Abstract
Despite effective new therapies, adaptive resistance remains the main obstacle in AML therapy. Autophagy induction is a key mechanism for adaptive resistance. Leukemic blasts at diagnosis express higher levels of the apical autophagy kinase ULK1 compared to normal hematopoietic cells. Exposure to chemotherapy and targeted agents upregulate ULK1, hence we hypothesize that developing ULK1 inhibitors may present the unique opportunity for clinical translation of autophagy inhibition. Accordingly, we demonstrate that ULK1 inhibition, by genetic and pharmacological means, suppresses treatment-induced autophagy, overcomes adaptive drug-resistance, and synergizes with chemotherapy and emerging anti-leukemia agents like venetoclax (ABT-199). The study next aims at exploring the underlying mechanisms. Mechanistically, ULK1 inhibition downregulates MCL1 anti-apoptotic gene, impairs mitochondrial function and downregulates components of the CD44-xCT system, resulting in impaired reactive oxygen species (ROS) mitigation, DNA damage and apoptosis. For further validation, several mouse models of AML were generated. In these mouse models, ULK1 deficiency impaired leukemic cell homing and engraftment, delayed disease progression and improved survival. Therefore, in the study we validated our hypothesis and identified ULK1 as an important mediator of adaptive resistance to therapy and an ideal candidate for combination therapy in AML. Therefore, we propose ULK1 inhibition as a therapeutically relevant treatment option to overcome adaptive drug-resistance in AML.
Keywords: ABT-199 (Venetoclax), Combination therapy, Drug-resistance, Mitochondria, Reactive Oxygen Species (ROS)
Introduction
Acute myelogenous leukemia (AML) is the most common hematological neoplasm diagnosed in adults, and the second-most common hematological disorder diagnosed in children. Cytarabine-based chemotherapy is the backbone of therapy, particularly for younger and fit patients, but inherent/acquired resistance and relapses are common. By mutational profiling, patients can be categorized into multiple molecular prognostic subtypes (1, 2), and treated with mutation-targeted therapies like FLT3, IDH, and other pathway-specific inhibitors (3–5). The Bcl2 inhibitor venetoclax (ABT-199) has expanded the therapeutic armamentarium for elderly or ‘unfit’ patients. Venetoclax-based combinations as frontline therapy have impressive response rates and extended survival, but most patients relapse due to adaptive resistance (6). Importantly, autophagy emerges as a putative adaptive resistance factor through molecular reprogramming (7–14).
Autophagy is an evolutionarily-conserved cellular process that maintains cellular homeostasis under nutrient deprivation, hypoxia, or chemotherapy-induced/metabolic/oncogenic stress. Therapy-resistant surviving cells are addicted to autophagy, presenting a targetable vulnerability (11–17). In AML, therapy-resistance usually resides in the leukemic stem cells (LSCs) (18, 19). Previous studies have shown the crucial role of autophagy in AML-LSC maintenance and survival, and that LSCs have higher basal autophagy (20). Mechanistically, LSCs are dependent on mitochondrial homeostasis including mitochondrial respiration (or oxidative phosphorylation, OXPHOS) (21, 22), competent mitophagy, mitochondrial fission and mitigation of reactive oxygen species (ROS) (23, 24). In addition, LSCs are more dependent on OXPHOS than hematopoietic stem cells (HSCs) (21, 22). Therefore, LSCs are more vulnerable to autophagy inhibition, oxidative stress, and disruption of mitochondrial functions (14, 17). We previously identified ATG7, a key autophagy ligase, as a therapeutic target to overcome adaptive resistance to AML therapies (7). Here we show that treatment with AraC and idarubicin (Ida) (standard AML chemotherapies), and ABT-199 (Bcl2 inhibitor) induce autophagy. Also, AraC and ABT-199-resistant cells have increased basal autophagy. However, translational work on autophagy inhibition is limited by paucity of inhibitors targeting autophagy-specific kinases (25). So, here we sought to target the apical kinase Unc51-like autophagy activating kinase 1 (ULK1). ULK1, a core autophagy-initiating component, is the only serine/threonine kinase in this pathway, and phosphorylates at least 15 downstream targets including Atg13, Beclin1, and Vps34 (26). This is a node where AMPK and mTOR, the two major metabolic sensing pathways converge, reciprocally regulating ULK1 (27).
Since autophagy addiction is a unique vulnerability in the AML cells compared to the normal cells, and also as we found from publicly available data that ULK1 is upregulated in AML, therefore, the inhibition of ULK1 presents a potential therapeutic window. With this rationale, in this study we have generated both genetic and pharmacologic systems to inhibit ULK1. These systems effectively synergize with AraC, Ida and ABT-199 in AML. We also demonstrate the molecular changes that are associated with ULK1 inhibition, facilitating apoptosis in in vitro systems and delay in disease progression in in vivo systems. The most relevant context in this case is the emergence of therapy-resistance, and to simulate this in vitro, we have generated several drug-resistant cells in this study. Finally, we have also generated in vivo model systems to demonstrate the anti-leukemic effect of ULK1 inhibition. Overall, we establish that ULK1-mediated autophagy inhibition impairs leukemia growth, and overcomes adaptive drug-resistance in AML. Therefore, this study highlights the potential of targeting ULK1 for further translational work and clinical benefit.
Materials and Methods
Cell Lines
HEK293T, HL-60 and MV4;11 were purchased from American Type Culture Collection (ATCC, Manassas, VA, USA); and OCI-AML2, OCI-AML3, MOLM13 and MOLM14 from Deutsche Sammlung von Mikroorganismen und Zellkulturen GmbH (DSMZ, Braunschweig, Germany). Cells were cultured in RPMI 1640 media supplemented with 10% fetal bovine serum (Millipore-Sigma) and 1% Penicillin-Streptomycin-L-Glutamine (Corning) and maintained at 37°C in a humidified incubator with 5% CO2. All cells were fingerprinted every 6 months at the institutional core facility and routinely confirmed to be mycoplasma-free.
Mouse Models
ULK1-WT and KO model:
Six-weeks old male NODscid-IL2Rγnull (NSG) mice (Jackson Laboratory) were intravenously injected with 106 control (WT) or ULK1-KO cells (n=13). Disease progression was monitored by bioluminescence imaging and estimating human CD45+ve cells in peripheral blood. Six mice from each group were euthanized on day 30, and tissues (bone, spleen and liver) were analyzed histologically. Disease burden in bone marrow (BM) and spleen was quantified by human CD45+ve cells. Remaining mice were monitored for survival and human CD45+vity at death.
AML-PDX model:
This model was generated by intravenous injection of AML patient sample (PDX#5) in 6-weeks old male NSG mice (more information in supplementary data).
Ex vivo model:
This model was generated by intravenous injection of luciferized-OCI-AML3 treated with DMSO, SBI-0206965 or ABT-199 alone, and in combination for 24 hours (viable cells in equal numbers), in 6-weeks old male NSG mice (more information in supplementary data).
Patient Samples
Mononuclear cells were isolated from peripheral blood or BM by Ficoll density centrifugation, grown in cytokine-enriched media, and maintained at 37°C/humidified incubator/5% CO2.
Genetically modified cell lines
In OCI-AML3, shRNA-mediated MCL1 (shMcl1) or ULK1 (shULK1) knockdown was performed, with respective vector-containing controls following institutional guidelines (28).
For ULK1 knockout, luciferized-OCI-AML3were lentivirally-transduced with CRISPR-Cas9-GFP-sgRNA (Applied Biological Materials Inc.), followed by single-cell sorting, clone expansion, and screening after 3–4 weeks.
Overexpression in OCI-AML3 were generated similarly for MCL1, ULK1, and CD44 using lentiviral transduction.
Drug-Resistant Cells
Cells were constantly exposed to increasing doses of drugs over 2–3 months until at least 10-fold higher IC50 values were achieved to generate drug-resistant cells (more information in supplementary data).
Drug sensitivity and combination synergy analysis
To calculate sensitivity to a single agent, cells were treated with the drug for 72 hours, and cell numbers and percent apoptosis were calculated based on Annexin V/DAPI assay. Similarly, to analyze the synergy in drug combinations, cells were treated with 2 or more drugs in a constant ratio for 72 hours, and cell numbers and percent apoptosis were calculated. Using the data from Annexin V assay, IC50 and ED50 values were calculated using the Calcusyn software. Further, for analysis of synergy, the combination index (CI) values were calculated following the Chou Talalay’s method. This is an established algorithm-based automated computer simulation where CI value may be 1 (additive), >1 (antagonistic), or <1 (synergistic).
Western blotting
Post-treatment cells were rinsed with PBS, and lysed in RIPA buffer (50mM Tris, 150mM NaCl, 1% Triton X-100, 1% Sodium Deoxycholate, 0.1% SDS, 1mM EDTA) supplemented with 1X protease and phosphatase inhibitor cocktail (Thermo Fisher Scientific). Protein concentration was determined using Pierce BCA Protein Assay kit (Thermo Fisher Scientific) and measuring absorbance at 550 nm in VictorX3 Multilabel plate reader (Perkin Elmer). 25, 50 or 100 ug of protein was diluted in 1X SDS lysis buffer (10mM Tris pH 7.5, 100nM NaCl, 1% SDS), separated using SDS-PAGE electrophoresis (Biorad, Hercules, CA, USA) and transferred onto PVDF membrane (Millipore). Membranes were blocked using 0.5% casein in PBS, incubated with primary antibodies overnight. Secondary antibodies IRDye 680 and 800 were used for scanning on the Odyssey Infrared Imaging System (LI-COR Bioscences). Quantitation was done using Image Studio Lite Version 5.2 and ImageJ softwares. Details of antibodies are available in supplementary data.
Flow cytometry-based autophagy detection (LC3-PE assay)
LC3B was quantified by intracellular staining with PE-conjugated antibody, following manufacturer’s protocol (Cell Signaling Technology), as described earlier (7).
Reactive Oxygen Species (ROS) assay
The total intracellular ROS levels were measured using the ROS-ID Total ROS detection kit (Enzo Life Sciences, Farmingdale, NY, USA) following the manufacturer’s protocol. Briefly, 106 cells were pelleted and resuspended in diluted Oxidative Stress Detection Reagent (Green) for 30 minutes at 37°C in dark, followed by washing once with wash buffer (supplied) containing 0.25ug/ml DAPI and resuspended in PBS. Live (DAPI-ve) cells were gated and analyzed at FL-1 channel in Gallios flow cytometer (Beckman Coulter). Data analysis was performed using Kaluza Software for Gallios (Beckman Coulter). For detection of mitochondrial ROS similar protocol was followed using MitoSOX Red Mitochondrial Superoxide Indicator (Thermo Fisher Scientific) and analyzed at FL-2 channel.
Glutathione Assay
The cellular GSH level was measured by Glutathione Assay kit (Cayman Chemical, Ann Arbor, MI, USA) as per the manufacturer’s protocol. Briefly, cell extracts were sonicated in 0.5X MES buffer (50mM MES or phosphate pH 6–7, containing 1mM EDTA) and deproteinated with metaphosphoric acid. The deproteinated supernatant was collected, neutralized with triethanolamine. The level of total GSH was detected by measuring the product of glutathionylated 5–5′-dithiobis-2-nitrobenzoic acid at 405 nm using the multilabel plate reader VictorX3. 2-vinylpyridine was added to the deproteinated samples and incubated for 1 hour at room temperature to estimate GSSG converted to GSH. Reduced glutathione amount was calculated based on GSH and GSSG values.
Mitochondria analysis
To quantify mitochondrial mass, cells were incubated with 125nM MitoTracker Green (MTG, Thermo Fisher Scientific) for 30 minutes/37°C/dark, washed (PBS-containing DAPI 0.25ug/ml) and analyzed on Gallios flow cytometer (Beckman Coulter).
To measure mitochondrial outer membrane potential (MOMP), cells were incubated with Tetramethylrhodamine, Ethyl Ester, Perchlorate (TMRE, Thermo Fisher Scientific) and analyzed similar to MTG.
Mitochondrial respiration was measured by Seahorse XF Cell Mito Stress Test Kit (Agilent Technologies) and analyzed using Seahorse Wave Desktop Software (Agilent Technologies). All results were normalized for numbers.
Microscopy
Cells were washed twice with PBS, fixed with 4% paraformaldehyde (10 minutes in dark), followed by three washes (PBS-T; PBS containing 0.1% Tween-20) and permeabilized with PBS containing 0.5% Triton X-100 (10 minutes), followed by three washes with PBS-T and blocking with 1% bovine serum albumin (BSA) in PBS for 1 hour. For DNA damage analysis, cells were then stained overnight with γH2AX antibody (1:1000; moist chamber at 4°C), and washed thrice with PBS-T (10 minutes each). Secondary Alexa Fluor 594 goat anti-rabbit IgG (1:2000) was used for 1 hour followed by two washes with PBS-T and a final wash with PBS containing DAPI (0.25ug/ml). Cells were mounted, visualized and images were captured using FV1000 confocal laser-scanning microscope (Olympus). For mitochondrial imaging, OCI-AML3 cells with GFP-labeled mitochondria were treated with SBI-0206965 and fixed, prior to mounting with DAPI. The Imaris Image Analysis Software (Bitplane, Zurich, Switzerland) was used for quantification and batch analysis was performed using the “Surfaces” application.
Electron Microscopy
Samples were fixed with a solution containing 3% glutaraldehyde plus 2% paraformaldehyde in 0.1M cacodylate buffer pH 7.3, washed in 0.1M sodium cacodylate buffer and treated with 0.1% millipore-filtered cacodylate buffered tannic acid, post-fixed with 1% buffered osmium, and stained en bloc with 1% millipore-filtered uranyl acetate. Samples were dehydrated in increasing concentrations of ethanol, infiltrated, and embedded in LX-112 medium, and polymerized in a 60°C oven for 3 days. Ultrathin sections were cut in a Leica Ultracut microtome (Leica, Deerfield, IL, USA), stained with uranyl acetate and lead citrate in a Leica EM Stainer, and examined in a JEM 1010 transmission electron microscope (JEOL Inc., Peabody, MA, USA) at an accelerating voltage of 80 kV. Digital images were obtained using AMT Imaging System (Advanced Microscopy Techniques Corp, Danvers, MA, USA).
RNA Isolation and qRT-PCR
Total RNA was extracted using PureLink RNA Mini Kit (Thermo Fisher Scientific) with on-column DNase I digestion (Thermo Fisher Scientific) following manufacturer’s instructions and quantified using Nanodrop 2000 UV-Vis Spectrophotometer (Thermo Scientific, Wilmington, DE, USA).1 ug RNA was used for reverse transcription with Verso cDNA Synthesis Kit (Thermo Fisher Scientific). Quantitative real time PCR was performed using PowerUp SYBR Green Master Mix or TaqMan Master Mix (Thermo Fisher Scientific) on a 7900HT Fast Real-Time PCR System with Fast 96-Well Block Module (Applied Biosystems, Thermo Fisher Scientific, Foster City, CA, USA). Cycle threshold (Ct) values were converted to fold change in relative expression (RQ) values (2-ΔΔCt). Primers were generated at Integrated DNA Technologies (Coralville, IA, USA) and sequences are available in supplementary data.
Genomic and Mitochondrial DNA Isolation
Total genomic DNA was extracted using the PureLink Genomic DNA Mini Kit (Thermo Fisher Scientific) and quantified using Nanodrop 2000 UV-Vis Spectrophotometer (Thermo Scientific). 50–100 ng DNA was used for assessment of genomic or mitochondrial DNA using specific primers (sequences available in supplementary data).
Statistical Analysis
Statistical analyses were conducted using GraphPad. Unpaired two-tailed Student’s t-test was used and p-values < 0.05 considered significant. Error bars represent standard deviation (SD). p-values and replicates are indicated in figure legends.
Study Approval
Animal studies were approved by MD Anderson Cancer Center Institutional Animal Care and Use Committee. Mice were housed at pathogen-free institutional facilities. Primary AML and normal BM specimens were collected from patients after written informed consent following institutional guidelines and declaration of Helsinki principles.
Data Availability
The microarray data analyzed in Figure 1A were obtained from publicly available datasets GSE13159, GSE15434, GSE61804, GSE14468, GSE42519 and The Cancer Genome Atlas. All data generated in this study are available within the article and its supplementary data files.
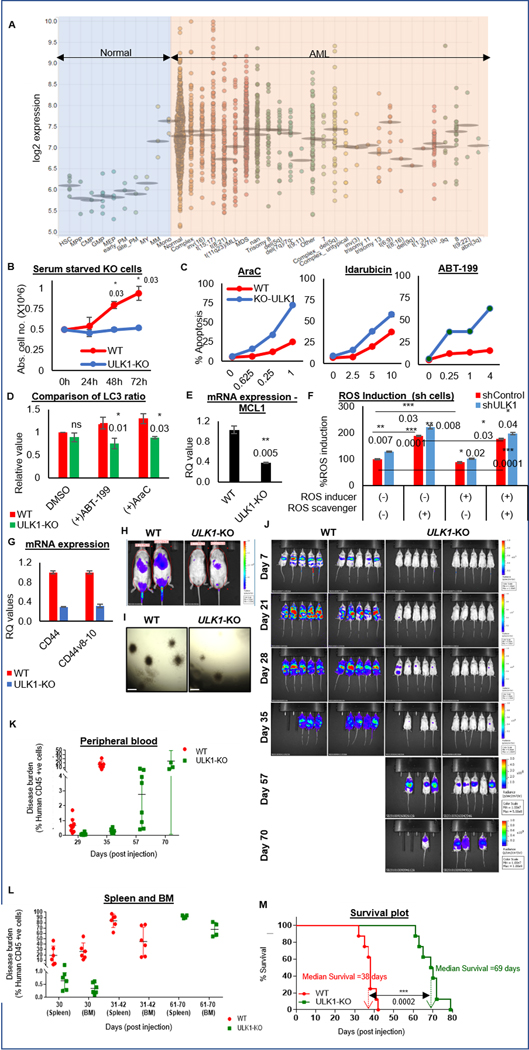
Figure 1. ULK1 is upregulated in AML, and its loss enhances therapy response in vitro and delays leukemia progression in vivo.
(A) Dot plot representation comparing ULK1 gene expression from curated microarray-based gene expression profiles of human AML (GSE13159, GSE15434, GSE61804, GSE14468, and The Cancer Genome Atlas) and FACS-sorted healthy hematopoietic cells in different stages of maturation (GSE42519), generated using the Bloodspot server and database. (B) Representative line graphs showing absolute cell numbers (from triplicates ± SD) measured by Trypan Blue staining using a Vi-Cell counter in control (WT) and ULK1-KO cells cultured simultaneously under serum starved conditions. Cells were collected every 24 hours. (C) Representative line graphs showing percent apoptosis (from triplicates ± SD) from Annexin V/DAPI assay in WT and ULK1-KO cells simultaneously treated with increasing doses as indicated of either AraC (left), Ida (middle) or ABT-199 (right) for 72 hours. (D) Representative bar graphs showing comparative analysis of LC3 ratio in whole cell lysates of control (WT) and ULK1-KO (KO) cells, treated with DMSO, AraC or ABT-199 (calculated from triplicates ± SD, in LC3-PE flow cytometry). Actin was used as the loading control and for densitometric measurements. (E) Representative bar graphs showing mean RQ values (from triplicates ± SD) from qRT-PCR assay performed for MCL1, on RNA isolated from control (WT) or ULK1-KO cells. (F) Representative bar graphs showing the percent ROS induced with respect to untreated shControl cells as 100 (from triplicates ± SD), in shControl and shULK1 cells, either untreated, or in the presence of ROS inducer (Pyocyanin) and/or ROS scavenger (NAC). (G) Representative bar graphs showing mean RQ values (from triplicates ± SD) from qRT-PCR assay performed for CD44 and its variant v8–10, on RNA isolated from control (WT) or ULK1-KO cells. All RQ values were generated from Ct values (2-ΔΔCt). (H) Representative images from bioluminescence imaging (BLI) performed 16 hours post-injection to analyze homing of the injected cells in mice. (I) Representative images of colony formation assay performed in control (WT) and ULK1-KO cells (4X magnification, 500 um scale bar). (J) BLI showing disease progression in 6-week old male NSG mice injected with luciferized control (WT) or ULK1-KO cells (106 cells each) via tail vein injection (n>10) over time. (K) Representative dot plot showing the percent of circulating human CD45+ve cells in individual mice at indicated time points by flow cytometry from peripheral blood, to measure the disease burden over the course of the experiment. (L) Representative dot plot showing the percentage of human CD45+ve cells in individual mice at indicated time points by flow cytometry from different organs (spleen and bone marrow), to measure the disease burden over the course of the experiment. (M) Kaplan-Meier survival plot showing survival benefit of ULK1-KO in comparison to control. The values were estimated by Mantel-Cox test showing a p-value of 0.0001 and median survival was 38 days for WT and 69 days for ULK1-KO group. Statistical significance of the experiments was calculated by standard Student’s t-test and p-values are indicated in respective graphs.
Results
ULK1 is upregulated in AML
Based on our previous work on targeting autophagy (7) and a strong rationale of targeting the apical autophagy kinase, we analyzed publicly available curated microarray-based gene expression profiles from human AML and healthy hematopoietic cells. AML cells have significantly higher ULK1 expression, compared to HSC counterparts, suggesting autophagy may be differentially induced by oncogenic stress in AML cells (Fig. 1A and Supplementary Table S1). This offers a potential therapeutic window to evaluate if targeting ULK1 can specifically eliminate AML cells.
shRNA-mediated knockdown (shULK1) or CRISPR/Cas9-mediated knockout (ULK1-KO) of ULK1 in OCI-AML3 cells (Supplementary Fig. S1A–C) did not alter viability, growth rate and cell cycle. It remained similar to the isogenic controls (WT) under normal culture conditions (Supplementary Fig. S1D–E). However, under stress (serum starvation or prolonged culture), there was inhibition of cell proliferation and loss of cell viability (Fig. 1B and S1E–F) suggesting that ULK1 is essential for the survival of leukemic cells under metabolic stress.
ULK1 loss enhances therapy response in AML cells by regulating apoptosis via MCL1 and ROS via CD44
Next, we asked if ULK1 plays a role in drug sensitivity of AML cells. Genetic targeting of ULK1 either in ULK1-KO or shULK1 resulted in significant increase in sensitivity to AML therapy-induced apoptosis (AraC, Ida and ABT-199; Fig. 1C and S1G). LC3 lipidation, a marker of autophagy induction, was lower in AraC or ABT-199-treated ULK1-KO cells than in WT (Fig. 1D and S1H). This indicates ULK1 deficiency increases sensitivity to AML drugs and decreases drug-induced autophagy.
To study the underlying mechanisms of enhanced apoptosis, we analyzed anti-apoptotic Bcl2 family proteins. ULK1-KO transcriptionally downregulated MCL1 (Fig. 1E). Protein levels of phospho-Erk1/2, an important upstream transcriptional regulator of MCL1, was also reduced upon ULK1 inhibition (Fig. S1I). To evaluate the effect of ULK1 inhibition on ROS, we performed total ROS detection assay. At the basal level, the control and shULK1 cells showed similar ROS levels, however, upon ROS induction with an inducer (pyocyanin, in this case), there was a significant increase in the levels of ROS in shULK1 cells compared to the control. Upon exposure to ROS scavenger, the ROS level decreases as expected, and the ROS induction increases again when ROS inducer is added in the presence of the ROS scavenger (Fig. 1F). This suggests that ULK1 plays a role in either increased production or reduced mitigation of ROS. Cellular ROS is maintained by intracellular antioxidants like reduced glutathione (GSH), and a redox system. GSH is in turn maintained by the cystine-glutamate antiporter system, consisting of solute carrier xCT (SLC7A11), glycoprotein CD98 and cell surface receptor CD44 (29, 30). To further dissect the mechanism of ULK1-mediated regulation of ROS, we analyzed the cellular ROS regulators, and found that ULK1 inhibition transcriptionally downregulated both WT and the v8–10 isoform of CD44 (Fig. 1G). Together, this data suggests that increase in ROS upon ULK1 inhibition may be a result of downregulated CD44 activity.
Targeting ULK1 delays leukemia progression
To examine the effect of ULK1 deficiency in vivo, we injected OCI-AML3 WT and ULK1-KO cells via tail vein and analyzed homing to spleen and BM by flow cytometry. As shown in figure 1H, ULK1-KO cells were significantly less abundant in spleen and BM compared to controls. To determine the engraftment potential, we performed colony formation assays with control (WT) and ULK1-KO cells. ULK1-KO cells showed reduced colony forming ability (Fig. 1I) suggesting that ULK1 deficiency hampers the proliferative capability of leukemic cells in vitro. There was a significant delay in disease progression in mice injected with ULK1-KO cells (Fig. 1J and S1J) compared to WT, accompanied with significantly reduced disease burden and prolonged survival in these mice (Fig. 2D–F; Supplementary Fig. S2K–L; and Supplementary Table S2). These data indicate that ULK1 is essential for homing, proliferation and self-renewal of AML cells.
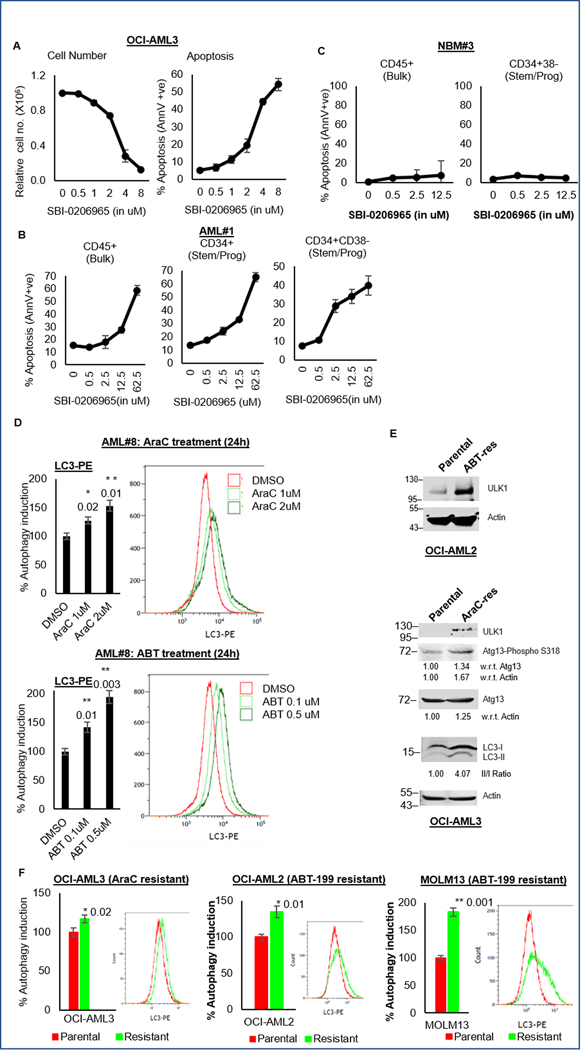
Figure 2. ULK1 inhibitor SBI-0206965 exhibits anti-leukemic effects and autophagic adaptation of drug resistant cells.
(A) Representative line graphs showing relative cell numbers (from triplicates ± SD; left) and percent apoptosis (from triplicates ± SD; right) from Annexin V/DAPI assay in OCI-AML3 cells treated with increasing doses as indicated of SBI-0206965 for 72 hours. (B-C) Representative line graphs showing percent apoptosis (from triplicates ± SD) from Annexin V/DAPI assay in primary AML blasts (B), and normal bone marrow cells (C) treated with increasing doses as indicated of SBI-0206965 for 72 hours. Cell pellets were additionally stained with CD45, CD34 and CD38 surface markers to identify different subpopulations [bulk (CD45+) and stem/progenitor (CD34+ or CD34+CD38-)] by flow cytometry. (D) Representative bar graphs showing percent autophagy induction (calculated from triplicates ± SD, in LC3-PE flow cytometry) with respect to DMSO treated control cells 100, in primary AML blasts treated with increasing doses as indicated of AraC (upper panel) or ABT-199 (lower panel) for 24 hours. Representative histograms corresponding to the autophagy induction are also shown. (E) Western blot analysis of whole cell lysates from parental and ABT-199 resistant OCI-AML2 cells (upper panel) and AraC resistant OCI-AML3 cells (lower panel) and, comparing autophagy markers. Actin was used as the loading control and for densitometric measurements. (F) Representative bar graphs showing basal autophagy (calculated from triplicates ± SD, in LC3-PE flow cytometry) with respect to parental cells as 100, with representative histograms, comparing parental and drug-resistant cells: AraC resistant OCI-AML3 cells (left), ABT-199 resistant OCI-AML2 cells (middle) and ABT-199 resistant MOLM13 cells (right). The statistical significance of the experiments was calculated by standard Student’s t-test and p-values are indicated in respective graphs.
ULK1 inhibitor, SBI-0206965 inhibits autophagy and exerts anti-leukemic effects
We next tried to replicate the findings above with an ULK1 inhibitor, SBI-0206965 (26). SBI-0206965 was effective as a single agent against multiple AML cells (n=5) (Fig. 2A, Supplementary Fig. S2A and Supplementary Table S3). SBI-0206965 reduced LC3 lipidation, and inhibited ULK1-specific target, phospho-Atg13 (Supplementary Fig. S2B–D). Patient-derived AML cells (n=8) also underwent apoptosis with SBI-0206965, in both bulk and stem/progenitor populations, with no or minimal cytotoxicity against normal bone marrow cells (n=4), indicating the clinical relevance of ULK1 inhibition (Fig. 2B–C; Supplementary Fig. S2E–N; and Supplementary Table S4). To further validate our findings and to rule out the potential FLT3 inhibitory activity of the compound on autophagy, we treated OCI-AML3 with AC220 (a potent and specific FLT3 inhibitor) and assessed the levels of LC3 by western blotting. As shown in figure S2O, AC220 showed no effect on autophagy, while SBI-0206965 could inhibit both basal and drug-induced autophagy. We further confirmed this data by demonstrating that ULK1-overexpression in OCI-AML3 cells impairs sensitivity to SBI-2026965, supporting the specificity of action (Supplementary Fig. S2P–Q).
Autophagy is an adaptation in chemotherapy and ABT-199 resistant AML cells
Chemotherapy and other targeted therapies are known to induce autophagy as a protective adaptation (11, 12, 15–17). Consistent with previous reports, we found that anti-leukemic drugs AraC, Ida and ABT-199 can induce autophagy in leukemic cells and primary AML blasts (Fig. 2D and S2R–U).
AraC and ABT-199- resistant cells generated in our laboratory (Supplementary Table S5 and Supplementary Fig. S2V) showed all hallmarks of autophagy upregulation autophagy upregulation. AraC-resistant cells had increased ULK1 expression, Atg13 phosphorylation, and LC3 lipidation. ABT-199-resistant cells also showed increased ULK1 expression (Fig. 2E. The resistant cells also had significantly higher basal autophagy compared to their parental counterparts (Fig. 2F). Together, these data indicate that the drug-resistant cells are dependent on autophagy for their survival and that autophagy inhibition can target this vulnerability.
ULK1 inhibition overcomes drug-resistance and is synergistic with AML therapies
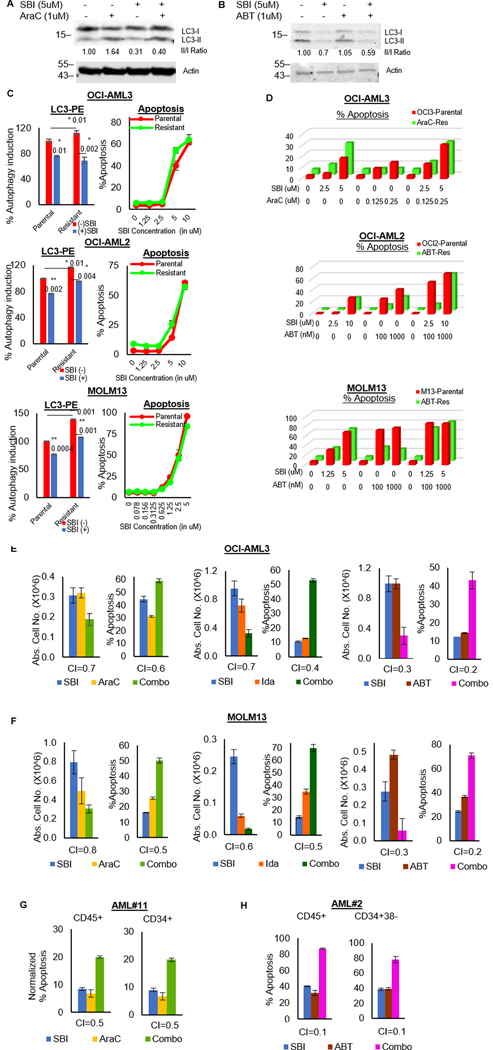
Next, we sought to evaluate the effect of ULK1 inhibition on autophagy-mediated drug resistance after therapy. First, we measured the levels of LC3 lipidation in OCI-AML3 after treatment with AraC or ABT-199. As shown in figure 3A–B, LC3 lipidation increased with treatment and decreased when combined with SBI-0206965. The increased basal autophagy in AraC or ABT-199-resistant cells was also reversed by SBI-0206965 (Fig. 3C). AraC or ABT-199 resistant cells were equally sensitive to ULK1 inhibition as their control counterparts (Supplementary Fig. S3A). Most importantly, SBI-0206965 could also restore sensitivity to AraC/ABT-199 in the respective resistant cells (Fig. 3D and S3B). Thus, ULK1 inhibition can effectively reverse drug-resistance.
Figure 3. ULK1 inhibition overcomes drug-resistance and synergizes with AML therapies.
(A-B) Western blot analysis for LC3 in whole cell lysates of OCI-AML3 cells treated with either SBI-0206965 alone or in combination with (A)AraC or (B) ABT-199, for 24 hours. Actin was used as the loading control and for densitometric measurements. (C) Representative bar graphs showing percent autophagy induction (calculated from triplicates ± SD, in LC3-PE flow cytometry) with respect to DMSO treated parental cells as 100 after 24 hours (left) and representative line graphs showing percent apoptosis (from triplicates ± SD) from Annexin V/DAPI assay after 72 hours (right); in parental and drug-resistant cells treated with/without SBI-0206965. (D) Representative bar graphs comparing percent apoptosis in parental versus drug-resistant cells. Data is based on Annexin V/DAPI assay 72 hours post-treatment with either SBI-0206965 or AML drugs alone and in combination (as indicated in the respective plots): OCI-AML3 (top), OCI-AML2 (middle) and MOLM13 (bottom). (E-F) Representative bar graphs showing absolute cell numbers (left) and percent apoptosis (right) from Annexin V/DAPI assay after 72 hours of treatment with either SBI-0206965 alone or AML drugs alone or in combination (as indicated in the respective plots), in cell lines OCI-AML3 (E) and MOLM13 (F). To assess the synergy in combinatorial treatments, combinatorial indices (CI-values) were generated according to Chou Talalay’s method (50) using the software Calcusyn. The drugs were treated over a dose range and bar graphs represent the best point of synergy. Values in the graphs represent triplicates ± SD. (G-H) Representative bar graphs showing percent apoptosis from Annexin V/DAPI assay in primary AML blasts, treated with indicated drug combinations for 72 hours. Cell pellets were additionally stained with CD45, CD34 and CD38 surface markers to identify different subpopulations [bulk (CD45+) and stem/progenitor (CD34+ or CD34+CD38)] by flow cytometry. CI-values were generated using software Calcusyn and bar graphs representing the best point of synergy were plotted. All graphs represent data from triplicates ± SD. The statistical significance of the experiments was calculated by standard Student’s t-test and p-values are indicated in respective graphs.
We next combined SBI-0206965 with the AML therapies. ULK1 inhibition was synergistic with both chemotherapy (AraC and Ida) and Bcl2 inhibition (ABT-199) across AML cell lines and patient samples (Fig. 3E–H; Supplementary Fig. S3C–H; and Supplementary Table S6). This corroborates our earlier observation where genetic abrogation of ULK1 enhanced sensitivity of AML cells to these therapeutic agents.
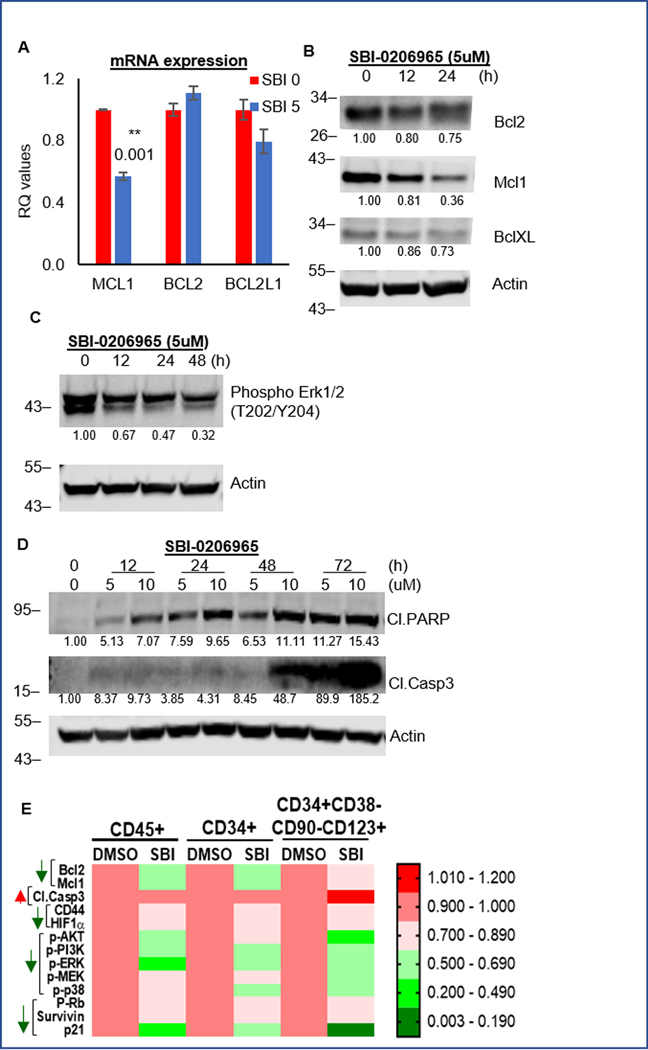
ULK1 inhibition downregulates Mcl1
Similar to ULK1-KO cells, SBI-0206965 decreased MCL1 transcriptionally, and at the protein level, decreased both Mcl1 and phospho-Erk1/2 (Fig. 4A–C and S4). Among the Bcl2 family members, Mcl1 downregulation was the most prominent. This was accompanied by increase in cleaved PARP and caspase 3 (Fig. 4D and S4). We further evaluated the effect of SBI on AML-PDX cells using CyTOF analysis of bulk and stem/progenitor populations. SBI-0206965 treatment significantly decreased anti-apoptotic Bcl2 family proteins and phospho-Erk1/2 in both populations (Fig. 4E). The concomitant increase in cleaved caspase 3, in the stem/progenitor compartment of PDX cells, further confirmed our data in cell lines.
Figure 4. SBI-0206965 downregulates Mcl1.
(A) Representative bar graphs showing mean RQ values (from triplicates ± SD) from qRT-PCR assay performed for Bcl2 family members, on RNA isolated from cells treated with either DMSO or SBI-0206965 for 24 hours. (B) Western blot analysis of whole cell lysates of OCI-AML3 cells treated with SBI-0206965 at different doses, for indicated time periods to analyze anti-apoptotic Bcl2 proteins: Bcl2, Mcl1 and BclXL. (C) Western blot analysis of whole cell lysates of OCI-AML3 cells treated 5uM of SBI-0206965 for indicated time periods. (D) Western blot analysis of whole cell lysates of OCI-AML3 cells treated with SBI-0206965 at different doses, for indicated time periods to analyze apoptosis markers: cleaved PARP and cleaved Caspase3. (E) Heat map representation of CyTOF experiment (generated using GraphPad) to show the changes in protein expression of signaling molecules, based on different functional clusters in different subpopulations: bulk (CD45+) and stem/progenitor populations (CD34+ and CD34+CD38-CD90-CD123+). Experiment was performed on primary AML blasts (PDX sample # 4) treated with 5uM of SBI-0206965 for 24 hours in vitro. Actin was used as loading control and for densitometric measurements for all western blots. The statistical significance of the experiments was calculated by standard Student’s t-test and p-values are indicated in respective graphs.
ULK1 inhibition impairs ROS mitigation
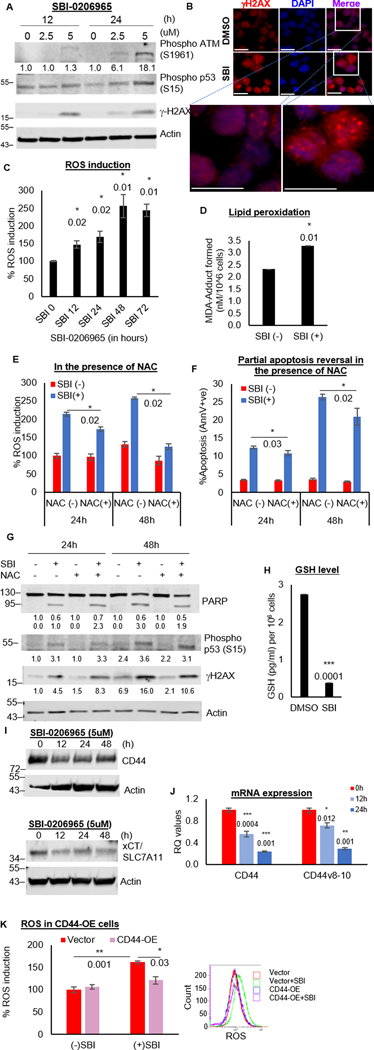
As CD44 and its isoforms were downregulated in ULK1-KO cells, we assessed ROS levels after treatment of OCI-AML3 cells with SBI-0206965. SBI-0206965 induced time-dependent increase in ROS and DNA damage, and lipid peroxidation (Fig. 5A–D and S5A). N-acetylcysteine (NAC), a known antioxidant, quenched SBI-0206965-induced ROS and partially reversed apoptosis, DNA damage and lipid peroxidation (Fig. 5E–G and S5B). This partial phenotypic reversal with NAC indicates that biological impact of ULK1 on DNA damage, extends beyond ROS induction.
Figure 5. ULK1 inhibition impairs ROS mitigation.
(A) Western blot showing DNA damage markers in whole cell lysates of OCI-AML3 cells treated with SBI-0206965. (B) Representative images from immunofluorescence in OCI-AML3 cells treated with DMSO or SBI-0206965 (24 hours): 1O−γH2AX, 2O−donkey anti-rabbit AF594, Nuclei–DAPI, 40X magnification, scale bar 10 um. (C) Representative bar graphs showing percent ROS induction with respect to DMSO treated control cells as 100 (from triplicates ± SD) in OCI-AML3 cells treated with SBI-0206965. (D) Representative bar graphs showing MDA-adduct formed in nM/106 cells (from triplicates ± SD) to quantify lipid peroxidation (colorimetric estimation based on generated standard curve) in OCI-AML3 cells treated with/without SBI-0206965 (24 hours). (E-G) OCI-AML3 cells were treated with/without SBI-0206965, in the presence/absence of NAC (1mM), for 24 or 48 hours. Representative bar graphs showing percent ROS induction with respect to DMSO treated control cells as 100 (from triplicates ± SD) (E) and percent apoptosis (from triplicates ± SD) from Annexin V/DAPI assay (F), and western blot analysis for DNA damage markers (G). (H) Representative bar graphs to estimate reduced glutathione (GSH) in OCI-AML3 cells treated with/without SBI-0206965 (5uM) for 24 hours in GSH/GSSG assay (colorimetric estimation based on generated standard curve). Bar graphs represent the amount in pg/ml/106 cells (from triplicates ± SD). (I) Western blot analysis for CD44 and xCT (SLC7A11 gene product) in whole cell lysates of OCI-AML3 cells treated with SBI-0206965 (5uM). Actin was used as the loading control and for densitometric measurements for all western blots. It should be noted that WB for xCT in fig 5I and pERK1/2 in fig 4C were performed on the same blot and hence, both have the same actin. (J) Representative bar graphs (mean RQ value from triplicates ± SD) from qRT-PCR assay performed for CD44 and its variant (v8–10) on RNA isolated from OCI-AML3 cells treated with SBI-0206965 (5uM). RQ values generated from Ct values (2-ΔΔCt). (K) Representative bar graphs showing percent ROS induction with respect to DMSO treated vector control cells as 100 (from triplicates ± SD; top) and corresponding representative histograms (bottom) in OCI-AML3 cells transiently overexpressing either vector or CD44, treated with/without SBI-0206965 (24 hours). The statistical significance of all experiments was calculated by standard Student’s t-test and p-values are indicated in respective graphs.
Increased ROS generation with SBI-0206965 was associated with significant decrease in the cellular levels of the antioxidant GSH (Fig. 5H). In addition, all 3 components of the cystine-glutamate antiporter system that are critical for glutathione reduction, especially xCT and CD44, were downregulated upon SBI-0206965 treatment (Fig. 5I, S5C and S4). Similar to ULK1-KO cells, the levels of CD44-WT and v8–10 were transcriptionally downregulated by SBI-0206965 (Fig. 5J).
Next, we overexpressed CD44 in OCI-AML3 (Supplementary Fig. S5D–F) and measured ROS levels after SBI-0206965 treatment. As shown in Fig. 5K, CD44 overexpression significantly mitigated SBI-0206965-induced ROS induction, indicating that CD44 levels are important in regulating ROS levels in this context. CyTOF analysis of SBI-0206965-treated AML-PDX cells further confirmed downregulation of CD44 in both bulk and stem/progenitor populations (Fig. 4E). Thus, ULK1 inhibition hampers ROS quenching in AML cells and contributes partly to ROS-mediated DNA damage.
ULK1 inhibition results in mitochondrial dysfunction
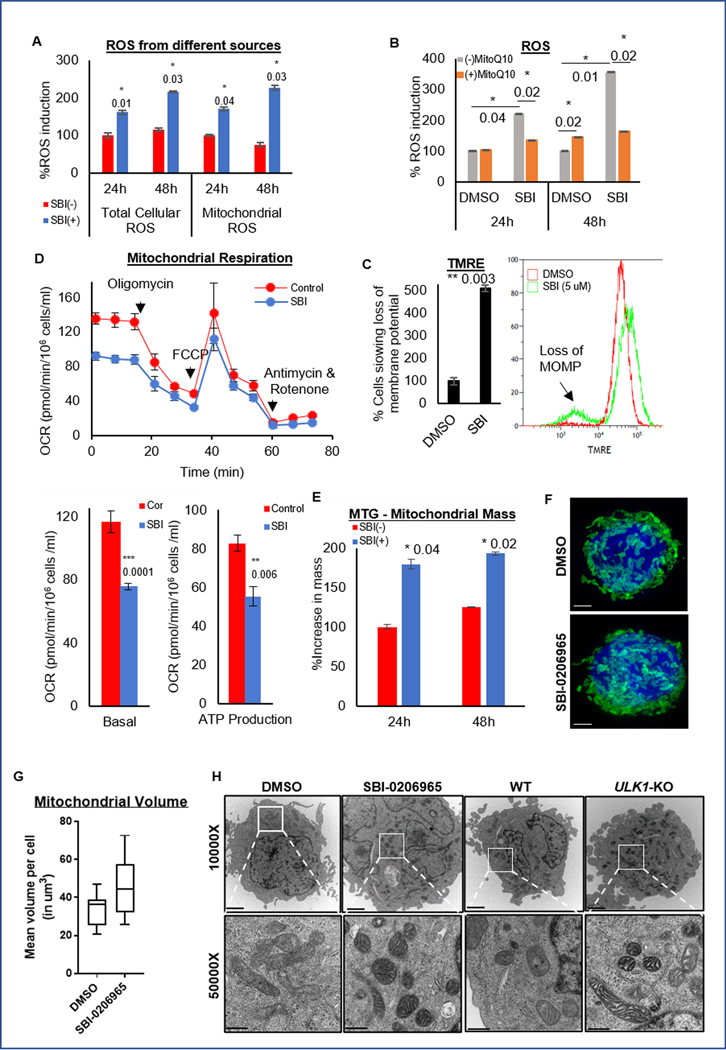
We next sought to investigate the source of excess ROS with ULK1 inhibition. To assess the source of ROS, we performed different ROS assays. Organelles like mitochondria are a major source of cell-intrinsic ROS (31), and we found that increased cellular ROS with SBI-0206965 was largely mitochondrial (Fig. 6A). Mitoquinone mesylate (MitoQ10), a mitochondria-specific antioxidant, almost completely abrogated ROS generation and partially reversed ULK1 inhibition-induced apoptosis (Fig. 6B and S6A), thus confirming the mitochondrial origin of ROS in this context.
Figure 6. ULK1 inhibition causes mitochondrial dysfunction.
(A) Representative bar graphs showing percent of total cellular ROS and mitochondrial ROS induced with respect to the DMSO treated control cells as 100 (from triplicates ± SD) in OCI-AML3 cells were treated with or without SBI-0206965 for 24 or 48 hours using ROS-ID and MitoSOX assays respectively. (B) Representative bar graphs showing the percent ROS induced with respect to the DMSO treated control cells as 100 (from triplicates ± SD) in OCI-AML3 cells were treated with or without SBI-0206965, in the presence or absence of mitochondria specific antioxidant MitoQ10 (100 nM), for 24 or 48 hours. (C) Representative bar graph (mean percentage from triplicates ± SD) (left), with representative histograms (right) of TMRE assay to quantify loss of membrane potential in OCI-AML3 cells were treated with SBI-0206965 for 24 hours. (D) Representative line graph (mean value from triplicates ± SD) from the Seahorse Mito Stress test showing Oxygen Consumption Rate (OCR) in different phases of the reaction, in OCI-AML3 cells treated with SBI-0206965 for 24 hours (top). Representative bar graphs (from triplicates ± SD) to quantify basal respiration and ATP production (bottom). (E) Representative bar graphs (percent mean fluorescence intensity from triplicates ± SD) to quantify mitochondrial mass in OCI-AML3 cells treated with SBI-0206965 for 24 and 48 hours and stained with MitoTracker Green (MTG) dye and analyzed by flow cytometry. (F) Representative images from Super-Resolution Microscopy of OCI-AML3 with GFP-labeled mitochondria treated with SBI-0206965 for 24 hours. Images were acquired using the DeltaVision OMX Blaze V4 at 100X magnification (scale bar 5 um). (G) Box plot representing the total volume of mitochondria per cell, quantified by Imaris software. (H) Representative images from Transmission Electron Microscopy acquired at 10000X (upper panel: scale bar 2 um) and 50000X (lower panel: scale bar 500 nm) magnifications from OCI-AML3 cells treated with DMSO or SBI-0206965 for 24 hours, and control (WT) or ULK1-KO cells. The statistical significance of the experiments was calculated by standard Student’s t-test and p-values are indicated in respective graphs.
Uncoupling of electron transport chain depolarizes mitochondrial membrane and leads to increased mitochondrial ROS levels (31, 32). As shown in Fig. 6C, SBI-0206965 treatment resulted in loss of mitochondrial outer membrane potential (MOMP). In addition, ULK1 inhibition resulted in distinct reductions in oxygen consumption rate, basal respiration and ATP production, reminiscent of mitochondrial dysfunction (Fig. 6D). Quantitation of mitochondrial content confirmed increased mitochondrial mass (Fig. 6E) as well as mitochondrial volume per cell with SBI-0206965 (Fig. 6F–G and S6B).
Increase in mitochondrial mass could be either due to increased mitochondrial biogenesis or reduced mitophagic clearance, and ULK1 deficiency is expected to inhibit mitophagic clearance (33, 34). SBI-0206965-treated and ULK1-KO cells showed increased mitochondrial numbers and typical morphology such as swollen mitochondrial cristae reminiscent of mitochondrial stress (Fig. 6H). Moreover, ULK1 inhibition did not change PPARGC1A expression (a marker of mitochondrial biogenesis) or mitochondrial DNA content suggesting that ULK1 inhibition has no effect on mitochondrial biogenesis (Supplementary Fig. S6C–D). Together, ULK1 inhibition impaired mitophagy, increased mitochondrial stress and ROS levels, decreased mitochondrial metabolism. Therefore, excess mitochondrial ROS production on one hand, and loss of ROS quenching on the other hand, skews the redox state in AML cells with ULK1 inhibition.
ULK1 inhibitor, SBI-0206965, delays leukemia progression
To determine if SBI-0206965 could recapitulate the colony forming and homing/engraftment effects of ULK1-KO, we performed colony formation assays with OCI-AML3 and primary AML blasts. Similar to ULK1-KO cells, SBI-0206965 treatment reduced colony forming ability (Supplementary Fig. S7A–I). In an AML-PDX model (Supplementary Fig. S7J), SBI-0206965 treatment resulted in reduced leukemic burden (Supplementary Fig. S7K–M) compared to the vehicle group. Corresponding CyTOF analysis on AML-PDX cells treated ex vivo with SBI-0206965, showed impaired proliferative/survival signaling (phosphoAkt, phosphoPI3K and phosphoMEK), decreased anti-apoptotic proteins (Bcl2 and Mcl1) and decreased microenvironmental factors like CD44 in both bulk (hCD45+) and LSC (CD34+CD38-CD123+) populations (Fig. 4E). These results indicate that ULK1 inhibition impairs AML LSC function.
ULK1 inhibition synergizes with chemotherapy increasing DNA damage
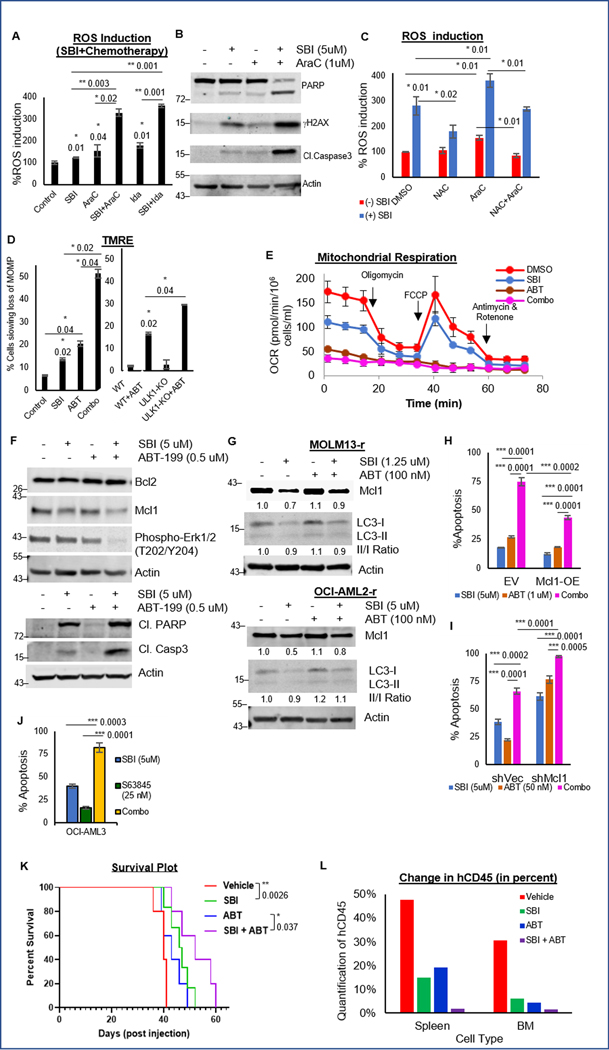
We first examined the synergy between ULK1 inhibition and chemotherapy. ULK1 inhibition synergistically enhanced the sensitivity of AML cells to chemotherapy (Fig. 1C, S1F, 3E–G and S3C–H). This effect was mediated through ROS induction by chemotherapy (AraC or Ida) and was significantly enhanced in combination with SBI-0206965 leading to increased DNA damage (Fig. 7A–B and S8A). The excess ROS production was partially reversed by NAC-mediated ROS quenching (Fig. 7C).
Figure 7. In vitro and in vivo synergy with ULK1 inhibition.
(A) Representative bar graphs showing percent ROS induction with respect to DMSO treated control cells as 100 (from triplicates ± SD) in OCI-AML3 cells treated with single agents AraC or Ida and in combination with SBI-0206965 (24 hours). (B) Western blot analysis for DNA damage markers in whole cell lysates of OCI-AML3 cells treated with single agents SBI-0206965 or AraC and in combination (24 hours). (C) Representative bar graphs showing percent ROS induction with respect to DMSO treated control cells as 100 (from triplicates ± SD) in OCI-AML3 cells treated with single agents SBI-0206965 or AraC and in combination, in presence/absence of NAC (24 hours). (D) Representative bar graphs (mean percentage from triplicates ± SD, TMRE assay) quantifying loss of membrane potential in OCI-AML3 cells treated with single agents SBI-0206965 or ABT-199, and in combination (24 hours, left); or in control (WT) and ULK1-KO cells treated with DMSO or ABT-199 (24 hours, right). (E) Representative line graph (mean value from triplicates ± SD, Seahorse Mito Stress test) showing Oxygen Consumption Rate (OCR) in OCI-AML3 cells treated with single agents SBI-0206965 (24 hours) or ABT-199 (2 hours), and in combination. (F) Western blot analysis in whole cell lysates of OCI-AML3 cells treated with single agents SBI-0206965 or ABT-199, and in combination (24 hours). (G) Western blot analysis for Mcl1 and LC3 in whole cell lysates of ABT-199 resistant cells treated with single agents SBI-0206965 or ABT-199, and in combination (24 hours). (H-I) Representative bar graphs showing percent apoptosis (from triplicates ± SD, Annexin V/DAPI assay) in empty vector or MCL1 overexpressing cells (H), and shControl or shMcl1 cells (I) treated with single agents SBI-0206965 or ABT-199, and in combination (72 hours). (J) Representative bar graphs showing percent apoptosis (from triplicates ± SD, Annexin V/DAPI assay) in OCI-AML3 cells treated with single agents SBI-0206965 or Mcl1 inhibitor (S63845), and in combination (72 hours). (K) Kaplan-Meier survival plot showing survival benefit of SBI-0206965 compared to control, and the benefit of adding SBI-0206965 to ABT-199. Indicated p-values were estimated by Mantel-Cox test. Median survival in days: vehicle (40), SBI-0206965 (47), ABT-199 (43) and combination (52). (L) Representative bar graph showing average percent human CD45+ve cells in spleen and bone marrow (flow cytometry), as a measure of tissue infiltration at the time of death. Actin was used as the loading control in all western blots. The statistical significance of all experiments (except in K) was calculated by standard Student’s t-test and p-values are indicated in respective graphs.
ULK1 inhibition synergizes with ABT-199/venetoclax through Mcl1 inhibition
Since Bcl2 family proteins regulate mitochondrial apoptosis by modulating MOMP and Bcl-2 inhibitors such as ABT-199 induce mitochondrial apoptosis in AML cells (28), we evaluated the effect of combining ULK1 inhibition and Bcl2 inhibition on apoptosis. ULK1 inhibition synergistically enhanced the sensitivity of AML cells to ABT-199 (Fig. 1C, S1F, 3E–F, 3H and S3C–H). This effect was due to synergistic impairment of mitochondrial functions, membrane potential and respiration (Fig. 7D–E and S8B–D). Mcl1 is important for leukemic cell survival and in sensitivity to ABT-199 (28). ULK1 inhibition downregulated Mcl1 (Fig. 1E and 4A). While Mcl1 level was increased in cells treated with ABT-199 alone, likely as acquired resistance, SBI-0206965 alone or in combination with ABT-199 reduced Mcl1, and increased PARP and Caspase 3 cleavage (Fig. 7F and S8E). Furthermore, ABT-199-resistant cells that have higher expression of Mcl1 at baseline, were sensitive to SBI-0206965 suggesting that ULK1 inhibition can overcome resistance to venetoclax via downregulation of Mcl1, inducing apoptosis (Fig. 7G and S8F).
To further validate our findings, we generated two isogenic cell lines, Mcl1 overexpression (Mcl1-OE) and Mcl1 knockdown (shMcl1). In Mcl1-OE cells, SBI-0206965 and ABT-199 combination remained synergistic, although significantly dampened (Fig. 7H and S8G–H). Strikingly, shMcl1 cells were extremely sensitive (20-fold) to SBI-0206965-mediated apoptosis, and showed significantly enhanced apoptotic synergy with ABT-199 (Fig. 7I and S8I–J). Mcl1 inhibitor S63845, was also highly synergistic in combination with SBI-0206965, highlighting additional possibilities of combination therapies (Fig. 7J and S8K–L). Together, these data identify Mcl1 downregulation as the main mechanism of synergy in combined Bcl2 and ULK1 inhibition to overcome ABT-199 resistance.
ULK1 inhibitor, SBI-0206965 synergizes with ABT-199 in an ex vivo model of AML
To verify the synergy of ULK1 inhibition with ABT-199 in vivo, we generated an ex vivo system, wherein AML cells were treated with drugs ex vivo and injected via tail vein (Supplementary Fig. S8M). ULK1 inhibition enhanced overall survival in the SBI-0206965-treated group of mice compared to control. While ABT-199 alone did not improve survival, its combination with SBI-0206965 extended survival (Fig. 7K and Supplementary Table S7). We confirmed corresponding reduction in disease burden with SBI-0206965 alone, and in combination with ABT-199 (Fig. 7L and S8N–P). This data further supports the clinical translation of combinatorial effects of ULK1 and Bcl-2 inhibition.
Discussion
Venetoclax-based combinations represent a good frontline option for elderly patients (35). However, despite high rates of initial remissions with existing therapies, the major concern is the persistence of quiescent and resistant LSCs (18, 19). This scenario can likely be improved with therapies targeting LSCs and mechanisms responsible for inherent/adaptive resistance. Autophagy presents a critical adaptive resistance in LSCs (17). Low ROS is crucial in retaining LSC ‘stemness’ (21), and is maintained by removal of damaged mitochondria by mitophagy and mitochondrial fission (23). Mitophagy and mitochondrial dynamics depend on autophagy-related mitochondrial proteins PINK1 and Parkin (36), upstream autophagy regulators LKB1 (37), and AMPK (38), mitochondrial fission/fusion proteins Mfns, Fis1, Drp1, etc. (21, 23, 39–41). Metabolic/stress sensing inputs from LKB1, AMPK, mTOR, and other regulators converge on the core apical autophagy kinase ULK1 (41–45). Accordingly, our experimental strategy evaluates how existing and emerging therapies activate autophagy in AML, and the effects of inhibiting ULK1 in this context.
We found that ULK1 is upregulated in leukemic blasts compared to normal hematopoietic cells, indicating a possible therapeutic window. We show that chemotherapies or VENETOCLAX induce autophagy through ULK1 activation as an adaptive mechanism of resistance in AML cells. High basal autophagy in these resistant cells further supports a therapeutic strategy targeting ULK1.
Chronic exposure to ROS may play a role in oncogenesis, but acute ROS induction has deleterious effects on proteins and lipids in cancer cells (43). Apoptosis in response to chemotherapy is associated with increase in cellular ROS generation and DNA damage (44). While we show that ULK1 inhibition alone caused DNA damage, its combination with chemotherapies further enhanced DNA damage, due to a combination of increased ROS generation and impaired ROS mitigation. Other potential mechanisms of synergy in DNA damage include limited nucleotide availability and/or impaired repair with ULK1 inhibition. DNA damage is closely followed by DNA damage response (DDR) ensuring genomic integrity restoration, particularly in malignant cells with numerous replication forks. Recent reports suggest autophagy can modulate DDR pathway proteins (45). ATP turnover is important for DDR and is also autophagy-dependent. Previous studies show that loss of autophagy impairs homologous recombination (46). Our data show, anti-oxidants such as NAC and MitoQ10 robustly reversed ROS generation and attenuated DNA damage. Mechanistically we found that ULK1 inhibition impaired ROS mitigation via CD44 downregulation. While CD44-mediated effects only partially account for the ROS mitigation strategy, in certain LSC populations this mechanism appears more relevant (47).
Mitophagy, the key mitochondrial clearance mechanism, maintains HSC/LSCs (48). We showed that ULK1-mediated autophagy inhibition increased mitochondrial mass without increasing mitochondrial biogenesis. Importantly, this effect was concomitant with compromised mitochondrial functions. Accumulation of damaged mitochondria leads to increased ROS generation while downregulation of the CD44-xCT and GSH axis impaired ROS mitigation and this in turn potentiates DNA damage and apoptosis. Interestingly, v8–10 isoform of CD44 is important in stabilizing the cystine-glutamate antiporter system to mitigate ROS (29). We found that along with WT, this variant (v8–10) was also downregulated by ULK1inhibition. Therefore, ULK1 inhibition can potentially target AML-LSCs by disabling cellular ROS mitigation.
In our in vivo data, we present striking prolongation of survival in mice bearing ULK1-KO AML without any additional therapy. ULK1-KO cells have impaired homing ability and ROS mitigation, leading to slower disease progression in these mice. In an AML-PDX model, lower disease burden in SBI-0206965-treated mice, supports this finding. The impact of ULK1 inhibition on progenitors and LSCs was further supported by impaired clonogenic growth of patient-derived AML cells and signaling changes in the LSC compartment. Therefore, our data using both genetic and pharmacologic ULK1 inhibition indicate the role of ULK1 in maintaining LSCs.
We previously reported that anti-apoptotic Mcl1 plays a crucial role in resistance to Bcl2 inhibition in AML (28). In addition, Erk signaling was reported to interact with autophagy proteins, and knocking-out autophagy proteins Atg5 or Atg7 also reduce Erk phosphorylation (49). Therefore, decreased Erk1/2 protein phosphorylation is possibly a sum effect of apical autophagy inhibition via ULK1. This in turn could lead to transcriptional downregulation of MCL1. Our data highlights the potential of combining ULK1 inhibitors with Bcl2 and/or Mcl1 inhibitors. Synergy of ULK1 inhibition with AraC also raises the possibility of improving efficacy of existing therapies with ULK1 inhibition.
SBI-0206965, used in our experiments as a tool ULK1 inhibitor, also has other targets, most prominently FLT3. According to the drug’s known FLT3 activity, FLT3-ITD cells were most sensitive to SBI-0206965. Therefore, to avoid the confounding impact of FLT3 activity, OCI-AML3 (a non-FLT3 mutant AML cell line) was used in all major experiments. Moreover, SBI-0206965 was used as a tool compound, and ULK1-KO cells were generated early on to carry out most experiments. Nonetheless, the effects seen with the genetic perturbation, were replicated while using the inhibitor.
Interestingly, in the context of overcoming drug-resistance (AraC and ABT-199 in this case), we show that not only does ULK1 inhibition reverse the drug-induced autophagic adaptation, but also induces apoptosis as a single agent in the drug-resistant cells.
In conclusion, our data clearly supports clinical benefit of ULK1 inhibitor in our AML models. This strategy can potentially synergize with current and emerging therapies to eradicate the persistent residual disease responsible for relapses. ULK1 inhibitors, for the first time, are being tested in early phase clinical trials and we will be working towards clinical translation of our data in AML.
Supplementary Material
Implications.
ULK1 drives a cell-intrinsic adaptive resistance in AML and targeting ULK1 mediated autophagy can synergize with existing and emerging AML therapies to overcome drug-resistance and induce apoptosis.
Acknowledgments
Mass cytometry by Duncan Mak, flow cytometry and cell sorting assisted by Wendy Schober and Nalini Patel, and confocal imaging assisted by Dr. Jared Burks, were performed in North Campus Flow Cytometry & Cellular Imaging Facility at The University of Texas MD Anderson Cancer Center under the supervision of Dr. Michael Andreeff and supported in part by the National Institutes of Health (NIH) through MD Anderson’s Cancer Center Support Grant (CCSG) CA016672 (to M.A). Electron microscopy was performed by Kenneth Dunner at the High-Resolution Electron Microscopy Facility at The University of Texas MD Anderson Cancer Center supported by CCSG NIH P30CA016672. Super-Resolution microscopy was performed assisted by Dr. Thomas Zal and Malgorzata A. Zal in the Advanced Microscopy Core at The University of Texas MD Anderson Cancer Center funded by CCSG NIH 1S10 RR029552 (to T.Z). All cell lines were authenticated at the CCSG Characterized Cell Line Core facility at The University of Texas MD Anderson Cancer Center supported by NCI grant CA16672. We thank Dr. Hong Mu, Dr. Bin Yuan, and Lauren Ostermann for assisting in mice maintenance and drug treatments. We thank Dr. Rodrigo Jacamo for generating OCI-AML3 with GFP-labeled mitochondria. Primary AML samples were procured and distributed by personnel in the Molecular Hematology and Therapy Section at The University of Texas MD Anderson Cancer Center and information regarding mutational backgrounds were collected by Zoe Alaniz and Ahmed Al Rawi. Dr. Nicholas Cosford is grateful to the Epstein Foundation and the Sanford Burnham Prebys NCI CCSG P30CA030199 for support. Dr. Gautam Borthakur is grateful for the Shirley Stein Scientific Endowed Research Award.
Footnotes
Conflict of interest statement:
The authors declare no potential conflicts of interest.
References
- 1.Papaemmanuil E, Gerstung M, Bullinger L, Gaidzik VI, Paschka P, Roberts ND, et al. Genomic Classification and Prognosis in Acute Myeloid Leukemia. New England Journal of Medicine. 2016;374(23):2209–21. doi: 10.1056/NEJMoa1516192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Marando L, Huntly BJP. Molecular Landscape of Acute Myeloid Leukemia: Prognostic and Therapeutic Implications. Current oncology reports. 2020;22(6):61-. doi: 10.1007/s11912-020-00918-7. PubMed PMID: . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Perl AE. The role of targeted therapy in the management of patients with AML. Blood Advances. 2017;1(24):2281. doi: 10.1182/bloodadvances.2017009829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yu J, Jiang PYZ, Sun H, Zhang X, Jiang Z, Li Y, et al. Advances in targeted therapy for acute myeloid leukemia. Biomarker Research. 2020;8(1):17. doi: 10.1186/s40364-020-00196-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cucchi DGJ, Polak TB, Ossenkoppele GJ, Uyl–De Groot CA, Cloos J, Zweegman S, et al. Two decades of targeted therapies in acute myeloid leukemia. Leukemia. 2021;35(3):651–60. doi: 10.1038/s41375-021-01164-x. [DOI] [PubMed] [Google Scholar]
- 6.Konopleva M, Pollyea DA, Potluri J, Chyla B, Hogdal L, Busman T, et al. Efficacy and Biological Correlates of Response in a Phase II Study of Venetoclax Monotherapy in Patients with Acute Myelogenous Leukemia. Cancer Discov. 2016;6(10):1106–17. Epub 2016/08/16. doi: 10.1158/2159-8290.CD-16-0313. PubMed PMID: 27520294; PubMed Central PMCID: PMC5436271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Piya S, Kornblau SM, Ruvolo VR, Mu H, Ruvolo PP, McQueen T, et al. Atg7 suppression enhances chemotherapeutic agent sensitivity and overcomes stroma-mediated chemoresistance in acute myeloid leukemia. Blood. 2016;128(9):1260–9. doi: 10.1182/blood-2016-01-692244. PubMed PMID: WOS:000384754100018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Heydt Q, Larrue C, Saland E, Bertoli S, Sarry JE, Besson A, et al. Oncogenic FLT3-ITD supports autophagy via ATF4 in acute myeloid leukemia. Oncogene. 2018;37(6):787–97. Epub 2017/10/23. doi: 10.1038/onc.2017.376. PubMed PMID: 29059168; PubMed Central PMCID: PMC5808073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jang JE, Eom J-I, Jeung H-K, Cheong J-W, Lee JY, Kim JS, et al. AMPK–ULK1-Mediated Autophagy Confers Resistance to BET Inhibitor JQ1 in Acute Myeloid Leukemia Stem Cells. Clinical Cancer Research. 2017;23(11):2781. doi: 10.1158/1078-0432.CCR-16-1903. [DOI] [PubMed] [Google Scholar]
- 10.Sehgal AR, Konig H, Johnson DE, Tang D, Amaravadi RK, Boyiadzis M, et al. You eat what you are: autophagy inhibition as a therapeutic strategy in leukemia. Leukemia. 2015;29(3):517–25. Epub 2014/11/26. doi: 10.1038/leu.2014.349. PubMed PMID: 25541151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li X, Zhou Y, Li Y, Yang L, Ma Y, Peng X, et al. Autophagy: A novel mechanism of chemoresistance in cancers. Biomedicine & Pharmacotherapy. 2019;119:109415. doi: 10.1016/j.biopha.2019.109415. [DOI] [PubMed] [Google Scholar]
- 12.Mele L, del Vecchio V, Liccardo D, Prisco C, Schwerdtfeger M, Robinson N, et al. The role of autophagy in resistance to targeted therapies. Cancer Treatment Reviews. 2020;88:102043. doi: 10.1016/j.ctrv.2020.102043. [DOI] [PubMed] [Google Scholar]
- 13.Usman RM, Razzaq F, Akbar A, Farooqui AA, Iftikhar A, Latif A, et al. Role and mechanism of autophagy-regulating factors in tumorigenesis and drug resistance. Asia-Pacific Journal of Clinical Oncology. 2021;17(3):193–208. doi: 10.1111/ajco.13449. [DOI] [PubMed] [Google Scholar]
- 14.Smith AG, Macleod KF. Autophagy, cancer stem cells and drug resistance. The Journal of Pathology. 2019;247(5):708–18. doi: 10.1002/path.5222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mizushima N, Komatsu M. Autophagy: Renovation of Cells and Tissues. Cell. 2011;147(4):728–41. doi: 10.1016/j.cell.2011.10.026. [DOI] [PubMed] [Google Scholar]
- 16.Mulcahy Levy JM, Thorburn A. Autophagy in cancer: moving from understanding mechanism to improving therapy responses in patients. Cell Death & Differentiation. 2020;27(3):843–57. doi: 10.1038/s41418-019-0474-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ianniciello A, Rattigan KM, Helgason GV. The Ins and Outs of Autophagy and Metabolism in Hematopoietic and Leukemic Stem Cells: Food for Thought. Frontiers in cell and developmental biology. 2018;6:120-. doi: 10.3389/fcell.2018.00120. PubMed PMID: 30320108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shlush LI, Mitchell A, Heisler L, Abelson S, Ng SWK, Trotman-Grant A, et al. Tracing the origins of relapse in acute myeloid leukaemia to stem cells. Nature. 2017;547(7661):104–8. Epub 20170628. doi: 10.1038/nature22993. PubMed PMID: 28658204. [DOI] [PubMed] [Google Scholar]
- 19.Eppert K, Takenaka K, Lechman ER, Waldron L, Nilsson B, van Galen P, et al. Stem cell gene expression programs influence clinical outcome in human leukemia. Nat Med. 2011;17(9):1086–93. Epub 20110828. doi: 10.1038/nm.2415. PubMed PMID: 21873988. [DOI] [PubMed] [Google Scholar]
- 20.Dykstra KM, Fay HRS, Massey AC, Yang N, Johnson M, Portwood S, et al. Inhibiting autophagy targets human leukemic stem cells and hypoxic AML blasts by disrupting mitochondrial homeostasis. Blood Advances. 2021;5(8):2087–100. doi: 10.1182/bloodadvances.2020002666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lagadinou ED, Sach A, Callahan K, Rossi RM, Neering SJ, Minhajuddin M, et al. BCL-2 inhibition targets oxidative phosphorylation and selectively eradicates quiescent human leukemia stem cells. Cell stem cell. 2013;12(3):329–41. Epub 2013/01/17. doi: 10.1016/j.stem.2012.12.013. PubMed PMID: 23333149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pollyea DA, Stevens BM, Jones CL, Winters A, Pei S, Minhajuddin M, et al. Venetoclax with azacitidine disrupts energy metabolism and targets leukemia stem cells in patients with acute myeloid leukemia. Nature Medicine. 2018;24(12):1859–66. doi: 10.1038/s41591-018-0233-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pei S, Minhajuddin M, Adane B, Khan N, Stevens BM, Mack SC, et al. AMPK/FIS1-Mediated Mitophagy Is Required for Self-Renewal of Human AML Stem Cells. Cell Stem Cell. 2018;23(1):86-100.e6. Epub 2018/06/14. doi: 10.1016/j.stem.2018.05.021. PubMed PMID: 29910151; PubMed Central PMCID: PMC6035102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nguyen TD, Shaid S, Vakhrusheva O, Koschade SE, Klann K, Thölken M, et al. Loss of the selective autophagy receptor p62 impairs murine myeloid leukemia progression and mitophagy. Blood. 2019;133(2):168–79. Epub 20181129. doi: 10.1182/blood-2018-02-833475. PubMed PMID: 30498063. [DOI] [PubMed] [Google Scholar]
- 25.Mauthe M, Orhon I, Rocchi C, Zhou X, Luhr M, Hijlkema K-J, et al. Chloroquine inhibits autophagic flux by decreasing autophagosome-lysosome fusion. Autophagy. 2018;14(8):1435–55. doi: 10.1080/15548627.2018.1474314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Egan DF, Chun MGH, Vamos M, Zou HX, Rong J, Miller CJ, et al. Small Molecule Inhibition of the Autophagy Kinase ULK1 and Identification of ULK1 Substrates. Molecular Cell. 2015;59(2):285–97. doi: 10.1016/j.molcel.2015.05.031. PubMed PMID: WOS:000362457000015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kim J, Kundu M, Viollet B, Guan K-L. AMPK and mTOR regulate autophagy through direct phosphorylation of Ulk1. Nature Cell Biology. 2011;13:132. doi: 10.1038/ncb2152 https://www.nature.com/articles/ncb2152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pan R, Hogdal LJ, Benito JM, Bucci D, Han L, Borthakur G, et al. Selective BCL-2 Inhibition by ABT-199 Causes On-Target Cell Death in Acute Myeloid Leukemia. Cancer Discovery. 2014;4(3):362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ishimoto T, Nagano O, Yae T, Tamada M, Motohara T, Oshima H, et al. CD44 Variant Regulates Redox Status in Cancer Cells by Stabilizing the xCT Subunit of System xc− and Thereby Promotes Tumor Growth. Cancer Cell. 2011;19(3):387–400. doi: 10.1016/j.ccr.2011.01.038. [DOI] [PubMed] [Google Scholar]
- 30.Piya S, Mu H, Bhattacharya S, Lorenzi PL, Davis RE, McQueen T, et al. BETP degradation simultaneously targets acute myelogenous leukemia stem cells and the microenvironment. J Clin Invest. 2019;130. Epub 2019/03/05. doi: 10.1172/JCI120654. PubMed PMID: 30829648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Holmström KM, Finkel T. Cellular mechanisms and physiological consequences of redox-dependent signalling. Nature Reviews Molecular Cell Biology. 2014;15:411. doi: 10.1038/nrm3801. [DOI] [PubMed] [Google Scholar]
- 32.Murphy MP. How mitochondria produce reactive oxygen species. The Biochemical journal. 2009;417(1):1–13. Epub 2008/12/12. doi: 10.1042/BJ20081386. PubMed PMID: 19061483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kundu M, Lindsten T, Yang C-Y, Wu J, Zhao F, Zhang J, et al. Ulk1 plays a critical role in the autophagic clearance of mitochondria and ribosomes during reticulocyte maturation. Blood. 2008;112(4):1493. doi: 10.1182/blood-2008-02-137398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Murakawa T, Okamoto K, Omiya S, Taneike M, Yamaguchi O, Otsu K. A Mammalian Mitophagy Receptor, Bcl2-L-13, Recruits the ULK1 Complex to Induce Mitophagy. Cell Rep. 2019;26(2):338–45 e6. Epub 2019/01/10. doi: 10.1016/j.celrep.2018.12.050. PubMed PMID: 30625316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.DiNardo CD, Pratz K, Pullarkat V, Jonas BA, Arellano M, Becker PS, et al. Venetoclax combined with decitabine or azacitidine in treatment-naive, elderly patients with acute myeloid leukemia. Blood. 2019;133(1):7–17. doi: 10.1182/blood-2018-08-868752. PubMed PMID: WOS:000454811300004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Youle RJ, Narendra DP. Mechanisms of mitophagy. Nature Reviews Molecular Cell Biology. 2011;12(1):9–14. doi: 10.1038/nrm3028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nakada D, Saunders TL, Morrison SJ. Lkb1 regulates cell cycle and energy metabolism in haematopoietic stem cells. Nature. 2010;468(7324):653–8. doi: 10.1038/nature09571. PubMed PMID: 21124450; PubMed Central PMCID: PMC3059717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Saito Y, Chapple Richard H, Lin A, Kitano A, Nakada D. AMPK Protects Leukemia-Initiating Cells in Myeloid Leukemias from Metabolic Stress in the Bone Marrow. Cell Stem Cell. 2015;17(5):585–96. doi: 10.1016/j.stem.2015.08.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chen H, Chan DC. Mitochondrial Dynamics in Regulating the Unique Phenotypes of Cancer and Stem Cells. Cell Metabolism. 2017;26(1):39–48. doi: 10.1016/j.cmet.2017.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pei S, Minhajuddin M, Adane B, Khan N, Stevens BM, Mack SC, et al. AMPK/FIS1-Mediated Mitophagy Is Required for Self-Renewal of Human AML Stem Cells. Cell Stem Cell. 2018;23(1):86–100 e6. Epub 2018/06/19. doi: 10.1016/j.stem.2018.05.021. PubMed PMID: 29910151; PubMed Central PMCID: PMC6035102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jacquel A, Luciano F, Robert G, Auberger P. Implication and Regulation of AMPK during Physiological and Pathological Myeloid Differentiation. International journal of molecular sciences. 2018;19(10):2991. doi: 10.3390/ijms19102991. PubMed PMID: 30274374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Egan D, Kim J, Shaw RJ, Guan K-L. The autophagy initiating kinase ULK1 is regulated via opposing phosphorylation by AMPK and mTOR. Autophagy. 2011;7(6):643–4. Epub 2011/06/01. doi: 10.4161/auto.7.6.15123. PubMed PMID: 21460621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Barrera G Oxidative stress and lipid peroxidation products in cancer progression and therapy. ISRN Oncol. 2012;2012:137289. Epub 20121017. doi: 10.5402/2012/137289. PubMed PMID: 23119185; PubMed Central PMCID: PMC3483701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Srinivas US, Tan BWQ, Vellayappan BA, Jeyasekharan AD. ROS and the DNA damage response in cancer. Redox Biology. 2019;25:101084. doi: 10.1016/j.redox.2018.101084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hewitt G, Korolchuk VI. Repair, Reuse, Recycle: The Expanding Role of Autophagy in Genome Maintenance. Trends in Cell Biology. 2017;27(5):340–51. doi: 10.1016/j.tcb.2016.11.011. PubMed PMID: WOS:000399554400004. [DOI] [PubMed] [Google Scholar]
- 46.Liu EY, Xu N, O’Prey J, Lao LY, Joshi S, Long JS, et al. Loss of autophagy causes a synthetic lethal deficiency in DNA repair. Proceedings of the National Academy of Sciences. 2015;112(3):773. doi: 10.1073/pnas.1409563112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Piya S, Yang Y, Bhattacharya S, Mu H, Ruvolo V, Baran N, et al. Targeting NOTCH1-MYC-CD44 Axis to Eliminate Leukemia Initiating Cells (LIC) in T-ALL. Clinical Lymphoma Myeloma and Leukemia. 2019;19:S196–S7. doi: 10.1016/j.clml.2019.07.046. [DOI] [Google Scholar]
- 48.Ito K, Turcotte R, Cui J, Zimmerman SE, Pinho S, Mizoguchi T, et al. Self-renewal of a purified Tie2+ hematopoietic stem cell population relies on mitochondrial clearance. Science. 2016;354(6316):1156–60. Epub 20161013. doi: 10.1126/science.aaf5530. PubMed PMID: 27738012; PubMed Central PMCID: PMC5164878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Martinez-Lopez N, Athonvarangkul D, Mishall P, Sahu S, Singh R. Autophagy proteins regulate ERK phosphorylation. Nature Communications. 2013;4(1):2799. doi: 10.1038/ncomms3799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chou TC. Drug combination studies and their synergy quantification using the Chou-Talalay method. Cancer Res. 2010;70(2):440–6. Epub 20100112. doi: 10.1158/0008-5472.can-09-1947. PubMed PMID: 20068163. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The microarray data analyzed in Figure 1A were obtained from publicly available datasets GSE13159, GSE15434, GSE61804, GSE14468, GSE42519 and The Cancer Genome Atlas. All data generated in this study are available within the article and its supplementary data files.
Figure 1. ULK1 is upregulated in AML, and its loss enhances therapy response in vitro and delays leukemia progression in vivo.
(A) Dot plot representation comparing ULK1 gene expression from curated microarray-based gene expression profiles of human AML (GSE13159, GSE15434, GSE61804, GSE14468, and The Cancer Genome Atlas) and FACS-sorted healthy hematopoietic cells in different stages of maturation (GSE42519), generated using the Bloodspot server and database. (B) Representative line graphs showing absolute cell numbers (from triplicates ± SD) measured by Trypan Blue staining using a Vi-Cell counter in control (WT) and ULK1-KO cells cultured simultaneously under serum starved conditions. Cells were collected every 24 hours. (C) Representative line graphs showing percent apoptosis (from triplicates ± SD) from Annexin V/DAPI assay in WT and ULK1-KO cells simultaneously treated with increasing doses as indicated of either AraC (left), Ida (middle) or ABT-199 (right) for 72 hours. (D) Representative bar graphs showing comparative analysis of LC3 ratio in whole cell lysates of control (WT) and ULK1-KO (KO) cells, treated with DMSO, AraC or ABT-199 (calculated from triplicates ± SD, in LC3-PE flow cytometry). Actin was used as the loading control and for densitometric measurements. (E) Representative bar graphs showing mean RQ values (from triplicates ± SD) from qRT-PCR assay performed for MCL1, on RNA isolated from control (WT) or ULK1-KO cells. (F) Representative bar graphs showing the percent ROS induced with respect to untreated shControl cells as 100 (from triplicates ± SD), in shControl and shULK1 cells, either untreated, or in the presence of ROS inducer (Pyocyanin) and/or ROS scavenger (NAC). (G) Representative bar graphs showing mean RQ values (from triplicates ± SD) from qRT-PCR assay performed for CD44 and its variant v8–10, on RNA isolated from control (WT) or ULK1-KO cells. All RQ values were generated from Ct values (2-ΔΔCt). (H) Representative images from bioluminescence imaging (BLI) performed 16 hours post-injection to analyze homing of the injected cells in mice. (I) Representative images of colony formation assay performed in control (WT) and ULK1-KO cells (4X magnification, 500 um scale bar). (J) BLI showing disease progression in 6-week old male NSG mice injected with luciferized control (WT) or ULK1-KO cells (106 cells each) via tail vein injection (n>10) over time. (K) Representative dot plot showing the percent of circulating human CD45+ve cells in individual mice at indicated time points by flow cytometry from peripheral blood, to measure the disease burden over the course of the experiment. (L) Representative dot plot showing the percentage of human CD45+ve cells in individual mice at indicated time points by flow cytometry from different organs (spleen and bone marrow), to measure the disease burden over the course of the experiment. (M) Kaplan-Meier survival plot showing survival benefit of ULK1-KO in comparison to control. The values were estimated by Mantel-Cox test showing a p-value of 0.0001 and median survival was 38 days for WT and 69 days for ULK1-KO group. Statistical significance of the experiments was calculated by standard Student’s t-test and p-values are indicated in respective graphs.