Abstract
A 100-fold increase in luciferase activity was observed in 293 cells, stably expressing Epstein-Barr nuclear antigen 1 (EBNA1; 293-EBNA1 cells), that had been transiently transfected with plasmids carrying Epstein-Barr virus (EBV) oriP sequences. This increase was observed in comparison to reporter gene activity obtained after transfection with a plasmid carrying no oriP sequences. The luciferase gene on these plasmids was under the control of either the cytomegalovirus immediate-early 1 gene enhancer-promoter (CMV IE1) or the Rous sarcoma virus promoter. The increase of reporter gene activity was not due to plasmid replication, since a similar enhancement was observed in the presence of aphidicolin, an inhibitor of replicative DNA synthesis, or after deletion of the dyad symmetry (DS) element within oriP. Luciferase production was not increased in the presence of only the DS element. Microinjection of plasmids carrying the CMV IE1 promoter-driven luciferase gene with or without oriP sequences into the nuclei of 293-EBNA1 cells resulted in a 17-fold increase in luciferase activity. Cytoplasmic injection of these plasmids led to an enhancement of luciferase activity of up to 100-fold. This difference in the factor of activation after nuclear or cytoplasmic injection could be ascribed to increased transport of plasmids carrying oriP from the cytoplasm to the nucleus in the presence of EBNA1. These data suggest the possibility of substantially increasing the apparent expression of a gene under the control of a strong constitutive promoter in the presence of oriP sequences and EBNA1. This improvement in expression is due to intranuclear enhancement of gene expression. oriP-specific transport of plasmid DNA from the cytoplasm of 293-EBNA1 cells to the nucleus seems to contribute to the observed effect.
The use of nonviral vectors, consisting of plasmid DNA and complexing synthetic compounds, for gene therapeutic applications is hampered by the low efficiency of gene transfer (23). Efficient gene delivery requires (i) entry of nonviral vectors into target cells, (ii) avoidance of or escape from the endosomes, (iii) stability of the plasmid DNA, and (iv) liberation of the plasmid DNA from complexing reagents before or after (v) the plasmid enters the nucleus. Maintenance of the plasmids in the nucleus and efficient expression of the therapeutic gene are also desired.
Investigation of the individual steps of cationic lipid-based nonviral vector delivery has identified the entrapment of DNA in the endosomal compartment as an important barrier to gene transfer and expression (27). In COS and HeLa cells, cationic lipid DNA complexes are mainly taken up by endocytosis, leading to the aggregation of the majority of the molecules in large perinuclear vesicles; only a very small fraction of plasmids reaches the nucleus (27). Entry of cytoplasmic DNA into the nucleus also seems to be a limiting step. Gold-labeled naked plasmid DNA, injected into the cytoplasm of postmitotic rat myotubes, was shown to be transported into the nucleus through the nuclear pore complexes (NPCs) with very low efficiency (7). Therefore, requirements for the improvement of nonviral vectors could be met not only by increasing the efficiency of transport of the plasmid DNA to the nucleus but also by further increasing the efficiency of transcription from the few DNA copies that have reached the nucleus and by replication of these plasmids.
We have studied the effect of Epstein-Barr virus (EBV) oriP sequences and Epstein-Barr nuclear antigen-1 (EBNA1) on apparent or finally detectable gene expression after transfection of cell lines with nonviral vectors.
The genome of the γ herpesvirus EBV is maintained in latently infected cells as an episome. Its persistence, or the persistence of engineered EBV-derived plasmids, requires EBNA1 and the origin of replication, oriP (25). Regulated to one round per cell cycle (26), replication is activated by multiple EBNA1 homodimers bound to oriP, which is composed of the following two functional subelements (19, 24): (i) the family of repeats (FR), comprising 20 copies of a 30-bp binding site motif for EBNA1, and (ii) 960 bp downstream of the FR element, the dyad symmetry element (DS), which consists of four copies of a 16-bp binding site for EBNA1. The DS region is the site for initiation of replication (11, 24), while the FR element acts as an EBNA1-dependent enhancer and participates in DNA replication (24). The phosphoprotein EBNA1 (641 amino acids) carries functional domains for DNA replication and transcriptional activation. A nuclear localization signal (NLS) is located in the vicinity of the DNA-binding domain in the C-terminal half of the protein (2), enabling the signal-mediated transport of EBNA1 from the cytoplasm into the nucleus through the NPC (for a review, see reference 14). That EBNA1 is capable of forming a complex with the nuclear transporter karyopherin α2 has been demonstrated (9). Transcriptional enhancer functions of EBNA1 and oriP or FR have been observed in an EBV context for the latent membrane protein promoter (12) and in a heterologous context for the EBV Cp promoter, herpes simplex virus thymidine kinase gene (HSV-tk) promoter, and simian virus 40 (SV40) minimal promoter (18, 20, 21).
We have analyzed luciferase activity in 293 cells stably expressing EBNA1 (293-EBNA1 cells) by transfection or microinjection of plasmids carrying the human cytomegalovirus (CMV) immediate-early 1 gene enhancer (IE1) promoter- or Rous sarcoma virus (RSV) long terminal repeat promoter-driven expression cassette for the reporter gene, with or without oriP sequences. We tried to characterize the contributions of replication, transcriptional enhancer functions, and potential stimulation of plasmid transport to the improvement of the finally detectable, apparent gene expression.
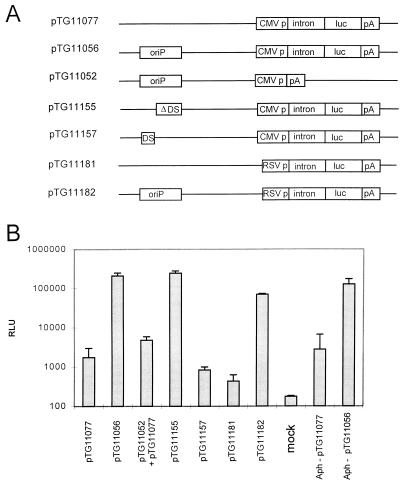
To analyze apparent luciferase expression in the presence of oriP and EBNA1, plasmid constructs derived from pCEP4 (Invitrogen) were used. The EBNA1-coding sequences (positions 8246 to 5519 in pCEP4) were deleted by restriction with NsiI and ClaI, T4 DNA polymerase treatment, and self-ligation (Fig. 1A) (plasmid pTG11052; oriP+, no reporter gene). The EBV genome fragment containing the oriP sequence (positions 7334 to 9519 according to the numbering system in reference 3) was retained. Insertion of the firefly luciferase gene (6) and the mouse HMG1 intron (13) into the empty expression cassette of pTG11052, consisting of CMV IE1 promoter and SV40 polyadenylation signal, resulted in plasmid pTG11056 (oriP+) (Fig. 1A). Deletion of the oriP sequences in pTG11056 led to pTG11077 (oriP−) (Fig. 1A). Deletion of only the DS elements within oriP (positions 8994 to 9134 according to reference 3) resulted in pTG11155 (ΔDS) (Fig. 1A). The insertion of the DS element (positions 9021 to 9133 according to reference 3) in pTG11077 (oriP−) led to the construct pTG11157 (DS+) (Fig. 1A).
FIG. 1.
(A) Schematic representation of the expression cassettes and oriP status of the tested plasmids. Plasmids were based on pCEP4 (Invitrogen) deleted for the EBNA1-encoding sequence. The plasmids pTG11056, pTG11052, and pTG11182 carry the wild-type oriP sequences (positions 7334 to 9519 according to reference 3), indicated by oriP. The DS element (positions 8994 to 9134 according to reference 3) within oriP was deleted (pTG11155; ΔDS), or oriP was replaced by the DS element only (positions 9021 to 9133 according to reference 3) (pTG11157). The luciferase (luc) expression cassette contains the mouse HMG1 intron (intron) and the SV40 polyadenylation signal (pA) and was under the control of either the CMV IE1 promoter (CMV p) or the RSV promoter (RSV p; pTG11181 and pTG11182). pTG11052 carries an empty expression cassette. (B) Representation of the relative light units (RLU) obtained after quantification of luciferase activity in 293-EBNA1 cells (Invitrogen) transfected with the indicated plasmids: 3 × 104 cells per well had been seeded on 48-well plates (Costar) and transfected with 30 ng of the indicated plasmid by using lipofectin (Gibco BRL) as described in the manufacturer’s protocol. Aphidicolin (Aph) treatment (1 μg/ml) of 293-EBNA1 cells before and during the transfection is indicated. Cells were harvested 16 h after transfection, and total cell protein was extracted (Promega lysis buffer). One-fifth of the material was used to quantify luciferase activity (Promega luciferase kit; Berthold Microlumat LB96P).
293-EBNA1 (Invitrogen) cells were transiently transfected with these plasmids. The 293-EBNA1 cell line had been established by stable transfection of 293 cells (16) with a CMV IE1 promoter-driven expression plasmid for EBNA1. Cells were seeded at a density of 3 × 104 cells/well. The next day, the cells were transfected with 30 ng of the respective plasmid by using lipofectin (Gibco BRL) in accordance with the manufacturer’s protocol. Cells were harvested 16 h after transfection, and total cell protein was extracted by using reporter lysis buffer (Promega). One-fifth of the extract was used to quantify luciferase activity (Promega kit). The relative light units (RLU) are shown in Fig. 1B. Transfection with pTG11056 (oriP+) led to a 100-fold increase in luciferase activity compared to that after transfection with pTG11077 (oriP−). This difference was not due to different transfection efficiencies since a cotransfected CMV IE1 promoter-driven β-galactosidase expression plasmid (pTG11078) gave rise to comparable numbers of blue cells (data not shown). The transfection of 293 cells (EBNA1 negative) with pTG11056 (oriP+) or pTG11077 (oriP−) led to comparable luciferase activities (see Table 1).
TABLE 1.
Fold increase in luciferase activity due to the presence of oriP sequences in EBNA1-positive or -negative cellsa
| Cell type | EBNA1 status | Fold increase in luciferase activity (oriP+/oriP−) |
|---|---|---|
| 293 | Negative | 0.6 |
| 293-EBNA1 | Positive | 100 |
| BJAB- K88 | Negative | 3 |
| BJAB- HR1K | Positive | 79 |
| BJAB- 95-8 | Positive | 126 |
| Daudi | Positive | 72 |
| NL03 | Negative | 1 |
| NL03/EBV | Positive | 61 |
The cell lines 293 and 293-EBNA1 were transfected with pTG11056 (oriP+) or pTG11077 (oriP−) as described in the legend to Fig. 1. In addition, the B- lymphoma cell lines BJAB- K88 (17), BJAB- HR1K (10), and BJAB- B95-8 (10), as well as Daudi cells (ATCC CCL-213), normal human lymphocytes (NL03), and an EBV-immortalized B-lymphoma cell line derived from NL03 (NL03/ EBV) were tested: 106 cells were transfected with 1 μg of pTG11056 (oriP+) or pTG11077 (oriP−) by using lipofectin. The cells were lysed 16 h after transfection, and the luciferase activity in one-fifth of the cell extract was quantified as described in the legend to Fig. 1.
The increase in luciferase activity was observed not only in 293-EBNA1 cells but also in EBNA1-positive B-lymphoblast cell lines: 106 cells of the B-lymphoblast cell lines Daudi (ATCC CCL-213), BJAB- K88 (17), BJAB- HR1K (10), and BJAB- B95-8 (10), and 106 cells of normal lymphocytes (NL03) and of the EBV-immortalized B-lymphoblast cell line EBV/NL03 derived therefrom were transfected with 1 μg of pTG11056 (oriP+) or pTG11077 (oriP−) by using lipofectin. The increase in luciferase activity expressed as a factor (oriP+/oriP−) was between 61 and 126 in EBNA1-positive cell lines (Table 1). In contrast, EBNA1-negative cell lines like BJAB- K88 and normal lymphocytes (NL03) led to comparable luciferase activities in the absence and presence of oriP sequences (oriP+/oriP− factor between 1 and 3).
In 293-EBNA1 cells, cotransfection with pTG11077 (oriP−) and pTG11052 (oriP+; no luciferase expression cassette) did not lead to a significant increase in luciferase activity (Fig. 1), indicating that oriP sequences have to be localized in cis to the luciferase expression cassette in order to increase luciferase activity. The large increase in oriP-EBNA1-dependent apparent gene expression was not restricted to plasmids carrying the CMV IE1 promoter, since its replacement by the RSV promoter (Fig. 1A) (pTG11181 [oriP−] and pTG11182 [oriP+]) also led to an increase in luciferase activity of more than 100-fold (Fig. 1B). These plasmids had been obtained by replacement of the CMV IE1 promoter within plasmid pTG11077 or pTG11056 by the RSV promoter derived from pREP4 (Invitrogen; positions 418 to 1041).
The increase in luciferase activity after transfection with the CMV IE1 promoter carrying plasmids was not influenced by the deletion of the DS elements (pTG11155; ΔDS, FR+), while the DS element alone (pTG11157; DS+, FR−) was not sufficient to obtain such a stimulatory effect (Fig. 1B).
We analyzed the effect of aphidicolin (Boehringer-Mannheim), an inhibitor of replicative DNA synthesis, on the oriP/EBNA1-mediated increase of luciferase activity. 293-EBNA1 cells were cultured for 24 h before transfection in the presence of aphidicolin (1 μg/ml), throughout the transfection with pTG11077 (oriP−) or pTG11056 (oriP+), and during subsequent culturing. The action of aphidicolin under these conditions had been confirmed by the reduction of thymidine incorporation by 293-EBNA1 cells to background levels (data not shown). The analysis of luciferase activity 16 h after transfection in the presence of aphidicolin (Fig. 1B, Aph-pTG11077 and Aph-pTG11056) still revealed a substantial 50-fold stimulatory effect (Fig. 1B). These data show that DNA replication did not contribute to the oriP-EBNA1-mediated increase of gene expression.
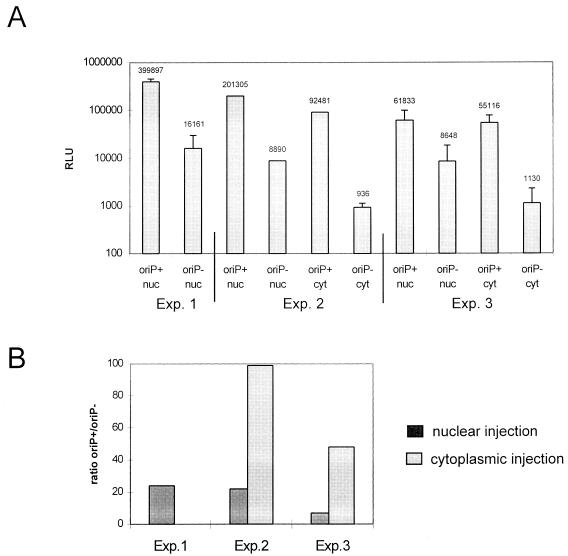
To further characterize the phenomenon, we microinjected pTG11056 (oriP+) or pTG11077 (oriP−) into the nucleus or cytoplasm of 293-EBNA1 cells. Briefly, 10-ng/μl solutions of pTG11056 (oriP+) or pTG11077 (oriP−) were injected into the nucleus or cytoplasm of 100 293-EBNA1 cells. These cells were harvested 16 h after injection, and total protein was extracted. The luciferase activity in one-fifth of the extract was measured (Fig. 2A). After nuclear injection in three independent experiments, a mean increase (oriP+/oriP−) in luciferase activity by a factor of 17 was observed (Fig. 2B). This effect could be ascribed to intranuclear enhancer functions, like transcriptional or posttranscriptional activation. Cytoplasmic injection, however, led to an increase of up to 100-fold in luciferase activity (Fig. 2B). We propose that increased nuclear import of plasmids carrying oriP sequences could be responsible for the stronger stimulatory effect after cytoplasmic injection.
FIG. 2.
(A) Representation of luciferase activity in 293-EBNA1 cells after microinjection. Plasmid pTG11056 (oriP+; 10 ng/μl) or pTG11077 (oriP−; 10 ng/μl) was injected into the nucleus (nuc) or cytoplasm (cyt) of 100 293-EBNA1 cells in three independent experiments. Cells were harvested 16 h after injection, and total protein extract was prepared. One-fifth of the material was used to quantify luciferase activity. Only nuclear injection was used in experiment 1. (B) Schematic representation of the ratio (oriP+/oriP−) in luciferase activity after nuclear or cytoplasmic injection of pTG11056 (oriP+) and pTG11077 (oriP−).
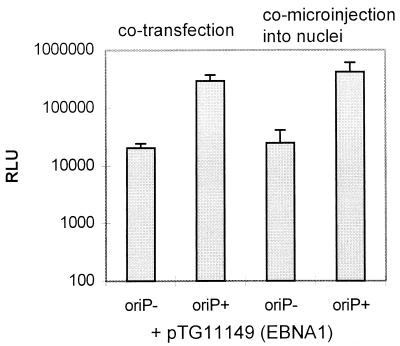
To evaluate the improvement of gene transfer and expression by oriP sequences and EBNA1 in cells lacking EBNA1, we constructed an EBNA1 expression plasmid (pTG11149). The EBNA1-coding sequence (positions 107940 to 109880 according to reference 3) was inserted into a CMV IE1 promoter-driven expression cassette carrying an SV40 polyadenylation signal, and its expression was verified by transient transfection of 293 cells (16) with pTG11149, followed by Western blot analysis (data not shown). The steady-state level of EBNA1 16 h after transfection was 10-fold higher than in the same number of 293-EBNA1 cells (data not shown). 293 cells (3 × 104) were cotransfected with 30 ng of pTG11149 (EBNA1) and 30 ng of either plasmid pTG11056 (oriP+) or plasmid pTG11077 (oriP−) by using lipofectin. Alternatively, solutions containing 10 ng of pTG11149 (EBNA1) per μl and 10 ng of pTG11056 (oriP+) or pTG11077 (oriP−) per μl were microinjected into 100 nuclei of 293 cells. Cells were harvested 16 h after transfection or microinjection, and total protein extract was analyzed for luciferase activity. The data shown in Fig. 3 indicate a 16-fold increase after transfection and a 17-fold increase after nuclear comicroinjection. This suggests that the oriP-EBNA1-mediated increase of reporter gene expression can be induced in cells that are lacking EBNA1. The main mechanism seems to be intranuclear enhancement of gene expression, since the effects after nuclear comicroinjection and after transfection were similar. This contrasts with the results obtained with 293-EBNA1 cells after transfection (100-fold increase) or nuclear injection (17-fold increase) with pTG11056 and pTG11077. The less pronounced fold increase after cotransfection of 293 cells compared to that after transfection of 293- EBNA1 cells might be due to the lack of preexisting EBNA1 at the moment of transfection, preventing the contribution of plasmid transport.
FIG. 3.
Representation of luciferase activity in 293 cells after cotransfection and nuclear comicroinjection with EBNA1 expression plasmid pTG11149 together with pTG11056 (oriP+) or pTG11077 (oriP−). A total of 3 × 104 293 cells were seeded per well on 48-well plates. The next day, the cells were transfected with 30 ng of pTG11149 together with 30 ng of pTG11077 or pTG11056. Alternatively, a solution of 10 ng of pTG11149 per μl together with 10 ng of pTG11056 or pTG11077 per μl was injected into the nuclei of 100 293 cells. At 16 h after cotransfection or comicroinjection, the cells were harvested and total cell protein was prepared. One-fifth of the material was analyzed with respect to luciferase activity. The obtained RLU are indicated.
So far, we have observed a strong oriP-EBNA1-mediated increase of apparent gene expression after transfection of cell lines using lipofectin as an example for nonviral vectors. We tried to better characterize the effect of oriP and EBNA1 on plasmid delivery, replication, and reporter gene expression.
The oriP-EBNA1-mediated increase of luciferase activity in the presence of aphidicolin or in the absence of the origin of replication, the DS element, rules out a significant contribution of replication.
Nuclear microinjection or comicroinjection of equal amounts of plasmids carrying identical luciferase expression cassettes allowed the detection of enhancement of transcriptional or posttranscriptional functions. By using this technique, a similar increase in luciferase activity was observed in 293-EBNA1 and in 293 cells of ca. 17-fold. Transcriptional enhancer effects by oriP-EBNA1 or FR-EBNA1 have already been described. Insertion of FR into a plasmid carrying an HSV-tk promoter-driven or an SV40 minimal promoter-driven expression cassette for chloramphenicol acetyl transferase (CAT) increased reporter gene activity by a factor of 66 or 35, respectively (20). In our work, it is noteworthy that the efficiency of the CMV IE1 promoter, described to be a very strong promoter-enhancer (4), can still be increased significantly in the presence of oriP and EBNA1.
When oriP-mediated enhancements of luciferase activity in 293-EBNA1 cells after cytoplasmic and nuclear injection were compared, we observed a significantly higher increase after cytoplasmic injection (up to 100-fold). Assuming that equal amounts of plasmids with and without oriP had been delivered into the respective cell compartments, we could explain the difference in luciferase activity by the different fate of plasmid DNA with or without oriP in the cytoplasm. We propose that plasmids with oriP sequences, injected into the cytoplasm, are tightly bound by EBNA1 molecules in the same way that the bacterially expressed C-terminal half of EBNA1 is bound tightly to its specific DNA target with a Kd for the consensus binding site of 1 × 10−11 to 2 × 10−11 M (1). The stable complex of plasmid and multiple EBNA1 protein molecules carrying NLSs would then be transported through the NPCs into the nucleus, thereby increasing the intranuclear number of copies of luciferase expression cassettes. The need for preexisting EBNA1 at the time point of transfection, like that given in 293-EBNA1 cells, might suggest that DNA is in a transportable status only for a limited period of time after transfection. We tried to monitor the localization of transfected pTG11056 (oriP+) and pTG11077 (oriP−) in 293-EBNA1 cells by fluorescence in situ hybridization. Plasmid DNA could be detected in the cytoplasm, while no plasmid DNA could be found in the nucleus, irrespective of which plasmid had been transfected (data not shown). This confirms that the majority of transfected plasmids are retained in the cytoplasm and suggests that the sensitivity of our assay is not sufficient to detect single copies of plasmids. Such detection would, however, be necessary for a direct proof of transport phenomena.
The microinjection experiments described herein were performed in analogy to work by Graessmann et al. (15), who compared differences in CAT activity after injection of plasmids with SV40 promoter- and HSV-tk promoter-driven expression cassettes into the nucleus or cytoplasm of T-antigen-negative cells. CAT activity was always higher after injection of the plasmid with the SV40 promoter-driven expression cassette. However, this difference was 5- to 10-fold higher after cytoplasmic injection than after nuclear injection. The authors ascribed this effect to improved transport of SV40-promoter-carrying plasmids into the nucleus. Work by Dean (5) also suggests that the SV40 early promoter increases the import of plasmids through the NPCs. The authors proposed that these sequences were recognized and bound by cellular proteins with NLSs, driving the plasmids through the NPC.
The potential to exploit the oriP-EBNA1-mediated increase of apparent gene expression in vivo, either by using plasmids that carry oriP sequences and an expression cassette for EBNA1 or by delivery of plasmids coupled to EBNA1, remains to be evaluated. EBNA1 has been shown to be tumorigenic in mice (22). It would be necessary to define nontransforming EBNA1 mutants or peptides that have retained the activation functions but not the tumorigenic potency. The potential exists to target EBNA1-positive tumors with plasmids that carry oriP sequences, as suggested by Evans et al. (8) as well as by our own work with B-lymphoblast cell lines.
With the goal of creating better nonviral vectors, we suggest that our understanding of the oriP-EBNA1 system described in this paper could help to improve apparent gene expression by intranuclear activation of therapeutic gene expression and to improve transport of plasmids from the cytoplasm to the nucleus.
Acknowledgments
This work was supported in part by the Association Française contre les Myopathies (AFM), Association Française de Lutte contre la Mucoviscidose (AFLM), and by the Ministère Française de l’Industrie.
We thank O. Keppler and P. Pawlita (DKFZ) for providing us with BJAB- derived cell lines. The preparation of normal lymphocytes (NL03) and the EBV immortalized cell line NL03/EBV were kindly provided by F. Oberling, DKFZ, Heidelberg. We thank Andrea Pavirani and Michael Courtney for critically reading the manuscript.
REFERENCES
- 1.Ambinder R F, Shah W A, Rawlins D R, Hayward G S, Hayward S D. Definition of the sequence requirements of the EBNA-1 protein to its palindromic target sites in Epstein-Barr virus DNA. J Virol. 1990;64:2369–2379. doi: 10.1128/jvi.64.5.2369-2379.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ambinder R F, Mullen M, Chang Y, Hayward G S, Hayward D S. Functional domains of Epstein-Barr virus nuclear antigen EBNA-1. J Virol. 1991;65:1466–1478. doi: 10.1128/jvi.65.3.1466-1478.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baer R, Bankier A T, Biggin M D, Deininger P L, Farrell P J, Gibson T J, Hatfull G, Hudson G S, Satchwell S C, Seguin C, Tuffnell P S, Barrell B G. DNA sequence and expression of the B95-8 Epstein-Barr virus genome. Nature. 1984;310:207–209. doi: 10.1038/310207a0. [DOI] [PubMed] [Google Scholar]
- 4.Boshart M, Weber F, Jahn G, Dorsch-Haesler K, Fleckenstein B, Schaffner W. A very strong enhancer is located upstream of an immediate early gene of human cytomegalovirus. Cell. 1958;41:521–530. doi: 10.1016/s0092-8674(85)80025-8. [DOI] [PubMed] [Google Scholar]
- 5.Dean D. Import of plasmid DNA into the nucleus is sequence specific. Exp Cell Res. 1997;230:293–302. doi: 10.1006/excr.1996.3427. [DOI] [PubMed] [Google Scholar]
- 6.DeWet J R, Wood K V, DeLuca M, Helsinki D R, Subramani S. Firefly luciferase gene: structure and expression in mammalian cells. Mol Cell Biol. 1987;7:725–737. doi: 10.1128/mcb.7.2.725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dowty M E, Williams P, Zhang G, Hagstrom J E, Wolff J A. Plasmid DNA entry into postmitotic nuclei of primary rat myotubes. Proc Natl Acad Sci USA. 1995;92:4572–4576. doi: 10.1073/pnas.92.10.4572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Evans T J, Brooks L, Farrell P J. A strategy for specific targeting of therapeutic agents to tumour cells of virus-associated cancers. Gene Ther. 1997;4:264–267. doi: 10.1038/sj.gt.3300392. [DOI] [PubMed] [Google Scholar]
- 9.Fischer N, Kremmer E, Lautscham G, Mueller-Lantzsch N, Grässer F A. Epstein-Barr virus nuclear antigen 1 forms a complex with the nuclear transporter Karyopherin α2. J Biol Chem. 1997;272:3999–4005. doi: 10.1074/jbc.272.7.3999. [DOI] [PubMed] [Google Scholar]
- 10.Fresen K-O, Merkt B, Bornkamm G W, zur Hausen H. Heterogeneity of Epstein-Barr virus originating from P3HR-1 cells. I. Studies on EBNA induction. Int J Cancer. 1977;19:317–323. doi: 10.1002/ijc.2910190306. [DOI] [PubMed] [Google Scholar]
- 11.Gahn T A, Schildkraut C L. The Epstein-Barr virus origin of plasmid replication, oriP, contains both the initiation and termination sites of DNA replication. Cell. 1989;58:527–535. doi: 10.1016/0092-8674(89)90433-9. [DOI] [PubMed] [Google Scholar]
- 12.Gahn T A, Sugden B. An EBNA-1-dependent enhancer acts from a distance of 10 kilobase pairs to increase expression of the Epstein-Barr virus LMP gene. J Virol. 1995;69:2633–2636. doi: 10.1128/jvi.69.4.2633-2636.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gautier C, Mehtali M, Lathe R. A ubiquitous mammalian expression vector, pHMG, based on a housekeeping gene promoter. Nucleic Acids Res. 1989;17:8389. doi: 10.1093/nar/17.20.8389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Görlich D, Mattaj I W. Nucleocytoplasmic transport. Science. 1996;271:1513–1518. doi: 10.1126/science.271.5255.1513. [DOI] [PubMed] [Google Scholar]
- 15.Graessmann M, Menne J, Liebler M, Graeber I, Graessmann A. Helper activity for gene expression, a novel function of the SV40 enhancer. Nucleic Acids Res. 1989;17:6603–6612. doi: 10.1093/nar/17.16.6603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Graham F L, Smiley J, Russel W C, Nairu R. Characteristics of a human cell line transformed by DNA from human adenovirus type 5. J Gen Virol. 1977;36:59–77. doi: 10.1099/0022-1317-36-1-59. [DOI] [PubMed] [Google Scholar]
- 17.Klein G, Sugden B, Leibold W, Menézes J. Infection of EBV-genome-negative and -positive human lymphoblastoid cell lines with biologically different preparations of EBV. Intervirology. 1974;3:232–244. doi: 10.1159/000149760. [DOI] [PubMed] [Google Scholar]
- 18.Puglielli M T, Woisetschlaeger M, Speck S H. oriP is essential for EBNA gene promoter activity in Epstein-Barr virus-immortalized lymphoblastoid cell lines. J Virol. 1996;70:5758–5768. doi: 10.1128/jvi.70.9.5758-5768.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Reisman D, Yates J, Sugden B. A putative origin of replication of plasmids derived from Epstein-Barr virus is composed of two cis-acting components. Mol Cell Biol. 1985;5:1822–1832. doi: 10.1128/mcb.5.8.1822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Reisman D, Sugden B. trans activation of an Epstein-Barr viral transcriptional enhancer by the Epstein-Barr viral nuclear antigen 1. Mol Cell Biol. 1986;6:3838–3846. doi: 10.1128/mcb.6.11.3838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sugden B, Warren N. A promoter of Epstein-Barr virus that can function during latent infection can be transactivated by EBNA1, a viral protein required for viral DNA replication during latent infection. J Virol. 1989;63:2644–2649. doi: 10.1128/jvi.63.6.2644-2649.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wilson J B, Bell J L, Levine A J. Expression of Epstein-Barr virus nuclear antigen-1 induces B cell neoplasia in transgenic mice. EMBO J. 1996;15:3117–3125. [PMC free article] [PubMed] [Google Scholar]
- 23.Wolff J A. Naked DNA transport and expression in mammalian cells. Neuromuscular Disorders. 1997;7:314–318. doi: 10.1016/s0960-8966(97)00055-2. [DOI] [PubMed] [Google Scholar]
- 24.Wysokenski D A, Yates J L. Multiple EBNA1-binding sites are required to form an EBNA1-dependent enhancer and to activate a minimal replicative origin within oriP of Epstein-Barr virus. J Virol. 1989;63:2657–2666. doi: 10.1128/jvi.63.6.2657-2666.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yates J L, Warren N, Sugden B. Plasmids derived from Epstein-Barr virus replicate stably in a variety of mammalian cells. Nature. 1985;313:812–815. doi: 10.1038/313812a0. [DOI] [PubMed] [Google Scholar]
- 26.Yates J L, Guan N. Epstein-Barr virus-derived plasmids replicate only once per cell cycle and are not amplified after entry into cells. J Virol. 1991;65:483–488. doi: 10.1128/jvi.65.1.483-488.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zabner J, Fasbender A J, Moninger T, Poellinger K A, Welsh M J. Cellular and molecular barriers to gene transfer by a cationic lipid. J Biol Chem. 1995;270:18997–19007. doi: 10.1074/jbc.270.32.18997. [DOI] [PubMed] [Google Scholar]