makes the popular vegetable an excellent source of supplemental dietary Se (Finley et al., 2004). The isolation of the BoSMT gene will allow up-regulation of BoSMT in broccoli to further enhance SeMSC production for improving its health-promoting properties. In addition, it provides an avenue for down-regulation of BoSMT activity to investigate the effect of alteration of SeMSC formation on the overall Se metabolism in this secondary Se accumulator.
MATERIALS AND METHODS
Plant Materials
Broccoli (Brassica oleracea var. italica) cv Green Comet (G30771) was obtained from the Plant Genetic Resources Unit at Geneva, New York, and used in this study. One-week-old seedlings germinated in a petri dish were transplanted to 2-L pots containing a modified Johnson's solution (Wang et al., 2002) and grown in a greenhouse at 24°C with 16-h daylength for 4 weeks. The nutrient solution was changed every 2 weeks. Three days after replacing with fresh solution at plant age of 4 weeks, Na2SeO4 or Na2SeO3 (Sigma, St. Louis) was added to give a final concentration of 10, 20, 40, 50, 75, or 100 μm. To study the effect of S on Se metabolism, additional MgSO4 was added to the nutrient solution containing 40 μm of selenate to achieve a concentration of 0.1, 1.0, and 10 mm of sulfate in addition to the 0.2 mm of sulfate found in the modified Johnson's solution. One week after the treatments, young leaf samples from at least 5 individual plants were combined, frozen in liquid nitrogen, ground to powder, and stored at −80°C for RNA and DNA extraction as well as for HPLC analysis. For analysis of total Se, the samples were dried at 65°C in an oven for over 3 d. The same treatments were repeated three to four more times. To avoid possible effects of phytovolatilized Se on nearby plants, nontreated control plants were grown in a separate room. For broccoli floret samples, plants were grown hydroponically until proper size of florets were formed, and then the plants were treated with Na2SeO4, Na2SeO3, and MgSO4 as described above.
Isolation of BoSMT Gene from Broccoli
A broccoli cDNA library was constructed using mRNA from selenate-treated florets with the BD SMART cDNA Library Construction kit (CLONTECH). The ligated cDNA in λTriplEx2 cloning vector was packaged with Gigapack II Gold packing extract (Stratagene, La Jolla, CA) according to the manufacturer's instructions. The cDNA library was screened by colony hybridization using radiolabeled AtHMT2 probe (Ranocha et al., 2000). Prehybridization, hybridization, and washing of the filters were performed at 65°C as described (Li et al., 2003). The putative positive clones were sequenced using an ABI Prizm 3100 Genetic Analyzer (Applied Biosystems, Foster City, CA). DNA sequence analysis was performed using the Lasergene program (DNASTAR, Madison, WI).
Functional Characterization of BoSMT by Heterologous Expression in Escherichia coli
The BoSMT coding sequence along with the other two putative positive clones were amplified from the original cDNA clones with TaKaRa Ex Taq polymerase (Panvera, Madison, WI) using primers containing KpnI and XbaI restriction enzyme sites. The amplified KpnI-XbaI fragments were cloned into pTriplEx2 and transformed into E. coli strain MTD123 (ΔyagD ΔmetE ΔmetH; Thanbichler et al., 1999), which lacks Met synthase and HMT activity and thus exhibits a very low background for SMT enzyme activity assay. In addition, the coding sequences of AtHMT1 and AtHMT2 from Arabidopsis (Arabidopsis thaliana; Ranocha et al., 2000) and AbSMT from Astragalus bisulcatus (Neuhierl et al., 1999) were also cloned in pTriplEx2 and transformed along with the vector only into MTD123 cells as positive and negative controls. All of the constructs were sequenced to verify the inserts and the nucleotide sequences at the cloning sites.
To test whether these clones function in the MTD123 cells, an individual colony from each construct was grown overnight in M9 medium (Sambrook and Russell, 2001) containing 0.8% Glc, 100 μm S-methylmethionine, and 100 μg mL−1 of ampicillin (Ranocha et al., 2000). The overnight culture was reinoculated in 3 mL of the same medium at an initial OD600 of 0.1 and grown for 16 h at 37°C. Cell growth was measured at OD600. The clones that complemented MTD123 cells gave an OD600 of >0.6, while those that did not complement the cells showed no changes in the initial OD600 value.
Enzyme Extraction and Semiquantitative and Quantitative Enzyme Activity Assay
BoSMT enzyme from E. coli cells was extracted using the method described by Ranocha et al. (2000) with some modifications. A single colony of BoSMT construct in MTD123 cells was inoculated in 5 mL of M9 medium containing 0.8% Glc, 100 μm l-Met, and 100 μg mL−1 of ampicillin and shaken at 37°C overnight. Two milliliters of the culture was added to 200 mL of the same medium and allowed to grow at 37°C with vigorous shaking until the OD600 reached 0.5 to 0.6. Bacteria cells were collected by centrifugation at 4,000g for 10 min and washed 3 times with a buffer containing 100 mm HEPES-KOH, pH 7.0, 1 mm dithiothreitol, and 10% glycerol. Cell pellets were resuspended in the buffer, broken using a chilled French Pressure Cell Press (Travenol Lab, Silver Spring, MD) at 1,000 psi, and sonicated with a Cell Disruptor 350 (Branson, Danbury, CT) on ice. After centrifugation at 10,000g for 15 min, the supernatant was concentrated with an Amicon Ultra filtration kit (10,000 MWCO, Millipore, Billerica, MA) and used either directly for enzyme activity assays or stored at −80°C until use.
BoSMT enzyme activity assays were conducted following the methods of Neuhierl and Böck (1996) and Böck and Neuhierl (2002) with slight modification. In the semiquantitative assay, a reaction mixture in a total volume of 50 μL was comprised of 50 mm sodium citrate, pH 7.0, 10 mm magnesium acetate, 5 mm dithiothreitol, 5 to 10 μL of enzyme extract, and 0.5 mm selenocysteine (freshly reduced from selenocysteine with 10 times molar excess of sodium borohydride at 25°C for at least 30 min). The reaction was started by the addition of S-methylmethionine. At different time points (i.e. 0, 10, 20, 30, and 60 min), 5 μL of samples were collected and spotted onto silica gel 60 thin-layer chromatography (TLC) plates (EM Science, Gibbstown, NJ). The TLC plates were developed in 1-butanol:acetic acid:water at 4:1:1 (v/v/v), stained with ninhydrin reagent (Sigma), and baked at 80°C until spots were clearly detected. SeMSC product was identified by comparing the spot mobility with that of a commercial standard and quantified using ImageQuant software (Amersham, Piscataway, NJ).
The quantitative BoSMT enzyme assay was carried out in a total volume of 15 μL in an AtmosBag (Aldrich, Milwaukee, WI) filled with N2 gas to avoid substrate oxidation. The reaction mixture was similar to that of the semiquantitative assay except that radiolabeled substrate [methyl-14C]adenosylmethionine (52.7 mCi mmol−1, NEN Life Science Products, Boston) was used as the methyl donor. Se-[14C]methylselenocysteine was detected using a Storm 840 Phosphoimager (Molecular Dynamics, Sunnyvale, CA). Quantification was done by spotting serial dilutions of [methyl-14C]adenosylmethionine onto the same TLC plates as described by Neuhierl and Böck (1996).
Bacterial Se Tolerance Analysis
To examine if the bacteria expressing BoSMT contained an enhanced level of Se tolerance, MTD123 cells expressing BoSMT, AbSMT, or the pTriplEx2 empty vector were grown overnight in M9 media containing 0.8% Glc, 100 μm l-Met, and 100 μg mL−1 of ampicillin. The overnight culture (20 μL) was reinoculated in 3 mL of the same medium, collected at an OD600 of approximately 0.4, and washed with 1 mL of 0.9% NaCl twice. The pretreated cells were adjusted to OD600 of 0.05 and grown in the presence or absence of 100 μm of selenate or selenite at 37°C for 16 h. Cell density (OD600) was then measured.
To examine if the bacteria expressing BoSMT contained an increased level of total Se accumulation, the overnight cultures of MTD123 cells expressing BoSMT, AbSMT, or the pTriplEx2 empty vector were adjusted to OD600 of 0.6 and grown in the same M9 medium in the presence of 100 μm Na2SeO4 at 37°C for 16 h. Cells were harvested, washed 5 times with 18.2 mΩ distilled water, pelleted, fresh weight measured, and then analyzed for total Se with ICP as described below.
Nucleic Acid Analysis
Genomic DNA (10 μg) isolated from leaf tissue of broccoli (Li et al., 2003) was digested with various restriction enzymes, separated on 0.8% agarose gels, and blotted onto Hybond-N+ nylon membranes (Amersham). 32P-labeled BoSMT probe was generated by PCR using a random priming DECAprime II kit (Ambion, Austin, TX) following the manufacturer's instruction. Prehybridization, hybridization, and washing of the membranes were performed at 65°C as described by Li et al. (2003).
Total RNA from combined young leaf and floret tissues of broccoli was extracted using Trizol reagent (Invitrogen, Carlsbad, CA). Messenger RNA was purified from 1 mg of total RNA using the PolyATtract mRNA Isolation System (Promega, Madison, WI). The mRNA (1 μg/sample) was separated and transferred onto Hybond-N+ filters. Equal loading of mRNA was verified by ethidium bromide staining and by probing with a cauliflower actin-8 gene. The filters were prehybridized in ULTRAhyb (Ambion) at 42°C for 1 h and hybridized with a 32P-labeled probe in the same solution overnight. The membranes were washed at 42°C for 2 × 5 min in 2× SSC, 2 × 5 min in 1× SSC, and 1 × 10 min in 0.5× SSC with 0.1% (w/v) SDS.
Quantification of SeMSC by HPLC
The procedures for extraction and analysis of SeMSC were performed according to the method described by Ellis et al. (2004) with some modifications. The plant tissue materials were extracted overnight at 4°C in 50 mm HCl (10:1, v/w) and centrifuged at 12,000g to remove cell debris. AccQTaq derivatives of extracted SeMSC and other amino acids were created using an AccQ-Fluor Reagent kit (Waters, Milford, MA). The samples were separated on a Waters Symmetry C18 reverse-phase column (4.6 × 250 mm) using a Dionex HPLC system (Sunnyvale, CA) equipped with an RF 2000 fluorescence detector. The Waters AccQTag Chemistry Package gradient profile was followed at a flow rate of 1.0 mL min−1 over a 60-min period. SeMSC and methylcysteine in the chromatogram were identified by coelution with the standards (Sigma). Concentration of SeMSC in the samples was calculated based on peak areas and a calibration curve generated with the commercial standard.
Analysis of Total Se by ICP
Dried tissues (approximately 100 mg) of leaves and florets were weighed and acid digested in 1.0 mL HNO3 with 1.5 mL HClO4 at 120°C for 1 h and then at 220°C until HClO4 fumes were observed. The samples were diluted with 18 mΩ water to 15 mL. Total Se in the samples was determined using an ICP trace analyzer emission spectrometer (model ICAP 61E trace analyzer, Thermo Electron, San Jose, CA). The instrument was calibrated with 10% HCIO4 as the low standard and 2.5 μg g−1 Se in a multi-element standard as the high standard. The Se was determined using the 196.0-nm line.
Novel Materials
Upon request, all novel materials described in this publication will be made available in a timely manner for noncommercial research purposes, subject to the requisite permission from any third-party owners of all or parts of the material. Obtaining any permission will be the responsibility of the requestor.
Sequence data from this article have been deposited with the EMBL/GenBank data libraries under accession number AY817737.
Acknowledgments
We gratefully acknowledge Dr. August Böck for kindly providing the bacterial strain MTD123 (ΔyagD ΔmetE ΔmetH). We thank Dr. Shan Lu for helpful suggestions and Ms. Kelly Cosman, Diano O'Halloran, and Mr. Robert Hung for their excellent technical assistance.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.104.056549.
References
- Arvy MP (1993) Selenate and selenite uptake and translocation in bean plants (Phaseolus vulgaris). J Exp Bot 44: 1083–1087 [Google Scholar]
- Banuelos GS, Ajwa HA, Wu L, Guo X, Akohoue S, Zambrzuski S (1997) Selenium-induced growth reduction in brassica land races considered for phytoremediation. Ecotoxicol Environ Saf 36: 282–287 [DOI] [PubMed] [Google Scholar]
- Beck MA, Levander OA, Handy J (2003) Selenium deficiency and viral infection. J Nutr 133: 1463S–1467S [DOI] [PubMed] [Google Scholar]
- Böck A, Neuhierl B (2002) Selenocysteine methyltransferase. Methods Enzymol 347: 203–207 [DOI] [PubMed] [Google Scholar]
- Bourgis F, Roje S, Nuccio ML, Fisher DB, Tarczynski MC, Li C, Herschbach C, Rennenberg H, Pimenta MJ, Shen TL, et al (1999) S-Methylmethionine plays a major role in phloem sulfur transport and is synthesized by a novel type of methyltransferase. Plant Cell 11: 1485–1497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown TA, Shrift A (1981) Exclusion of selenium from proteins of selenium tolerant Astragalus species. Plant Physiol 67: 1051–1053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown TA, Shrift A (1982) Selenium: toxicity and tolerance in higher plants. Biol Rev 57: 59–84 [Google Scholar]
- Cai XJ, Block E, Uden PC, Zhang X, Quimby BD, Sullivan JJ (1995) Allium chemistry: identification of selenoamino acids in ordinary and selenium-enriched garlic, onion, and broccoli using gas-chromatography with atomic-emission detection. J Agric Food Chem 43: 1754–1757 [Google Scholar]
- Clark LC, Combs GF, Turnbull BW, Slate EH, Chalker DK, Chow J, Davis LS, Glover RA, Graham GF, Gross EG, et al (1996) Effects of selenium supplementation for cancer prevention in patients with carcinoma of the skin: a randomized controlled trial. JAMA 276: 1957–1963 [PubMed] [Google Scholar]
- Combs GF Jr, Gray WP (1998) Chemopreventive agents: selenium. Pharmacol Ther 79: 179–192 [DOI] [PubMed] [Google Scholar]
- Davis CD, Zeng HW, Finley JW (2002) Selenium-enriched broccoli decreases intestinal tumorigenesis in multiple intestinal neoplasia mice. J Nutr 132: 307–309 [DOI] [PubMed] [Google Scholar]
- De Souza MP, Pilon-Smits EA, Lytle CM, Hwang S, Tai J, Honma TS, Yeh L, Terry N (1998) Rate-limiting steps in selenium assimilation and volatilization by indian mustard. Plant Physiol 117: 1487–1494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Draize JH, Beath OA (1935) Observations on the pathology of “blind staggers” and “alkali disease”. Am Vet Med Assoc J 86: 753–763 [Google Scholar]
- Driscoll DM, Copeland PR (2003) Mechanism and regulation of selenoprotein synthesis. Annu Rev Nutr 23: 17–40 [DOI] [PubMed] [Google Scholar]
- Ellis DR, Salt DE (2003) Plants, selenium and human health. Curr Opin Plant Biol 6: 273–279 [DOI] [PubMed] [Google Scholar]
- Ellis DR, Sors TG, Brunk DG, Albrecht C, Orser C, Lahner B, Wood KV, Harris HH, Pickering IJ, Salt DE (2004) Production of Se-methylselenocysteine in transgenic plants expressing selenocysteine methyltransferase. BMC Plant Biol 4: 1–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finley JW (2003) Reduction of cancer risk by consumption of selenium-enriched plants: Enrichment of broccoli with selenium increases the anticarcinogenic properties of broccoli. J Med Food 6: 19–26 [DOI] [PubMed] [Google Scholar]
- Finley JW, Davis CD (2001) Selenium (Se) from high-selenium broccoli is utilized differently than selenite, selenate and selenomethionine, but is more effective in inhibiting colon carcinogenesis. Biofactors 14: 191–196 [DOI] [PubMed] [Google Scholar]
- Finley JW, Davis CD, Feng Y (2000) Selenium from high selenium broccoli protects rats from colon cancer. J Nutr 130: 2384–2389 [DOI] [PubMed] [Google Scholar]
- Finley JW, Grusak MA, Keck AS, Gregoire BR (2004) Bioavailability of selenium from meat and broccoli as determined by retention and distribution of 75Se. Biol Trace Elem Res 99: 191–209 [DOI] [PubMed] [Google Scholar]
- Finley JW, Ip C, Lisk DJ, Davis CD, Hintze KJ, Whanger PD (2001) Cancer-protective properties of high-selenium broccoli. J Agric Food Chem 49: 2679–2683 [DOI] [PubMed] [Google Scholar]
- Fleming J, Chose A, Harrison PR (2001) Molecular mechanisms of cancer prevention by selenium compounds. Nutr Cancer 40: 42–49 [DOI] [PubMed] [Google Scholar]
- Foresta C, Flohe L, Garolla A, Roveri A, Ursini F, Maiorino M (2002) Male fertility is linked to the selenoprotein phospholipid hydroperoxide glutathione peroxidase. Biol Reprod 67: 967–971 [DOI] [PubMed] [Google Scholar]
- Ganther HE (1999) Selenium metabolism, selenoproteins and mechanisms of cancer prevention: complexities with thioredoxin reductase. Carcinogenesis 20: 1657–1666 [DOI] [PubMed] [Google Scholar]
- Hamilton JW (1975) Chemical examination of seleniferous cabbage Brassica oleracea capitata. J Agric Food Chem 23: 1150–1152 [DOI] [PubMed] [Google Scholar]
- Hopper JL, Parker DR (1999) Plant availability of selenite and selenate as influenced by the competing ions phosphate and sulfate. Plant Soil 210: 199–207 [Google Scholar]
- Ip C (1998) Lessons from basic research in selenium and cancer prevention. J Nutr 128: 1845–1854 [DOI] [PubMed] [Google Scholar]
- Ip C, Hayes C, Budnick RM, Ganther HE (1991) Chemical form of selenium, critical metabolites, and cancer prevention. Cancer Res 51: 595–600 [PubMed] [Google Scholar]
- Ip C, Thompson HJ, Zhu ZJ, Ganther HE (2000) In vitro and in vivo studies of methylseleninic acid: evidence that a monomethylated selenium metabolite is critical for cancer chemoprevention. Cancer Res 60: 2882–2886 [PubMed] [Google Scholar]
- Kahakachchi C, Boakye HT, Uden PC, Tyson JF (2004) Chromatographic speciation of anionic and neutral selenium compounds in Se-accumulating Brassica juncea (Indian mustard) and in selenized yeast. J Chromatogr 1054: 303–312 [PubMed] [Google Scholar]
- Kryukov GV, Castellano S, Novoselov SV, Lobanov AV, Zehtab O, Guigo R, Gladyshev VN (2003) Characterization of mammalian selenoproteomes. Science 300: 1439–1443 [DOI] [PubMed] [Google Scholar]
- Larsen PR, Berry MJ (1995) Nutritional and hormonal regulation of thyroid hormone deiodinases. Annu Rev Nutr 15: 323–352 [DOI] [PubMed] [Google Scholar]
- LeDuc DL, Tarun AS, Montes-Bayon M, Meija J, Malit MF, Wu CP, AbdelSamie M, Chiang CY, Tagmount A, deSouza M, et al (2004) Overexpression of selenocysteine methyltransferase in Arabidopsis and Indian mustard increases selenium tolerance and accumulation. Plant Physiol 135: 377–383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leustek T (2002) Sulfate metabolism. In CR Somerville, EM Meyerowitz, eds, The Arabidopsis Book, American Society of Plant Biologists, Rockville, MD, pp 1–17 [DOI] [PMC free article] [PubMed]
- Li L, Lu S, O'Halloran DM, Garvin DF, Vrebalov J (2003) High-resolution genetic and physical mapping of the cauliflower high-β-carotene gene Or (orange). Mol Genet Genomics 270: 132–138 [DOI] [PubMed] [Google Scholar]
- McKenzie RC, Rafferty TS, Beckett GJ, Arthur JR (2001) Effects of selenium on immunity and aging. In DL Hatfield, ed, Selenium, Its Molecular Biology and Role in Human Health. Kluwer Academic Publishers, Boston, pp 257–272
- Medina D, Thompson H, Ganther H, Ip C (2001) Se-methylselenocysteine: a new compound for chemoprevention of breast cancer. Nutr Cancer 40: 12–17 [DOI] [PubMed] [Google Scholar]
- Neuhierl B, Böck A (1996) On the mechanism of selenium tolerance in selenium-accumulating plants. Purification and characterization of a specific selenocysteine methyltransferase from cultured cells of Astragalus bisculatus. Eur J Biochem 239: 235–238 [DOI] [PubMed] [Google Scholar]
- Neuhierl B, Thanbichler M, Lottspeich F, Böck A (1999) A family of S-methylmethionine-dependent thiol/selenol methyltransferases. Role in selenium tolerance and evolutionary relation. J Biol Chem 274: 5407–5414 [DOI] [PubMed] [Google Scholar]
- Pickering IJ, Wright C, Bubner B, Ellis D, Persans MW, Yu PE, George GN, Prince RC, Salt DE (2003) Chemical form and distribution of selenium and sulfur in the selenium hyperaccumulator Astragalus bisulcatus. Plant Physiol 131: 1460–1467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranocha P, Bourgis F, Ziemak MJ, Rhodes D, Gage DA, Hanson AD (2000) Characterization and functional expression of cDNAs encoding methionine-sensitive and -insensitive homocysteine S-methyltransferases from Arabidopsis. J Biol Chem 275: 15962–15968 [DOI] [PubMed] [Google Scholar]
- Rotruck JT, Pope AL, Ganther HE, Hafeman DG, Hoekstra WG (1973) Selenium biochemical role as a component of glutathione peroxidase. Science 179: 588–590 [DOI] [PubMed] [Google Scholar]
- Sambrook J, Russell DW (2001) Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY
- Schwarz K, Foltz CM (1957) Selenium as an integral part of factor 3 against dietary necrotic liver degeneration. J Am Chem Soc 70: 3292–3293 [PubMed] [Google Scholar]
- Shibagaki N, Rose A, McDermott JP, Fujiwara T, Hayashi H, Yoneyama T, Davies JP (2002) Selenate-resistant mutants of Arabidopsis thaliana identify Sultr1;2, a sulfate transporter required for efficient transport of sulfate into roots. Plant J 29: 475–486 [DOI] [PubMed] [Google Scholar]
- Soriano-Garcia M (2004) Organoselenium compounds as potential therapeutic and chemopreventive agents: a review. Curr Med Chem 11: 1657–1669 [DOI] [PubMed] [Google Scholar]
- Stadtman TC (1996) Selenocysteine. Annu Rev Biochem 65: 83–100 [DOI] [PubMed] [Google Scholar]
- Sugihara S, Kondo M, Chihara Y, Yuji M, Hattori H, Yoshida M (2004) Preparation of selenium-enriched sprouts and identification of their selenium species by high-performance liquid chromatography-inductively coupled plasma mass spectrometry. Biosci Biotechnol Biochem 68: 193–199 [DOI] [PubMed] [Google Scholar]
- Tagmount A, Berken A, Terry N (2002) An essential role of S-adenosyl-L-methionine : L-methionine S-methyltransferase in selenium volatilization by plants. Methylation of selenomethionine to selenium-methyl-L-selenium-methionine, the precursor of volatile selenium. Plant Physiol 130: 847–856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura T, Stadtman TC (1996) A new selenoprotein from human lung adenocarcinoma cells: purification, properties, and thioredoxin reductase activity. Proc Natl Acad Sci USA 93: 1006–1011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terry N, Zayed AM, De Souza MP, Tarun AS (2000) Selenium in higher plants. Annu Rev Plant Physiol Plant Mol Biol 51: 401–432 [DOI] [PubMed] [Google Scholar]
- Thanbichler M, Neuhierl B, Böck A (1999) S-methylmethionine metabolism in Escherichia coli. J Bacteriol 181: 662–665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Garvin DF, Kochian LV (2002) Rapid induction of regulatory and transporter genes in response to phosphorus, potassium, and iron deficiencies in tomato roots. Evidence for cross talk and root/rhizosphere-mediated signals. Plant Physiol 130: 1361–1370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whanger PD (2002) Selenocompounds in plants and animals and their biological significance. J Am Coll Nutr 21: 223–232 [DOI] [PubMed] [Google Scholar]
- Whanger PD (2004) Selenium and its relationship to cancer: an update. Br J Nutr 91: 11–28 [DOI] [PubMed] [Google Scholar]
- White PJ, Bowen HC, Parmaguru P, Fritz M, Spracklen WP, Spiby RE, Meacham MC, Mead A, Harriman M, Trueman LJ, et al (2004) Interactions between selenium and sulphur nutrition in Arabidopsis thaliana. J Exp Bot 55: 1927–1937 [DOI] [PubMed] [Google Scholar]
- Zayed A, Lytle CM, Terry N (1998) Accumulation and volatilization of different chemical species of selenium by plants. Planta 206: 284–292 [Google Scholar]