Abstract
Five site-specific adeno-associated virus integrants generated in a model system with an Epstein-Barr virus- based shuttle vector have been characterized. The results suggest a deletion-substitution mechanism of recombination.
Adeno-associated virus (AAV) type 2 has the unique property of integrating at a specific site (19q13.3-qter) in the human genome (6–8, 12). To study the mechanism of site-specific integration, a model system with an Epstein-Barr virus (EBV)-based shuttle vector (Fig. 1) that carries the preintegration site from 19q (AAVS1) (Fig. 1) was developed (3). Human cell lines containing the shuttle vector, with inserts from AAVS1 of either 0.51, 1.6, or 8.2 kb, were infected with AAV, and the recombinants produced were analyzed (4). These studies demonstrated that site specificity was determined by DNA sequence. Detailed genetic studies with the model system have demonstrated that sequences in AAVS1 corresponding to the Rep-binding site (RBS) and the terminal resolution site (TRS) are required for site-specific integration (9). AAV integration in the model system resembled integration into chromosome 19 in several ways: integration was site specific, the integration event was associated with disruption and rearrangement of the AAVS1 target, and the AAV integrants in several instances were in a head-to-tail tandem array similar to those observed for chromosomal integrants. A majority of the recombinants generated in the model system had junctions between AAV and AAVS1 clustered around the RBS of AAVS1. This has also been observed for several chromosomal integrants (2, 13).
FIG. 1.
The substrates for site-specific integration. (A) Schematic representation of the AAV genome; (B) schematic representation of the EBV-based shuttle vector with the AAVS1 insert of 0.51 or 1.6 kb. The AAVS1 sequences show the putative TRS and an RBS. BamHI and HindIII were the restriction sites used for cloning the AAVS1 inserts. The genes of the vector backbone in which the recombinants had deletions are indicated; hygR, hygromycin resistance gene; EBNA-1, EBV-encoded nuclear antigen; oriP, EBV origin; ITR, inverted terminal repeat.
Integration of AAV into the human genome is associated with extensive rearrangements of viral and cellular sequences flanking the integration site (2, 5, 6, 8, 11, 13). These rearrangements include deletion of AAV sequences and the disruption of the AAVS1 preintegration site, and they have also been observed in the model system (4). In addition, the provirus structures in chromosome 19 show rather complex rearrangements, such as inversions and translocations of AAV and AAVS1 sequences. Recombinants generated in the episomal model system have not been characterized previously with respect to such rearrangements. To characterize rearrangements associated with AAV integration into the episome, a set of five recombinants from the model system were chosen and their structures were analyzed in detail.
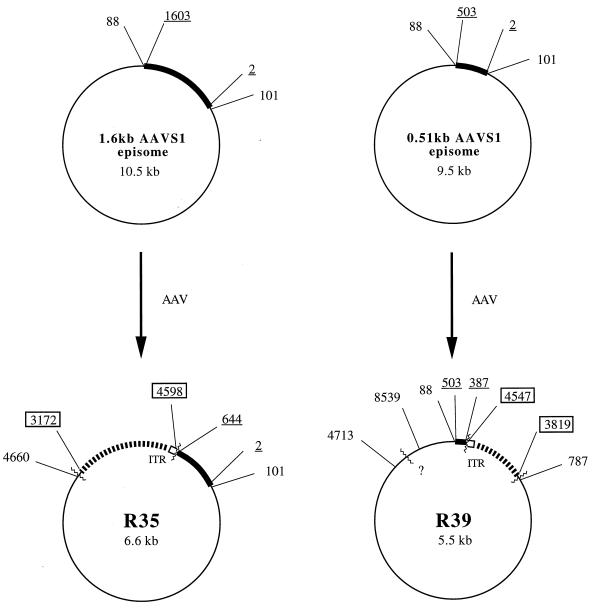
Five recombinants derived from cell lines carrying either the 0.51- or the 1.6-kb AAVS1 insert were selected and subjected to detailed restriction mapping. Recombinants were digested with restriction endonucleases (AvaI, PstI, and SmaI) that give characteristic restriction patterns. After electrophoresis and transfer to a nylon membrane, the digests were hybridized with the following three probes: AAV, AAVS1, and the backbone sequence of the EBV shuttle vector. By this approach, additional crossover points were identified. The newly identified junctions were confirmed by detailed sequencing. Overall structures of the five recombinants were determined and are shown in Fig. 2 and 3. The substrates used to produce the recombinants in the model system are shown in Fig. 1. The five recombinants showed considerable deletions of AAV, as well as of AAVS1 and of vector backbone sequences, and are summarized in Table 1. Table 2 shows that there is no correlation between the sizes of the deletions and the insertions found in the recombinants. Deletions of AAV DNA have been characterized previously (4).
FIG. 2.
Structures of the recombinants R35 and R39 derived from cell lines carrying the 1.6-kb AAVS1 and the 0.51-kb AAVS1 episomes. ▮▮▮▮▮▮, AAV sequences (boxed numbers); ▪, AAVS1 sequences (underlined numbers); ——, vector backbone sequences (plain numbers); ······, unknown sequences; □, ITR, AAV terminal repeats; ∧∧∧∧∧∧, recombinant junctions; ?, unknown position of a junction within the vector sequence.
FIG. 3.
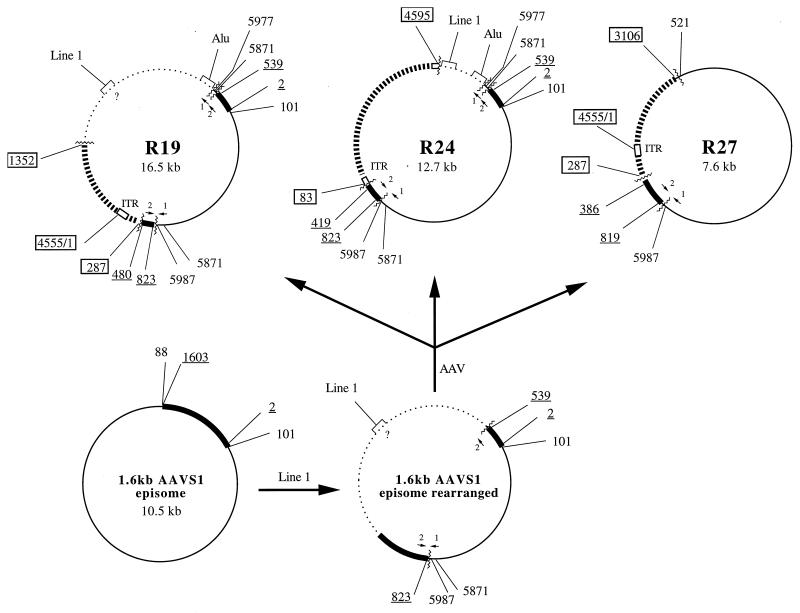
Structures of the recombinants R19, R24, and R27 derived from a cell line carrying the 1.6-kb AAVS1 episome. The recombinants most likely arose from an 1.6-kb AAVS1 episome that had recombined with a Line-1 element prior to AAV infection. R19, R24, and the rearranged AAV-negative episome were isolated from the same cell line and the same experiment. R27 was retrieved from the same cell line as well, but at a later passage. ▮▮▮▮▮▮, AAV sequences (boxed numbers); ▪, AAVS1 sequences (underlined numbers); ——, vector backbone sequences (plain numbers); ······, unknown sequences; □, ITR, AAV terminal repeats; ∧∧∧∧∧∧, recombinant junctions;  , duplicated vector sequence (nt 5871 to 5977);
, duplicated vector sequence (nt 5871 to 5977);  , duplicated AAVS1 sequence (nt 480 to 539); ?, unknown position of the Line-1 sequence within the unrelated sequence of R19 and the rearranged EBV episome.
, duplicated AAVS1 sequence (nt 480 to 539); ?, unknown position of the Line-1 sequence within the unrelated sequence of R19 and the rearranged EBV episome.
TABLE 1.
Summary of the deletions of AAV, AAVS1, and vector backbone sequences found in the recombinants
| Recombinant (AAVS1)a | AAV deletion (nt) | AAVS1 deletion upstream of RBS | AAVS1 deletion downstream of RBS (nt) | 220.2 deletion (nt) |
|---|---|---|---|---|
| R19 (1.6) | 288– 1351 | 824–1603 | 1– 88; 5988– 8952 | |
| R24 (1.6) | 1– 82; 4595– 4680 | 824– 1603 | 1– 88; 5988– 8952 | |
| R27 (1.6) | 288– 3105 | 1– 386 | 820– 1603 | 1– 88; 101– 520; 5988– 8952 |
| R35 (1.6) | 1– 3171; 4598– 4680 | 645– 1603 | 1– 88; 4667– 8952 | |
| R39 (0.51) | 1– 3818; 4547– 4680 | 1– 387 | 101– 786; 4713– 8539 |
Recombinants R19, R24, R27, and R35 were isolated from the same cell line carrying the 1.6-kb AAVS1 sequence. Recombinants R19 and R24, R27, and R35 were retrieved after AAV infection of the cell line at passages 10, 11, and 21, respectively. Recombinant R39 was isolated from a cell line carrying the 0.51-kb AAVS1 sequence. Numbers in parentheses are sizes in kilobases.
TABLE 2.
Comparison of the lengths of deleted episome sequences and the lengths of inserted sequencesa
| Recom- binant | Original size of episome (kb) | Size of recom- binant (kb) | Length of deletion (kb) | Length of insertion (kb) | Length of inserted AAV (kb) |
|---|---|---|---|---|---|
| R19 | 10.5 | 16.5 | 3.8 | 9.8 | 3.5 |
| R24 | 10.5 | 12.7 | 3.8 | 6.0 | 4.5 |
| R27 | 10.5 | 7.6 | 4.6 | 1.7 | 1.7 |
| R35 | 10.5 | 6.6 | 5.3 | 1.4 | 1.4 |
| R39 | 9.5 | 5.5 | 4.7 | 0.7 | 0.7 |
On account of the inherent instability of the AAVS1-shuttle vector, it was not possible to distinguish between deletions occurring prior to or during the AAV integration event.
All recombinants had large regions of the EBV EBNA-1 (nucleotides [nt] 4162 to 6772) and oriP (nt 6773 to 8952) genes deleted from the vector sequences. One recombinant (R39) also showed deletion within the hygromycin resistance gene (nt 635 to 1661). The Escherichia coli plasmid genes were necessarily retained in recombinants by virtue of their cloning in E. coli.
Similarly, extensive deletions of AAVS1 sequences were found either upstream (R39) or downstream (R19, R24, and R35) or both upstream and downstream (R27) of the RBS. Apart from the region surrounding the RBS and the TRS, there seems to be no tendency to maintain any part of the AAVS1 locus after AAV integration. This finding confirms earlier models of the AAV integration event that are based on the hypothesis that the RBS and TRS play an important role in this recombination event (1, 9, 10).
In addition to deletions, integration into human chromosome 19 is associated with complex rearrangements which lead to duplications and inversions of both AAV and cellular sequences. In a recent model for AAV integration based on replication, Rep binds to the RBS and introduces a nick into the TRS (9). The rearrangements are postulated to arise as a consequence of template switching by the replication complex. To find out whether strand switching also leads to such rearrangements during the process of AAV integration into the EBV episome, the selected recombinants were analyzed for sequence duplications and/or inversions. With one exception, restriction analysis showed that the recombinants did not contain any rearrangements other than the deletions noted above. A short stretch of AAVS1 and a short stretch of shuttle vector DNA were duplicated in two recombinants (R19 and R24). Detailed sequencing showed that the duplications were identical in both recombinants and had the following features. The duplicated vector sequence had a second junction with an unknown sequence which was homologous to the Alu sequence and was identical in the two recombinants. The duplicated sequences were in orientations opposite to one another; while the AAVS1 duplications were in the same orientation, the shuttle vector duplications were inverted with respect to one another. The duplications of AAVS1 and vector sequences were identical in both R19 and R24. This finding suggested that the rearrangements had occurred before the AAV integration event. Another observation confirmed this suggestion. The two recombinants R19 and R24 and an AAV-negative episome had been isolated from the same experiment. Dot blot hybridization and restriction mapping showed that the AAV-negative isolate and the two AAV recombinants contained the same Line- 1 sequence and the same duplications of AAVS1 (reference 3 and data not shown). Therefore, it is likely that prior to AAV integration, the shuttle vector plasmid carrying the AAVS1 insert underwent recombination with an extrachromosomal Line-1 element that led to the duplications of AAVS1 and vector sequences. AAV integration as a second event led to different junctions, with the AAVS1 sequence in each of the two recombinants (R19 and R24) reflecting the independence of the AAV integrations. In the recombinant R24, AAV integration was associated with deletion of a part of the unrelated sequence containing Line-1. If this scenario is true, then the formation of R27, which was isolated from the same cell line (but at a later passage), can also readily be explained. In this case, integration of AAV into the rearranged shuttle vector resulted not only in deletion of AAV sequences, but also in removal of the Line-1-containing sequences as well as the bordering duplicated stretches of AAVS1 and the vector.
Elucidation of the overall structures of five recombinants (Fig. 2 and 3) allowed us to evaluate the extent of rearrangements occurring in association with AAV integration into the shuttle vector. Fundamentally, these are deletions of AAV, AAVS1, and vector sequences. A total of 11 proviral structures of AAV in chromosome 19 have been analyzed in detail thus far (2, 5, 8, 14). The findings show that in chromosomal integration, inversions and/or translocations of both AAV and cellular sequences occur, in addition to deletion of AAV sequences. The more complex rearrangements seen at the chromosomal level may reflect postintegration instability. This would not be detected in the model system because of the short time between insertion and isolation of recombinant structures. We suggest, therefore, that AAV integrates into the AAVS1-shuttle vector via a simpler process involving a deletion-substitution mechanism.
In the AAVS1-shuttle vector, a region responsible for the instability of the plasmid during cell passage was localized to a 300-bp region near the 5′ end of AAVS1 (3, 9). Genetic analysis has shown that this region is not required for AAV integration (9). AAVS1-shuttle vectors which lacked a short sequence reputed to enhance recombination within the 300-bp region were stable. Therefore, the observed loss of the EBNA sequences could have been caused by either the AAVS1 sequence or the integration event but do not appear to have been caused by the inherent instability of the shuttle vector itself. Deletion of oriP is most likely to have been caused by the integration event, because earlier loss would lead to loss of the shuttle vector by dilution. As suggested above, the translocations in R19 and R24 seem to have arisen by recombination of the AAVS1-shuttle vector with the Line-1 sequence. This apparently happened prior to AAV integration. The Line-1 and the Alu element have also been found at AAV and cellular junctions in chromosomal proviral structures (2, 13), giving rise to the possibility that the instability region within the AAVS1 target promotes recombination with extrachromosomal DNAs as well as AAV integration. Thus, in analyzing the mechanism of AAV integration, it is essential to distinguish between rearrangements that are related to AAV integration and those that represent AAV-independent recombination events. With respect to the use of AAV for gene therapy, it would be an advantage if one could avoid unnecessary rearrangements. How ever, it is possible that the proclivity to rearrangement inherent in 19q13.3 actually enhances AAV integration and, thus, the high efficiency of the recombination event seen in vivo.
Although only a small number of recombinants retrieved from the model system have been investigated, the data are entirely consistent with integration by a deletion-substitution mechanism.
Acknowledgments
We thank Catherine Giraud for providing the recombinants. We thank Catherine Giraud, Peter Ward, and Patrick Menesis for helpful discussions and critical reading of the manuscript. We thank Nenita Cortez for excellent technical support.
This work was supported by grant GM50032 from the National Institutes of Health.
REFERENCES
- 1.Chiorini J A, Wiener S M, Yang L, Smith R H, Safer B, Kilcoin N P, Liu Y, Urcelay E, Kotin R M. The roles of AAV Rep proteins in gene expression and targeted integration. Curr Top Microbiol Immunol. 1996;218:25–33. doi: 10.1007/978-3-642-80207-2_2. [DOI] [PubMed] [Google Scholar]
- 2.Dutheil N, Deprez A, Begue A, Delobel B, Montpellier C, Walz C, Schlehofer J R, Duppressoir T. Proceedings of the VIIth International Parvovirus Workshop. 1997. Molecular characterization of AAV-2 integration sites in latently infected HeLa cells, abstr. P8. [Google Scholar]
- 3.Giraud C, Winocour E, Berns K I. Site-specific integration by adeno-associated virus is directed by a cellular DNA sequence. Proc Natl Acad Sci USA. 1994;91:10039–10043. doi: 10.1073/pnas.91.21.10039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Giraud C, Winocour E, Berns K I. Recombinant junctions formed by site-specific integration of adeno-associated virus into an episome. J Virol. 1995;69:6917–6924. doi: 10.1128/jvi.69.11.6917-6924.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kotin R M, Berns K I. Organization of adeno-associated virus DNA in latently infected Detroit 6 cells. Virology. 1989;170:460–467. doi: 10.1016/0042-6822(89)90437-6. [DOI] [PubMed] [Google Scholar]
- 6.Kotin R M, Linden R M, Berns K I. Characterization of a preferred site on human chromosome 19q for integration of adeno-associated virus by non-homologous recombination. EMBO J. 1992;11:5071–5078. doi: 10.1002/j.1460-2075.1992.tb05614.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kotin R M, Menninger J C, Ward D C, Berns K I. Mapping and direct visualization of a region-specific viral DNA integration site on chromosome 19q13-qter. Genomics. 1991;10:831–834. doi: 10.1016/0888-7543(91)90470-y. [DOI] [PubMed] [Google Scholar]
- 8.Kotin R M, Siniscalco M, Samulski R J, Zhu X D, Hunter L, Laughlin C A, McLaughlin S, Muzyczka N, Rocchi M, Berns K I. Site-specific integration by adeno-associated virus. Proc Natl Sci USA. 1990;87:2211–2215. doi: 10.1073/pnas.87.6.2211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Linden R M, Winocour E, Berns K I. The recombination signals for adeno-associated virus site-specific integration. Proc Natl Sci USA. 1996;93:7966–7972. doi: 10.1073/pnas.93.15.7966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Linden R M, Ward P, Giraud C, Winocour E, Berns K I. Site-specific integration by adeno-associated virus. Proc Natl Sci USA. 1996;93:11288–11294. doi: 10.1073/pnas.93.21.11288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.McLaughlin S K, Collis P, Hermonat P L, Muzyczka N. Adeno-associated virus general transduction vectors: analysis of proviral structures. J Virol. 1988;62:1963–1973. doi: 10.1128/jvi.62.6.1963-1973.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Samulski R J, Zhu X, Xiao X, Brook J D, Housman D E, Epstein N, Hunter L A. Targeted integration of adeno-associated virus (AAV) into human chromosome 19. EMBO J. 1991;10:3941–3950. doi: 10.1002/j.1460-2075.1991.tb04964.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Surosky R T, Urabe M, Godwin S G, McQuiston S A, Kurtzman G J, Ozawa K, Natsoulis G. Adeno-associated virus Rep proteins target DNA sequences to a unique locus in the human genome. J Virol. 1997;71:7951–7959. doi: 10.1128/jvi.71.10.7951-7959.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yang C C, Xiao X, Zhu X, Ansardi D C, Epstein N D, Frey M R, Matera A G, Samulski R J. Cellular recombination pathways and viral terminal repeat hairpin structures are sufficient for adeno-associated virus integration in vivo and in vitro. J Virol. 1997;71:9231–9247. doi: 10.1128/jvi.71.12.9231-9247.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]