Abstract
cDNAs encoding a high-affinity sulfate transporter and an adenosine 5′-phosphosulfate reductase from potato (Solanum tuberosum L. cv Désirée) have been cloned and used to examine the hypothesis that sulfate uptake and assimilation is transcriptionally regulated and that this is mediated via intracellular O-acetylserine (OAS) pools. Gas chromotography coupled to mass spectrometry was used to quantify OAS and its derivative, N-acetylserine. Treatment with external OAS increased sulfate transporter and adenosine 5′-phosphosulfate reductase gene expression consistent with a model of transcriptional induction by OAS. To investigate this further, the Escherichia coli gene cysE (serine acetyltransferase EC 2.3.1.30), which synthesizes OAS, has been expressed in potato to modify internal metabolite pools. Transgenic lines, with increased cysteine and glutathione pools, particularly in the leaves, had increased sulfate transporter expression in the roots. However, the small increases in the OAS pools were not supportive of the hypothesis that this molecule is the signal of sulfur (S) nutritional status. In addition, although during S starvation the content of S-containing compounds decreased (consistent with derepression as a mechanism of regulation), OAS pools increased only following extended starvation, probably as a consequence of the S starvation. Taken together, expression of these genes may be induced by a demand-driven model, via a signal from the shoots, which is not OAS. Rather, the signal may be the depletion of intermediates of the sulfate assimilation pathway, such as sulfide, in the roots. Finally, sulfate transporter activity did not increase in parallel with transcript and protein abundance, indicating additional posttranslational regulatory mechanisms.
Higher plants use inorganic sulfate as their major source of sulfur (S), which is reduced and assimilated to Cys. Sulfate is actively taken up into the root by high-affinity sulfate transporters, and transport of sulfate around the plant depends upon a gene family of sulfate transporters (Hawkesford, 2003). The cloning of high-affinity (Smith et al., 1997; Takahashi et al., 2000), low-affinity (Smith et al., 1995; Takahashi et al., 2000), and organelle-specific (Kataoka et al., 2004b) sulfate transporters has provided molecular probes with which to study the regulation of S uptake and the control of S assimilation.
Reductive sulfate assimilation is a multistep pathway in which sulfate is activated, reduced to sulfide, and incorporated into Cys, which may be used for the synthesis of other S-containing compounds (Leustek and Saito, 1999; Hawkesford and Wray, 2000; Leustek et al., 2000). Reductive assimilation occurs in most tissues, but predominantly in plastids of green tissue where the supply of the required ATP and redox equivalent is plentiful. The final step of Cys biosynthesis is catalyzed by the Ser acetyltransferase (SAT; EC 2.3.1.30)/O-acetylserine (thiol) lyase (OASTL; EC 4.2.99.8) bienzyme complex (Bogdanova and Hell, 1997). SAT acetylates l-Ser using acetyl-CoA to form O-acetylserine (OAS), which in a reaction catalyzed by OASTL is then combined with sulfide to form l-Cys (Saito, 1999). Activity of SAT is regulated indirectly by OAS, as SAT is active only when complexed with OASTL, and this complex is disrupted by free OAS (Droux et al., 1998). OASTL is virtually inactive when complexed; however, the concentration of OASTL is far in excess of SAT, and the free active OASTL is responsible for the production of Cys (Ruffet et al., 1994; Droux et al., 1998).
The control of expression of genes for sulfate transporters and several components of the S-assimilatory pathway may be mediated by feedback loops involving key metabolites of Cys biosynthesis. Sulfate transporter expression has been shown to be influenced by both S availability and by the exogenous addition of OAS (Smith et al., 1997). Sulfate uptake and transport to the shoots of tobacco (Nicotiana tabacum) plants was repressed by glutathione and l-Cys (Herschbach and Rennenberg, 1994). Similar observations have been made for ATP sulfurylase and adenosine 5′-phosphosulfate (APS) reductase in Lemna minor (Neuenschwander et al., 1991). Cys (Bolchi et al., 1999) or glutathione (Lappartient et al., 1999) have been suggested to repress expression of ATP-sulfurylase and sulfate uptake. APS reductase also has a major influence on the control of flux through the pathway (Tsakraklides et al., 2002; Vauclare et al., 2002).
From such observations a model of a regulatory circuit controlling sulfate uptake and assimilation has been proposed (Hawkesford and Smith, 1997). In this model, expression of genes involved in uptake and assimilation are under both a positive regulation by OAS, which accumulates when insufficient sulfide is available to utilize the OAS for Cys synthesis, and under a negative feedback control from some product of sulfate assimilation when S supply is in excess. Evidence for this model is however limited by the inability to distinguish between OAS acting as an indirect signal of S availability or as a limiting substrate for Cys synthesis. Increasing OAS availability will increase the demand for the supply of reduced S by depleting pools of reduction pathway intermediates, any of which could be signals of demand.
The roles of both OASTL and SAT in determining Cys synthesis have been examined by transgenic approaches. Chloroplastic OASTL has been overexpressed in tobacco, and although enzyme activity increased, there was no effect on Cys content. However, when isolated chloroplasts were incubated with OAS or OAS together with sulfite or sulfate, there was an increase in Cys production. Addition of sulfate or sulfite alone had no effect (Saito et al., 1994). This suggests that SAT rather than OASTL may be rate limiting for Cys biosynthesis and, hence, S assimilation. When SAT has been overexpressed in tobacco (Błaszczyk et al., 1999; Wirtz and Hell, 2003) or in potato (Solanum tuberosum; Harms et al., 2000), both Cys and glutathione content of plant tissue were increased, further supporting the position of this enzyme as a limiting step. Excess synthesis of OAS, at least in the cytosol, is prevented by the feedback inhibition of cytosolic SAT by Cys (Noji and Saito, 2002).
In this paper, we report the cloning of a high-affinity sulfate transporter in potato roots and investigate the possible role of OAS on the control of expression of the sulfate uptake and assimilation using plants overexpressing cysE (Harms et al., 2000).
RESULTS
Characteristics of cDNA Encoding a Potato Sulfate Transporter
A cDNA encoding a sulfate transporter has been isolated from potato roots by reverse transcription (RT)-PCR (AF309643). The cDNA (StST1) was sequenced and found to be 2,439 nucleotides in length and contained a single long open reading frame that encoded a 657-amino acid polypeptide. The polypeptide encoded by StST1 has significant identity (E < 0.01; Pearson, 2000) with sequences of other plant H+/sulfate cotransporters. The highest identity is with the high-affinity plant H+/sulfate cotransporter from tomato (Lycopersicon esculentum), LeST1 (97% amino acid identity), indicating that this transporter belongs to the designated Group 1 type of transporter (Hawkesford, 2003; Howarth et al., 2003) and is highly likely to be of the high-affinity type. A partial cDNA fragment (987 bp; 329 amino acids representing >73% of the polypeptide sequence) for a potato APS reductase (AJ506751) was also isolated (StAPR).
Effect of External OAS Supply
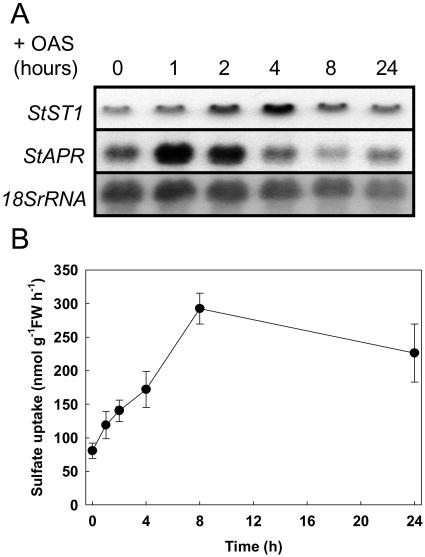
Application of OAS to the roots of hydroponically grown plants resulted in increased abundance of StST1 and StAPR (Fig. 1A). In both cases, increased abundance was transient with maximal mRNA being observed at 4 and 1 h after OAS addition for StST1 and StAPR, respectively. Concomitant with the increased sulfate transporter transcript, measurable influx capacity increased more than 3-fold after 8 h (Fig. 1B).
Figure 1.
Effect of external OAS supply (0.2 mm) on sulfate transporter and APS reductase mRNA abundance in root and on the sulfate uptake capacity of the roots. A, Northern-blot analysis of the mRNA abundance of StST1 and StAPR in roots; 10 μg of total RNA was loaded in each lane from four pooled plants. Equal loading was verified using an 18SrRNA probe. B, Sulfate uptake capacity of roots measured during a 10-min incubation period. Data are the means ± se of five separate plant samples.
Effect of Overexpressing cysE in Potato
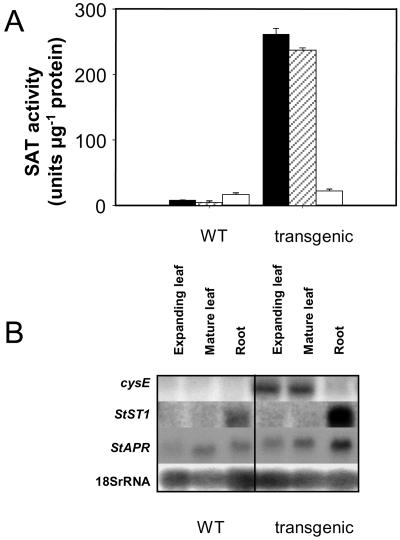
The effect of overexpressing cysE on SAT activity and expression was determined. The SAT activity in leaf tissue of transgenic plants (line 26; Harms et al., 2000) was higher than that of control plants (Fig. 2A). This increase can be correlated directly to the expression of cysE mRNA in the leaves of transgenic plants (Fig. 2B). Although the cysE gene is under the control of the constitutive 35S cauliflower mosaic virus promoter, only a slight increase was seen in the SAT activity and in cysE expression in the roots of the transgenics compared to wild-type plants (Fig. 2, A and B). Differential activity of this constitutive promoter has been noted previously (Holtorf et al., 1995). However, an increase in the expression of StST1, in the roots of the transgenic line compared to wild-type plants, was observed (Fig. 2B). StAPR was also induced to a small extent, particularly in the roots (Fig. 2B). These results were confirmed in an independent transgenic line (line 48; Harms et al., 2000; data not shown).
Figure 2.
SAT activity and mRNA abundance of cysE and StST1 in leaves and roots of nontransformed (WT) and transgenic potato overexpressing cysE. A, SAT activity (unit μg−1 protein), where one unit catalyzes the acetylation of 1 pmol of l-Ser. Data are the means ± se of five separate plant samples. Young (solid bar) and mature (hatched) leaves and roots (white) were sampled from plants grown with 1 mm sulfate for 3 weeks. B, Northern-blot analysis of the mRNA abundance of cysE, StST1, and StAPSR in leaves and roots; 10 μg of total RNA was loaded in each lane. Equal loading was verified using an 18SrRNA probe.
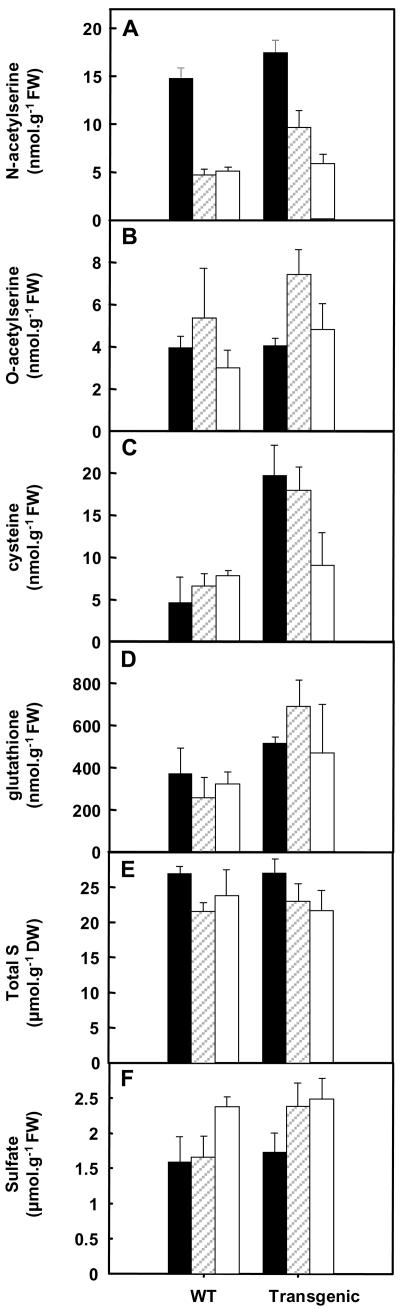
The impact of cysE overexpression on key metabolites of S assimilation was examined (Fig. 3). Steady-state extractable pools of OAS and N-acetylserine (NAS) were similar in both the wild type and the transgenic line (Fig. 3, A and B). This may be expected as under the experimental conditions of adequate S nutrition, all the OAS was converted to Cys and glutathione, which were found to be higher in the transgenic line compared to wild type in all tissues (Fig. 3, C and D), and supports the idea that OAS may limit S assimilation (Saito et al., 1994). No overall differences were observed in the total S and sulfate pools, with the possible exception of an increase in sulfate pools in mature leaves of the transgenic line (Fig. 3, E and F). The free amino acid pools (data not shown) of the transgenic line compared to wild-type plants were also similar.
Figure 3.
Metabolite pools in leaves and roots of nontransformed (WT) and transgenic potato overexpressing cysE. Young (solid bar) and mature (hatched) leaves and roots (white) were sampled from plants grown with 1 mm sulfate for 3 weeks. NAS (A), OAS (B), Cys (C), glutathione (D), total S (E), and sulfate (F) were determined. Data are the means ± se of five separate plant samples except OAS and NAS, which are the means ± se of three separate plant samples.
Effect of S Starvation
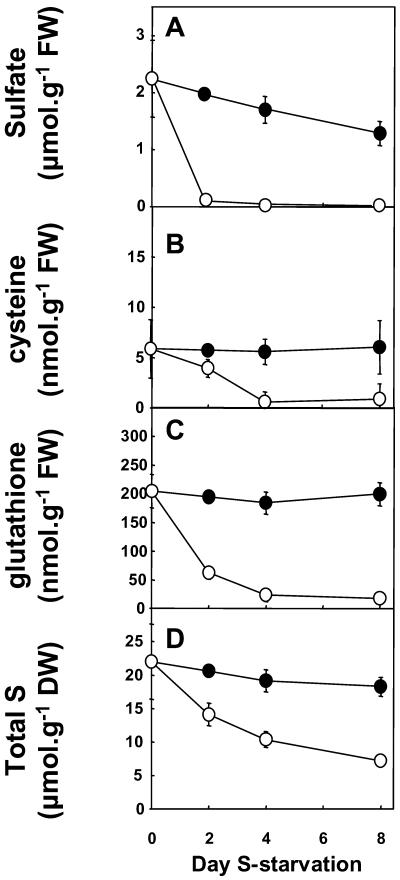
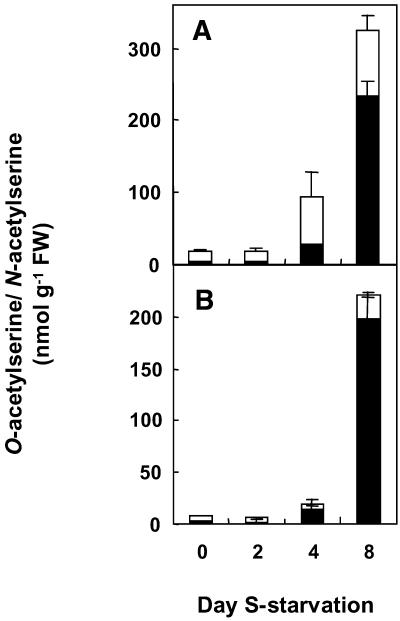
Metabolite pools of the S-assimilatory pathway were examined in roots of the wild type over a time course of S deficiency (Fig. 4). Sulfate, Cys, glutathione, and total S content decreased within 2 d of the imposed S limitation (Fig. 4). Decreased total S content (Fig. 4D) was due mainly to the decrease in sulfate-S content (Fig. 4A). Leaf tissue metabolite pools were also examined and gave similar patterns of reduction in response to S limitation (data not shown). OAS and NAS were measured during the 8-d time course of S starvation (Fig. 5). Both OAS and NAS increased during S starvation in both leaves and roots. OAS and NAS remained low in mature leaf tissues (data not shown). In roots, accumulation of OAS and NAS occurred only at late stages (day 8) of S starvation (Fig. 5B).
Figure 4.
Effect of S starvation on S pools in root tissue of potato. Root tissue was sampled from plants grown with 1 mm sulfate for 3 weeks and then either maintained on 1 mm sulfate (black circles) or transferred to minimal sulfate (20 μm) for 0, 2, 4, or 8 d (white circles). Sulfate (A), Cys (B), glutathione (C), and total S (D) were determined. Data are the means ± se of five separate plant samples.
Figure 5.
Effect of S starvation on the OAS and NAS concentrations in leaf and root tissue of potato. Young leaves (A) and roots (B) were sampled from plants grown with 1 mm sulfate for 3 weeks and then on minimal sulfate (20 μm) for 0, 2, 4, or 8 d. The OAS (black) and NAS (white) concentrations were determined by GC-MS. Data are the means ± se of three separate plant samples.
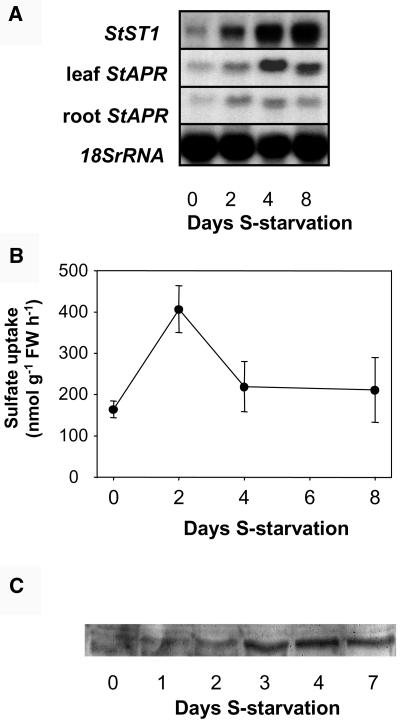
The effect of S starvation on StST1 and StAPR expression, S uptake capacity, and StST1 protein abundance was examined (Fig. 6). As previously seen for other plant species, S starvation leads to an increase in StST1 (in the roots) and StAPR (in both roots and leaves) mRNA abundance (Fig. 6A) and an increase of the S uptake capacity of the roots (Fig. 6B). The increase in the transporter activity was consistently only transient in nature. Using an antibody produced to a StST1-specific oligopeptide, the abundance of the StST1 protein was examined in the plasma membrane fractions isolated from the roots (Fig. 6C). Little or no protein was detected by western blotting until day 3. Approximately equal detectable levels were evident after this time point.
Figure 6.
Effect of S starvation on sulfate uptake capacity, StST1 mRNA abundance, and sulfate transporter protein in potato. Roots were sampled from wild-type plants grown with 1 mm sulfate for 3 weeks and then minimal sulfate (20 μm) for 0, 2, 4, or 8 d. A, Northern-blot analysis of StST1 and StAPR mRNA abundance in roots and StAPR abundance in leaves; 10 μg of total RNA was loaded in each lane. Equal loading was verified using an 18SrRNA probe (leaf data not shown). B, Sulfate uptake capacity of roots measured during a 10-min incubation period. Data are the means ± se of five separate plant samples. C, Western blot of isolated plasma membranes of root tissues probed with the anti-StST antibody. Part of this data was presented in a meeting report (Hawkesford et al., 2003).
Essentially similar data for the effect of S starvation on the cysE overexpressing line were obtained (data not shown).
DISCUSSION
A regulatory model has been proposed that suggests that S starvation leads to an accumulation of OAS and that this accumulation positively regulates ST gene expression (Hawkesford and Smith, 1997; Smith et al., 1997). Recent analytical advances have enabled the measurement of OAS in tissue samples to test this regulatory model. While OAS increased with S starvation (Fig. 5), this occurred after the observed increase in StST1 mRNA abundance in the roots (Fig. 6).
OAS spontaneously converts to NAS in a nonenzymatic reaction at a rate of about 1% per minute at neutral pH (Flavin and Slaughter, 1965), and NAS has been suggested to be the inducer in prokaryotic systems (Ostrowski and Kredich, 1989). In plant feeding experiments, only exogenously applied OAS, and not NAS, induced expression of regulated S-assimilatory pathway enzymes (Neuenschwander et al., 1991; Hawkesford et al., 1995). In the data reported here, the ratio of extracted OAS/NAS varied between tissues and treatments, and neither molecule appeared to be a candidate signal molecule based on bulk-tissue analysis. The OAS-to-NAS conversion may occur during extraction. The model would be valid only if there were localized pools of OAS, which were masked by measurements of bulk tissue OAS. It is possible that the large increase in OAS pool size, which occurs only following prolonged S deprivation, is a consequence of a metabolic imbalance for which the plant can no longer compensate. As such, it is not a good candidate for an early sensitive signal of S deficiency.
In this report, OAS pools were manipulated by expressing the Escherichia coli gene cysE (SAT), the enzyme responsible for OAS production. The data reported here (Fig. 3) show a small increase in OAS and NAS content in root tissues (59% and 14%, respectively) of the transgenic line compared to the wild type in the presence of an adequate S supply. It is questionable as to whether these increases would be sufficient to initiate changes in gene expression. Most of the OAS produced as a consequence of the cysE expression would be immediately utilized for the production of Cys and subsequently incorporated into the glutathione pool, both of which are increased. Under circumstances of enhanced flux through the assimilatory pathway, pool sizes of other assimilatory-pathway intermediates would be expected to fluctuate. One candidate for a regulatory metabolite would be sulfide as it may be substantially depleted if OAS supply were enhanced. Sulfide has been suggested to act antagonistically to NAS in the prokaryotic regulatory circuit (Kredich, 1993).
An increased sulfate transporter expression (Fig. 2) was seen in the roots of the transgenic lines. This observation would be expected and consistent with the original model if OAS pools were enhanced by this overexpression; however, only a limited OAS increase was seen in the root tissues. The SAT expression was most apparent in leaf tissues where the presumed increased OAS synthesis resulted in an increase in Cys and glutathione pools. This situation may have triggered a demand signal from the shoots to the roots. Root-specific regulatory circuits would be responsive to phloem-translocated signals from the shoot caused by the cysE overexpression. However, this signal could not be reduced levels of Cys or glutathione, as previously suggested (Lappartient and Touraine, 1996; Lappartient et al., 1999), as root Cys and glutathione pools were elevated in the transgenic line (Fig. 3, C and D), either from elevated biosynthesis in the root or from biosynthesis in the shoot followed by translocation to the root. Rather than a predicted repression, an increase in sulfate transporter expression was observed (Fig. 2). Sulfate is probably also not a signal because the pool in the transgenic plants is not changed.
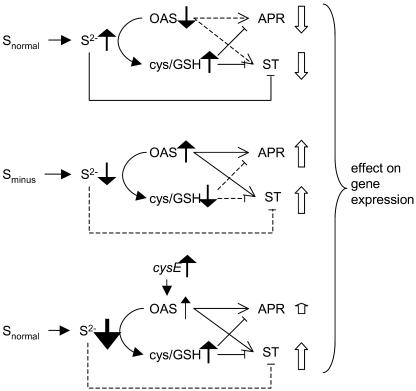
The current status of the regulatory model is presented (Fig. 7). With an adequate S supply (Fig. 7A), gene expression may be repressed by Cys, glutathione, or sulfide; little OAS is present for inducing gene expression. During initial stages of S deficiency (Fig. 7B), gene expression is derepressed as a result of decreased Cys, glutathione, or sulfide pools; accumulating OAS may induce expression, however accumulation is modest at first. With cysE expression (Fig. 7C), a small increase in OAS and larger increases in Cys and glutathione were observed; sulfide is likely to be depleted and no longer represses expression. Derepression is the most likely mechanism for inducing gene expression during S limitation, and the local root concentration of a sulfate assimilatory-pathway intermediate such as sulfide is a candidate for this role. While OAS may have an additional positive regulatory role, and the data reported here do not definitively prohibit this possibility, an alternative explanation may be that in feeding experiments it supplies additional substrate, which otherwise would limit Cys production. This enhanced Cys synthesis (which may be observable only as an increase in the glutathione pool) increases the demand for reduced S in the form of sulfide, which depletes this repressive metabolite pool and induces sulfate transporter expression.
Figure 7.
Possible regulatory links between metabolite pools and transcriptional control of expression of a sulfate transporter (ST) and APS reductase (APR). Positive and negative regulatory signals are indicated by solid lines if operational and broken lines when ineffective. Changes in pool sizes are indicated by vertical arrows to the right of the metabolite. Changes in mRNA pools are indicated by the open arrows adjacent to APR and ST.
Particularly evident is the lack of correlation between mRNA abundance and protein level as compared to measurable sulfate uptake activity (Fig. 6). Large changes in mRNA abundance paralleling modest changes in sulfate transporter activity were also observed in barley (Hordeum vulgare; Smith et al., 1997) and Arabidopsis (Arabidopsis thaliana; Takahashi et al., 2000). Such studies suggest that control of sulfate uptake is at the transcriptional level. Here, only a small and transient increase in uptake capacity was observed upon removal of an external supply of S. It is noteworthy that in a previous study on barley (Smith et al., 1997), the increase in capacity reached a maximum after 4 d of S starvation and then decreased slightly (see figure 6B in Smith et al., 1997), in contrast to mRNA abundance which continued to increase. In this study, the availability of an antibody has enabled visualization of the sulfate transporter protein. As the transporter protein in the plasma membrane increased in abundance compared to the S-replete condition while uptake capacity was comparable, the only possible conclusion is that there are additional posttranslational control mechanisms in operation. A precedent for such regulation has been made by recent reports of the involvement of phosphorylation/dephosphorylation and CRE1/WOL/AHK4 in the regulation of high-affinity sulfate transporters in Arabidopsis (Maruyama-Nakashita et al., 2004a, 2004b). Additionally, maximal activity of another sulfate transporter (SULTR2;1) requires coexpression of another sulfate transporter (SULTR3;5), raising the possibility of further posttranslational regulatory mechanisms (Kataoka et al., 2004a) in addition to the well-established transcriptional regulation.
MATERIALS AND METHODS
Plant Material and Growth Conditions
Potato (Solanum tuberosum L. cv Désirée) plants overexpressing the Escherichia coli gene cysE (SAT EC 2.3.1.30; Harms et al., 2000) were grown hydroponically in nutrient solution containing 7.0 mm KNO3, 5.0 mm Ca(NO3)2, 1.0 mm KH2PO4, 2.0 mm CaCl2, 1.7 mm Mg(NO3)2, 1.0 mm MgCl2, 1.0 mm MgSO4, 0.1 mm NaCl, 0.05 mm FeNaEDTA, 25 μm H3BO3, 10 μm Mn(NO3)2, 1 μm Zn(CH3COO)2, 0.8 μm (NH4)6Mo7O24, and 0.64 μm Cu(NO3)2. Plants were grown on 1 mm sulfate for 3 weeks in a controlled environment chamber with a 16-h photoperiod at 20/10°C. Leaf (young, expanding and mature, fully expanded) and root tissues were harvested for analysis. OAS (0.2 mm) was added as required. For S starvation experiments, plants were grown for 3 weeks with 1 mm sulfate, transferred to 20 μm sulfate (20 μm MgSO4, 0.98 mm MgCl2), and leaf and root tissue was harvested 0, 2, 4, and 8 d after transfer onto minimal sulfate.
Isolation of a Potato Sulfate Transporter cDNA
Total RNA was extracted from root tissue according to Verwoerd et al. (1989). The central region of the sulfate transporter was obtained from RNA by RT-PCR, using degenerate primers designed to sequences from domains conserved between other sulfate transporters (antisense 5′-AARTTCATGTGMGGBTTACG-3′ [first-strand cDNA synthesis and PCR]; sense 1, 5′-CTCACCATYGCHAGYCTYTGYAT-3′ [PCR1]; sense 2, 5′-CCTCAGGAYMTYGSWTATGC-3′ [PCR2]). The amplified fragment of 950 bp was completely sequenced on both strands. Using this sequence, gene-specific primers (GSP) were synthesized for 5′- and 3′-RACE. The 5′-RACE system (Invitrogen, Carlsbad, CA) was used according to the manufacturer's instructions. The first-strand cDNA was synthesized from 1 μg of total root RNA using GSP1 (5′-AGGATCAAGTTCTGGCTGAAGC-3′) and reamplified using nested primer GSP2 (5′-TCCAAGCAAGAGAGACACCACA-3′). The DNA was cloned into pGEM-T Easy vector (Promega, Madison, WI). The 3′-RACE system (Invitrogen) was also used according to the manufacturer's instructions. The first-strand cDNA was synthesized from 1 μg of total RNA using GSP3 (5′-CAGCAGTCAACTATATGGCTGG-3′). The PCR product was also purified and cloned into pGEM-T Easy vector (Promega). DNA sequences were determined on an ABI PRISM 310 genetic analyzer.
Isolation of a Potato APS Reductase Gene Fragment
The central region of an APS reductase was also obtained from RNA by RT-PCR, using degenerate primers designed to sequences conserved between other APS reductases (antisense 5′-CATCTCTMTKYTCWGAHGGRTAC-3′; sense 5′-ATYATGGAYAARGCTCTYGAG-3′). The APS reductase fragment was cloned and sequenced as described above.
SAT Enzyme Assay
SAT activity was assayed using a procedure adapted from Kredich and Tomkins (1966). Frozen plant tissue was homogenized in 100 μL of ice-cold 0.1 m Tris, pH 7.6, and centrifuged (14,000g, 10 min, 4°C). The assay was performed consistently 2 h after extraction. The assay was carried out in a final volume of 500 μL, which contained 63 mm Tris, pH 7.6; 1.25 mm Na2EDTA; 1.25 mm DTNB; 0.1 mm acetyl-CoA; 1 mm l-Ser; and 10 μL of extract. The rate of reaction was followed at 412 nm for 3 min. The reaction rate from a blank sample containing all material except l-Ser was subtracted from the reaction rate obtained with l-Ser. The assay was done in duplicate from three independent isolations. A unit of enzyme is defined as that amount of enzyme that catalyzes the acetylation of 1 pmol of l-Ser per min in the conditions of the assay.
Sulfur Pools
Total S was determined by digesting 100 mg of lyophilized plant material in a mixture of concentrated HNO3 and HClO4 (85:15, v/v). The digested material was resuspended in 5% (v/v) HCl and S determined by inductively coupled plasma-atomic emission spectroscopy (Applied Research Laboratories, Accuris, Ecublens, Switzerland) at 182 nm (Blake-Kalff et al., 2000). Sulfate was measured by extracting 100 mg of frozen, homogenized plant material in 1 mL of deionized water at 80°C for 30 min, after which the extract was filtered through a 0.45-μm filter. Sulfate concentrations in the extracts were determined by ion chromatography (Dionex 2000i/sp) using an AS9SC separation column fitted with an AS9G guard column (Dionex, Sunnyvale, CA). The eluent solution consisted of 1.8 mm Na2CO3, 1.7 mm NaHCO3. Thiols were determined by HPLC using a Zorbax OSD 5-μm column (Jones Chromatography, Mid Glamorgan, UK), following derivatization with mono-bromobimane (Zhao et al., 1996) and following the method described by Newton et al. (1981).
Sulfate Uptake Assay
Rates of sulfate uptake by plant roots were determined for five replicate plants held on a support frame (Clarkson et al., 1989) for a 10-min incubation period in nutrient solution containing 1 mm sulfate with 50 μCi L−1 35SO42− (Amersham Biosciences, Little Chalfont, UK). Roots were rinsed for two 30-s washes in nutrient solution without tracer, blotted dry, and weighed. Individual roots were then extracted in 1 mL of 0.1 n HCl at 95°C for 10 min and allowed to cool before adding 9 mL of Ultima Gold scintillation fluid (Packard Bioscience B.V., Groningen, The Netherlands) and counted. Uptake is expressed as per gram fresh weight of roots.
Determination of NAS and OAS
NAS and OAS were determined using gas chromotography coupled to mass spectrometry (GC-MS). Plant material (150–200 mg fresh weight) was frozen in liquid nitrogen immediately after harvest. The frozen material was ground to a fine powder and subsequently 1,400 μL of methanol, pH 3; 50 μL of ribitol (0.2 mg/mL); and 50 μL of dH2O were added and the sample incubated at 70°C for 15 min. The extract was centrifuged (15,000g, 3 min), and the supernatant transferred to a vial containing 1,500 μL of dH2O. The pellet was washed with 750 μL of CHCl3, incubated at 37°C for 5 min, centrifuged (15,000g, 3 min), and the resulting supernatant added to the previous supernatant/dH2O mixture. This supernatant mixture was vortexed, centrifuged (2,250g, 10 min), and 1 mL of the upper phase transferred to a new vial and dried under vacuum. Samples were prepared for GC-MS and analyzed as described by Fiehn et al. (2000).
RNA Isolation and Northern-Blot Analysis
Total RNA was extracted from frozen material according to Verwoerd et al. (1989). The RNA was separated on agarose/formaldehyde gels and blotted onto positive-charged nylon membrane. The northern hybridization was performed according to Sambrook et al. (1989) with prehybridization and hybridization at 65°C. Blots were hybridized with random-primed 32P probes, washed sequentially with 2× SSC and 1× SSC (150 mm NaCl, 15 mm C6H5Na3O7, pH 7) containing 0.1% (w/v) SDS, and exposed to Kodak BioMax MS film (Eastman-Kodak, Rochester, NY).
Antibody Production and Western Blotting
A rabbit antisera against a 20mer oligopeptide situated near to the N-terminal region of StST1 (NMATDISRVASSRRHSENGL) coupled to keyhole limpet hemocyanin was prepared (Bioworld, Dublin, OH). Plasma membranes were prepared by two-phase partitioning (5 mm KCl and 6.6% [w/w] dextran T-500 [Amersham Biosciences] and 6.6% [w/w] PEG3350 [Sigma, Poole, UK]) from hydroponically grown roots and resolved by SDS-PAGE (Hawkesford and Belcher, 1991). Western blotting used immunogold detection system (British Biocell, Cardiff, UK). Antibody concentrations were 1:1,000 for the primary antibody and 1:200 for the goat anti-rabbit gold-labeled secondary antibody.
Sequence data from this article have been deposited with the EMBL/GenBank data libraries under accession numbers AF309643 and AJ506751.
Acknowledgments
We thank O. Fiehn, V. Nikiforova, and K. Riedel (MP-MPP Golm) for help with the OAS determinations.
This work was supported by Framework IV and V of the European Union (grant nos. BIO4–CT97–2182, QLRT–2000–00103, and QLRT–2001–02928), the European Molecular Biology Organization (grant no. 9303 to A.B.), and Max-Planck Society (grants to H.H., R.H.). Rothamsted Research receives grant-aided support from the Biotechnology and Biological Sciences Research Council of the United Kingdom.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.104.057521.
References
- Blake-Kalff MMA, Hawkesford MJ, Zhao FJ, McGrath SP (2000) Diagnosing sulphur deficiency in field-grown oilseed rape (Brassica napus L.) and wheat (Triticum aestivum L.). Plant Soil 225: 95–107 [Google Scholar]
- Błaszczyk A, Brodzik R, Sirko A (1999) Increased resistance to oxidative stress in transgenic tobacco plants overexpressing bacterial serine acetyltransferase. Plant J 20: 237–243 [DOI] [PubMed] [Google Scholar]
- Bogdanova N, Hell R (1997) Cysteine synthesis in plants: protein-protein interactions of serine acetyltransferase from Arabidopsis thaliana. Plant J 11: 251–262 [DOI] [PubMed] [Google Scholar]
- Bolchi A, Petrucco S, Tenca PL, Foroni C, Ottonello S (1999) Coordinate modulation of maize sulfate permease and ATP sulfurylase mRNAs in response to variations in sulfur nutritional status: stereospecific down-regulation by L-cysteine. Plant Mol Biol 39: 527–537 [DOI] [PubMed] [Google Scholar]
- Clarkson DT, Saker LR, Purves JV (1989) Depression of nitrate and ammonium transport in barley plants with diminished sulphate status: evidence of co-regulation of nitrogen and sulphate intake. J Exp Bot 40: 953–963 [Google Scholar]
- Droux M, Ruffet M-L, Douce R, Job D (1998) Interactions between serine acetyltransferase and O-acetylserine (thiol) lyase in higher plants: structural and kinetic properties of the free and bound enzymes. Eur J Biochem 255: 235–245 [DOI] [PubMed] [Google Scholar]
- Fiehn O, Kopka J, Dormann P, Altmann T, Trethewey RN, Willmitzer L (2000) Metabolite profiling for plant functional genomics. Nat Biotechnol 18: 1157–1161 [DOI] [PubMed] [Google Scholar]
- Flavin M, Slaughter C (1965) Synthesis of the succinic ester of homoserine, a new intermediate in the bacterial synthesis of methionine. Biochemistry 4: 1370–1375 [DOI] [PubMed] [Google Scholar]
- Harms K, von Ballmoos P, Brunold C, Höfgen R, Hesse H (2000) Expression of a bacterial serine acetyltransferase in transgenic potato plants leads to increased levels of cysteine and glutathione. Plant J 22: 335–343 [DOI] [PubMed] [Google Scholar]
- Hawkesford MJ (2003) Transporter gene families in plants: the sulphate transporter gene family—redundancy or specialization? Physiol Plant 117: 155–165 [Google Scholar]
- Hawkesford MJ, Belcher AR (1991) Differential protein synthesis in response to sulphate and phosphate deprivation: identification of possible components of plasma membrane systems in cultured tomato roots. Planta 185: 323–329 [DOI] [PubMed] [Google Scholar]
- Hawkesford MJ, Buchner P, Hopkins L, Howarth JR (2003) The plant sulfate transporter family: specialised function and integration with whole plant nutrition. In J-C Davidian, D Grill, LJ De Kok, I Stulen, MJ Hawkesford, E Schnug, H Rennenberg, eds, 5th Workshop on Sulfur Transport and Assimilation: Regulation, Interaction, Signalling. Backhuys Publishers, Leiden, The Netherlands, pp 1–10
- Hawkesford MJ, Schneider A, Belcher AR, Clarkson DT (1995) Regulation of enzymes involved in the sulfur-assimilatory pathway. Z Pflanzenernähr Bodenk 158: 55–57 [Google Scholar]
- Hawkesford MJ, Smith FW (1997) Molecular biology of higher plant sulphate transporters. In WJ Cram, LJ de Kok, I Stulen, C Brunold, H Rennenberg, eds, Sulphur Metabolism in Higher Plants. Backhuys Publishers, Leiden, The Netherlands, pp 13–25
- Hawkesford MJ, Wray JL (2000) Molecular genetics of sulfate assimilation. Adv Bot Res 33: 160–208 [Google Scholar]
- Herschbach C, Rennenberg H (1994) Influence of glutathione (GSH) on net uptake of sulphate and sulphate transport in tobacco plants. J Exp Bot 45: 1069–1076 [Google Scholar]
- Holtorf S, Apel K, Bohlmann H (1995) Comparison of different constitutive and inducible promoters for the overexpression of transgenes in Arabidopsis thaliana. Plant Mol Biol 29: 637–646 [DOI] [PubMed] [Google Scholar]
- Howarth JR, Fourcroy P, Davidian J-C, Smith FW, Hawkesford MJ (2003) Cloning of two contrasting sulfate transporters from tomato induced by low sulfate and infection by the vascular pathogen Verticillium dahliae. Planta 218: 58–64 [DOI] [PubMed] [Google Scholar]
- Kataoka T, Hayashi N, Yamaya T, Takahashi H (2004. a) Root-to-shoot transport of sulfate in Arabidopsis: evidence for the role of SULTR3;5 as a component of low-affinity sulfate transport system in the root vasculature. Plant Physiol 136: 4198–4204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kataoka T, Watanabe-Takahashi A, Hayashi N, Ohnishi M, Mimura T, Buchner P, Hawkesford MJ, Yamaya T, Takahashi H (2004. b) Vacuolar sulfate transporters are essential determinants controlling internal distribution of sulfate in Arabidopsis. Plant Cell 16: 2693–2704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kredich NM (1993) Gene regulation of sulfur assimilation. In LJ de Kok, I Stulen, H Rennenberg, C Brunold, WE Rauser, eds, Sulfur Nutrition and Sulfur Assimilation in Higher Plants. SPB Academic Publishing, The Hague, The Netherlands, pp 37–47
- Kredich KM, Tomkins GM (1966) The enzymic synthesis of L-cysteine in Escherichia coli and Salmonella typhimurium. J Biol Chem 241: 4955–4965 [PubMed] [Google Scholar]
- Lappartient AG, Touraine B (1996) Demand-driven control of root ATP sulfurylase activity and SO42− uptake in intact canola: the role of phloem-translocated glutathione. Plant Physiol 111: 147–157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lappartient AG, Vidmar JJ, Leustek T, Glass ADM, Touraine B (1999) Inter-organ signaling in plants: regulation of ATP sulfurylase and sulfate transporter genes expression in roots mediated by phloem-translocated compound. Plant J 18: 89–95 [DOI] [PubMed] [Google Scholar]
- Leustek T, Martin MN, Bick J-A, Davies JP (2000) Pathways and regulation of sulfur metabolism revealed through molecular and genetic studies. Annu Rev Plant Physiol Plant Mol Biol 51: 141–165 [DOI] [PubMed] [Google Scholar]
- Leustek T, Saito K (1999) Sulfate transport and assimilation in plants. Plant Physiol 120: 637–643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maruyama-Nakashita A, Nakamura Y, Watanabe-Takahashi A, Yamaya T, Takahashi H (2004. a) Induction of SULTR1;1 sulfate transporter in Arabidopsis roots involves protein phosphorylation/dephosphorylation circuit for transcriptional regulation. Plant Cell Physiol 45: 340–345 [DOI] [PubMed] [Google Scholar]
- Maruyama-Nakashita A, Nakamura Y, Yamaya T, Takahashi H (2004. b) A novel regulatory pathway of sulfate uptake in Arabidopsis roots: implication of CRE1/WOL/AHK4-mediated cytokinin-dependent regulation. Plant J 38: 779–789 [DOI] [PubMed] [Google Scholar]
- Neuenschwander U, Suter M, Brunold C (1991) Regulation of sulfate assimilation by light and O-acetyl-L-serine in Lemna minor L. Plant Physiol 97: 253–258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newton GL, Dorian R, Fahey RC (1981) Analysis of biological thiols: derivatization with monobromobimane and separation by reverse-phase high-performance liquid chromatography. Anal Biochem 114: 383–387 [DOI] [PubMed] [Google Scholar]
- Noji M, Saito K (2002) Molecular and biochemical analysis of serine acetyltransferase and cysteine synthase towards sulfur metabolic engineering in plants. Amino Acids 22: 231–243 [DOI] [PubMed] [Google Scholar]
- Ostrowski J, Kredich NM (1989) Molecular characterization of the cysJIH promoters of Salmonella typhimurium and Escherichia coli: regulation by cysB protein and N-acetyl-L-serine. J Bacteriol 171: 130–140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson WR (2000) Flexible sequence similarity searching with the FASTA3 program package. Methods Mol Biol 132: 185–219 [DOI] [PubMed] [Google Scholar]
- Ruffet M-L, Droux M, Douce R (1994) Purification and kinetic properties of serine acetyltransferase free of O-acetylserine(thiol)lyase from spinach chloroplasts. Plant Physiol 104: 597–604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito K (1999) Regulation of sulfate transport and synthesis of sulfur-containing amino acids. Curr Opin Plant Biol 3: 188–195 [PubMed] [Google Scholar]
- Saito K, Kurosawa M, Tatsuguchi K, Takagi Y, Murakoshi I (1994) Modulation of cysteine biosynthesis in chloroplasts of transgenic tobacco overexpressing cysteine synthase [O-acetylserine(thiol)-lyase]. Plant Physiol 106: 887–895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T (1989) Molecular Cloning: A Laboratory Manual, Ed 2. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY
- Smith FW, Ealing PM, Hawkesford MJ, Clarkson DT (1995) Plant members of a family of sulfate transporters reveal functional subtypes. Proc Natl Acad Sci USA 92: 9373–9377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith FW, Hawkesford MJ, Ealing PM, Clarkson DT, Vandenberg PJ, Belcher AR, Warrilow AGS (1997) Regulation of expression of a cDNA from barley roots encoding a high affinity sulfate transporter. Plant J 12: 875–884 [DOI] [PubMed] [Google Scholar]
- Takahashi H, Watanabe-Takahashi A, Smith FW, Blake-Kalff M, Hawkesford MJ, Saito K (2000) The roles of three functional sulfate transporters involved in uptake and translocation of sulfate in Arabidopsis thaliana. Plant J 23: 171–182 [DOI] [PubMed] [Google Scholar]
- Tsakraklides G, Martin M, Chalam R, Tarczynski MC, Schmidt A, Leustek T (2002) Sulfate reduction is increased in transgenic Arabidopsis thaliana expressing 5′-adenylylsulfate reductase from Pseudomonas aeruginosa. Plant J 32: 879–889 [DOI] [PubMed] [Google Scholar]
- Vauclare P, Kopriva S, Fell D, Suter M, Sticher L, von Ballmoos P, Krähenbühl U, Op den Camp R, Brunold C (2002) Flux control of sulphate assimilation in Arabidopsis thaliana: adenosine 5′-phosphosulphate reductase is more susceptible than ATP sulphurylase to negative control by thiols. Plant J 31: 729–740 [DOI] [PubMed] [Google Scholar]
- Verwoerd TC, Dekker BM, Hoekema A (1989) A small-scale procedure for the rapid isolation of plant RNAs. Nucleic Acids Res 17: 2362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wirtz M, Hell R (2003) Production of cysteine for bacterial and plant biotechnology: application of cysteine feedback-insensitive isoforms of serine acetyltransferase. Amino Acids 24: 195–203 [DOI] [PubMed] [Google Scholar]
- Zhao FJ, Hawkesford MJ, Warrilow AGS, McGrath SP, Clarkson DT (1996) Responses of two wheat varieties to sulfur addition and diagnosis of sulfur deficiency. Plant Soil 181: 317–327 [Google Scholar]