Abstract
We have defined a 160-nucleotide region, Mψ, from the 5′ leader region of the Rous sarcoma virus genome that is sufficient to direct the packaging of a heterologous RNA. Mψ contains the putative O3 stem structure that has previously been shown, and that has been confirmed in this study, to be important for the efficient packaging of avian leukosis-sarcoma virus RNA. Analyses of several O3 stem mutants revealed that other regions within Mψ can interfere with the proper folding of altered sequences which are predicted to form a wild-type O3 stem.
The packaging signal, or ψ, is the cis-acting signal in retroviral RNA necessary for encapsidation of the RNA into the virion. In an early attempt to identify the avian leukosis-sarcoma virus (ALSV) packaging signal, deletions were constructed in the 5′-untranslated region of a cloned avian sarcoma provirus. Several mutants which had packaging defects were identified. These mutants contained a deletion in an overlapping region of 31 bp located between the primer binding site (PBS) and the Gag initiation codon (8). Subsequently, a 150-bp deletion that encompasses this region was engineered in an avian leukosis virus provirus and stably expressed in a quail cell line to produce the Q2bn-4D line (13). While particles produced from these cells are unable to package their own genomes, they are able to package viral RNAs lacking viral structural genes, indicating that they retain trans-acting packaging functions. Using the Q2bn packaging cell line, we previously identified a 270-nucleotide (nt) packaging signal from the 5′ end of the Rous sarcoma virus (RSV) genome. This sequence, termed Aψ, is sufficient for the packaging of a heterologous RNA (1). Aψ is located just downstream of the primer binding site and continues to the 5′ major splice site, located in the 5′ end of gag coding sequences. Aψ contains the last of three upstream open reading frames (uORF-3) located in the leader regions of ALSVs, which may play a role in packaging (3, 4, 12). Additionally, Aψ contains two 8-nt inverted repeats that are also conserved among several ALSVs and that are predicted to form the stem of the O3 stem-loop (5).
In this report we define Mψ, a 160-nt region from within Aψ sufficient for packaging in our heterologous system. Mψ retains uORF-3 and the O3 stem inverted repeats from Aψ. We also describe the results of our initial experiments to determine the secondary structures within Mψ necessary for packaging. In agreement with studies by Knight et al. (9), we find that the structure of the O3 stem, but not the sequence composing the stem, is necessary for packaging.
In order to further define the regions necessary for packaging, we tested sequences from within Aψ for their abilities to confer packaging in our heterologous system. Aψ deletion mutants were placed 3′ of the coding sequence for the neomycin phosphotransferase gene (neo) in a derivative of pCMVneo (2, 10). This plasmid, pASY185, has an MluI linker inserted at the unique SmaI site (Fig. 1). The constructs were then transfected into the Q2bn packaging cell line (13), and mass cultures of drug G418-resistant cells were obtained (1). In initial experiments, Western blot analyses were performed on viral supernatants concentrated by high-speed centrifugation through a 20% sucrose cushion and quantitated, as previously described (1), in order to determine the relative amounts of viral particles released by the cells. In later experiments, the more quantitative radioimmunoprecipitation assays (RIPAs) were performed. For these analyses, cells were plated at a density of 107 cells per 100-mm-diameter plate at least 18 h before metabolic labeling. The cells were then labeled with [35S]methionine for 5 h, and the label was chased overnight by the addition of complete medium. Supernatants were collected, and labeled viral particles were concentrated by high-speed centrifugation through a 20% sucrose cushion. One-half of the collected particles were set aside for RNA analysis; the remaining half were precipitated in the presence of a mixture consisting of antibody buffer, 5 μl of α-PrB (11), and protein A-Sepharose beads. The antigen-antibody complexes were washed as previously described (14). Bound proteins were eluted and separated on a sodium dodecyl sulfate–12.5% polyacrylamide gel. Following electrophoresis, the gel was dried and, after an overnight exposure, scanned by a Molecular Dynamics PhosphorImager. Radioactive bands corresponding to the CA protein were quantitated, in machine units, with ImageQuant software.
FIG. 1.
Constructs used to assay regions of the genome necessary for ψ function. Portions of the RSV Prague C genome were cloned into pASY185, a pCMVneo derivative. Numbering is from the first nucleotide of the 5′ R region of the RSV Prague C genome (GenBank accession no., J02342). The S1 and S2 inverted repeats are boxed.
To determine the relative amounts of neo-specific RNA contained in the viruses, RNase protection assays were performed on concentrated virus with the Direct Protect kit (Ambion Inc.). RNAs were probed with antisense neo RNA either 170 or 250 nt in length. Protected RNA bands were scanned and quantitated as described above. Packaging efficiency was determined by calculating the ratio of the amount of neo RNA in the particles to the amount of CA protein, as measured by Western blotting or RIPA. The ratios obtained for the deletion mutants were then normalized to that of the pCMVneo-Mψ construct.
The deletions were obtained by using Aψ as the template in the Erase-a-base system (Promega, Inc.). The sizes and locations of the deletions obtained are shown in Fig. 1. AψΔ3′ contains a deletion of 80 nt from the 3′ end of Aψ. Δ5′Aψ contains a deletion of 30 nt from the 5′ end of Aψ. By Western and RNase protection analyses (data not shown) we found that the packaging efficiencies of AψΔ3′ and Δ5′Aψ were similar to that of Aψ. These results are summarized in Table 1. Next, the packaging efficiency of the 160-base Mψ, which contains the deletions found in both AψΔ3′ and Δ5′Aψ, was determined. Again, packaging levels were similar to that of Aψ (Table 1).
TABLE 1.
Lengths and packaging efficienciesa of viral sequences from the 5′ region of RSV
| Clone | Length (nt) | Packaging efficiency |
|---|---|---|
| Aψ | 270 | +++ |
| AψΔ3′ | 190 | +++ |
| Δ5′Aψ | 240 | +++ |
| Mψ | 160 | +++ |
| Δ5′Mψ | 144 | + |
| MψΔ3′ | 105 | ++ |
Packaging efficiency was calculated as the ratio of the relative amount of neo-specific RNA packaged in particles, as measured by RNase protection analysis, to the relative number of particles, as measured by Western analysis. Packaging efficiencies were normalized to that of Mψ. For more detail see the text. Each plus sign represents approximately 1 order of magnitude.
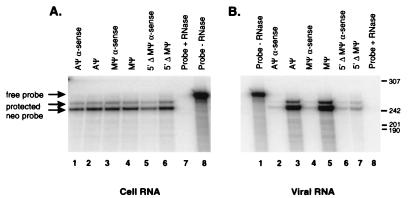
We next deleted either end of Mψ to determine the effect on packaging. Δ5′Mψ contains a deletion of 16 nt from the 5′ end of Mψ, whereas MψΔ3′ contains a deletion of 55 nt from the 3′ end of Mψ (Fig. 1). These Mψ deletions were placed 3′ of neo in the sense and antisense orientations. Similar numbers of viral particles were released from the cells transfected with the mutants, as measured by Western blot analysis (data not shown). RNase protection analysis of cell RNA indicated that similar levels of neo RNA are present in the cells (Fig. 2A; data not shown for MψΔ3′ constructs). RNase protection analysis of RNA from viral particles released from the cells showed similar levels of packaging of neo RNA containing Aψ and Mψ in the sense orientation (Fig. 2B, lanes 3 and 5). In contrast, the level of packaging of Δ5′Mψ RNA was reduced over 18-fold compared to that of Mψ (Fig. 2B, lanes 5 and 7). The efficiency of MψΔ3′ was reduced over sixfold compared to that of Mψ (Fig. 5C, lanes 3 and 4). These results are summarized in Table 1. The RNAs containing ψ in the antisense orientation were packaged at very low levels (Fig. 2B, lanes 2, 4, and 6).
FIG. 2.
(A) RNase protection analysis to determine the amount of neo RNA in cells. Total cell RNAs were extracted from G418-resistant mass cultures of Q2bn-4D cells transfected with plasmids, as indicated above each lane, and annealed to the antisense neo probe. The sample in lane 7 had no cell RNA added. The sample in lane 8 was not treated with RNase. The arrows show the expected locations of the free probe and the neo-protected band, seen as a doublet. (B) RNase protection analysis to determine the amount of neo RNA packaged in virions released from the cells described in the legend for panel A. Virus was collected, and RNAs were extracted and annealed to the antisense neo probe. The sample in lane 1 had no RNase added. The sample in lane 8 had no viral RNA added.
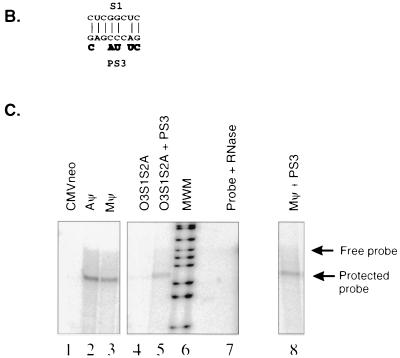
FIG. 5.
(A) The sequences of O3S1A and O3S2A are shown above and below, respectively, the wt O3 stem. Nucleotides differing from those of the wt sequence are highlighted. (B) RIPA to determine the amount of viral particles released from G418-resistant mass cultures of Q2bn-4D cells transfected with plasmids, as indicated above each lane, and precipitated with the α-PrB antibody. The expected size of the capsid (CA) band is indicated. (C) RNase protection analysis of the RNA from virus released from cells described for panel B. The expected sizes of the free probe and the protected neo probe are indicated by arrows. The sample in lane 1 had no RNase added. The sample in lane 2 had no viral RNA added.
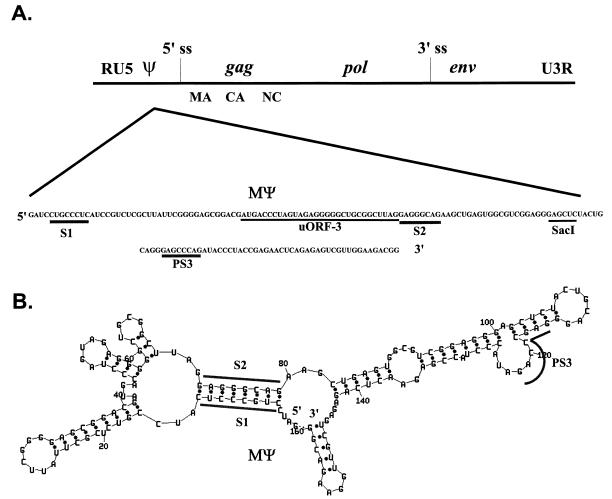
Mψ is the shortest sequence which can confer a high level of packaging of a heterologous RNA that we have identified. The primary sequence of Mψ is given in Fig. 3A. Mψ does not contain any sequences from the Gag open reading frame. Nor are sequences from the primer binding site and 5′ major splice site included in the 160-nt region. Mψ does contain the 31-nt sequence shown by Katz et al. to be involved in packaging (8). Additionally, the entire uORF-3 is located within Mψ (Fig. 3A). The Mψ sequence was placed in the M-Fold RNA folding program to determine the predicted secondary structure (6, 7, 15). The most stable structure is shown in Fig. 3B. The O3 stem-loop structure, which is conserved in the computer-generated folding models of the leader region of several ALSV strains (5) and which has been shown to play a role in packaging (9), is also largely conserved in our model of Mψ. The inverted repeats composing the stem, which we have named S1 and S2, are underlined in Fig. 3B.
FIG. 3.
(A) The RSV genome showing the location and sequence of the minimal packaging region, Mψ. The S1 and S2 inverted repeats composing the O3 stem, as well as the downstream PS3 region, are underlined in gray. The SacI restriction site and uORF-3 are underlined in black. R, repeated sequence; U5, unique 5′ sequences; gag, gene encoding viral structural proteins; MA, matrix protein; CA, capsid protein; NC, nucleocapsid protein; pol, gene encoding reverse transcriptase and integrase; env, gene encoding glycoproteins; U3, unique 3′ sequences; 5′ ss, 5′ splice donor site; 3′ ss, 3′ splice acceptor site. (B) Predicted secondary structure of Mψ. The M-Fold RNA folding program based on the computer algorithm of Zuker et al. (6, 7, 15) was used. The folding energy obtained was −59.0 kcal. The S1, S2, and PS3 regions are outlined.
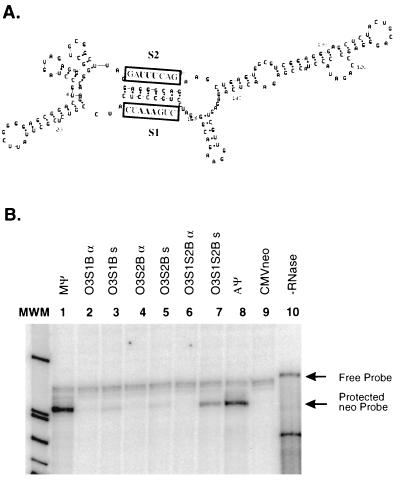
The importance of the O3 stem in packaging is supported by our Δ5′Mψ mutant. As described above, the deletion of just 16 nt, which includes the S1 region, from the 5′ end of Mψ results in an 18-fold reduction in packaging over wild-type (wt) Mψ. To more directly study the role of the O3 stem in packaging, we made a series of mutations in S1 and S2 which disrupt the correct formation of the stem. These point mutations were made by standard PCR techniques. We constructed the same S1 and S2 mutations made by Knight et al. (9) for use in our heterologous system. In mutant O3S1B, three contiguous nucleotides from S1, CCC, have been changed to AAA. In mutant O3S2B, three contiguous nucleotides from S2, GGG, have been changed to UUU. Finally, O3S1S2B contains both mutations, restoring the complementarity between S1 and S2. These sequences are summarized in Table 2 and in Fig. 4A. The mutated packaging signals were inserted in pASY185 in both the sense and antisense orientations. To measure the number of particles released from cell lines transfected with the mutants, RIPAs were performed. These studies indicated that similar numbers of particles were released from cells. RNase protection analysis of RNA from particles released from the cells showed that the packaging of O3S1B and O3S2B sense RNAs is reduced 16- and 30-fold, respectively, over that of wt Mψ RNA (Fig. 4B, lanes 1, 3, and 5). In particles released from the compensatory mutant, O3S1S2B, packaging cells, the amount of neo-containing RNA is similar to that of wt Mψ (Fig. 4B, lanes 1 and 7). These packaging efficiencies are summarized in Table 2. The antisense mutants had greatly reduced packaging levels compared to wt Mψ (Fig. 4B, lanes 1, 2, 4, and 6). From these data, we concluded that the O3 stem structure plays a role in packaging, while the primary sequence of the stem is not important. The regions of the Gag polyprotein that bind to the packaging signal during encapsidation may directly interact with the O3 stem, or O3 may stabilize a region directly involved in packaging. Regions downstream of S2 may also be involved in packaging, as our MψΔ3′ mutant, in which all Mψ sequences downstream of the SacI site were deleted, had a sixfold reduction in packaging compared to wt Mψ.
TABLE 2.
Sequences and packaging efficiencies of O3 stem mutants
| Clone | Sequence (5′ to 3′)a of:
|
Packaging efficiencyb | ||
|---|---|---|---|---|
| S1 | S2 | PS3 | ||
| wt Mψ | CUGCCCUC | GAGGGCAG | GAGCCCAG | 1.00 |
| O3S1B | CUGAAAUC | GAGGGCAG | GAGCCCAG | 0.06 |
| O3S2B | CUGCCCUC | GAUUUCAG | GAGCCCAG | 0.03 |
| O3S1S2B | CUGAAAUC | GAUUUCAG | GAGCCCAG | 1.10 |
| O3S1A | CUCGGCUC | GAGGGCAG | GAGCCCAG | 0.18 |
| O3S2A | CUGCCCUC | GAGCCGAG | GAGCCCAG | 0.09 |
| O3S1S2A | CUCGGCUC | GAGCCGAG | GAGCCCAG | 0.02 |
| O3S1S2A+PS3 | CUCGGCUC | GAGCCGAG | CAGAUCUC | 0.29 |
| wt Mψ+PS3 | CUGCCCUC | GAGGGCAG | CAGAUCUC | 1.09 |
Sequence differing from that of the wt is shown in boldface.
Packaging efficiency was calculated as the ratio of the relative amount of neo-specific RNA packaged in particles, as measured by RNase protection analysis, to the relative number of particles, as measured by RIPA. Packaging efficiencies were normalized to that of wt Mψ. For more details, see the text.
FIG. 4.
(A) The sequences of O3S1B and O3S2B are shown above and below, respectively, the wt O3 stem. Nucleotides differing from those of the wt sequence are highlighted. (B) RNase protection analysis to determine the amount of neo RNA packaged in virions released from G418-resistant mass cultures of Q2bn-4D cells transfected with plasmids, as indicated above each lane, and annealed to the antisense neo probe. The expected locations of free probe and the protected neo band are indicated by arrows. No RNase was added to the sample in lane 10.
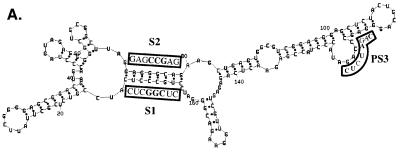
To confirm and extend these results, we made another set of mutations which altered the O3 stem sequence at different nucleotides. In mutant O3S1A, three contiguous nucleotides from the S1 stem, GCC, have been changed to CGG. In mutant O3S2A, 3 nt from the S2 stem, GGC, have been changed to CCG. O3S1S2A contains both mutations, restoring the complementarity between S1 and S2. These sequences are summarized in Table 2 and Fig. 5A. The numbers of particles released by the cell lines were similar as measured by RIPA (Fig. 5B). As in the case of the O3S1B and O3S2B mutants, the levels of O3S1A and O3S2A RNAs in the particles, as measured by RNase protection analysis, were reduced compared to that of wt Mψ (Fig. 5C, lanes 3, 5, and 6). However, unlike the packaging of the O3S1S2B compensatory mutant RNA, the packaging of O3S1S2A RNA was greatly reduced compared to that of wt Mψ (Fig. 5C, lanes 3 and 7). The packaging efficiencies are summarized in Table 2. This was a surprising result, as the mutations were expected to restore the complementarity of the inverted repeat, presumably restoring the secondary structure of the O3 stem.
An examination of the sequences surrounding the S1 and S2 inverted repeats revealed a region, PS3 (Fig. 3), 36 nt downstream of S2, that is complementary to S1 in 7 of 8 nt in the O3S1S2A mutant (Fig. 6A and B). To test whether PS3 was preventing the proper folding of the mutated O3 stem by interacting with S1 in the O3S1S2A mutant, we made an additional mutant, O3S1S2A+PS3, that contains the O3S1S2A mutations as well as a mutation in PS3 that destroys complementarity to the S1 mutation. These sequences are summarized in Table 2 and Fig. 6A and B. By measuring the number of particles released from these cells (data not shown) and measuring the amount of neo RNA in these particles (Fig. 6C, lanes 3, 4, and 5) we found that the packaging efficiency of O3S1S2A+PS3 is increased over 10-fold compared to that of O3S1S2A. The PS3 region itself does not appear to play a role in the folding of wt Mψ. When the PS3 mutation was placed in the context of wt Mψ, it did not affect packaging (Fig. 6C, lanes 3 and 8). These packaging efficiencies are summarized in Table 2. These experiments highlight the complexity of RNA structural analysis. Mutation in one region can have an effect on the folding of regions which appear noncontiguous in computer-generated RNA folding models.
FIG. 6.
(A) The sequence of O3S1S2A+PS3. The sequences that differ from that of the wt O3 stem are highlighted. The PS3 region is outlined, and the sequences mutated in O3S1S2A+PS3 are indicated. (B) The predicted base pairing between S1 and PS3 in the O3S1S2A mutant. The nucleotide changes in the O3S1S2A+PS3 mutant are shown in boldface below the PS3 sequence. (C) RNase protection analysis of the RNA from virus released from G418-resistant mass cultures of Q2bn-4D cells transfected with plasmids, as indicated above each lane, and annealed to the neo probe. The expected sizes of the free probe and the protected neo probe are indicated by arrows. The sample in lane 7 had no viral RNA added.
Despite the identification of Mψ, there exists a paradox in our understanding of RSV packaging. In mammalian retroviruses and the avian spleen necrosis virus, ψ is located downstream of the 5′ major splice site. As a result, the packaging signal is not found on subgenomic species. In contrast, Mψ is located upstream of the 5′ major splice site and therefore is found on both full-length and env and src subgenomic species. Yet the full-length message is preferentially packaged (1). One explanation is that there are regions in the intron that augment packaging in the full-length RNA. Similarly, there may be sequences in env that in some way inhibit packaging when inserted next to Mψ in the spliced RNAs. This model, in which additional sequences play a role in packaging, is supported by our observation that the rate of packaging of Aψ and Mψ heterologous RNAs is down 10-fold compared to the rate of packaging of the full-length RSV genome (data not shown). This possibility points out a limitation of our heterologous assay. In wt RSV infection, the subgenomic RNAs may compete with genomic RNA for binding to the Gag motifs that specifically recognize the packaging region. Since this competition does not exist in our assay, we may be overestimating the precise efficiency of Mψ. Thus, while we have identified sequences sufficient for selective encapsidation of our heterologous RNA, there may be additional sequences that further select the encapsidation of the full-length viral genome. We are currently performing experiments to investigate this possibility.
With this short packaging signal identified, we are presently performing a more extensive investigation of the secondary structures involved in packaging. Another important aspect of RSV packaging that has yet to be elucidated is the role, if any, of uORF-3. This open reading frame has been shown to be efficiently translated (3, 4, 12), and when the translation of uORF3 is altered, the packaging efficiency is reduced (3). Donze et al. (3) have suggested that uORF-3 contains an RNA secondary structure that inhibits the packaging of the RNA. During translation of the open reading frame, this inhibitory structure is removed and packaging can proceed. The predicted fold of Mψ does show stem-loop structures in these sequences. We are currently testing the effect of mutating the AUG of uORF-3 on packaging in our heterologous system.
Acknowledgments
J.D.B. and A.Y. contributed equally to this work.
This work was supported by a grant from the National Cancer Institute (CA 18282) to M.L.L., A.Y. was supported by a Leukemia Society of America postdoctoral fellowship, and J.D.B. was supported by a National Science Foundation graduate fellowship.
We thank Julie Overbaugh for critical review of the manuscript.
REFERENCES
- 1.Aronoff R, Hajjar A M, Linial M L. Avian retroviral RNA encapsidation: reexamination of functional 5′ RNA sequences and the role of nucleocapsid Cys-His motifs. J Virol. 1993;67:178–188. doi: 10.1128/jvi.67.1.178-188.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aronoff R, Linial M L. Specificity of retroviral RNA packaging. J Virol. 1991;65:71–80. doi: 10.1128/jvi.65.1.71-80.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Donze O, Damay P, Spahr P F. The first and third uORFs in RSV leader RNA are efficiently translated: implications for translational regulation and viral RNA packaging. Nucleic Acids Res. 1995;23:861–868. doi: 10.1093/nar/23.5.861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Donze O, Spahr P F. Role of the open reading frames of Rous sarcoma virus leader RNA in translation and genome packaging. EMBO J. 1992;11:3747–3757. doi: 10.1002/j.1460-2075.1992.tb05460.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hackett P B, Dalton M W, Johnson D P, Petersen R B. Phylogenetic and physical analysis of the 5′ leader RNA sequences of avian retroviruses. Nucleic Acids Res. 1992;19:6929–6934. doi: 10.1093/nar/19.24.6929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jaeger J A, Turner D H, Zuker M. Improved predictions of secondary structures for RNA. Proc Natl Acad Sci USA. 1989;86:7706–7710. doi: 10.1073/pnas.86.20.7706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jaeger J A, Turner D H, Zuker M. Predicting optimal and suboptimal secondary structure for RNA. Methods Enzymol. 1990;183:281–306. doi: 10.1016/0076-6879(90)83019-6. [DOI] [PubMed] [Google Scholar]
- 8.Katz R A, Terry R W, Skalka A M. A conserved cis-acting sequence in the 5′ leader of avian sarcoma virus RNA is required for packaging. J Virol. 1986;59:163–167. doi: 10.1128/jvi.59.1.163-167.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Knight J B, Si Z H, Stoltzfus C M. A base-paired structure in the avian sarcoma virus 5′ leader is required for efficient encapsidation of RNA. J Virol. 1994;68:4493–4502. doi: 10.1128/jvi.68.7.4493-4502.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Linial M. Creation of a processed pseudogene by retroviral infection. Cell. 1987;49:93–102. doi: 10.1016/0092-8674(87)90759-8. [DOI] [PubMed] [Google Scholar]
- 11.Linial M, Fenno J, Burnette W B, Rohrschneider L. Synthesis and processing of viral glycoproteins in two nonconditional mutants of Rous sarcoma virus. J Virol. 1980;36:280–290. doi: 10.1128/jvi.36.1.280-290.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Moustakas A, Sonstegard T S, Hackett P B. Alterations of the three short open reading frames in the Rous sarcoma virus leader RNA modulate viral replication and gene expression. J Virol. 1993;67:4337–4349. doi: 10.1128/jvi.67.7.4337-4349.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stoker A W, Bissell M J. Development of avian sarcoma and leukosis virus-based vector-packaging cell lines. J Virol. 1988;62:1008–1015. doi: 10.1128/jvi.62.3.1008-1015.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yu S F, Linial M L. Analysis of the role of the bel and bet open reading frames of human foamy virus by using a new quantitative assay. J Virol. 1993;67:6618–6624. doi: 10.1128/jvi.67.11.6618-6624.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zuker M. On finding all suboptimal foldings of an RNA molecule. Science. 1989;244:48–52. doi: 10.1126/science.2468181. . (Review.) [DOI] [PubMed] [Google Scholar]