Abstract
We have used viruslike particles (VLPs) of human papillomaviruses to study the structure and assembly of the viral capsid. We demonstrate that mutation of either of two highly conserved cysteines of the major capsid protein L1 to serine completely prevents the assembly of VLPs but not of capsomers, whereas mutation of all other cysteines leaves VLP assembly unaffected. These two cysteines form intercapsomeric disulfides yielding an L1 trimer. Trimerization comprises about half of the L1 molecules in VLPs but all L1 molecules in complete virions. We suggest that trimerization of L1 is indispensable for the stabilization of intercapsomeric contacts in papillomavirus capsids.
Papillomaviruses are widespread, highly species- and tissue-specific viruses of higher vertebrates. They infect exclusively the skin or the oral and genital mucosae, inducing warts (or papillomas), condylomata, and other benign or malignant intraepithelial lesions. To date, over a hundred types of human papillomaviruses (HPV) have been identified, and some of them are strongly associated with invasive cervical carcinoma, the second most frequent cancer of women worldwide (25).
Due to the low virus content of many lesions and our incapacity to develop a tissue culture system for the large-scale propagation of papillomaviruses, only few HPVs have been purified in quantities sufficient for structural analysis. Recently, however, several groups have obtained viruslike particles (VLPs) by expressing the major capsid protein L1 either alone or together with the minor capsid protein L2 by using vaccinia virus (6, 24) or baculovirus expression systems (11, 15, 22). VLPs do not contain a viral genome but are otherwise indistinguishable from authentic virions according to cryoelectron microscopy and three-dimensional image reconstruction at 3.5- nm resolution (7). Moreover, VLPs are amenable to genetic analysis. Therefore VLPs offer a promising approach to investigating the structure and the still-unknown mechanisms of assembly, cellular uptake, and disassembly of papillomaviruses.
Papillomaviruses and the genetically unrelated polyomaviruses are grouped together into the family Papovaviridae because they share a number of structural features, including a circular double-stranded DNA genome, associated with histones to form a minichromosome, and a spherical capsid of icosahedral symmetry (T = 7). Capsids consist of 72 capsomers, each a pentamer of the major capsid protein L1 or VP1 (1, 16). In addition, polyomavirus capsids contain one copy of either of the minor capsid proteins, VP2 or VP3, per capsomer, and papillomavirus capsids probably contain 12 copies of the minor capsid protein L2 located at the 12 pentavalent capsomers (21).
Although reducing agents are required to disassemble simian virus 40 (SV40) (3) and polyomavirus (23) in vitro, disulfide bonds were, at best, considered to contribute somewhat to virion stability and were proposed to form only after particle assembly or even after viral release (9). Therefore, they should play no role in viral assembly. In contrast, we have shown previously that reduction of the disulfide bonds in VLPs of papillomaviruses always leads to disassembly into capsomers (18). This indicates that disulfide formation could be essential for the assembly of papillomavirus capsids. To address this issue, we have constructed L1 cysteine mutants and have analyzed their capacity for VLP assembly, using as the prototype HPV type 33 (HPV-33), which has been cloned from an invasive cervical carcinoma and is closely related to HPV-16 and other genital HPVs (2).
L1 trimers cross-linked by disulfides are present in VLPs and virions.
Initially, we analyzed the degree of disulfide cross-linking in VLPs and virions. VLPs of HPV-16, -18, and -33 were obtained by synthesis of the major capsid proteins L1 by the baculovirus expression system. VLPs purified by cesium chloride density gradient centrifugation banded at a density of 1.29 g/cm3 and were subsequently sedimented into a cesium chloride cushion (1.29 g/cm3) to remove capsomers and free L1 molecules. Virions were isolated from vulgar warts collected from the hands of a patient and banded at a density of 1.34 g/cm3 in cesium chloride. VLPs and virions were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) followed by Western blot analysis. Monoclonal antibody 33L1-7 served as the primary antibody (17). For SDS-PAGE under nonreducing conditions β-mercaptoethanol was omitted from the sample buffer and 8% polyacrylamide gels were used. As previously described for HPV-11 and HPV-33 (14, 22), all VLPs yielded two broad bands when analyzed under nonreducing conditions: about 50% of the L1 molecules migrated as monomers with an apparent molecular mass of 50 kDa (Fig. 1A, left panel) and about 50% migrated as 150- to 160-kDa oligomers. Careful calibration using various molecular mass standards indicates that this oligomer corresponds to L1 trimers (Fig. 1A, left panel). In contrast to VLPs, all of the L1 proteins of virions from warts migrated as oligomers (Fig. 1A). The oligomeric L1 protein from virions (the HPV type of which has not been determined) had an apparent molecular mass of 180 kDa. Since a mass of 60 kDa was determined for the monomer (Fig. 1B), the oligomeric L1 in virions should also correspond to trimers. Under reducing conditions all of the L1 proteins migrated as monomers, which demonstrates that trimerization of L1 proteins occurs through disulfide bonding in both virions and VLPs.
FIG. 1.
Analysis of disulfide bonds in VLPs and virions. VLPs obtained by using the baculovirus expression system and virions extracted from a hand wart were purified by cesium chloride density gradient centrifugation and subsequently analyzed by nonreducing (A) and reducing (B) SDS-PAGE. L1 proteins were visualized by immunoblotting using the cross-reactive monoclonal antibody 33L1-7 (17). The apparent molecular masses of marker proteins are indicated. Lanes 16, 18, and 33 contain HPV-16, -18, and -33 VLPs, respectively.
L1 trimer formation in VLPs is mediated by only two cysteines.
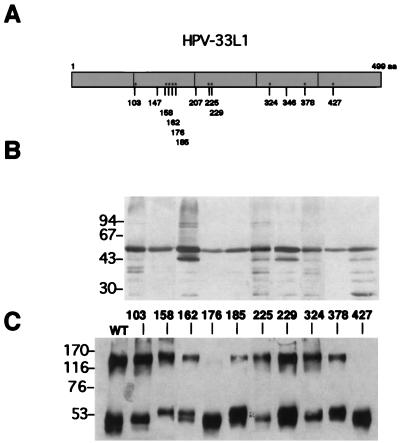
To analyze if specific cysteines were involved in the L1 trimer formation, single point mutations, substituting serine for cysteine, were constructed. HPV-33, which is related to HPV- 16, was used for this analysis. There are 13 cysteine residues in the 499-amino-acid sequence of the HPV-33 L1 protein (4) (Fig. 2A), 10 of which occur frequently in other HPVs as well. These were mutated one by one with an oligonucleotide-directed mutagenesis kit obtained from Amersham (Sculptor in vitro mutagenesis system). In all cases TGT was mutated to AGT, and TGC was mutated to AGC. The mutations were confirmed by sequencing, and the 10 mutants obtained were expressed in Sf9 insect cells by using recombinant baculoviruses, as previously described (22). Each of the mutants yielded the expected 55-kDa protein, as shown by Western blotting of cellular lysates (Fig. 2B). A few proteolytic breakdown products were always observed in all preparations. To isolate VLPs, nuclei of infected cells were subjected to mild sonication and fractionated by cesium chloride density gradient centrifugation. Mutant and wild-type L1 proteins were obtained at 1.29 g/cm3. To probe for disulfide cross-linking in these proteins, they were analyzed by SDS-PAGE in the absence of a reducing agent. Wild-type L1 and eight of the L1 mutants migrated in two bands, one of them corresponding to monomers with an apparent molecular mass of 50 to 55 kDa and the other corresponding to a protein of about 150 kDa corresponding to trimers, as shown above (Fig. 2C). The amount of L1 protein recovered as trimers never exceeded 50% of the total L1 protein. Surprisingly, none of our L1 preparations, mutant or wild type, ever contained any disulfide-linked dimers. Two of the mutants, the mutants carrying serine instead of cysteine 176 or cysteine 427 (C176S and C427S, respectively) failed to form trimers (Fig. 2C). This demonstrates that a single point mutation of either C176 or C427 in HPV-33 L1 converts all trimers in VLPs into monomers, whereas all other cysteine mutants leave the trimerization unaffected. This is best explained by exclusive disulfide bonds of C176 with C427 in neighboring L1 molecules. The most plausible structure for a trimer formed by disulfides between only two cysteines is a covalently closed, cyclical arrangement of three L1 molecules around a threefold axis, in which each molecule donates two cysteines to a total of three intermolecular disulfides. However, a linear arrangement of three L1 molecules cannot be excluded on the basis of our data.
FIG. 2.
Analysis of disulfide bonds in cysteine mutants and wild-type L1 protein by SDS-PAGE under reducing and nonreducing conditions. (A) Schematic diagram of the positions of cysteine residues within the L1 protein of HPV-33. ∗, cysteines conserved among papillomaviruses; aa, amino acids. (B) SDS-PAGE under reducing conditions. Lysates of Sf9 insect cells infected with recombinant baculoviruses are shown. In addition to the full-length L1 protein, some degradation products are seen. (C) SDS-PAGE under nonreducing conditions. L1 proteins extracted from nuclei of infected Sf9 cells were partially purified by cesium chloride density gradient centrifugation. Fractions corresponding to a buoyant density of 1.29 g/cm3 were subjected to gel electrophoresis. Numbers between the two gels indicate the positions of the mutated cysteines in the L1 protein. The apparent molecular masses of marker proteins are indicated to the left of each gel (in kilodaltons). WT, wild type.
L1 trimer formation is critical for VLP assembly.
If trimer formation is a critical step in the assembly of viral capsids, then C176S and C427S should be unable to form VLPs. To examine the effect of the mutations on the capacity of L1 proteins to assemble into VLPs, wild-type and mutant L1 proteins from the cesium chloride fractions were sedimented through sucrose gradients. Whereas the wild type and the eight trimer-forming mutants had high sedimentation coefficients, as expected for VLPs, the C176S and C427S proteins sedimented only with about 9 S, consistent with assembly into capsomers (not shown). The structure of the L1 complexes was then examined by electron microscopy. Aliquots from the fractions of the cesium chloride density gradient centrifugation corresponding to 1.29 g/cm3 were dialyzed against phosphate-buffered saline, pH 7.3, and spotted onto carbon-coated copper grids. They were negatively stained with 5% ammonium molybdate, pH 7.0, containing 1% trehalose and with 2% phosphotungstic acid, pH 7.0, respectively, as previously described (8). Samples were examined under a Zeiss EM900 transmission electron microscope at an instrumental magnification of ×30,000 and ×50,000, respectively. The wild type and the eight fast sedimenting mutants formed mainly spherical VLPs with diameters of 50 to 60 nm and some tubular structures, but the C176S and C427S proteins only formed capsomers (Fig. 3), in agreement with the sedimentation analysis. This demonstrates that the assembly of viral capsids requires disulfide bonds involving either of these two cysteines.
FIG. 3.
Electron micrographs of particles assembled from cysteine mutant or wild-type (wt) L1 protein. L1 proteins partially purified by cesium chloride density gradient centrifugation were dialyzed against phosphate-buffered saline, spotted onto carbon-coated copper grids, and negatively stained with 5% ammonium molybdate containing 1% trehalose (for wild type, C162S, C185S, and C378S) or with 2% phosphotungstic acid (for C176S and C427S). Photographs were taken with a Zeiss EM900 transmission electron microscope at an instrumental magnification of ×30,000 (for wild type, C162S, C185S, and C378S) or ×50,000 (for C176S and C427S). The bars represent 100 nm.
We have previously shown that reduction of disulfides in VLPs leads to disassembly into capsomers, not L1 monomers (18). Now we have demonstrated that the cysteine mutants that do not form trimers assemble into capsomers but not into capsids. The L1 molecules present in these capsomers are not disulfide bonded. Clearly, disulfides forming L1 trimers are intercapsomeric; i.e., the three L1 molecules are from separate capsomers.
It is interesting to analyze whether intermolecular disulfide bonds between C176 and C427 of L1 molecules located in separate capsomers are sterically compatible with the structure expected for L1. Until now the structure of papillomaviruses has only been studied by cryoelectron microscopy, the resolution of which is at the capsomer level. However, the crystal structures of the related SV40 and polyomavirus have been resolved at 3.8 Å (13) and 3.65 Å (19), respectively, and the SV40 structure has recently been refined to a 3.1-Å resolution (20). Although there is only moderate sequence similarity between the L1 and VP1 proteins of papillomaviruses and polyomaviruses, the arrangement of β-strands found for VP1 and that predicted for the L1 sequences of HPVs by using various secondary-structure predictions and CLUSTAL-W for alignment (10) are similar. According to these predictions, the two essential cysteines C176 and C427 are located in a loop and in the C-terminal region, respectively. In SV40 both the corresponding loop and the C terminus are located on the same side of the VP1 structure. In HPV-33 both the potential loop (amino acids 160 to 189) and the C-terminal arm (amino acids 348 to 499) are considerably larger than in SV40, providing ample flexibility for disulfide bonding.
Cysteine mutant of HPV-18.
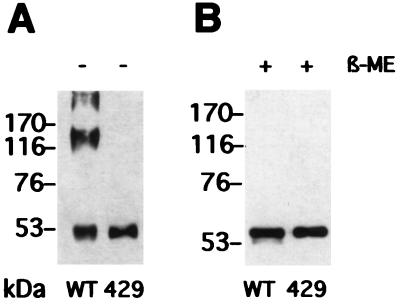
To probe for the generality of the importance of these cysteines for the assembly of papillomavirus capsids, we introduced a single point mutation corresponding to C427S into the L1 protein of HPV-18. HPV-18 is a more distantly related HPV type, a representative of the high-risk viruses most often associated with genital carcinoma. Since C427 of HPV-33 corresponds to C429 in HPV-18, the C429S mutant of HPV-18 L1 was constructed and used to obtain a recombinant baculovirus, as described above. When expressed in Sf9 cells, the mutant formed neither trimers (Fig. 4) nor VLPs (not shown), in contrast to wild-type HPV-18 L1 (Fig. 4). This demonstrates that L1 trimer formation using the two cysteines identified is not a privilege of HPV-33. Indeed, C176 and C427 are conserved in all of the 103 L1 sequences published to date, including even the distantly related animal papillomaviruses. In addition, the corresponding C424 in HPV- 11 is essential for virion stability and assembly (12). Taken together, these data suggest that the formation of trimers of L1 by way of disulfides between these two cysteines is a critical step towards capsid assembly of all papillomaviruses.
FIG. 4.
Analysis of disulfide bonds in cysteine mutant 18L1-C429S (lanes 429) and wild-type HPV-18 L1 protein (lanes WT) by SDS-PAGE under reducing (B) and nonreducing (A) conditions. The apparent molecular masses of marker proteins are indicated. β-ME, β-mercaptoethanol.
It is interesting that only half of the L1 molecules in VLPs produced in insect cells are disulfide bonded into trimers, whereas all of the L1 molecules in virions isolated from warts are disulfide bonded, indicating structural differences between VLPs and virions that are not detectable by electron microscopy. Complete cross-linking of L1 proteins through disulfides has previously been observed for HPV-1a virions, even though the molecular weight of the complex was not determined (5). Clearly, disulfide-linked L1 trimers are an important structural feature of the papillomavirus capsid. The difference in the degree of L1 cross-linking observed between virions and VLPs cannot be explained by the presence of L2 in virions, since incorporation of L2 into VLPs does not alter L1 cross-linking (not shown, but see references 18 and 22). Possibly DNA packaging or different reducing environments in keratinocytes and insect cells can explain the additional disulfide bonds found in virions.
Acknowledgments
M. Sapp and C. Fligge contributed equally to this work.
We thank G. Orth for HPV clones, J. T. Schiller for the HPV-16 L1/ L2 recombinant baculovirus K-L1/L2, T. Dandekar for help with alignment and secondary-structure predictions, and C. Purtak and M. Werr for expert assistance.
This work was supported by grants from the Deutsche Forschungsgemeinschaft to M.S. and R.E.S.
REFERENCES
- 1.Baker T S, Newcomb W W, Olson N H, Cowsert L M, Olson C, Brown J C. Structures of bovine and human papillomaviruses. Biophys J. 1991;60:1445–1456. doi: 10.1016/S0006-3495(91)82181-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Beaudenon S, Kremsdorf D, Croissant O, Jablonska S, Wain-Hobson S, Orth G. A novel type of human papillomavirus associated with genital neoplasias. Nature. 1986;321:246–249. doi: 10.1038/321246a0. [DOI] [PubMed] [Google Scholar]
- 3.Christiansen G, Landers T, Griffith J, Berg P. Characterization of components released by alkali disruption of simian virus 40. J Virol. 1977;21:1079–1084. doi: 10.1128/jvi.21.3.1079-1084.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cole S T, Streeck R E. Genome organization and nucleotide sequence of human papillomavirus type 33, which is associated with cervical cancer. J Virol. 1986;58:991–995. doi: 10.1128/jvi.58.3.991-995.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Doorbar J, Gallimore P H. Identification of proteins encoded by the L1 and L2 open reading frames of human papillomavirus 1a. J Virol. 1987;61:2793–2799. doi: 10.1128/jvi.61.9.2793-2799.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hagensee M E, Yaegashi N, Galloway D A. Self-assembly of human papillomavirus type 1 capsids by expression of the L1 protein alone or by coexpression of the L1 and L2 capsid proteins. J Virol. 1993;67:315–322. doi: 10.1128/jvi.67.1.315-322.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hagensee M E, Olson N H, Baker T S, Galloway D A. Three-dimensional structure of vaccinia virus-produced human papillomavirus type 1 capsids. J Virol. 1994;68:4503–4505. doi: 10.1128/jvi.68.7.4503-4505.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Harris J R, Gebauer W, Markl J. Keyhole limpet haemocyanin: negative staining in the presence of trehalose. Micron. 1995;26:25–33. [Google Scholar]
- 9.Harrison S C, Skehel J J, Wiley D C. Virus structure. In: Fields B N, Knipe D M, Howley P M, et al., editors. Virology. 3rd ed. Vol. 1. New York, N.Y: Lippincott-Raven; 1996. pp. 59–99. [Google Scholar]
- 10.Higgins D G, Thompson J D, Gibson T J. Using CLUSTAL for multiple sequence alignment. Methods Enzymol. 1996;266:383–402. doi: 10.1016/s0076-6879(96)66024-8. [DOI] [PubMed] [Google Scholar]
- 11.Kirnbauer R, Booy F, Cheng N, Lowy D R, Schiller J T. Papillomavirus L1 major capsid protein self-assembles into virus-like particles that are highly immunogenic. Proc Natl Acad Sci USA. 1992;89:12180–12184. doi: 10.1073/pnas.89.24.12180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li M, Beard P, Estes P A, Lyon M K, Garcea R L. Intercapsomeric disulfide bonds in papillomavirus assembly and disassembly. J Virol. 1998;72:2160–2167. doi: 10.1128/jvi.72.3.2160-2167.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liddington R C, Yan Y, Moulai J, Sahli R, Benjamin T L, Harrison S C. Structure of simian virus 40 at 3.8-Å resolution. Nature. 1991;354:278–284. doi: 10.1038/354278a0. [DOI] [PubMed] [Google Scholar]
- 14.McCarthy M P, White W I, Palmer-Hill F, Koenig S, Suzich J A. Quantitative disassembly and reassembly of human papillomavirus type 11 viruslike particles in vitro. J Virol. 1998;72:32–41. doi: 10.1128/jvi.72.1.32-41.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rose R C, Bonnez W, Reichman R C, Garcea R L. Expression of human papillomavirus type 11 L1 protein in insect cells: in vivo and in vitro assembly of viruslike particles. J Virol. 1993;67:1936–1944. doi: 10.1128/jvi.67.4.1936-1944.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Salunke D M, Caspar D L D, Garcea R L. Self-assembly of purified polyomavirus capsid protein VP1. Cell. 1986;46:895–904. doi: 10.1016/0092-8674(86)90071-1. [DOI] [PubMed] [Google Scholar]
- 17.Sapp M, Kraus U, Volpers C, Snijders P J, Walboomers J M, Streeck R E. Analysis of type-restricted and cross-reactive epitopes on virus-like particles of human papillomavirus type 33 and in infected tissues using monoclonal antibodies to the major capsid protein. J Gen Virol. 1994;75:3375–3383. doi: 10.1099/0022-1317-75-12-3375. [DOI] [PubMed] [Google Scholar]
- 18.Sapp M, Volpers C, Müller M, Streeck R E. Organization of the major and minor capsid proteins in human papillomavirus type 33 virus-like particles. J Gen Virol. 1995;76:2407–2412. doi: 10.1099/0022-1317-76-9-2407. [DOI] [PubMed] [Google Scholar]
- 19.Stehle T, Harrison S C. Crystal structure of murine polyomavirus in complex with straight-chain and branched-chain sialyloligosaccharide receptor fragments. Structure. 1996;4:183–194. doi: 10.1016/s0969-2126(96)00021-4. [DOI] [PubMed] [Google Scholar]
- 20.Stehle T, Gamblin S J, Yan Y, Harrison S C. The structure of simian virus 40 refined at 3.1 Å resolution. Structure. 1996;4:165–182. doi: 10.1016/s0969-2126(96)00020-2. [DOI] [PubMed] [Google Scholar]
- 21.Trus B L, Roden R B S, Greenstone H L, Vrhel M, Schiller J T, Booy F P. Novel structural features of bovine papillomavirus revealed by a three-dimensional reconstruction to 9 Å resolution. Nat Struct Biol. 1997;4:413–420. doi: 10.1038/nsb0597-413. [DOI] [PubMed] [Google Scholar]
- 22.Volpers C, Schirmacher P, Streeck R E, Sapp M. Assembly of the major and the minor capsid protein of human papillomavirus type 33 into virus-like particles and tubular structures in insect cells. Virology. 1994;200:504–512. doi: 10.1006/viro.1994.1213. [DOI] [PubMed] [Google Scholar]
- 23.Walter G, Deppert W. Intermolecular disulfide bonds: an important structural feature of the polyomavirus capsid. Cold Spring Harbor Symp Quant Biol. 1974;39:255–257. doi: 10.1101/sqb.1974.039.01.033. [DOI] [PubMed] [Google Scholar]
- 24.Zhou J, Sun X Y, Stenzel D J, Frazer I H. Expression of vaccinia recombinant HPV 16 L1 and L2 ORF proteins in epithelial cells is sufficient for assembly of HPV virion-like particles. Virology. 1991;185:251–257. doi: 10.1016/0042-6822(91)90772-4. [DOI] [PubMed] [Google Scholar]
- 25.zur Hausen H, de Villiers E M. Human papillomaviruses. Annu Rev Microbiol. 1994;48:427–447. doi: 10.1146/annurev.mi.48.100194.002235. [DOI] [PubMed] [Google Scholar]