Abstract
The endosperm of Golden Rice (Oryza sativa) is yellow due to the accumulation of β-carotene (provitamin A) and xanthophylls. The product of the two carotenoid biosynthesis transgenes used in Golden Rice, phytoene synthase (PSY) and the bacterial carotene desaturase (CRTI), is lycopene, which has a red color. The absence of lycopene in Golden Rice shows that the pathway proceeds beyond the transgenic end point and thus that the endogenous pathway must also be acting. By using TaqMan real-time PCR, we show in wild-type rice endosperm the mRNA expression of the relevant carotenoid biosynthetic enzymes encoding phytoene desaturase, ζ-carotene desaturase, carotene cis-trans-isomerase, β-lycopene cyclase, and β-carotene hydroxylase; only PSY mRNA was virtually absent. We show that the transgenic phenotype is not due to up-regulation of expression of the endogenous rice pathway in response to the transgenes, as was suggested to be the case in tomato (Lycopersicon esculentum) fruit, where CRTI expression resulted in a similar carotenoid phenomenon. This means that β-carotene and xanthophyll formation in Golden Rice relies on the activity of constitutively expressed intrinsic rice genes (carotene cis-trans-isomerase, α/β-lycopene cyclase, β-carotene hydroxylase). PSY needs to be supplemented and the need for the CrtI transgene in Golden Rice is presumably due to insufficient activity of the phytoene desaturase and/or ζ-carotene desaturase enzyme in endosperm. The effect of CRTI expression was also investigated in leaves of transgenic rice and Arabidopsis (Arabidopsis thaliana). Here, again, the mRNA levels of intrinsic carotenogenic enzymes remained unaffected; nevertheless, the carotenoid pattern changed, showing a decrease in lutein, while the β-carotene-derived xanthophylls increased. This shift correlated with CRTI-expression and is most likely governed at the enzyme level by lycopene-cis-trans-isomerism. Possible implications are discussed.
Golden Rice (Oryza sativa) denotes a genetically modified rice capable of biosynthesizing and accumulating β-carotene (provitamin A) in the endosperm, yielding a characteristic yellow color in the polished grains. Golden rice was developed to help cope with vitamin A deficiency, a problem that prevails in developing countries, affecting millions (Underwood, 2000; UNICEF, 2000). Vitamin A deficiency causes mortality and growth retardation in children and frequently impairs vision, leading to blindness. It also contributes indirectly to anemia by interfering with iron bioavailability (for review, see Haschke and Javaid, 1991).
β-Carotene and some related carotenoids, collectively termed provitamin A, are converted to retinal through symmetric oxidative cleavage at the central C15-C15′ double bond. The corresponding enzyme,β-β-carotene-15,15′-oxygenase, has been identified from Drosophila melanogaster (von Lintig and Vogt, 2000), chicken (Wyss et al., 2000), and mammals (Redmond et al., 2001; Yan et al., 2001).
The first Golden Rice (Ye et al., 2000) and more recent versions (Datta et al., 2003; Hoa et al., 2003; Paine et al., 2005) all rely on the expression of two transgenes (see Fig. 1). The first encodes phytoene synthase (PSY), which utilizes the endogenously synthesized geranygeranyl-diphosphate to form phytoene, a colorless carotene with a triene chromophore (Burkhardt et al., 1997). The second encodes the bacterial CRTI, a carotene desaturase that introduces conjugation by adding four double bonds. The combined activity of PSY and CRTI should lead to the formation of lycopene, which is red, due to its undecaene chromophore. Lycopene, however, has never been observed in any transformant. Instead, α- andβ-carotene are found as well as variable amounts of oxygenated derivatives such as lutein and zeaxanthin (Fig. 1). In other words, in diverse genetic backgrounds of rice (Taipei 309, Cocodrie, Kaybonnet, IR64, Asanohikari, MTL 250, Nang Hzong Cho Dao, Mot Bui, BR 29), combined expression of PSY-CRTI in the endosperm led to a carotenoid pattern that revealed that the pathway proceeded beyond the end point that would be predicted based on the enzymatic function of the transferred gene combination.
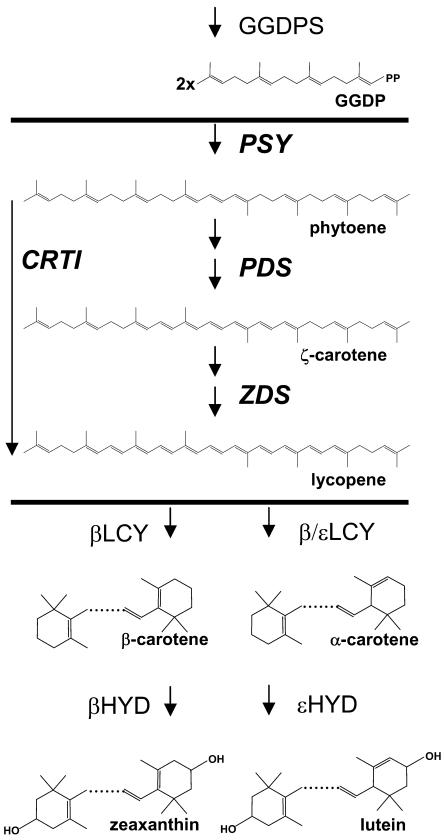
Figure 1.
Carotenoid biosynthesis in transgenic rice endosperm. The precursor molecule geranylgeranyl-diphosphate (GGDP) is synthesized in wild-type endosperm. The enzymatic activities between horizontal bars are supplemented by transformation. This can be done either by using the two plant-type desaturases, PDS and ZDS, or by using the bacterial carotene desaturase, CrtI. However, lycopene does not appear as a product; instead, the carotenoids shown below the bottom bar are found in transgenic endosperm, among which β-carotene is predominant.
There are two possible explanations to account for this observation. The first is that enzymes downstream in the pathway, such as lycopene cyclases (LCYs) and α- and β-carotene hydroxylases (HYDs), are expressed in an enzymatically active form in wild-type rice endosperm, while PSY and one or both of the plant carotene desaturases, phytoene desaturase (PDS) and ζ-carotene desaturase (ZDS), are not. Synthesis of lycopene by PSY and CRTI in the transgenic plant provides the substrate for these downstream enzymes and consequently enables the formation of downstream products. The observation that the expression of PSY alone led to phytoene accumulation but not to desaturated products (Burkhardt et al., 1997) is evidence for the absence of at least one active desaturase, namely PDS. Similarly, the expression of CRTI alone did not produce any color in rice endosperm (A. Klöti and P. Beyer, unpublished data) because of the lack of PSY activity.
The second explanation involves the feedback induction of endogenous carotenoid biosynthetic genes as a result of the presence of transgenes. This was shown to be the case in tomato (Lycopersicon esculentum) fruit upon expression solely of CRTI. Again,β-carotene rather than lycopene increased and endogenous carotenoid genes were shown to be up-regulated, except for PSY, which was repressed (Römer et al., 2000). Such CRTI-dependent up-regulation may be based on the fact that the plant desaturases, PDS and ZDS, together produce a tetra-cis configured form of lycopene, termed prolycopene (Bartley et al., 1999), which is subsequently isomerized to the trans form by a recently identified isomerase, carotene cis-trans-isomerase (CRTISO; Isaacson et al., 2002, 2004; Park et al., 2002). In contrast, the bacterial CRTI leads to the exclusive formation of the all-trans form of lycopene. Likewise, an increased formation of specific carotenogenic mRNAs and proteins was consistently observed in daffodil flowers upon artificial accumulation of all-trans lycopene after addition of the LCY inhibitor chlorophenylthiotriethylamine, and the total carotenoid content was elevated (Al-Babili et al., 1999).
To distinguish between these two conceivable explanations, we examined Golden Rice integration events to detect potential alterations in the expression of the endogenous carotenoid biosynthetic genes in response to the transgenes, using TaqMan real-time PCR. We utilized transgenic lines that contained CrtI under the control of the constitutive cauliflower mosaic virus (CaMV) 35S promoter, which allowed us to investigate both leaves and the endosperm of developing seeds. We further substantiated our results by using plant model systems expressing CRTI. The results obtained enabled us to explain the Golden Rice phenotype, as described below.
RESULTS
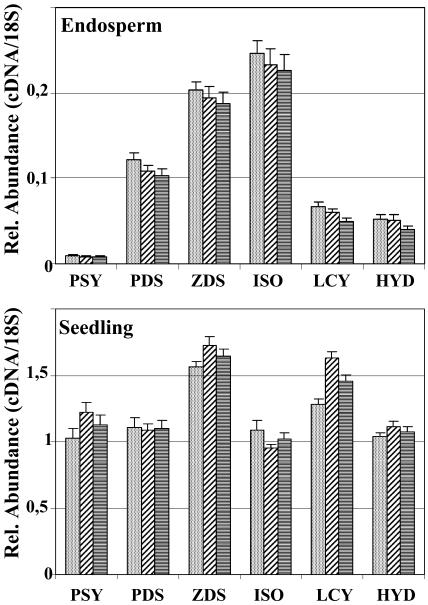
Previously described Golden Rice transformation events expressing CRTI constitutively and PSY in the endosperm (Ye et al., 2000; Hoa et al., 2003) were used to carry out expression analyses. The high specificity of TaqMan real-time PCR allowed us to measure the intrinsic rice Psy mRNA levels without interference by the expression of the Psy transgene. Homozygosity of the materials was confirmed by inverse PCR and by the absence of color segregation in T3 seeds. Transgenic and wild-type plants were sown in a randomized plot in the greenhouse, and RNA was extracted from immature milky-stage endosperm. Leaf material was harvested from 16-d-old seedlings. TaqMan real-time PCR was used to characterize mRNA levels in the wild type or to monitor potential changes in rice endogenous mRNA levels in the transgenic samples. The pathway from phytoene to α-carotene and β-carotene and further on to zeaxanthin was surveyed by monitoring mRNA levels of rice PSY, PDS, ZDS, CRTISO, β-LCY, ɛ-LCY, and β-HYD. Figure 2 shows the results obtained with wild-type materials.
Figure 2.
TaqMan real-time PCR analysis of the expression of rice genes involved in carotenoid biosynthesis in wild-type rice plants. The expression of PSY, PDS, ZDS, CRTISO, β-LCY, and β-HYD were assayed using RNA preparations from immature endosperm and from leaves, as detailed in “Materials and Methods.” Differently shaded bars each represent RNA pools of six individuals.
All carotenoid biosynthetic genes investigated were found to be expressed in rice endosperm, albeit at very low levels. Psy mRNA was the least abundant, 130-fold lower than in leaves, followed by mRNAs for LCY (25-fold), HYD (22-fold), PDS (10-fold), ZDS (8-fold), and CRTISO (4-fold). The barely detectable amount of Psy mRNA found in endosperm explains the need for the Psy transgene to produce carotenoids in rice grains. However, the remaining transcripts, all required to produce β-carotene and hydroxylated xanthophylls, were present. This raises questions about the necessity of the CrtI transgene in Golden Rice and may suggest either one or both of the two desaturases, PDS and ZDS, are inactive or that their respective protein levels are too low.
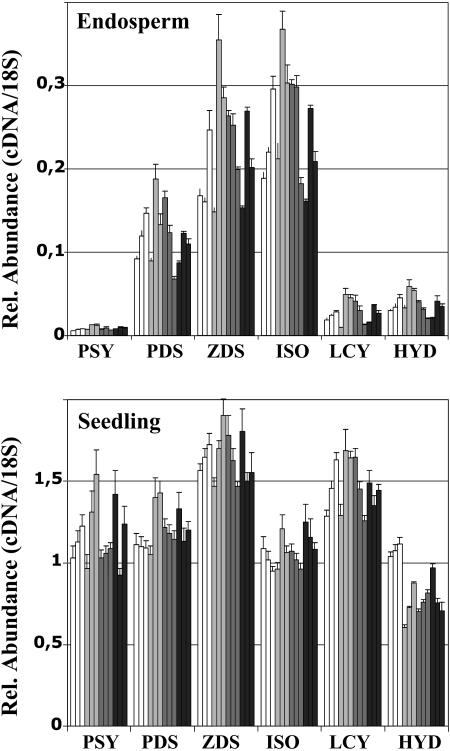
To investigate whether carotenoid accumulation in Golden Rice endosperm is due to a modification in this expression pattern of the rice carotenoid biosynthetic genes, a similar set of experiments was carried out using the transgenic plants (Fig. 3). However, no significant changes in expression of the six rice genes investigated was observed, neither in the endosperm nor in leaves from three independent transformation events. These data indicate that the transgenes (especially CrtI) did not cause feedback regulation to alter the mRNA levels of these endogenous genes, suggesting that, in fact, rice LCY and rice HYD are functional in the endosperm and reveal their activity with the availability of lycopene produced by the transgenic PSY and CRTI. Further experiments were necessary to verify the functional presence of rice PDS, ZDS, and CRTISO (see below).
Figure 3.
TaqMan real-time PCR analysis of the expression of rice genes involved in carotenoid biosynthesis in transgenic rice plants. The expression of PSY, PDS, ZDS, CRTISO, β-LCY, and β-HYD were assayed using RNA preparations from immature endosperm and from leaves as detailed in “Materials and Methods.” White bars, wild-type; light gray bars, T3 siblings of a Golden Rice event obtained with the vector pCaCar (Hoa et al., 2003); dark gray bars, T3 siblings of a different Golden Rice event obtained with the vector pCaCar; black bars, T4 siblings of a Golden Rice event obtained with the vector pB19hpc (Ye et al., 2000). Each bar represents the analysis on a RNA pool extracted from six siblings of the subsequent T-generation.
Carotenoid Analysis
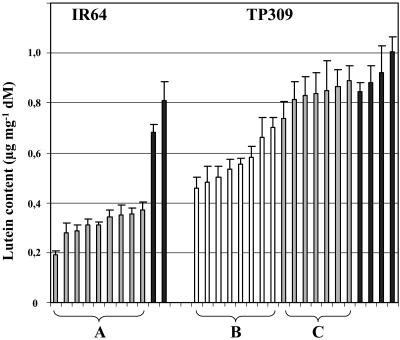
Wild-type leaves, in contrast to endosperm, possess an active carotenoid biosynthetic pathway. Pigment changes could be provoked by the constitutive expression of CRTI, but presumably not by PSY, the expression of which was driven by the endosperm-specific gt1 promoter (pCaCar and pB19hpc; see Ye et al., 2000; Hoa et al., 2003). CRTI is known to be enzymatically active in chloroplasts, as shown by its capability to confer norflurazon resistance in tobacco (Nicotiana tabacum; Misawa et al., 1993) and rice (P. Lucca, unpublished data). Our analyses revealed that in many events, the xanthophyll pattern had shifted in favor of the β-carotene-derived xanthophylls, mainly of violaxanthin and neoxanthin. Some selected examples are shown in Figure 4. A decrease in lutein (as the predominant α-carotene-derived xanthophyll) was frequently observed. The total carotenoid amount, however, did not decrease accordingly, but was partially compensated by an equivalent increase of β-carotene-derived xanthophylls (data not shown). It must be assumed that CRTI activity exerts a hitherto unknown influence on carotenoid biosynthesis in leaves, leading to the observed altered pattern.
Figure 4.
Lutein content in leaves of selected Golden Rice events carrying CrtI under control of the CaMV 35S promoter. A, Lutein content in transgenic IR64 events (siblings of one event; 37B-2b; Hoa et al., 2003). B, Lutein content in transgenic Taipei 309 events (Man 2.2, Man 2.5, and hpc 6.3.1; used in TaqMan real-time PCR analysis). C, Lutein content in Taipei 309 48–67 events showing no effect. Black bars represent the lutein content in the respective wild-type samples. Each bar represents a pool of extracts made from six siblings and represents the mean of three measurements.
The Level of CRTI Expression Correlates with the Decrease in Lutein Content in Leaves
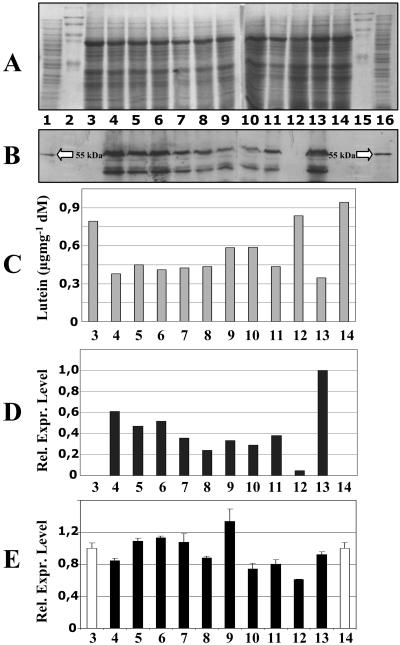
Western blots were carried out with leaf extracts to investigate CRTI protein levels. Figure 5 shows a band of the expected molecular mass of 55 kD, with varying intensities for the different transgenic events investigated. A second cross-reactive band was always observed, which was absent from the wild-type controls and may represent a degradation product. A clear correlation was observed between the level of RNA and protein of CrtI and the decrease in lutein abundance. For example, the event in lane 13 expresses CRTI strongly and has a lower lutein content, and the event expressing CRTI weakly, shown in lane 12, has approximately wild-type lutein levels (lane 3). The signals of all other lines more or less mirror this inverse relationship.
Figure 5.
CrtI RNA and protein expression and lutein content in transgenic rice leaves. A, Coomassie Blue-stained SDS-polyacrylamide gel. B, Western blot conducted with anti-CRTI antibodies. C, Lutein content. D, Relative RNA expression levels of CrtI measured by TaqMan real-time PCR (the strongest expressing line 13 was set to 1). E, Relative RNA expression levels of the ɛ-LCY measured by TaqMan real-time PCR (Taipei 309 and IR 64 wild types in lanes 3 and 14, respectively, were set to 1). Lanes 1 and 16, E. coli-expressing CRTI without transit peptides as positive controls; lanes 2 and 15, prestained marker proteins; lanes 3 and 14, wild-type Taipei 309 and IR 64, respectively; lanes 4 to 12, transgenic Taipei 309 rice events from 4 independent events (lanes 4–6, Man 2.2; lanes 7 and 8, Man 2.5; lanes 9–11, hpc 6.3.1; lane 12, TP 48–67); lane 13, transgenic IR 64 event (37B-2b).
An evident explanation for this loss of lutein could be a down-regulation of ɛ-LCY. When examined by TaqMan real-time PCR, the mRNA changes observed were found to be only minor and did not correlate with the observed lutein-phenotype (Fig. 5E).
Regulation of Lutein Content by CRTI: A Common Principle?
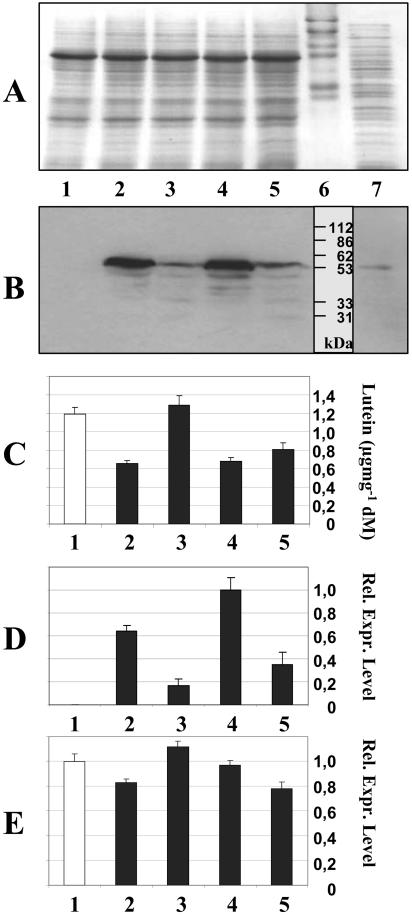
To investigate the effects of CRTI expression in a green tissue from a different species, we transformed Arabidopsis (Arabidopsis thaliana) with the vector pSToff. This experiment is analogous to rice transformation with pCaCar or pB19hpc (Ye et al., 2000; Hoa et al., 2003), which both contain CrtI under the control of the CaMV 35S promoter. Four homozygous transgenic Arabidopsis lines were investigated in detail in the T3 generation. Western-blot analyses shown in Figure 6B revealed high- and low-expressing lines, as mirrored by the respective CrtI mRNA abundance (Fig. 6D). These differentially expressing lines showed the same variation in lutein accumulation found in rice leaves (Fig. 6C), with low levels of CRTI expression corresponding to wild-type lutein content (lane 3), while strongly expressing lines were significantly lower in lutein (lanes 2 and 4). As with rice, the decreased lutein content was partially compensated by an increase of β-carotene-derived xanthophylls (data not shown). Similarly, no changes in the expression of ɛ-LCY were detected that correlated with the observed changes in lutein abundance. A complete set of TaqMan real-time PCR experiments was also conducted for PSY, PDS, ZDS, CRTISO, β-LCY, and the two known β-HYDs (Tian et al., 2003) to account for the full pathway. However, no obvious changes in specific RNA abundance were ever observed (data not shown).
Figure 6.
CrtI RNA and protein expression and lutein content in transgenic Arabidopsis leaves. A, Coomassie Blue-stained SDS-polyacrylamide gel. B, Western blot conducted with anti-CRTI antibodies. C, Lutein content. D, Relative RNA expression levels of CrtI measured by TaqMan real-time PCR (the strongest expressing line 4 was set to 1). E, Relative RNA expression levels of the ɛ-LCY measured by TaqMan real-time PCR (wild type in lane 1 was set to 1). Lane 7, E. coli-expressing CRTI as positive control; lane 6, prestained marker proteins; lane 1, wild-type leaves; lanes 2 to 5, leaves from transgenic Arabidopsis lines. Each bar represents a pool of extracts from 20 siblings. See “Materials and Methods” for details.
Transformation experiments with carotenoid biosynthetic genes targeting photosynthetically inactive tissues often produce more pronounced effects than green tissues, probably because the latter have a tighter regulation at the protein level. We therefore used the transgenic Arabidopsis lines to generate photosynthetically inactive root calli (Banno et al., 2001) and investigated their carotenoid patterns. We observed an analogous effect, i.e. a decrease of lutein from 36% in wild-type samples to 24% in line 2, 27% in line 3, 16% in line 4, and 18% in line 5 (line numbering as in Fig. 6).
Why Is Golden Rice Yellow?
The experiments described above show that CRTI expression in rice, Arabidopsis, and tobacco BY2 cells (see supplemental data) does not alter the expression of the investigated carotenoid biosynthetic genes. This implies that the carotenoid pattern in Golden Rice is the result of functional endogenous ɛ/β-LCYs and α/β-HYDs enabling the formation of α- and β-carotene and derived xanthophylls in the endosperm. Additionally, PDS and ZDS are apparently inactive or their expression levels are too low in the wild-type background, necessitating substitution with CRTI in Golden Rice.
By analysis of the carotenoids in Golden Rice, it is not possible to judge the activity of the rice CRTISO, because the CRTI pathway produces all-trans carotene precursors that do not require cis-to-trans isomerization. The investigation of CRTISO in rice endosperm is only possible in a transgenic background expressing PDS and ZDS, which delivers the plant-specific tetra-cis-lycopene, termed prolycopene.
Transgenic rice producing carotenoid was made using the cultivar Asanohikari expressing PSY, PDS, and ZDS, all from daffodil (Narcissus pseudonarcissus) and each individually under the control of the endosperm-specific Glu-2-promoter with a Ubi1-hpt expression cassette as the selectable marker. A total of 23 events were regenerated and analyzed. Western blots revealed signals for all three proteins that were not detectable in wild-type endosperm (data not shown). The carotenoid content in the resulting yellow grains ranged from 0.2 to 0.8 μg g−1. This is the same range as in Golden Rice, which relies on the same daffodil PSY but on CRTI as the desaturase (Hoa et al., 2003). In addition, HPLC analysis revealed a similar carotenoid pattern, with β-carotene as the main component, accompanied by α-carotene and varying amounts of zeaxanthin and lutein. However, a detailed HPLC analysis of the colored carotenoids revealed the occurrence of small amounts of ζ-carotene isomers and of prolycopene. These intermediates are absent from CRTI-expressing lines and are indicative for PDS and ZDS catalysis. This confirms that the introduced plant-type carotene desaturation pathway operated mechanistically as expected.
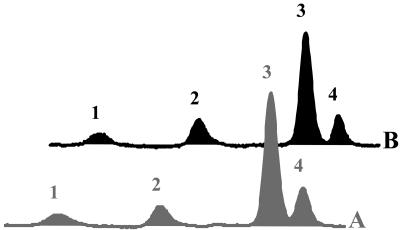
Analysis of the isomer pattern of the β-carotene formed revealed it to be indistinguishable from the one in CRTI-expressing lines (Fig. 7). This shows that the rice CRTISO is enzymatically active in rice endosperm, leading to the identical isomer equilibrium of cyclic carotenes, regardless of the poly-cis or all-trans configuration of the lycopene precursor.
Figure 7.
Separation of isomers of α- and β-carotene of Golden Rice integration events relying on the bacterial desaturase CRTI (A) and the plant (PDS, ZDS) desaturation system (B). 1, 15-cis-β-carotene; 2, all-trans-α-carotene; 3, all-trans-β-carotene; 4, 9-cis-β-carotene. Both A and B show the same distinct isomer distribution.
DISCUSSION
We have here investigated the effects of introduced bacterial CRTI on the expression of critical endogenous carotenoid biosynthesis genes in rice leaves, rice endosperm, Arabidopsis leaves, and tobacco BY2 cells (see supplemental data). Based on published observations (Al-Babili et al., 1999; Römer et al., 2000), it was conceivable that expression of the CrtI transgene provokes transcriptional activation of the carotenoid biosynthetic genes in the target tissue by a feedback regulatory loop. This could be initiated by either the absence of the tetra-cis-lycopene, normally produced by the two plant desaturases (PDS, ZDS), or the constitutive presence of all-trans lycopene, mediated by CRTI. Along these lines of thinking, lycopene isomers would not only represent orthodox intermediates but would also exert a regulatory role. Consequently, the newly identified carotenoid pathway enzyme of plants and cyanobacteria, CRTISO (Isaacson et al., 2002; Park et al., 2002), might play an additional role as regulator. In an extreme scenario, Golden Rice could be yellow due to the transcriptional activation of the entire carotenoid biosynthetic pathway. Alternatively, downstream enzymes, such as cyclases and hydroxylases, might be expressed in wild-type rice in an active form. Their activity would be enabled upon delivery of lycopene mediated by the introduced upstream transgenes.
Wild-type rice endosperm displays low levels of all mRNAs required for xanthophyll formation, i.e. PSY, PDS, ZDS, CRTISO, β-LCY, ɛ-LCY, β-HYD, and ɛ-HYD. Among these, Psy expression is lowest. Given the sensitivity of the PCR method used, the Psy transcript is effectively absent and this is consistent with the fact that introduction of PSY is mandatory, although alone not sufficient for the golden phenotype. The sole expression of PSY resulted in the accumulation of only phytoene (Burkhardt et al., 1997). The essential requirement for CRTI apparently conflicts with the presence of PDS and ZDS transcripts in wild-type endosperm as shown by TaqMan real-time PCR. Perhaps this is because the explanation lies in the level of protein expression and enzymatic activity rather than with mRNA. Due to the low expression, the complicated reaction mechanisms of these enzymes and the unavailability of radioactive carotene substrates, the respective investigations were not possible in vitro and a transgenic approach was chosen instead.
The endosperm-specific expression of the PDS/ZDS system instead of CRTI in rice endosperm resulted in the formation of comparable levels of colored carotenoids. Thus, the rice endosperm provides the complex requirements for the activity of the plant desaturases. PDS requires a redox chain, employing quinones, a quinone-reductase, and molecular oxygen as a terminal electron acceptor (Beyer et al., 1989; Mayer et al., 1990; Nievelstein et al., 1995; Norris et al., 1995) to which it is linked via an oxidase identified through the immutans mutation of Arabidopsis (for review, see Kuntz, 2004). This redox pathway is especially important in non-green carotenoid-bearing tissues like endosperm, while the photosynthetic electron transport is thought to play an analogous role in chloroplasts. Therefore, in one interpretation, colored carotenoids do not form in the endosperm with Psy as the only transgene because the expression of the rice PDS/ZDS system is too low.
It has been shown that, in contrast to PSY, CRTI is not rate limiting and is capable of desaturating large amounts of phytoene, thereby increasing β-carotene accumulation (Paine et al., 2005). In its primary sequence, CRTI is unrelated to the plant-type desaturases; it may have simpler requirements and may therefore be more effective in rice endosperm than the plant-type desaturases. However, this remains to be investigated in greater detail.
By homology, CRTISO is related to CRTI, and it appears that in evolution CrtIso originated from CrtI (Isaacson et al., 2002; Park et al., 2002). In fact, CRTISO is enzymatically active in rice endosperm, as we show. PSY, PDS, and ZDS expression in rice endosperm installed the poly-cis pathway of carotene desaturation, but the β-carotene formed was predominantly in the all-trans form, accompanied by the typical pattern of cis-isomers, among which the 9-cis form is the most abundant. Furthermore, endosperm from transgenic plants relying on CRTI-mediated desaturation yielded the identical isomer ratio of β-carotenes, indicating that rice CRTISO catalyzes the formation of this equilibrium of specific isomers. This is consistent with the recently published characterization of CRTISO in vitro (Isaacson et al., 2004). The activity of CRTISO is also crucial with respect to the formation of cyclic end groups. In its absence, cyclic carotenoids and derived xanthophylls, as observed in these transgenic rice integration events, would not form in non-green tissues, e.g. as shown with the tangerine mutation in tomato fruit and in etioplasts from the Arabidopsis ccr2 mutant, both lacking functional CRTISO (Isaacson et al., 2002; Park et al., 2002).
Clearly, LCY activities and the activities of the divergent class of β-HYDs (for review, see Tian and DellaPenna, 2004) rely on the expression of the respective rice genes in the endosperm. In all rice cultivars transformed so far, the complementation with these activities by transformation has been found to be unnecessary. Moreover, the activity of the rice LCYs never proved to be rate limiting, since lycopene did not accumulate. Thus, Golden Rice is yellow because of the activity of these intrinsic rice cyclases.
TaqMan real-time PCR analyses with RNA isolated from transgenic rice endosperm did not give any indication of a feedback regulatory loop that may affect the expression of rice carotenoid biosynthetic genes because all were unchanged in level when compared to the wild type. The same was true in leaves of rice and Arabidopsis, both expressing CRTI constitutively, despite a change in the xanthophyll composition. In both cases, lutein decreased, partially compensated by an increase in β-carotene and its derived xanthophylls. The same was also true in tobacco BY2 cells transformed with CaMV 35S CrtI, but this time, however, no analogous change in carotenoid composition was found, probably because BY2 cells do not accumulate lutein and zeaxanthin (data not shown).
The change in the xanthophyll ratio observed in rice and Arabidopsis leaves varied in an event-dependent manner and was inversely correlated to the CRTI expression level. The flux of substrate into either branch of xanthophyll formation is controlled by the two LCYs, converting lycopene into β-carotene (β,β-carotene) and α-carotene (β,ɛ-carotene). However, no significant changes in transcript levels of the cyclases were found that would mirror the change in carotenoid composition. It must therefore be assumed that the state of geometric isomerism in lycopene has a certain impact on the probability of β- or ɛ-ring formation. It is interesting to note that the lack of CRTISO in mutants of tomato and Arabidopsis resulted in just the same change toward a preponderance of the β-carotene-derived xanthophylls in leaves (Isaacson et al., 2002; Park et al., 2002). Thus, the observed effect is most likely at the level of enzymatic catalysis and geometric isomerism of lycopene, but not at the level of gene expression.
The effect described is evident only in tissues exhibiting carotenoid biosynthesis in the wild type, such as in leaves. It cannot occur in transformed endosperm lacking carotenoids in the wild type. However, the decrease in lutein in leaves, as described, may have a negative impact on photosynthetic performance. Lutein, the most abundant xanthophyll, is primarily attached to the light-harvesting complex II. Using mutants of Arabidopsis, it has been shown that decreased lutein content leads to a reduction of PSII antenna size (Lokstein et al., 2002). This suggests that lutein has the ability to optimize antenna structure and stability to ensure efficient light harvesting. Therefore, it might be preferable to express CrtI under the control of a tissue-specific promoter; the use of a constitutive promoter has consequently been abandoned in newer versions of golden rice (Paine et al., 2005).
Taken together, we show that Golden Rice grains show no changes in the level of the carotenogenic mRNAs investigated. To search for any potentially occurring transcriptional alteration on a broader scale, a gene expression analysis using a rice gene chip experiment is being completed.
MATERIALS AND METHODS
Vector Construction
Transformation of tobacco (Nicotiana tabacum) BY2 cells and Arabidopsis (Arabidopsis thaliana) plants was performed using the binary vector pSToff containing CrtI under the control of the constitutive CaMV 35S promoter. The vector pSToff was constructed by transferring the CrtI expression cassette (CaMV 35S promoter CrtI-nos terminator) from pBaal2 (Ye et al., 2000) into the binary vector pCAMBIA 1300 (CAMBIA, Canberra, Australia). The resulting vector was then transformed into Agrobacterium tumefaciens strain GV3101 (Koncz and Schell, 1986).
To establish the plant carotene desaturation pathway, PCR was employed to make expression cassettes using the daffodil (Narcissus pseudonarcissus) Psy, Pds, and Zds cDNAs with the rice (Oryza sativa) Gt1 promoter. The native 5′ untranslated region was included in all 3 cases (as given in the EMBL database) and 141, 151, and 350 nucleotides, respectively, of the 3′ untranslated region. The terminators used were from CaMV 35S (for Psy), potP1-11 (for Pds), and nos (for Zds). The resulting 3 expression cassettes were ligated into pJH0104h, a Bin19-based binary vector containing the hpt marker gene under the control of the Ubi-1 promoter and nos terminator.
Growth Conditions and Transformation
Tobacco BY2 cells were cultivated in 100 mL Murashige and Skoog medium (Murashige and Skoog, 1962) supplemented with KH2PO4 (200 μg mL−1), 2,4-dichlorophenoxyacetic acid (2 μg mL−1), thiamine-HCl (10 μg mL−1), and myoinositol (100 μg mL−1) at 27°C under constant shaking (125 rpm) in the dark. Subcultivation was done weekly by transferring 1.5 mL to fresh medium. For transformation, 6-d-old cultures were cocultivated with A. tumefaciens GV3101 cells containing pSToff for 4 d at 27°C in the dark. After 6 to 8 weeks under hygromycin selection (60 μg mL−1), transformed cell lines were recovered. For maintaining the transformed lines, cells were cultivated in the presence of hygromycin. For analytical purposes, cells were subcultured in antibiotic-free media and harvested after 4 d.
Transformation of Arabidopsis plants was done according to the protocol of Bechtold and Pelletier (1998). Transgenic seedlings were identified by germination on Murashige and Skoog agar plates containing hygromycin (30 μg mL−1). Expression of the transgene was monitored by TaqMan real-time PCR.
Seeds of Arabidopsis wild type and 4 CRTI-expressing lines (L11-5, L12-5, L14-2, and L27-3) were germinated on Murashisge and Skoog agar plates containing 30 μg mL−1 hygromycin for 10 d. Twenty seedlings from each line were transferred to soil and grown at 65 μmol m−2 s−1 irradiance and an 8-h/16-h day/night cycle at 21°C for 5 weeks. For each line, 5 leaves of each of the 20 seedlings were harvested and combined to produce RNA pools (see below).
For callus generation, 10-d-old seedlings were transferred to callus induction medium agar plates (Banno et al., 2001) and incubated in the dark at 22°C for 6 weeks. Calli derived from individual lines were combined for analytical purposes.
The method used for rice transformation was based on previous protocols (Tanaka et al., 1990; Hiei et al., 1994) using cultivar Asanohikari with the RecA− supervirulent A. tumefaciens AGL1 (Zhang et al., 1997) with the following modifications. Embryogenic calli of 3 to 4 mm were incubated with A. tumefaciens, spread onto R2COMAS (R2 Micro, 0.5 R2 Macro, B5 vitamins, 20 g L−1 Suc, 10 g L−1 Glc, 1 g L−1 casein hydrolysate, 2 mg L−1 2,4-dichlorophenoxyacetic acid, and 100 μm acetosyringone, pH 5.2) and placed in the dark at 26°C. After selection, surviving embryogenic calli were transferred to regeneration medium (0.5 N6 Macro, N6 Micro, and vitamins, AA amino acids, 20 g L−1 Suc, 1 g L−1 casein hydrolysate, 0.2 mg L−1 naphthyleneacetic acid, 1 mg L−1 kinetin, 50 mg L−1 hygromycin B, pH 5.8, gelrite 6 g L−1) to form transgenic plantlets.
Wild-type seedlings from 2 rice cultivars, TP309 and IR64, as well as several homozygous Golden Rice transformation events, were grown in an incubator (Rumed, Laatzen, Germany) at 50 to 70 μmol m−1 s−2 irradiance and a 16-h/8-h day/night cycle at 28°C for 16 d. Six siblings of each event were harvested and combined for analysis. To minimize variations, seedlings of comparable size (25–30 cm) were used.
Determination of Homozygosity
Homozygosity of rice transformants was determined either by the segregation of color of propagated grains or by PCR. Using inverse PCR, the integration loci of the T-DNA within the rice genome were identified. Subsequently, homozygosity was investigated by PCR using primers deduced from the flanking genomic DNA and from the T-DNA sequence.
For inverse PCR, circular genomic DNA molecules were produced according to the following protocol: 10 μg genomic DNA were digested with HindIII overnight, purified using the GFX-DNA purification kit (Amersham Biosciences, Freiburg, Germany), and eluted with 100 μL 60°C prewarmed water. Then 40 μL of the digested DNA were ligated using T4-ligase (New England Biolabs, Frankfurt) in a total volume of 300 μL. Before adding the enzyme, the reaction mixture was heated for 5 min at 50°C and chilled on ice. The ligation reaction was then incubated for 2 h at room temperature followed by an overnight incubation at 16°C. The enzyme was then heat inactivated for 10 min at 70°C and DNA was precipitated by adding 30 μL NaAc (3 m, pH 4.7), 800 μL ethanol, and by incubating for 2 h at −20°C. After centrifugation, the pellet of circular genomic DNA was washed with 70% ethanol, dried, and dissolved in 100 μL of distilled water. The inverse PCR reactions were performed in a Mastercycler gradient (Eppendorf, Hamburg, Germany) using the inverse primers SalII, SalII-rev for pCaCar transformants (Hoa et al., 2003) and BsalII, BsalII-rev for pB19hpc-lines (Ye et al., 2000). The primers used (given in Supplemental Table I) were deduced from the 3′ region of the T-DNAs and allowed the identification of the right-side flanking sequences. Reactions were performed in a total volume of 25 μL containing 1.5 μL circular genomic DNA, 5 pmol of each primer, 0.5 μL dNTPs (10 mm), and 0.3 μL Advantage polymerase mix (CLONTECH, Heidelberg) in the provided buffer. An annealing temperature gradient covering a range between 45.1°C and 65.1°C was applied, and the reactions were performed as follows: initial denaturation for 2 min at 96°C, followed by 37 cycles of denaturation for 30 s at 94°C, annealing for 30 s, and polymerization for 4 min 30 s at 68°C. After a final step (5 min at 68°C), reaction mixtures were subjected to gel electrophoresis and PCR products were purified using the GFX-DNA purification kit (Amersham Biosciences) and ligated into the vector pCR2.1 (Invitrogen, Karlsruhe, Germany). Sequencing of the obtained products and BLASTN searches allowed the identification of the integration loci with the lines Man 2.2, Man 2.5, and pB19hpc15.b.2. Subsequently, PCR reactions were performed using a primer pair corresponding to either the wild-type or the transgenic alleles. Homozygosity was shown by the absence of products corresponding to the wild-type alleles.
TaqMan Real-Time PCR Assays
Total RNA was isolated using Trizol (Invitrogen) according to the manufacturer's protocol. Equal amounts of total RNA from at least three independent samples per event were combined and subjected to RNA purification and on-column DNaseI digestion using the Qiagen RNeasy mini kit (Qiagen, Hilden, Germany). First-strand cDNA synthesis was performed using the TaqMan reverse transcription reagents (Applied Biosystems, Darmstadt, Germany), according to the manufacturer's protocol. MGB-TaqMan probes and primers (Supplemental Table 2) were designed based on cDNA sequences from tobacco, Arabidopsis, and rice, using Primer Express 2.0 software (Applied Biosystems). Specific mRNA levels were quantified by TaqMan real-time PCR (ABIPrism 7000) using 18S rRNA for normalization (Human 18S rRNA PDAR; Applied Biosystems). 6FAM and VIC reporter (5′ end) dyes were used for the MGB-TaqMan probes and 18S rRNA, respectively. The relative quantities of the transcripts were calculated by using the standard curve method (Livak, 1997).
The following transgenic events were used in this study: IR64 37B-2b, TP 309 48–67 (transformed with pCaCar; Hoa et al., 2003); Man 2.2 A 28, Man 2.5 A67 (transformed with pCaCar), and pB19hpc 6.3.1. (Ye et al., 2000).
Carotenoid Extraction and Analysis
Samples were ground to a fine powder under liquid nitrogen and lyophilized. Lyophilized samples (5 mg for Arabidopsis and rice leaf samples, 150 mg for BY2 and Arabidopsis callus samples) were incubated with 4 mL ethanol/1% 2,6-di-tert-butyl-4-methylphenol (Sigma, Taufkirchen, Germany) at 85°C for 10 min, vigorously mixed once at 5 min. After chilling on ice, 6 mL 1% (w/v) NaCl solution and 3 mL petroleum ether:diethyl ether (2:1, v/v) were added to the sample, followed by brief vigorous mixing. The organic phase was recovered after centrifugation for 10 min at 1,400g. The aqueous phase was extracted a second time as described above. The combined organic phases were dried and dissolved in 50 μL chloroform. Ten microliters were subjected to quantitative analysis using HPLC with a C30 reversed-phase column (YMC Europe GmbH, Schermbeck, Germany) and a gradient system, as described (Hoa et al., 2003). For quantitative analyses, 200 μg α-tocopherol acetate (Sigma) were added to each sample as an internal standard prior to extraction. For normalization and quantification, all peaks were normalized relative to the internal α-tocopherol acetate standard to correct for extraction and injection variability. An external standard, echinenone, run separately, was then used to calculate carotenoid amounts. All carotenoid peaks were integrated at their individual λmax and underwent a second normalization to correct for their individual molar extinction coefficients relative to echinenone (=1), using violaxanthin/neoxanthin (0.896), lutein (0.969), zeaxanthin (0.891), other xanthophylls (0.911), chlorophyll a (1.311), chlorophyll b (0.885), α-carotene (0.791), β-carotene (0.887), phytoene (1.745), and phytofluene (1.622). Carotenoids were identified by their absorption spectra, monitored using a photodiode array detector (PDA 996; Waters, Eschborn, Germany).
cis-Isomers of β-carotene were analyzed and identified with the help of published data (Sander et al., 1994) using a gradient system consisting of solvent A, MeOH:tert-butylmethyl ether, 1:1 (v/v), and solvent B, MeOH:H2O, 20:1 (v/v). The column (C30, as above) was developed as follows: 70% B, isocratic for 20 min, followed by a linear gradient from 70% B to 0% B within 20 min. These final conditions were maintained for another 20 min. The solvent flow was constant at 1.5 mL min−1.
Western-Blot Analysis
Ground leaf material was incubated at 98°C in sample buffer (65 mm Tris-HCl, pH 6.75, 4% (w/v) SDS, 10% (v/v) β-mercaptoethanol, 20% (v/v) glycerol) for 10 min. After centrifugation at 21,000g for 5 min, the supernatant was subjected to SDS-PAGE (Laemmli, 1970) using 10% polyacrylamide gels and subsequent western blotting. Applied protein amounts were verified by Coomassie Brilliant Blue staining of a gel run in parallel. For immunodetection, anti-CRTI antibodies were raised in mice against a purified His-tagged CRTI fusion protein expressed in Escherichia coli cells. The antibodies were then affinity purified according to the method of Smith and Fisher (1984). For detection, horseradish peroxidase-coupled secondary anti-mouse antibodies and the ECL system (Amersham Biosciences) were employed according to the manufacturer's protocol.
Distribution of Materials
Materials described in this publication are available to members of the Golden Rice licensee network within the research and development strategies of the Golden Rice Humanitarian Board. Proposals may be addressed to P.B., in the first instance, for consideration by the Golden Rice Humanitarian Board.
Supplementary Material
Acknowledgments
We thank Jorge Mayer for valuable discussions and Randy Cassada for critically reading the manuscript.
This work was supported by the Rockefeller Foundation, New York, and by HarvestPlus (www.harvestplus.org).
The online version of this article contains Web-only data.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.104.057927.
References
- Al-Babili S, Hartung W, Kleinig H, Beyer P (1999) CPTA modulates levels of carotenogenic proteins and their mRNAs and affects carotenoid and ABA content as well as chromoplast structure in Narcissus pseudonarcissus flowers. Plant Biol 1: 607–612 [Google Scholar]
- Banno H, Ikeda Y, Niu QW, Chua NH (2001) Overexpression of Arabidopsis ESR1 induces initiation of shoot regeneration. Plant Cell 13: 2609–2618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartley GE, Scolnik PA, Beyer P (1999) Two Arabidopsis thaliana carotene desaturases, phytoene desaturase and zeta-carotene desaturase, expressed in Escherichia coli, catalyze a poly-cis pathway to yield pro-lycopene. Eur J Biochem 259: 396–403 [DOI] [PubMed] [Google Scholar]
- Bechtold N, Pelletier G (1998) In planta Agrobacterium-mediated transformation of adult Arabidopsis thaliana plants by vacuum infiltration. Methods Mol Biol 82: 259–266 [DOI] [PubMed] [Google Scholar]
- Beyer P, Mayer M, Kleinig H (1989) Molecular oxygen and the state of geometric isomerism are essential in the carotene desaturation and cyclization reactions in daffodil chromoplasts. Eur J Biochem 184: 141–150 [DOI] [PubMed] [Google Scholar]
- Burkhardt PK, Beyer P, Wünn J, Kloeti A, Armstrong GA, Schledz M, v. Lintig J, Potrykus I (1997) Transgenic rice (Oryza sativa) endosperm expressing daffodil (Narcissus pseudonarcissus) phytoene synthase accumulates phytoene, a key intermediate of provitamin A biosynthesis. Plant J 11: 1071–1078 [DOI] [PubMed] [Google Scholar]
- Datta K, Baisakh N, Oliva N, Torrizo L, Abrigo E, Tan J, Rai M, Rehana S, Al-Babili S, Beyer P, et al (2003) Bioengineered “golden” indica rice cultivars with β-carotene metabolism in the endosperm with hygromycin and mannose selection systems. Plant Biotech J 1: 81–90 [DOI] [PubMed] [Google Scholar]
- Haschke F, Javaid N (1991) Nutritional anemias. Acta Paediatr Scand 374: 38–44 [DOI] [PubMed] [Google Scholar]
- Hiei Y, Ohta S, Komari T, Kumashiro T (1994) Efficient transformation of rice (Oryza sativa L.) mediated by Agrobacterium and sequence analysis of the boundaries of the T-DNA. Plant J 6: 271–282 [DOI] [PubMed] [Google Scholar]
- Hoa TTC, Al-Babili S, Schaub P, Potrykus I, Beyer P (2003) Golden indica and japonica rice lines amenable to deregulation. Plant Physiol 113: 161–169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isaacson T, Ohad I, Beyer P, Hirschberg J (2004) Analysis in vitro of the enzyme CRTISO establishes a poly-cis-carotenoid biosynthesis pathway in plants. Plant Physiol 136: 4246–4255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isaacson T, Ronen G, Zamir D, Hirschberg J (2002) Cloning of tangerine from tomato reveals a carotenoid isomerase essential for the production of beta-carotene and xanthophylls in plants. Plant Cell 14: 333–342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koncz C, Schell J (1986) The promoter of TL-DNA gene 5 controls the issue specific expression of chimeric genes carried by a novel type of Agrobacterium binary vector. Mol Gen Genet 204: 383–396 [Google Scholar]
- Kuntz M (2004) Plastid terminal oxidase and its biological significance. Planta 218: 896–899 [DOI] [PubMed] [Google Scholar]
- Laemmli UK (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227: 680–685 [DOI] [PubMed] [Google Scholar]
- Livak KJ (1997) User Bulletin No. 2: ABI PRISM 7700 Sequence Detection System. PE Applied Biosystems, Foster City, CA, pp 11–15
- Lokstein H, Tian L, Polle JE, DellaPenna D (2002) Xanthophyll biosynthetic mutants of Arabidopsis thaliana: altered nonphotochemical quenching of chlorophyll fluorescence is due to changes in photosystem II antenna size and stability. Biochim Biophys Acta 1553: 309–319 [DOI] [PubMed] [Google Scholar]
- Mayer MP, Beyer P, Kleinig H (1990) Quinone compounds are able to replace molecular oxygen as terminal electron acceptor in phytoene desaturation in chromoplasts of Narcissus pseudonarcissus L. Eur J Biochem 191: 359–363 [DOI] [PubMed] [Google Scholar]
- Misawa N, Yamano S, Linden H, de Felipe MR, Lucas M, Ikenaga H, Sandmann G (1993) Functional expression of the Erwinia uredovora carotenoid biosynthesis gene crtl in transgenic plants showing an increase of beta-carotene biosynthesis activity and resistance to the bleaching herbicide norflurazon. Plant J 4: 833–840 [DOI] [PubMed] [Google Scholar]
- Murashige T, Skoog K (1962) A revised medium for rapid growth and bioassays with tobacco tissue cultures. Physiol Plant 15: 473–497 [Google Scholar]
- Nievelstein V, Vandekerckhove J, Tadros MH, v. Lintig J, Nitschke W, Beyer P (1995) Carotene desaturation is linked to a respiratory redox pathway in N. pseudonarcissus chromoplasts membranes. Involvement of a 23 kDa oxygen-evolving-complex-linked protein. Eur J Biochem 233: 864–872 [DOI] [PubMed] [Google Scholar]
- Norris SR, Barrette TR, DellaPenna D (1995) Genetic dissection of carotenoid synthesis in Arabidopsis defines plastoquinone as an essential component of phytoene desaturation. Plant Cell 12: 2139–2149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paine JA, Shipton CA, Chaggar S, Howells RM, Kennedy MJ, Vernon G, Wright SY, Hinchliffe E, Drake R (2005) Improving the nutritional value of golden rice through increased pro-vitamin A content. Nat Biotechnol (in press) [DOI] [PubMed]
- Park H, Kreunen SS, Cuttriss AJ, DellaPenna D, Pogson BJ (2002) Identification of the carotenoid isomerase provides insight into carotenoid biosynthesis, prolamellar body formation, and photomorphogenesis. Plant Cell 14: 321–332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redmond TM, Gentleman S, Duncan T, Yu S, Wiggert B, Gantt E, Cunningham FX Jr (2001) Identification, expression, and substrate specificity of a mammalian beta-carotene 15,15′-dioxygenase. J Biol Chem 276: 6560–6565 [DOI] [PubMed] [Google Scholar]
- Römer S, Fraser PD, Kiano JW, Shipton CA, Misawa N, Schuch W, Bramley PM (2000) Elevation of the provitamin A content of transgenic tomato plants. Nat Biotechnol 18: 666–669 [DOI] [PubMed] [Google Scholar]
- Sander LC, Sharpless KE, Craft NE, Wise SA (1994) Development of engineered stationary phases for the separation of carotenoid isomers. Anal Chem 66: 1667–1674 [DOI] [PubMed] [Google Scholar]
- Smith DE, Fisher PA (1984) Identification, developmental regulation, and response to heat shock of two antigenically related forms of a major nuclear envelope protein in Drosophila embryos: application of an improved method for affinity purification of antibodies using polypeptides immobilized on nitrocellulose blots. J Cell Biol 99: 20–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka A, Mita S, Ohta S, Kyozuka J, Shimamoto K, Nakamura K (1990) Enhancement of foreign gene expression by a dicot intron in rice but not in tobacco is correlated with an increased level of mRNA and an efficient splicing of the intron. Nucleic Acids Res 11: 6767–6770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian L, DellaPenna D (2004) Progress in understanding the origin and functions of carotenoid hydroxylases in plants. Arch Biochem Biophys 430: 22–29 [DOI] [PubMed] [Google Scholar]
- Tian L, Magallanes-Lundback M, Musetti V, DellaPenna D (2003) Functional analysis of beta- and epsilon-ring carotenoid hydroxylases in Arabidopsis. Plant Cell 15: 1320–1332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Underwood BA (2000) Overcoming micronutrient deficiencies in developing countries: Is there a role for agriculture? In NS Scrimshaw, ed, Food and Nutrition Bulletin, Vol 21, No 4. United Nations University Press, New York, pp 356–360
- UNICEF (2000) The State of World's Children: Statistical Tables. UNICEF, New York
- von Lintig J, Vogt K (2000) Filling the gap in vitamin A research. Molecular identification of an enzyme cleaving beta-carotene to retinal. J Biol Chem 275: 11915–11920 [DOI] [PubMed] [Google Scholar]
- Wyss A, Wirtz G, Woggon W, Brugger R, Wyss M, Friedlein A, Bachmann H, Hunziker W (2000) Cloning and expression of beta,beta-carotene 15,15′-dioxygenase. Biochem Biophys Res Commun 271: 334–336 [DOI] [PubMed] [Google Scholar]
- Yan W, Jang GF, Haeseleer F, Esumi N, Chang J, Kerrigan M, Campochiaro M, Campochiaro P, Palczewski K, Zack DJ (2001) Cloning and characterization of a human beta,beta-carotene-15,15′-dioxygenase that is highly expressed in the retinal pigment epithelium. Genomics 72: 193–202 [DOI] [PubMed] [Google Scholar]
- Ye X, Al-Babili S, Klöti A, Zhang J, Lucca P, Beyer P, Potrykus I (2000) Engineering the provitamin A (β-carotene) biosynthetic pathway into (carotenoid-free) rice endosperm. Science 287: 303–305 [DOI] [PubMed] [Google Scholar]
- Zhang J, Xu RJ, Elliott MC, Chen DF (1997) Agrobacterium-mediated transformation of elite indica and japonica rice cultivars. Mol Biotechnol 8: 223–231 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.