Abstract
Human immunodeficiency virus type 1 (HIV-1) variants resistant to protease inhibitors have been shown to contain a mutation in the p1/p6 Gag precursor cleavage site. At the messenger RNA level, this mutation generates a U UUU UUU sequence that is reminiscent of the U UUU UUA sequence required for ribosomal frameshifting and Gag-Pol synthesis. To test whether the p1/p6 cleavage site mutation was generating a novel frameshift site, HIV sequences were inserted in translation vectors containing a chloramphenicol acetyltransferase (CAT) reporter gene requiring −1 frameshifting for expression. All sequences containing the original HIV frameshift site supported the synthesis of CAT but expression was increased 3- to 11-fold in the presence of the mutant p1/p6 sequence. When the original frameshift site was abolished by mutation, expression remained unchanged when using constructs containing the mutant p1/p6 sequence, whereas it was decreased 2- to 4.5-fold when using wild-type p1/p6 constructs. Similarly, when introduced into HIV molecular clones, the p1/p6 mutant sequence supported Gag-Pol synthesis and protease activity in the absence of the original frameshift site, indicating that this sequence could also promote ribosomal frameshifting in virus-expressing cells.
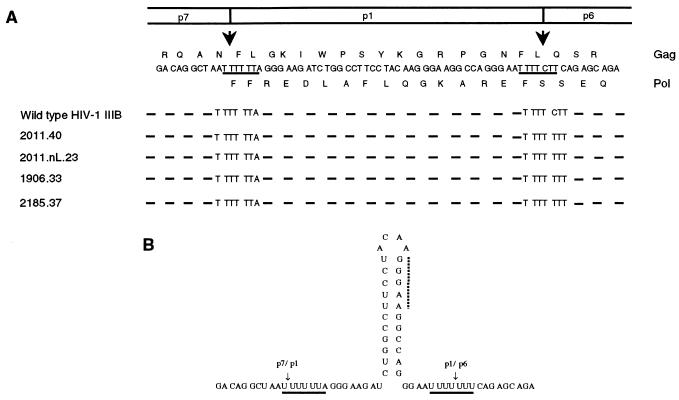
To overcome the antiviral effects of protease inhibitors in culture or in vivo, human immunodeficiency virus type 1 (HIV-1) accumulates mutations in its protease gene and in Gag precursor cleavage sites (reviewed in reference 21). Two cleavage sites were shown to be mutated in resistant variants: the p1/p6 cleavage site (3, 6, 25) and the NC(p7)/p1 cleavage site (6, 25). These mutations improve peptide hydrolysis by the protease in vitro and improve polyprotein processing in virions (6). In all mutants analyzed, the p1/p6 mutation involves an L→F modification at the p1′ position of the scissile bond (6). In the DNA, this mutation is a C-to-T transition of the first base of the leucine codon, replacing the wild-type AAT TTT CTT sequence in this region with the AAT TTT TTT sequence (Fig. 1A). When this sequence is transcribed into RNA, the resulting mutant stretch of nucleotides, AAU UUU UUU, is quite reminiscent of the AAU UUU UUA sequence required for ribosomal frameshifting and Gag-Pol synthesis (Fig. 1B) (13). Interestingly, this mutant sequence is also located in close proximity to the original frameshift site in HIV, which itself overlaps the p7/p1 cleavage site sequence in Gag (Fig. 1A). This therefore suggested not only that the p1/p6 cleavage site mutation was improving the processing of precursors at the protein level but also that the mutant sequence could constitute a novel slippery site promoting ribosomal frameshifting during mRNA translation.
FIG. 1.
Nucleic acid sequences of the p1/p6 cleavage site mutation in HIV-1 protease inhibitor-resistant variants. (A) Variants obtained in the presence of protease inhibitors were sequenced in the p7/p1/p6 region, and DNA sequences were compared to that of the HIV-1 IIIB strain (5, 6, 17). The portion of the DNA sequence from HIV-1 IIIB is shown in its entirety as well as the deduced amino acid sequences (indicated in single-letter codes), read either in the Gag frame (top) or in the Pol frame (bottom). The arrows indicate the scissile bonds of the p7/p1 and p1/p6 cleavage sites. The sequences of variants obtained in the presence of palinavir (2011.40 and 2011.nL.23), BILA 1906 BS (1906.33), and BILA 2185 BS (2185.37) are shown, with sequence identity illustrated by a dash. All mutants contain a C-to-T transition at the p1/p6 junction. (B) Transcribed into RNA, the p7/p1/p6 sequence is predicted to give a stem-loop structure, with the p7/p1 and p1/p6 potential slippery sites (underlined) lying on either side. The dotted line shows a sequence possibly involved in transient pairing with 18S rRNA (see the text).
In HIV, as in many other retroviruses (7, 9, 13, 14), frameshifting is required to synthesize two polyproteins (Gag and Gag-Pol in HIV) starting from the same initiation codon of an mRNA. Translation of the HIV Gag terminates at the carboxy-terminal end of the p6 protein, around codon 500 of the mRNA, whereas synthesis of Gag-Pol requires a shift of the reading frame in the 5′ direction (−1 shift) at the p7/p1 junction, around codon 432 of the mRNA (13). Translation of Gag-Pol then proceeds in this new reading frame until a stop codon is reached, about 3,000 nucleotides later. Ribosomal −1 frameshifting is a very controlled event requiring both a heptameric X XXY YYZ consensus slippery sequence (U UUU UUA in HIV) and a downstream secondary RNA structure which causes the ribosome to pause (a stem-loop in HIV; Fig. 1B) (4, 8, 9, 13). Under optimal conditions, however, frameshifting is a rare event, occurring only for 1 of 10 to 20 ribosomes. This controlled frequency ensures that the synthesis of Gag and Gag-Pol occurs in the correct ratio, which is required for optimal enzyme activation and virus assembly (12, 18).
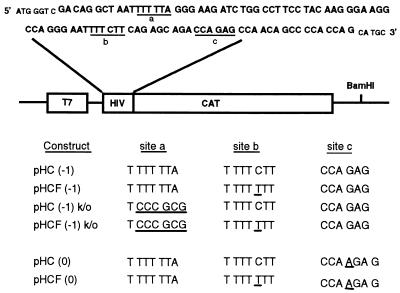
Since protease inhibitor-resistant variants have impaired protease activity due to mutations (5, 10, 20), they could benefit from an increased level of Gag-Pol frameshifting that would increase the level of enzyme proteins in the virus. To determine if the p1/p6 mutation observed in resistant HIV was indeed creating a novel frameshift site, in vitro translation vectors were constructed. A plasmid construct in which a 93-bp DNA sequence encompassing the HIV p7/p1/p6 region was inserted at the beginning of the chloramphenicol acetyltransferase (CAT) coding sequence of pHC(−1), a derivative of plasmid bluescript SK− (Stratagene), was made (Fig. 2). The HIV sequence encodes the original frameshift site (site a), the putative stem-loop structure, and the p1/p6 cleavage site sequence (site b). Expression of the CAT reporter gene is driven by a T7 promoter, and requires a −1 frameshift in the HIV sequence, in the absence of which an in-frame stop codon terminates translation at codon 45 of the CAT coding sequence. Construct pHC(−1) has the wild-type HIV sequence at the beginning of the CAT gene sequence, while construct pHCF(−1) has an HIV sequence in which a C-to-T transition was inserted at the p1/p6 junction (site b). As controls, constructs in which no frameshift was required for CAT expression were used. These constructs [pHC(0) and pHCF(0)] contain an additional base pair four codons downstream of the putative second frameshift site, at site c, which renders the HIV gene sequence in frame with the CAT gene sequence. All constructs were linearized at the unique BamHI site located 3′ of the CAT coding sequence, and 1 μg of DNA was used for in vitro RNA transcription by the T7 RNA polymerase. RNA was then translated in a rabbit reticulocyte lysate. In assays using [35S]methionine, messengers from both pHC(0) and pHCF(0) constructs were efficiently translated into a protein whose molecular mass, 28 kDa, corresponded to the expected molecular mass of the HIV-CAT fusion protein. In contrast, constructs pHC(−1) and pHCF(−1), requiring a −1 frameshift for expression, gave only weak bands of 28 kDa, as expected (data not shown).
FIG. 2.
Construction of a CAT expression vector for in vitro translation. A 93-bp DNA sequence encompassing the HIV p7/p1/p6 region was inserted in the beginning of the coding sequence of a CAT reporter gene which is under the control of a T7 promoter. The HIV and CAT sequences are not in the same reading frame, so a −1 frameshift in the HIV sequence is required to produce the CAT protein. The inserted HIV sequence is shown in boldface, with the original frameshift site (site a), the p1/p6 junction (site b), and the region in which an additional base pair was added to make the HIV and CAT sequences in frame (site c) underlined. The sequences of sites a, b, and c of all constructs used in this study are indicated in the bottom half of the figure, with added or modified nucleotides underlined. Apart from these three sites, all constructs were identical and were used in similar in vitro transcription and translation experiments.
To quantitate the frequency of frameshifting generated with these constructs, the level of CAT expression was assessed by an immunological CAT enzyme-linked immunosorbent assay (ELISA). As shown in Table 1, the wild-type HIV sequence [pHC(−1)] gave a frameshift of from 5 to 12.9% in rabbit reticulocyte lysates with respect to the in-frame pHC(0) construct, a result similar to that already reported for this sequence (19, 24). The presence of a mutation at the p1/p6 junction in pHCF(−1) clones, however, gave a frameshift of from 15 to 30% with respect to the in-frame control constructs [pHCF(0)], an approximately threefold increase compared to that for the wild-type HIV sequence. Both capped (experiment 3) and uncapped (experiments 1 and 2) RNAs were assayed in these experiments, with no significant difference found. CAT levels determined by the method of chloramphenicol acetylation (CAT assays) (22) also gave results comparable to those in Table 1 (data not shown). Therefore, these experiments suggest that the mutation in the HIV p1/p6 cleavage site sequence was enhancing the extent of ribosomal frameshifting in vitro.
TABLE 1.
The p1/p6 cleavage site mutation promotes ribosomal frameshifting leading to CAT expression in vitroa
| Expt | Results for construct containing wild-type site a
|
Results for construct containing a nonslippery site a
|
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CAT expression (pg/ml)b
|
% Frameshiftc
|
CAT expression (pg/ml)b
|
Decrease in CAT expression (fold)d
|
|||||||||
| pHC(0) | pHC(−1) | pHCF(0) | pHCF(−1) | pHC(−1)/ pHC(0) | pHCF(−1)/ pHCF(0) | pHC(−1) | pHC(−1)k/o | pHCF(−1) | pHCF(−1)k/o | pHC(−1)/ pHC(−1)k/o | pHCF(−1)/ pHCF(−1)k/o | |
| 1 | 104,245 | 13,537 | 121,226 | 36,745 | 12.9 | 30 | 11,622 | 4,353 | 31,992 | 24,386 | 2.6 | 1.3 |
| 2 | 96,164 | 8,322 | 136,111 | 21,375 | 8.6 | 15 | 7,156 | 1,570 | 14,029 | 12,139 | 4.5 | 1.1 |
| 3 | 86,214 | 4,338 | 61,357 | 11,974 | 5.0 | 19.5 | 4,338 | 1,166 | 12,778 | 11,046 | 3.7 | 1.1 |
The three independent experiments shown for each assay used either uncapped RNA (experiments 1 and 2 for wild-type site a and experiment 1 for nonslippery site a) or capped RNA (experiment 3 for wild-type site a and experiments 2 and 3 for nonslippery site a). Site a is the original HIV frameshift site which can be either wild type (T TTT TTA) or mutated to the unslippery sequence T CCC GCG.
CAT ELISA values were determined according to a CAT standard curve.
Calculated by dividing the ELISA result for pHC(−1) or pHCF(−1) by that for pHC(0) or pHCF(0), respectively, and multiplying by 100.
Calculated by dividing the ELISA result for pHC(−1) or pHCF(−1) by that for pHC(−1)k/o or pHCF(−1)k/o, respectively.
The effect of the p1/p6 mutation could be due to the generation of a novel frameshift site in the pHCF(−1) sequence. However, there is a possibility that the increase in frameshifting could also be due to the presence of novel or more complex secondary structures in the mutant mRNA. To distinguish between these two possibilities, constructs pHC(−1)k/o and pHCF(−1)k/o were generated (Fig. 2). In these constructs, the original T TTT TTA slippery sequence at site a was replaced by site-directed mutagenesis with the nonslippery sequence T CCC GCG, thereby preventing frameshifting at this site. This mutation also generated a novel SacII restriction site in the DNA, used for easy screening. Table 1 shows the levels of CAT expression found when comparing these pHC(−1)k/o and pHCF(−1)k/o constructs with their parental pHC(−1) and pHCF(−1) constructs. Abolishing the original frameshift site in pHC(−1)k/o reduced CAT expression by 2.6- to 4.5-fold with respect to the wild-type HIV sequence. Interestingly, residual CAT levels were consistently observed with pHC(−1)k/o constructs, suggesting that ribosomal slippage at the original frameshift site was not the only mechanism generating CAT expression in this system. In assays using mutant pHCF(−1) and pHCF(−1)k/o constructs on the other hand, there was no significant difference in CAT expression (1.1- to 1.3-fold reduction) suggesting that the p1/p6 cleavage site mutation could compensate for the change in the original frameshift site. Again, uncapped RNAs (experiment 1), capped RNAs (experiments 2 and 3), and CAT assays (data not shown) gave similar results. Since ribosomal slippage still efficiently occurs in the absence of the original frameshift site in pHCF(−1)k/o constructs, the p1/p6 mutation must therefore generate a novel frameshift site.
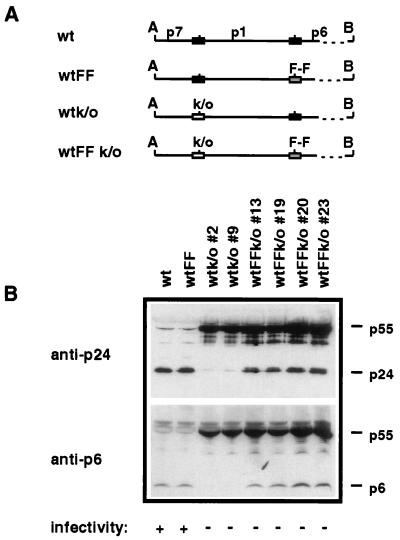
To determine the relevance of these results for virions, HIV molecular clones were constructed (Fig. 3A). These clones, derived from a modified pNL4.3 (1) plasmid called 2.12 (6), contained all the NL4.3 HIV genetic information except for a 930-bp sequence taken from the HIV-I IIIB strain. This 930-bp sequence contained part of the p7 gene, the entire p1 and p6 genes, and 3′ sequences leading into the reverse transcriptase gene (6). Clone wt contained all wild-type HIV-1 sequences, whereas site-directed mutagenesis was conducted to introduce the C-to-T transition at the p1/p6 junction of clone wtFF. Mutagenesis was also carried out to modify the original frameshift site from T TTT TTA to T CCC GCG in constructs wtk/o and wtFFk/o. Proviral DNA (2 μg) from these clones was transfected in human embryonic kidney cell line 293 to produce virions, as described previously (6). Western blot analysis of recovered virions showed that whereas both wt and wtFF virions produce mature p24 protein by processing the p55gag precursor, wtk/o viruses, in which the original frameshift site was abolished by mutation, produce very low levels of the p24 protein (Fig. 3B). These wtk/o viruses did, however, contain high levels of precursor polyprotein p55gag, suggesting that processing by an active protease, which requires ribosomal frameshifting to synthesize Gag-Pol, was deficient in the absence of a functional slippery sequence. In contrast, wtFFk/o viruses containing a C-to-T mutation at the p1/p6 junction produced significant amounts of mature p24 protein, although the original slippery sequence had been abolished, indicating that Gag-Pol frameshifting had occurred in these viruses. In all wtFFk/o clones examined, large amounts of unprocessed p55gag precursors were detected, suggesting that protease activity, although obviously present in these viruses, must be suboptimal. An analysis of p6 protein expression by Western blotting confirmed the presence of active protease in wtFFk/o clones but not in wtk/o clones (Fig. 3B). These results therefore indicate that the C-to-T mutation at the p1/p6 junction gives rise to a functional frameshift site in HIV, in the absence of the original slippery sequence.
FIG. 3.
Construction and analysis of HIV frameshift mutant molecular clones. (A) A 930-bp DNA fragment containing part of the p7 gene, the entire p1 gene, and 3′ sequences leading into the reverse transcriptase gene from HIV-1 strain IIIB was inserted between the unique ApaI (A) and BST1107 (B) restriction sites of a modified NL4.3 vector called 2.12 to generate clone wt. Site-directed mutagenesis was then used as described previously (11) to introduce the p1/p6 cleavage site mutation in clone wtFF and to abolish the original frameshift site in clones wtk/o and wtFFk/o. The original frameshift site is represented by a solid box, the abolished frameshift site is represented by an open box, the wild-type p1/p6 cleavage site is also represented by a solid box, and the mutated p1/p6 cleavage site is represented by a shaded box. (B) Viral particles harvested upon transfection of molecular clones were analyzed by Western blotting using monoclonal antibodies directed either against the p24 protein (clone 39/5.1.23; ID Labs Inc.) or the p6 protein (23). Proteins of 55, 41, 24, and 6 kDa representing the p55gag and p41 precursors and the mature p24 and p6 proteins, respectively, were detected. Two independent wtk/o clones and four independent wtFFk/o clones are shown. Immune reactivity was detected by a chemiluminescent substrate detection assay. The infectivities of viruses, determined by infection of C8166 cells with transfection supernatants, are shown as positive (+) or negative (−) at the bottom of the figure.
Surprisingly, although Gag-Pol synthesis seemed possible in wtFFk/o viruses and high levels of mature p24 and p6 proteins were produced in these mutant viruses, no productive infection of T cells could be obtained. Indeed, 10 days following infection of T-cell line C8166 with wtFFk/o virus, no p24 protein could be detected in culture supernatants by a sensitive ELISA and no cytopathic effect could be observed (Fig. 3B, bottom). This lack of infectivity could be due to the mutations introduced to abolish the original frameshift site.
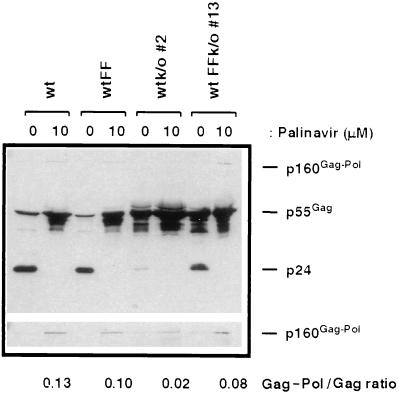
Finally, to unambiguously demonstrate that Gag-Pol synthesis had occurred in wtFFk/o viruses and to confirm the presence of an active protease in these mutants, proviral DNA was transfected in the presence of the protease inhibitor palinavir (15) (Fig. 4). High concentrations of this protease inhibitor should block all protease activity and lead to the accumulation of unprocessed precursors p55gag and p160gag-pol in virions (15). Figure 4 shows that in the presence of 10 μM palinavir, wt viruses indeed did not contain mature p24 protein and that there was an accumulation of the polyprotein precursors p55gag and p160gag-pol due to inactivation of the protease. Quantification of precursor bands by density integration indicated that the Gag-Pol/Gag ratio in these viruses is 0.13. Similar results were obtained with wtFF viruses, where there are about 10 times more Gag than Gag-Pol precursors (ratio of 0.10). In wtk/o viruses, the very low level of mature p24 protein observed previously disappeared in the presence of palinavir, and there was also a slight accumulation of the p160gag-pol precursor, giving a Gag-Pol/Gag ratio of 0.02, about sixfold lower than that observed for wt viruses. In wtFFk/o viruses, p24 production was also completely blocked by the presence of palinavir and significant levels of the p160gag-pol precursor were observed. The Gag-Pol/Gag ratio was estimated to be 0.08 for these mutant viruses, about fourfold higher than that observed for wtk/o viruses. Therefore, Gag-Pol synthesis does occur quite efficiently in wtFFk/o viruses in the absence of the original slippery sequence.
FIG. 4.
Blocking of protease activity in mutant viral clones leads to the accumulation of polyprotein precursors. Viral particles were produced in the presence of the protease inhibitor palinavir (10 μM) and then harvested and analyzed by Western blotting using an anti-p24 monoclonal antibody. Proteins of 24, 55, and 160 kDa, representing the mature p24 protein and the p55gag and p160gag-pol precursors, respectively, were detected. Longer exposure was required for efficient p160gag-pol detection (bottom panel). Gag-Pol/Gag ratios were estimated by comparing results obtained from density integration of the 160- and 55-kDa bands.
These results therefore demonstrate that the U UUU UUU p1/p6 mutant sequence found in HIV protease inhibitor-resistant variants can promote ribosomal frameshifting both in vitro and in virus-expressing cells. This is somewhat surprising because although the mutant sequence does contain a consensus slippery heptamer, the latter is not in close proximity to any 3′ secondary structure, as determined by RNA substructure search analysis (data not shown). The major function of secondary structures is to cause the ribosome to pause, a process during which frameshifting is thought to be more likely to occur (8, 9). Two mechanisms could explain how frameshifting could occur in the absence of 3′ secondary structures. First, as the mutant p1/p6 sequence is a stretch of seven consecutive U’s, it is considered superslippery (2, 24) and may not require ribosomal pausing at all for frameshifting. Second, frameshifting could occur following 5′ base pairing between viral and ribosomal RNAs, a frameshift-enhancing mechanism already described for the Escherichia coli dnax gene (16). Interestingly, analysis of mRNA sequences around the HIV p1/p6 junction reveals 5′ sequences that could pair with eukaryotic small-unit rRNA (bases 2113 to 2117 in the mRNA [Fig. 1B] and bases 1115 to 1119 in the 18S rRNA). Experiments using constructs in which both the original frameshift site and 5′ flanking sequences are altered by mutagenesis could give more insight into these possibilities.
Acknowledgments
The M35/2F8 anti-p6 monoclonal antibody (23) was a generous gift from M. G. Samgadharan (Advanced BioScience Laboratories Inc., Kensington, Md.). We thank M. G. Cordingley for critical review of the manuscript.
L.D. is a recipient of a University-Industry fellowship award from the Medical Research Council of Canada.
REFERENCES
- 1.Adachi A, Gendelman H E, Koenig S, Folks T, Willey R, Rabson A, Martin M A. Production of acquired immunodeficiency syndrome-associated retrovirus in human and nonhuman cells transfected with an infectious molecular clone. J Virol. 1986;59:284–291. doi: 10.1128/jvi.59.2.284-291.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brierly I, Jenner A J. Mutational analysis of the “slippery-sequence” component of a coronavirus ribosomal frameshifting signal. J Mol Biol. 1992;227:463–479. doi: 10.1016/0022-2836(92)90901-U. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Carrillo A, Sham H, Norbeck D, Kempf D, Kohlbrenner W, Plattner J, Leonard J, Molla A. Abstracts of the Fourth Conference on Retroviruses and Opportunistic Infections 1997. Washington. D.C: Infectious Diseases Society of America; 1997. Selection and analysis of HIV-1 variants with increased resistance to ABT-378, a novel protease inhibitor, abstr. 462. [Google Scholar]
- 4.Chamorro M, Parkin N, Varmus H E. An RNA pseudoknot and an optimal heptameric shift site are required for highly efficient ribosomal frameshifting on a retroviral messenger RNA. Proc Natl Acad Sci USA. 1992;89:713–717. doi: 10.1073/pnas.89.2.713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Croteau G, Doyon L, Thibeault D, McKercher G, Pilote L, Lamarre D. Impaired fitness of human immunodeficiency virus type 1 variants with high-level resistance to protease inhibitors. J Virol. 1997;71:1089–1096. doi: 10.1128/jvi.71.2.1089-1096.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Doyon L, Croteau G, Thibeault D, Poulin F, Pilote L, Lamarre D. Second locus involved in human immunodeficiency virus type 1 resistance to protease inhibitors. J Virol. 1996;70:3763–3769. doi: 10.1128/jvi.70.6.3763-3769.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Farabaugh P J. Alternative readings of the genetic code. Cell. 1993;74:591–596. doi: 10.1016/0092-8674(93)90507-M. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Farabaugh P J. Programmed translational frameshifting. Annu Rev Genet. 1996;30:507–528. doi: 10.1146/annurev.genet.30.1.507. [DOI] [PubMed] [Google Scholar]
- 9.Gesteland R F, Atkins J F. Recoding: dynamic reprogramming of translation. Annu Rev Biochem. 1996;65:741–768. doi: 10.1146/annurev.bi.65.070196.003521. [DOI] [PubMed] [Google Scholar]
- 10.Gulnik S V, Suvorov L I, Liu B, Yu B, Anderson B, Mitsuya H, Erickson J W. Kinetic characterization and cross-resistance patterns of HIV-1 protease mutants selected under drug pressure. Biochemistry. 1995;34:9282–9287. doi: 10.1021/bi00029a002. [DOI] [PubMed] [Google Scholar]
- 11.Higuchi R, Krummel B, Saiki R K. A general method of in vitro preparation and specific mutagenesis of DNA fragments: study of protein and DNA interaction. Nucleic Acids Res. 1988;16:7351–7367. doi: 10.1093/nar/16.15.7351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hunter E. Macromolecular interactions in the assembly of HIV and other retroviruses. Semin Virol. 1994;5:71–83. [Google Scholar]
- 13.Jacks T, Power M D, Masiarz F R, Luciw P A, Barr P J, Varmus H E. Characterization of ribosomal frameshifting in HIV-1 gag-pol expression. Nature. 1988;331:280–283. doi: 10.1038/331280a0. [DOI] [PubMed] [Google Scholar]
- 14.Jacks T, Madhani H D, Masiarz F R, Varmus H E. Signals for ribosomal frameshifting in the Rous sarcoma virus gag-pol region. Cell. 1988;55:447–458. doi: 10.1016/0092-8674(88)90031-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lamarre D, Croteau G, Wardrop E, Bourgon L, Thibeault D, Clouette C, Vaillancourt M, Cohen E, Pargellis C, Yoakim C, Anderson P C. Antiviral properties of palinavir, a potent inhibitor of the human immunodeficiency virus type 1 protease. Antimicrob Agents Chemother. 1997;41:965–971. doi: 10.1128/aac.41.5.965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Larsen B, Wills N M, Gesteland R F, Atkins J F. rRNA-mRNA base pairing stimulates a programmed −1 ribosomal frameshift. J Bacteriol. 1994;176:6842–6851. doi: 10.1128/jb.176.22.6842-6851.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Myers G, Korber B, Foley B, Jeang K T, Mellors J W, Wain-Hobson S. Human retroviruses and AIDS: a compilation and analysis of nucleic acid and amino acid sequences. Los Alamos, N.Mex: Theoretical Biology and Biophysics, Los Alamos National Laboratory; 1996. [Google Scholar]
- 18.Park J, Morrow C D. Overexpression of the gag-pol precursor from human immunodeficiency virus type 1 proviral genomes results in efficient proteolytic processing in the absence of virion production. J Virol. 1991;65:5111–5117. doi: 10.1128/jvi.65.9.5111-5117.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Parkin N T, Chamorro M, Varmus H E. Human immunodeficiency virus type 1 gag-pol frameshifting is dependent on downstream mRNA secondary structure: demonstration by expression in vivo. J Virol. 1992;66:5147–5151. doi: 10.1128/jvi.66.8.5147-5151.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pazhanisamy S, Stuver M, Cullinan A B, Margolin N, Rao B G, Livingston D J. Kinetic characterization of human immunodeficiency virus type-1 protease-resistant variants. J Biol Chem. 1996;271:17979–17985. doi: 10.1074/jbc.271.30.17979. [DOI] [PubMed] [Google Scholar]
- 21.Schinazi R F, Larder B A, Mellors J W F. Mutations in retroviral genes associated with drug resistance. Int Antivir News. 1997;5:129–142. [Google Scholar]
- 22.Seed B, Sheen J Y. A simple phase-extraction assay for chloramphenicol acetyletransferase activity. Gene. 1988;67:271–277. doi: 10.1016/0378-1119(88)90403-9. [DOI] [PubMed] [Google Scholar]
- 23.Veronese F, Rahman R, Copeland T D, Oroszlan S, Gallo R C, Sarngadharan M G. Immunological and chemical analysis of p6, the carboxyl-terminal fragment of HIV p15. AIDS Res Hum Retroviruses. 1987;3:253–264. doi: 10.1089/aid.1987.3.253. [DOI] [PubMed] [Google Scholar]
- 24.Wilson W, Braddock M, Adams S E, Rathjen P D, Kingsman S M, Kingsman A J. HIV expression strategies: ribosomal frameshifting is directed by a short sequence in both mammalian and yeast systems. Cell. 1988;55:1159–1169. doi: 10.1016/0092-8674(88)90260-7. [DOI] [PubMed] [Google Scholar]
- 25.Zhang Y-M, Imamichi H, Imamichi T, Lane H C, Falloon J, Vasudevachari M B, Salzman N P. Drug resistance during Indinavir therapy is caused by mutations in the protease gene and in its Gag substrate cleavage sites. J Virol. 1997;71:6662–6670. doi: 10.1128/jvi.71.9.6662-6670.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]