Abstract
HIV-1 cores, which contain the viral genome and replication machinery, must disassemble (uncoat) during viral replication. However, the viral and host factors that trigger uncoating remain unidentified. Recent studies show that infectious cores enter the nucleus and uncoat near the site of integration. Here, we show that efficient uncoating of nuclear cores requires synthesis of a double-stranded DNA (dsDNA) genome >3.5 kb and that the efficiency of uncoating correlates with genome size. Core disruption by capsid inhibitors releases viral DNA, some of which integrates. However, most of the viral DNA is degraded, indicating that the intact core safeguards viral DNA. Atomic force microscopy and core content estimation reveal that synthesis of full-length genomic dsDNA induces substantial internal strain on the core to promote uncoating. We conclude that HIV-1 cores protect viral DNA from degradation by host factors and that synthesis of long double-stranded reverse transcription products is required to trigger efficient HIV-1 uncoating.
Synthesis of long viral DNA is needed to disassemble HIV-1 cores, a requirement for integration into host chromosomes.
INTRODUCTION
HIV-1 virions contain a capsid shell (core) that encloses the viral genome and must disassemble (uncoat) so that the newly synthesized viral double-stranded DNA (dsDNA) can integrate into the host chromosomes. Although uncoating is essential for virus infection, it remains one of the most poorly understood steps in HIV-1 replication. Studies of uncoating have been difficult in part because methods to quantify the amount of capsid protein (CA) associated with viral complexes have not been available and because it has been difficult to identify the rare infectious viral cores from the vast majority of noninfectious viral cores. Consequently, previous studies have drawn differing conclusions regarding the intracellular location, timing, and molecular triggers of uncoating [reviewed in (1, 2)]. Historically, it was thought that HIV-1 cores must uncoat in the cytoplasm because their size exceeds the diameter of the nuclear pore complex and that uncoating is necessary to allow reverse transcription of the viral genome [reviewed in (3, 4)]. Viral core uncoating in the cytoplasm has been observed (5, 6), which can lead to efficient proteasomal degradation of viral complexes (7) and detection of viral DNA by host immune sensors (8, 9). However, our recent studies indicate that the rare viral cores that lead to productive infection are intact when they enter the nucleus and uncoat near their integration sites <1.5 hours before integration (10, 11). Consistent with our results, cryo–electron tomography studies have revealed that the diameter of nuclear pores is large enough for import of intact cores and that conical-shaped capsids enter the nuclei of infected cells (12–14). This argument is bolstered by the observation that interactions between the viral core and host cleavage and polyadenylation specificity factor 6 (CPSF6), which require a capsid lattice, are essential for the nuclear import of viral cores (15, 16) and integration into speckle-associated chromatin domains, the preferred sites of proviral integration (16–18). Recent studies have also shown that capsids remain largely intact and play an indispensable role in supporting efficient reverse transcription (19) and that reverse transcription is completed in the nucleus (10, 20–22).
Previous studies that sought to identify the molecular triggers of uncoating did not consider the uncoating of viral cores in the nucleus and focused on uncoating events in the cytoplasm or near the nuclear envelope. Binding of a CPSF6 mutant to cytoplasmic cores prevents their nuclear import and leads to substantial loss of infectivity, suggesting that viral cores that uncoat in the cytoplasm do not lead to productive infection (23). Some studies concluded that uncoating occurs in the cytoplasm soon after initiation of reverse transcription (24) and that reverse transcription destabilizes cytoplasmic viral cores (25). Another study suggested that completion of reverse transcription is not sufficient to induce uncoating and that a central polypurine tract needed for the formation of a central DNA flap is required to induce uncoating at the nuclear envelope before nuclear entry (26). A study from our lab (10) and a recent study (27) showed that reverse transcription is required to induce efficient uncoating of nuclear viral cores, but the extent of reverse transcription needed to induce uncoating was not determined. Another recent study showed that inhibition of reverse transcription did not affect the CA levels that accumulated in the nucleus, but it was not determined whether the CA was associated with nuclear viral cores or if reverse transcription induced uncoating of nuclear viral cores (22). It was also recently observed that capsid stiffness transiently increases ~7 hours after initiation of reverse transcription, suggesting that the capsid stiffness changes correlate with the initiation of capsid disassembly (28). However, these observations were made in the absence of inositol hexakisphosphate (IP6), which results in unstable capsids and low reverse transcription efficiency (19); consequently, the capsid stiffness changes could not be directly correlated with the progression of reverse transcription or uncoating. The lack of consensus in the field for >20 years reflects the difficulties in studying the uncoating mechanism in vitro and in infected cells. Here, we used a live-cell imaging assay to directly visualize the uncoating of nuclear HIV-1 cores and complementary biochemical assays to identify the viral and host factors that play a critical role in HIV-1 uncoating.
RESULTS
Reverse transcription is required for efficient uncoating
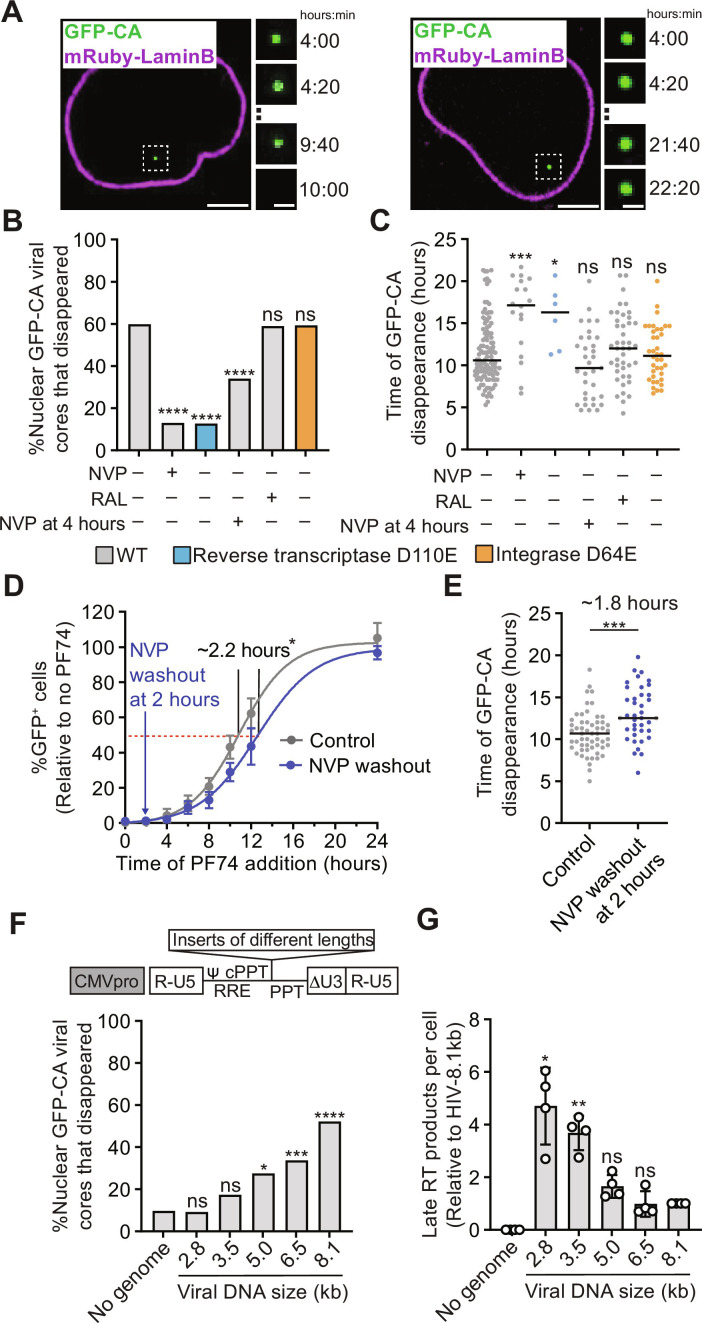
We determined HIV-1 uncoating efficiency by live-cell imaging of HIV-1 cores labeled with green fluorescent protein (GFP)–tagged capsid protein (GFP-CA; fig. S1, A and B) in HeLa cells expressing mRuby-LaminB. We determined the percentage of GFP-CA–labeled nuclear cores that disappeared between ~4 and 22 hours after infection, which was defined as uncoating (Fig. 1A). In untreated cells, in cells treated with the HIV-1 integrase inhibitor raltegravir (RAL), or in cells infected with HIV-1 carrying the integrase catalytic site mutation D64E, ~60% of the nuclear HIV-1 cores uncoated (Fig. 1B). However, uncoating was significantly reduced to ~13% in cells treated with reverse transcriptase inhibitor nevirapine (NVP) or infected with HIV-1 carrying the reverse transcriptase catalytic site mutation D110E. HIV-1 uncoating occurred ~10.6 hours after infection in untreated cells or when integration was inhibited, whereas the few cores that uncoated in the absence of reverse transcription did so much later at ~16 to 17 hours after infection (Fig. 1C). The observation that RAL treatment had no effect on uncoating indicated that uncoating does not require integrase activity (Fig. 1, B and C). We also assessed the time of HIV-1 core uncoating by addition of the capsid binding inhibitor PF-3450074 (PF74) at different times and determining the infectivity of an HIV-1–based vector that expresses a GFP reporter. The average loss of sensitivity to PF74 occurred ~10.8 hours after infection (Fig. 1D), which is similar to the time of uncoating of nuclear cores measured by live-cell imaging assays (~10.6 hours; Fig. 1C). These data agree with previous studies (10, 27) and indicate that reverse transcription is required for efficient HIV-1 uncoating.
Fig. 1. Effect of reverse transcription, integration, and genome size on uncoating of nuclear HIV-1 cores.
(A) Live-cell imaging of aphidicolin-treated HeLa cells expressing mRuby-LaminB infected with GFP-CA–labeled HIV-1. Examples of GFP-CA disappearance in the nucleus (left) or remaining in the nucleus until the end of a movie (right). Scale bars, 10 μm (large) and 2 μm (small). (B) Percentage of nuclear HIV-1 cores that disappeared during the movies for the indicated virus and cell treatments. All vector genomes are 10.3 kb. (C) Time (hours) after infection of GFP-CA disappearance. (D) PF74 time-of-addition experiment in untreated cells (control) or in cells treated with NVP at time of infection followed by washout 2 hours later. The percentage of GFP reporter–expressing cells was determined ~1 day after infection and is shown relative to untreated control cells (set to 100%; means ± SD, n = 6 biological replicates). (E) Time (hours) after infection of GFP-CA disappearance in untreated control cells or in cells treated with NVP at time of infection followed by washout 2 hours later. (F) Top: Schematic of different-sized lentiviral vectors by addition of non–HIV-1 sequence (see "Synthesis of long dsDNA reverse transcription products is required for efficient uncoating"). Bottom: Percentage of nuclear HIV-1 cores containing no viral RNA genome or different-sized genomes that disappeared during the movies. Expected size (kilobase) of genome after reverse transcription is indicated for each vector. cPPT, central polypurine tract; PPT, polypurine tract; RRE, rev-response element. (G) Late RT products for indicated viruses 6 hours after infection of aphidicolin-treated HeLa cells with p24-normalized virus input (means ± SD, Welch’s t test, n = 4 biological replicates). For (B), (E), and (F), the number of HIV-1 cores analyzed for each sample ranged from 51 to 187 (average = ~94). Fisher’s exact test. For (C) and (E), lines = median; Mann-Whitney U test. *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001; ns, not significant.
Uncoating occurs ~10 hours after reverse transcription initiation
To gain insight into the effect of delaying reverse transcription initiation on the kinetics of uncoating, we treated cells with NVP at the time of infection, washed out the NVP 2 hours later, and then determined when viruses lost sensitivity to PF74. We found that, when initiation of reverse transcription was delayed by 2 hours, viruses lost sensitivity to PF74 ~2 hours later than viruses in untreated cells (~10.8 hours versus ~13.0 hours), indicating that uncoating was also delayed by ~2 hours (Fig. 1D). A similar 2-hour NVP treatment delayed the uncoating of nuclear viral cores by ~2 hours, as determined by the loss of the GFP-CA signals in the imaging assays (~10.7 versus ~12.5 hours; Fig. 1E). These results indicate that the kinetics of uncoating are closely correlated with the timing of reverse transcription and that uncoating of infectious viral cores occurs ~10 hours after reverse transcription initiation.
Synthesis of long dsDNA reverse transcription products is required for efficient uncoating
Previous studies did not determine the extent of reverse transcription required for efficient uncoating. To address this question, we first determined the kinetics of reverse transcription by measuring early and late reverse transcription products (referred to as early RT and late RT products, respectively, hereafter) at different times after infection (fig. S1C). Next, we added NVP at a point when ~75% of the late RT products had been synthesized (4 hours after infection) and then assessed the uncoating efficiency of nuclear HIV-1 cores. Treatment of cells with NVP after reverse transcription progressed to a late stage significantly reduced uncoating efficiency from ~60 to ~34%, indicating that efficient uncoating requires synthesis of long dsDNA products. To further determine how the length reverse transcription products affects uncoating efficiency, we constructed a series of lentiviral vectors of varying size by addition of non–HIV-1 sequences and determined the effect of genome size on uncoating efficiency (Fig. 1F and fig. S2A). We found that all viral cores entered the nucleus with similar efficiency irrespective of genome size (fig. S2, B and C), and their uncoating efficiency positively correlated with genome size (Fig. 1F). Furthermore, the uncoating efficiency of cores containing genomes <3.5 kb was not significantly different from the cores that did not contain any genome, indicating that synthesis of a genome >3.5 kb is required to induce uncoating above background levels of reverse transcription-independent capsid disassembly (Fig. 1F). Viral cores containing small genomes (≤3.5 kb) efficiently produced late RT products (Fig. 1G), indicating that the low uncoating efficiency of viral cores containing the small genomes was not due to a failure to synthesize viral DNA. We conclude that HIV-1 uncoating efficiency is directly influenced by the size of the viral DNA genome and that efficient uncoating requires a genome >3.5 kb.
Intact cores protect viral DNA from degradation
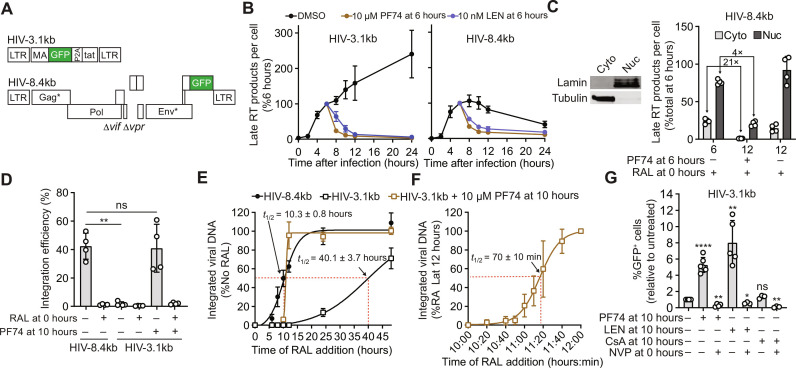
Treatment of infected cells with 10 μM PF74 disrupts most nuclear GFP-CA–labeled HIV-1 cores (10, 11). To determine whether PF74-induced capsid disruption can release short viral DNA that is capable of integration and reporter gene expression, we constructed HIV-1–based vectors HIV-3.1kb and near-full length HIV-8.4kb that both express a GFP reporter (Fig. 2A and fig. S3; see Supplementary Text). The nuclear import efficiency of cores containing these genomes was similar (fig. S4A) and both efficiently produced late RT DNA (fig. S4B). Similar to the other short vectors ≤3.5 kb (Fig. 1F), the cores containing the HIV-3.1kb genome were defective for uncoating, exhibiting a 6.3-fold lower efficiency of uncoating of nuclear cores compared to the cores containing the HIV-8.4kb genome (fig. S4C). Treatment of cells 6 hours after infection with 10 μM of PF74 or 10 nM of the capsid binding inhibitor lenacapavir led to the rapid (<2 hours) loss of both HIV-3.1kb and HIV-8.4kb viral DNA (Fig. 2B). To determine whether viral DNA released from cores in the cytoplasm or the nucleus was degraded, we isolated the cytoplasmic and nuclear fractions before DNA extraction (Fig. 2C, left). The levels of cytoplasmic and nuclear viral DNA were decreased by 21-fold and 4-fold, respectively, 6 hours after treatment with PF74 (PF74 was added at 6 hours after infection) (Fig. 2C). Thus, while most viral DNA released from the HIV-1 cores in the cytoplasmic and nuclear fractions was degraded, the viral DNA in the nuclear fractions, which potentially included DNA from viral cores associated with the nuclear envelope, was degraded less efficiently than in the cytoplasmic fraction. Moreover, because we inhibited integration with RAL in these experiments, the less efficient degradation of DNA released from the nuclear cores cannot be attributed to a protective effect of viral DNA integration. These findings indicate that capsid inhibitors PF74 and lenacapavir can disrupt HIV-1 cores, exposing viral DNA to the host environment where it can be degraded. Our observation that cytoplasmic HIV-1 cores protect viral DNA from degradation is consistent with previous studies showing that HIV-1 cores shield viral DNA from cytoplasmic nucleases (29–32) and innate immune sensors (8, 33). We extend these findings and show that HIV-1 cores in the nucleus also protect viral DNA from degradation by host factors. These results also suggest that HIV-1 cores shield the viral DNA from detection by nuclear DNA sensors [reviewed in (34)].
Fig. 2. Effect of CA inhibitor-induced disruption of HIV-1 cores containing short genomes on viral DNA levels, integration, and expression of GFP reporter.
(A) Schematic of short (HIV-3.1kb) and near-full length (HIV-8.1kb) HIV-1–based vectors encoding a GFP reporter. LTR, long terminal repeat. (B) Measurement of Late RT products for HIV-3.1kb (left) and HIV-8.4kb (right) at different times after infection of aphidicolin-treated HeLa cells (means ± SD, n = 3 biological replicates). DMSO, dimethyl sulfoxide. (C) Left: Western blot analysis of cytoplasmic and nuclear fractions of HeLa cells, representative of four experiments. Right: Measurement of cytoplasmic and nuclear late RT products before (6 hours) and after (12 hours) PF74 treatment of aphidicolin- and RAL-treated HeLa cells (means ± SD, n = 4 biological replicates). (D) Measurement of integration efficiency, defined as the percentage of late RT products detected at 24 hours after infection that was integrated by 7 days after infection, for indicated samples (means ± SD, Welch’s t test, n = 4 biological replicates). Integrated viral DNA was detected using the late RT primer and probe set at 7 days after infection. (E and F) Measurement of integrated viral DNA for indicated samples after RAL addition at different time points (means ± SD, n = 3 biological replicates). (G) Fold change in the percentage of GFP reporter–expressing cells measured 1 day after infection of HeLa cells with HIV-3.1kb; data shown relative to untreated control (set to 1; means ± SD, Welch’s t test, n = 3 to 6 biological replicates). The percentage of GFP reporter–expressing cells in untreated cells was set to 1. *P < 0.05, **P < 0.01, and ****P < 0.0001.
The late RT products of the HIV-3.1kb genome continued to increase from 8 to 24 hours after infection, whereas the late RT products of the HIV-8.4kb genome decreased over the same time period (Fig. 2B). This difference in late RT product accumulation is consistent with the observation that viral cores containing the 3.1-kb genome do not uncoat, and, as a result, the late RT products continue to accumulate. In contrast, the viral cores containing the 8.4-kb genome begin to uncoat as reverse transcription is completed, leading to degradation of most of the viral DNA released from the uncoated cores, while some DNA integrates; as a result, the late RT products of the 8.4-kb genome decrease between 8 and 24 hours after infection.
Short vector DNA integrates after core disruption with capsid inhibitors
Although most of the short HIV-3.1kb DNA was degraded after PF74-induced capsid disassembly (Fig. 2B), the integration efficiency of the short HIV-3.1kb DNA that survived degradation, defined as the percentage of late viral DNA detected at 24 hours after infection that had integrated, was high and similar to that of the near full-length HIV-8.4kb DNA in untreated cells (~38 versus ~43%, respectively; Fig. 2D). The integration efficiency of HIV-3.1kb in cells not treated with PF74 was only ~2%, presumably because most short viral DNAs remained trapped inside intact HIV-1 capsids. As expected, RAL treatment of cells resulted in low integration efficiency (Fig. 2D). The integration kinetics of the short viral DNA released from PF74-disrupted capsids was determined by adding RAL at different times and measuring integrated viral DNA. The viral DNA released from PF74-disrupted capsids integrated into the host genome <2 hours after PF74 addition (Fig. 2E). Additional experiments using more frequent time points within the first 2 hours of PF74 addition indicated that the short HIV-3.1kb DNA integrates ~77 min after PF74 addition (Fig. 2F), which is consistent with our previous findings that the full-length viral DNA integrates <1.5 hours after uncoating of nuclear HIV-1 cores (10). We also determined the kinetics of integration of the ~2% of HIV-3.1kb viral DNAs that integrated without PF74 treatment (Fig. 2D); we found that integration of the HIV-3.1kb DNA in the absence of PF74 treatment was substantially delayed compared to that of HIV-8.4kb (Fig. 2E). Analysis of infected HeLa cells 1 to 4 days after infection indicated that most of the viral cores that fail to uncoat within the first 24 hours of infection remain stable for at least 4 days after infection (fig. S5; see Supplementary Text). Treatment of cells with PF74 or lenacapavir 10 hours after infection of HeLa cells or T cell lines with HIV-3.1kb led to a ~5.3- to 8.0-fold increase in the percentage of GFP reporter–expressing cells compared to that of untreated control cells (Fig. 2G and fig. S6; see Supplementary Text). Overall, these results indicated that the viral DNA that was released by disruption of intact cores with PF74 or lenacapavir treatment and survived degradation by host nucleases could integrate and express the GFP reporter.
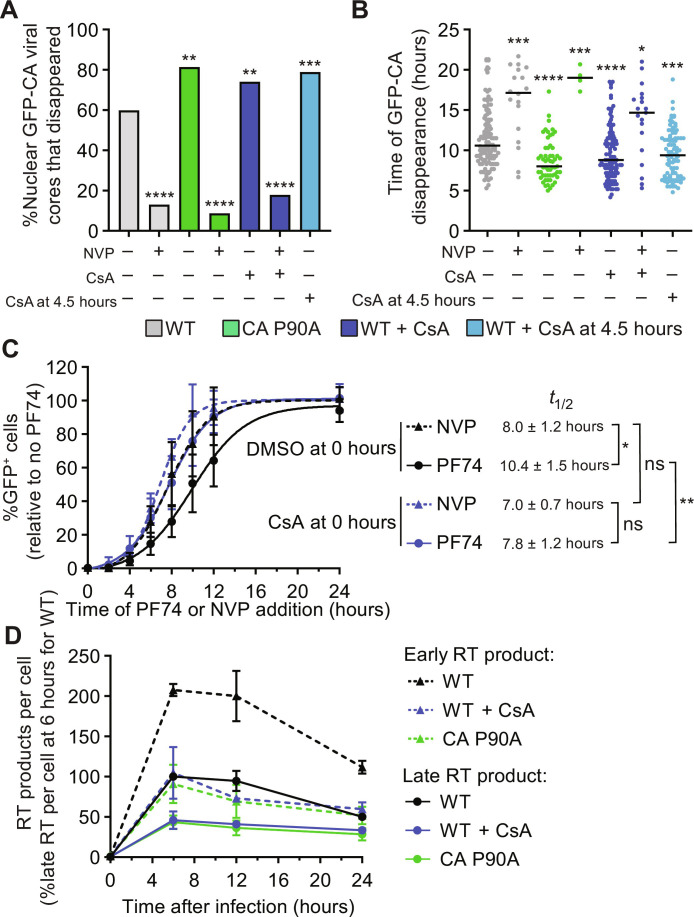
CypA binding to nuclear viral cores delays uncoating
Host factor cyclophilin A (CypA) directly binds to HIV-1 cores (35–38), protects them from TRIM5α restriction (39), and potentially stabilizes them (36, 40). However, the effect of CypA exerts on nuclear HIV-1 cores is not well understood [reviewed in (41)]. Inhibiting CypA-CA interactions using the CA mutation P90A (42) or treatment of target cells with cyclosporin A (CsA), a competitive inhibitor of the CypA-CA interaction, increased the uncoating efficiency of nuclear HIV-1 cores and resulted in faster uncoating kinetics (Fig. 3, A and B), indicating that CypA binding stabilizes viral cores and delays uncoating. CsA addition after HIV-1 cores entered the nucleus (~4.5 hours after infection) also resulted in increased uncoating efficiency and faster uncoating kinetics (Fig. 3, A and B), suggesting that CypA, or another CsA-sensitive cyclophilin, binds to nuclear HIV-1 cores. NVP treatment significantly decreased the uncoating efficiency of the P90A mutant cores or wild-type (WT) cores in CsA-treated cells (Fig. 3, A and B). These findings indicate that CypA-CA interactions influence uncoating efficiency and kinetics, but reverse transcription remains essential for efficient uncoating of nuclear HIV-1 cores. Consistent with this observation, the addition of the CsA 10 hours after infection of HeLa cells with HIV-3.1kb did not increase the percentage of GFP reporter–expressing cells (Fig. 2G). These results indicated that, unlike PF74 or lenacapavir treatment, CsA treatment was not sufficient to disrupt viral cores and release the short viral DNA that could express the GFP reporter.
Fig. 3. Effect of reverse transcription, CypA-CA interaction, and CA mutations on uncoating of nuclear HIV-1 cores.
(A) HeLa cells were challenged with GFP-CA–labeled virus and live-cell microscopy was performed. The percentage of nuclear HIV-1 cores that disappeared during the movies for the indicated virus and cell treatments is shown. An average of 101 HIV-1 cores was analyzed for sample (Fisher’s exact test). (B) Time (hours) after infection of GFP-CA disappearance (line = median; Mann-Whitney U test was used to compare statistical significance from WT HIV-1 cores in untreated cells). For (A) and (B), WT virus with and without NVP treatment is same as in Fig. 1 (B and C) and is shown as reference. (C) HeLa cells were infected with an HIV-1–based GFP reporter virus, cells were treated with CsA at the time of infection, NVP or PF74 was added at different time points after infection, and the percentage of GFP reporter–expressing cells was determined ~1 day after infection. Data are shown relative to infected cells not treated with PF74 (means ± SD, n = 4 biological replicates). The time of NVP or PF74 addition when the percentage of GFP reporter expressing cells was half of the percentage of GFP reporter expressing untreated or CsA-treated cells (t1/2) was determined by nonlinear curve fitting and compared using Welch’s t test. (D) Quantitation of early RT and late RT products for indicated viruses at different times after infection of aphidicolin-treated HeLa cells with p24-normalized virus input (means ± SD, n = 3 biological replicates). *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001.
Next, we assessed the timing of reverse transcription and uncoating by determining the loss of sensitivity to NVP and PF74, respectively, in the absence or presence of CsA (Fig. 3C). As we previously observed (10), the average loss of sensitivity to NVP occurred at ~8.0 hours after infection, whereas the average loss of sensitivity to PF74 occurred at ~10.4 hours after infection, indicating that uncoating occurred ~2.4 hours after the completion of reverse transcription. In contrast, the average loss of sensitivity to NVP and PF74 in CsA-treated cells occurred at similar times (~7.0 and ~7.8 hours after infection; P > 0.05). These observations indicate that, in the absence of CypA binding to viral cores, uncoating occurs at approximately the same time that reverse transcription is completed. Moreover, the average loss of sensitivity to PF74 in CsA-treated cells occurred ~2.6 hours earlier than in untreated control cells (~7.8 hours versus ~10.4 hours), which is consistent with the faster uncoating of nuclear HIV-1 cores when the CypA-CA interaction is inhibited (Fig. 3B). We also measured early RT and late RT products at different time points after infection and observed a ~2-fold decrease in viral DNA levels when the CypA-CA interaction was inhibited (Fig. 3D), which is consistent with a previous report showing that CypA promotes reverse transcription (43). Overall, these results suggest that CypA promotes reverse transcription by stabilizing nuclear HIV-1 cores to ensure that reverse transcription is completed before uncoating. It is important to note that although CypA has been shown to be localized to the nucleus (44, 45), there are several CsA-sensitive nuclear cyclophilins (46), and the identity of the cyclophilin that binds to nuclear viral cores and delays uncoating remains an open question at this time.
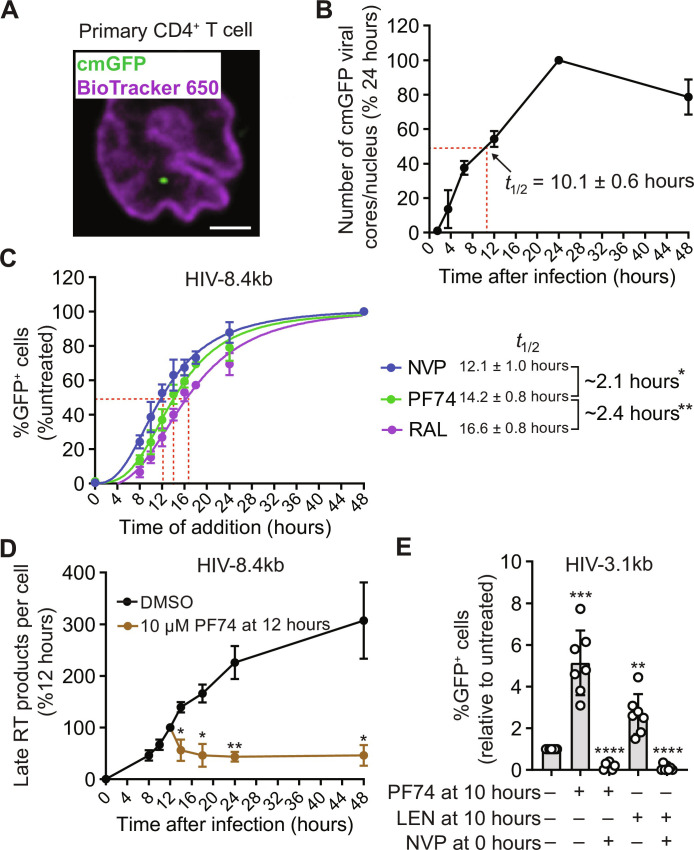
Viral cores with short vector DNA uncoat inefficiently in primary CD4+ T cells
Next, we performed imaging and time-of-addition experiments in primary CD4+ T cells, the natural target cells of HIV-1 infection, to determine the kinetics of nuclear import, reverse transcription, uncoating, and integration. We labeled viral cores with GFP that is passively trapped inside intact cores during virion maturation [content marker GFP (cmGFP)]. The cmGFP is released from the viral cores when core integrity is lost and serves as a marker for intact viral cores (fig. S7) (11). Nuclear import of viral cores labeled with cmGFP occurred ~10.1 hours after infection (Fig. 4, A and B). In the infected CD4+ T cells, HIV-8.4kb GFP reporter viruses remained sensitive to NVP, PF74, and RAL until ~12.1, ~14.2, and ~16.6 hours after infection, respectively (Fig. 4C). In HeLa cells, the loss of sensitivity to NVP occurred 8.0 hours after infection (Fig. 3C), indicating that reverse transcription was delayed in the CD4+ T cells compared to that in the HeLa cells. Despite the delay in reverse transcription, loss of sensitivity to PF74 occurred ~2.1 hours after loss of sensitivity to NVP, indicating that uncoating occurred ~2 hours after completion of reverse transcription. Furthermore, integration occurred ~2.4 hours after loss of sensitivity to PF74. These results, which are consistent with our previous findings in HeLa cells (10), imply that, in CD4+ T cells, reverse transcription is completed within the nucleus ~12 hours after infection, uncoating occurs ~2 hours after completion of reverse transcription, and integration occurs ~2 to 2.5 hours after uncoating. Infection of CD4+ T cells with the HIV-8.4kb vector and treatment with PF74 12 hours later resulted in a rapid ~50% decrease in viral DNA levels (Fig. 4D), indicating that HIV-1 cores protect viral DNA from degradation in the CD4+ T cells. Moreover, infection of CD4+ T cells with HIV-3.1kb and treatment ~10 hours later with PF74 or lenacapavir resulted in a ~2.7- to 5.1-fold increase in the percentage of GFP reporter–expressing cells compared to that of untreated control cells (Fig. 4E). These findings demonstrate that, in primary CD4+ T cells, reverse transcription of the short HIV-3.1kb DNA does not induce efficient uncoating, that short viral DNA can be released upon capsid inhibitor-induced disruption of viral cores, and that intact HIV-1 capsids safeguard the viral DNA from degradation.
Fig. 4. Kinetics of nuclear import, reverse transcription, uncoating, and integration in primary activated CD4+ T cells, and effect of CA inhibitor-induced disruption of HIV-1 cores on viral DNA levels and expression of GFP reporter.
(A) HIV-1 core labeled with content marker GFP (cmGFP) in the nucleus of a primary activated CD4+ T cell 12 hours after infection; nucleus stained using BioTracker 650 Dye. Scale bar, 2 μm. (B) Number of nuclear cmGFP-labeled HIV-1 cores per cell at the indicated time points (means ± SD, n = 3 biological replicates, two donors). An average of 2.6 nuclear HIV-1 cores per cell was detected 24 hours after infection (set to 100%). (C) Percentage of GFP-expressing cells measured 48 hours after infection after addition of NVP, PF74, and RAL at different time points (means ± SD, n = 4 biological replicates, four donors). (D) Quantitation of late RT products at different times after infection with HIV-8.4kb (means ± SD, Welch’s t test, n = 4 biological replicates, four donors). An average of 0.4 late RT products per cell was detected 12 hours after infection (set to 100%). (E) Fold change in the percentage of GFP reporter–expressing cells measured 1 day after infection with HIV-3.1kb; data shown relative to untreated control (set to 1; means ± SD, Welch’s t test, n = 7 biological replicates). *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001.
HIV-1 NC condenses viral DNA
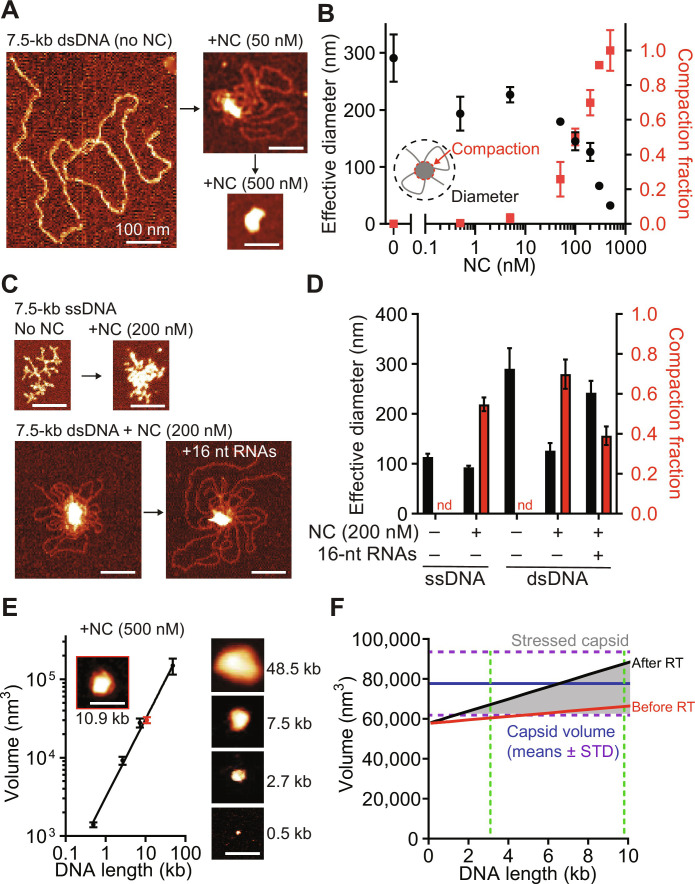
Reverse transcription of flexible single-stranded RNA into a stiff dsDNA that resists compression has been proposed to increase internal pressure within the core that is counteracted by HIV-1 nucleocapsid (NC)–mediated dsDNA condensation (28, 47, 48). Previous studies, however, have not measured condensation as a function of dsDNA length, which increases as reverse transcription progresses or as genome length is altered as in the experiments described above. They also did not consider the conformations of single-stranded DNA (ssDNA) regions transiently formed during reverse transcription, the interplay between the degraded RNA template and the full length proviral dsDNA product, and the relative geometric proportions of these molecules in comparison with the capsid, as described here. Uncoating is proposed to occur when the amount of NC is insufficient to fully condense the newly synthesized dsDNA. However, testing the role of NC in cell-based assays is complicated by the fact that NC plays a critical role in genomic RNA packaging (49) and in facilitating reverse transcription [reviewed in (50)]. Thus, mutations in NC may lead to a decrease or increase in uncoating if the net effect of the mutations is inhibition of reverse transcription or inhibition of dsDNA condensation, respectively. Because the results of cell-based assays may be difficult to interpret, we opted to use in vitro assays to study the role of NC in HIV-1 uncoating. We measured the size of NC-dsDNA complexes by atomic force microscopy (AFM) at increasing levels of dsDNA saturation with NC (Fig. 5, A and B). At 50 nM NC, the dsDNA is only partially condensed in a large central globule surrounded by a single layer of DNA, whereas, at 500 nM NC, all the DNA is fully condensed into a single globule. These results indicate that dsDNA in viral cores, in which NC concentration is estimated to be ~40 mM (see Materials and Methods), should be completely condensed unless the amount of NC available for binding to dsDNA is limiting. During reverse transcription, reverse transcriptase switches templates between the two co-packaged genomic RNA strands, resulting in synthesis of one dsDNA molecule (51). As a result, RNA fragments equivalent to at least one full genome (and perhaps more) would remain in the core and bind to NC. Adding equimolar amounts of 16-nucleotide (nt) RNA oligonucleotides, which represent the single-stranded RNA fragments that remain in the capsid after reverse transcription (52), reduced dsDNA condensation (Fig. 5, C and D). Although the amount of NC in the capsid is sufficient to compact full-length genomic RNAs or dsDNA, it may not be sufficient to compact the amount of nucleic acid in the capsid at the end of reverse transcription, which increases as much as ~150 to 200%. Consequently, the full-length dsDNA would not be fully condensed, increasing the internal pressure on the viral core, leading to uncoating. HIV-1 plus-strand DNA synthesis occurs in multiple segments that are separated by gaps (53, 54). The ssDNA regions increase local flexibility by two orders of magnitude [dsDNA is rigid at lengths up to 50 nm, while ssDNA bends at sub-nanometer lengths; (55)] and vastly decrease the forces required to keep the DNA contained in the capsid (fig. S8A). Thus, reverse transcription may need to progress to a stage where long stretches of dsDNA are synthesized to increase the internal pressure on the core and induce uncoating.
Fig. 5. AFM measurement of nucleic acid compaction by HIV-1 NC and modeling of capsid contents.
(A) Atomic force microscopy (AFM) images of 7.5-kb dsDNA incubated with or without NC (same lateral scale). (B) Diameter of dsDNA and fraction of dsDNA in central globule after incubation of 7.5-kb dsDNA with different NC concentrations (means ± SEM, n = 3 biological replicates). (C) AFM images of ssDNA with or without NC (top) and dsDNA with or without equimolar free 16-nt RNA oligos before NC incubation (bottom). (D) Diameter and fraction of DNA in central globule for the indicated conditions; nd, not determined (means ± SEM, n = 3 biological replicates). (E) Minimal bounding volume (left) and AFM images (right) of NC-condensed dsDNA of different lengths (means ± SEM, n = 3 biological replicates). NC-condensed HIV-1 DNA (red). (F) Total volume of capsid contents (nucleic acid plus protein) as a function of packaged viral RNA length before (red) and after RT [black, partial (33%) dsDNA condensation by a conserved quantity of NC present in capsid]; dotted green lines indicate sizes of HIV-3.1kb and full-length HIV-1; internal capsid volumes (blue; mean + SD) are plotted in comparison; stressed capsids (gray; volume of capsid contents > internal capsid volume). See "HIV-1 NC condenses viral DNA" for details of volume measurements and estimates. Scale bars, 100 nm.
Our measurements show that the volumes of completely condensed dsDNA/NC globule with dsDNA length ranging between 0.5 and 48.5 kb increase proportionally to dsDNA length (Fig. 5E and fig. S8, B and C). As expected (56–58), NC did not show a noteworthy sequence preference and equally condensed different dsDNA substrates, including HIV-1 DNA. Assuming the total viral RNA and dsDNA packaged into the capsid increase in proportion with genome length and the major proteins in the capsid (NC, RT, IN, and Vpr; fig. S8D) remain constant, we estimated the entire nucleic acid and protein contents of the capsid as a function of genome length (details of the model are described in Materials and Methods). Before reverse transcription, the packaged viral RNA and proteins are predicted to occupy ~75% of the total capsid volume (Fig. 5F and fig. S9). Completely condensed full-length viral dsDNA could fit inside the remaining capsid space. However, there is insufficient NC to condense the full-length HIV-1 dsDNA (~9.8 kb), and the volume of the dsDNA exceeds the available space in the capsid (Fig. 5F); as a result, a large fraction of capsids of different sizes experience strain following reverse transcription, which leads to uncoating. These predictions are consistent with our findings that the efficiency of uncoating increases with genome length and that short genomes ≤3.5 kb do not induce efficient uncoating (Fig. 1F).
DISCUSSION
Our results show that reverse transcription is required for uncoating, that uncoating occurs ~10 hours after reverse transcription initiation, and that synthesis of long double-stranded reverse transcription products is required for efficient uncoating. Some previous studies showed that inhibiting reverse transcription with NVP inhibited uncoating of cytoplasmic (24) as well as nuclear viral cores (10, 27), but the temporal link between reverse transcription initiation and uncoating was not established and other studies concluded that uncoating occurred soon after initiation of reverse transcription (5, 6, 59). Most previous studies did not examine the uncoating of nuclear viral cores, which are likely to be infectious, and biochemical assays for uncoating measure the behavior of a population of viral cores, most of which were defective and did not lead to productive infection. Here, we used two separate assays to study uncoating. The first assay used a live-cell imaging assay to directly visualize the uncoating of nuclear HIV-1 cores, and the second assay used time-of-addition assays with an inhibitor of reverse transcriptase (NVP) and capsid binding inhibitor (PF74) to determine the kinetics of reverse transcription and uncoating of infectious viral cores, respectively.
Our studies of viral cores containing different-sized genomes show that synthesis of long double-stranded reverse transcription products > 3.5 kb is required for efficient HIV-1 uncoating. Furthermore, the genome size positively correlated with uncoating efficiency. These observations suggest that synthesis of dsDNA, which is less flexible than genomic RNA and resists compression, exerts internal pressure on the capsid, resulting in uncoating. AFM measurements showed that NC can condense dsDNA to reduce the pressure on the viral capsid and ensure completion of reverse transcription. The size of the NC-condensed dsDNA and estimation of viral core protein and nucleic acid contents reveal that synthesis of full-length reverse transcription products induces substantial internal strain on the cores to promote uncoating. We also show that the CypA-CA interactions can stabilize the viral cores, reducing uncoating efficiency and delaying the kinetics of uncoating of nuclear viral cores. Reverse transcription was still required to induce uncoating of viral cores not bound to CypA, indicating that synthesis of long dsDNA products remains an essential requirement for uncoating. On the basis of these findings, we propose a model in which HIV-1 uses at least two different mechanisms to ensure that reverse transcription is completed before uncoating. First, NC-mediated condensation of viral dsDNA regulates the internal pressure exerted by the viral dsDNA genome to ensure that reverse transcription is completed inside the intact capsid until sufficient internal pressure is exerted to induce uncoating. Second, CypA binding stabilizes nuclear HIV-1 cores to ensure that reverse transcription does not induce premature uncoating (fig. S9). The completion of reverse transcription before uncoating provides several advantages for the virus, including evading host nucleases and avoiding detection by innate DNA sensors (8, 33), ensuring efficient reverse transcription [this study and (43)], and capsid-mediated delivery of the viral preintegration complex to gene-rich speckle-associated chromatin domains, the preferred sites of HIV-1 integration (16–18).
Although our studies indicate that synthesis of long double-stranded reverse transcription products is required for uncoating of nuclear viral cores, the observed ~2.1-hour delay between reverse transcription completion and uncoating in primary CD4+ T cells (Fig. 4C) suggests that additional steps after DNA synthesis involving viral and/or host factors are needed to trigger HIV-1 uncoating. Alternatively, internal strain imposed by long dsDNA may gradually weaken the viral core structural integrity and induce uncoating. We observed that the ~2.4-hour delay between reverse transcription completion and uncoating in HeLa cells is abolished when the CypA-CA interaction is inhibited, which is consistent with the hypothesis that CypA or another CsA-sensitive nuclear cyclophilin binds to nuclear HIV-1 cores and delays uncoating to ensure that reverse transcription is completed before capsid disassembly. The role of CypA in post-nuclear entry steps of HIV-1 replication is not well understood [reviewed in (41)]. The inhibition of the CypA-CA interaction has been shown to alter proviral integration site preference (60), suggesting a role for CypA in the nucleus of infected cells. In addition, low levels of a fluorescently tagged oligomeric form of CypA (CypA-DsRed), which binds to capsid more efficiently than endogenous CypA, were found to be associated with nuclear viral complexes (61); however, the levels of endogenous CypA or other cyclophilins binding to nuclear viral cores is not known. Although CypA is not required for efficient in vitro endogenous reverse transcription reactions (19), the IP6 levels in the reactions may stabilize cores and overcome any potential requirement for CypA. Previous studies have shown that CypA binding protects the viral cores from host restriction factor TRIM5α (39), promotes reverse transcription (43), and delays nuclear import (62). On the basis of the observations in this study, we propose that CypA-CA interactions increase capsid stability to ensure that reverse transcription is completed before uncoating.
It is not known whether uncoating involves complete or substantial loss of the capsid lattice or whether large fragments of capsid remain associated with the viral preintegration complex. We previously observed a rapid loss of viral core-associated GFP-CA signal below the limit of detection ~1 to 3 min after loss of capsid integrity (10, 11). The uncoating kinetics of GFP-CA–labeled viral cores measured by imaging (Fig. 1C) and unlabeled viral cores measured by PF74 time-of-addition assay (Figs. 1D and 3C) were nearly identical, indicating that the GFP-CA label does not affect uncoating kinetics. Loss of NC from viral cores after rupture of the capsid lattice could further accelerate decondensation of viral DNA (47), suggesting a rapid uncoating event. However, partially disassembled capsids were observed after an endogenous reverse transcription reaction (19), and release of viral DNA from virus core-like structures was observed in the nuclei of infected cells (14). Another study observed varying degrees of capsid disassembly after endogenous reverse transcription, ranging from no fractures to large fractures with large missing pieces of the capsid lattice (63). The degree to which the capsid ruptures and disassembles likely depends on both IP6 levels and host factor binding. Additional studies involving CypA and IP6 are needed to fully elucidate the role of these factors in stabilizing the capsid during reverse transcription and the dynamics of capsid disassembly.
Last, the uncoating and integration kinetics in primary CD4+ T cells determined by using time-of-addition assays with PF74 and RAL, respectively, indicate that there is likely a short window (<2.4 hours) for the released viral DNA to interact with nuclear host factors like LEDGF/p75 (64) to facilitate the formation of a competent preintegration complex and ensure proper integration targeting.
In summary, our findings support a model in which HIV-1 capsid evolved to remain intact until reverse transcription is completed, protect the viral DNA from degradation by host factors, shield the viral DNA from detection by innate immune sensors, and deliver the viral DNA to its preferred genomic sites for integration (60, 65).
MATERIALS AND METHODS
Cells and reagents
HeLa [American Type Culture Collection (ATCC) CCL-2], HeLa-based cell lines (described below), and human embryonic kidney 293T cells (ATCC CRL-3216) were maintained in Dulbecco’s modified Eagle’s medium supplemented with 10% fetal calf serum (FCS; HyClone, Logan, UT) and 1% penicillin-streptomycin (50 U and 50 μg/ml, respectively; Lonza, Walkersville, MD). CEM-SS cells [NIH HIV Reagent Program, Division of AIDS, National Institute of Allergy and Infectious Diseases (NIAID), National Institutes of Health (NIH); contributed by P. L. Nara; ARP-776) and Jurkat cells (clone E6-1; ATCC TIB-152) were maintained in RPMI 1640 medium (CellGro, Manassas, VA) supplemented with 10% FCS and 1% penicillin-streptomycin. Peripheral blood mononuclear cells (PBMCs) were isolated from the blood of anonymous, healthy donors using Ficoll-Paque Plus (Cytiva, 17-1440-02). PBMCs were activated using Dynabeads Human T-activator CD3/CD28 (Gibco, 11161D) for ~3 to 5 days, and CD4+ T cells were isolated using the Dynabeads CD4 Positive Isolation Kit (Invitrogen, 11331D). The purity (>99%) and activation status (>70%) of isolated CD4+ T cells were assessed by flow cytometry after staining with anti–CD4–fluorescein isothiocyanate (BD Pharmingen, 555346) and anti–CD25-phycoerythrin (BD Pharmingen; 555432) antibodies, respectively. PBMCs and CD4+ T cells were maintained in RPMI 1640 supplemented with 10% FCS, 1% penicillin-streptomycin, and interleukin-2 (50 to 100 U/ml; Sigma-Aldrich). All cells were maintained in a humidified incubator at 37°C with 5% CO2.
A HeLa-based cell line that constitutively expresses mRuby-LaminB fusion protein [HeLa:mRuby-LaminB cells; (11)] was primarily used for live-cell imaging. However, other HeLa-based cell lines were occasionally used for live-cell imaging (described below). The HeLa:mRuby-LaminB(sin) cell line, which also constitutively expresses mRuby-LaminB fusion protein, was generated by transduction of HeLa cells with pLVXsin-CMV-mRuby-LaminB-P2A-Puro, a bicistronic lentiviral vector that contains a partial deletion in the U3 of the 3′ long terminal repeat and expresses an mRuby-LaminB fusion protein and puromycin resistance gene under the control of the cytomegalovirus (CMV) promoter. mRuby-LaminB–expressing cells were selected for resistance to puromycin (1 μg/ml). The HeLa-Bgl cell line expresses truncated bacterial protein BglG that is fused to mCherry at the C terminus and contains a nuclear localization signal (Bgl-mCherry) from a doxycycline-inducible promoter and was previously described (10).
The HeLa-Bgl:Tat-Rev cell line was produced by transduction of HeLa-Bgl cells with a murine leukemia virus–based vector that expresses codon-optimized HIV-1 Tat and Rev separated by an in-frame self-cleaving peptide from porcine teschovirus 2A (P2A) upstream of an internal ribosome entry site, hygromycin resistance gene, and was previously described (10). In this study, the HeLa-Bgl:Tat-Rev is simply referred to as HeLa:Tat-Rev because the expression of Bgl-mCherry was not induced with doxycycline for the infectivity experiments.
NVP and RAL were obtained through the NIH AIDS Reagent Program and were used at final concentrations of 5 and 10 μM, respectively. PF74 (Sigma-Aldrich) and lenacapavir (MedChemExpress) were used at a final concentration of 10 and 10 nM, respectively, unless indicated otherwise. CsA (CalBioChem) was used at a final concentration of 5 μM. Aphidicolin (MilliporeSigma) was used at a final concentration of 10 μg/ml. Nuclei were stained with 4′,6-diamidino-2-phenylindole (DAPI; MilliporeSigma) for fixed-cell assays or BioTracker 650 Red Nuclear Dye (Millipore) for live-cell imaging assays.
Vectors
The HIV-1–based vectors pHGFP, which contains a gfp reporter gene in place of nef and does not express env, and pHGFP-GFPCA, which is similar to pHGFP except that GFP was inserted between MA and CA and proteolytic processing generates a GFP-CA fusion protein during virus maturation, were previously described (10). CA mutant P90A was generated by site-directed mutagenesis of CA in pHGFP and pHGFP-CA (QuikChange II Site-directed mutagenesis kit; Agilent), resulting in pHGFP(P90A) and pHGFP-GFPCA(P90A), respectively. RT mutant D110E was generated by swapping a 653–base pair (bp) fragment containing a portion of the pol by digestion of pHDVEGFP-D110E (62) with Sbf I and Age I into pHGFP and pHGFP-GFPCA, generating pHGFP(D110E) and pHGFP-GFPCA(D110E), respectively. Integrase mutant D64E was generated by cloning a synthesized (Genewiz) 723-bp fragment containing a portion of pol into pHGFP and pHGFP-GFPCA, generating pHGFP(D64E) and pHGFP-GFPCA(D64E), respectively. HIV Gag-iGFP ΔEnv vector (NIH AIDS Reagent Program, Division of AIDS, NIAID, NIH, from B. Chen; catalog no. 12455) is an HIV-1–based vector that contains an internal GFP (iGFP) between MA and CA and, after proteolytic processing, expresses free GFP and CA (11). pcHELP is an HIV-1 helper construct that lacks a packaging signal and primer-binding site; the helper construct expresses all the viral proteins except Nef and Env (66). To generate pcHELP-GFPCA, a helper construct that expresses GFP-CA fusion protein during virion maturation, a 2885-bp fragment digested from pHGFP-GFPCA using Bss HII and Sbf I was cloned into pcHELP.
A frameshift mutation in gag that results in a premature stop codon was introduced in pHGFP-GFPCA by Sph I digestion and Klenow fill-in, generating an HIV-1–based vector (HIV-GFP-GFP) that contains gfp in the 5′ end of the viral genome (gfp in between MA and CA) and gfp in the 3′ end of the viral genome (gfp in nef orf). To generate HIV-11.1kb, the gfp in the nef open reading frame was replaced with mNeonGreen (mNG). HIV-2.7kb was generated by digestion of HIV-GFP-GFP with Pfo I (cuts within gfp) and removal of the sequence between the 5′ gfp and 3′ gfp and ligation of the vector backbone. HIV-3.1kb, which expresses a matrix (MA)–GFP fusion protein and codon-optimized Tat separated by an in-frame self-cleaving P2A peptide, was generated by the synthesis of a gBlock (Integrated DNA Technologies) containing P2A-Tat sequence and cloning of the fragment into HIV-2.7kb using Xho I. HIV-8.4kb was generated by introducing a frameshift mutation leading to a premature stop codon in gag by Sph I digestion and Klenow fill-in of HDV-EGFP (67), an HIV-1–based vector that expresses a gfp reporter in the nef open reading frame and contains deletions in vpr and vif. pHCMV-G (68) expresses the VSV-G protein.
The different-sized lentiviral vectors were generated by deleting or adding various non–HIV-1, nonfunctional sequences. HIV-2.8kb, a small lentiviral vector that contains essential elements for packaging genomic RNA into virus particles and reverse transcription, including the packaging signal, rev-response element, central polypurine tract, and the polypurine tract, was generated by digestion of the lentiviral vector CMV13p>eGFP (Protein Expression Laboratory at the National Cancer Institute-Frederick) with Eco RI and Cla I and removal of a fragment containing the CMV promoter-gfp–woodchuck hepatitis virus posttranscriptional regulatory element (WPRE), blunting the ends with mung bean nuclease [New England Biolabs (NEB)], and ligation of the vector backbone. To generate intermediate vectors HIV-6.7kb and HIV-8.2kb, a 1520- or 2995-bp fragment of lacZ from XLong-BSL (69) (lacZ does not contain start codon and is not expressed), respectively, was cloned into a lentiviral vector that contains a CMV promoter-gfp-WPRE cassette. HIV-3.5kb was generated by removal of a 3181-bp fragment (lacZ-CMVpromoter-gfp) from HIV-6.7kb by digestion with Psp OMI and Xho I, blunting the ends with the Quick Blunting Kit (NEB), and ligation of the vector backbone. HIV-5.0kb was generated by removal of a 1636-bp fragment (CMVpromoter-gfp) from HIV-6.7kb by digestion with Eco RI and Xho I, blunting the ends with the Quick Blunting Kit (NEB), and ligation of the vector backbone. HIV-6.5kb was generated by the removal of a 1711-bp fragment (CMV promoter-GFP) from HIV-8.2kb by digestion with Eco RI and Xho I, blunting of ends with Quick Blunting Kit (NEB), and ligation of the vector backbone. HIV-8.1kb was generated by cloning a 3071-bp fragment of the ouabain-resistance gene from XLong-BSL into vector named Protein Expression Lab 5.0kb (PEL 5.0kb) by digestion with Xba I (ouabain resistance gene did not contain a start codon and was cloned in reverse orientation). The vector size in the name indicates the expected length (kilobase) of the DNA genome after plus-strand transfer of the R-U5 to the 5′ end and completion of reverse transcription. All constructs were verified by sequencing.
Virus production and infection
Infectious virions labeled with GFP-CA were prepared by co-transfection of 293T cells with pHGFP and pHGFP-GFPCA (or corresponding plasmids expressing RT, IN, or CA mutants) at a 1:10 plasmid ratio and pHCMV-G. Virions labeled with GFP-CA that contain different-sized lentiviral genomes (HIV-2.8kb to HIV-8.1kb), genomes for studying large and small vectors (HIV-11.1kb and HIV-2.7kb), or HIV-3.1kb and HIV-8.4kb, were prepared by co-transfection of 293T cells with pCHELP-GFPCA and pCHELP at a 1:10 plasmid ratio (total of 3 μg of DNA), lentiviral genome (up to 7 μg), and pHCMV-G. For some experiments, unlabeled viruses were generated by omitting pCHELP-GFPCA. Equal molar concentrations of plasmids expressing lentiviral genome were transfected, and pGEM-3Zf (Promega; up to 7 μg) was used to normalize plasmid DNA input for transfection. Virions containing no viral genome were prepared by co-transfection of 293T cells with pCHELP-GFPCA and pCHELP at a 1:10 plasmid ratio (total of 3 μg of DNA), pGEM-3Zf (7 μg), and pHCMV-G. Infectious virions that were labeled with the GFP content marker (cmGFP) were prepared by co-transfection of 293T cells with Gag-iGFP ΔEnv and pHGFP at a 1:2 plasmid ratio and pHCMV-G. Supernatants from the transfected 293T cells were collected ~24 hours later and filtered, and the HIV-1 particles were concentrated by ultracentrifugation (100,000g) for 1.5 hours at 4°C through a 20% sucrose cushion (w/v) in 1× phosphate-buffered saline. For some experiments, virus levels were determined by HIV-1 p24 ELISA assay (XpressBio).
For live-cell and fixed-cell imaging experiments (described below), HeLa and HeLa-based cell lines were seeded in ibiTreated μ-slides (4 × 104 cells per well; ibidi) 1 day before infection. For measuring RT products or infectivity, HeLa cells were seeded in 48-well plates (3 × 104 cells per well) 1 day before infection. For live-cell imaging experiments, HeLa cell lines were challenged with GFP-CA–labeled viruses at a low multiplicity of infection (~1 nuclear particle per cell) with polybrene (10 μg/ml; Sigma-Aldrich) via spinoculation (1200g, 1 hour) at 15°C, which permitted virion binding to cell membranes but prevented virion endocytosis (70). For fixed-cell imaging experiments, the number of virus particles in each virus preparation was determined by single virion analysis, and an equal number of GFP-labeled virus particles was used for infections. After spinoculation, the medium was replaced with prewarmed medium to allow internalization of the virus (defined as the 0-hour time point). For live-cell imaging experiments, aphidicolin (10 μg/ml) was also added at the time of infection to prevent cell division during long movies. The infected cells were maintained at 37°C in a humified incubator. Time-lapse images of the infected cells were acquired by spinning disk confocal microscopy (described below). For NVP washout experiments, 5 μM NVP was added immediately after spinoculation and removed 2 hours later by washing cells three times with complete medium (3 min per wash). For time-of-addition experiments, medium was replaced with medium containing NVP, RAL, or PF74 at different times after infection. Cells were collected at indicated time points (1 to 4 days after infection) by Trypsin (Gibco) or TrypLE (Gibco), and virus infectivity was determined by measuring the percentage of GFP reporter–expressing cells by flow cytometry (LSRFortessa; BD Biosciences).
For live-cell imaging of activated CD4+ T cells (1 × 105 cells per well), VSV-G-pseudotyped HIV-1 labeled with cmGFP, polybrene (5 μg/ml), aphidicolin (10 μg/ml; Sigma-Aldrich), and BioTracker 650 Red Nuclear Dye (1 μl/ml; Sigma-Aldrich) were briefly mixed and centrifuged onto ibiTreated μ-slides (1200g, 1 hour, 15°C). The amount of virus used resulted in ~2 to 3 nuclear cmGFP particles per cell at the 24-hour time point. For measuring RT products or infectivity, activated CD4+ T cells (1 × 105 cells per well), virus (10 ng p24), polybrene (5 μg/ml), and aphidicolin (10 μg/ml) were briefly mixed and centrifuged onto ibiTreated μ-slides (1200g, 1 hour, 15°C). For time-of-addition experiments, complete medium containing 2× NVP, PF74, and RAL (final concentration of 5, 10, and 10 μM, respectively) was added at various times after infection, and the percentage of GFP reporter–expressing cells was determined 2 days after infection by flow cytometry. To measure GFP reporter expression after disruption of HIV-1 cores with PF74 or LEN, activated CD4+ T cells were infected with HIV-3.1kb (10 ng p24), complete medium containing 2× PF74 or LEN (final concentration of 10 and 10 nM, respectively) was added 10 hours after infection, and the percentage of GFP reporter–expressing cells was determined 1 day after infection by flow cytometry.
Cell fractionation and Western blots
HeLa cells were collected using trypsin or TrypLE at the indicated time point and the cytoplasm and nuclear fractions were obtained using NE-PER Nuclear and Cytoplasmic Extraction Reagents (Thermo Fisher Scientific). Viral and cellular DNA was extracted from the cytoplasmic and nuclear fractions using the QIAamp Blood Mini Kit (Qiagen). To determine the quality of the fractionation, the fractions were subjected to SDS–polyacrylamide gel electrophoresis and Western blot analysis using rabbit anti-Lamin A/C antibody (Sigma-Aldrich) and mouse anti–α-tubulin antibody (Sigma-Aldrich) followed by goat anti-rabbit antibody (IRDye-800CW; LI-COR) and goat anti-mouse antibody (IRDye-680RD; LI-COR), respectively. Western blots were imaged and quantitated using the Odyssey infrared imaging system (LI-COR).
Detection of reverse transcription products and integrated viral DNA
Virus stocks were treated with deoxyribonuclease I (Invitrogen) for 1 hour at 37°C to degrade contaminating plasmid DNA. Viral and cellular DNA was extracted from HeLa cells (collected by trypsin) or activated primary CD4+ T cells (collected by gently pipetting cells from the well) using the QIAamp Blood Mini Kit (Qiagen). Viral and cellular DNA was detected by quantitative polymerase chain reaction (PCR) using LightCycler 480 Probes Master (Roche), the LightCycler 480 Real-Time PCR System (Roche), and the following primer/probe sets (71): early RT products (RU5: forward primer hRU5-F2 5′-GCCTCAATAAAGCTTGCCTTGA-3′ and reverse primer hRU5-R 5′-TGACTAAAAGGGTCTGAGGGATCT-3′), late RT products (U5Ψ: forward primer MH531 5′-TGTGTGCCCGTCTGTTGTGT-3′ and reverse primer MH532 (5′-GAGTCCTGCGTCGAGAGATC-3′), and probe P-HUS-SS1 (5′-FAM-TAGTGTGTGCCCGTCTGTTGTGTGAC-IOWA BLK FQ-3′). To detect late RT products for some lentiviral vectors (HIV-2.8kb to HIV8.1kb), the reverse primer MH532mod (5′-CGCTTCAGCAAGCCGAGTC-3′) was used instead of MH532. Host CCR5, which was used to determine the cell count (assumed two copies per cell), was detected using CCR5-For (5′-CCAGAAGAGCTGAGACATCCG-3′, CCR5-Rev (5′-GCCAAGCAGCTGAGAGGTTACT-3′), and P-CCR5-01 (5-FAM-TCCCCTACAAGAAACTCTCCCCGG-IOWA BLK FQ-3′). To detect integrated viral DNA, the infected cells were allowed to divide for 7 days following infection (to dilute unintegrated DNA), collected, and proviruses were detected using the late RT primer and probe set.
Microscopy and image processing
Confocal images were acquired using a Nikon Eclipse Ti-E microscope equipped with a Yokogawa CSU-X1 spinning disk unit and a Plan-Apochromat 100× numerical aperture (NA) 1.49 oil objective, using 405-nm (DAPI), 488-nm (GFP), 561-nm (mRuby), and 647-nm (Cy5 dye) lasers for illumination. Images were captured using a TwinCam system (Cairn) equipped with a 565-nm splitter and two iXon Ultra (Andor) cameras. For some experiments, confocal images were acquired using a Nikon Eclipse Ti-E microscope equipped with a Yokogawa CSU-W1 spinning disk unit and a Plan-Apochromat 100× NA 1.49 oil objective, using 405-nm (DAPI), 488-nm (GFP), 561-nm (mRuby), and 647-nm (Cy5 dye) lasers for illumination. Images were captured using a 565-nm splitter and two ORCA-fusion BT cameras (Hamamatsu). A Tokai Hit microscope stage top incubator (Tokai) was used for all live-cell imaging experiments. Time-lapse images of the infected cells (described below) were examined using Nikon Elements or ImageJ (72). For display, a pixel-averaging filter was applied to the images and the contrast was adjusted.
HeLa cells were infected with GFP-CA–labeled virions and time-lapse images of HeLa cells starting ~4 hours after infection were acquired every 20 min for 18 hours using spinning disk confocal microscopy (13 slices with 0.4-μm step size). The GFP-CA spots were manually tracked, and the time of disappearance was recorded. Primary activated CD4+ T cells were infected with virus labeled with cmGFP and imaged (31 slices with 0.4-μm step size) at various time points after infection. The number of cmGFP spots per nucleus (identified using BioTracker 650 Red Nuclear Dye) was manually determined.
To determine the nuclear import efficiency of GFP-labeled virus particles in fixed HeLa cells, the number of fluorescently labeled virus particles for each virus preparation was first quantified. GFP-labeled virus particles were centrifuged onto ibiTreated μ-slides (1200g for 1 hour) and then immediately imaged by spinning disk confocal microscopy. The diffraction-limited spots were detected in each image using Localize (73) and quantified using a custom MATLAB program (10). A custom MATLAB program (10) was used to determine the percentage of total GFP signals inside the nucleus of infected HeLa cells 6 hours after infection. First, the GFP signals were detected using Localize (73). Colocalization of the fluorescently labeled virus particles with a mask of the nucleus interior (based on immunofluorescence staining using an anti-Lamin A/C antibody; Sigma-Aldrich) followed by detection using a Cy5-labeled secondary antibody (Thermo Fisher Scientific) or DAPI staining was determined using a custom-written MATLAB program as previously described (10). The percentage of GFP spots that colocalized with the nucleus mask was determined.
Atomic force microscopy
Double-stranded phage vector M13mp18 (NEB) was linearized by digestion with restriction enzyme Bam HI (NEB) and diluted to a concentration of 0.5 ng/μl (~100 pM) in a buffer containing 150 mM NaCl, 10 μM spermidine, and 10 mM Hepes (pH 7.5). Recombinant WT NL4-3 HIV-1 NCp7, a 55–amino acid protein, was produced and purified as previously described (74, 75). The DNA was incubated with varying concentrations of NC at 37°C for 1 min (longer incubation times leads to aggregation of DNA substrates) in a volume of 5 μl and then deposited on a freshly cleaved mica surface. The sample was rinsed with deionized water, blown dry under a stream of argon gas, and then imaged by a MultiMode 8 AFM and Nanoscope V controller (Bruker) using peak force tapping mode. Images were analyzed using Gwyddion software with low and high height thresholds (0.25 and 1 nm) used to differentiate the DNA from background and NC condensate from free DNA, respectively. For each individual DNA molecule, the effective diameter was calculated using its maximum span along both the x and y axes, and the compaction fraction was calculated by the ratio of the integrated volume of the NC condensate to the volume of the entire DNA molecule. For RNA competitor experiments, a 16 nt (UCAGUCAGUCAGUCAG) RNA oligo (Integrated DNA Technologies) was mixed with the DNA at an equimolar ratio before incubation with NC. A single-stranded form of M13mp18 was used in place of the dsDNA for ssDNA experiments. For experiments on different length substrates, 48.5–kilo–base pair (kbp) lambda phage DNA (Roche), 2.7-kbp linearized pUC19 (NEB), and a 500-bp PCR product from pUC19 were each diluted to 0.5 ng/μl before incubation and deposition. To isolate HIV-1 DNA, plasmid pHGFP(D110E) was digested with Bsr BI (NEB) and a 10.9-kbp fragment containing HIV-1 DNA (10.3 kbp) and genomic sequences flanking the 5′ and 3′ ends of the viral DNA (~0.3 kbp each) was isolated by gel purification (Qiagen). A minimum of three experimental samples (biological replicates) were prepared for each condition, with multiple individual DNA molecules imaged for each sample.
Estimating nucleic acid and protein contents of the capsid
The volume of the capsid contents was calculated by summing the volumes of the RNA genomes, packaged proteins, and the reverse-transcribed DNA product. RNA volume is calculated using established values of diameter 1 nm and length 0.55 nm/nt. The proportionality of protein molecular weight and size has been previously established using AFM imaging (76) and confirmed on our system using previously studied proteins T4 gp32 (77), Escherichia coli SSB (78), and LINE-1 ORF1p (79) with a proportionality constant of 2.09 nm3/kDa. A similar value for average protein density has been previously reported (80). The capsid is assumed to contain 100 molecules of RT (117 kDa), 100 molecules IN (32 kDa) (5% of the Gag-Pol of the average number of Gag molecules per virion), and 200 molecules viral protein R (14 kDa) (81). We assumed 2000 molecules of NC (6.6 kDa) in the mature capsid based on the average quantity of Gag polyprotein per virion, from which NC is cleaved, in the immature virion.
It has been established that HIV-1 RT switches templates between copackaged RNA templates 8 to 10 times to form one dsDNA composed of genetic information from each of the genomes (51). As a result, only about 50% of the genomic RNA is reverse-transcribed, while the other 50% remains as RNA fragments with an average length of approximately 1 kb. The genomic RNA that is reverse-transcribed is degraded by the RNase H domain of RT into smaller fragments of undefined lengths, with fragments of 8 to 9 nt often observed during in vitro RT assays (52). It is not known whether some of the RNA fragments generated by RNase H may be too short to bind to NC; therefore, we estimate that the total amount of nucleic acid in the viral core is increased by 150 to 200%. Therefore, the level of NC inside the capsid is enough to saturate the originally packaged RNA, but, after the completion of RT, similar binding affinities between NC and RNA or DNA results in only one-third of the DNA being fully condensed, while two-thirds are uncondensed due to NC binding residual RNA fragments that remain after the cleaving of the RNA genome (52). From the substrate length dependence, we measured condensed DNA, including bound NC, to have a volume of 3000 nm3/kbp. In comparison, the excluded diameter of the uncondensed dsDNA has been measured to be 3.1 nm (82, 83), substantially larger than the crystallographic B DNA diameter of 2 nm due to the two layers of structured water molecules on the highly polar and patterned dsDNA surface. Taking into the account that the rise per bp of B DNA duplex is 0.34 nm/bp, the volume of tightly packed uncondensed dsDNA is nearly equal to the volume of the tight NC-dsDNA globule. However, the globule also contains approximately one NC molecule per 5 bp of condensed dsDNA, leading to the overall capsid content volume reduction by the net volume of all NC molecules within the dsDNA condensate.
The internal volume of the capsid was calculated by approximating its cross-sectional area as a “coffin” shape (an elongated hexagon with ends perpendicular to its long axis). The average length and maximum width of mature capsids were previously measured to be 120 ± 22 and 60 ± 8 nm, respectively (84), and the minimum width of the capsid as it tapers at its end was measured to be approximately 20 nm (85). Using a capsid wall width of 6 nm and by rotating this cross section around its long central axis to form a truncated cone shape, the internal volume of the capsid is calculated to be 78,000 nm3 on average using these external dimensions. This results in an internal capsid NC concentration of ~40 mM (2000 molecules) and a ribonucleotide triphosphate concentration of ~420 mM (dimer of 9.8 knt RNA), with an additional ~420 mM of deoxynucleotide triphosphates after the completion of RT.
Data analysis and statistics
Welch’s unpaired t tests were used to analyze parametric data. Mann-Whitney U tests were used to analyze nonparametric data. Fisher’s exact test was used to analyze 2 × 2 contingency tables. All statistical tests were performed in Prism 9 (GraphPad Software). P values of <0.05 were considered significant. The time when 50% of the viruses lost sensitivity to the antiviral compounds (t1/2) was determined by nonlinear (sigmoidal) curve fitting (Prism 9).
Acknowledgments
We thank E. Freed and S. Hughes for valuable discussions and suggestions during manuscript preparation. We thank R. J. Gorelick, Frederick National Laboratory for Cancer Research, for preparing HIV-1 NCp7 used in these studies. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the US government.
Funding: This work was supported by the Intramural Research Program of the National Institutes Health, National Cancer Institute, Center for Cancer Research ( Z1A BC011436 to V.K.P. and Z1A BC010504 to W.-S.H.) and supplemental funding provided to support collaboration with the B-HIVE Group funded by U54A1170855; Office of AIDS Research (V.K.P. and W.-S.H.); and National Institutes Health grant R56AI167700 (M.C.W.).
Author contributions: Conceptualization: All authors. Investigation: R.C.B., M.M., M.C.W., W.-S.H., and V.K.P. Supervision: M.C.W. and V.K.P. Writing—original draft: All authors. Writing—review and editing: All authors.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. All reagents will be made available upon request.
Supplementary Materials
This PDF file includes:
Supplementary Text
Figs. S1 to S9
Legend for data file S1
Other Supplementary Material for this manuscript includes the following:
Data file S1
REFERENCES AND NOTES
- 1.Ambrose Z., Aiken C., HIV-1 uncoating: Connection to nuclear entry and regulation by host proteins. Virology 454–455, 371–379 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Campbell E. M., Hope T. J., HIV-1 capsid: The multifaceted key player in HIV-1 infection. Nat. Rev. Microbiol. 13, 471–483 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arhel N., Revisiting HIV-1 uncoating. Retrovirology 7, 96 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dvorin J. D., Malim M. H., Intracellular trafficking of HIV-1 cores: Journey to the center of the cell. Curr. Top. Microbiol. Immunol. 281, 179–208 (2003). [DOI] [PubMed] [Google Scholar]
- 5.Cosnefroy O., Murray P. J., Bishop K. N., HIV-1 capsid uncoating initiates after the first strand transfer of reverse transcription. Retrovirology 13, 58 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mamede J. I., Cianci G. C., Anderson M. R., Hope T. J., Early cytoplasmic uncoating is associated with infectivity of HIV-1. Proc. Natl. Acad. Sci. U.S.A. 114, E7169–E7178 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Francis A. C., Melikyan G. B., Single HIV-1 imaging reveals progression of infection through CA-dependent steps of docking at the nuclear pore, uncoating, and nuclear transport. Cell Host Microbe 23, 536–548.e6 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rasaiyaah J., Tan C. P., Fletcher A. J., Price A. J., Blondeau C., Hilditch L., Jacques D. A., Selwood D. L., James L. C., Noursadeghi M., Towers G. J., HIV-1 evades innate immune recognition through specific cofactor recruitment. Nature 503, 402–405 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yoh S. M., Mamede J. I., Lau D., Ahn N., Sanchez-Aparicio M. T., Temple J., Tuckwell A., Fuchs N. V., Cianci G. C., Riva L., Curry H., Yin X., Gambut S., Simons L. M., Hultquist J. F., Konig R., Xiong Y., Garcia-Sastre A., Bocking T., Hope T. J., Chanda S. K., Recognition of HIV-1 capsid by PQBP1 licenses an innate immune sensing of nascent HIV-1 DNA. Mol. Cell 82, 2871–2884.e6 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Burdick R. C., Li C., Munshi M., Rawson J. M. O., Nagashima K., Hu W. S., Pathak V. K., HIV-1 uncoats in the nucleus near sites of integration. Proc. Natl. Acad. Sci. U.S.A. 117, 5486–5493 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li C., Burdick R. C., Nagashima K., Hu W. S., Pathak V. K., HIV-1 cores retain their integrity until minutes before uncoating in the nucleus. Proc. Natl. Acad. Sci. U.S.A. 118, e2019467118 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schifferdecker S., Zila V., Müller T. G., Sakin V., Anders-Össwein M., Laketa V., Kräusslich H.-G., Müller B., Direct capsid labeling of infectious HIV-1 by genetic code expansion allows detection of largely complete nuclear capsids and suggests nuclear entry of HIV-1 complexes via common routes. mBio 13, e0195922 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zila V., Margiotta E., Turonova B., Muller T. G., Zimmerli C. E., Mattei S., Allegretti M., Borner K., Rada J., Muller B., Lusic M., Krausslich H. G., Beck M., Cone-shaped HIV-1 capsids are transported through intact nuclear pores. Cell 184, 1032–1046.e18 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Müller T. G., Zila V., Peters K., Schifferdecker S., Stanic M., Lucic B., Laketa V., Lusic M., Müller B., Kräusslich H.-G., HIV-1 uncoating by release of viral cDNA from capsid-like structures in the nucleus of infected cells. eLife 10, e64776 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Burdick R. C., Deleage C., Duchon A., Estes J. D., Hu W. S., Pathak V. K., Intranuclear positions of HIV-1 proviruses are dynamic and do not correlate with transcriptional activity. mBio 13, e0325621 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wei G., Iqbal N., Courouble V. V., Francis A. C., Singh P. K., Hudait A., Annamalai A. S., Bester S., Huang S. W., Shkriabai N., Briganti L., Haney R., KewalRamani V. N., Voth G. A., Engelman A. N., Melikyan G. B., Griffin P. R., Asturias F., Kvaratskhelia M., Prion-like low complexity regions enable avid virus-host interactions during HIV-1 infection. Nat. Commun. 13, 5879 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Achuthan V., Perreira J. M., Sowd G. A., Puray-Chavez M., McDougall W. M., Paulucci-Holthauzen A., Wu X., Fadel H. J., Poeschla E. M., Multani A. S., Hughes S. H., Sarafianos S. G., Brass A. L., Engelman A. N., Capsid-CPSF6 interaction licenses nuclear HIV-1 trafficking to sites of viral DNA integration. Cell Host Microbe 24, 392–404.e8 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Francis A. C., Marin M., Singh P. K., Achuthan V., Prellberg M. J., Palermino-Rowland K., Lan S., Tedbury P. R., Sarafianos S. G., Engelman A. N., Melikyan G. B., HIV-1 replication complexes accumulate in nuclear speckles and integrate into speckle-associated genomic domains. Nat. Commun. 11, 3505 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Christensen D. E., Ganser-Pornillos B. K., Johnson J. S., Pornillos O., Sundquist W. I., Reconstitution and visualization of HIV-1 capsid-dependent replication and integration in vitro. Science 370, eabc8420 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dharan A., Bachmann N., Talley S., Zwikelmaier V., Campbell E. M., Nuclear pore blockade reveals that HIV-1 completes reverse transcription and uncoating in the nucleus. Nat. Microbiol. 5, 1088–1095 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rensen E., Mueller F., Scoca V., Parmar J. J., Souque P., Zimmer C., Di Nunzio F., Clustering and reverse transcription of HIV-1 genomes in nuclear niches of macrophages. EMBO J. 40, e105247 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Selyutina A., Persaud M., Lee K., KewalRamani V., Diaz-Griffero F., Nuclear import of the HIV-1 core precedes reverse transcription and uncoating. Cell Rep. 32, 108201 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lee K., Ambrose Z., Martin T. D., Oztop I., Mulky A., Julias J. G., Vandegraaff N., Baumann J. G., Wang R., Yuen W., Takemura T., Shelton K., Taniuchi I., Li Y., Sodroski J., Littman D. R., Coffin J. M., Hughes S. H., Unutmaz D., Engelman A., KewalRamani V. N., Flexible use of nuclear import pathways by HIV-1. Cell Host Microbe 7, 221–233 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hulme A. E., Perez O., Hope T. J., Complementary assays reveal a relationship between HIV-1 uncoating and reverse transcription. Proc. Natl. Acad. Sci. U.S.A. 108, 9975–9980 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yang Y., Fricke T., Diaz-Griffero F., Inhibition of reverse transcriptase activity increases stability of the HIV-1 core. J. Virol. 87, 683–687 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Arhel N. J., Souquere-Besse S., Munier S., Souque P., Guadagnini S., Rutherford S., Prevost M. C., Allen T. D., Charneau P., HIV-1 DNA Flap formation promotes uncoating of the pre-integration complex at the nuclear pore. EMBO J. 26, 3025–3037 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gifford L. B., Melikyan G. B., HIV-1 capsid uncoating is a multistep process that proceeds through defect formation followed by disassembly of the capsid lattice. ACS Nano 18, 2928–2947 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rankovic S., Varadarajan J., Ramalho R., Aiken C., Rousso I., Reverse transcription mechanically initiates HIV-1 capsid disassembly. J. Virol. 91, e00289-17 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kumar S., Morrison J. H., Dingli D., Poeschla E., HIV-1 Activation of innate immunity depends strongly on the intracellular level of TREX1 and sensing of incomplete reverse transcription products. J. Virol. 92, e00001-18 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yan N., Regalado-Magdos A. D., Stiggelbout B., Lee-Kirsch M. A., Lieberman J., The cytosolic exonuclease TREX1 inhibits the innate immune response to human immunodeficiency virus type 1. Nat. Immunol. 11, 1005–1013 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Papa G., Albecka A., Mallery D., Vaysburd M., Renner N., James L. C., IP6-stabilised HIV capsids evade cGAS/STING-mediated host immune sensing. EMBO Rep. 24, e56275 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Siddiqui M. A., Saito A., Halambage U. D., Ferhadian D., Fischer D. K., Francis A. C., Melikyan G. B., Ambrose Z., Aiken C., Yamashita M., A novel phenotype links HIV-1 capsid stability to cGAS-mediated DNA sensing. J. Virol. 93, e00706-19 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gao D., Wu J., Wu Y. T., Du F., Aroh C., Yan N., Sun L., Chen Z. J., Cyclic GMP-AMP synthase is an innate immune sensor of HIV and other retroviruses. Science 341, 903–906 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Diner B. A., Lum K. K., Cristea I. M., The emerging role of nuclear viral DNA sensors. J. Biol. Chem. 290, 26412–26421 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gamble T. R., Vajdos F. F., Yoo S., Worthylake D. K., Houseweart M., Sundquist W. I., Hill C. P., Crystal structure of human cyclophilin A bound to the amino-terminal domain of HIV-1 capsid. Cell 87, 1285–1294 (1996). [DOI] [PubMed] [Google Scholar]
- 36.Liu C., Perilla J. R., Ning J., Lu M., Hou G., Ramalho R., Himes B. A., Zhao G., Bedwell G. J., Byeon I. J., Ahn J., Gronenborn A. M., Prevelige P. E., Rousso I., Aiken C., Polenova T., Schulten K., Zhang P., Cyclophilin A stabilizes the HIV-1 capsid through a novel non-canonical binding site. Nat. Commun. 7, 10714 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Luban J., Bossolt K. L., Franke E. K., Kalpana G. V., Goff S. P., Human immunodeficiency virus type 1 Gag protein binds to cyclophilins A and B. Cell 73, 1067–1078 (1993). [DOI] [PubMed] [Google Scholar]
- 38.Thali M., Bukovsky A., Kondo E., Rosenwirth B., Walsh C. T., Sodroski J., Göttlinger H. G., Functional association of cyclophilin A with HIV-1 virions. Nature 372, 363–365 (1994). [DOI] [PubMed] [Google Scholar]
- 39.Kim K., Dauphin A., Komurlu S., McCauley S. M., Yurkovetskiy L., Carbone C., Diehl W. E., Strambio-De-Castillia C., Campbell E. M., Luban J., Cyclophilin A protects HIV-1 from restriction by human TRIM5α. Nat. Microbiol. 4, 2044–2051 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shah V. B., Shi J., Hout D. R., Oztop I., Krishnan L., Ahn J., Shotwell M. S., Engelman A., Aiken C., The host proteins transportin SR2/TNPO3 and cyclophilin A exert opposing effects on HIV-1 uncoating. J. Virol. 87, 422–432 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Padron A., Prakash P., Pandhare J., Luban J., Aiken C., Balasubramaniam M., Dash C., Emerging role of cyclophilin A in HIV-1 infection: From producer cell to the target cell nucleus. J. Virol. 97, e0073223 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yoo S., Myszka D. G., Yeh C., McMurray M., Hill C. P., Sundquist W. I., Molecular recognition in the HIV-1 capsid/cyclophilin A complex. J. Mol. Biol. 269, 780–795 (1997). [DOI] [PubMed] [Google Scholar]
- 43.De Iaco A., Luban J., Cyclophilin A promotes HIV-1 reverse transcription but its effect on transduction correlates best with its effect on nuclear entry of viral cDNA. Retrovirology 11, 11 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhong Z., Ning J., Boggs E. A., Jang S., Wallace C., Telmer C., Bruchez M. P., Ahn J., Engelman A. N., Zhang P., Watkins S. C., Ambrose Z., Cytoplasmic CPSF6 regulates HIV-1 capsid trafficking and infection in a cyclophilin a-dependent manner. mBio 12, e03142-20 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bannon J. H., O'Donovan D. S., Kennelly S. M., Mc Gee M. M., The peptidyl prolyl isomerase cyclophilin A localizes at the centrosome and the midbody and is required for cytokinesis. Cell Cycle 11, 1340–1353 (2012). [DOI] [PubMed] [Google Scholar]
- 46.Rajiv C., Davis T. L., Structural and functional insights into human nuclear cyclophilins. Biomolecules 8, 161 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gien H., Morse M., McCauley M. J., Kitzrow J. P., Musier-Forsyth K., Gorelick R. J., Rouzina I., Williams M. C., HIV-1 nucleocapsid protein binds double-stranded DNA in multiple modes to regulate compaction and capsid uncoating. Viruses 14, 235 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rouzina I., Bruinsma R., DNA confinement drives uncoating of the HIV Virus. Eur. Phys. J. Spec. Top. 223, 1745–1754 (2014). [Google Scholar]
- 49.Berkowitz R. D., Ohagen A., Hoglund S., Goff S. P., Retroviral nucleocapsid domains mediate the specific recognition of genomic viral RNAs by chimeric Gag polyproteins during RNA packaging in vivo. J. Virol. 69, 6445–6456 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rein A., Henderson L. E., Levin J. G., Nucleic-acid-chaperone activity of retroviral nucleocapsid proteins: Significance for viral replication. Trends Biochem. Sci. 23, 297–301 (1998). [DOI] [PubMed] [Google Scholar]
- 51.Delviks-Frankenberry K., Galli A., Nikolaitchik O., Mens H., Pathak V. K., Hu W. S., Mechanisms and factors that influence high frequency retroviral recombination. Viruses 3, 1650–1680 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wisniewski M., Balakrishnan M., Palaniappan C., Fay P. J., Bambara R. A., Unique progressive cleavage mechanism of HIV reverse transcriptase RNase H. Proc. Natl. Acad. Sci. U.S.A. 97, 11978–11983 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Thomas D. C., Voronin Y. A., Nikolenko G. N., Chen J., Hu W. S., Pathak V. K., Determination of the ex vivo rates of human immunodeficiency virus type 1 reverse transcription by using novel strand-specific amplification analysis. J. Virol. 81, 4798–4807 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Charneau P., Clavel F., A single-stranded gap in human immunodeficiency virus unintegrated linear DNA defined by a central copy of the polypurine tract. J. Virol. 65, 2415–2421 (1991). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Smith S. B., Cui Y., Bustamante C., Overstretching B-DNA: The elastic response of individual double-stranded and single-stranded DNA molecules. Science 271, 795–799 (1996). [DOI] [PubMed] [Google Scholar]
- 56.Bazzi A., Zargarian L., Chaminade F., De Rocquigny H., Rene B., Mely Y., Fosse P., Mauffret O., Intrinsic nucleic acid dynamics modulates HIV-1 nucleocapsid protein binding to its targets. PLOS ONE 7, e38905 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kanevsky I., Chaminade F., Chen Y., Godet J., René B., Darlix J.-L., Mély Y., Mauffret O., Fossé P., Structural determinants of TAR RNA-DNA annealing in the absence and presence of HIV-1 nucleocapsid protein. Nucleic Acids Res. 39, 8148–8162 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Krishnamoorthy G., Roques B., Darlix J.-L., Mély Y., DNA condensation by the nucleocapsid protein of HIV-1: A mechanism ensuring DNA protection. Nucleic Acids Res. 31, 5425–5432 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hulme A. E., Kelley Z., Foley D., Hope T. J., Complementary assays reveal a low level of ca associated with viral complexes in the nuclei of HIV-1-infected cells. J. Virol. 89, 5350–5361 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Schaller T., Ocwieja K. E., Rasaiyaah J., Price A. J., Brady T. L., Roth S. L., Hue S., Fletcher A. J., Lee K., KewalRamani V. N., Noursadeghi M., Jenner R. G., James L. C., Bushman F. D., Towers G. J., HIV-1 capsid-cyclophilin interactions determine nuclear import pathway, integration targeting and replication efficiency. PLOS Pathog. 7, e1002439 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Francis A. C., Marin M., Shi J., Aiken C., Melikyan G. B., Time-resolved imaging of single HIV-1 uncoating in vitro and in living cells. PLOS Pathog. 12, e1005709 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Burdick R. C., Delviks-Frankenberry K. A., Chen J., Janaka S. K., Sastri J., Hu W. S., Pathak V. K., Dynamics and regulation of nuclear import and nuclear movements of HIV-1 complexes. PLOS Pathog. 13, e1006570 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yu A., Lee E. M. Y., Briggs J. A. G., Ganser-Pornillos B. K., Pornillos O., Voth G. A., Strain and rupture of HIV-1 capsids during uncoating. Proc. Natl. Acad. Sci. U.S.A. 119, e2117781119 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Cherepanov P., Maertens G., Proost P., Devreese B., Van Beeumen J., Engelborghs Y., De Clercq E., Debyser Z., HIV-1 integrase forms stable tetramers and associates with LEDGF/p75 protein in human cells. J. Biol. Chem. 278, 372–381 (2003). [DOI] [PubMed] [Google Scholar]
- 65.Sowd G. A., Serrao E., Wang H., Wang W., Fadel H. J., Poeschla E. M., Engelman A. N., A critical role for alternative polyadenylation factor CPSF6 in targeting HIV-1 integration to transcriptionally active chromatin. Proc. Natl. Acad. Sci. U.S.A. 113, E1054–E1063 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Mochizuki H., Schwartz J. P., Tanaka K., Brady R. O., Reiser J., High-titer human immunodeficiency virus type 1-based vector systems for gene delivery into nondividing cells. J. Virol. 72, 8873–8883 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Unutmaz D., KewalRamani V. N., Marmon S., Littman D. R., Cytokine signals are sufficient for HIV-1 infection of resting human T lymphocytes. J. Exp. Med. 189, 1735–1746 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Yee J. K., Miyanohara A., LaPorte P., Bouic K., Burns J. C., Friedmann T., A general method for the generation of high-titer, pantropic retroviral vectors: Highly efficient infection of primary hepatocytes. Proc. Natl. Acad. Sci. U.S.A. 91, 9564–9568 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Nikolaitchik O. A., Dilley K. A., Fu W., Gorelick R. J., Tai S. H., Soheilian F., Ptak R. G., Nagashima K., Pathak V. K., Hu W. S., Dimeric RNA recognition regulates HIV-1 genome packaging. PLOS Pathog. 9, e1003249 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.O'Doherty U., Swiggard W. J., Malim M. H., Human immunodeficiency virus type 1 spinoculation enhances infection through virus binding. J. Virol. 74, 10074–10080 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Mbisa J. L., Delviks-Frankenberry K. A., Thomas J. A., Gorelick R. J., Pathak V. K., Real-time PCR analysis of HIV-1 replication post-entry events. Methods Mol. Biol. 485, 55–72 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Schneider C. A., Rasband W. S., Eliceiri K. W., NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 9, 671–675 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zenklusen D., Larson D. R., Singer R. H., Single-RNA counting reveals alternative modes of gene expression in yeast. Nat. Struct. Mol. Biol. 15, 1263–1271 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Carteau S., Gorelick R. J., Bushman F. D., Coupled integration of human immunodeficiency virus type 1 cDNA ends by purified integrase in vitro: Stimulation by the viral nucleocapsid protein. J. Virol. 73, 6670–6679 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Cruceanu M., Urbaneja M. A., Hixson C. V., Johnson D. G., Datta S. A., Fivash M. J., Stephen A. G., Fisher R. J., Gorelick R. J., Casas-Finet J. R., Rein A., Rouzina I., Williams M. C., Nucleic acid binding and chaperone properties of HIV-1 Gag and nucleocapsid proteins. Nucleic Acids Res. 34, 593–605 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Fuentes-Perez M. E., Dillingham M. S., Moreno-Herrero F., AFM volumetric methods for the characterization of proteins and nucleic acids. Methods 60, 113–121 (2013). [DOI] [PubMed] [Google Scholar]
- 77.Cashen B. A., Morse M., Rouzina I., Karpel R. L., Williams M. C., Dynamic structure of T4 gene 32 protein filaments facilitates rapid noncooperative protein dissociation. Nucleic Acids Res. 51, 8587–8605 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Naufer M. N., Morse M., Moller G. B., McIsaac J., Rouzina I., Beuning P. J., Williams M. C., Multiprotein E. coli SSB–ssDNA complex shows both stable binding and rapid dissociation due to interprotein interactions. Nucleic Acids Res. 49, 1532–1549 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Cashen B. A., Naufer M. N., Morse M., Jones C. E., Williams M. C., Furano A. V., The L1-ORF1p coiled coil enables formation of a tightly compacted nucleic acid-bound complex that is associated with retrotransposition. Nucleic Acids Res. 50, 8690–8699 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Fischer H., Polikarpov I., Craievich A. F., Average protein density is a molecular-weight-dependent function. Protein Sci. 13, 2825–2828 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.J. M. Coffin, S. H. Hughes, H. Varmus, in Retroviruses (Cold Spring Harbor Laboratory Press, 1997), pp. xv–843. [PubMed] [Google Scholar]
- 82.Brandariz-Nunez A., Robinson S. J., Evilevitch A., Pressurized DNA state inside herpes capsids-A novel antiviral target. PLOS Pathog. 16, e1008604 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Rau D. C., Lee B., Parsegian V. A., Measurement of the repulsive force between polyelectrolyte molecules in ionic solution: Hydration forces between parallel DNA double helices. Proc. Natl. Acad. Sci. U.S.A. 81, 2621–2625 (1984). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Briggs J. A., Wilk T., Welker R., Krausslich H. G., Fuller S. D., Structural organization of authentic, mature HIV-1 virions and cores. EMBO J. 22, 1707–1715 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Briggs J. A., Grunewald K., Glass B., Forster F., Krausslich H. G., Fuller S. D., The mechanism of HIV-1 core assembly: Insights from three-dimensional reconstructions of authentic virions. Structure 14, 15–20 (2006). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Text
Figs. S1 to S9
Legend for data file S1
Data file S1