Abstract
Many insect pests, including the brown planthopper (BPH), undergo windborne migration that is challenging to observe and track. It remains controversial about their migration patterns and largely unknown regarding the underlying genetic basis. By analyzing 360 whole genomes from around the globe, we clarify the genetic sources of worldwide BPHs and illuminate a landscape of BPH migration showing that East Asian populations perform closed-circuit journeys between Indochina and the Far East, while populations of Malay Archipelago and South Asia undergo one-way migration to Indochina. We further find round-trip migration accelerates population differentiation, with highly diverged regions enriching in a gene desert chromosome that is simultaneously the speciation hotspot between BPH and related species. This study not only shows the power of applying genomic approaches to demystify the migration in windborne migrants but also enhances our understanding of how seasonal movements affect speciation and evolution in insects.
Genome-wide analyses are used to reconstruct the evolution and global migration pattern of a windborne migratory pest.
INTRODUCTION
Migration is a common strategy among animals in response to changes in seasonal habitats. Migratory animals avoid adverse circumstances and might travel up to thousands of kilometers to locate appropriate environments during changing seasons. Insects are the most abundant group of terrestrial migrants (1). Migration in most insects differs from migration in vertebrates due to their short life cycle, tiny body size, and limited self-propelled flight capability. Perhaps the most well-known migratory insect species is the monarch butterfly (Danaus plexippus), which travels up to 4000 km from its eastern North American breeding grounds to overwintering sites in central Mexico (2, 3). However, the vertebrate-like, self-powered migratory behavior often observed in monarchs is uncommon in insects. Most migratory insects undertake multigenerational migration, during which they cannot reach the ultimate destination within a single generation and instead mate and reproduce along the journey. Moreover, many small insects such as hemipteran migrants fly at high altitudes, where their migration is largely wind-driven. While migration has been extensively studied in the monarch butterfly and some vertebrate migrants for navigation mechanisms and “migratory genes” (4–7), it remains largely unknown whether the migration in other insects relies on similar functional modules and leads to a similar evolutionary consequence. Methodological developments and extensive field studies have enabled the tracking of migratory pests at a macroscale (8–10) and even at individual level for large insects (11). However, it remains challenging to definitively determine the movement dynamics of small windborne migrants.
The brown planthopper (BPH), Nilaparvata lugens (Stål) (Hemiptera: Delphacidae), is a highly destructive rice pest worldwide. These specialized pests pose a serious threat to rice production by extracting liquid nutrients from phloem and transmitting various rice viruses in Asian regions (12–14). BPHs have a wide distribution ranging from Asia to northern Australia (15). They are distributed in tropical regions, including the major rice-planting areas in South Asia (SA) and Southeast Asia (SEA), all the year round and seasonally present in temperate and subtropical regions of East Asia (EA), reaching as far north as the Far East (16, 17). BPHs do not undergo diapause and hence cannot survive the cold winter in EA; instead, they migrate southward to overwintering sites in tropical regions and then remigrate northward the following spring (16, 18–22). Because of their small body size (2 to 3.5 mm in length) and low flight speed (approximately 0.3 m/s), BPHs are considered classic windborne migrants capable of long-distance migration with the assistance of high-altitude air flows (monsoons), but with no effective control over displacement speed and heading direction (18, 19, 23). In the past few decades, ecological studies based on field traps and radar observations have provided predictions on the overall migration scope, as well as overwintering sites, for EA populations (16, 21, 22, 24). However, many aspects of BPH migration are still unknown or controversial, such as whether most EA migrants return or further travel to SA or SEA.
Molecular evidence has been extensively used to trace the evolutionary and colonization history of animals and plants (5, 25, 26). Previous molecular-level studies on BPH migration based on either scattered markers or incomplete sampling could not yield enough resolution to completely differentiate BPH populations (27–30). In this study, we sampled worldwide BPH populations for whole-genome sequencing and pioneer to genetically track their migratory routes. We also aim to investigate the existence of migratory genes in windborne insect migrants and to explore the evolutionary consequences of these migrations.
RESULTS
Characterize the genetic relationship across BPHs around the globe
We collected BPH populations from 90 geographic locations from across their worldwide distribution, including almost all cultivated rice-planting areas across EA, SA, and SEA, from 2009 to 2018 (Fig. 1A and data S1). We also sampled an Australian population of BPH and two individuals of the sister species, Nilaparvata muiri, from the wild rice species, Leersia hexandra Swart (data S1). A total of 360 planthoppers were subjected to whole-genome sequencing, with an average of 10.9-fold coverage per sample, yielding 5.05 Tb of clean data in total (data S1). By mapping to the chromosomal-level assembly of BPH (31), a total of 25.23 million high-quality variants were identified, equaling approximately 26 variants/kb. The dense variation dataset allows for a high level of resolution in population genetic studies.
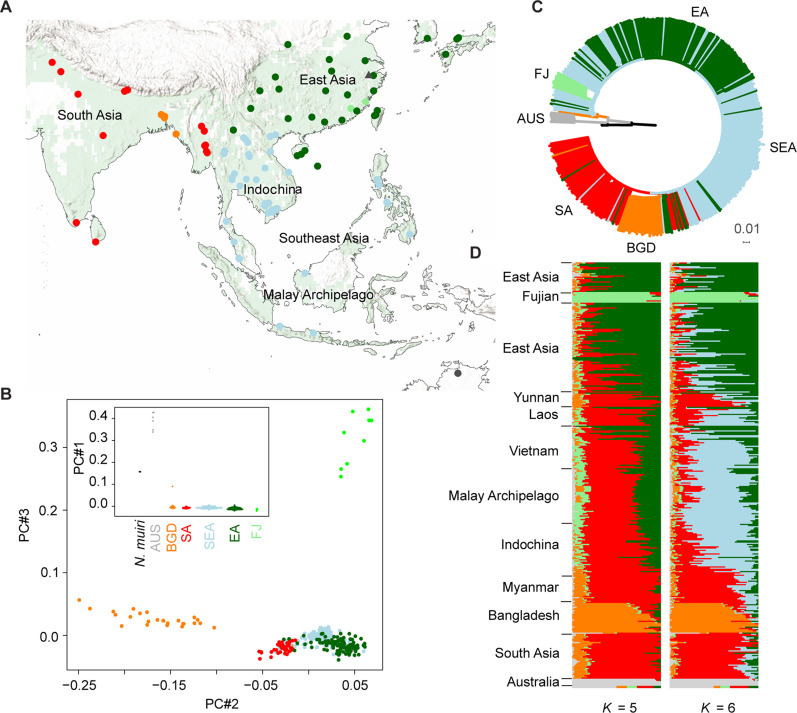
Fig. 1. Geographic and genetic relationship across worldwide BPHs.
(A) Sampling locations of BPH and two N. muiri individuals across the majority of worldwide distribution regions. The sampling region covers more than 90% of the worldwide rice planting areas [green background in the inset map; data from Monfreda (87)]. The sampling map was generated using ArcMap. Dark green points, EA populations; blue points, SEA populations; red points, SA populations; light green points, FJ populations; orange points, BGD population; gray points, AUS populations; triangle, N. muiri. (B) PCA of all sequenced individuals based on whole-genome SNPs. The first three components are displayed, with the first component being shown as inset. (C) Neighbor-joining phylogeny of all sequenced individuals based on whole-genome SNPs using N. muiri as outgroup. Branch colors indicate the information of geographic distribution: Green, EA; blue, SEA; red, SA; orange, BGD; light green, FJ; gray, AUS; black, N. muiri. See the cladogram format of this tree in fig. S1. (D) Ancestry analysis of all sequenced individuals. The colors in each column represent the ancestry proportion of presumed genetic groups ranging from K = 5 to 6. The presented result was inferred by sNMF (71). See alternative results, inferred by admixture (72), in fig. S2.
We first analyzed the genetic relationship among all sequenced individuals to characterize population structures of existing BPHs. Both the principal components analysis (PCA; the first principal component) and the inferred neighbor-joining (NJ) phylogeny evidently differentiated Australia (AUS) population from Asian BPHs (Fig. 1, B and C, and fig. S1). This deep split might be attributed to geographic barrier and host difference, given that no cultivated rice is planted in North Australia. A previous hybridization experiment showed that the F1 and F2 generations of Australian and Philippine populations developed normally (32), suggesting divergence between Australian and Asian populations has not reached the speciation level. AUS population was thus mainly used as the outgroup in subsequent analyses.
The Asian clade exhibited weak bootstrap support for most internal nodes and short branches between them (Fig. 1C and fig. S1), suggesting a generally low level of population differentiation across Asian BPHs. Among the 90 sampled geographic populations, only 12 populations were identified as independent lineages with robust bootstrap support (>90%; fig. S1). On the other hand, most genetically distinguished sublineages consisted of samples with unrelated geographic origins (fig. S1). Some samples showed the most proximity to those sampled as distantly as 2600 km away, e.g., a BPH sampled in Vietnam (YN3B-4) was tightly clustered with three samples from Korea (KOR) (fig. S1). The collapsed pattern of spatial structures at continental scales suggests massive movements across Asian BPH populations.
Relatively speaking, BPHs from SA and neighboring regions, encompassing Myanmar and the southwest border of China (referred to as SA), were found evidently distinguishable from East Asian (EA) populations based on phylogeny, PCA (component #2), and the model-based ancestry analysis, whereas Southeast Asian (SEA) samples were largely intermixed with SA or EA samples (Fig. 1, B to D, and figs. S2 and S3A). Further increase of presumed ancestry (k = 6), the analysis achieved the lowest cross-validation error (0.630) and considerably differentiated most Southeast Asian (SEA) samples from EA and SEA samples (Fig. 1D).
Two populations from Fujian (FJ), China, and three populations from Bangladesh (BGD) were found to form two distinct monophyletic lineages with long branches (Fig. 1C). These deep splits were further supported by PCA analyses (components #2 and 3), ancestry analysis (at k = 5) (Fig. 1D and fig. S2), and mitochondrion-based phylogeny (fig. S4). In addition, no individuals sampled from these populations were genetically placed outside the sublineages. These groups are likely to be resident populations, independent from their geographically nearby populations.
Determination of the distribution and movement scope of different BPH groups
We have characterized three genetic groups (EA, SEA, and SA) for most Asian BPHs, each covering a vast distributional area (fig. S5). They are very likely to engage in long-range migration but relatively distinct from each other, implying that the migration scope of each group is relatively closed and unique. We aimed to determine the main movement scope of each group based on the geographic populations they were involved with. To achieve this, we analyzed the geographic populations by assigning each involving sample to a designated main group based on its estimated ancestry, while the individual without dominant ancestry was alternatively classified to an admixed group (Materials and Methods). This approach genetically characterized 80 EA BPHs, 96 SEA BPHs, 70 SA BPHs, and 74 admixed BPHs (data S1). According to the involving individual ancestry, each geographic population was in turn classified as one of the main groups (EA, SEA, or SA) or considered admixed (Materials and Methods and data S1). Thus, the movement scope of each main group could be determined based on the spatial distribution of all included populations (Fig. 2A and fig. S5).
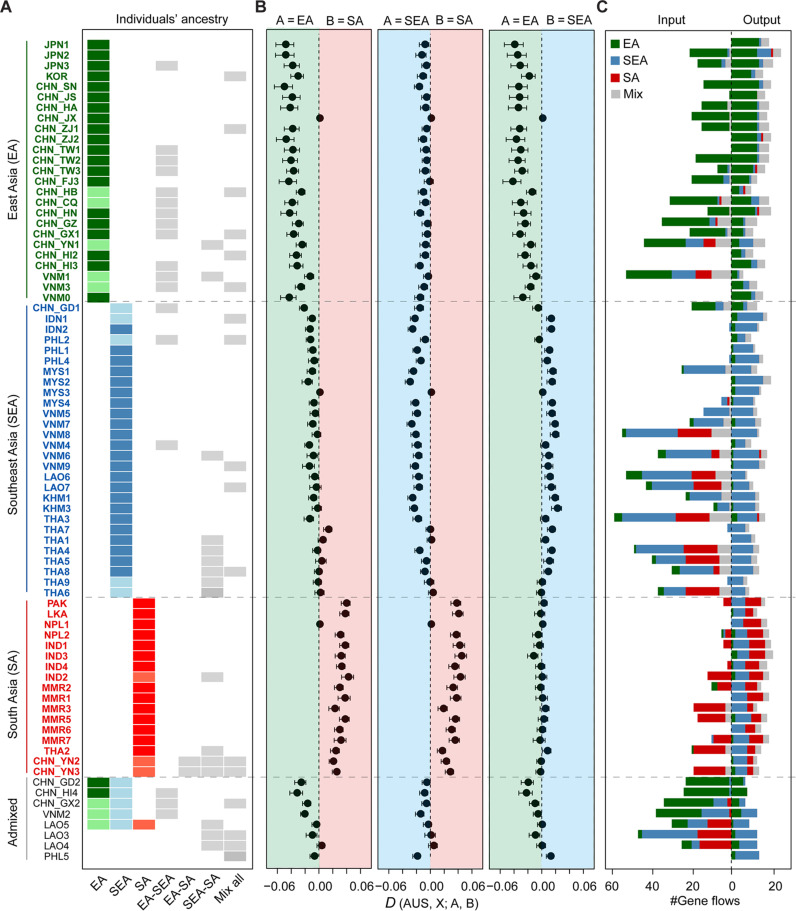
Fig. 2. Genetic sources of and gene flows worldwide BPHs.
(A) Genetic sources of each geographic population along with its genetic relationship to one of the main genetic groups. The genetic sources of all sampled individuals of the focal population (names shown on the left of each row, and individuals included in each population were listed in data S1) are shown with colored cells, i.e., what main groups the sampled individuals of this population were genetically assigned to. Each individual was defined to a main genetic group or an admixed group based on the inferred ancestry component (see Materials and Methods). Admixed populations are displayed in gray cells. (B) D statistics are shown to support the genetic assignment results. D indicates the comparison between the affinity of each geographic population with one of the main groups A and that with B (names shown on the top). D < 0 supports the affinity of the target population with the main group A, while D > 0 supports that with group B. Australian populations were used as outgroup. (C) Bar plots illustrating the inferred gene flows on focal populations. Left: Gene flows from other populations of certain groups (in color) to the focal population. Right: Gene flows from the focal population to other populations.
As a result, 25 geographic populations, comprising either EA individuals only or additionally including admixed individuals, were genetically classified as EA populations (Fig. 2A). In addition to the geographic locations of EA, three populations from north-central Vietnam were also identified as EA populations, signifying the likely southernmost border of the EA group (Fig. 2A and fig. S5). This genetically defined border agrees with previous field surveys that propose the north-central Vietnam region as a potential source of EA migrants (20, 21, 33) and elucidates the limited influx of immigrants from other regions into EA.
We also defined 28 and 17 geographic populations as SEA and SA populations, respectively (Fig. 2A). The involved geographic locations indicate that SEA BPHs travel throughout the western Pacific islands and the majority of the Indochina peninsula, whereas SA BPHs travel through SA, spanning from Pakistan in the west to northwestern Thailand and southwestern China in the east. The eight remaining admixed populations are primarily distributed in Indochina and the southern coastal regions of China (Fig. 2A and fig. S5).
To assess the robustness of grouping, we additionally identified “core” samples to respectively represent the extreme ancestry of EA, SEA, and SA (see Materials and Methods, fig. S3B, and data S1) and conducted D statistics to compare the genetic proximity of each geographic population to these core ancestries. As expected, all defined EA populations exhibited closer genetic proximity to the core EA ancestry than to SEA or SA ancestry; likewise, defined SEA and SA populations showed genetic affinity to their respective SEA and SA ancestries when compared with other ancestries (Fig. 2B).
Reconstruction of the evolutionary history of BPH groups
The maximum-likelihood phylogenetic analyses across core samples and across 21 nonadmixed populations both placed EA-SEA and SA populations as sister clades (Fig. 3A and fig. S6A), suggesting a relatively ancient split between SA and SEA-EA. In addition, this population-level phylogenetic analysis revealed the origins of two derived populations. The BGD group was clustered with SA populations (Fig. 3A), suggesting a derived history from the local adaptation of passing SA populations. Although geographically surrounded by nearby EA populations, the FJ group was genetically clustered with SEA populations, indicative of an SEA origin. This nonprimary route from the Philippines to the eastern coast of China is possibly due to the west Pacific typhoons, as suggested in previous studies (34–36). Main population characteristics support the founder effects of these two local populations, including higher levels of linkage disequilibrium, less fractions of negative Tajima’s D, and increased minor allele frequency (fig. S6, B to D).
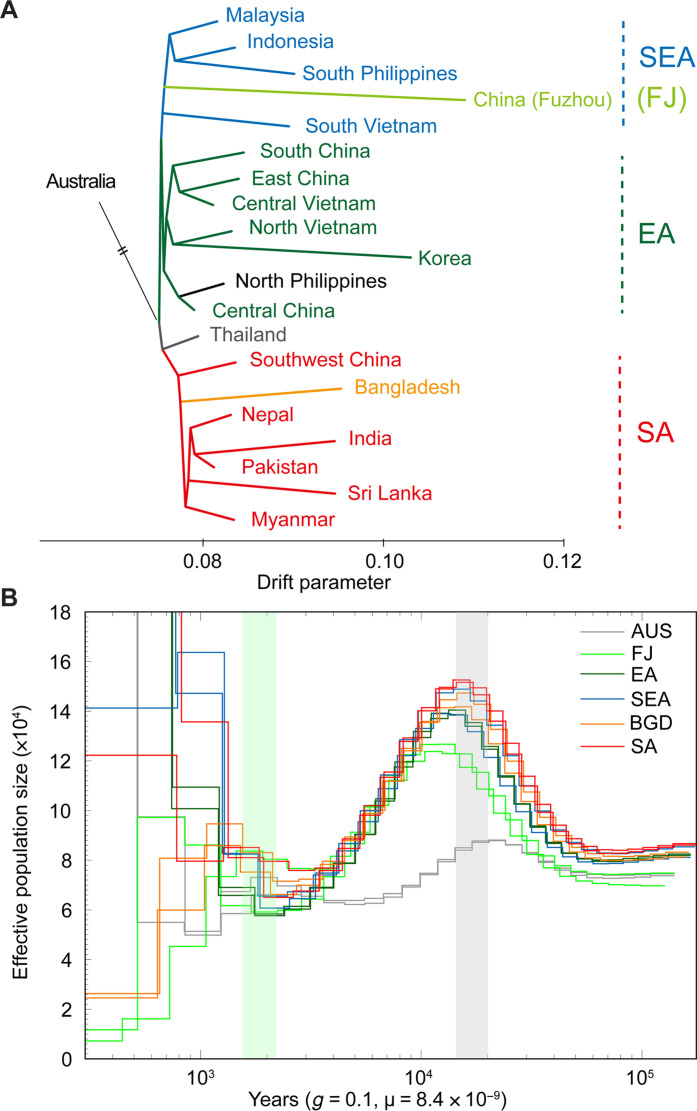
Fig. 3. Inferred evolutionary history of main BPH groups.
(A) Inferred phylogeny among nonadmixed populations using TreeMix. From top to bottom, populations used as follows: MYS2, IDN1, PHL5, CHN_FJ2, VNM4, CHN_HI3, CHN_ZJ2, VNM0, VNM3, KOR, PHL2, CHN_HB, THA7 + THA1 + THA9, CHN_YN2, BGD1, NPL1, IND3, PAK, LKA, and MMR6. (B) Demographic history of main BPH groups inferred by the PSMC model. “Core” samples from each main group were selected as representatives to show the effective size and divergence times. Gray shadow indicates the period Asia BPHs began to decline, while the green shadow indicates the period when different Asia groups fully diverged. The mutation rate (8.4 × 10−9) was set based on Drosophila (76).
We further estimated the dynamic population history using the pairwise sequentially Markovian coalescent (PSMC) model (37). This analysis revealed a concordant history of population expansion among all Asian groups, which diverged from the Australian population since ~30,000 years ago (Fig. 3B). Approximately 20,000 years ago, around the Last Glacial Maximum (LGM), all groups began to decline (Fig. 3B), presumably due to the shortage of host plants (38, 39). The adverse host conditions might have driven host shifts in herbivores, leading to founder effects (bottlenecks) and population differentiation. The multiple sequentially Markovian coalescent (MSMC) (40) model recovered these progressive split events overlapped with the concordant population decline in PSMC (fig. S7). Furthermore, both PSMC and MSMC models inferred that the main BPH groups completely separated from each other and independently evolved since approximately 2000 years ago (Fig. 3B and fig. S7), coinciding with the extensive cultivation of rice in Asia. It is thus possible that the increased paddy cultivation throughout Asia allowed BPHs to expand territories via long-range migrations, leading to population recovery. These patterns jointly indicate an important role of host condition changes in driving the global dispersal of BPHs. Similarly, the ancient split between SA and EA/SEA was possibly another consequence of host shift, as indica cultivars were domesticated and extensively planted in SA, in comparison to japonica in EA and SEA (41).
Inference of the migration routes within each BPH group
Characterizing the dynamic processes within the scope is more challenging than defining the movement boundary, especially for small animals such as BPHs that are difficult to track. Here, we attempted to infer the main movement routes based on pairwise gene flows and the genetic differences across traveling sites. To do this, we used the three-population test to check whether the target population was an admixture between two tested populations. We conservatively defined a gene flow between two populations (from A to X) only when the target population (X) showed signatures of mixture [Z < −3 of (X; A, B)] and B was not defined as the source (see Materials and Methods).
Several lines of evidence suggest that the inferred gene flows are reliable in the relevant geographic context. First, none of the genetically distinct populations (i.e., AUS, BGD, and FJ) showed mixture signatures (Fig. 4). Second, we found predominant output flows in populations distant from Indochina (as donor), while populations within or approaching Indochina showed much more input flows than output (as recipient) (Fig. 2C). Assembly of these progressive flows reconstructs three main migratory trajectories from various remote locations of EA, Malay Archipelago, and SA to Indochina (Fig. 4 and figs. S8 to S11), agreeing with the directions of documented prevailing winds that largely influence these windborne migrants (fig. S12). More specifically, the two central Vietnam populations (VNM0 and VNM3) sampled in early spring exhibited evident gene flows to EA populations (fig. S8), agreeing with the fact that EA migration starts from north-central Vietnam every early spring (22). By contrast, we detected admixed input gene flows in the further north Vietnamese population sampled in late summer when EA migrants leave (fig. S8), possibly due to population recolonization by nearby tropical populations (22). Nevertheless, we note that this conservative approach might overlook intermediate mixtures between populations, i.e., potential links between “internal” breeding area along the migratory route could be underestimated, and might be biased by the sampling seasons.
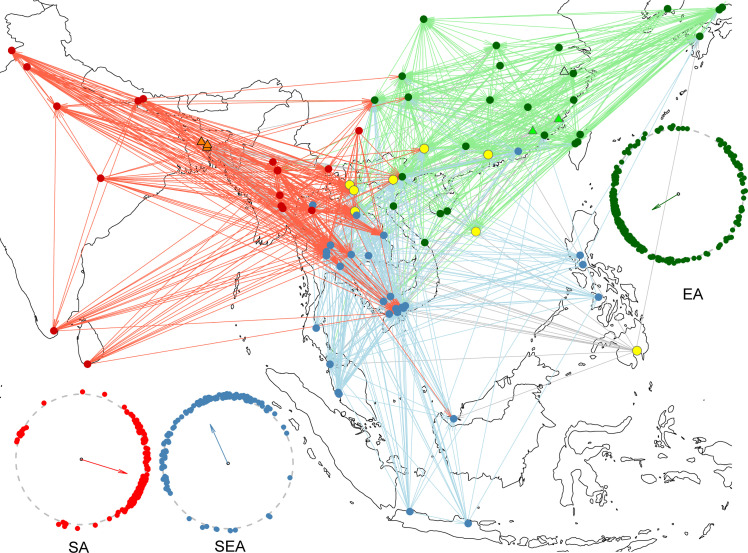
Fig. 4. Inferred migratory trajectories of worldwide BPHs.
Inferred gene flows between any pair of geographic populations (see Materials and Methods). Colored circles indicate geographic populations: green, EA populations; blue, SEA populations; red, SA populations; yellow, admixed populations. Lines with the same colors represented genes flows started from the corresponding populations. Gray lines showed gene flows started from admixed populations. Light green and orange triangles represented FJ and BGD populations. The colored clocks summarize the calculated angles, based on the GPS information, of all inferred gene flows within the corresponding group. The arrow direction indicates the mean angle representing the prevailing flows. To highlight the main pattern, only long-range gene flows (>1000 km) are plotted here (see figs. S8 to S11 for detailed information of all gene flows and fig. S12 for associated wind patterns).
As a result, we inferred 628 and 280 gene flows within and between main groups (EA, SEA, and SA), respectively (Fig. 4). In EA, we observed considerable northeastward gene flows from Vietnam to various EA locations and more reverse gene flows from the remote populations, such as Japan, Korea, and coastal regions of China, stepwise toward populations proximal to or within Indochina (Fig. 4 and fig. S9). In SEA, massive gene flows were found from the archipelagos of insular SEA toward central and southern Indochina, as well as short-range flows from southern Vietnam to Thailand and Laos (Fig. 4 and fig. S10). In SA, the prevailing gene flows were eastward, extending from as far west as Pakistan to central-south Indochina (Fig. 4 and fig. S11).
Overall, we found gene flows toward Indochina were evident in all three main groups, while reverse gene flows were present in EA but sparsely observed within SEA and SA (Fig. 4). Thus, we propose two distinct seasonal movement patterns for global BPHs: a multigenerational, round-trip migration in EA and two respective one-way migrations to Indochina in SEA (Malay Archipelago) and SA. These patterns can be attributed to the climate conditions of the involved locations. Most remote destinations of EA migrants are temperate regions where BPHs cannot overwinter, necessitating the remigration of EA populations southward for overwintering in the tropics. By contrast, most stopover areas of SA and SEA BPHs are in the tropics, where the host plant (rice) is grown year-round for breeding.
EA migratory populations are highly diverged from other populations
It is noteworthy that Indochina serves as both the overwintering site of EA migrants and the destination of SA and SEA migrations (fig. S5), enabling the potential of genetic exchanges among geographically proximate locations. However, we observed very few outliers with SEA or SA ancestry being sampled in any remote locations of EA (Fig. 2A). We further found that many EA populations, particularly those distant from Indochina, exhibit relatively high levels of genetic differentiation (dXY) from all other populations, whereas SEA and SA populations within or surrounding Indochina showed relatively low levels of dXY from each other (fig. S13). Some EA populations even showed higher levels of dXY with each other than with SEA populations (fig. S13). Considering the recent split between EA and SEA groups (Fig. 3A), it is likely that the long-distance, round-trip migration of EA not only maintains the original divergence from other groups but also has further triggered within-group differentiation, resulting in the observed high dXY.
Highly diverged regions occurred on a single chromosome
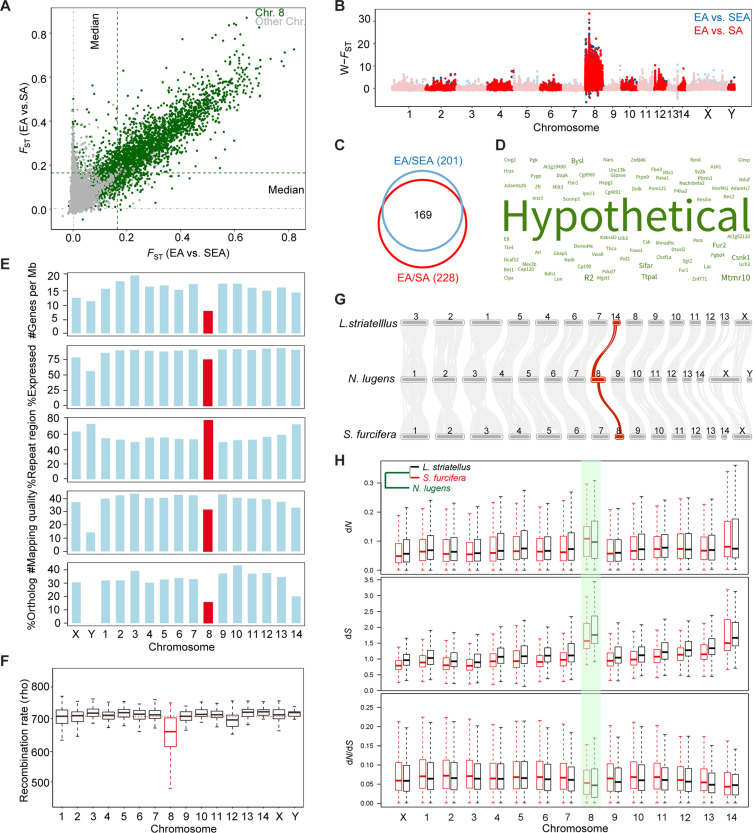
Genetic divergence is typically caused by either natural selection or genetic drift. We were interested in identifying the highly diverged regions between EA and other groups and addressing whether these regions might account for the unique adaptation to the long-distance migration in EA. Many genomic regions show correlated genetic differentiation between EA versus SEA and EA versus SA (Fig. 5A). To our surprise, the vast majority of these highly diverged genomic regions are located on a single chromosome, chromosome 8, rather than being interspersed throughout the genome (Fig. 5A). By isolating genomic segments with the highest 1% FST, we found that 95.1% (between EA and SEA) and 93.2% (between EA and SA) of these segments were located on chromosome 8 (data S3). In comparison with other chromosomes, we observed significantly higher population differentiation statistics between EA and other groups on chromosome 8, including FST (P < 2.2 × 10−16, one-sided Wilcoxon rank-sum test; Fig. 5B) and XP-CLR [the cross-population composite likelihood ratio; (42)] (P < 6.09 × 10−64, one-sided Wilcoxon rank-sum test; fig. S14 and data S2).
Fig. 5. Genome-wide selection and divergence of EA from other migratory groups.
(A) Window-based genomic divergence (FST) between EA and groups (dots). Median values of windows on chromosome 8 and the rest of genomes are indicated by green and gray dashed lines, respectively. (B) W-transformed FST between EA and SEA (red dots) and between EA and SA (blue dots) along chromosomes. (C) Gene counts with the most evident signatures of population differentiation. (D) Key word frequency for annotation of 169 overlapped gene as shown in (C). (E) Genomic features across different BPH chromosomes. From top to bottom, the number of protein-coding genes per megabase genome, the percentage of expressed genes out of all located protein-coding genes, the percentage of repeat elements of each chromosome, the mapping quality of reads, and the percentage of three-way orthologs among three planthoppers out of all protein-coding genes. Chromosome 8 is in red. (F) Distribution of recombination rates across different chromosomes for EA populations in 4Ne*r (where Ne is the effective size and r is the recombination rate). The red dashed line indicates the mean value of rho across all chromosomes. The two horizontal lines above and below the box represented the quartiles, the horizontal line in the middle represented the median, and the two whiskers showed the 1.5× interquartile expression on each chromosome. (G) Chromosome-scale synteny blocks between N. lugens and related species L. striatellus and S. furcifera using MCScanX (88). (H) Evolutionary rates of ortholog genes among planthopper species. The phylogenetic tree is based on (48). The distribution of dN, dS, and dN/dS on each chromosome are shown in black boxes for N. lugens versus L. striatellus and red boxes for N. lugens versus S. furcifera. Chromosome 8 is highlighted in green.
The top highly diverged regions encompass 201 genes (between EA and SEA) and 228 genes (between EA and SA); the majority (169) of these genes are overlapped (Fig. 5C). However, we observed that most of these genes had extremely low expression levels across 55 representative developmental stages and tissues of BPH (fig. S15). Furthermore, we found that a great proportion of these genes are either specific to BPH (38) or annotated with unknown function (44) (Fig. 5D and data S4). There were only 32 genes annotated with biological pathways, but no pathways were significantly enriched (corrected P < 0.05, hypergeometric test).
The highly diverged chromosome is a gene desert
What factors have caused chromosome 8 to be the differentiation outlier across all chromosomes? It was first ruled out as a sexual chromosome based on the sequencing coverage between males (X-) and females (XX) (31). Chromosome 8 encodes 512 protein-coding genes, exhibiting the lowest gene density among all chromosomes (7.5 genes/Mb, 31 to 62% lower than the other 15 chromosomes; Fig. 5E and data S2). Through profiling the spatial-temporal expression across 55 representative transcriptomes, we observed a significantly lower level of expression in genes of chromosome 8 in comparison to any other nonsexual chromosomes (P < 4.34 × 10−9, one-sided Wilcoxon rank-sum test) (fig. S16 and data S2). In addition, this chromosome has a much higher ratio of unexpressed genes (25.4%), which is much higher than other autosomes ranging from 6.8 to 14.3% (Fig. 5E and data S2). Combined, chromosome 8 shows apparent features of a gene desert (43).
On the other hand, we found that chromosome 8 is of remarkably high level of repeat content (79.4%, higher than any other chromosomes; Fig. 5E), which often causes improper mapping and false-positive variation calling. The overall mapping quality on chromosome 8 was indeed lower than most other chromosomes (Fig. 5E). We thus reanalyzed the population differentiation statistics by excluding variations within repetitive regions and found extreme divergence outliers still enriched in chromosome 8 (fig. S17, A and B). To avoid Wahlund effect by pooling multiple populations within a group, the comparisons were also performed between representative single populations, which still revealed the most highly diverged signatures on chromosome 8 (fig. S18, A and B). Together, the correlation between the overwhelmingly diverged signatures and the gene desert chromosome is unlikely to be caused by technical issues.
Chromosome 8 reconciles population differentiation and speciation
Population differentiation is considered to be associated with local recombination rate (44, 45). Correspondingly, we observed significantly lower recombination rates, especially in the EA group, in chromosome 8 than in the rest of genome (P < 5.19 × 10−4, one-sided Wilcoxon rank-sum test; Fig. 5F). A reduced recombination rate is thought to impede gene flow and create species barriers (46, 47), prompting us to investigate the role of chromosome 8 in speciation. We compared the BPH genome with those of two other closely related species, the white-backed planthopper Sogatella furcifera and the small BPH Laodelphax striatellus, which diverged from BPH approximately 64.4 million years (48). We characterized 5542 three-way orthologs that share the consistent chromosomal assignment across the three planthopper species and found only 80 of them located on chromosome 8, which (15.6% of all protein-coding genes) is significantly underrepresented compared to other chromosomes (33.41%; Chi-square test, P = 1.9 × 10−10; Fig. 5E and data S2). Chromosome 8 also exhibited the lowest density of genomic synteny blocks across all chromosomes except Y (Fig. 5G). We further estimated the synonymous (dS) and nonsynonymous (dN) substitution rates between BPH and the other two planthoppers (S. furcifera and L. striatellus), respectively, for orthologous genes. As a result, orthologs on chromosome 8 present significantly higher dN and dS than those on any other chromosomes, except chromosome 14 (Fig. 5H). These lines of evidence jointly suggest that chromosome 8 has undergone rapid evolution in terms of both gene turnover and sequence variation along with the speciation of planthoppers.
The comparison between SEA and SA groups identified multiple peaks with relatively even divergence signatures, including those from chromosome 8 (fig. S17C). However, the degree of divergence is much lower than the comparisons with EA (fig. S17). Coupled with the proposed history that SA first split from EA/SEA and then EA and SEA separated, we hypothesize that the speciation hotspot on chromosome 8 has been anciently maintained along with the within-species differentiation of BPH and further promoted along with the evolved round-trip migration in EA.
DISCUSSION
In this study, we applied population genomic approaches to characterize the genetic architectures and evolutionary history of worldwide BPHs and pioneered the inference of the main seasonal movement patterns for these windborne migrants based on gene flows. We propose that BPHs of SEA and EA first diverged from those in SA and that EA populations further diverged from tropical SEA populations along with the evolved northward migration to colonize temperate regions. Our results reveal that BPHs in EA undergo a round-trip seasonal migration between Indochina and EA, while those in SA and Malay Archipelago seem to exhibit one-way migration, from their respective remote ends into Indochina (Fig. 4). Notably, the inferred massive migration in SEA and SA is unexpected to some degree, as relatively short-distance movement is expected for BPHs living in the tropical continuous-breeding areas (22, 49). Correspondingly, previous studies showed a population sampled in Philippines is genetically distinct from populations in SEA and EA (29). It is noteworthy that the inferred gene flows from Philippines to Indochina in our study were based on five other different populations that were all sampled in February and March (data S1), when the monsoon wind from the western Pacific Ocean prevails (fig. S12). A likely scenario reconciling the disparity would be the existence of multiple populations with different migratory properties in Philippines, including both local populations as we found in FJ and migratory populations moving with the winds.
Because most Asian BPHs travel toward Indochina, there is a geographic potential for interbreeding among populations of EA, SEA, and SA in this region. Correspondingly, we found relatively evident spatial population genetic structures in remote populations of SEA and SA, with a gradual lack of structure signatures as they approach Indochina (Fig. 1 and fig. S1). Although the round-trip migration reduces genetic structuring among most populations in EA (Fig. 1 and fig. S1), admixed signatures were frequently detected in southern populations proximal to Indochina (Fig. 2 and figs. S5, S10, and S11). Previous studies have suggested frequent interbreeding of rice planthoppers across the areas of Greater Mekong subregion and EA migrants overwinter in central-south areas of Indochina (22, 50, 51). Thus, a progressive gene introgression from SA and SEA to EA should occur during the overwintering periods. On the other hand, we found that EA and SEA populations are genetically differentiable (Fig. 1D and fig. S3B) and unexpectedly observed ongoing rapid differentiation within EA populations (fig. S13). These patterns, in turn, indicate an existing “mechanism” that helps maintain the genetic ancestry and traits in EA populations.
We observed EA ancestry dominated in north-central Vietnam populations sampled on planted rice in January to March (data S1), differing from the SEA ancestry in south Vietnam populations and even from the admixed ancestry in further north populations sampled in late summer (Fig. 2A and figs. S5 and S8). The unexpected genetic disparities among Vietnamese populations could be explained either by an assumed scenario that the short-range movements of planthoppers in the Greater Mekong are seasonal or by assumed constraints on interbreeding. Further behavioral assays and genomic studies on more samples from additional locations and seasons might help resolve the disparity. Nevertheless, this finding supports the possibility of north-central Vietnam being an optional overwintering area for EA migrants. Moreover, recent studies have identified 23°N to 25°N as the potential northern overwintering boundary for BPH, encompassing southern areas of China such as Hainan, Guangdong, Guangxi, and FJ (16, 24, 52). Respective colonization of dispersed overwintering sites might help EA populations maintain their relatively unique genetic structures.
It is also unexpected that there are no SEA or SA outliers in central-north sampling sites of EA (Fig. 2A and fig. S5), given that the direction of BPH migration largely depends on winds. Why do SEA and SA immigrants arriving in Indochina not travel north together with other EA remigrants? It is unlikely due to a memory-based migratory trajectory similar to birds, as it cannot be contemplated in such small insects. A plausible explanation is that outlier migrants have been eliminated during the northward journey, which is partially compatible with the theory of “contemporary within-season selection” proposed by Drake and Reynolds (53). This theory suggests that the contemporary selection is progressively occurring in long-distance migration that successively advances the populations migrating northeastward (53). We indeed observed accelerated signatures of population subdifferentiation within EA (fig. S13). To test whether long-flying populations have been subjected to “contemporary selection,” we tried to compare the population reaching Japan, the most northeast site of EA migration, with other EA populations from representative sites and tropical populations from SEA and SA (fig. S18). As expected, we observed evident outliers on chromosome 8 when comparing with populations from SEA and SA (fig. S18). In addition, we found multiple genomic regions to be highly diverged between Japan and other EA populations, further supporting the observed ongoing differentiation among them. However, none of the most diverged regions encode biologically meaningful genes related to presumed traits such as flight, cold tolerance, or metabolism. Instead, most of them encode hypothetical proteins, except a gene on chromosome X encoding the solute carrier (SLC) family protein that is highly diverged with populations of Hainan (south China) and Shaanxi (northwest China) (fig. S18C). This divergence also occurred when comparing with an India population of SA (fig. S18A). Coupled with its potential role in xenobiotic metabolism, the repeated evolution of this gene is possibly subjected to the selection of insecticide or host races. Together, our findings support the further differentiation along with the long-distance migration in EA, while this ongoing isolation is more likely to be driven by genetic drift, which is commonly expected in “founder” populations exploiting territories. As multigenerational migrants, partial journeys of EA BPHs act as territory colonization or lead to the extinction of a subpopulation within a single generation. Consequently, the continued “founder effect” and range expansion contributed to the accelerated population differentiation within the EA group.
A possible alternative scenario may be that the long-distance migration in EA BPHs is highly selective in functional constraints for other immigrants. The highly diverged regions between EA migrants and tropical populations were unexpectedly limited to a single chromosome (chromosome 8). Simultaneously, there were remarkable differences in this chromosome between BPH and its closely related species, S. furcifera and L. striatellus, in terms of the densities of orthologous genes and syntenic blocks (Fig. 5, E and G). We further found that the high level of divergence on this chromosome was anciently maintained across the main groups of BPH and further augmented along with the evolution of EA. Multiple lines of evidence indicated that this chromosome resembles a gene desert (Fig. 5), enriched in genes with unknown functions and/or of low expression levels (Fig. 5E and fig. S16). Genes with low levels of expression are generally considered less important for phenotypes and are subject to selection relaxation (54–56). Thus, variations could escape the purging effects of selection and accumulate in this chromosome through genetic drift. A similar pattern has been observed in X chromosomes of aphids and explained by the relaxation of purifying selection on X-linked genes (57) despite that chromosome 8 of BPH is not a sexual chromosome. The little functional evidence questions whether migratory genes truly exist in windborne migrating insects. Nevertheless, we still found that several genes encoding Ras-related proteins were included in the highly diverged gene list (data S4). The Ras signaling pathway widely involves in hypoxia stress, ultraviolet irradiation, and heat response (58–60), which may contribute to the adaptation of BPH to adverse environmental conditions during long-distance migration. We also identified a single genomic region from the unplaced scaffold (scaffold001) showing a high level of divergence (data S4). The encompassing genes encode myotubularin-related protein 10-B, alpha-tocopherol transfer protein, and nucleolar protein 6, respectively, which were suggested associated with muscle weakness, infertility, or innate immunity in mammals or nematodes (61–64). Whether these genes, independent of the divergence hotspot, confer the unique adaptations to long-distance migration in EA deserves further verifications.
In summary, our study shows the power of applying genomics methodology in dissecting migration in insects, particularly in windborne insects whose migration is challenging to track directly. On the basis of the genome-wide variations, the complex population structures and evolutionary history of global BPHs are resolved at high resolution. In addition, gene flow–based assembly of trajectories clarifies the migration patterns of BPH, differentiating round-trip and one-way journeys between temperate and tropical populations. The insights gained in this study not only advance our understanding of windborne migration in insects but also benefit the control and warning of migratory pests.
MATERIALS AND METHODS
Sampling
BPHs were collected from worldwide distributed regions from 2009 to 2018. A total of 358 female adult BPHs from 92 geographic locations were lastly subjected for sequencing according to the geographic representativeness, including 122 samples from EA, 139 samples from SEA, 91 samples from SA, and 6 samples from AUS (Fig. 1A and data S1). We also sampled the closely related species of BPH, N. muiri, from Zhejiang, China as the outgroup to polarize some analyses. Two fifth instar nymphs were subjected to sequencing. All the samples were maintained in absolute ethanol before DNA extraction.
Sequencing
At least 1.0 μg of genomic DNA per sample was extracted by the SDS method. DNA purity and integrity were analyzed by gel electrophoresis. Sequencing libraries were constructed by the NEBNext Ultra II DNA Library Prep Kit following the manufacturer’s instruction. The entire library was prepared by randomly fragmenting genomic DNA with Covaris ultrasonic disruptors, followed by end repair, A-tailing, add adaptors, purification, and polymerase chain reaction (PCR) amplification. All BPH samples were sequenced on the Illumina HiSeq X platform, generating paired-end reads of 150 base pair with an average depth of 10× per sample. Two N.muiri individuals were sequenced with an approximate depth of 40×.
Quality control
Raw sequencing data were filtered for low-quality bases and adapters using fastp (65). The read pair was discarded if (i) more than 10% of bases were ambiguous; (ii) more than 50% of bases were of low quality (Phred quality score ≤ 5); (iii) aligned to any adapters. As a result, approximately 5 T of clean bases (33.7 billion reads) were retained for subsequent analyses (data S1).
SNP calling
Clean reads were mapped against the BPH reference genome (31) using Burrows-Wheeler Alignment (BWA) Tool (66) v0.7.17-r1188 with parameters “mem -t 8 -k 32 -M –R”. The alignment results were sorted by SAMtools v1.9 (67) and retained in SAM format. Variant calling was performed following the best practice of GATK using the version 4.1.8.1 (68). Briefly, potential PCR duplicates were first removed by the module “MarkDuplicates”, and then the module “HaplotypeCaller” was applied to characterize variants for each sample. After all 360 BPH samples being called with individual variants, the module “GenomicsDBImport” was used to combine all individual files and the module “GenotypeGVCFs” was used to genotyping all samples at each variant locus. To control the error rate in single-nucleotide polymorphism (SNP) calling due to low-quality mapping or low-coverage sequencing, a set of stringent thresholds was applied to filter the raw variant sets, including the overall variant quality (50), the maximum missing rate across all samples (50%), the minor allele frequency (1%), the maximum mean sequencing depth at each locus (20×), and the minimum mean sequencing depth (3×).
Genetic relationship across individuals
Bi-allelic SNPs were retained for inferring the phylogenetic relationship across individuals. Low-sequencing coverage might cause underestimation of heterozygous variations. Thus, genotypes of loci with sequencing coverage lower than 8× were masked. A total of 5,684,172 SNP loci with at least 120 nonmasked individuals and minor allele frequency higher than 0.01 were further retained to infer the phylogenomic tree. The genetic distance matrix across all 360 individuals was calculated on the basis of the average pairwise distance as described (5). The distance matrix was provided for MEGA 7 (69) to infer the NJ phylogenetic relationship. Bootstrap analysis was performed by randomly selecting loci 5,684,172 times and recalculating the matrix 100 times. EIGENSOFT v5.0.2 (70) was also used to perform the PCA analysis using default settings. Ancestry analyses were performed using sNMF v2.0 (71) and admixture v1.3.0 (72). Ancestors (K) were predefined from 2 to 6, and cross-validation analysis was conducted to compare the reliability of each K.
Assignment of genetic groups
The BPH individual was assigned to a designated group based on its estimated ancestry frequency by sNMF, when K = 6. To do this, the ancestry was sorted on the basis of probability from high to low, and then each ancestry was assigned to a given individual if the probability of this ancestry was estimated higher than 1/6, unless the first ancestry had a dominant probability higher than 2/3, or its probability was lower than half of the previous ancestry. Individuals assigned with more than one kind of ancestry were considered admixed; otherwise, they were assigned to a main group such as EA, SEA, SA, BGD, FJ, or AUS accordingly. On the basis of the ancestry assignment results of the involved samples, each geographic population was further classified into either a main group if no more than one main kinds of individuals or an admixed group.
To test the reliability of grouping, f4 statistics were applied using qpDstat v900 of AdmixTools (73). In each D statistics (AUS, X; A, B), the affinity of a given geographic population X to a pairwise main group A and B was tested, when using AUS as the outgroup. Each main group was represented by core samples, which were selected considering the proportion of dominant ancestry (>80%) and the genetic divergence from Indochina samples in PCA.
Gene flow between populations
The gene flow between each possible pairwise population was inferred on the basis of the iterative three-population test (f3 statistics). The test can provide clear evidence of admixture, i.e., a significantly negative f3 (X; A, B) implies that X is admixed with populations close to A and B. f3 statistics were applied across all possible combinations of geographic populations. In the case of the significantly negative f3 (X; X1, X2) (Z < −3), X was considered admixed with X1 or X2 based on the comparison between the count number of significantly negative f3 (X; X1, Y) and that of f3 (X; X2, Y), in which Y refers to any other populations of the same main group besides X, X1/X2. In other words, the gene flow from X1 to X was considered when most f3 (X; X1, Y) has a significantly negative value (Z < −3), unless Y has been determined as the donor of X. The genetic differentiation (dXY) was calculated using custom scripts between each pair of populations.
Demographic history
The population-level phylogenetic tree was inferred on the basis of frequency alleles using TreeMix v1.13 (74) with parameter “-global.” The analyses were performed using “core” samples and all involved samples, respectively. Dynamic demographic history of each main group was inferred by the PSMC model (37). “Core” samples from each group were selected as representatives. First, the read alignment results were transformed to the diploid consensus sequence using SAMtools, bcftools, and vcfutils.pl scripts (75). Then, consensus sequences were transformed to fasta format by “fq2psmcfa -q 20”. Consequently, parameters “psmc -N25 -t15 -r5 -p “4+25*2+4+6” were used for the PSMC model. The generation time g was set as 1/12 year(s), and the mutation rate μ was 8.4 × 10−9 as referred Drosophila (76). MSMC v1.1.0 (40) was also applied to infer the demographic history using phased genotypes. We transformed the BWA alignment results to vcf format using SAMtools mpileup command and then phased the data implemented BEAGLE v4.1 (77) with default settings. Linkage disequilibrium was estimated for “core” samples of each main group on PopLDdecay (78) by calculating the squared correlation coefficient (r2). The max distances between two SNPs were less than 300 kb. Sites with minor allele frequency less than 0.005 and missing allele ratio great than 0.25 were filtered. Tajima’s D and nucleotide diversity (π) were calculated along 10-kb sliding windows using custom Perl scripts. Cross-population comparison was performed on the basis of a balanced size of sampling, for which samples were randomly selected from the group with larger sample size. Minor allele frequency was measured with plink v1.9 (79) by “--freq” command.
Characterization of highly diverged regions and selective sweeps
Variants with more than two alleles across samples and individuals having first and second degree relation were further removed from the analysis (data S5). For population differentiation, Wright’s fixation index (FST) (80) and allele frequency differentiation (XP-CLR) (42) were applied for cross-validation. FST was calculated for each locus using VCFtools v0.1.17 (81) and subsequently estimated window boundaries based on the smoothing spline model using GenWin v0.1 (82). Window-based FST was normalized across the genome by transforming to W statistics (82). XP-CLR was calculated for sliding 10-kb windows using a python module (github.com/hardingnj/xpclr). To characterize the most highly diverged regions between EA and other groups, the genomic windows with the highest 1% W-transformed FST between EA and SEA and between EA and SA were considered. To limit the selection in EA, additional reduction of π in EA is required.
Selective sweeps were identified by applying several independent approaches. Selection signatures of a single population (group) were considered for each 10-kb sliding window based on the reduction of π. For haplotype-based analyses, we first phased genotypes using BEAGLE v25Nov19.28d (77). Recombination rates of each chromosome were calculated for each main group using an R package FastEPRR2.0 (83). To do this, the value of rho (rho = 4Ne × r, where Ne is the effective population size and r is the recombination rate) was calculated in 1-Mb genomic windows based on the phased VCF file as described above.
Annotations of gene models and pathways were referenced to the previous study (31). Expression profiles of genes were determined on the basis of available transcriptome data of 55 representative developmental stages and tissues (84). Pathway enrichment analysis was applied using the online platform OmicShare (www.omicshare.com/tools). Pathway with q value < 0.05 was considered as a significantly enriched pathway.
Comparative genomics among three planthopper species
Genome sequence and gene annotations of another two planthopper species were generated as described. The longest transcript was retained for each single gene model. BLASTP was conducted for any pairs of the three planthopper gene set and reciprocal best BLASTP hits were considered as 1 versus 1 ortholog genes between species. Three-way ortholog genes were determined across the three species by combining the consensus 1 versus 1 orthologs of each pairwise comparison. Protein sequences of each three-way ortholog were multi-aligned using muscle v3.8.31 (85). Corresponding nucleotide sequences were retrieved using a Perl script, pal2nal v14 (86). The synonymous (dS) and nonsynonymous (dN) substitution rates were calculated using the codeml module of PAML v4.8 (86) under the phylogenetic context as described (48).
Acknowledgments
We thank M. Ashfaque (PARC, Pakistan), S. Zakaria (SKU, Indonesia), J. Pilianto (Brawijaya University, Indonesia), Y.H. Song (GNU, Korea), H. Noda (NIAS, Japan), S. Sanada (NARO, Japan), C. Huy Nguyen (Vietnam Academy of Agricultural Sciences, Vietnam), N. Kado (Plant Protection Office, Shimane Prefecture, Japan), G. Bellis (Department of Agriculture, Fisheries and Forestry, Australia), S. Sarathbabu (Mizoran University Aizawl, India), W. Sriratanasak (Division of Rice Research and Development, Thailand), W. Janlapha (Prachinburi Rice Research Center, Thailand), S.H. Gu (Taiwan Museum of Natural Science), X.-D. Li (Yunnan Academy of Agricultural Sciences), D.-W. Zhang (Zunyi Normal University), B. Chen (Chongqing Normal University), D. Wang (Northwest A&F University), F.-K. Huang (Guangxi Academy of Agricultural Sciences), Y.-K. Li (Hainan University), Y.-Z. Li (Hunan Agricultural University), C.-Y. Niu (Huazhong Agricultural University), L.-H. Lu (Guangdong Academy of Agricultural Sciences), T.-Y. Wei and G. Yang (Fujian A&F University), J.-l. Peng (Protection Station of Xiamen, Fujian), Z.-L. Shu (Zhenjiang Institute of Agricultural Science, Jiangsu), S.-L. Jin (Xinyang Normal University), H.-J. Xiao (Jiangxi Agricultural University), H. Ma (Shandong Academy of Agricultural Sciences), Z.-Y. Liu, M.-X. Jiang (Zhejiang University), W.-Q. Zhang (Sun Yat-sen University), Z.-X. Lu and H.-X. Xu (Zhejiang Academy of Agricultural Sciences), X.-Y. Zhou and K.-M. Wu (Institute of Plant Protection, CAAS) for the kind help in BPH collections.
Funding: This work was supported by the National Natural Science Foundation of China (32230086, 31630057, 32225008, and 32021001), National key R & D plan in the 14th five year plan (2021YFD1401100), and Chinese Academy of Sciences (XDB27040205, Project for Young Scientists in Basic Research).
Author contributions: Conceptualization: C.-X.Z. and S.Z. Formal analysis: S.Z., Q.-L.H., G.-Q.F., Y.-X.Y., Y.-X.Z., and A.-D.C. Investigation: C.-X.Z., J.-C.Z., J.-B.L., and Q.-L.H. Software: S.Z., Q.-L.H., Y.-X.Y., G.-Q.F., and Y.X.Z. Resources: C.-X.Z., Q.-L.H., J.-B.L., J.-C.Z., D.-T.L., Y.-H.L., X.-Y.Z., X.C., S.-L.W., Z.-C.W., N.M., S.S.O., T.T., P.N.S., J.J., I.H.S., M.T.I., S.M.M.R., N.A.A., Z.-R.Z., K.L.H., G.L., H.-J.H., J.-M.L., and J.-P.C. Methodology: S.Z. Validation: S.Z. and Q.-L.H. Visualization: S.Z. and Q.-L.H. Data curation: C.-X.Z. and Q.-L.H. Writing—original draft: C.-X.Z., S.Z., and Q.-L.H. Writing—review and editing: C.-X.Z., S.Z., Q.-L.H., P.N.S., M.T.I., K.L.H., N.M., N.A.A., S.M.M.R., Z.-R.Z., and I.H.S. Supervision: C.-X.Z. Funding acquisition: C.-X.Z. and S.Z. Project administration: C.-X.Z. and S.Z.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: All genomic sequencing data has been deposited in NCBI under the BioProject PRJNA749496. All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials.
Supplementary Materials
This PDF file includes:
Figs. S1 to S18
Legends for data S1 to S5
Other Supplementary Material for this manuscript includes the following:
Data S1 to S5
REFERENCES AND NOTES
- 1.Chapman J. W., Reynolds D. R., Wilson K., Long-range seasonal migration in insects: Mechanisms, evolutionary drivers and ecological consequences. Ecol. Lett. 18, 287–302 (2015). [DOI] [PubMed] [Google Scholar]
- 2.Brower L. P., Understanding and misunderstanding the migration of the monarch butterfly (Nymphalidae) in North America: 1857-1995. J. Lepid. Soc. 49, 304–385 (1995). [Google Scholar]
- 3.Vidal O., Rendon-Salinas E., Dynamics and trends of overwintering colonies of the monarch butterfly in Mexico. Biol. Conserv. 180, 165–175 (2014). [Google Scholar]
- 4.Reppert S. M., Gegear R. J., Merlin C., Navigational mechanisms of migrating monarch butterflies. Trends Neurosci. 33, 399–406 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhan S., Zhang W., Niitepold K., Hsu J., Haeger J. F., Zalucki M. P., Altizer S., de Roode J. C., Reppert S. M., Kronforst M. R., The genetics of monarch butterfly migration and warning colouration. Nature 514, 317–321 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lundberg M., Liedvogel M., Larson K., Sigeman H., Grahn M., Wright A., Åkesson S., Bensch S., Genetic differences between willow warbler migratory phenotypes are few and cluster in large haplotype blocks. Evol. Lett. 1, 155–168 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gu Z., Pan S., Lin Z., Hu L., Dai X., Chang J., Xue Y., Su H., Long J., Sun M., Ganusevich S., Sokolov V., Sokolov A., Pokrovsky I., Ji F., Bruford M. W., Dixon A., Zhan X., Climate-driven flyway changes and memory-based long-distance migration. Nature 591, 259–264 (2021). [DOI] [PubMed] [Google Scholar]
- 8.Huestis D. L., Dao A., Diallo M., Sanogo Z. L., Samake D., Yaro A. S., Ousman Y., Linton Y. M., Krishna A., Veru L., Krajacich B. J., Faiman R., Florio J., Chapman J. W., Reynolds D. R., Weetman D., Mitchell R., Donnelly M. J., Talamas E., Chamorro L., Strobach E., Lehmann T., Windborne long-distance migration of malaria mosquitoes in the Sahel. Nature 574, 404–408 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stefanescu C., Soto D. X., Talavera G., Vila R., Hobson K. A., Long-distance autumn migration across the Sahara by painted lady butterflies: Exploiting resource pulses in the tropical savannah. Biol. Lett. 12, 20160561 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Knight S. M., Pitman G. M., Flockhart D. T. T., Norris D. R., Radio-tracking reveals how wind and temperature influence the pace of daytime insect migration. Biol. Lett. 15, 20190327 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Menz M. H. M., Scacco M., Bürki-Spycher H. M., Williams H. J., Reynolds D. R., Chapman J. W., Wikelski M., Individual tracking reveals long-distance flight-path control in a nocturnally migrating moth. Science 377, 764–768 (2022). [DOI] [PubMed] [Google Scholar]
- 12.J. A. Cheng, "Rice planthopper problems and relevant causes in China" in Planthoppers: New threats to the sustainability of intensive rice production systems in Asia, H. K. L., H. B., Eds. (International Rice Research Institute, 2009), pp. 157–178. [Google Scholar]
- 13.Bottrell D. G., Schoenly K. G., Resurrecting the ghost of green revolutions past: The brown planthopper as a recurring threat to high-yielding rice production in tropical Asia. J. Asia Pac. Entomol. 15, 122–140 (2012). [Google Scholar]
- 14.Lu M. H., Chen X., Liu W. C., Zhu F., Lim K. S., McInerney C. E., Hu G., Swarms of brown planthopper migrate into the lower Yangtze River Valley under strong western Pacific subtropical highs. Ecosphere 8, e01967 (2017). [Google Scholar]
- 15.V. Dyck, B. Thomas, "The brown planthopper problem" in Brown planthopper: Threat to rice production in Asia (International Rice Research Institute, 1979), pp. 3–17. [Google Scholar]
- 16.Cheng X. N., Chen R. C., Xi X., Yang L. M., Yang J. S., Studies on the migrations of brown planthopper Nilaparvata lugens (Stål). Acta Entom. Sin. 22, 1–21 (1979). [Google Scholar]
- 17.K. Sogawa, in Rice Planthoppers: Ecology, Management, Socio Economics and Policy, K. L. Heong, J. Cheng, M. M. Escalada, Eds. (Springer, 2015), pp. 33–63. [Google Scholar]
- 18.Riley J. R., Cheng X. N., Zhang X. X., Reynolds D. R., Xu G. M., Smith A. D., Cheng J. Y., Bao A. D., Zhai B. P., The Long-Distance Migration of Nilaparvata lugens(Stål) (Delphacidae) in China: Radar Observations of Mass Return Flight in the Autumn. Ecol. Entomol. 16, 471–489 (1991). [Google Scholar]
- 19.Riley J. R., Reynolds D. R., Smith A. D., Rosenberg L. J., Cheng X. N., Zhang X. X., Xu G. M., Cheng J. Y., Bao A. D., Zhai B. P., Wang H. K., Observations on the Autumn Migration of Nilaparvata lugens(Homoptera: Delphacidae) and Other Pests in East Central China. Bull. Entomol. Res. 84, 389–402 (1994). [Google Scholar]
- 20.Otuka A., Matsumura M., Watanabe T., Dinh T. V., A migration analysis for rice planthoppers, Sogatella furcifera (Horváth) and Nilaparvata lugens (Stål) (Homoptera: Delphacidae), emigrating from northern Vietnam from April to May. Appl. Entomol. Zool. 43, 527–534 (2008). [Google Scholar]
- 21.Hu G., Lu M. H., Tuan H. A., Liu W. C., Xie M. C., McInerney C. E., Zhai B. P., Population dynamics of rice planthoppers, Nilaparvata lugens and Sogatella furcifera (Hemiptera, Delphacidae) in Central Vietnam and its effects on their spring migration to China. Bull. Entomol. Res. 107, 369–381 (2017). [DOI] [PubMed] [Google Scholar]
- 22.Wu Q. L., Hu G., Tuan H. A., Chen X., Lu M. H., Zhai B. P., Chapman J. W., Migration patterns and winter population dynamics of rice planthoppers in Indochina: New perspectives from field surveys and atmospheric trajectories. Agric. For. Meteorol. 265, 99–109 (2019). [Google Scholar]
- 23.Chen R., Wu J., Zhu S., Zhang J., Flight capacity of the brown planthopper Nilaparavata lugens Stål. Acta Entom. Sin. 27, 121–126 (1984). [Google Scholar]
- 24.Ju L., Yu L., Yi-Fei G., Xia-Nian C., Qiang F. U., Gao H. U., Investigation of the overwintering of three species of rice pest, Nilaparvata lugens,Sogatella furcifera and Cnaphalocrocis medinalis in China. Chin. J. Appl. Entomol. 50, 253–260 (2013). [Google Scholar]
- 25.Xiang H., Liu X. J., Li M. W., Zhu Y. N., Wang L. Z., Cui Y., Liu L. Y., Fang G. Q., Qian H. Y., Xu A. Y., Wang W., Zhan S., The evolutionary road from wild moth to domestic silkworm. Nat. Ecol. Evol. 2, 1268–1279 (2018). [DOI] [PubMed] [Google Scholar]
- 26.Huang X., Kurata N., Wei X., Wang Z. X., Wang A., Zhao Q., Zhao Y., Liu K., Lu H., Li W., Guo Y., Lu Y., Zhou C., Fan D., Weng Q., Zhu C., Huang T., Zhang L., Wang Y., Feng L., Furuumi H., Kubo T., Miyabayashi T., Yuan X., Xu Q., Dong G., Zhan Q., Li C., Fujiyama A., Toyoda A., Lu T., Feng Q., Qian Q., Li J., Han B., A map of rice genome variation reveals the origin of cultivated rice. Nature 490, 497–501 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mun J. H., Song Y. H., Heong K. L., Roderick G. K., Genetic variation among Asian populations of rice planthoppers,Nilaparvata lugens and Sogatella furcifera (Hemiptera: Delphacidae): Mitochondrial DNA sequences. Bull. Entomol. Res. 89, 245–253 (1999). [Google Scholar]
- 28.Matsumoto Y., Matsumura M., Sanada-Morimura S., Hirai Y., Sato Y., Noda H., Mitochondrial cox sequences of Nilaparvata lugens and Sogatella furcifera (Hemiptera, Delphacidae): Low specificity among Asian planthopper populations. Bull. Entomol. Res. 103, 382–392 (2013). [DOI] [PubMed] [Google Scholar]
- 29.Hereward J. P., Cai X. H., Matias A. M. A., Walter G. H., Xu C. X., Wang Y. M., Migration dynamics of an important rice pest: The brown planthopper (Nilaparvata lugens) across Asia-Insights from population genomics. Evol. Appl. 13, 2449–2459 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tyagi S., Narayana S., Singh R. N., Srivastava C. P., Twinkle S., Das S. K., Jeer M., Migratory behaviour of Brown planthopper, Nilaparvata lugens (Stål) (Hemiptera: Delphacidae), in India as inferred from genetic diversity and reverse trajectory analysis. 3 Biotech 12, 266 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ye Y. X., Zhang H. H., Li D. T., Zhuo J. C., Shen Y., Hu Q. L., Zhang C. X., Chromosome-level assembly of the brown planthopper genome with a characterized Y chromosome. Mol. Ecol. Resour. 21, 1287–1298 (2021). [DOI] [PubMed] [Google Scholar]
- 32.Claridge M., Hollander J. D., Morgan J., Variation in courtship signals and hybridization between geographically definable populations of the rice brown planthopper, Nilaparvata lugens (Stål). Biol. J. Linn. Soc. 24, 35–49 (1985). [Google Scholar]
- 33.Bao Y., Cang X., Yang S., Chen C., Xie X., Lu M., Liu W., Impact of atmospheric low temperature stress on the initial immigration of Nilaparvata lugens (Stål) in a year in China. Acta Ecol. Sin. 40, 7519–7533 (2020). [Google Scholar]
- 34.Otuka A., Watanabe T., Suzuki Y., Matsumura M., Furuno A., Chino M., A migration analysis of the rice planthopper Nilaparvata lugens from the Philippines to East Asia with three-dimensional computer simulations. Popul. Ecol. 47, 143–150 (2005). [Google Scholar]
- 35.Otuka A., Huang S. H., Sanada-Morimura S., Matsumura M., Migration analysis of Nilaparvata lugens (Hemiptera: Delphacidae) from the Philippines to Taiwan under typhoon-induced windy conditions. Appl. Entomol. Zool. 47, 263–271 (2012). [Google Scholar]
- 36.Liu C.-H., Study on the long-distance migration of the brown planthopper in Taiwan. Chin. J. Entomol. 4, 49–54 (1984). [Google Scholar]
- 37.Li H., Durbin R., Inference of human population history from individual whole-genome sequences. Nature 475, 493–496 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Huang P., Schaal B. A., Association between the geographic distribution during the last glacial maximum of Asian wild rice, Oryza rufipogon (Poaceae), and its current genetic variation. Am. J. Bot. 99, 1866–1874 (2012). [DOI] [PubMed] [Google Scholar]
- 39.Ray N., Adams J., A GIS-based vegetation map of the world at the last glacial maximum (25,000-15,000 BP). Internet Archaeol. 11, 1–44 (2001). [Google Scholar]
- 40.Schiffels S., Wang K., MSMC and MSMC2: The multiple sequentially markovian coalescent. Methods Mol. Biol. 2090, 147–166 (2020). [DOI] [PubMed] [Google Scholar]
- 41.Gutaker R. M., Groen S. C., Bellis E. S., Choi J. Y., Pires I. S., Bocinsky R. K., Slayton E. R., Wilkins O., Castillo C. C., Negrao S., Oliveira M. M., Fuller D. Q., Guedes J. A. D., Lasky J. R., Purugganan M. D., Genomic history and ecology of the geographic spread of rice. Nat. Plants 6, 492–502 (2020). [DOI] [PubMed] [Google Scholar]
- 42.Chen H., Patterson N., Reich D., Population differentiation as a test for selective sweeps. Genome Res. 20, 393–402 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Venter J. C., Adams M. D., Myers E. W., Li P. W., Mural R. J., Sutton G. G., Smith H. O., Yandell M., Evans C. A., Holt R. A., Gocayne J. D., Amanatides P., Ballew R. M., Huson D. H., Wortman J. R., Zhang Q., Kodira C. D., Zheng X. H., Chen L., Skupski M., Subramanian G., Thomas P. D., Zhang J., Miklos G. L. G., Nelson C., Broder S., Clark A. G., Nadeau J., McKusick V. A., Zinder N., Levine A. J., Roberts R. J., Simon M., Slayman C., Hunkapiller M., Bolanos R., Delcher A., Dew I., Fasulo D., Flanigan M., Florea L., Halpern A., Hannenhalli S., Kravitz S., Levy S., Mobarry C., Reinert K., Remington K., Abu-Threideh J., Beasley E., Biddick K., Bonazzi V., Brandon R., Cargill M., Chandramouliswaran I., Charlab R., Chaturvedi K., Deng Z., Di Francesco V., Dunn P., Eilbeck K., Evangelista C., Gabrielian A. E., Gan W., Ge W., Gong F., Gu Z., Guan P., Heiman T. J., Higgins M. E., Ji R. R., Ke Z., Ketchum K. A., Lai Z., Lei Y., Li Z., Li J., Liang Y., Lin X., Lu F., Merkulov G. V., Milshina N., Moore H. M., Naik A. K., Narayan V. A., Neelam B., Nusskern D., Rusch D. B., Salzberg S., Shao W., Shue B., Sun J., Wang Z., Wang A., Wang X., Wang J., Wei M., Wides R., Xiao C., Yan C., Yao A., Ye J., Zhan M., Zhang W., Zhang H., Zhao Q., Zheng L., Zhong F., Zhong W., Zhu S., Zhao S., Gilbert D., Baumhueter S., Spier G., Carter C., Cravchik A., Woodage T., Ali F., An H., Awe A., Baldwin D., Baden H., Barnstead M., Barrow I., Beeson K., Busam D., Carver A., Center A., Cheng M. L., Curry L., Danaher S., Davenport L., Desilets R., Dietz S., Dodson K., Doup L., Ferriera S., Garg N., Gluecksmann A., Hart B., Haynes J., Haynes C., Heiner C., Hladun S., Hostin D., Houck J., Howland T., Ibegwam C., Johnson J., Kalush F., Kline L., Koduru S., Love A., Mann F., May D., McCawley S., McIntosh T., McMullen I., Moy M., Moy L., Murphy B., Nelson K., Pfannkoch C., Pratts E., Puri V., Qureshi H., Reardon M., Rodriguez R., Rogers Y. H., Romblad D., Ruhfel B., Scott R., Sitter C., Smallwood M., Stewart E., Strong R., Suh E., Thomas R., Tint N. N., Tse S., Vech C., Wang G., Wetter J., Williams S., Williams M., Windsor S., Winn-Deen E., Wolfe K., Zaveri J., Zaveri K., Abril J. F., Guigó R., Campbell M. J., Sjolander K. V., Karlak B., Kejariwal A., Mi H., Lazareva B., Hatton T., Narechania A., Diemer K., Muruganujan A., Guo N., Sato S., Bafna V., Istrail S., Lippert R., Schwartz R., Walenz B., Yooseph S., Allen D., Basu A., Baxendale J., Blick L., Caminha M., Carnes-Stine J., Caulk P., Chiang Y. H., Coyne M., Dahlke C., Mays A. D., Dombroski M., Donnelly M., Ely D., Esparham S., Fosler C., Gire H., Glanowski S., Glasser K., Glodek A., Gorokhov M., Graham K., Gropman B., Harris M., Heil J., Henderson S., Hoover J., Jennings D., Jordan C., J. Jordan, J. Kasha, Kagan L., Kraft C., Levitsky A., Lewis M., Liu X., Lopez J., Ma D., Majoros W., McDaniel J., Murphy S., Newman M., Nguyen T., Nguyen N., Nodell M., Pan S., Peck J., Peterson M., Rowe W., Sanders R., Scott J., Simpson M., Smith T., Sprague A., Stockwell T., Turner R., Venter E., Wang M., Wen M., Wu D., Wu M., Xia A., Zandieh A., Zhu X., The sequence of the human genome. Science 291, 1304–1351 (2001). [DOI] [PubMed] [Google Scholar]
- 44.Geraldes A., Basset P., Smith K. L., Nachman M. W., Higher differentiation among subspecies of the house mouse (Mus musculus) in genomic regions with low recombination. Mol. Ecol. 20, 4722–4736 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nachman M. W., Payseur B. A., Recombination rate variation and speciation: Theoretical predictions and empirical results from rabbits and mice. Philos. Trans. R. Soc. B-Biol. Sci. 367, 409–421 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Martin S. H., Davey J. W., Salazar C., Jiggins C. D., Recombination rate variation shapes barriers to introgression across butterfly genomes. PLoS Biol. 17, e2006288 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Slotman M. A., Reimer L. J., Thiemann T., Dolo G., Fondjo E., Lanzaro G. C., Reduced recombination rate and genetic differentiation between the M and S forms of Anopheles gambiae s.s. Genetics 174, 2081–2093 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ma W., Xu L., Hua H., Chen M., Guo M., He K., Zhao J., Li F., Chromosomal-level genomes of three rice planthoppers provide new insights into sex chromosome evolution. Mol. Ecol. Resour. 21, 226–237 (2021). [DOI] [PubMed] [Google Scholar]
- 49.Otuka A., Sakamoto T., Chien H. V., Matsumura M., Sanada-Morimura S., Occurrence and short-distance migration of Nilaparvata lugens (Hemiptera: Delphacidae) in the Vietnamese Mekong Delta. Appl. Entomol. Zool. 49, 97–107 (2014). [Google Scholar]
- 50.Li X.-Y., Chu D., Yin Y.-Q., Zhao X.-Q., Chen A.-D., Khay S., Douangboupha B., Kyaw M. M., Kongchuensin M., Ngo V. V., Possible source populations of the white-backed planthopper in the Greater Mekong Subregion revealed by mitochondrial DNA analysis. Sci. Rep. 6, 39167 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yin Y., Li X., Chu D., Zhao X., Sathya K., Douangboupha B., Kyaw M. M., Kongchuensin M., Somrith A., Ngo V. V., Extensive gene flow of white-backed planthopper in the Greater Mekong Subregion as revealed by microsatellite markers. Sci. Rep. 7, 15905 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Xue J., Zhou X., Zhang C.-X., Yu L.-L., Fan H.-W., Wang Z., Xu H.-J., Xi Y., Zhu Z.-R., Zhou W.-W., Pan P.-L., Li B.-L., Colbourne J. K., Noda H., Suetsugu Y., Kobayashi T., Zheng Y., Liu S., Zhang R., Liu Y., Luo Y.-D., Fang D.-M., Chen Y., Zhan D.-L., Lv X.-D., Cai Y., Wang Z.-B., Huang H.-J., Cheng R.-L., Zhang X.-C., Lou Y.-H., Yu B., Zhuo J.-C., Ye Y.-X., Zhang W.-Q., Shen Z.-C., Yang H.-M., Wang J., Wang J., Bao Y.-Y., Cheng J.-A., Genomes of the rice pest brown planthopper and its endosymbionts reveal complex complementary contributions for host adaptation. Genome Biol. 15, 521 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.V. Drake, D. Reynolds, "Migrations of pest and beneficial insects" in Radar entomology: Observing insect flight and migration (CABI, 2012), pp. 312–346. [Google Scholar]
- 54.Duret L., Mouchiroud D., Determinants of substitution rates in mammalian genes: Expression pattern affects selection intensity but not mutation rate. Mol. Biol. Evol. 17, 68–74 (2000). [DOI] [PubMed] [Google Scholar]
- 55.Nabholz B., Ellegren H., Wolf J. B., High levels of gene expression explain the strong evolutionary constraint of mitochondrial protein-coding genes. Mol. Biol. Evol. 30, 272–284 (2013). [DOI] [PubMed] [Google Scholar]
- 56.Nabholz B., Sarah G., Sabot F., Ruiz M., Adam H., Nidelet S., Ghesquière A., Santoni S., David J., Glémin S., Transcriptome population genomics reveals severe bottleneck and domestication cost in the African rice (Oryza glaberrima). Mol. Ecol. 23, 2210–2227 (2014). [DOI] [PubMed] [Google Scholar]
- 57.Mathers T. C., Wouters R. H. M., Mugford S. T., Swarbreck D., van Oosterhout C., Hogenhout S. A., Chromosome-scale genome assemblies of aphids reveal extensively rearranged autosomes and long-term conservation of the X chromosome. Mol. Biol. Evol. 38, 856–875 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Li Y., Zhang L., Kang M., Guo X., Xu B., AccERK2, a map kinase gene from Apis cerana cerana, plays roles in stress responses, developmental processes, and the nervous system. Arch. Insect Biochem. Physiol. 79, 121–134 (2012). [DOI] [PubMed] [Google Scholar]
- 59.Inoue H., Tateno M., Fujimura-Kamada K., Takaesu G., Adachi-Yamada T., Ninomiya-Tsuji J., Irie K., Nishida Y., Matsumoto K., A Drosophila MAPKKK, D-MEKK1, mediates stress responses through activation of p38 MAPK. EMBO J. 20, 5421–5430 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wang L., Cui S., Liu Z., Ping Y., Qiu J., Geng X., Inhibition of mitochondrial respiration under hypoxia and increased antioxidant activity after reoxygenation of Tribolium castaneum. PLOS ONE 13, e0199056 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sarikaya E., Sabha N., Volpatti J., Pannia E., Maani N., Gonorazky H. D., Celik A., Liang Y., Onofre-Oliveira P., Dowling J. J., Natural history of a mouse model of X-linked myotubular myopathy. Dis. Model. Mech. 15, dmm049342 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lim Y., Traber M. G., Alpha-tocopherol transfer protein (alpha-TTP): Insights from alpha-tocopherol transfer protein knockout mice. Nutr. Res. Pract. 1, 247–253 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Buj-Bello A., Furling D., Tronchère H., Laporte J., Lerouge T., Butler-Browne G. S., Mandel J. L., Muscle-specific alternative splicing of myotubularin-related 1 gene is impaired in DM1 muscle cells. Hum. Mol. Genet. 11, 2297–2307 (2002). [DOI] [PubMed] [Google Scholar]
- 64.Fuhrman L. E., Goel A. K., Smith J., Shianna K. V., Aballay A., Nucleolar proteins suppress Caenorhabditis elegans innate immunity by inhibiting p53/CEP-1. PLOS Genet. 5, e1000657 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Chen S. F., Zhou Y. Q., Chen Y. R., Gu J., fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 34, i884–i890 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Li H., Durbin R., Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics 26, 589–595 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., Durbin R., Proc G. P. D., The sequence alignment/Map format and SAMtools. Bioinformatics 25, 2078–2079 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.McKenna A., Hanna M., Banks E., Sivachenko A., Cibulskis K., Kernytsky A., Garimella K., Altshuler D., Gabriel S., Daly M., DePristo M. A., The Genome Analysis Toolkit: A MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 20, 1297–1303 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kumar S., Stecher G., Tamura K., MEGA7: Molecular Evolutionary Genetics Analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33, 1870–1874 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Patterson N., Price A. L., Reich D., Population structure and eigenanalysis. PLos Genet. 2, e190 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Frichot E., Mathieu F., Trouillon T., Bouchard G., Francois O., Fast and efficient estimation of individual ancestry coefficients. Genetics 196, 973–983 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Alexander D. H., Novembre J., Lange K., Fast model-based estimation of ancestry in unrelated individuals. Genome Res. 19, 1655–1664 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Patterson N., Moorjani P., Luo Y. T., Mallick S., Rohland N., Zhan Y. P., Genschoreck T., Webster T., Reich D., Ancient admixture in human history. Genetics 192, 1065–1093 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Pickrell J. K., Pritchard J. K., Inference of population splits and mixtures from Genome-wide allele frequency data. PLOS Genet. 8, e1002967 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Danecek P., Bonfield J. K., Liddle J., Marshall J., Ohan V., Pollard M. O., Whitwham A., Keane T., McCarthy S. A., Davies R. M., Twelve years of SAMtools and BCFtools. GigaScience 10, giab008 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Haag-Liautard C., Dorris M., Maside X., Macaskill S., Halligan D. L., Houle D., Charlesworth B., Keightley P. D., Direct estimation of per nucleotide and genomic deleterious mutation rates in Drosophila. Nature 445, 82–85 (2007). [DOI] [PubMed] [Google Scholar]
- 77.Browning S. R., Browning B. L., Rapid and accurate haplotype phasing and missing-data inference for whole-genome association studies by use of localized haplotype clustering. Am. J. Hum. Genet. 81, 1084–1097 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Zhang C., Dong S. S., Xu J. Y., He W. M., Yang T. L., PopLDdecay: A fast and effective tool for linkage disequilibrium decay analysis based on variant call format files. Bioinformatics 35, 1786–1788 (2019). [DOI] [PubMed] [Google Scholar]
- 79.Purcell S., Neale B., Todd-Brown K., Thomas L., Ferreira M. A. R., Bender D., Maller J., Sklar P., de Bakker P. I. W., Daly M. J., Sham P. C., PLINK: A tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 81, 559–575 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Hudson R. R., Slatkin M., Maddison W. P., Estimation of levels of gene flow from DNA sequence data. Genetics 132, 583–589 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Danecek P., Auton A., Abecasis G., Albers C. A., Banks E., DePristo M. A., Handsaker R. E., Lunter G., Marth G. T., Sherry S. T., The variant call format and VCFtools. Bioinformatics 27, 2156–2158 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Beissinger T. M., Rosa G. J. M., Kaeppler S. M., Gianola D., de Leon N., Defining window-boundaries for genomic analyses using smoothing spline techniques. Genet. Sel. Evol. 47, 30 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Gao F., Ming C., Hu W., Li H., New software for the fast estimation of population recombination rates (FastEPRR) in the genomic era. G3 6, 1563–1571 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Xue J., Bao Y.-Y., Li B.-L., Cheng Y.-B., Peng Z.-Y., Liu H., Xu H.-J., Zhu Z.-R., Lou Y.-G., Cheng J.-A., Transcriptome analysis of the brown planthopper Nilaparvata lugens. PLOS ONE 5, e14233 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Edgar R. C., MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32, 1792–1797 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Suyama M., Torrents D., Bork P., PAL2NAL: Robust conversion of protein sequence alignments into the corresponding codon alignments. Nucleic Acids Res. 34, W609–W612 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Monfreda C., Ramankutty N., Foley J. A., Farming the planet: 2. Geographic distribution of crop areas, yields, physiological types, and net primary production in the year 2000. Glob. Biogeochem. Cycle 22, GB1022 (2008). [Google Scholar]
- 88.Wang Y., Tang H., Debarry J. D., Tan X., Li J., Wang X., Lee T. H., Jin H., Marler B., Guo H., Kissinger J. C., Paterson A. H., MCScanX: A toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 40, e49 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S18
Legends for data S1 to S5
Data S1 to S5