Abstract
Voltage imaging enables high-throughput investigation of neuronal activity, yet its utility is often constrained by a low signal-to-noise ratio (SNR). Conventional denoising algorithms, such as those based on matrix factorization, impose limiting assumptions about the noise process and the spatiotemporal structure of the signal. While deep learning based denoising techniques offer greater adaptability, existing approaches fail to fully exploit the fast temporal dynamics and unique short- and long-range dependencies within voltage imaging datasets. Here, we introduce CellMincer, a novel self-supervised deep learning method designed specifically for denoising voltage imaging datasets. CellMincer operates on the principle of masking and predicting sparse sets of pixels across short temporal windows and conditions the denoiser on precomputed spatiotemporal auto-correlations to effectively model long-range dependencies without the need for large temporal denoising contexts. We develop and utilize a physics-based simulation framework to generate realistic datasets for rigorous hyperparameter optimization and ablation studies, highlighting the key role of conditioning the denoiser on precomputed spatiotemporal auto-correlations to achieve 3-fold further reduction in noise. Comprehensive benchmarking on both simulated and real voltage imaging datasets, including those with paired patch-clamp electrophysiology (EP) as ground truth, demonstrates CellMincer’s state-of-the-art performance. It achieves substantial noise reduction across the entire frequency spectrum, enhanced detection of subthreshold events, and superior cross-correlation with ground-truth EP recordings. Finally, we demonstrate how CellMincer’s addition to a typical voltage imaging data analysis workflow improves neuronal segmentation, peak detection, and ultimately leads to significantly enhanced separation of functional phenotypes.
1. Introduction
Voltage imaging utilizes fluorescent reporters, either small-molecule dyes or genetically encoded proteins, to measure the membrane potential of electrically active cells. Compared to traditional patch-clamp electrophysiology (EP), voltage imaging offers higher throughput and is less invasive. This technique has been used to monitor neuronal electrical activity during behavioral assays in vivo [1], as well as to characterize the functional effects of pharmacological and genetic perturbations in primary and iPSC-derived mammalian neurons in vitro [2, 3]. The increased throughput, control, and flexibility of voltage imaging have enabled significant advances in our understanding of biology. Recent developments have improved the brightness of both voltage-sensitive dyes [4] and heterologously expressed voltage-sensitive proteins [5]. However, the achievable signal-to-noise ratio (SNR) remains limited compared to conventional patch-clamp techniques due to factors such as dye quantum yield, short exposure times (<2 ms) needed to capture neuronal action potentials, and constraints on excitation intensity to prevent sample damage.
Limitations on SNR have two practical effects. First, small-magnitude electrical events of interest, such as subthreshold post-synaptic potentials, could be lost in the background temporal noise. Previous work powered by tracking the timing of action potential firing between neurons provided some insight to how neurons might wire together in small circuits. However, the site of inter-neuronal communication is the synapse. A measure of synaptic connectivity, demonstrated as subthreshold activity measured at the soma, would provide a greater understanding of how neuronal circuits are formed and how synaptic connections are modified during different forms of plasticity. Second, cells expressing relatively low amounts of fluorescent reporters can be lost in a comparatively high autofluorescent background, lowering the effective throughput of voltage imaging.
These technical challenges have motivated the development of data denoising algorithms to computationally enhance the SNR and enable the recovery of obscured and subtle fluorescent signals. Matrix factorization is an effective class of algorithms for fluorescence image denoising [6] [7], as the sparse and static signal sources (e.g. neurites) in these imaging assays create an ideal setting for approximating entire fluorescence recordings as low-rank decompositions. Principal component analysis (PCA), non-negative matrix factorization (NMF), and penalized matrix decomposition (PMD) [8] are popular implementations of this concept. These approaches, while being highly efficient and effective at data denoising, suffer from a number of caveats. These include: (1) implicit parametric assumptions on the nature of the noise that are theoretical approximation of the actual complex data generating process; (2) usage of spatiotemporal regularizations to encourage robustness and model identifiability, such as total variation penalty or temporal continuity, that are often violated (e.g. spike events, spatially heterogeneous expression of the fluorescent reporter); (3) making strong assumptions about the background fluorescence component to allow their approximate subtraction as a simple data preprocessing step. These modeling assumptions, while laying a strong foundation, ultimately hamper the expressivity of conventional denoising algorithms.
We envision that the ideal denoising algorithm should minimize the explicit assumptions made about the noise process while maximizing the potential to learn the complex spatiotemporal relationships that govern the signal. Deep neural networks (DNNs), which have no theoretical limit to complexity, can in principle solve the issue of denoising model expressivity. However, deep learning denoising models pretrained on large datasets of natural images [9] are ill-equipped to operate in the low-SNR regime of fluorescence imaging [10], requiring a suitable model to be trained from scratch for this data domain. This immediately poses a challenge for supervised learning approaches which require clean images as a learning target, which are not available in voltage imaging. A powerful recent training strategy, called self-supervised denoising, circumvents the requirement of having clean ground truth data by exploiting a key property of many noise processes: by appropriately partitioning the raw data into compartments, and predicting one compartment from the other, it is often possible to eliminate predictors of noise while retaining the ability to predict the underlying signal. Noise2Noise (N2N) [11] and Noise2Self (N2S) [12] are prominent examples of self-supervised denoising techniques proposed for images. These methods have consistently been shown to produce state-of-the-art results, including in fluorescence imaging, even compared to counterparts that are trained on pairs of noisy and clean data [10]. In particular, the Noise2Self algorithm, which we use as a foundation to build upon here, operates on the following elegant and simple principle: suppose a sparse set of pixels are masked out from a noisy image, and a neural network is trained to predict the value of the sparsely masked pixels from the rest of the image, i.e. the majority of pixels. Assuming that the noise in the masked pixels is uncorrelated with the rest of the pixels, the optimal predictor can at best predict the noiseless signal component; in practice, it can excel at this task given the strong spatial correlations and redundancy in biological images. It follows that the optimal masked pixel predictor in turn behaves as an optimal pixel denoiser. Noisy data itself provides the needed evidence for teasing out the signal component, circumventing the need for clean training data.
One of the main challenges in extending self-supervised image denoising approaches to spatiotemporal data, such as voltage imaging recordings, is that these datasets contain thousands of frames, and that each frame is individually too signal-deficient to self-supervise its own denoising. At the same time, GPU hardware memory constraints and efficient training considerations prevent us from ingesting and processing entire voltage imaging movies with neural networks to exploit frame-to-frame correlations. The middle ground strategy adopted by several authors is to process the movie in overlapping and truncated local denoising temporal contexts, i.e. chunks of adjacent frames. For instance Li et al. [13] developed DeepCAD, a Noise2Self-like denoising method based on reconstructing whole masked frames from temporally downsampled movie chunks, and demonstrated its capacity to restore a high imaging SNR from low-SNR calcium imaging recordings. Lecoq et al. [14] developed DeepInterpolation, a Noise2Noise-like whole-frame interpolation-based deep learning method acting on small temporal windows which also allowed them to increase the SNR and retrieve a significantly higher fraction of neuronal segments from calcium imaging.
These existing approaches are sub-optimal for two important reasons. (1) As we will show in later sections, while such leave-frame-out approaches work remarkably well for calcium imaging, the denoising performance degrades strikingly when the same methods are applied to voltage imaging data (see Sec. 2.3). The much faster temporal dynamics of voltage imaging data compared to calcium imaging imply that each movie frame contains unique and valuable signal that cannot be entirely inferred from the adjacent frames. For instance, the evidence for a neuronal spike is most prominently present in a single frame. (2) Unless the local denoising context is impractically large and contains hundreds of movie frames, the neural network is incapable of estimating long-range pixel-to-pixel temporal correlations that are arguably key to effective signal extraction and noise removal. As we will show in later sections, explicitly precomputing and supplementing the short-context local denoiser with such information results in a striking boost in the denoising performance.
In this work, we introduce CellMincer, a self-supervised deep learning method specifically designed for denoising voltage imaging datasets based on the Noise2Self denoising framework. CellMincer introduces several key refinements over the currently existing self-supervised movie denoising methods to address the aforementioned caveats. The key methodological contributions of CellMincer include: (1) development of an efficient and expressive two-stage spatiotemporal data processing deep neural network architecture, comprising a frame-wise 2D U-Net module for spatial feature extraction, followed by a pixelwise 1D convolutional module for temporal data post-processing; (2) replacing the common task of whole-frame prediction with masking and predicting a sparse set of pixels from a small number of adjacent frames; this training methodology allows the denoiser to have access to the unique information contained in any individual frame as well as the supporting context in its neighboring frames; (3) precomputing spatiotemporal auto-correlations at multiple length scales, and providing such precomputed statistics as a conditioner to the denoiser neural network (that otherwise processes smaller spatiotemporal regions of the movie at a time); (4) developing and leveraging a physics-based simulation framework to generate highly realistic pairs of clean ground truth and noisy recording realization for hyperparameter optimization and performing ablation studies to tease apart the roles of various modeling choices in a controlled setting.
Using benchmarking experiments performed on simulated data and real voltage imaging data with paired patch-clamp EP recordings as a proxy for ground truth, we show that CellMincer yields state-of-the-art results as measured in terms of several practical metrics. These include a peak signal-to-noise ratio (PSNR) average gain of 24 dB compared to the raw data (an increase of 2 dB over the next best benchmarked method), a 14 dB reduction in high-frequency (>100 Hz) noise (a further reduction of 10.5 dB from the next best method), a 5–10 percentage point increase of -score in detecting sub-threshold events compared to the other algorithms and across all voltage magnitudes in the 1–10 mV range (in which the baseline -score ranges from 5–14%), and more than 20% increase in the cross-correlation between low-noise EP recordings and voltage imaging. A striking result from our ablation study is the pivotal role of conditioning the denoiser on precomputed global features, resulting in a nearly 5 dB (or approximately 3-fold) boost in average PSNR gain, as well as a highly-concentrated distribution of PSNR gain across all frames and electrical stimulation amplitudes (see Fig. 1e). Finally, to demonstrate the utility of CellMincer to real end-to-end biological hypothesis testing, we compare the voltage imaging of chronically tetrodotoxin-treated and unperturbed cultured hPSC-derived neurons, and demonstrate that CellMincer denoising enables reliable identification and segmentation of nearly 2-fold as many neurons as in the raw data, improved identification of spiking events, and ultimately significantly enhanced statistical separation between the two functional phenotypes.
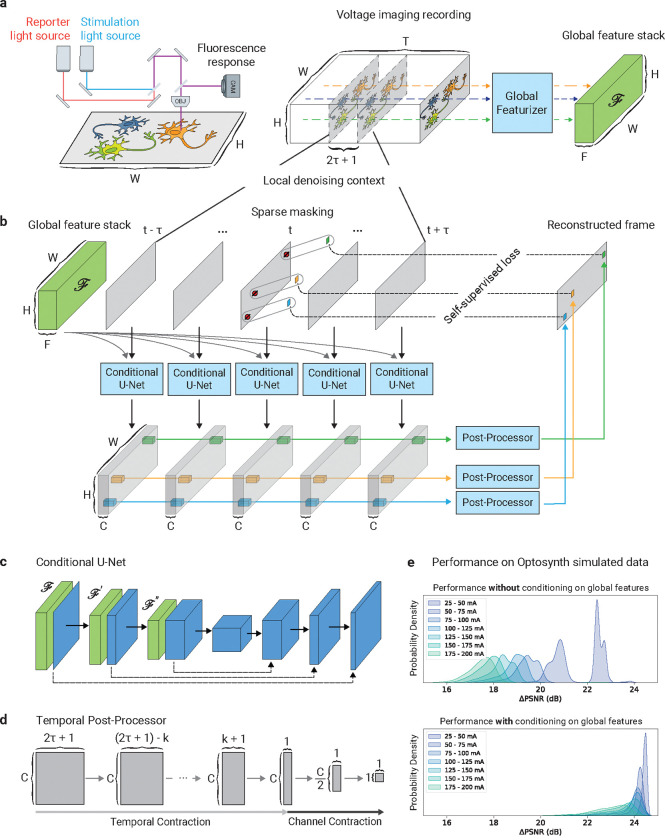
Figure 1:
Overview of voltage imaging data and CellMincer denoising model. (a) A simplified schematic diagram of a typical optical voltage imaging experiment (left). The spatially resolved fluorescence response is recorded over time to produce a voltage imaging movie. A key component of CellMincer’s preprocessing pipeline is the computation of spatial summary statistics and various auto-correlations from the entire recording, which are concatenated into a stack of global features (right). (b) An overview of CellMincer’s deep learning architecture. (c) The conditional U-Net convolutional neural network (CNN). At each step in the contracting path, the precomputed global feature stack is spatially downsampled in parallel and concatenated to the intermediate spatial feature maps. (d) The temporal post-processor neural network. The sequence of pixel embeddings are convolved with a 1D kernel along the time dimension, producing a single vector of length . A multilayer perceptron subsequently reduces this vector to a single value. (e) A compaxrison of model performance on simulated data before and after introducing global features as a U-Net conditioner. The distributions of PSNR gain are binned by stimulation amplitude. Using global features confers an average increase of 5 dB to the denoiser, roughly corresponding to a 3-fold noise reduction.
2. Results
2.1. CellMincer self-supervised denoising framework
The CellMincer denoising pipeline involves three stages: (1) data preprocessing and global feature extraction; (2) self-supervised pretraining of the denoising neural network; (3) inference of denoised movie. In the preprocessing step, we take a recording represented as a three-dimensional tensor with shape . We treat each pixel as a separate time series of samples, fit a low-order polynomial function to each, and thereby decompose the movie as a sum of smooth trend and detrended residual tensors. The trend tensor primarily represents the background fluorescence, whereas the residual detrended tensor represents a noisy measurement of the electrical activity. Going forward, we perform self-supervised denoising only over the detrended residual component, and add the smooth trend component back after the inference step. To set the stage for self-supervised pretraining, we calculate various summary statistics for each pixel, including temporal mean, temporal variance, and all bidirectionally lagged spatiotemporal auto-correlations with adjacent pixels. These statistics are computed separately for both the slow moving average and the fast residual components of the detrended movie, and at two different spatially downsampled resolutions to account for auto-correlations with longer spatial lags. Finally, we concatenate these precomputed statistics as a tensor of shape to represent pixelwise global statistics, where is the total number of computed statistics per pixel. Supplemental Sec. S.3 fully describes our preprocessing and feature extraction stage. This step is schematically referred to as global featurizer in Fig. 1a.
We present the denoising strategy we employ in CellMincer in two stages for clarity. First, we describe the architecture of the DNN that we purport to be capable to performing efficient denoising. Next, we describe the self-supervised training strategy a la Noise2Self that allows the denoiser to train without clean targets.
Our proposed denoising DNN takes as input a series of consecutive frames, corresponding to time points , from the detrended movie and aims to predict a denoised reconstruction of the frame in the middle, at time point . We refer to as the temporal order, and to as the context size of the local denoiser. Crucially, the DNN additionally takes the precomputed global feature stack as a conditioner to supplement the local denoiser with long-range spatiotemporal statistics. The architecture of the denoising DNN consists of a single-frame spatial feature extractor followed by a temporal post-processor (see Fig. 1b). The spatial component is implemented as a U-Net convolutional neural network (CNN) with a small but consequential modification: to condition the convolutional operations on , we concatenate an appropriately spatially downsampled copy of prior to each convolution block on the contracting path (see Fig. 1c). The conditional U-Net extracts deep, native-resolution -channel single-frame embeddings from each of the consecutive frames (see Fig. 1b). The resulting embeddings are concatenated into a 4D tensor of shape :
| (1) |
where denotes concatenation along the time dimension. This intermediate tensor is routed to the temporal post-processor, which consists of a series of temporal convolutional layers, reducing each set of pixel embeddings across all frames to a final output pixel. The output of the temporal post-processor represents a denoised reconstruction of the middle frame:
| (2) |
Note that the temporal post-processor treats pixels as independent, only operating on time and U-Net feature channels (see Fig. 1d). This two-stage constrained network design enables efficient spatiotemporal data processing by logically compartmentalizing the flow of information; the U-Net facilitates information mixing across pixels within individual frames, while the post-processor convolves information across frames for individual pixels. Refer to Supplemental Sec. S.4 for architectural details.
We train the CellMincer denoiser in a self-supervised fashion as follows. At the beginning of each training iteration, pixels chosen at random in the frame at time are replaced with Gaussian noise with pixel-specific in-distribution mean and variance before the frame is fed into the network:
| (3) |
Here, is a binary mask containing a sparse number of ones, and and correspond to the temporal mean and standard deviation of the detrended movie at position . These masked pixels are then used as the training targets, where the loss is computed between the network’s predicted values and their pre-masked values with the following loss function:
| (4) |
where (see Fig. 1b). Here, , refer to the preceding and the following frames surrounding the frame at time , respectively. As for a choice of pixel loss function, we have experimented with both and 2 and found the latter to result in higher PSNR (see Supplemental Sec. S.6). Even though these pre-masked target values do not represent actual ground truth but noisy realizations, their noise contribution cannot be predicted by the neural network provided that the masking mechanism decorrelates the pixel noise between masked and unmasked compartments (-invariance, see Ref. [12]). In fluorescence imaging, the main source of noise is pixelwise Poisson-Gaussian noise, which is uncorrelated across pixels, allowing us to satisfy the -invariance condition as a matter of masking individual pixels. CellMincer’s implementation additionally permits the use of alternate masking mechanisms (e.g. inclusion of margin around each masked pixel) if needed to account for correlated noise. Crucially, since the identities of the masked pixels are not revealed to the denoiser (e.g. implicitly by using a special masked pixel value, as is typically done), the network is incentivised to denoise not only the sparse subset of masked pixels but the frame in its entirety. This training strategy allows CellMincer to operate very efficiently at inference time, when we feed the noisy detrended movie in -length overlapping sliding windows to the network and denoise each window’s entire middle frame. To avoid the typical practice of producing truncated results, we augment all model inputs with appropriate spatial padding at training and inference time, and we pad the beginning and end of the denoised movie with copies of its first and last frame respectively.
Our implementation of CellMincer can jointly train on many datasets across multiple GPUs to produce a highly generalizable model, but satisfactory results can be achieved by training on a single voltage imaging dataset with as few as 5000 frames. Because of the model’s self-supervised training scheme, the dataset to be denoised can also serve as the model’s only training data.
2.2. Architecture optimization of CellMincer via realistic physics-based simulations
To optimize the architecture and hyperparameters of CellMincer and study the impact of various design choices on the baseline denoising performance, one needs noiseless ground truth voltage imaging data. While experimental sourcing of true noiseless data is impractical due to technical limitations (e.g. the trade-off between signal-to-noise ratio and sampling rate, photo-bleaching and sample heating at higher illumination), we can aim to generate such ground truth recordings and their noisy realizations via carefully crafted simulations. These simulated data can then be used to study and optimize the model architecture and serve as a benchmark to evaluate the performance of CellMincer compared to other denoising methods.
To these ends, we developed Optosynth, a methodology for generating physics-based synthetic optical voltage imaging data using single-neuron morphological reconstructions and paired EP measurements from the Allen Brain Patch-seq dataset [15–17]. In brief, Optosynth simulates a noiseless voltage imaging readout by sampling neurons from a Patch-seq dataset, arranging them on a synthetic imaging field, and modeling the fluorescence signal density as an appropriate conversion function of the measured membrane potential. To produce realistic voltage imaging readouts, we additionally include low-passing of EP to the fluorescence sensor sampling rate, action potential wavefront propagation and decay, variability in fluorescent reporter expression, point spread function (PSF), and static and dynamics background autofluorescence. We generate noisy readings from noiseless simulations by adding Poisson shot noise and Gaussian sensor thermal noise. Optosynth’s simulations are highly customizable, enabling generation of synthetic datasets that can represent a wide range of experimental conditions, noise levels, and magnifications. A detailed description of Optosynth is provided in Supplemental Sec. S.5.
The CellMincer model is specified by a large set of hyperparameters which determine the architecture of the underlying DNNs, the self-supervised training parameters, and the optimizer scheduling. To optimize over this hyperparameter space, we first identified a baseline configuration that specifies a design empirically capable of denoising our Optosynth datasets and training it to sufficient convergence. We then constructed a series of single-hyperparameter variations of the baseline configuration and evaluated their performance on Optosynth data. Our hyperparameter variations included the inclusion or exclusion of global features , the length of the denoising window , the U-Net parameters (depth, number of channels), the temporal post-processor architecture, the loss function, and the rate of pixel masking during self-supervised training. Our evaluation procedure consisted of training the model on a subset of our Optosynth data, denoising both the training data and unseen data (biological replicates generated using Optosynth) with our trained model, and computing the denoised imaging’s peak signal-to-noise ratio (PSNR) with respect to the ground truth. These results determined our final selection of hyperparameters used in subsequent benchmarking experiments.
Foremost, we found that conditioning the U-Net on global features produced the most significant improvement by wide margins, up to 5 dB gain in PSNR (see Supplementary Fig. S 2b, rows 1–3). Without inclusion of global features, model performance gains relied heavily on increasing the local denoising temporal context windows (see Supplementary Fig. S 2b, rows 1, 4–7; the context size is varied from 5 to 21 frames ~ 10–42 ms). We note that such large temporal context sizes exceed the observed temporal correlation lengths in voltage imaging (see lagged cross-correlations in Fig. 2d and Fig. 3c), suggesting that the unconditioned denoiser is taking advantage of large context sizes to infer pixel-to-pixel spatial correlations rather than temporal correlations. To further underscore this point, we note that PSNR gains of a denoiser explicitly conditioned on precomputed global auto-correlations saturate between 5 and 9 frames ~ 10–18 ms, which coincides with the typical temporal correlation length in neuronal activity (see Supplementary Fig. S 2e, rows 1–5). Clearly, precomputing global features and conditioning the denoiser is a much more effective and computationally efficient alternative to using longer denoising context windows.
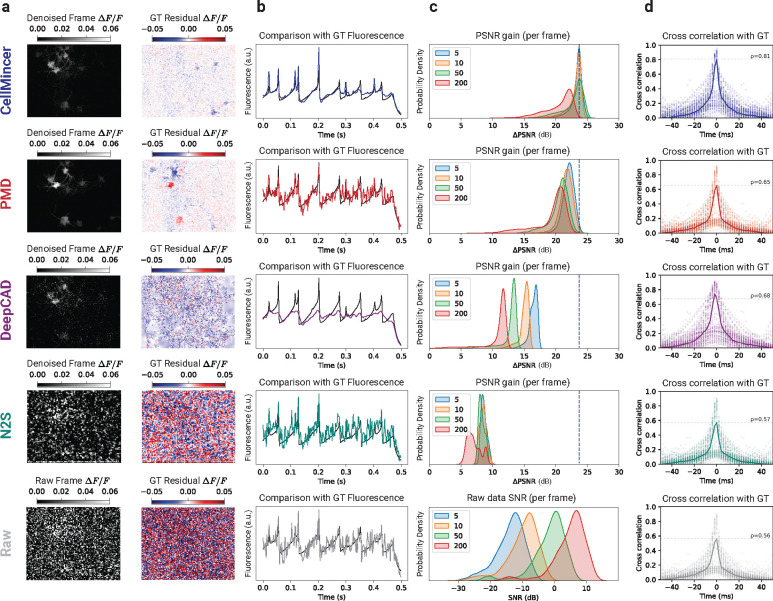
Figure 2:
Benchmarking CellMincer and three other denoising methods on simulated voltage imaging. (a) Sample denoised frame visualizations (grayscale images) and their residuals with respect to simulated ground truth imaging (red/blue images). Both the denoised and residual images are shown as relative change in fluorescence with respect to a frame-averaged polynomial regression of the baseline (see Supplemental Sec. S.3). (b) Sample denoised ROI-averaged neuron traces (color), overlaid with the ground truth (black). (c) Distributions of single-frame PSNR gain achieved through denoising. Each distribution corresponds to a different value of simulated photon-per-fluorophore count (shown in the legend), which is the measure of raw data SNR in Optosynth simulations (see Supplemental Sec. S.5). The dashed vertical line over the top four rows is a guide for the eye and indicates the mode of CellMincer’s PSNR gain distribution for the lowest SNR data (corresponding to ). The plot at the bottom row shows the SNR distributions of the raw datasets at different levels. (d) Distributions of lagged cross-correlations between denoised single-neuron traces and their ground truths. Their medians are overlaid with peak correlations at labeled. Abbreviations: GT (ground truth).
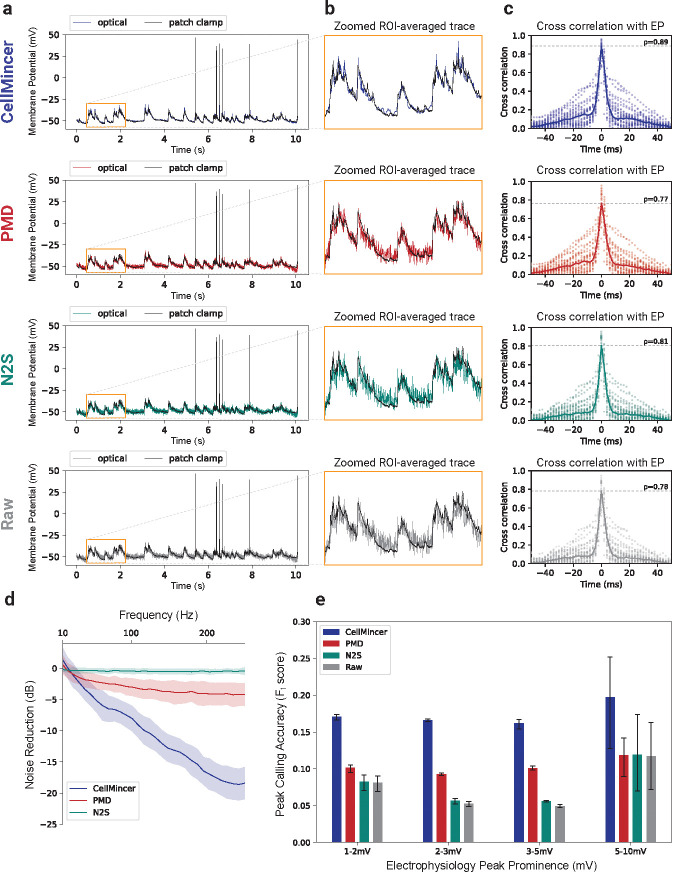
Figure 3:
Benchmarking CellMincer and two other denoising methods on paired optical and patch clamp datasets. (a) Sample denoised ROI-averaged neuron traces (color), aligned to the EP-derived ground truth (black). (b) Inlays of subthreshold activity as indicated in the previous column, magnified. (c) Distributions of lagged cross-correlations between denoised single-neuron traces and their corresponding aligned EP signals. Their medians are overlaid with peak correlations at labeled. (d) Average noise reductions at varying frequency ranges achieved through denoising. (e) Peak-calling accuracy F1-scores over a range of EP peak prominence levels, using the EP signal as ground truth. Abbreviations: ROI (region of interest).
Another advantage of conditioning the denoiser on precomputed global features is achieving more robust PSNR gain characteristics across different stimulation amplitudes. This can be seen by comparing the violin plots of PSNR gain distributions for unconditioned and conditioned denoisers in Supplementary Fig. S 2. The PSNR gain distribution of the unconditioned denoiser (baseline, first row, panel b-c)) varies from +16 dB to +23 dB, and is highly variable in particular for recordings at lower electrical stimulation amplitudes (shown in blue). This observation further underscores that the performance of an unconditioned denoiser relies on its ability to infer correlations solely from the local context, which can be unreliable when neurons are not active under low stimulation. In contrast, the PSNR gain distributions of all conditioned model variants (rows 2–2, panel b-c, and all rows of panel e-f) are tightly concentrated from +22 dB to +24 dB.
Besides the crucial importance of conditioning the denoiser on global features, we find that other design decisions (like U-Net depth, number of channels, loss function selection, and the amount of masked pixels) have a surprisingly small effect on model performance. This indicates that CellMincer works reliably and does not require extensive parameter fine-tuning when used with other voltage imaging datasets. A more detailed description of these optimization experiments and their results are provided in Supplemental Sec. S.6.
2.3. CellMincer outperforms existing methods at denoising simulated voltage imaging data
For our benchmark evaluations, we include a selection of denoising algorithms applicable to movie-like data. Importantly, the algorithms share the precondition that clean reference data for training is not needed. DeepCAD [13] is a self-supervised deep learning algorithm for denoising calcium imaging data. Penalized matrix decomposition (PMD) [8] is a training-free algorithm based on a regularized, low-rank factorization of the data. As a baseline, we also include the original implementation of Noise2Self (N2S) [12] for images, which denoises movie frames individually.
To cover a range of noise conditions, we chose four different SNRs by varying the number of photons-per-fluorophore in simulations (see Supplemental Sec. S.5). For each level of SNR, denoted in increasing order as , we generated five synthetic datasets using Optosynth with associated ground truth. We then trained CellMincer and each of the other training-based denoising methods on three of the five datasets and subsequently used them to denoise all five datasets. PMD, which is a single-sample denoising algorithm, was used to individually denoise the five datasets instead. With these denoised datasets along with the original noisy datasets, we conducted a series of evaluations centered on comparing them to our ground truth imaging. We present our Optosynth benchmarks in Fig. 2.
To visualize imaging quality, our first benchmark evaluation compared the results of denoising a single movie frame in both absolute intensity and residual intensity with respect to ground truth, of which CellMincer and PMD exhibit notably cleaner residual frames (Fig. 2a). However, PMD retained significant error at neuron locations, while DeepCAD and N2S were less effective at removing noise throughout the frame. Our next evaluation explored the resolution of single-neuron signals from the imaging. To extract these signals, we inferred single-neuron masks from the raw data (Supplemental Sec. S.8) and used them to produce ROI-averaged traces. We overlaid these traces with their respective ground truth for a sample neuron (Fig. 2b) and observed significantly less noise from CellMincer and DeepCAD. However, we noted that DeepCAD, unlike the other methods, does not fully reconstruct the spiking events. After establishing a visual evaluation of CellMincer and the compared methods, we sought to quantify these differences with the PSNR metric. In column c, we compared the distributions of single-frame PSNR gain achieved by denoising. Each frame PSNR was computed over the union of pixels contained in the neuron ROIs gathered from all five datasets, and only frames during stimulation periods were considered. These restrictions mitigated the influence of background pixels in our performance metric. We found that CellMincer demonstrates a consistent lead in PSNR gain over the other algorithms. Additionally, CellMincer is more consistent across our range of input SNRs, while the other methods yield a clear reduction in PSNR gain for higher-quality datasets. For reference, the SNR distribution of the raw datasets are given at the bottom of column c for each of the four simulated scenarios. Finally, we aimed to show that CellMincer does indeed reconstruct the activity exhibited in the ground truth single-neuron traces without temporal bias. In column d, we computed the distributions of lagged cross-correlations between the denoised and ground truth traces over the Optosynth neurons at and overlaid the median. All cross-correlations sharply peaked at , and CellMincer exhibited a zero-lag median cross-correlation of , significantly higher than of DeepCAD (0.68) and PMD (0.65).
2.4. CellMincer improves the detection of subthreshold events in real voltage imaging data with paired electrophysiology
To extend our evaluation of CellMincer to real data, we further evaluated CellMincer on 26 external datasets from a previously published study with simultaneous voltage imaging with chemically-synthesized voltage-sensitive fluorophore, BeRST [18, 19], and patch-clamp EP recordings, the latter of which can be repurposed as a high-confidence source of ground truth. BeRST is a chemically-synthesized far-red voltage-sensitive fluorophore. Previously, we showed that BeRST can be used in cultured rat hippocampal neurons to track changes in neuronal activity in models of development and disease [20]. Data from that study contained simultaneous recordings of BeRST fluorescence (voltage imaging) and single-cell patch clamp recordings (EP) that could serve as a ground truth for benchmarking of CellMincer. Please refer to Supplemental Sec. S.2 for the details of the experimental procedure.
Our aim in the following benchmarking experiments was to extract single-neuron denoised imaging traces and compare them to their associated patch-clamp EP signals. While both modalities operate on the same underlying neural activity, they differ substantially in sampling rate, noise characteristics, and artifacts. It is thus necessary for us to minimally resolve these data modality incompatibilities by applying a series of common filters and transformations to map them onto a shared scale. These include removing a slowly-varying trend from both measurements, temporal alignment of the two recordings, and performing a global affine transformation on the detrended fluorescence recordings to make them comparable in scale to EP recordings (in mV). See Supplemental Sec. S.8 for a detailed description of our alignment procedure.
Using an analogous approach to that presented in our simulated data benchmarking, we trained the deep learning based models on as many as all 26 of the available joint datasets, depending on the capabilities of the model implementations. DeepCAD was excluded from this benchmark due to difficulties with training it on large quantities of data, as well as its previously noted poor performance in reconstructing spiking events (see Fig. 2). With these trained models, we identified a subset of 22 datasets exhibiting discernible activity suitable for benchmarking purposes. Each model was used to denoise these benchmarking datasets, while PMD, as before, denoised each dataset individually. From the resulting denoised datasets, we extracted ROI-averaged traces and aligned them to their corresponding EP signals. The concordance between these aligned traces to the underlying EP activity forms the basis of our benchmark results, shown in Fig. 3.
Our first step was to visualize the quality of our imaging traces after mapping them to the EP scale. From plotting these traces against their corresponding EP signals (Fig. 3a), we found that CellMincer again exhibited significantly less noise than the other algorithms. The improvement is particularly apparent when examining the baseline trace relative to subthreshold activity (Fig. 3b). To characterize this noise reduction, we computed the spectral power, binned by frequency, of the residual signal before and after denoising for each algorithm (Supplemental Sec. S.9). We plot the reduction in power of these residuals, measured in dB, across the frequency range (Fig. 3d). At frequencies above 100Hz, CellMincer achieves an average noise reduction of 14.3 dB, far greater than that of PMD (3.8) and N2S (0.5). Using a process analogous to that in our simulated data benchmark, we also compared the distributions of lagged cross-correlations between the voltage imaging and EP data (Fig. 3c). CellMincer similarly exhibited a higher median cross-correlation (), while the other algorithms remained on par with the raw data (0.78).
After demonstrating that CellMincer, by way of its enhanced noise reduction, could potentially resolve signals of a smaller magnitude than that which can be seen by the other algorithms, we sought an approach to quantify this small-signal reconstruction fidelity. Due to difficulties with reliably processing these imaging traces with tools designed for EP signals, we devised an analytic method based on peak-calling (Supplemental Sec. S.9). From this analysis, we plotted the peak-calling accuracy of each algorithm as an F1-score, binned over several ranges of peak magnitudes (Fig. 3e), and found that CellMincer exhibited a 1.7 to 3-fold increase in F1-score over the other benchmarked algorithms and the raw data across all peak magnitude ranges. CellMincer was the only algorithm to maintain F1 score > 0.15 across all ground truth mV changes (from 1 mV to 10 mV). At the 5–10 mV range, in contrast to CellMincer, the other two denoising methods (PMD, N2S) do not show any statistically significant improvement over the raw data. At changes below 5 mV, only CellMincer and PMD improve the accuracy over the raw data while N2S does not. Furthermore, CellMincer outperforms the accuracy of PMD significantly in this regime. These evaluations demonstrated that on real voltage imaging, CellMincer produces significant quantitative improvements in the reconstruction of single-neuron traces when compared to traces derived from the raw data and from data denoised by standard algorithms. These improvements have meaningful implications for the potential uses of CellMincer denoising to recover underlying subthreshold activity using voltage imaging data.
2.5. CellMincer improves neuron segmentation and detection of subtle changes in neural activity
To demonstrate the utility of CellMincer in a representative end-to-end biological hypothesis testing workflow, we present a complete such analysis with and without CellMincer as a data-denoising component, and we quantify the impact of CellMincer on improving the detection of subtle phenotypic changes. Specifically, we compare the spiking activity of unperturbed and chronically tetrodotoxin (TTX)-treated cultured hPSC-derived neurons via Optopatch voltage imaging. TTX is a voltage-gated sodium channel blocker which, when used to treat cultured neurons, prevents them from firing action potentials. Prolonged silencing with TTX increases intrinsic excitability of neurons [21]. This homeostatic plasticity is also displayed in hPSC-derived neurons [3]. We incubated hPSC-derived neuronal cultures in 500 nM TTX for 48 hours and washed it out prior to Optopatch recordings. Parallel unperturbed cultures were incubated in TTX-free media. In both cases, we subjected the neurons to eight stimulation periods in increasing intensity and measured their action potential via Optopatch voltage imaging. Please refer to Supplemental Sec. S.1 for the details of the experimental procedure.
We analysed the obtained recordings as follows. We performed a pixelwise detrending preprocessing step on both raw and denoised datasets, computed independent spatial components using a PCA/ICA decomposition approach [22], and identified neuronal components through careful comparative review of the obtained components and the activity traces. We finally derived an ROI-averaged trace from each identified neuron for downstream analysis, which focused on counting and comparing the statistics of high-amplitude action potential spikes. Additional details are provided in Supplemental Sec. S.10.
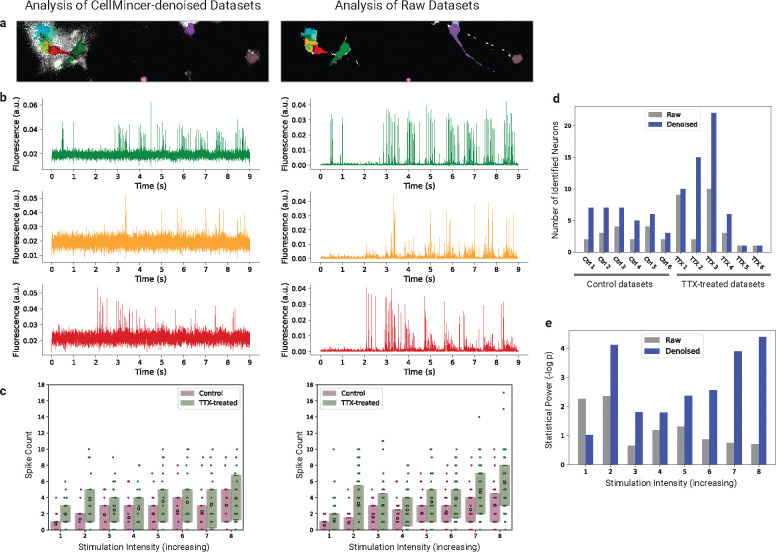
Fig. 4a showcases a segmentation of identifiable neurons on a sample frame from the raw and CellMincer-denoised analysis. It is evident that: (1) among the neurons that are reliably identifiable in both the raw and denoised dataset, CellMincer more clearly delineates their boundaries. This is particularly evident among the cluster of overlapping neurons shown on the left side of the field of view in Fig. 4a); (2) CellMincer enables better separation and detection of more neuronal components, in particular neurons with fainter fluorescence signal, as well as more reliable spike-counting. To substantiate the latter, we plotted the ROI-averaged traces from three neurons side-by-side in Fig. 4b (color-matched to the neuron components shown in Fig. 4a). As explored in the previous experiments, the most salient improvement brought about by CellMincer is in the form of a significant reduction in the background noise (see Fig. 3c). In the present context, this has the effect of highlighting subtler spiking events and enabling them to be called with greater confidence (compare the raw and denoised traces shown in Fig. 4b). The total number of confidently detected neurons are shown in Fig. 4d and establishes that in most recordings, CellMincer denoising allows identification of twice or more as many neurons with distinct spiking patterns. As a result of improved neuron segmentation and spike counting following denoising, aggregating spike statistics over more neurons results in a larger statistical separation between the control and chronically TTX-treated populations. This can be visualized by comparing the boxplots shown in Fig. 4c. To further quantify this finding, we performed a Wilcoxon rank sum test, the result of which is shown in Fig. 4e. Notably, CellMincer denoising yields significantly greater statistical power to separate the two conditions, with this separation increasing at higher stimulation intensities, as evidenced in the tail-end of Fig. 4e. Interestingly, the lowest stimulation intensity shows a deviation from this trend, yet it still aligns with the overall conclusion that chronically TTX-treated neurons exhibit heightened excitability.
Figure 4:
Comparing the spiking activity of chronically tetrodotoxin (TTX)-treated vs. control hPSC-derived neurons with raw and CellMincer-denoised Optopatch voltage imaging data. (a) Raw and denoised versions of a sample frame, colored with the neuron components identified in their corresponding datasets. (b) Corresponding ROI-averaged single-neuron traces detected in both versions of the above frame. (c) Spike count distributions, separated by neuron population and stimulation intensity. Spikes were identified in each detected neuron’s trace and binned by their stimulation intensity. (d) Detected neuron counts in the raw and denoised versions of each dataset. (e) Statistical power of the Wilcoxon Rank Sum test applied to the neuron population differentiation hypothesis, reported as the negative logarithm of its p-value.
3. Discussion
We introduced CellMincer, a self-supervised deep learning method specifically designed for denoising voltage imaging datasets, and discussed several key methodological refinements over the existing approaches. These include: (1) an efficient and expressive two-stage spatiotemporal data processing deep neural network architecture, comprising a frame-wise 2D U-Net module for spatial feature extraction, followed by a pixelwise 1D convolutional module for temporal data post-processing; (2) performing self-supervised training by masking a sparse set of pixels rather than entire frames, allowing the model to access both intra- and inter-frame information as needed for effective denoising of voltage imaging datasets; (3) conditioning local denoisers on a set of precomputed spatiotemporal auto-correlations at multiple length scales, resulting in a significant boost in denoising accuracy; (4) introducing a physics-based simulation framework to generate highly realistic pairs of clean and noisy voltage imaging movies for the purpose of hyperparameter optimization and ablation studies. We evaluated CellMincer’s performance on both simulated and real datasets, including an external previously published dataset comprising 30 voltage imaging experiments with simultaneous patch-clamp EP recordings [20], and established that CellMincer outperforms the existing denoising approaches. Finally, we demonstrated the utility of CellMincer in downstream analyses, resulting in a more robust identification of neurons and spiking events and ultimately a higher statistical power for separating neuron populations based on functional phenotypes.
CellMincer denoising holds the potential to advance the study of complex neuronal communication through multiple avenues. Firstly, traditional methods often involve measuring postsynaptic potentials at the cell body to understand synaptic transmission. However, the inherent biophysical properties of synapses, coupled with the intricate dendritic morphology, such as shape, branching, and diameter, can distort electrical signals originating at the synapse. Consequently, the activity recorded at the soma may not accurately depict events at the synapse. This discrepancy poses a challenge in electrophysiological techniques, which predominantly require recordings at the soma. CellMincer, however, presents a promising solution by facilitating the direct examination of electrical activity at the synapse through voltage imaging techniques and computational SNR enhancement. Secondly, CellMincer denoising enables the collection of usable data even from low magnification recordings with low signal-to-noise ratios (SNRs). For instance, enough data was acquired in seconds to clearly separate the TTX-treated groups (Fig. 4). This is in contrast to single cell recordings either by patch-clamp EP or high magnification voltage imaging, which would take multiple recording days.
CellMincer’s improved performance over similar denoising methods largely stems from precomputing global movie features and using these features as a conditioner for the denoiser network that otherwise operates on small temporal contexts. The inclusion of global spatial features alongside CellMincer’s localized context processing allows the algorithm to exploit persistent long-range correlations in the data without directly ingesting the entire dataset with a neural network, an intractable computational operation. These carefully crafted features (see Supplemental Sec. S.3) meaningfully contribute to data modeling because the neuron signal sources are generally fixed in space, making the behavior of individual pixels, as well as pixel-pixel relationships, highly consistent. Our ablation studies reveal that providing global features to the denoiser results in a striking 3-fold boost in the PSNR (or approximately a 5 dB gain). While increasing the denoising context size endows the local denoiser with more global information and improves its performance, we notice that there is still a ~ 4 dB gap between an unconditioned denoiser with a context size of 21 frames and a conditioned denoiser with a context size of only 9 frames, see Supplementary Fig. S 2.
Another advantage of leveraging precomputing global features in conditioning short-context denoisers is computational efficiency. Existing denoising methods that rely on long contexts to achieve satisfactory denoising performance will inevitably demand relatively larger computational resources. For instance, when extended to a training corpora of 26 datasets (Sec. 2.4), DeepCAD’s computational resource demands exceeded our limits and we had to exclude it from the benchmarking. Likewise, DeepInterpolation [14], another self-supervised deep neural network denoiser, could not be made to process our datasets and was thus also not included in benchmarking. In contrast, we were able to efficiently train CellMincer on our largest training corpora using widely-available commodity GPUs.
The pre-training approach we employed to train the CellMincer model on voltage imaging data might not be optimally tailored to handle other functional imaging modalities, such as calcium imaging, characterized by substantially different spatiotemporal dynamics. A characteristic difference in the dynamics of voltage imaging and calcium imaging is the presence of single spiking events occurring within 5 to 10 frames. The performance gap between CellMincer and DeepCAD on voltage imaging suggests that the most effective fluorescence imaging denoisers are highly specific to their target domains. CellMincer’s architecture uses a context window length on par with the timescale of a typical voltage imaging spiking event, and its training scheme maximizes the utility of this context by enabling inference from same-frame pixels. Conversely, DeepCAD predicts whole frames from a large, temporally downsampled neighborhood of frames, a strategy which foregoes mutual information carried by proximal pixels in exchange for training scalability. Our hypothesis of the specificity of the model (and self-supervision task) to the data domain is further reinforced by an experiment in which we compare the performance of CellMincer and DeepCAD on calcium imaging data (see Supplementary Fig. S 5). Both algorithms were trained on seven low-SNR calcium imaging datasets [13], and their denoised outputs were compared to high-SNR versions of the same datasets. We noticed a significant drop in the performance of CellMincer on denoising calcium imaging data, and improved performance of DeepCAD, which is opposite to our findings on fast voltage imaging data. We concluded that CellMincer’s capacity to model short-term fluctuations becomes a hindrance when the underlying signal has inherently slow dynamics, whereas DeepCAD’s whole-frame masking and the implicit bias of slow dynamics becomes advantageous.
A notable difficulty in conducting the analysis of neuron imaging traces in relation to corresponding EP activity is the lack of available tools for analyzing waveforms that diverge from the highly specific characteristics of EP. The Electrophys Feature Extraction Library (EFEL) [23], one such tool, can extract a variety of EP features such as spike half-widths but is much less conclusive when the input signals were adapted from fluorescence imaging. Our solution, prominence-thresholded peak calling, is motivated by the biological significance of partial and total depolarization events, indicative of subthreshold activity and action potentials respectively. Thus, identifying peaks in the EP represents a sensible first-order approximation for the locations of these events and can function as a task to which we subject our imaging traces. While most action potentials stand in such stark contrast to the surrounding baseline that they are evident in any form of the trace, the elevated baseline in the traces produced by PMD, N2S, and the raw data is likely to hide less pronounced EP events and introduce more false positives. Although the absolute peak-calling performance across all methods is low, primarily owing to the inherent incompatibilities between the 50 kHz EP signal and its derived 500 Hz imaging, our assessment is that for EP peaks between 2–5 mV, there is indeed information in the raw data that corresponds to this activity but is not immediately visible, and CellMincer is distilling this information to allow for more confident judgements.
A limitation of CellMincer’s default self-supervised training scheme is that in uniformly sampling random crops of the training data, CellMincer spends an overwhelming majority of its computation time on static background pixels as opposed to pixels containing meaningful neuronal activity. We introduced an option in CellMincer to increase its sampling efficiency without introducing network bias by oversampling such meaningful data crops, defined as exceeding the top of average luminosity across all crops in the dataset for chosen between 0.1 and 1, to 50% of each training batch (importance sampling). We can then correct the loss calculation knowing the constructed ratio of meaningful samples. While this feature was not incorporated into the models used in our main benchmarking experiments, we found that it reduced the performance gap between CellMincer and DeepCAD on calcium imaging. We expect further exploration of this direction, namely adaptive sampling and hard sample mining in the context of self-supervised training, will find application beyond the present domain.
Precomputation of global features should be carried out with additional considerations in situations where either neuron locations or noise characteristics could be non-stationary (e.g. certain in vivo recordings). In such cases, the temporal distributional shift along a long recording interval may render any one set of precomputed global features less relevant to variable local contexts of the recording. Since CellMincer’s objective function is unbiased (see Eq. 4), conditioning on poor (or even irrelevant) global features will not degrade the performance of the method. However, to take full advantage of the notion of feature-conditioning, we stipulate that a more effective strategy in denoising non-stationary data would be to pre-segment the movie into approximately stationary intervals and denoising each section separately using its own precomputed features. This is streamlined by CellMincer’s ability to pre-train on and denoise an arbitrary collection of recordings.
We believe that CellMincer’s architecture is adequately powered for denoising many forms of fluorescence imaging modalities of electrically active cells. Optimizing the hyperparameters and training schedule of CellMincer for related data domains (e.g. calcium imaging) would be a natural avenue for future work. While our analysis shows that CellMincer satisfactorily operates on a single dataset (both for training and denoising), we hypothesize that training a generalist large-scale CellMincer foundation model on a large and diverse biomedical imaging corpora (and perhaps using more scalable architectures such as the Vision Transformer [24]) is another promising area of future research. Intriguingly, inspecting and building on the saliency and attention maps underlying the Vision Transformer could lay a novel roadmap for segmenting a wide range of functional imaging datasets into functional units, much like the recently demonstrated utility of self-supervised models of natural images (e.g. DINOv2 [25]) in segmenting natural images and performing various other image-based downstream tasks.
We have made available separate pre-trained CellMincer models on synthetic Optosynth data from Sec. 2.3, the BeRST voltage imaging data from Sec. 2.4, and the Optopatch voltage imaging data from Sec. 2.5. Even though training a CellMincer model from scratch can take 10–12 hours on a typical dataset and publicly available commodity GPU (see Supplemental Sec. S.4), using one of the pre-trained models as is or fine-tuning it presents a faster and less computationally intensive approach to the adoption of our method. In the future, we hope that the availability pre-trained CellMincer foundation models on a large and diverse voltage imaging dataset, combined with efficient model selection and fine-tuning strategies, will further reduce the computational cost of using CellMincer.
Supplementary Material
4. Acknowledgements
This work was supported by a grant from SFARI (#890477, SLF, RN), by the National Institute of Mental Health (R01 MH128366-01A1, SLF, RN; R21 MH120423, SLF; and U01MH115727, MB, RN), by the National Institute of Health (R01 NS098088, ASW, EWM), by the Stanley Center for Psychiatric Research, by a SPARC award from Broad Institute (year 2019–2020, TM, MB, SLF), and by a BroadIgnite award from Broad Institute (2021–2022, BW, MB). The template HT076 was a gift from Dr. Adam Cohen. The authors thank Luca D’Alessio for insightful comments during various stages of the development and testing of CellMincer.
Footnotes
5 Code Availability
The code repository containing the CellMincer pipeline can be found at https://github.com/cellarium-ai/CellMincer. The repository containing the Optosynth simulation framework can be found at https://github.com/cellarium-ai/Optosynth. An auxiliary repository with reproductions of the analysis used to produce the figures can be found at https://github.com/cellarium-ai/CellMincerPaperAnalysis.
6. Data Availability
All data used to conduct the benchmarking experiments with Optosynth data, BeRST voltage imaging with paired EP data, and Optopatch datasets for chronically TTX-treated and unperturbed hPSC-derived neurons can be accessed either directly from the Google Cloud bucket found at gs://broad-dsp-cellmincer-data, or through notebooks in the aforementioned paper analysis repository.
References
- [1].Adam Yoav, Kim Jeong J, Lou Shan, Zhao Yongxin, Xie Michael E, Brinks Daan, Wu Hao, Mostajo-Radji Mohammed A, Kheifets Simon, Parot Vicente, Chettih Selmaan, Williams Katherine J, Gmeiner Benjamin, Farhi Samouil L, Madisen Linda, Buchanan E Kelly, Kinsella Ian, Zhou Ding, Paninski Liam, Harvey Christopher D, Zeng Hongkui, Arlotta Paola, Campbell Robert E, and Cohen Adam E. Voltage imaging and optogenetics reveal behavior-dependent changes in hippocampal dynamics. Nature, 569(7756):413–417, May 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].St-Pierre François, Marshall Jesse D, Yang Ying, Gong Yiyang, Schnitzer Mark J, and Lin Michael Z. High-fidelity optical reporting of neuronal electrical activity with an ultrafast fluorescent voltage sensor. Nature neuroscience, 17(6):884–889, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Hochbaum Daniel R, Zhao Yongxin, Farhi Samouil L, Klapoetke Nathan, Werley Christopher A, Kapoor Vikrant, Zou Peng, Kralj Joel M, Maclaurin Dougal, Smedemark-Margulies Niklas, et al. All-optical electrophysiology in mammalian neurons using engineered microbial rhodopsins. Nature methods, 11(8):825–833, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Kulkarni Rishikesh U and Miller Evan W. Voltage imaging: pitfalls and potential. Biochemistry, 56(39):5171–5177, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Lin Michael Z and Schnitzer Mark J. Genetically encoded indicators of neuronal activity. Nature neuroscience, 19(9):1142–1153, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Huang Shaosen, Zhao Yong, and Qin Binjie. Two-hierarchical nonnegative matrix factorization distinguishing the fluorescent targets from autofluorescence for fluorescence imaging. Biomed. Eng. Online, 14:116, December 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Aonishi Toru, Maruyama Ryoichi, Ito Tsubasa, Miyakawa Hiroyoshi, Murayama Masanori, and Ota Keisuke. Imaging data analysis using non-negative matrix factorization. Neurosci. Res., 179:51–56, June 2022. [DOI] [PubMed] [Google Scholar]
- [8].Witten Daniela M, Tibshirani Robert, and Hastie Trevor. A penalized matrix decomposition, with applications to sparse principal components and canonical correlation analysis. Biostatistics, 10(3):515–534, July 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Zhang Kai, Zuo Wangmeng, Chen Yunjin, Meng Deyu, and Zhang Lei. Beyond a gaussian denoiser: Residual learning of deep cnn for image denoising. IEEE transactions on image processing, 26(7):3142–3155, 2017. [DOI] [PubMed] [Google Scholar]
- [10].Zhang Yide, Zhu Yinhao, Nichols Evan L, Wang Qingfei, Zhang Siyuan, Smith Cody J, and Howard S. A Poisson-Gaussian denoising dataset with real fluorescence microscopy images. Proc. IEEE Comput. Soc. Conf. Comput. Vis. Pattern Recognit., pages 11702–11710, December 2018. [Google Scholar]
- [11].Lehtinen Jaakko, Munkberg Jacob, Hasselgren Jon, Laine Samuli, Karras Tero, Aittala Miika, and Aila Timo. Noise2Noise: Learning image restoration without clean data. March 2018. [Google Scholar]
- [12].Batson Joshua and Royer Loic. Noise2Self: Blind denoising by Self-Supervision. January 2019.
- [13].Li Xinyang, Li Yixin, Zhou Yiliang, Wu Jiamin, Zhao Zhifeng, Fan Jiaqi, Deng Fei, Wu Zhaofa, Xiao Guihua, He Jing, Zhang Yuanlong, Zhang Guoxun, Hu Xiaowan, Chen Xingye, Zhang Yi, Qiao Hui, Xie Hao, Li Yulong, Wang Haoqian, Fang Lu, and Dai Qionghai. Real-time denoising enables high-sensitivity fluorescence time-lapse imaging beyond the shot-noise limit. Nat. Biotechnol., 41(2):282–292, February 2023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Lecoq Jérôme, Oliver Michael, Siegle Joshua H, Orlova Natalia, Ledochowitsch Peter, and Koch Christof. Removing independent noise in systems neuroscience data using DeepInterpolation. Nat. Methods, 18(11):1401–1408, November 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Gouwens Nathan W, Sorensen Staci A, Baftizadeh Fahimeh, Budzillo Agata, Lee Brian R, Jarsky Tim, Alfiler Lauren, Baker Katherine, Barkan Eliza, Berry Kyla, Bertagnolli Darren, Bickley Kris, Bomben Jasmine, Braun Thomas, Brouner Krissy, Casper Tamara, Crichton Kirsten, Daigle Tanya L, Dalley Rachel, de Frates Rebecca A, Dee Nick, Desta Tsega, Lee Samuel Dingman, Dotson Nadezhda, Egdorf Tom, Ellingwood Lauren, Enstrom Rachel, Esposito Luke, Farrell Colin, Feng David, Fong Olivia, Gala Rohan, Gamlin Clare, Gary Amanda, Glandon Alexandra, Goldy Jeff, Gorham Melissa, Graybuck Lucas, Gu Hong, Hadley Kristen, Hawrylycz Michael J, Henry Alex M, Hill Dijon, Hupp Madie, Kebede Sara, Kim Tae Kyung, Kim Lisa, Kroll Matthew, Lee Changkyu, Link Katherine E, Mallory Matthew, Mann Rusty, Maxwell Michelle, McGraw Medea, McMillen Delissa, Mukora Alice, Ng Lindsay, Ng Lydia, Ngo Kiet, Nicovich Philip R, Oldre Aaron, Park Daniel, Peng Hanchuan, Penn Osnat, Pham Thanh, Pom Alice, Popović Zoran, Potekhina Lydia, Rajanbabu Ramkumar, Ransford Shea, Reid David, Rimorin Christine, Robertson Miranda, Ronellenfitch Kara, Ruiz Augustin, Sandman David, Smith Kimberly, Sulc Josef, Sunkin Susan M, Szafer Aaron, Tieu Michael, Torkelson Amy, Trinh Jessica, Tung Herman, Wakeman Wayne, Ward Katelyn, Williams Grace, Zhou Zhi, Ting Jonathan T, Arkhipov Anton, Sümbül Uygar, Lein Ed S, Koch Christof, Yao Zizhen, Tasic Bosiljka, Berg Jim, Murphy Gabe J, and Zeng Hongkui. Integrated morphoelectric and transcriptomic classification of cortical GABAergic cells. Cell, 183(4):935–953.e19, November 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Lee Brian R, Budzillo Agata, Hadley Kristen, Miller Jeremy A, Jarsky Tim, Baker Katherine, Hill Dijon, Kim Lisa, Mann Rusty, Ng Lindsay, Oldre Aaron, Rajanbabu Ram, Trinh Jessica, Vargas Sara, Braun Thomas, Dalley Rachel A, Gouwens Nathan W, Kalmbach Brian E, Kim Tae Kyung, Smith Kimberly A, Soler-Llavina Gilberto, Sorensen Staci, Tasic Bosiljka, Ting Jonathan T, Lein Ed, Zeng Hongkui, Murphy Gabe J, and Berg Jim. Scaled, high fidelity electrophysiological, morphological, and transcriptomic cell characterization. Elife, 10, August 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Berg Jim, Sorensen Staci A, Ting Jonathan T, Miller Jeremy A, Chartrand Thomas, Buchin Anatoly, Bakken Trygve E, Budzillo Agata, Dee Nick, Ding Song-Lin, Gouwens Nathan W, Hodge Rebecca D, Kalmbach Brian, Lee Changkyu, Lee Brian R, Alfiler Lauren, Baker Katherine, Barkan Eliza, Beller Allison, Berry Kyla, Bertagnolli Darren, Bickley Kris, Bomben Jasmine, Braun Thomas, Brouner Krissy, Casper Tamara, Chong Peter, Crichton Kirsten, Dalley Rachel, de Frates Rebecca, Desta Tsega, Lee Samuel Dingman, D’Orazi Florence, Dotson Nadezhda, Egdorf Tom, Enstrom Rachel, Farrell Colin, Feng David, Fong Olivia, Furdan Szabina, Galakhova Anna A, Gamlin Clare, Gary Amanda, Glandon Alexandra, Goldy Jeff, Gorham Melissa, Goriounova Natalia A, Gratiy Sergey, Graybuck Lucas, Gu Hong, Hadley Kristen, Hansen Nathan, Heistek Tim S, Henry Alex M, Heyer Djai B, Hill Dijon, Hill Chris, Hupp Madie, Jarsky Tim, Kebede Sara, Keene Lisa, Kim Lisa, Kim Mean-Hwan, Kroll Matthew, Latimer Caitlin, Levi Boaz P, Link Katherine E, Mallory Matthew, Mann Rusty, Marshall Desiree, Maxwell Michelle, McGraw Medea, McMillen Delissa, Melief Erica, Mertens Eline J, Mezei Leona, Mihut Norbert, Mok Stephanie, Molnar Gabor, Mukora Alice, Ng Lindsay, Ngo Kiet, Nicovich Philip R, Nyhus Julie, Olah Gaspar, Oldre Aaron, Omstead Victoria, Ozsvar Attila, Park Daniel, Peng Hanchuan, Pham Trangthanh, Pom Christina A, Potekhina Lydia, Rajanbabu Ramkumar, Ransford Shea, Reid David, Rimorin Christine, Ruiz Augustin, Sandman David, Sulc Josef, Sunkin Susan M, Szafer Aaron, Szemenyei Viktor, Thomsen Elliot R, Tieu Michael, Torkelson Amy, Trinh Jessica, Tung Herman, Wakeman Wayne, Waleboer Femke, Ward Katelyn, Wilbers René, Williams Grace, Yao Zizhen, Yoon Jae-Geun, Anastassiou Costas, Arkhipov Anton, Barzo Pal, Bernard Amy, Cobbs Charles, de Witt Hamer Philip C, Ellenbogen Richard G, Esposito Luke, Ferreira Manuel, Gwinn Ryder P, Hawrylycz Michael J, Hof Patrick R, Idema Sander, Jones Allan R, Keene C Dirk, Ko Andrew L, Murphy Gabe J, Ng Lydia, Ojemann Jeffrey G, Patel Anoop P, Phillips John W, Silbergeld Daniel L, Smith Kimberly, Tasic Bosiljka, Yuste Rafael, Segev Idan, de Kock Christiaan P J, Mansvelder Huibert D, Tamas Gabor, Zeng Hongkui, Koch Christof, and Lein Ed S. Human neocortical expansion involves glutamatergic neuron diversification. Nature, 598(7879):151–158, October 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Huang Yi-Lin, Walker Alison S, and Miller Evan W. A photostable silicon rhodamine platform for optical voltage sensing. Journal of the American Chemical Society, 137(33):10767–10776, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Milosevic Milena M, Jang Jinyoung, McKimm Eric J, Zhu Mei Hong, and Antic Srdjan D. In vitro testing of voltage indicators: Archon1, arclightd, asap1, asap2s, asap3b, bongwoori-pos6, berst1, flicr1, and chi-vsfp-butterfly. eneuro, 7(5), 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Walker Alison S, Raliski Benjamin K, Karbasi Kaveh, Zhang Patrick, Sanders Kate, and Miller Evan W. Optical spike detection and connectivity analysis with a far-red voltage-sensitive fluorophore reveals changes to network connectivity in development and disease. Frontiers in neuroscience, 15:643859, 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Desai Niraj S, Rutherford Lana C, and Turrigiano Gina G. Plasticity in the intrinsic excitability of cortical pyramidal neurons. Nature neuroscience, 2(6):515–520, 1999. [DOI] [PubMed] [Google Scholar]
- [22].Mukamel Eran A, Nimmerjahn Axel, and Schnitzer Mark J. Automated analysis of cellular signals from large-scale calcium imaging data. Neuron, 63(6):747–760, September 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Ranjan Creators, Van Geit Rajnish1, Moor Werner1, Rössert Ruben1, Riquelme Christian1, Damart Juan Luis1, Jaquier Tanguy1, Tuncel Aurélien1, Anil1 Show affiliations 1. Blue Brain Project, EPFL. eFEL. [Google Scholar]
- [24].Dosovitskiy Alexey, Beyer Lucas, Kolesnikov Alexander, Weissenborn Dirk, Zhai Xiaohua, Unterthiner Thomas, Dehghani Mostafa, Minderer Matthias, Heigold Georg, Gelly Sylvain, Uszkoreit Jakob, and Houlsby Neil. An image is worth 16×16 words: Transformers for image recognition at scale. October 2020.
- [25].Oquab Maxime, Darcet Timothée, Moutakanni Théo, Vo Huy, Szafraniec Marc, Khalidov Vasil, Fernandez Pierre, Haziza Daniel, Massa Francisco, Alaaeldin El-Nouby Mahmoud Assran, Ballas Nicolas, Galuba Wojciech, Howes Russell, Huang Po-Yao, Li Shang-Wen, Misra Ishan, Rabbat Michael, Sharma Vasu, Synnaeve Gabriel, Xu Hu, Jegou Hervé, Mairal Julien, Labatut Patrick, Joulin Armand, and Bojanowski Piotr. DINOv2: Learning robust visual features without supervision. April 2023.
- [26].Nehme Ralda, Zuccaro Emanuela, Ghosh Sulagna Dia, Li Chenchen, Sherwood John L, Pietilainen Olli, Barrett Lindy E, Limone Francesco, Worringer Kathleen A, Kommineni Sravya, et al. Combining ngn2 programming with developmental patterning generates human excitatory neurons with nmdar-mediated synaptic transmission. Cell reports, 23(8):2509–2523, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Berryer Martin H, Tegtmeyer Matthew, Binan Loïc, Valakh Vera, Nathanson Anna, Trendafilova Darina, Crouse Ethan, Klein Jenny A, Meyer Daniel, Pietiläinen Olli, et al. Robust induction of functional astrocytes using ngn2 expression in human pluripotent stem cells. Iscience, 26(7):106995, 2023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Fan Linlin Z, Nehme Ralda, Adam Yoav, Jung Eun Sun, Wu Hao, Eggan Kevin, Arnold Don B, and Cohen Adam E. All-optical synaptic electrophysiology probes mechanism of ketamine-induced disinhibition. Nature methods, 15(10):823–831, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Loshchilov Ilya and Hutter Frank. SGDR: Stochastic gradient descent with warm restarts. August 2016.
- [30].Izmailov Pavel, Podoprikhin Dmitrii, Garipov Timur, Vetrov Dmitry, and Wilson Andrew Gordon. Averaging weights leads to wider optima and better generalization. March 2018.
- [31].Allen Institute for Brain Science. Allen software development kit (allen sdk). https://allensdk.readthedocs.io, 2023.
- [32].Gouwens Nathan W, Sorensen Staci A, Baftizadeh Fahimeh, Budzillo Agata, Lee Brian R, Jarsky Tim, Alfiler Lauren, Baker Katherine, Barkan Eliza, Berry Kyla, et al. Integrated morphoelectric and transcriptomic classification of cortical gabaergic cells. Cell, 183(4):935–953, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Lee Brian R, Budzillo Agata, Hadley Kristen, Miller Jeremy A, Jarsky Tim, Baker Katherine, Hill DiJon, Kim Lisa, Mann Rusty, Ng Lindsay, et al. Scaled, high fidelity electrophysiological, morphological, and transcriptomic cell characterization. Elife, 10:e65482, 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Berg Jim, Sorensen Staci A, Ting Jonathan T, Miller Jeremy A, Chartrand Thomas, Buchin Anatoly, Bakken Trygve E, Budzillo Agata, Dee Nick, Ding Song-Lin, et al. Human neocortical expansion involves glutamatergic neuron diversification. Nature, 598(7879):151–158, 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Stuart Greg, Spruston Nelson, Sakmann Bert, and Häusser Michael. Action potential initiation and backpropagation in neurons of the mammalian cns. Trends in neurosciences, 20(3):125–131, 1997. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data used to conduct the benchmarking experiments with Optosynth data, BeRST voltage imaging with paired EP data, and Optopatch datasets for chronically TTX-treated and unperturbed hPSC-derived neurons can be accessed either directly from the Google Cloud bucket found at gs://broad-dsp-cellmincer-data, or through notebooks in the aforementioned paper analysis repository.