Abstract
T helper 17 (TH17) cells are implicated in autoimmune diseases, and several metabolic processes are shown to be important for their development and function. In this study, we report an essential role for sphingolipids synthesized through the de novo pathway in TH17 cell development. Deficiency of SPTLC1, a major subunit of serine palmitoyl transferase enzyme complex that catalyzes the first and rate-limiting step of de novo sphingolipid synthesis, impaired glycolysis in differentiating TH17 cells by increasing intracellular reactive oxygen species (ROS) through enhancement of nicotinamide adenine dinucleotide phosphate oxidase 2 activity. Increased ROS leads to impaired activation of mammalian target of rapamycin C1 and reduced expression of hypoxia-inducible factor 1–alpha and c-Myc–induced glycolytic genes. SPTLCI deficiency protected mice from developing experimental autoimmune encephalomyelitis and experimental T cell transfer colitis. Our results thus show a critical role for de novo sphingolipid biosynthetic pathway in shaping adaptive immune responses with implications in autoimmune diseases.
Sphingolipid synthesis regulates metabolic rewiring underlying TH17 differentiation by influencing ROS production.
INTRODUCTION
T helper 17 (TH17) cells are a subset of CD4+ T helper cells that characteristically produce interleukin-17A (IL-17A) and mediate protective immunity against extracellular bacteria and fungi (1–3). However, aberrant TH17 cells are also associated with the pathogenesis of many inflammatory and autoimmune diseases like multiple sclerosis, inflammatory bowel diseases, rheumatoid arthritis, and psoriasis, suggesting that fine-tuning of their differentiation and function is required for proper health. Naïve T cells undergo programmed activation and differentiation to form mature T helper cells. Following antigenic stimulation, transforming growth factor–β1 (TGF-β1) along with IL-6 initiates TH17 differentiation by inducing the expression of retinoic acid–related orphan receptor gamma t (RORγt), a master transcription factor that controls the production of TH17–specific cytokines (4, 5). Further, under in vivo conditions, TH17 cells are stabilized by environmental factors including cytokines like IL-1β and IL-23, hypoxia, glucose, salt, and microbiota, indicating the role of intrinsic and extrinsic factors in TH17 cell development and function (4, 6–13).
The quiescent naïve T cells derive their energy mainly from mitochondrial oxidative phosphorylation (OXPHOS) for maintaining their minimal functions. However, upon activation, T cells increase their bioenergetic and biosynthetic demands for supporting rapid cell growth and proliferation. To meet this increased energy demand and increased need for the metabolic intermediates for synthesis of macromolecules (nucleic acid, lipids, and proteins), T cells up-regulate OXPHOS and aerobic glycolysis (14–17). Increased glycolysis in these cells is mediated by mammalian target of rapamycin (mTOR), a metabolic sensor, which induces the expression of genes related to glycolysis via transcription factors c-Myc and hypoxia-inducible factor 1–alpha (HIF-1α) (18–20). Further, studies have shown requirement for distinct energy metabolic programs during differentiation of T helper cell subsets. Glycolysis is important for TH17 development, whereas regulatory T (Treg) cell development depends on OXPHOS. Blocking glycolysis inhibits TH17 cell development in vitro and inhibits the development of experimental autoimmune encephalomyelitis (EAE) (10, 21, 22). Growing evidence also highlights the need for other metabolic processes like fatty acid and cholesterol biosynthesis and amino acid metabolism (glutaminolysis) in TH17 cell differentiation (23–27). However, the role of other metabolites/metabolic processes in regulating the energy metabolism and, hence, metabolic rewiring during TH17 differentiation is still emerging (28–30).
Sphingolipids, a class of lipids with sphingoid base as backbone, are generally membrane components and are essential for the survival of all eukaryotic cells from yeast to mammals (31–33). They constitute around 10 to 15% of total membrane lipids and regulate the biophysical properties of the membranes including the organization of proteins for cell recognition and signaling (33–37). Apart from their structural function, sphingolipids also play important roles as signaling molecules in many cell types including immune cells (33, 38–43). Sphingolipid-mediated biological outcomes are determined by the levels of sphingolipids in the cells, which, in turn, are regulated by a balance maintained between their synthesis and catabolism (31–33).
De novo sphingolipid biosynthetic pathway is the major contributor of cellular sphingolipids. Their synthesis begins in the endoplasmic reticulum (ER) with the condensation of serine and palmitoyl–coenzyme A to form 3-ketosphinganine (3-KDS), catalyzed by the rate-limiting enzyme serine palmitoyltransferase (SPT). In subsequent reactions, 3-KDS is converted to ceramide, which is then transported to Golgi complex for the synthesis of complex sphingolipids including sphingomyelin and glycosphingolipids (31). Mammalian SPT consists of two large subunits (SPTLC1 and SPTLC2 or SPTLC3) and one small subunit ssSPTa (small subunit a of SPT) or ssSPTb (small subunit b of SPT). The hetero oligomer of SPTLC1 and SPTLC2 contribute to the catalytic site, and, hence, activity and small subunits enhance the SPT activity and confer substrate specificity. In humans, missense mutations in genes encoding for SPTLC1 and SPTLC2 are associated with hereditary sensory neuropathy type 1 (HSAN-I) and a form of amyotrophic lateral sclerosis (44–46). Sphingolipid biosynthesis induces a conformational change in the murine norovirus receptor and facilitates viral infection since cells deficient in SPTLC2 are resistant to norovirus infection (47). In dendritic cells, sphingolipid biosynthesis is critical for phagocytosis of Candida albicans, and SPTLC2 deficiency in CD8+ T cell impairs its response to viral infection (41, 42). However, the role of de novo sphingolipid biosynthesis in CD4+ T helper cell differentiation and function is largely unknown.
Here, we report a critical role for de novo sphingolipid biosynthesis in regulating redox balance and metabolic rewiring during TH17 cell differentiation. By deleting SPTLC1 specifically in CD4+ T cells, we demonstrate that sphingolipids synthesized by this pathway are essential for regulating reactive oxygen species (ROS) production from nicotinamide adenine dinucleotide phosphate (NADPH) oxidase. SPTLC1 deficiency increased the intracellular ROS and inhibited mTORC1 activity leading to reduced glycolysis and TH17 differentiation. ROS scavenger N-acetyl cysteine (NAC) and the NADPH oxidase (NOX) inhibitor diphenyleneiodonium chloride (DPI) rescued the defects in SPTLC1-deficient TH17 cells. A similar rescue was observed upon treatment with 3-KDS, the product of SPT reaction. Supplementation with downstream products ceramide or glucosylceramide but not sphingomyelin or galactosylceramide rescued the cytokine profile of TH17 cells, indicating a specific requirement for glucosylceramide in TH17 differentiation. Further, SPTLC1 deficiency showed defective TH17 development in vivo and showed attenuated response in EAE and colitis. Thus, our data position de novo sphingolipid biosynthesis as a metabolic checkpoint, which can be explored as a therapeutic target in TH17-mediated autoimmune diseases.
RESULTS
SPTLC1 modulates CD4+ T helper subset differentiation
Although the role of sphingolipids synthesized via de novo pathway is appreciated in many cellular functions of immune cells including T cells, its role in CD4+ T helper cell differentiation and function is yet to be explored in detail (40–42, 48). We examined the potential role of the de novo sphingolipid biosynthetic pathway in CD4+ T cell biology by using Sptlc1 conditional knockout (KO) mice, as the germline deletion of Sptlc1 is embryonic lethal (34). For deleting Sptlc1 specifically in T cells, mice with loxP-flanked Sptlc1 alleles (Sptlc1floxl/flox) were crossed with CD4-Cre transgenic mice. Sptlc1 deletion was confirmed in CD4+ T cells by quantitative polymerase chain reaction (qPCR) (fig. S1A) and by mass spectrometry analysis of sphingolipid content. Sptlc1 deletion significantly reduced the levels of sphingomyelin, ceramide, and hexosylceramide in naïve cells (fig. S1B). Under homeostatic conditions, in thymus, SPTLC1-deficient mice showed equal frequency and number of CD4+ CD8+ double-positive (DP) and CD8+ single-positive (SP) thymocytes and slightly reduced frequency and number of CD4+ SP thymocytes compared to wild-type (WT) mice (fig. S1C). In peripheral lymphoid compartments, CD4+ T cell frequency and numbers were slightly reduced in spleen, while these were not affected in lymph nodes (fig. S1, D and E). In both spleen and lymph nodes, CD8+ T cell frequency and numbers were drastically reduced (fig. S1, D and E). However, the naïve, effector, and memory CD4+ T cell frequencies were comparable between WT and KO mice in spleen (fig. S1F). These data suggest a possible role for SPTLC1 in CD4+ T cell development in thymus and for the peripheral CD4+ and CD8+ T cell homeostasis.
Next, we explored the role of SPTLC1 in CD4+ T helper cell differentiation. Naïve CD4+ T cells from WT and Sptlc1-deficient mice were differentiated into major T helper subsets [TH1, TH2, TH17, and induced Treg (iTreg) cells], with different cytokine and neutralizing antibodies as described in Materials and Methods. After 4 days, the cells were analyzed for subset-specific intracellular cytokines and transcription factors in viable cells (fig. S2A) [interferon-γ (IFN-γ) for TH1 cells, IL-4 for TH2 cells, and IL-17A for TH17 cells and forkhead box protein 3 (FOXP3) for iTreg cells]. SPTLC1 deficiency increased the frequency of IFN-γ+ cells under TH1 differentiating conditions (fig. S2B). However, under TH2 differentiating conditions, SPTLC1 deficiency decreased IL-4+ cells and increased IFN-γ+ cells (fig. S2B). SPTLC1-deficient cells showed defective TH17 differentiation, characterized by reduced frequency of IL-17A+ cells and an unanticipated increase in frequency of IFN-γ+ cells (Fig. 1A). We also found that SPTLC1 deficiency enhanced the iTreg differentiation as evidenced by increased frequency of FOXP3-positive cells (fig. S2C) and increased Foxp3 mRNA levels (fig. S2D). Together, these data suggest that SPTLC1 differentially regulate CD4+ T cell subset differentiation.
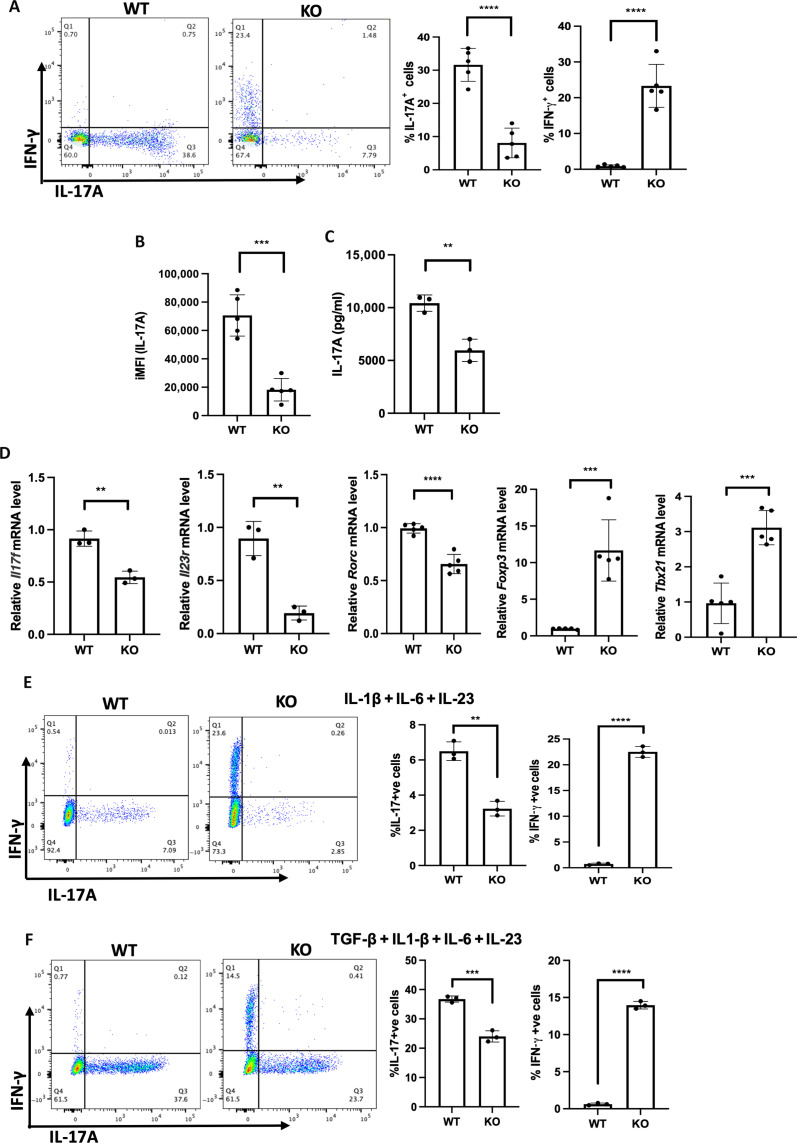
Fig. 1. SPTLC1 is essential for TH17 differentiation in vitro.
(A) Naïve CD4+ T cells from WT or KO mice were differentiated into TH17 cells (as described in Materials and Methods) for 4 days, and intracellular cytokines were scored by flow cytometry. The left panel is a representative flow figure showing frequency of IL-17A+ and IFN-γ+ cells, and the right panel shows the cumulative data of the same. n = 5 biologically independent samples, representative of five experiments. (B) iMFI of IL-17A. iMFI = (MFI)(P) where, MFI is the median fluorescence intensity of cytokine-positive cells, and P is the percentage of cytokine-positive cells. (C) WT and KO naïve T cells were differentiated under TH17 conditions for 4 days and reactivated with anti-CD3 for 24 hours, and the culture supernatant was analyzed for IL-17A by bioplex assay. n = 3 biologically independent samples. (D) WT and KO naïve T cells were differentiated into TH17 cells for 3 days and RNA quantified by reverse transcription qPCR. n = 3 to 5 biologically independent samples. (E and F) WT and KO naïve CD4+ T cells were differentiated into TH17 cells with IL-1β + IL-6 + IL-23 (E) or TGF-β1 + IL1-β + IL-6 + IL-23 (F) for 4 days and analyzed for frequency of IL-17A+ and IFN-γ+ cells by flow cytometry. The left panel is a representative flow figure, and the right panel shows the cumulative data for the same. n = 3 biologically independent samples. Each dot represents an individual mouse. All data presented as means ± SEM: **P < 0.01; ***P < 0.001; ****P < 0.0001; ns, not significant.
TH17 cells are implicated in many autoimmune and inflammatory disorders. TH17 cells maintain a distinct metabolic profile, alterations of which could impair their immune response. Increasing evidence suggests that the distinct metabolic state in TH17 cells is indeed contributed and maintained by changes in various metabolic processes during its differentiation and function (23, 27–29). Since sphingolipid flux through de novo synthetic pathway is shown to modulate the metabolic fitness of immune cells and inhibiting the SPT enzyme reduced the inflammation in many autoimmune settings, we hypothesize a role for de novo sphingolipid pathway in TH17 differentiation and function (32, 41, 42, 49). Hence, we sought to delineate the role of SPTLC1 in TH17 cells and evaluate molecular interactions that integrate de novo sphingolipid pathway to determination of T cell fates. We estimated ceramide, sphingomyelin, and hexosylceramide levels in the different subsets by mass spectrometry. We found that levels of hexosylceramide in TH17 cells were trending higher than in other subsets (fig. S2E). Further analysis of SPTLC1-deficient TH17 cells showed reduced intracellular levels of IL-17A per cell [integrated mean fluorescence intensity (iMFI)] and reduced amount of secreted IL-17A in SPTLC1-deficient TH17 cells (Fig. 1, B and C). In addition, we observed a significant decrease in the expression levels of TH17 lineage–specific genes (Fig. 1D). SPTLC1-deficient cells also showed increased expression of mRNA for FOXP3 under TH17 polarizing conditions (Fig. 1D). The defect in IL-17A production was also seen under pathogenic TH17 cell differentiating conditions: (i) IL-1β, IL-6, and IL-23 (Fig. 1E) and (ii) TGF-β1 + IL-1β + IL-6 + IL-23 (Fig. 1F) (9, 11, 50). Defective IL-17A production is not attributed to reduced T cell activation, as SPTLC1-deficient CD4+ T cells under TH17 polarizing conditions showed similar levels of surface activation markers, CD25 and CD69 (fig. S3A). Further, analysis of WT and KO CD4+ T cell activated under unbiased conditions also showed similar levels of these activation markers and similar levels of phosphorylated Zeta-chain–associated protein kinase-70 (ZAP70), an early T cell receptor (TCR) signaling marker (fig. S3, A and B). Cell proliferation and cell numbers were also reduced in SPTLC1-deficient CD4+ T cells under TH17 conditions (fig. S3, C and D). QVD-OPh, a pan-caspase inhibitor, restored cell viability but failed to rescue IL-17A production, indicating that the defect in cytokine production is due to intrinsic defects of cells and not due to cell death (fig. S3E). Moreover, Ki-67hi +ve (proliferating) cells in SPTLC1-deficient CD4+ T cells under TH17 polarizing conditions also showed reduced frequency of IL-17A+ cells and increased frequency in IFN-γ+ cells compared to the WT counterpart, indicating that the differentiation defect is independent of proliferation (fig. S3F). Since SPTLC1-deficient TH17 cells showed increased frequency of IFN-γ+ cells (Fig. 1A), and since IFN-γ can inhibit the TH17 cell differentiation (51), we investigated whether IFN-γ has a role in the observed phenotype. IFN-γ signals through the interferon-gamma receptor (IFNGR), which is a heterodimer composed of IFNGR1 and IFNGR2 (52). We analyzed the expression of IFNGR1 in WT and KO TH17 cells as a measure of IFNGR expression. We noted that SPTLC1-deficient TH17 cells had reduced expression of IFNGR1 compared to WT TH17 cells (fig. S4A). We then knocked down the expression of IFNGR1 in differentiating TH17 cells using SMARTpool Accell mouse Ifngr1 as described in Materials and Methods. We confirmed that this resulted in a reduction of cell surface expression of IFNGR1 (about 45%) in TH17 cells (fig. S4B). Accell nontargeting control pool small interfering RNA (siRNA) is used as negative control. Knockdown of IFNGR1 in WT cells did not alter the frequency of IL-17A+ cells (fig. S4C). Knockdown of IFNGR1 in SPTLC1-deficient cells did not rescue the defect, and they showed similar frequencies of IL-17A+ cells compared to SPTLC1-deficient cells (fig. S4C). Of note, Ifngr1 siRNA further increased the IFN-γ+ cells in KO SPTLC1-deficient TH17 cells, which was also observed upon control siRNA addition to these cells (fig. S4C). The nonspecific response of SPTLC1 KO cells to siRNA is currently not understood. To further rule out the role of IFN-γ in the observed defect in the SPTLC1-deficient TH17 cells, we differentiated naïve CD4 T cells from WT and IFNGR1 KO mice into TH17 cells in the presence of myriocin, a potent inhibitor of SPT (53). Myriocin-treated WT TH17 cells showed defects in TH17 differentiation evidenced by reduced frequency of IL-17A+ cells compared to WT TH17 cells (fig. S4D). IFNGR1 KO TH17 cells showed slightly reduced frequency of IL-17A+ cells compared to WT TH17 cells, and myriocin-treated IFNGR1 KO cells showed similar frequency of reduction in IL-17A+ cells compared to myriocin-treated WT TH17 cells (fig. S4D). IFNGR1 KO was confirmed by surface staining for IFNGR1 in TH1 cells (fig. S4E). TH17 culture condition also has neutralizing antibody to IFN-γ. Together, these data indicate that defects in the TH17 differentiation observed in SPLCT1-deficient cells are independent of increased IFN-γ production in these cells.
T cell–specific deletion of SPTLC1 affects TH17 cell function in vivo
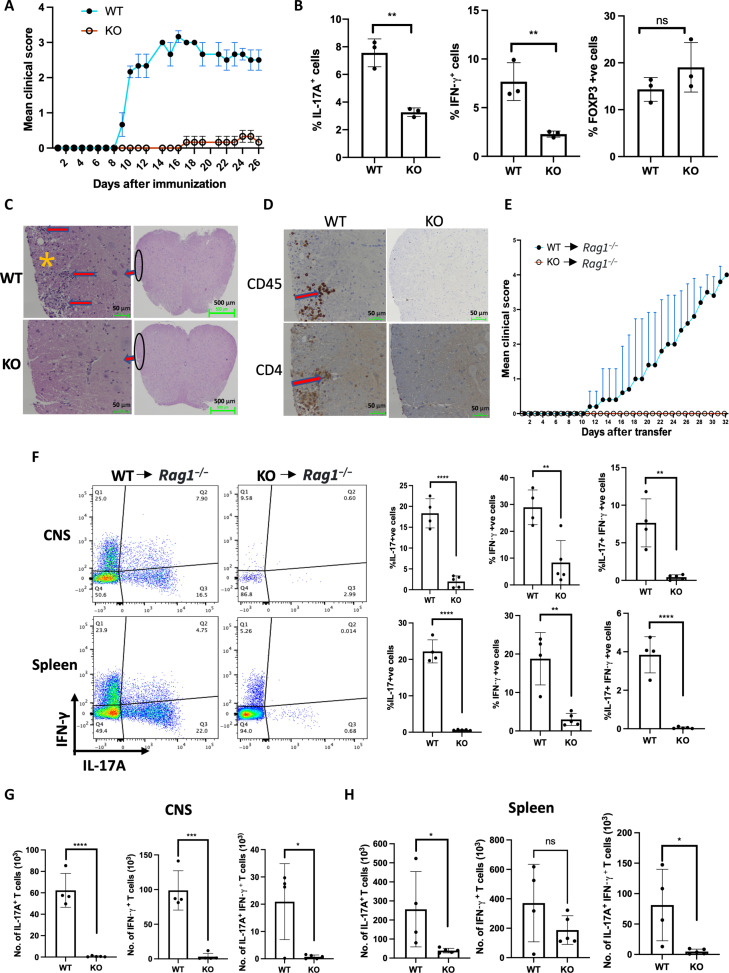
TH17 cells are required for mediating protective immunity against extracellular bacterial and fungal infections, but aberrant activation of these cells is also implicated in many autoimmune diseases including multiple sclerosis, a demyelinating disease in humans (3, 54). To examine the role of SPTLC1 in TH17 cell differentiation and function in vivo, we induced EAE, an animal model of human multiple sclerosis (55). Mice were immunized with the myelin oligodendrocyte glycoprotein (MOG)33-55 peptide in complete Freund’s adjuvant (CFA) followed by pertussis toxin and monitored daily for 28 days. WT mice displayed the disease symptoms at day 9, which peaked at day 15 after the initial immunization with a maximum score of around 3 (Fig. 2A). However, the KO mice had mild symptoms that were delayed, and the disease did not progress beyond a score of 0.5 (Fig. 2A). Analysis of the splenic CD4+ T cell population showed reduced IL-17A+ cells in KO mice compared to WT (Fig. 2B). Unlike in vitro differentiation studies, the IFN-γ+ cells were also reduced in this condition. The frequency of FOXP3+ cells did not differ between WT and KO spleen (Fig. 2B). Histopathological analysis of spinal cords showed reduced infiltration of leukocytes and demyelination in the KO mice compared to the WT (Fig. 2C). Further, immunohistochemical analysis confirmed fewer infiltration of CD45+ and CD4+ T cells in the spinal cord of KO mice (Fig. 2D). Since SPTLC1-deficient cells showed increased FOXP3 expression under in vitro TH17 differentiating conditions, we analyzed FOXP3 in splenic CD4+ T cells of immunized mice at disease preonset time point to assess the possibility of TH17 plasticity in vivo. On day 8 after immunization (disease preonset time point), analysis of splenic CD4+ T cells from SPTLC1-deficient mice showed similar frequency of FOXP3-positive cells compared to the WT counterpart (fig. S5A). SPTLC1-deficient mice showed reduced frequency of IL-17A+ cells in the spleen at this time (fig. S5A). Since SPTLC1-deficient mouse showed slightly reduced CD4+ T cells in spleen and lymph nodes under homeostatic conditions (fig. S1, D and E), we induced EAE by adapting an adoptive transfer protocol (fig. S5B) (55). In these experiments, cells from spleen and lymph nodes of immunized mice were cultured ex vivo under TH17 differentiating conditions, and CD4+ T cells were isolated for adoptive transfer. The cultured cells were assessed for intracellular cytokines before the transfer. CD4+ T cells from SPTLC1-deficient mice showed reduced frequency of IL-17A+, IFN-γ+, and IL-17A+IFN-γ+ DP cells compared to WT (fig. S5C). Equal numbers of ex vivo–differentiated cells were transferred to Rag−/− mice and scored daily for EAE symptoms. Mice receiving WT TH17 cells showed the symptoms of EAE from day 10, and, on the other hand, mice receiving KO TH17 cells did not show signs of EAE throughout the study (Fig. 2E). We examined CD4+ T cells from the central nervous system (CNS) by harvesting spinal cord and brain at the peak of the disease. We found significantly reduced frequency and numbers of infiltrating IL-17A+, IFN-γ+, and IL-17A+IFN-γ+ DP cells in mice receiving KO cells compared to WT cells receiving mice (Fig. 2, F and G). Further, analysis of spleen also showed reduced frequency and numbers of IL-17A+, IFN-γ+, and IL-17A+IFN-γ+ DP cells in mice receiving KO cells compared to their counterpart (Fig. 2, F and H).
Fig. 2. SPTLC1 is required for TH17 differentiation in vivo.
(A) Mean clinical EAE scores of WT and KO mice immunized with MOG35–45 peptide. n = 3 mice per experimental group, representative of two independent experiments. (B) Frequency of IL-17A+, IFN-γ+, and FOXP3-positive cells in splenocytes of immunized mice at day 28. n = 3 biologically independent samples. (C) Representative hematoxylin and eosin staining of spinal cord sections obtained from indicated immunized mice at day 28. Red arrows and asterisks indicate infiltrated inflammatory cells and demyelinated foci, respectively. Scale bars, 50 μm (left) and 500 μm (right). n = 3 biologically independent samples. (D) Immunohistochemical staining for CD4+ and CD45+ cells in spinal cord sections from immunized WT and KO mice at day 28. The red arrow indicates positive cells. Scale bar, 50 μm. n = 3 biologically independent samples. (E) Mean clinical scores for EAE in RAG−/− mice transferred with equal number of ex vivo differentiated CD4+ T cells from MOG35–45 immunized WT and KO mice. RAG−/− mice receiving WT cells (n = 4); RAG−/− mice receiving KO cells (n = 5). (F to H) CNS and spleen were isolated from the RAG−/− mice receiving WT cells at the peak of the disease and from corresponding RAG−/− mice receiving KO cells and scored for frequency (F) and absolute number (G and H) of IL-17A+, IFN-γ+, and IL-17A+IFN-γ+ DP cells by flow cytometry. (F) The left panel is a representative flow figure, and the right panel shows the cumulative data of the same. The top panel is from CNS and the bottom panel is from spleen. (G and H) CNS (G) and spleen (H). RAG−/− mice receiving WT cells (n = 4); RAG−/− mice receiving KO cells (n = 5). Each dot represents an individual mouse. All data presented as means ± SEM: *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001; ns, not significant.
We additionally evaluated the intrinsic role of SPTLC1 in the T cell transfer model of colitis, a mouse model for inflammatory bowel disease mediated by TH17 cells (56), by adoptive transfer of sorted naïve (CD4+ CD45RBhi) T cells from WT and KO mice into Rag−/− mice (fig. S5D). Rag−/− mice that received WT naïve CD4+ T cells showed significantly reduced body weight and increased colon weight-to-length ratio compared to Rag−/− mice that received KO CD4+ T cells (fig. S5, E and F). Further, histopathological examination of distal colon of Rag−/− mice that received WT naïve CD4+ T cells showed severe histiocytic colitis with hyperplastic crypt epithelium, whereas Rag−/− mice that received KO CD4+ T cells showed no signs of colitis (fig. S5G). Consistent with this finding, analysis of CD4+ T cells in the colon of Rag−/− mice receiving the KO CD4+ T cells showed reduced IL-17A+–positive cells (fig. S5H). These data suggest that SPTLC1 is required for TH17 differentiation and function in vivo.
SPTLC1 is required for metabolic reprograming in TH17 differentiation
To investigate the mechanism underlying the defective TH17 differentiation in SPTLC1-deficient cells, we first checked IL-2 signaling by measuring phosphorylation of signal transducer and activator of transcription 5 (p-STAT5) by flow cytometry. IL-2 is known to suppress TH17 differentiation and enhance iTreg differentiation through STAT5 signaling (57–59). SPTlC1-deficient TH17 cells showed reduced p-STAT5 phosphorylation compared to WT cells (Fig. 3A), ruling out IL-2 as the causative factor.
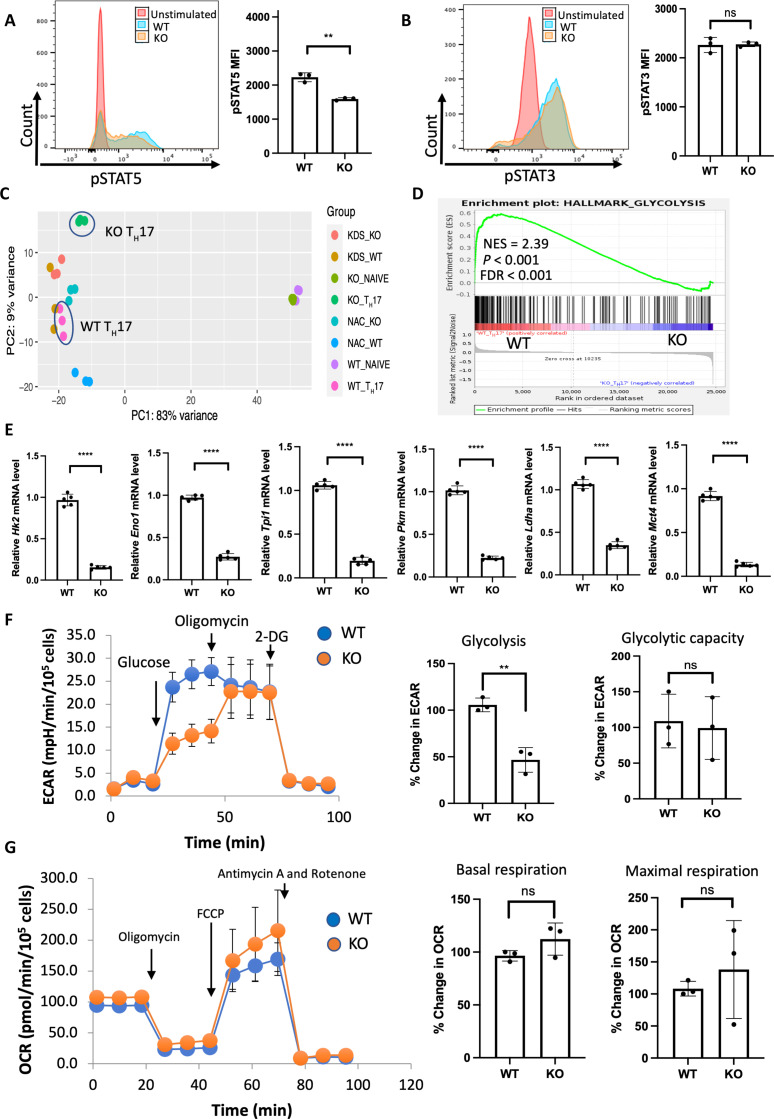
Fig. 3. SPTLC1 is required for metabolic reprograming in TH17 cells.
(A) WT and KO naïve cells were differentiated under TH17 conditions for 12 hours and scored for phosphorylated STAT5 by flow cytometry. The left panel is a representative flow figure, and the right panel shows the cumulative data of the pSTAT5 MFI. n = 3 biologically independent samples. (B) WT and KO naïve cells were differentiated under TH17 conditions for 12 hours and scored for phosphorylated STAT3 by flow cytometry. The left panel is a representative flow figure, and the right panel shows the cumulative data of the pSTAT3 MFI. n = 3 biologically independent samples. (C) Principal components analysis (PCA) of WT and KO TH17 cell RNA-seq data. n = 3 biological replicates per group. (D) GSEA result of HALLMARK_GLYCOLYSIS gene sets between WT and KO TH17 cells. (E) qPCR analysis of indicated glycolytic genes in WT and KO TH17 cells (naïve CD4+ T cells cultured under TH17 conditions for 3 days). n = 5 biologically independent samples. (F) WT and KO naïve T cells were differentiated under TH17 conditions for 3 days and ECAR measured in equal number of viable cells by Seahorse analyzer. The left panel shows the representative data, and the right panel shows the percentage change in ECAR for glycolysis and glycolytic capacity. n = 3 biologically independent samples. (G) TH17 cells were differentiated as in (F) and OCR measured in equal number of viable cells by Seahorse analyzer. The left panel shows the representative data, and the right panel shows the cumulative data for basal and maximal respiration. n = 3 biologically independent samples. Each dot represents an individual mouse. All data presented as means ± SEM: **P < 0.01; ****P < 0.0001; ns, not significant.
TGF-β induces the expression of both RORγt and FOXP3. Under TH17 conditions, IL-6, through STAT3 signaling, inhibits FOXP3 expression by TGF-β and enhances differentiation of TH17 cells. Therefore, we assessed the phosphorylation status of STAT3 by flow cytometry and found no change in the levels of STAT3 phosphorylation between WT and KO TH17 cells, indicating that SPTLC1 does not regulate the IL-6 signaling pathway in TH17 cells (Fig. 3B). Our recent report documented ER stress in SPTLC1-deficient bone marrow cells (34). Therefore, we checked for markers of ER stress in differentiated TH17 cells. The ER stress marker, BiP, did not differ between WT and KO TH17 cells at both mRNA and protein levels (fig. S6, A and B). The mRNA levels of XBP1s were reduced in KO TH17 cells (fig. S6C), and the protein levels of PERK did not differ between WT and KO TH17 cells (fig. S6D). These results indicate that SPTLC1 deficiency did not induce ER stress in the KO TH17 cells.
To further investigate how SPTLC1 regulates TH17 differentiation, we performed bulk RNA sequencing (RNA-seq) in WT and KO TH17 cells. The gene expression profiles of WT and KO TH17 cells were clearly distinct (Fig. 3C) with 5683 differentially expressed genes (DEGs), which include 2944 up-regulated and 2739 down-regulated genes (fig. S6E). Next, we performed unbiased pathway enrichment analysis and compared RNA-seq data of WT and KO TH17 cells. Notably, among enriched pathways, most of them belonged to carbohydrate and related metabolic processes (fig. S6F). When T cells are activated, they enhance aerobic glycolysis (19). Among the T cell subsets, TH17 cells show maximum glycolysis, and its inhibition affects TH17 differentiation (10, 54). Gene set enrichment analysis (GSEA) revealed that transcripts from KO TH17 cells negatively correlated with glycolytic genes, suggesting a potential metabolic alteration during TH17 differentiation (Fig. 3D and fig. S7A). To confirm, we performed qPCR for the genes related to glycolysis in WT and KO TH17 cells and found that SPTLC1 deficiency reduced the expression of glycolytic genes (Fig. 3E). To analyze glycolysis in real time, we measured extracellular acidification rate (ECAR) in equal numbers of viable WT and KO TH17 cells by an extracellular flux analyzer. SPTLC1 deficiency showed significantly reduced glycolysis after addition of glucose without altering the glycolytic capacity (Fig. 3F). In Cell Mito Stress Test (Agilent), basal oxygen consumption rate (OCR) and maximal respiration did not differ in SPTLC1-deficient cells compared to WT (Fig. 3G). To assess the detailed bioenergetic state of these cells, we calculated rates of adenosine triphosphate (ATP) production by glycolysis, JATPglyc, and oxidative metabolism, JATPox, using methods developed by Brand and Mookerjee that do not use exogenous supplementation of substrates under basal conditions and use only glucose as a substrate for assessing ATP production (60). For this, OCR and ECAR were measured simultaneously in TH17 cells under the basal condition, followed by sequential addition of glucose, oligomycin, carbonyl cyanide-p-trifluoromethoxyphenylhydrazone (FCCP), and rotenone plus antimycin A (fig. S7B). ATP production from glycolysis and OXPHOS in basal and all injections is presented in stacked column graph (fig. S7C). Under the basal conditions, cells are assumed to produce ATP mainly by OXPHOS from glycogen. This process is compromised in SPTLC1-deficient TH17 cells (fig. S7D). When glucose is added in WT TH17, JATPglyc increased, and JATPox decreased (fig. S7, E and D). Under this condition, SPTLC1-deficient TH17 cells showed reduced JATPglyc compared to WT cells; however, JATPox increased more than in WT cells (fig. S7, E and D). Further, SPTLC1-deficient TH17 cells continued to show reduced ATP production by glycolysis during subsequent addition of oligomycin (which inhibited the mitochondrial ATP synthesis) and FCCP (under maximal respiration) (fig. S7E). However, FCCP addition increased the ATP production via OXPHOS in SPTLC1-deficient cells (fig. S7D). The total ATP production did not significantly differ between WT and SPTLC1-deficient TH17 cells in glucose and FCCP addition (fig. S7F). Our data suggest that while SPTLC1 deficiency impairs glycolysis in TH17 cells, and mitochondria retain the ability to sustain ATP production through OXPHOS when supplemented with exogenous substrate, they have defective OXPHOS under basal conditions.
SPTLC1 deficiency reduces c-Myc, HIF-1α, and mTOR signaling
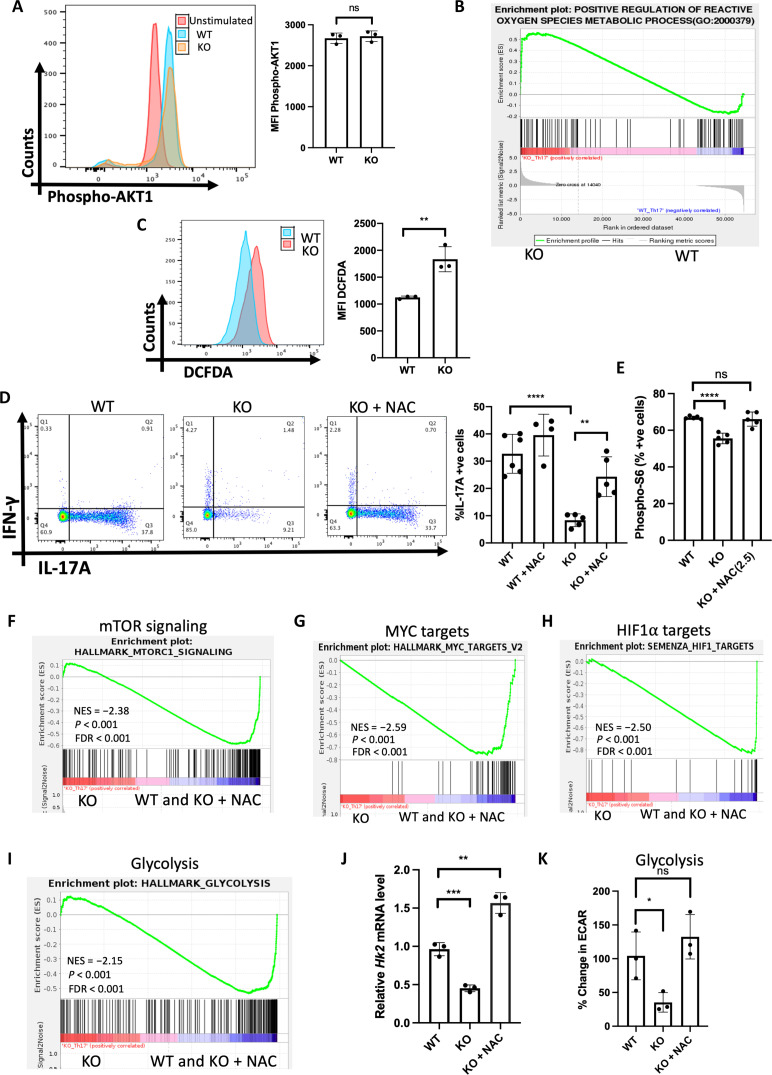
Metabolic rewiring in activated T cells require coordination between different signaling pathways and transcription factors (19). In TH17 cells, phosphatidylinositol 3-kinase (PI3K)–Akt–mediated mTORC1 signaling integrates the metabolic rewiring toward glycolysis by regulating c-Myc and HIF-1α–mediated glycolytic gene expression (10, 15, 18, 20). To understand why SPTLC1 initiated sphingolipid flux is integral to TH17 cell development, we explored the relationship between SPTLC1 and the three key metabolic regulators during TH17 differentiation. We checked mTORC1 activity in WT and SPTLC1-deficient CD4+ T cells under TH17 conditions by measuring phosphorylation of S6 subunit (S6) and 4E-BP1(p4E-BP1), two well-established downstream targets of mTORC1. SPTLC1-deficient TH17 cells showed significant reduction in the phosphorylation of the two proteins compared to control cells (Fig. 4, A and B). In concordance with these observations, the GSEA of RNA-seq data showed negative enrichment of mTORC1 pathway genes in the mutant (Fig. 4C and fig. S8A). In differentiating TH17 cells, mTORC1 regulates glycolysis through c-Myc and HIF-1α. Therefore, we measured the mRNA expression levels of Myc and Hif1a and found that compared to the WT, SPTLC1-deficient TH17 cells showed reduced expression of both Myc and Hif1a (Fig. 4, D and F). Correspondingly, protein levels of Myc and HIF-1α were also reduced in SPTLC1-deficient cells (Fig. 4, E and G). Furthermore, RNA-seq analysis revealed that SPTLC1 deficiency impaired the expression of c-Myc and HIF-1α target genes in TH17 cells (Fig. 4, H and I, and fig. S8, B and C). These data indicate the importance of SPTLC1 in regulating mTORC1 activity and metabolic rewiring in developing TH17 cells.
Fig. 4. SPTLC1 deficiency reduces c-Myc, HIF-1α, and mTORC1 signaling.
(A) Naïve CD4+T cells from WT and KO were differentiated under TH17 conditions for 12 hours and scored for frequency of phospho-S6+ cells by flow cytometry. The left panel is a representative flow figure, and the right panel shows the cumulative percent pS6-positive cells. n = 5 biologically independent samples. (B) Immunoblot of p-4E-BP1 in WT and KO naïve CD4+ T cells differentiated for 12 hours under TH17 polarizing conditions. The left panel shows representative Western blot, and the right panel is the quantitative data for the same. n = 5 biologically independent samples from two independent experiments. (C) GSEA for mTORC1 signaling genes between WT and KO TH17 cells. (D) WT and KO naïve T cells were differentiated into TH17 cells for 3 days and Hif1a mRNA quantified by qPCR. n = 5 biologically independent samples. (E) Immunoblot of HIF-1α in WT and KO naïve CD4+ T cells differentiated for 3 days under TH17 polarizing conditions. The left panel shows representative Western blot, and the right panel is the quantitative data for the same. n = 3 biologically independent samples. (F) Myc mRNA quantified by qPCR in cells differentiated as in (D). n = 5 biologically independent samples. (G) Immunoblot of c-Myc in WT and KO naïve CD4+ T cells differentiated for 12 hours under TH17 polarizing conditions. The left panel shows representative Western blot, and the right panel is the quantitative data for the same. n = 5 biologically independent samples from two independent experiments. (H and I) GSEA for hallmark gene sets (H) c-Myc target genes and (I) HIF-1α target genes between WT and KO TH17 cells. Each dot represents an individual mouse. All data presented as means ± SEM: **P < 0.01; ***P < 0.001; ****P < 0.0001; ns, not significant.
SPT enzyme product 3-KDS rescues the defects in SPTLC1-deficient TH17 cells
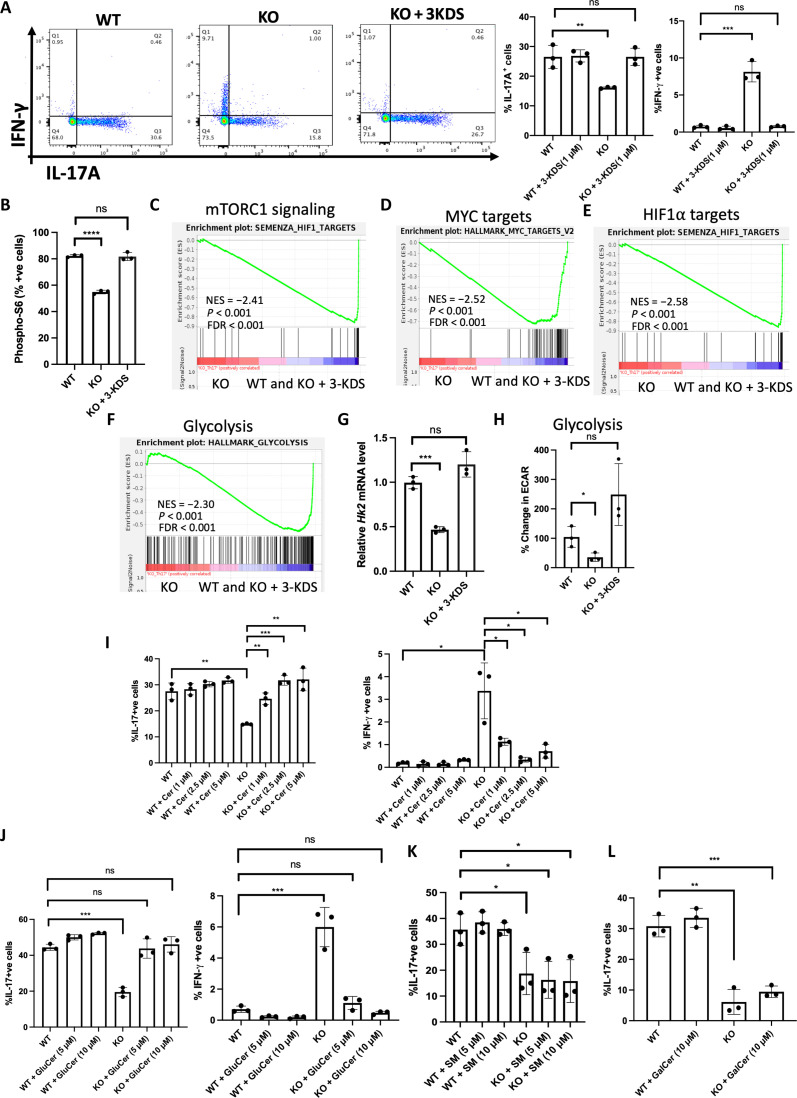
SPTLC1 is a component of the catalytic core of SPT enzyme complex whose activity is required for maintaining cellular levels of sphingolipids. To test whether SPT enzymatic activity is necessary for TH17 differentiation and to examine the specificity of the de novo sphingolipid biosynthetic pathway defect in the observed phenotype, we performed rescue experiments with 3-KDS, the product of SPT reaction. For this, naïve CD4+ T cells were pretreated with 3-KDS and then differentiated under TH17 conditions. The addition of 3-KDS rescued all observed defects of SPTLC1-deficient TH17 cells. 3-KDS increased the frequency of IL-17A+ cells and reduced the frequency of IFN-γ + cells to the WT levels (Fig. 5A). 3-KDS also rescued the mTORC1 signaling defect in SPTLC1-deficient TH17 cells. 3-KDS restored the phospho-S6–positive cells to the WT level (Fig. 5B) and enriched mTORC1 signaling genes (Fig. 5C and fig. S9A). Expression of c-Myc and HIF-1α target genes were also restored back to WT levels in SPTLC1-deficient cells in the presence of 3-KDS (Fig. 5, D and E, and fig. S9, B and C). Further, 3-KDS rescued the defect in glycolysis in SPTLC1-deficient TH17 cells. 3-KDS supplementation up-regulated the glycolysis-related genes in SPTLC1-deficient TH17 cells as evidenced by GSEA (Fig. 5F and fig. S9D), which is further confirmed by qPCR analysis of Hk2 mRNA (Fig. 5G). 3-KDS also significantly restored ECAR, a measure of glycolysis (Fig. 5H). De novo sphingolipid biosynthesis pathway produces various biologically important sphingolipid species. 3-KDS, formed in the first reaction by SPT enzyme, in subsequent reactions is converted to ceramide, which serves as a building block for the synthesis of complex sphingolipids like glucosylceramide, galactosylceramide, and sphingomyelin. To investigate which of these downstream products could influence TH17 differentiation, we preincubated SPTLC1-deficient naïve CD4+ T cells with ceramide or glucosylceramide or galactosylceramide or sphingomyelin and then differentiated them under TH17 conditions. Supplementation of ceramide and glucosylceramide rescued the defects in the cytokine production in SPTLC1-deficient TH17 cells (Fig. 5, I and J). However, supplementation of sphingomyelin or galactosylceramide did not rescue the defect (Fig. 5, K and L), indicating the specific requirement of glucosylceramide for TH17 differentiation. Together, these data suggest a critical role for sphingolipid synthesis via the de novo synthetic pathway for TH17 differentiation and implies that the physiological regenerative flux of sphingolipids via salvage pathway in the absence of de novo pathway is not sufficient for TH17 differentiation.
Fig. 5. SPT enzyme product 3-KDS rescues the defects in SPTLC1-deficient TH17 cells.
(A) WT and KO naïve cells were differentiated under TH17 conditions with or without 3-KDS (1 μM) for 4 days and scored for frequency of IL-17A+ and IFN-γ+ cells by flow cytometry. The left panel is a representative flow figure, and the right panel shows the cumulative data of the same. n = 3 biologically independent samples. (B) WT and KO naïve cells were differentiated under TH17 conditions for 12 hours with or without 3-KDS and scored for frequency of phospho-S6+ cells by flow cytometry. n = 3 biologically independent samples. (C to F) GSEA for (C) mTOR signaling genes, (D) c-Myc target genes, (E) HIF-1α target genes, and (F) glycolytic genes between KO and (WT and KO + 3-KDS) TH17 cells. (G) WT and KO naïve cells were differentiated under TH17 conditions with or without 3-KDS (1 μM) for 3 days and Hk2 mRNA quantified by qPCR. n = 3 biologically independent samples. (H) WT and KO naïve T cells were differentiated under TH17 conditions with or without 3-KDS for 3 days and ECAR measured in equal number of viable cells by Seahorse analyzer. n = 3 biologically independent samples. (I to L) WT and KO naïve cells were differentiated under TH17 conditions for 4 days with or without indicated concentrations of (I) C6 ceramide (Cer), (J) glucosylceramide (GluCer), (K) sphingomyelin (SM) (L), and galactosylceramide (GalCer), and reactivated with phorbol 12-myristate 13-acetate/ionomycin for 6 hours and scored for frequency of IL-17A+ and IFN-γ+ cells by flow cytometry. (I and J) Left: %IL-17A+ cells; right: % IFN-γ+ cells. n = 3 biologically independent samples. Each dot represents an individual mouse. All data presented as means ± SEM: *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001; ns, not significant.
SPTLC1 deficiency impairs TH17 differentiation by increasing intracellular ROS
In differentiating T cells, mTORC1 activity is regulated by PI3K–protein kinase B (AKT) (PI3K-AKT) signaling. Since mTORC1 activity is reduced in SPTLC1-deficient TH17 cells, we hypothesized that PI3K-AKT signaling might be compromised in these cells. We examined the phosphorylation status of AKT in differentiating TH17 cells as an index of PI3K-AKT signaling and found no difference between WT and KO cells (Fig. 6A). These data indicate that PI3K-AKT signaling is not compromised in these cells. Adenosine monophosphate (AMP)–activated protein kinase (AMPK), a negative regulator of mTOR activity, was unexpectedly decreased in the mutant cells indicating that AMPK activity was not responsible for reduced mTOR activity (fig. S10A). RNA-seq data showed increased expression of genes that positively regulate reactive ROS in SPTLC1-deficient TH17 cells, suggesting a possibility of oxidative stress in these cells (Fig. 6B and fig. S10B). Since ROS can modulate mTOR activity, we measured the intracellular ROS in WT and SPTLC1-deficient differentiating TH17 cells using 2′,7′-dichlorofluorescin diacetate (DCFDA), which measures total intracellular ROS. SPTLC1-deficient TH17 cells showed significantly increased intracellular ROS validating the RNA-seq data (Fig. 6C). To ascertain whether ROS was involved in the observed phenotype, we evaluated the benefits of an antioxidant in SPTLC1-deficient TH17 cells. We differentiated naive WT and SPTLC1-deficient cells into TH17 cells in the presence of antioxidant NAC. NAC rescued the defect in cytokine production in SPTLC1-deficient TH17 cells. NAC restored the frequency of IL-17A+ cells and reduced the frequency of IFN-γ+ cells (Fig. 6D and fig. S10C). NAC also corrected the defect in mTORC1 activity in the SPTLC1-deficient TH17 cells (Fig. 6, E and F, and fig. S11A) and restored the c-Myc and HIF-1α target gene levels (Fig. 6, G and H, and fig. S11, B and C). NAC improved glycolysis defects in SPTLC1-deficient TH17 cells. NAC supplementation up-regulated the glycolysis-related genes in SPTLC1-deficient TH17 cells as evidenced by GSEA (Fig. 6I and fig. S11D), which is further confirmed by qPCR analysis of Hk2 mRNA (Fig. 6J). Its treatment significantly restored ECAR, a measure of glycolysis (Fig. 6K). NAC also significantly reduced ROS production in SPTLC1-deficient TH17 cells (fig. S12A). Our data suggest that SPTLC1 deficiency increases intracellular ROS that disrupts the mTOR activity and its downstream targets.
Fig. 6. SPTLC1 deficiency impairs TH17 differentiation by increasing intracellular ROS.
(A) WT and KO naïve cells were differentiated under TH17 conditions for 12 hours and scored for phospho-Akt(S473) by flow cytometry. The left panel is a representative flow figure, and the right panel shows cumulative p-Akt MFI. n = 3 biologically independent samples. (B) GSEA for positive regulation of ROS genes between WT and KO TH17 cells. (C) WT and KO naïve cells were differentiated under TH17 conditions for 12 hours and intracellular ROS measured using DCFDA by flow cytometry. The left panel is a representative flow figure, and the right panel shows the cumulative data of DCFDA MFI. n = 3 biologically independent samples. (D) WT and KO naïve cells were differentiated under TH17 conditions for 4 days with or without NAC and scored for intracellular cytokine. The left panel is a representative flow figure, and the right panel shows the cumulative data of the same. n = 4 to 6 biologically independent samples. (E) WT and KO naïve cells were differentiated under TH17 conditions with or without NAC for 12 hours and scored for frequency of phospho-S6+ cells by flow cytometry. n = 5 biologically independent samples. (F to I) GSEA for (F) mTOR signaling genes, (G) c-Myc target genes, (H) HIF-1α target genes, and (I) glycolytic genes KO and (WT and KO + NAC) TH17 cells. n = 3 biologically independent samples. (J) Hk2 mRNA quantification by qPCR of TH17 under the indicated differentiation conditions. n = 3 biologically independent samples. (K) WT and KO naïve T cells were differentiated under TH17 conditions with or without NAC for 3 days and ECAR was measured in equal number of viable cells by Seahorse analyzer. n = 3 biologically independent samples. Each dot represents an individual mouse. All data presented as means ± SEM: *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001; ns, not significant.
SPTLC1 deficiency increases intracellular ROS by enhancing NOX activity
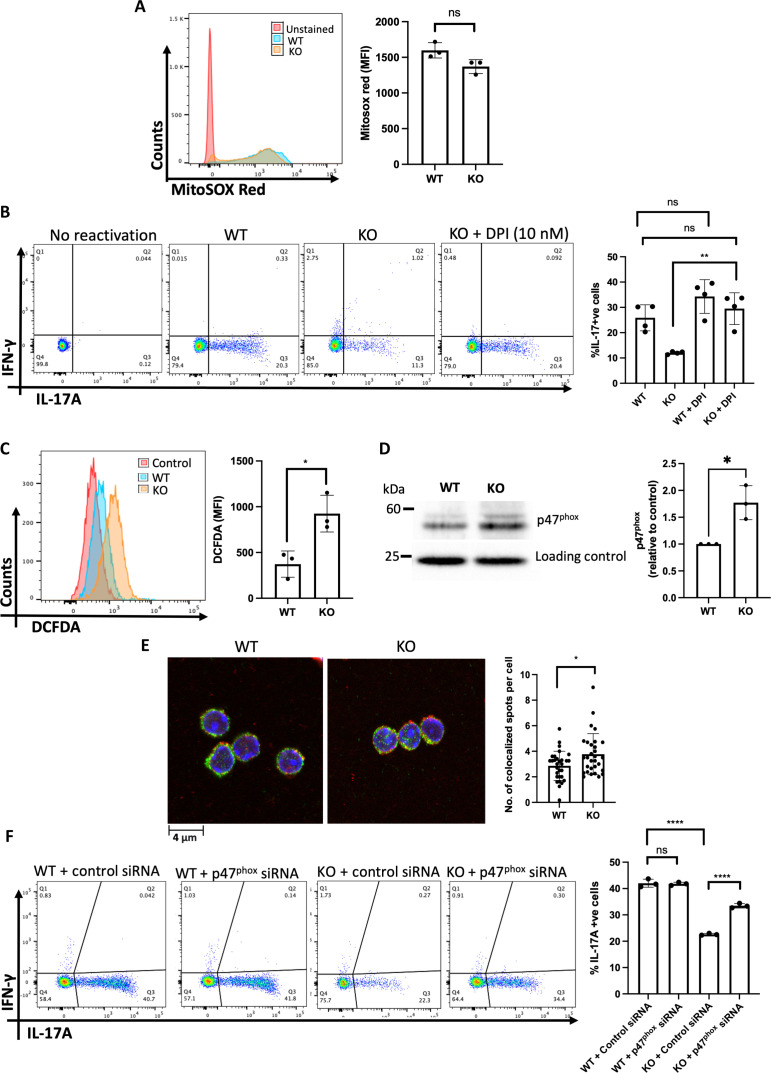
In T cells, intracellular ROS are mainly generated by mitochondria and NOX (61–64). Therefore, we next investigated the source of increased ROS in SPTLC1-deficient T cells. We measured mitochondrial ROS in WT and SPTLC1-deficient TH17 cells using MitoSOX red. There was no significant difference in mitochondrial ROS between WT and SPTLC1-deficient T cells (Fig. 7A). Next, we investigated the involvement of NOX using DPI, a NOX inhibitor. DPI preincubation, like NAC, corrected the defect in cytokine production in SPTLC1-deficient TH17 cells (Fig. 7B). DPI also significantly reduced ROS production in SPTLC1-deficient TH17 cells (fig. S12B). In T cells, TCR cross-linking produces intracellular ROS within minutes by activation of membrane NOX2 (63, 65). To check whether SPTLC1 influences NOX2-mediated ROS production, we measured total intracellular ROS using DCFDA in WT and KO in naïve CD4+ T cells 15 min after activation with anti-CD3. SPTLC1-deficient CD4+ T cells showed significantly increased ROS production compared to WT (Fig. 7C). Preincubation with DPI and NAC also significantly reduced the ROS production at this instance (fig. S12, C and D). The de novo sphingolipid biosynthetic pathway consumes NADPH at two steps while synthesizing ceramide (31, 66). Therefore, it is reasonable to posit that SPTLC1 deficiency could increase the NADPH levels and fuel the NOX resulting in increased ROS. To consider this possibility, we measured the cellular NADPH levels in TH17 cells and found no significant difference between WT and SPTLC1-deficient TH17 cells, thus ruling out elevated NADPH levels driving increased NOX activity (fig. S12E). NOX2 has membrane (gp91phox and p22phox) and cytosolic components (p47phox, p67phox, and p40phox). TCR cross-linking activates NOX2 by recruiting the cytosolic components to the membrane (67). Therefore, we measured the translocation of cytosolic p47phox to membrane by Western blot analysis and colocalization of p47phox and gp91phox as an index of NOX2 activity. We activated the naïve CD4+ T cells from WT and KO mice with anti-CD3 for 15 min; membrane fractions were isolated by ultracentrifugation and probed for p47phox. Compared to WT, KO cells showed increased levels of p47phox in the membrane fraction (Fig. 7D). In addition, we measured the colocalization of p47phox and gp91phox by confocal microscopy. Our analysis of confocal images showed increased colocalization p47phox and gp91phox in SPTLC1-deficient cells (Fig. 7E). However, we found that the total levels of p47phox did not differ between WT and SPTLC1-deficient naive CD4+ T cells after CD3 activation (fig. S12F). Last, to probe the involvement of NOX2 activity in the observed defect in SPTLC1-deficient TH17 cells, we knocked down p47phox, a component of NOX2, in naïve CD4+ T cells and differentiated them to TH17 cells. Knockdown of p47phox was confirmed by qPCR (fig. S12G). Knocking down p47phox significantly rescued the frequency of IL-17A+cells in SPTLC1 KO mutants. However, knocking down p47phox in WT cells did not alter the frequency of IL-17A+ cells compared to control siRNA-treated WT cells (Fig. 7F). These results together indicate higher levels of NOX2 activity in Sptlc1-deficient CD4+ T cells emphasizing the importance of Sptlc1 in maintaining appropriate levels of sphingolipids that modulate subcellular localization of protein complexes such as NOX2 in differentiating CD4+ T cells. An imbalance in NOX2 activity in Sptlc1 KO cells results in increased ROS production that compromises mTOR, HIF-1α, and c-Myc signaling pathways that ultimately results in defective TH17 differentiation.
Fig. 7. SPTLC1 deficiency increases intracellular ROS by enhancing NOX activity.
(A) WT and KO naïve CD4+ T cells were differentiated under TH17 conditions for 12 hours and mitochondrial ROS measured using Mitosox red. The left panel is a representative histogram, and the right panel shows the cumulative data (MFI) from n = 3 biologically independent samples. (B) WT and KO naïve CD4+ T cells were differentiated under TH17 conditions for 4 days with or without DPI (10 nM) and scored for intracellular cytokines. The left panel is a representative flow figure, and the right panel shows the cumulative data from n = 4 biologically independent samples. (C) WT and KO naïve CD4+ T cells cross-linked with anti-CD3 for 15 min with DCFDA and read in flow cytometry. The left panel is a representative histogram, and the right panel shows the cumulative data (MFI) from n = 3 biologically independent samples. (D) WT and KO naïve CD4+ T cells cross-linked as in (C) and membrane fraction immunoblotted for p47 phox. Each lane is a pooled sample from four to eight mice. The left panel shows representative blot, and the right panel is the quantitative data from three independent experiments. (E) CD4+ T cells cross-linked as in (C) and stained for gp91phox (green) and p47 phox (red) and analyzed using confocal microscopy. The left panel shows the representative confocal picture, and the right panel shows the number of colocalization spots per cell. n = 3 biologically independent samples. (F) WT and KO Naïve CD4+ T cells were nucleofected with control or p47phox siRNA, differentiated into TH17 cells, and scored for intracellular cytokines. The left panel is a representative flow figure, and the right panel shows the cumulative data. Each dot represents cells pooled from three mice. n = 3 independent experiment. All data presented as means ± SEM: *P < 0.05; **P < 0.01; ****P < 0.0001; ns, not significant.
DISCUSSION
Aberrant TH17 cells are associated with many autoimmune and inflammatory disorders and curbing their development or function remains an attractive therapeutic mode (3, 54, 68–70). In this study, we demonstrated that SPTLC1 deficiency impaired TH17 cell differentiation and attenuated EAE and colitis.
Activated T cells enhance glycolysis. Increasing evidence shows the requirement of glycolysis for TH17 differentiation (10, 12, 21). Here, we report an essential function for SPTLC1 in metabolic rewiring of differentiating TH17 cells. SPTLC1 deficiency impaired glycolysis. TCR activation along with CD28 costimulation activates the metabolic sensor mTORC1, which, in coordination with HIF-1α and c-Myc, rewires the metabolism to glycolysis (19). Growing evidence shows that perturbation of this axis impairs TH17 development and that different metabolites/metabolic pathways regulate this metabolic shift by modulating several key players of this pathway. Deletion of one-carbon metabolism enzyme methylenetetrahydrofolate dehydrogenase 2 suppresses mTORC1 signaling and alters histone methylation, impairing TH17 differentiation (29). Further, short-chain fatty acids are shown to activate mTOR (71). In this study, we show that SPTLC1 deficiency impaired mTORC1 activity and reduced the expression of its downstream targets, HIF-1α, c-Myc, and glycolytic genes. Further, supplementation of an SPT enzyme product, 3-KDS, rescued the mTORC1 activity, indicating a specific role for the de novo sphingolipid biosynthetic pathway in maintaining the mTORC1 activity during TH17 differentiation. Our result contrasted with a recent study where they have shown that specific deletion of SPTLC2, another major subunit of SPT enzyme, in T cells prolonged the activation of mTORC1 in CD8+ T cells (42). The observed differences could be attributed to the cell types and the specific conditions, where sphingolipid flux through the de novo pathway could regulate mTORC1 signaling differently.
Further, our study shows that SPTLC1 is essential for maintaining the redox balance in activated T cells, and its deficiency increases intracellular ROS. The absence of SPTLC1 exaggerates the ROS production via NOX. ROS once considered only as harmful metabolic by-products are now appreciated to play essential roles as second messengers in T cell biology (61–63, 72, 73). Recent evidence documented ROS as a negative regulator of TH17 differentiation (72, 74, 75). However, the multiple sclerosis drug dimethyl fumarate (DMF) can activate the transcription factor nuclear factor (erythroid-derived 2)–like 2 (Nrf2) and is believed to act as an antioxidant in T and other cells. Their precise mechanism of action in MS and T cells is still emerging (76, 77). In addition, in mice, DMF’s immune regulatory function is independent of NRF2 expression as it can modulate both innate and adaptive immune system in Nrf2 KO animals (78). Here, we show that the NAC, a ROS scavenger, reversed all the defects in SPTLC1-deficient TH17 cells. NAC corrected the defects in cytokine production, glycolysis, and mTORC1 activity. ROS can inhibit or activate mTOR signaling depending on cell types and conditions (79). Restoration of mTORC1 activity by NAC to WT levels places ROS upstream of mTORC1 and indicates that it inhibits mTORC1 activity in SPTLC1-deficient TH17 cells. In activated T cells, ROS is mainly produced by mitochondria and NOX, and both are implicated in T cell function (61, 65). SPTLC1 deficiency did not increase mitochondrial ROS as detected by Mitosox red. NOX2 is the main isoform expressed in mouse T cells, and TCR activation produces ROS via NOX2 within minutes (63). We detected increased ROS in SPTLC1-deficient CD4+ T cells as early as 15 min after TCR cross-linking and increased colocalization of p47phox and gp91phox. Further, DPI reversed the cytokine defects in SPTLC1-deficient TH17 cells, indicating NOX2 as a possible source of increased ROS in SPTLC1-deficient TH17 cells. In line with our data, a recent study in neuroblastoma cells showed that exogenous addition of glucosylceramide or overexpression of glucosylceramide synthase impaired NOX2-mediated ROS production and conferred chemoresistance. Glucosylceramide modulates membrane environment by inducing positive curvature that could affect NOX by influencing subunit-subunit and subunit-membrane interactions (80). Thus, our study highlights the importance of sphingolipid flux through de novo pathway in regulating NOX2 activity in T cells. It will be further interesting to study how various sphingolipids produced via de novo sphingolipid pathway could influence NOX assembly and, subsequently, ROS production.
In vitro culturing of SPTLC1-deficient naïve CD4+ T cells under TH17 conditions showed a defect in IL-17A production but showed increased IFN-γ and FOXP3 production. Further, SPTLC1 deficiency enhanced TH1 differentiation by increasing IFN-γ production and enhanced iTreg cells by increasing FOXP3 production. In the EAE model, SPTLC1 cKO mice showed delayed onset and reduced clinical symptoms of EAE compared to WT mice. This is accompanied by reduced IL-17A–positive cells in the CNS and spleen. Further, IFN-γ+ cells were also reduced in this condition, which was different from our in vitro culture where we observed an increase in IFN-γ+ cells under TH1 and TH17 differentiating conditions. This difference could be due to the difference in the activation and costimulation signals under in vivo conditions. When cells for adoptive transfer were differentiated using splenocytes (from MOG peptide primed mice) with MOG peptide and TH17 polarizing cytokines (as opposed to plate-bound anti-CD3 and -CD28 activation for in vitro experiments), CD4+ T cells from KO mice showed reduced IL-17A+ and IFN-γ+ cells as seen in active EAE. Further, we also noted no change in FOXP3+ Treg cells in the SPTLC cKO mice after EAE induction, indicating that specific deletion of SPTLC1 in T cells selectively impaired pathogenic TH17 and TH1 cells’ differentiation in vivo.
Sphingolipids are an important class of lipids having structural and functional roles in eukaryotic cells. Growing evidence indicates a definitive role for sphingolipid metabolism in T cell function. Loss of SPTLC2 impairs CD8+ T cell responses to infection, and age-dependent changes in sphingolipid composition alter CD4+ T cell function (39, 42). Further, the role of sphingolipid metabolism in T helper differentiation and function is still emerging. A recent report on a genome-scale metabolic modeling of human CD4+ T cells has documented a role for the SPT complex in TH17 cell cytokine production; however, the molecular details were undefined (40). Another study showed the importance of ceramide accumulation in Treg cell function (81). Consistently, the expression of sphingomyelin synthase 1, which converts ceramide into sphingomyelin, was found to be reduced in Treg cells (81). They showed that ceramide accumulation inhibited mTORC1 activity by activating protein phosphatase 2. However, another study showed the opposing effect where the pharmacological inhibition of acid sphingomyelinase or its genetic deficiency increased Treg cells in mice (82), indicating that the source of ceramide could have a different effect on cellular function. Sphingosine-1-phosphate (S1P) is shown to induce TH17 cells (83, 84), and reducing S1P ameliorates TH17-mediated alcoholic steatohepatitis in mice (85). In line with this, another study has shown that genetic ablation of sphingosine kinases affected expression of IL-17 in human T lymphocytes (86). Our study gains importance since we demonstrate the existence of a protein interaction network that connects homeostatic sphingolipid flux to glucose metabolism in differentiating TH17 cells.
MATERIALS AND METHODS
Mice and reagents
All animal studies were performed according to National Institutes of Health (NIH) guidelines for the use and care of live animals and were approved by the Animal Care and Use Committees of the National Cancer Institute (NCI) under the protocol 20-073. C57BL/6 CD4-Cre transgenic, RAG−/− mice and Ifngr KO (JAX:003288) mice were obtained from the Jackson Laboratory and bred in our institute animal facility under specific pathogen–free conditions. Sptlc1foxl/flox mice were generated in our laboratory as previously described (34). Sptlc1foxl/floxCD4-Cre (KO) conditional KO mice were generated by crossing Sptlc1floxl/flox mice to CD4-Cre transgenic mice. Sptlc1foxl/flox or Sptlc1+/+CD4-Cre were used as control mice (WT). All mice used for experiments were aged 8 to 12 weeks, and both female and male mice were used for experiments. Primers for qPCR were obtained from IDT (Integrated DNA Technologies, USA). Cytokines mIL-4, mIL-12, hTGF-β1, IL-7, and IL-1β were purchased from PeproTech, USA; hIL-2 was obtained from R&D Systems; and IL-6 and IL-23 were purchased from BioLegend, USA. Antibodies for activation and cytokine neutralization were procured from Bio X Cell, USA. All chemicals were purchased from Sigma-Aldrich unless otherwise mentioned.
Mouse cell isolation and in vitro differentiation of T helper cell subsets
Naïve CD4+ T cells were isolated from spleen and peripheral lymph nodes of 8- to 12-week-old mice by negative selection, using the Mojosort mouse CD4+ naïve T cell isolation kit (BioLegend, catalog no. 480006), according to the manufacturer’s protocol. Cell purity was assessed by fluorescence-activated cell sorting (FACS) and was consistently higher than 95%. Isolated cells were cultured in RPMI 1640 supplemented with 10% fetal bovine serum (FBS), penicillin (100 U/ml), streptomycin (100 mg/ml), and 50 mM 2-mercaptoethanol referred hereafter as complete medium (CM). For TH1 and TH2 differentiation, naïve CD4+ T cells at 1 million cells/ml concentration were activated by plate-bound anti-CD3 (1 μg/ml, Bio X Cell, catalog no. BP0001-1) and anti-CD28 (2 μg/ml, Bio X Cell, catalog no. BE0015-1) for 4 days along with IL-12 (10 ng/ml, PeproTech, catalog no. 210-12) and anti–IL-4 (10 μg/ml, Bio X Cell, catalog no. BP0045) for the TH1 condition, and IL-4 (10 ng/ml, PeproTech, catalog no. 214-14) and anti–IFN-γ (10 μg/ml, Bio X Cell, catalog no. BP0055) for the TH2 condition. For TH17 differentiation, naïve CD4+ T cells at 1 million cells/ml concentration were activated by plate-bound anti-CD3 (5 μg/ml) and soluble anti-CD28 (2 μg/ml) for 4 days along with hTGF-β1 (5 ng/ml, PeproTech, catalog no. 100-21C), IL-6 (40 ng/ml, BioLegend, catalog no. 575704), anti–IL-4 (10 μg/ml), and anti–IFN-γ (10 μg/ml). For pathogenetic TH17 differentiation, the naïve CD4+ T cells were activated with IL-1β (20 ng/ml, PeproTech, catalog no. 211-11B), IL-23 (20 ng/ml, BioLegend, catalog no. 589002), IL-6, anti–IL-4 (10 μg/ml), and anti–IFN-γ (10 μg/ml). For chemical treatments, naïve CD4+ T cells were preincubated with chemicals for 60 to 120 min before activation and differentiation. For myriocin treatment, naïve CD4+ T cells were incubated with myriocin (5 μM) for 12 hours and differentiated under TH17 conditions for 4 days.
Immunoblotting
Differentiated cells were collected at different time points and washed once with ice-cold phosphate-buffered saline (PBS) and lysed using radioimmunoprecipitation assay (RIPA) lysis buffer, containing protease and phosphatase inhibitors (Thermo Fisher Scientific). Proteins were quantified by the Bradford method (Bio-Rad reagent), and equal amounts of protein were fractionated by 12% SDS–polyacrylamide gel electrophoresis, electro-transferred onto polyvinylidene difluoride membranes, blocked with 5% bovine serum albumin (BSA) in tris-buffered saline supplemented with Tween 20, and incubated with primary antibodies overnight at 4°C. Primary antibodies, HIF-1α, phospho-4E-BP1 (Thr37/46), c-Myc, BiP, PERK, and AMPKα, p-AMPKα1(S485)/AMPKα2(S491) (all from Cell Signaling Technology, CST) were used at 1:1000 dilution. Membranes were washed and incubated with horseradish peroxidase–conjugated secondary antibodies (1:10,000, CST) for 1 hour at room temperature. The bands were detected using SuperSignal West Pico PLUS Chemiluminescent Substrate (Thermo Fisher Scientific). Membranes were stripped by Restore Western blot stripping buffer (Thermo Fisher Scientific) and reprobed for total proteins or β-actin (Abcam or Proteintech). The intensity of the bands was quantitated using Fiji ImageJ software. The intensity of each band normalized to the corresponding loading control.
For detecting membrane p47phox, 20 million WT or KO naïve cells were cross-linked with anti-CD3 for 15 min as described elsewhere and subjected to membrane fractionation. Cells were washed with ice-cold PBS and lysed with 1 ml of hypotonic lysis buffer [10 mM tris (pH 7.4), 1 mM EDTA, 25 mM NaF, and 1 mM dithiothreitol with protease and phosphatase inhibitors]. After 10 min of incubation on ice, the cells were homogenized using dounce homogenizer on ice (30 strokes) and then centrifuged for 10 min at 400g, 4°C to pellet nuclei and unbroken cells. Resulting supernatants were loaded to ultracentrifuge tubes and centrifuged at 40,000 rpm for 30 min at 4°C. Pellets were washed twice with ice-cold PBS and resuspended in RIPA lysis buffer containing protease and phosphatase inhibitors and subjected to Western blot analysis using monoclonal antibody to p47phox (Santa Cruz Biotechnology, sc-17845, clone D-10). Nonspecific band around 25 kDa was used as loading control for quantification. Three independent experiments were performed, and in each experiment, fold change was calculated relative to WT.
Real-time qPCR
Total RNA was extracted from in vitro differentiated T helper subsets using TRIzol reagent (Ambion catalog no. 15596026), and DNA was removed with amplification grade deoxyribonuclease (DNase) I (Sigma-Aldrich) before cDNA synthesis. First-strand cDNA synthesis was carried out with the Transcriptor First-Strand cDNA synthesis kit (Roche, catalog no. 04896866001) using Oligo (dT) primers. Reverse-transcribed cDNA was quantified by real-time PCR using iTag universal SYBR green supermix (Bio-Rad, catalog no. 172-5124) on Applied Biosystems QuantStudio 5 Real-Time PCR System. Actb was used as a reference gene to normalize for total RNA in each sample. Fold change was calculated by using the 2−ΔΔct method.
Oligonucleotides
The following primer pairs were used for analysis of gene expression by qPCR: Rorc, 5′-TGAGGCCATTCAGTATGTGG-3′(forward) and 5′-ACACCACCGTATTTGCCTTC-3′ (reverse); Il17f, 5′-CC-CCATGGGATTACAACATCAC-3′ (forward) and 5′-CATTGAT-GCAGCCTGAGTGTCT-3′ (reverse); Il23r, 5′-AACATGACATGC-ACCTGGAA-3′ (forward) and 5′-TCCATGCCTAGGGAATT-GAC-3′ (reverse); Ldha, 5′-CATTGTCAAGTACAGTCCACACT-3 (forward) and 5′-TTCCAATTACTCGGTTTTTGGGA-3′ (reverse); Pkm, 5′-GCCGCCTGGACATTGACTC-3′ (forward) 5′-CCATGAGAGAAATTCAGCCGAG-3 (reverse); Mct4, 5′-TCA-CGGGTTTCTCCTACGC-3′ (forward) and 5′-GCCAAAGCGGTT-CACACAC-3′ (reverse); Gpi, 5′-TCAAGCTGCGCGAACTTT-TTG-3′ (forward) and 5′-GGTTCTTGGAGTAGTCCACCAG-3′ (reverse); Tpi1, 5′-CCAGGAAGTTCTTCGTTGGGG-3′ (forward) and 5′-CAAAGTCGATGTAAGCGGTGG-3′ (reverse); Eno1, 5′-TGCGTCCACTGGCATCTAC-3′ (forward) and 5′-CAGAGCA-GGCGCAATAGTTTTA-3′ (reverse); Hif1a, 5′-ACCTTCATCGG-AAACTCCAAAG-3′ (forward) and 5′-CTGTTAGGCTGGG-AAAAGTTAGG-3′ (reverse); Myc, 5′-ATGCCCCTCAACGTGAA-CTTC-3′ (forward) and 5′-CGCAACATAGGATGGAGAGCA-3′ (reverse); Foxp3, 5′-GGTACACCCAGGAAAGACAGC-3′ (forward) and 5′-AAGACCTTCTCACAACCAGGC-3′ (reverse); Hk2, 5′-TGATCGCCTGCTTATTCACGG-3′ (forward) and 5′-AACCG-CCTAGAAATCTCCAGA-3′ (reverse); Glut1, 5′-CTCTGTCGGCC-TCTTTGTTAAT-3′ (forward) and 5′-CCAGTTTGGAGAAGCCC-ATAAG-3′ (reverse); Tbet, 5′-AGCAAGGACGGCGAATGTT-3′ (forward) and 5′-GGGTGGACATATAAGCGGTTC-3′ (reverse); Gata3, 5′-GAGGTGGTGTCTGCATTCCAA-3′ (forward) and 5′-TTTCACAGCACTAGAGACCCTGTTA-3′ (reverse); Sptlc1, 5′-ACGAGGCTCCAGCATACCAT-3′ (forward) 5′-TCAGAACG-CTCCTGCAACTTG-3′ (reverse); Actb, 5′-GGCTGTATTCCCCT-CCATCG-3′ (forward) and 5′-CCAGTTGGTAACAATGCCATG-T-3′ (reverse). All the primers were from IDT, USA.
Flow cytometry
For surface staining, cells were incubated with fluorescently labeled antibodies in FACS buffer (PBS with 2% FBS) for 30 min at 4°C in the dark. For detecting intracellular cytokines, 1 × 106 differentiated T helper cells or cells isolated from spleen, lymph nodes, small intestine, colon, spinal cord, or brain were reactivated with phorbol 12-myristate 13-acetate (PMA, 50 ng/ml) and ionomycin (1 μM) for 6 to 8 hours in the presence of brefeldin A (BFA) at 10 μg/ml concentration during the last 4 to 6 hours. After washing, the cells were stained with violet, fluorescent reactive dye in PBS for discriminating dead cells and fixed and permeabilized with Cytofix/Cytoperm fixation/permeabilization kit (BD, catalog no. 554714). Intracellular cytokine levels per cell were determined by iMFI. iMFI = (MFI)(P), where MFI is the median fluorescence intensity of cytokine-positive cells and P is the percentage of cytokine-positive cells. For staining p47phox, cells were fixed with BD Cytofix/Cytoperm fixation/permeabilization kit and stained with primary antibody for p47phox (600 ng, Santa Cruz Biotechnology, catalog no. sc-17845) followed by secondary antibody Alexa Flour goat anti-mouse immunoglobulin (IgG) (H + L) (1:1000; Invitrogen, catalog no. A32723). Transcription factor staining was performed using FOXP3-staining buffer set (eBioscience) according to the manufacturer’s instructions. Cells were incubated with conjugated antibodies for 1 hour at 4°C, washed twice in FACS buffer, and read using a flow cytometer.
For intracellular phosphoprotein staining, unstimulated or stimulated T helper cells were fixed with formaldehyde (final concentration of 1.5%) for 30 min at room temperature, permeabilized with ice-cold methanol, and stained with conjugated antibodies in staining media (PBS with 1% BSA) for 1 hour at room temperature. Data were acquired on a BD LSR Fortessa II or FACSCalibur and analyzed with FACSDiva or CellQuest Pro Software (BD, San Jose, CA, USA) or FlowJo 10.0.8 (Tree Star, Ashland, OR, USA). Antibody used for the flow cytometry are as follows: Alexa Flour 647 anti-mouse IL-17A (BD, catalog no. 560184); allophycocyanin (APC)–Cy7 anti-mouse IFN-γ (BD, catalog no. 561479); phycoerythrin (PE) anti-mouse IL-4 (BD, catalog no. 554435); PE anti–Hu/Mo phosphor-AKT1(Ser473, eBioscience, catalog no. 17-9715-42); Alexa Fluor 647 mouse anti-Stat5 (pY694, BD, catalog no. 612599); PE mouse anti-Stat3 (pY705, BD, catalog no. 612569); PE anti Hu/Mo phosphor-S6 (Ser235, Ser236, eBioscience, catalog no. 12-9007-42); PerCP-Cyanine5.5 anti-Mo/Rt FOXP3 (eBioscience, catalog no. 45-5773-82); BV786 Rat anti-mouse CD4 (BD, catalog no. 563331); PE anti-mouse CD8a (BioLegend, catalog no. 100708); PE anti-mouse CD4 (eBioscience, catalog no. 12-0043-82); APC-eFluor 780 (eBioscience, catalog no. 47-0691-82); fluorescein isothiocyanate (FITC) anti-Mo/Rt Ki-67 (eBioscience 12-5698-82); Percp-Cyanine5.5 anti-Mo CD62L (eBioscience, catalog no. 45-0621-82); PE mouse anti-Zap70 (Y319, BD, catalog no. 557881); PE anti-Mo CD119 (IFNGR1, eBioscience, catalog no. 12-1191-82); PE Armenian hamster IgG isotype (eBioscience, catalog no. 12-4888-81); FITC rat anti-mouse CD44 (BD, catalog no. 561859); Alexa Fluor 647 anti-Mo CD25 (eBioscience, catalog no. 51-0251-82); FITC anti-Mo CD3 (eBioscience, catalog no. 11-0032-82).
Seahorse metabolism assay
ECAR and OCR were measured using the Agilent Seahorse XFe24 bioanalyzer. The Seahorse XF glycolysis stress test kit (catalog no. 103020-100) or in-house chemicals were used to measure ECAR. Briefly, naïve CD4+ T cells were isolated from WT and KO and activated on αCD3/CD28-coated plates under TH17 differentiating conditions as described above for 3 days. Then, cells were harvested and washed in assay medium (XF RPMI 1640 with 2 mM glutamine), and 0.3 million cells in assay medium were spun onto a 24-well plate coated with Cell-Tak (BD Bioscience, catalog no. 354240). Then, ECAR was measured under basal conditions and in response to glucose (10 mM), oligomycin (1.0 μM), and 2-deoxyglucose (50 mM). ECAR values expressed as percentage change relative to WT (WT ECAR values defined as 100% after glucose or oligomycin addition for calculating glycolysis or glycolytic capacity, respectively). The Seahorse XF Cell Mito Stress Test Kit (catalog no. 103015-100) was used to measure the OCR according to the manufacturer’s instruction. A total of 250,000 cells were used per well. The OCR was measured under basal conditions and in response to 1 μM oligomycin, 1 μM FCCP, and 0.5 μM rotenone/antimycin A. Quantification of rate of intracellular ATP production from OXPHOS and glycolysis was estimated using methods developed by Mookerjee et al. (60). Briefly, cells were washed in Krebs-Ringer phosphate Hepes (KRPH) medium [2 mM Hepes, 136 mM NaCl, 2 mM NaH2PO4, 3.7 mM KCl, 1 mM MgCl2, 1.5 mM CaCl2, and 0.1% (w/v) fatty acid–free BSA (pH 7.4) at 37°C], and 0.3 million cells were seeded on a Cell-Tak–coated XFe24 Cell Culture microplate in 100 μl of KRPH buffer. Cells were centrifuged and incubated for 30 min at 37°C with atmospheric CO2. OCR and ECAR were measured under basal conditions and in response to 10 mM glucose (port A), 1 μM oligomycin (port B), 1 μM FCCP (port C), 1 μM rotenone, and 1 μM antimycin A (port D). Three measurement cycles of 1-min mix, 1-min wait, and 3-min measure were carried out. Calculations of JATP were performed using the spreadsheet provided in Mookerjee et al. (60).
Cytokine measurement by Luminex assays
For measuring secreted cytokines during recall response, 0.5 × 106 differentiated WT and KO viable TH17 cells were reactivated with plate-bound anti-CD3 (5 μg/ml) for 24 hours and supernatants were collected. Culture supernatants were collected and sent to Eve Technologies, Canada for measuring cytokine using Luminex technology.
ROS detection
For detecting total cellular ROS, naïve T cells were differentiated under TH17 conditions as mentioned above for 12 hours and incubated with 5 μM DCFDA for 30 min before the harvest. The reaction was terminated with ice-cold CM, washed with FACS buffer, and read using a flow cytometer. For detecting mitochondrial ROS, differentiating cells were incubated with 5 μM MitoSOX for 30 min before the harvest at 37°C in a cell culture incubator, washed, and read using a flow cytometer. For detecting anti-CD3–induced ROS generation, naïve T cells (1 × 106 cells/ml) were precoated with anti-CD3 (2C11) at 10 μg/ml for 30 min on ice, washed, and resuspended in ice-cold CM, and maintained on ice at 1 × 106 cells/0.1 ml. Cells were then transferred to tubes containing rabbit anti-hamster antibody (5 μg/ml; to cross-link 2C11) and DCFDA at 5 μM concentration in CM at 37°C and incubated for 15 min. The reaction was terminated by adding ice-cold FACS buffer, and the cells were washed before FACS measurement. In experiments using inhibitors, the cells were preincubated with drugs during the antibody incubation at 4°C and in the subsequent steps. To show the effect of NAC and DPI on ROS production in TH17 cells, naïve T cells were preincubated in NAC (5 mM) and DPI (10 nM) for 1 to 2 hours and differentiated under TH17 conditions for 3 days. Then, cells were harvested and CD3 cross-linked for 15 min in the presence of DCFDA, washed, and analyzed by flow cytometry.
RNA interference
Naive CD4+ T cells isolated from WT and SPTLC1-deficient mice were differentiated into TH17 cells in Accell siRNA delivery medium (Dharmacon, catalog no. B-005000-500) supplemented with 2.5% FBS. On day 1, cells were treated with SMARTPool Accell Mouse Ifngr1 siRNA (1 μm, Dharmacon, catalog no. E-043702-00-0005) or Accell nontargeting control pool (1 μm, Dharmacon, catalog no. D-001910-10-05). On day 5, cells were reactivated to analyze intracellular cytokines by flow cytometry.
Nucleofection
Naive CD4+ T cells (10 million) were transfected with 300 nM SMARTPool Accell mouse siRNA for p47phox (Ncf1, neutrophil cytosolic factor; Dharmacon, catalog no. E-057350-00-0010) or with 300 nM Accell nontargeting control pool using Amaxa P3 Primary Cell 4D-Nucleofector X Kit L (Lonza, catalog no. V4XP-3024) and program DN-100 (for mouse T cells) in a 4D-Nucleofector unit (Lonza) as per the manufacturer’s instructions. After nucleofection, the cells were transferred immediately to prewarmed CM and incubated for 24 hours at 37°C and differentiated into TH17 cells. Four days later, cells were reactivated to analyze intracellular cytokines by flow cytometry.
NADPH measurement
The intracellular NADPH levels in TH17 cells were determined by bioluminescent assay using the NADP/NADPH-Glo assay kit (Promega, catalog no. G9081) according to the manufacturer’s protocol. Briefly, naïve CD4+ T cells from WT and SPTLC1-deficient mice were differentiated under TH17 differentiating conditions for 4 days. A total of 0.5 million cells were used for the assay. The intensity of light detected by a luminometer is proportional to the amount of NADPH, and data were presented as relative light units.
Sphingolipid analysis
For analyzing sphingolipids in the T helper cell subset, naïve CD4+ T cells isolated from WT mice were differentiated under TH0, TH1, TH2, TH17, and iTreg conditions as mentioned above for 4 days. Cells were washed in cold PBS and pellets were stored at −80°C until extraction. Lipids were prepared from the respective cells by chloroform and methanol extraction. The internal standards, 500 pmol (20 μl) of Cer/Sph mixture I (Avanti Polar Lipids LM-6002), were added to the cells before extraction. Three biologically independent samples were taken for each subset. Lipids were normalized to the carbon content (100 μg/ml). Sphingolipids were analyzed by supercritical fluid chromatography coupled with mass spectrometry (SFC/MS/MS) using a Nexera UC system (Shimadzu Corp.) coupled to an LCMS-8060 system (Shimadzu Corp.) as previously described (87). The SFC analysis conditions were as follows: column, ACQUITY UPC2 Torus DEA Column (100 mm by 3.0 mm inner diameter; particle size, 1.7 μm, (Waters Co.); column temperature, 50°C; mobile phase A, supercritical carbon dioxide; mobile phase B, methanol/water (95:5, v/v) with 0.1% (w/v) ammonium acetate; flow rate of mobile phase, 1.0 ml/min; flow rate of make-up pump, 0.1 ml/min; and back-pressure regulator, 10 MPa. The gradient conditions were as follows: 1% B, 1 min; 1 to 76% B, 1 to 24 min; 76% B, 24 to 26 min; and 1% B, 26 to 30 min. The MS/MS operating conditions were as follows: polarity, positive ionization; electrospray voltage, 4.0 kV; desolvation line temperature, 250°C; heat block temperature, 400°C; nebulizing gas flow rate, 3.0 liters/min; drying gas flow rate, 10.0 liters/min; collision-induced dissociation gas pressure, 0.23 MPa; detector voltage, 2.16 kV; dwell time, 2 ms; and pause time, 2 ms. Data processing was performed using Skyline software v21.2 (MacCoss Laboratories).
Active and adoptive T cell transfer EAE induction
For active EAE induction, on day 0, 8- to 10-week-old age-matched Sptlc1+/+CD4-Cre (WT) and Sptlc1fl/flCD4-Cre (KO) mice were immunized subcutaneously in the hind flank with 400 μg of MOG35–55 (AnaSpec; AS-60130; ≥95%) emulsified in 100 μl of CFA containing Mycobacterium tuberculosis H37Ra (400 μg, BD Difco Laboratories; 231141) at a final volume of 200 μl in PBS. On days 0 and 2, the animals were injected with 200 ng of pertussis toxin (Sigma-Aldrich; P2980) in 100 μl of PBS intraperitoneally. Mice were observed daily for clinical signs of EAE up to 26 days after immunization and scored on a scale of 0 to 5: 0, no disease; 1, complete tail paralysis; 2, hindlimb weakness; 3, complete hindlimb paralysis; 4, hind- and forelimb paralysis; and 5, moribund/death. For the adoptive T cell transfer EAE model, WT (n = 10) and KO (n = 18) mice were primed with MOG peptide and CFA. At day 11, spleen and draining lymph nodes were isolated, and single-cell suspension was prepared after RBC lysis. The cells were then differentiated with MOG peptide (20 μg/ml) under TH17 differentiating conditions (IL-23, 8 ng/ml; IL-1a, 10 ng/ml; anti–IFN-γ, 10 μg/ml; anti–IL-4, 10 μg/ml). Four days later, CD4+ T cells were isolated by magnetic sorting as described above, and 5 million purified cells were transferred to female RAG−/− mice intraperitonially and scored daily for EAE up to 32 days after transfer as described above. Mice were euthanized when the scores reached 4 for two consecutive days or when body weight drops below 20% of the initial weight. A score of 4 is retained for these mice for the remainder of the time course of the experiment. During the experiment, the animals are managed according to the NCI Laboratory of Animal Sciences clinical assessment and management guidelines.
Isolation of mononuclear cells from CNS
Both brain and spinal cords were used for analyzing mononuclear cells infiltrating the CNS after EAE induction. After intracardial perfusion with cold PBS, brains were dissected, and spinal cords were obtained by flushing the spinal canal with cold PBS. Single-cell suspensions were prepared by pressing the tissues through a 70-μm cell strainer and mononuclear cells were obtained by 80/40% Percoll gradient centrifugation. Cells were removed from the interphase, washed, counted, and reactivated with PMA/ion with BFA for 6 hours and stained for intracellular cytokines.
Adoptive T cell transfer colitis model
For inducing colitis, naive CD4+D45RBhi T cells were purified from the spleen and lymph node of Sptlc1+/+CD4-Cre (WT) and Sptlc1fl/flCD4-Cre (KO) by FACS. Briefly, CD4+ T cells were enriched using the Mojosort mouse CD4+ T cell isolation kit, and after staining with BV786-conjugated CD4 and PE-conjugated CD45RB, naive CD4+CD45RBhi T cells were purified by cell sorting using a FACSAria II cell sorter (BD). Sorted cells were washed in sterile PBS, and 5 × 106 cells were transferred to Rag1−/− recipient mice intraperitoneally. Recipient mice were assessed two times a week for weight loss and other symptoms of disease for 8 weeks. At the end of the experiment, mice were euthanized, and spleen, mesenteric lymph nodes, small intestine, and colon were collected for immune cell isolation and histopathogy analysis. For isolating lamina propria lymphocytes, small intestine and colon were washed twice with Hanks’ balanced salt solution (HBSS, Gibco, Ca2+ and Mg2+ free), and intraepithelial cells were dissociated using HBSS supplemented with 5 mM EDTA by shaking the tissues at 37°C, 200 rpm for 30 min. After removing the epithelial cells, the tissues were minced and digested with 10% FBS-supplemented RPMI 1640 containing collagenase D (1 mg/ml, Roche, catalog no. 11088858001) and DNase I (100 μg/ml, Roche, catalog no. 10104159001) for 45 min at 37°C with constant shaking at 155 rpm. Digested tissues were filtered through a 70-μm cell strainer, and lymphocytes were isolated by Percoll gradient (40/80%) centrifugation.
Bulk RNA-seq
For RNA-seq, naïve CD4+ T cells from WT and KO mice were differentiated for 3 days under TH17 differentiating conditions in the presence or absence of NAC or 3-KDS. Samples from three independent biological replicates were processed for each condition. Total RNA was extracted using TRIzol, further treated with DNase I, and purified using the RNA Clean and Concentrator-5 kit (Zymoresearch). RNA (1.3 μg) was submitted to Novogene for total RNA-seq. The data obtained were further subjected to differential gene expression analysis using the DESeq2 R package. Principal components analysis (PCA) plots were generated using the plotPCA function. The DEGs were ranked on the basis of the log2 fold change and false discovery rate (FDR)–corrected P values. We used variance-stabilized transformation gene count values for downstream pathway analysis using the R package GAGE and GSEA with the commonly used gene set data based on Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways and Gene Ontology (GO) terms. The gene sets specific for the analysis were downloaded from http://bioinformatics.org/go2msig/ and modified with additional genes downloaded from https://gsea-msigdb.org/gsea/index.jsp. Gene sets having normalized P value <0.001 and an FDR cutoff of <0.1 were considered significant.
Confocal imaging
Naïve CD4 T cells from WT and KO mice were cross-linked with anti-CD3. One million cells were fixed and permeabilized with the Cytofix/Cytoperm fixation/permeabilization kit from BD according to the manufacturer’s instruction. Then, they were blocked with 5% normal goat serum (NGS) for 1 hour and incubated with primary antibody NOX2 [Proteintech (19013-1-AP), 1:100 dilution] and p47phox (Santa Cruz Biotechnology, 600 ng) in 5% NGS in permeabilization buffer for 1 hour. Cells were washed with 5% NGS in permeabilization buffer and incubated with secondary antibody (1:1000 dilution in 5% NGS in permeabilization buffer) overnight for 1 hour. Cells were washed and stained with 4′,6-diamidino-2-phenylindole (DAPI) (300 nM DAPI in PBS) for 1 to 5 min. After washing, cells were concentrated onto microscopic slide by cytospin method and mounted with VECTASHIELD (H-1000) mounting medium. The slides were imaged on ZEISS confocal laser scanning microscope LSM880. Confocal images were processed with Imaris software, wherein images were first smoothed by applying a Gaussian filter to each channel. Subsequently, all the punctate structures in each channel of a three-dimensional z-stack were traced with the spots tool by setting up appropriate thresholds. Spot-to-spot colocalization was performed by overlying spots from two different channels and applying the shortest distance (0.5 μm) between the two spots. Three independent experiments were performed for each treatment, and about 10 images from each experiment were analyzed. Each image contained at least three cells.
Statistics
Statistical analysis was performed using GraphPad Prism 9 software. No statistical analysis was used to predetermine the sample size, and no data were excluded from the analyses. Error bars present the means ± SEM. All data are representative of at least two independent experiments. Comparison between two groups was analyzed using an unpaired two-tailed Student’s t test. Comparison of more than two groups was analyzed by ordinary one-way analysis of variance (ANOVA) with Tukey’s multiple comparisons test. P < 0.05 was considered as statistically significant.
Acknowledgments
We thank I. Daar, S. Mackem, M. Lewandoski, T. Yamaguchi, and E. Davies for the scientific discussion. We thank A. Hackley for animal maintenance and help during animal experiments. We thank D. Lj. Jankovic and H. A. Young for providing us with additional Ifngr KO mice.
Funding: This work was supported by the intramural division of the National Cancer Institute, National Institutes of Health (Division of Health and Human Services). The sphingolipid analysis is partly supported by the Medical Research Center Initiative for High Depth Omics and the Cooperative Research Project Program of the Medical Institute of Bioregulation, Kyushu University (T.B.).
Author contributions: T.Ab., U.A., and J.K.A. conceived the study and designed the experiments; T.Ab. and V.P. performed the experiments; N.V. and D.B. analyzed the RNA-seq data; S.-H.L., T.B., S.D.F., and T.An. performed mass spectrometric analysis of sphingolipids; G.K. conceptualized the analysis of confocal data; B.K. conceptualized and performed the histopathological analysis; T.Ab. and J.K.A. drafted the original manuscript; and U.A., D.B., and T.B. reviewed and edited the manuscript. All authors reviewed and approved the manuscript.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. RNA-seq data have been deposited in Gene Expression Omnibus (GEO) under the accession number GSE254602. The mutant mice used in the study can be provided by the National Cancer Institute to academic and nonprofit requestors pending scientific review and a completed material transfer agreement substantially in the form of the NIH Material Transfer Agreement for the Transfer of Organisms (MTA-TO) to academic/not-for-profit organizations (https://techtransfer.nih.gov/resources#MTACTA). Commercial requestors may be required to take a license to the mutant mice substantially in the form of the NIH Biological Materials License Agreement (https://techtransfer.nih.gov/resources#MTACTA). Requests for the mutant mice should be submitted to J.K.A.
Supplementary Materials
This PDF file includes:
Figs. S1 to S12
REFERENCES AND NOTES
- 1.Harrington L. E., Hatton R. D., Mangan P. R., Turner H., Murphy T. L., Murphy K. M., Weaver C. T., Interleukin 17-producing CD4+ effector T cells develop via a lineage distinct from the T helper type 1 and 2 lineages. Nat. Immunol. 6, 1123–1132 (2005). [DOI] [PubMed] [Google Scholar]
- 2.Park H., Li Z., Yang X. O., Chang S. H., Nurieva R., Wang Y. H., Wang Y., Hood L., Zhu Z., Tian Q., Dong C., A distinct lineage of CD4 T cells regulates tissue inflammation by producing interleukin 17. Nat. Immunol. 6, 1133–1141 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tesmer L. A., Lundy S. K., Sarkar S., Fox D. A., Th17 cells in human disease. Immunol. Rev. 223, 87–113 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bettelli E., Carrier Y., Gao W., Korn T., Strom T. B., Oukka M., Weiner H. L., Kuchroo V. K., Reciprocal developmental pathways for the generation of pathogenic effector TH17 and regulatory T cells. Nature 441, 235–238 (2006). [DOI] [PubMed] [Google Scholar]
- 5.Mangan P. R., Harrington L. E., O'Quinn D. B., Helms W. S., Bullard D. C., Elson C. O., Hatton R. D., Wahl S. M., Schoeb T. R., Weaver C. T., Transforming growth factor-β induces development of the TH17 lineage. Nature 441, 231–234 (2006). [DOI] [PubMed] [Google Scholar]
- 6.Dang E. V., Barbi J., Yang H. Y., Jinasena D., Yu H., Zheng Y., Bordman Z., Fu J., Kim Y., Yen H. R., Luo W., Zeller K., Shimoda L., Topalian S. L., Semenza G. L., Dang C. V., Pardoll D. M., Pan F., Control of T(H)17/T(reg) balance by hypoxia-inducible factor 1. Cell 146, 772–784 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ivanov I. I., Atarashi K., Manel N., Brodie E. L., Shima T., Karaoz U., Wei D., Goldfarb K. C., Santee C. A., Lynch S. V., Tanoue T., Imaoka A., Itoh K., Takeda K., Umesaki Y., Honda K., Littman D. R., Induction of intestinal Th17 cells by segmented filamentous bacteria. Cell 139, 485–498 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kleinewietfeld M., Manzel A., Titze J., Kvakan H., Yosef N., Linker R. A., Muller D. N., Hafler D. A., Sodium chloride drives autoimmune disease by the induction of pathogenic TH17 cells. Nature 496, 518–522 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Langrish C. L., Chen Y., Blumenschein W. M., Mattson J., Basham B., Sedgwick J. D., McClanahan T., Kastelein R. A., Cua D. J., IL-23 drives a pathogenic T cell population that induces autoimmune inflammation. J. Exp. Med. 201, 233–240 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shi L. Z., Wang R., Huang G., Vogel P., Neale G., Green D. R., Chi H., HIF1alpha-dependent glycolytic pathway orchestrates a metabolic checkpoint for the differentiation of TH17 and Treg cells. J. Exp. Med. 208, 1367–1376 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wu C., Yosef N., Thalhamer T., Zhu C., Xiao S., Kishi Y., Regev A., Kuchroo V. K., Induction of pathogenic TH17 cells by inducible salt-sensing kinase SGK1. Nature 496, 513–517 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang D., Jin W., Wu R., Li J., Park S. A., Tu E., Zanvit P., Xu J., Liu O., Cain A., Chen W., High glucose intake exacerbates autoimmunity through reactive-oxygen-species-mediated TGF-β cytokine activation. Immunity 51, 671–681.e5 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhao Y., Liu Z., Qin L., Wang T., Bai O., Insights into the mechanisms of Th17 differentiation and the Yin-Yang of Th17 cells in human diseases. Mol. Immunol. 134, 109–117 (2021). [DOI] [PubMed] [Google Scholar]
- 14.Chapman N. M., Boothby M. R., Chi H., Metabolic coordination of T cell quiescence and activation. Nat. Rev. Immunol. 20, 55–70 (2020). [DOI] [PubMed] [Google Scholar]
- 15.O'Neill L. A. J., Kishton R. J., Rathmell J., A guide to immunometabolism for immunologists. Nat. Rev. Immunol. 16, 553–565 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Warburg O., On the origin of cancer cells. Science 123, 309–314 (1956). [DOI] [PubMed] [Google Scholar]
- 17.Ma E. H., Verway M. J., Johnson R. M., Roy D. G., Steadman M., Hayes S., Williams K. S., Sheldon R. D., Samborska B., Kosinski P. A., Kim H., Griss T., Faubert B., Condotta S. A., Krawczyk C. M., DeBerardinis R. J., Stewart K. M., Richer M. J., Chubukov V., Roddy T. P., Jones R. G., Metabolic profiling sing stable isotope tracing reveals distinct patterns of glucose utilization by physiologically activated CD8(+) T cells. Immunity 51, 856–870.e5 (2019). [DOI] [PubMed] [Google Scholar]
- 18.Chapman N. M., Chi H., Hallmarks of T-cell exit from quiescence. Cancer Immunol. Res. 6, 502–508 (2018). [DOI] [PubMed] [Google Scholar]
- 19.Shyer J. A., Flavell R. A., Bailis W., Metabolic signaling in T cells. Cell Res. 30, 649–659 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Waickman A. T., Powell J. D., mTOR, metabolism, and the regulation of T-cell differentiation and function. Immunol. Rev. 249, 43–58 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Damasceno L. E. A., Prado D. S., Veras F. P., Fonseca M. M., Toller-Kawahisa J. E., Rosa M. H., Publio G. A., Martins T. V., Ramalho F. S., Waisman A., Cunha F. Q., Cunha T. M., Alves-Filho J. C., PKM2 promotes Th17 cell differentiation and autoimmune inflammation by fine-tuning STAT3 activation. J. Exp. Med. 217, (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gerriets V. A., Kishton R. J., Nichols A. G., Macintyre A. N., Inoue M., Ilkayeva O., Winter P. S., Liu X., Priyadharshini B., Slawinska M. E., Haeberli L., Huck C., Turka L. A., Wood K. C., Hale L. P., Smith P. A., Schneider M. A., MacIver N. J., Locasale J. W., Newgard C. B., Shinohara M. L., Rathmell J. C., Metabolic programming and PDHK1 control CD4+ T cell subsets and inflammation. J. Clin. Invest. 125, 194–207 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Berod L., Friedrich C., Nandan A., Freitag J., Hagemann S., Harmrolfs K., Sandouk A., Hesse C., Castro C. N., Bahre H., Tschirner S. K., Gorinski N., Gohmert M., Mayer C. T., Huehn J., Ponimaskin E., Abraham W. R., Muller R., Lochner M., Sparwasser T., De novo fatty acid synthesis controls the fate between regulatory T and T helper 17 cells. Nat. Med. 20, 1327–1333 (2014). [DOI] [PubMed] [Google Scholar]
- 24.Cai F., Jin S., Chen G., The effect of lipid metabolism on CD4+ T cells. Mediators Inflamm. 2021, 6634532 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Endo Y., Asou H. K., Matsugae N., Hirahara K., Shinoda K., Tumes D. J., Tokuyama H., Yokote K., Nakayama T., Obesity drives Th17 cell differentiation by inducing the lipid metabolic kinase, ACC1. Cell Rep. 12, 1042–1055 (2015). [DOI] [PubMed] [Google Scholar]
- 26.Hu X., Wang Y., Hao L. Y., Liu X., Lesch C. A., Sanchez B. M., Wendling J. M., Morgan R. W., Aicher T. D., Carter L. L., Toogood P. L., Glick G. D., Sterol metabolism controls TH17 differentiation by generating endogenous RORγ agonists. Nat. Chem. Biol. 11, 141–147 (2015). [DOI] [PubMed] [Google Scholar]
- 27.Johnson M. O., Wolf M. M., Madden M. Z., Andrejeva G., Sugiura A., Contreras D. C., Maseda D., Liberti M. V., Paz K., Kishton R. J., Johnson M. E., de Cubas A. A., Wu P., Li G., Zhang Y., Newcomb D. C., Wells A. D., Restifo N. P., Rathmell W. K., Locasale J. W., Davila M. L., Blazar B. R., Rathmell J. C., Distinct regulation of Th17 and Th1 cell differentiation by glutaminase-dependent metabolism. Cell 175, 1780–1795.e19 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Puleston D. J., Baixauli F., Sanin D. E., Edwards-Hicks J., Villa M., Kabat A. M., Kaminski M. M., Stanckzak M., Weiss H. J., Grzes K. M., Piletic K., Field C. S., Corrado M., Haessler F., Wang C., Musa Y., Schimmelpfennig L., Flachsmann L., Mittler G., Yosef N., Kuchroo V. K., Buescher J. M., Balabanov S., Pearce E. J., Green D. R., Pearce E. L., Polyamine metabolism is a central determinant of helper T cell lineage fidelity. Cell 184, 4186–4202.e20 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sugiura A., Andrejeva G., Voss K., Heintzman D. R., Xu X., Madden M. Z., Ye X., Beier K. L., Chowdhury N. U., Wolf M. M., Young A. C., Greenwood D. L., Sewell A. E., Shahi S. K., Freedman S. N., Cameron A. M., Foerch P., Bourne T., Garcia-Canaveras J. C., Karijolich J., Newcomb D. C., Mangalam A. K., Rabinowitz J. D., Rathmell J. C., MTHFD2 is a metabolic checkpoint controlling effector and regulatory T cell fate and function. Immunity 55, 65–81.e9 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wagner A., Wang C., Fessler J., DeTomaso D., Avila-Pacheco J., Kaminski J., Zaghouani S., Christian E., Thakore P., Schellhaass B., Akama-Garren E., Pierce K., Singh V., Ron-Harel N., Douglas V. P., Bod L., Schnell A., Puleston D., Sobel R. A., Haigis M., Pearce E. L., Soleimani M., Clish C., Regev A., Kuchroo V. K., Yosef N., Metabolic modeling of single Th17 cells reveals regulators of autoimmunity. Cell 184, 4168–4185.e21 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gault C. R., Obeid L. M., Hannun Y. A., An overview of sphingolipid metabolism: From synthesis to breakdown. Adv. Exp. Med. Biol. 688, 1–23 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Maceyka M., Spiegel S., Sphingolipid metabolites in inflammatory disease. Nature 510, 58–67 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hannun Y. A., Obeid L. M., Sphingolipids and their metabolism in physiology and disease. Nat. Rev. Mol. Cell Biol. 19, 175–191 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Parthibane V., Acharya D., Srideshikan S. M., Lin J., Myerscough D. G., Abimannan T., Vijaykrishna N., Blankenberg D., Bondada L., Klarmann K. D., Fox S. D., Andresson T., Tessarollo L., Acharya U., Keller J. R., Acharya J. K., Sptlc1 is essential for myeloid differentiation and hematopoietic homeostasis. Blood Adv. 3, 3635–3649 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Parthibane V., Lin J., Acharya D., Abimannan T., Srideshikan S. M., Klarmann K., Yang A., Soheilian F., Nagashima K., Fox S. D., Andresson T., Tessarollo L., Keller J. R., Acharya U., Acharya J. K., SSSPTA is essential for serine palmitoyltransferase function during development and hematopoiesis. J. Biol. Chem. 296, 100491 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kunduri G., Le S. H., Baena V., Vijaykrishna N., Harned A., Nagashima K., Blankenberg D., Yoshihiro I., Narayan K., Bamba T., Acharya U., Acharya J. K., Delivery of ceramide phosphoethanolamine lipids to the cleavage furrow through the endocytic pathway is essential for male meiotic cytokinesis. PLOS Biol. 20, e3001599 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kunduri G., Turner-Evans D., Konya Y., Izumi Y., Nagashima K., Lockett S., Holthuis J., Bamba T., Acharya U., Acharya J. K., Defective cortex glia plasma membrane structure underlies light-induced epilepsy in cpes mutants. Proc. Natl. Acad. Sci. U.S.A. 115, E8919–E8928 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Albeituni S., Stiban J., Roles of ceramides and other sphingolipids in immune cell function and inflammation. Adv. Exp. Med. Biol. 1161, 169–191 (2019). [DOI] [PubMed] [Google Scholar]
- 39.Molano A., Huang Z., Marko M. G., Azzi A., Wu D., Wang E., Kelly S. L., Merrill A. H. Jr., Bunnell S. C., Meydani S. N., Age-dependent changes in the sphingolipid composition of mouse CD4+ T cell membranes and immune synapses implicate glucosylceramides in age-related T cell dysfunction. PLOS ONE 7, e47650 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sen P., Andrabi S. B. A., Buchacher T., Khan M. M., Kalim U. U., Lindeman T. M., Alves M. A., Hinkkanen V., Kemppainen E., Dickens A. M., Rasool O., Hyotylainen T., Lahesmaa R., Oresic M., Quantitative genome-scale metabolic modeling of human CD4(+) T cell differentiation reveals subset-specific regulation of glycosphingolipid pathways. Cell Rep. 37, 109973 (2021). [DOI] [PubMed] [Google Scholar]
- 41.Tafesse F. G., Rashidfarrokhi A., Schmidt F. I., Freinkman E., Dougan S., Dougan M., Esteban A., Maruyama T., Strijbis K., Ploegh H. L., Disruption of sphingolipid biosynthesis blocks phagocytosis of Candida albicans. PLOS Pathog. 11, e1005188 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wu J., Ma S., Sandhoff R., Ming Y., Hotz-Wagenblatt A., Timmerman V., Bonello-Palot N., Schlotter-Weigel B., Auer-Grumbach M., Seeman P., Löscher W. N., Reindl M., Weiss F., Mah E., Weisshaar N., Madi A., Mohr K., Schlimbach T., Velasco Cardenas R. M.-H., Koeppel J., Grünschläger F., Müller L., Baumeister M., Brügger B., Schmitt M., Wabnitz G., Samstag Y., Cui G., Loss of neurological disease HSAN-I-associated gene SPTLC2 impairs CD8+ T cell responses to infection by inhibiting T cell metabolic fitness. Immunity 50, 1218–1231.e5 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhu Y., Gumlaw N., Karman J., Zhao H., Zhang J., Jiang J. L., Maniatis P., Edling A., Chuang W. L., Siegel C., Shayman J. A., Kaplan J., Jiang C., Cheng S. H., Lowering glycosphingolipid levels in CD4+ T cells attenuates T cell receptor signaling, cytokine production, and differentiation to the Th17 lineage. J. Biol. Chem. 286, 14787–14794 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bejaoui K., Wu C., Scheffler M. D., Haan G., Ashby P., Wu L., de Jong P., Brown R. H. Jr., SPTLC1 is mutated in hereditary sensory neuropathy, type 1. Nat. Genet. 27, 261–262 (2001). [DOI] [PubMed] [Google Scholar]
- 45.Penno A., Reilly M. M., Houlden H., Laura M., Rentsch K., Niederkofler V., Stoeckli E. T., Nicholson G., Eichler F., Brown R. H. Jr., von Eckardstein A., Hornemann T., Hereditary sensory neuropathy type 1 is caused by the accumulation of two neurotoxic sphingolipids. J. Biol. Chem. 285, 11178–11187 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rotthier A., Auer-Grumbach M., Janssens K., Baets J., Penno A., Almeida-Souza L., Van Hoof K., Jacobs A., De Vriendt E., Schlotter-Weigel B., Loscher W., Vondracek P., Seeman P., De Jonghe P., Van Dijck P., Jordanova A., Hornemann T., Timmerman V., Mutations in the SPTLC2 subunit of serine palmitoyltransferase cause hereditary sensory and autonomic neuropathy type I. Am. J. Hum. Genet. 87, 513–522 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Orchard R. C., Wilen C. B., Virgin H. W., Sphingolipid biosynthesis induces a conformational change in the murine norovirus receptor and facilitates viral infection. Nat. Microbiol. 3, 1109–1114 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Solomon J. C., Sharma K., Wei L. X., Fujita T., Shi Y. F., A novel role for sphingolipid intermediates in activation-induced cell death in T cells. Cell Death Differ. 10, 193–202 (2003). [DOI] [PubMed] [Google Scholar]
- 49.Miller L. G. Jr., Young J. A., Ray S. K., Wang G., Purohit S., Banik N. L., Dasgupta S., Sphingosine toxicity in EAE and MS: Evidence for ceramide generation via serine-palmitoyltransferase activation. Neurochem. Res. 42, 2755–2768 (2017). [DOI] [PubMed] [Google Scholar]
- 50.Ghoreschi K., Laurence A., Yang X.-P., Tato C. M., McGeachy M. J., Konkel J. E., Ramos H. L., Wei L., Davidson T. S., Bouladoux N., Grainger J. R., Chen Q., Kanno Y., Watford W. T., Sun H.-W., Eberl G., Shevach E. M., Belkaid Y., Cua D. J., Chen W., O'Shea J. J., Generation of pathogenic TH17 cells in the absence of TGF-β signalling. Nature 467, 967–971 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lazarevic V., Chen X., Shim J. H., Hwang E. S., Jang E., Bolm A. N., Oukka M., Kuchroo V. K., Glimcher L. H., T-bet represses TH17 differentiation by preventing Runx1-mediated activation of the gene encoding RORγt. Nat. Immunol. 12, 96–104 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ivashkiv L. B., IFNγ: Signalling, epigenetics and roles in immunity, metabolism, disease and cancer immunotherapy. Nat. Rev. Immunol. 18, 545–558 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wadsworth J. M., Clarke D. J., McMahon S. A., Lowther J. P., Beattie A. E., Langridge-Smith P. R. R., Broughton H. B., Dunn T. M., Naismith J. H., Campopiano D. J., The chemical basis of serine palmitoyltransferase inhibition by myriocin. J. Am. Chem. Soc. 135, 14276–14285 (2013). [DOI] [PubMed] [Google Scholar]
- 54.Dong C., Targeting Th17 cells in immune diseases. Cell Res. 24, 901–903 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Miller S. D., Karpus W. J., Experimental autoimmune encephalomyelitis in the mouse. Curr. Protoc. Immunol. Chapter 15, 15.1.1–15.1.18 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Jiang P., Zheng C., Xiang Y., Malik S., Su D., Xu G., Zhang M., The involvement of TH17 cells in the pathogenesis of IBD. Cytokine Growth Factor Rev. 69, 28–42 (2023). [DOI] [PubMed] [Google Scholar]
- 57.Laurence A., Tato C. M., Davidson T. S., Kanno Y., Chen Z., Yao Z., Blank R. B., Meylan F., Siegel R., Hennighausen L., Shevach E. M., O'shea J. J., Interleukin-2 signaling via STAT5 constrains T helper 17 cell generation. Immunity 26, 371–381 (2007). [DOI] [PubMed] [Google Scholar]
- 58.Sakaguchi S., Sakaguchi N., Asano M., Itoh M., Toda M., Immunologic self-tolerance maintained by activated T cells expressing IL-2 receptor alpha-chains (CD25). Breakdown of a single mechanism of self-tolerance causes various autoimmune diseases. J. Immunol. 155, 1151–1164 (1995). [PubMed] [Google Scholar]
- 59.Zorn E., Nelson E. A., Mohseni M., Porcheray F., Kim H., Litsa D., Bellucci R., Raderschall E., Canning C., Soiffer R. J., Frank D. A., Ritz J., IL-2 regulates FOXP3 expression in human CD4+CD25+ regulatory T cells through a STAT-dependent mechanism and induces the expansion of these cells in vivo. Blood 108, 1571–1579 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mookerjee S. A., Gerencser A. A., Nicholls D. G., Brand M. D., Quantifying intracellular rates of glycolytic and oxidative ATP production and consumption using extracellular flux measurements. J. Biol. Chem. 292, 7189–7207 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Franchina D. G., Dostert C., Brenner D., Reactive oxygen species: Involvement in T cell signaling and metabolism. Trends Immunol. 39, 489–502 (2018). [DOI] [PubMed] [Google Scholar]
- 62.Belikov A. V., Schraven B., Simeoni L., T cells and reactive oxygen species. J. Biomed. Sci. 22, 85 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Jackson S. H., Devadas S., Kwon J., Pinto L. A., Williams M. S., T cells express a phagocyte-type NADPH oxidase that is activated after T cell receptor stimulation. Nat. Immunol. 5, 818–827 (2004). [DOI] [PubMed] [Google Scholar]
- 64.Tse H. M., Thayer T. C., Steele C., Cuda C. M., Morel L., Piganelli J. D., Mathews C. E., NADPH oxidase deficiency regulates Th lineage commitment and modulates autoimmunity. J. Immunol. 185, 5247–5258 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Devadas S., Zaritskaya L., Rhee S. G., Oberley L., Williams M. S., Discrete generation of superoxide and hydrogen peroxide by T cell receptor stimulation: Selective regulation of mitogen-activated protein kinase activation and fas ligand expression. J. Exp. Med. 195, 59–70 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hla T., Dannenberg A. J., Sphingolipid signaling in metabolic disorders. Cell Metab. 16, 420–434 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Taylor J. P., Tse H. M., The role of NADPH oxidases in infectious and inflammatory diseases. Redox Biol. 48, 102159 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Schnell A., Littman D. R., Kuchroo V. K., T(H)17 cell heterogeneity and its role in tissue inflammation. Nat. Immunol. 24, 19–29 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Yamagata T., Skepner J., Yang J., Targeting Th17 effector cytokines for the treatment of autoimmune diseases. Arch. Immunol. Ther. Exp. 63, 405–414 (2015). [DOI] [PubMed] [Google Scholar]
- 70.Yasuda K., Takeuchi Y., Hirota K., The pathogenicity of Th17 cells in autoimmune diseases. Semin. Immunopathol. 41, 283–297 (2019). [DOI] [PubMed] [Google Scholar]
- 71.Park J., Kim M., Kang S. G., Jannasch A. H., Cooper B., Patterson J., Kim C. H., Short-chain fatty acids induce both effector and regulatory T cells by suppression of histone deacetylases and regulation of the mTOR-S6K pathway. Mucosal Immunol. 8, 80–93 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Abimannan T., Peroumal D., Parida J. R., Barik P. K., Padhan P., Devadas S., Oxidative stress modulates the cytokine response of differentiated Th17 and Th1 cells. Free Radic. Biol. Med. 99, 352–363 (2016). [DOI] [PubMed] [Google Scholar]
- 73.Alfadda A. A., Sallam R. M., Reactive oxygen species in health and disease. J. Biomed. Biotechnol. 2012, 936486 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Fu G., Xu Q., Qiu Y., Jin X., Xu T., Dong S., Wang J., Ke Y., Hu H., Cao X., Wang D., Cantor H., Gao X., Lu L., Suppression of Th17 cell differentiation by misshapen/NIK-related kinase MINK1. J. Exp. Med. 214, 1453–1469 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kaufmann U., Kahlfuss S., Yang J., Ivanova E., Koralov S. B., Feske S., Calcium signaling controls pathogenic Th17 cell-mediated inflammation by regulating mitochondrial function. Cell Metab. 29, 1104–1118.e6 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Mills E. A., Ogrodnik M. A., Plave A., Mao-Draayer Y., Emerging understanding of the mechanism of action for dimethyl fumarate in the treatment of multiple sclerosis. Front. Neurol. 9, 5 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Yadav S. K., Soin D., Ito K., Dhib-Jalbut S., Insight into the mechanism of action of dimethyl fumarate in multiple sclerosis. J. Mol. Med. 97, 463–472 (2019). [DOI] [PubMed] [Google Scholar]
- 78.Schulze-Topphoff U., Varrin-Doyer M., Pekarek K., Spencer C. M., Shetty A., Sagan S. A., Cree B. A. C., Sobel R. A., Wipke B. T., Steinman L., Scannevin R. H., Zamvil S. S., Dimethyl fumarate treatment induces adaptive and innate immune modulation independent of Nrf2. Proc. Natl. Acad. Sci. U.S.A. 113, 4777–4782 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Li M., Zhao L., Liu J., Liu A., Jia C., Ma D., Jiang Y., Bai X., Multi-mechanisms are involved in reactive oxygen species regulation of mTORC1 signaling. Cell. Signal. 22, 1469–1476 (2010). [DOI] [PubMed] [Google Scholar]
- 80.Barth B. M., Gustafson S. J., Young M. M., Fox T. E., Shanmugavelandy S. S., Kaiser J. M., Cabot M. C., Kester M., Kuhn T. B., Inhibition of NADPH oxidase by glucosylceramide confers chemoresistance. Cancer Biol. Ther. 10, 1126–1136 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Apostolidis S. A., Rodriguez-Rodriguez N., Suarez-Fueyo A., Dioufa N., Ozcan E., Crispin J. C., Tsokos M. G., Tsokos G. C., Phosphatase PP2A is requisite for the function of regulatory T cells. Nat. Immunol. 17, 556–564 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Wiese T., Dennstädt F., Hollmann C., Stonawski S., Wurst C., Fink J., Gorte E., Mandasari P., Domschke K., Hommers L., Vanhove B., Schumacher F., Kleuser B., Seibel J., Rohr J., Buttmann M., Menke A., Schneider-Schaulies J., Beyersdorf N., Inhibition of acid sphingomyelinase increases regulatory T cells in humans. Brain Commun. 3, fcab020 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Eken A., Duhen R., Singh A. K., Fry M., Buckner J. H., Kita M., Bettelli E., Oukka M., S1P(1) deletion differentially affects TH17 and regulatory T cells. Sci. Rep. 7, 12905 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Huang M. C., Watson S. R., Liao J. J., Goetzl E. J., Th17 augmentation in OTII TCR plus T cell-selective type 1 sphingosine 1-phosphate receptor double transgenic mice. J. Immunol. 178, 6806–6813 (2007). [DOI] [PubMed] [Google Scholar]
- 85.Chu S., Sun R., Gu X., Chen L., Liu M., Guo H., Ju S., Vatsalya V., Feng W., McClain C. J., Deng Z., Inhibition of sphingosine-1-phosphate-induced Th17 cells ameliorates alcohol-associated steatohepatitis in mice. Hepatology 73, 952–967 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Barra G., Lepore A., Gagliardi M., Somma D., Matarazzo M. R., Costabile F., Pasquale G., Mazzoni A., Gallo C., Nuzzo G., Annunziato F., Fontana A., Leonardi A., De Palma R., Sphingosine kinases promote IL-17 expression in human T lymphocytes. Sci. Rep. 8, 13233 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Takeda H., Izumi Y., Takahashi M., Paxton T., Tamura S., Koike T., Yu Y., Kato N., Nagase K., Shiomi M., Bamba T., Widely-targeted quantitative lipidomics method by supercritical fluid chromatography triple quadrupole mass spectrometry. J. Lipid Res. 59, 1283–1293 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S12