Abstract
Phosphorylation of the expressed NS5A protein of hepatitis C virus (HCV), a member of the Hepacivirus genus of the family Flaviviridae, has been demonstrated in mammalian cells and in a cell-free assay by an associated kinase activity. In this report, phosphorylation is also shown for the NS5A and NS5 proteins, respectively, of bovine viral diarrhea virus (BVDV) and yellow fever virus (YF), members of the other two established genera in this family. Phosphorylation of BVDV NS5A and YF NS5 was observed in infected cells, transient expression experiments, and a cell-free assay similar to the one developed for HCV NS5A. Phosphoamino acid analyses indicated that all three proteins were phosphorylated by serine/threonine kinases. Similarities in the properties of BVDV NS5A, YF NS5, and HCV NS5A phosphorylation in vitro further suggested that closely related kinases or the same kinase may phosphorylate these viral proteins. Conservation of this trait among three quite distantly related viruses representing three separate genera suggests that phosphorylation of the NS5A/NS5 proteins or their association with cellular kinases may play an important role in the flavivirus life cycle.
The family Flaviviridae is currently comprised of three genera, Flavivirus, Pestivirus, and Hepacivirus. Several newly identified human and primate viruses, GBV-A, GBV-B, and GBV-C or hepatitis G virus, are also likely to be classified in this family. The Flaviviridae include numerous human and animal pathogens: agents of global importance include the human flaviviruses Japanese encephalitis virus, dengue virus, and yellow fever virus (YF); the animal pestiviruses classical swine fever virus, border disease virus, and bovine viral diarrhea virus (BVDV); and the hepacivirus hepatitis C virus (HCV). All Flaviviridae family members have a single-stranded, positive-sense RNA genome that is translated as a long viral polyprotein and processed by a combination of host and viral proteases into individual structural and nonstructural (NS) proteins (see reference 37 for a review of Flaviviridae features). Although members of the three Flaviviridae genera are only distantly related, their polyproteins are organized similarly (Fig. 1), with the structural proteins located in the N-terminal portion, followed by the NS proteins. Another common feature is the location of serine protease and nucleoside triphosphatase/helicase activities in the NS3 region and an RNA-dependent RNA polymerase activity near the C terminus of the polyprotein of viruses from all three genera. However, other features of the polyprotein differ among the three genera, such as the existence of an additional cleavage site in the NS5 region of HCV and pestiviruses, but not flaviviruses, that separates the N-terminal portion (NS5A) from the viral polymerase (NS5B).
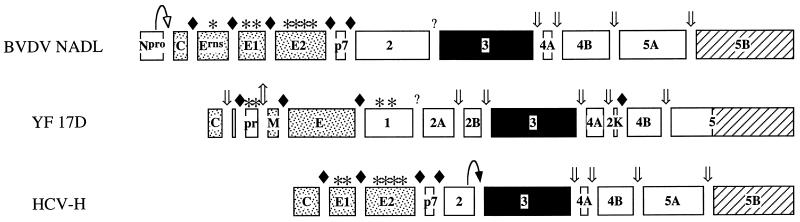
FIG. 1.
Features of the BVDV, YF, and HCV polyproteins. C, capsid; E, E1, E2, and Erns, envelope proteins; Npro, N-terminal autoprotease; prM, membrane precursor protein; p7 and 2K, small polypeptides of unknown function; 1, 2, 2A, 2B, 3, 4A, 4B, 5, 5A, and 5B, NS proteins. Stippled boxes, structural proteins; solid boxes, NS proteins containing the serine protease and nucleoside triphosphatase/helicase activities; striped boxes, polymerase domains. Asterisks indicate glycosylation sites; solid diamonds indicate host signalase cleavage sites; straight arrows mark the Golgi furin-like protease cleavage site in YF 17D (⇕) and the viral serine protease cleavage sites (⇓). The HCV NS2-3 and BVDV N-terminal autoproteases are indicated by curved arrows with solid and open arrowheads, respectively.
Phosphorylation of the NS5 protein has been demonstrated in cells infected with dengue virus type 2 (DEN-2) (23) and in extracts of cells infected with tick-borne encephalitis virus (TBE) (31). Phosphorylation of the HCV NS5A protein has also been shown in transiently transfected mammalian cells (22, 36). Phosphoamino acid analyses of transiently expressed DEN-2 NS5 and HCV NS5A and of in vitro-phosphorylated TBE NS5 have indicated that all three proteins are phosphorylated preferentially on serine residues (22, 23, 31, 36). The sites of phosphorylation in the NS5 proteins of DEN-2 and TBE are unknown, but site-directed mutagenesis experiments have suggested that phosphorylation of HCV NS5A may occur on Ser-2197, Ser-2210, and Ser-2204, as well as on serines in the C-terminal region of the protein (45). Phosphorylation of DEN-2 NS5 and a 56-kDa form of HCV NS5A has been observed in the absence of other viral proteins (23, 36, 45), but NS4A has been implicated in the production of a 58-kDa form of HCV NS5A (1, 22, 45). The effects of other viral proteins on the phosphorylation of DEN-2 or TBE NS5 have not been further examined. However, subcellular fractionation and immunoprecipitation (IP) experiments with DEN-2-infected cells have indicated that the phosphorylation state of NS5 correlates with its subcellular localization and ability to associate with NS3 (23), suggesting that NS5 phosphorylation may regulate viral replication and/or the expression of host genes, among other possibilities.
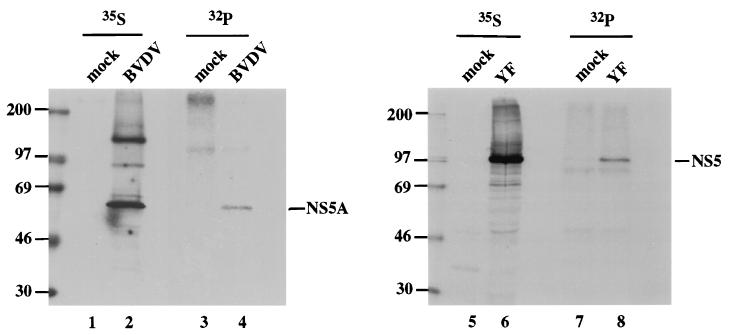
Reports that the DEN-2 and TBE NS5 proteins are phosphorylated suggested that YF NS5 is also likely to be phosphorylated. More generally, observations that viruses from two of the three Flaviviridae genera are phosphorylated within the NS5 region suggested that this characteristic may be conserved throughout the family and, furthermore, that despite the weak similarity between the NS5A region of HCV or pestiviruses and the NS5 region of flaviviruses, these proteins might share a common function related to their phosphorylation. To investigate the distribution of this trait among the Flaviviridae, we labeled BVDV- or YF-infected cells with [32P]orthophosphate and immunoprecipitated the BVDV NS5A or YF NS5 protein with region-specific antisera. As shown in Fig. 2, 32P labeling of both proteins was observed in infected cells (lanes 4 and 8), although the YF NS5 signal was somewhat stronger than that of BVDV NS5A. Phosphorylation of HCV NS5A was not examined in infected cells due to the current lack of an efficient cell culture system for its propagation.
FIG. 2.
Phosphorylation of BVDV NS5A and YF NS5 in virus-infected cells. Monolayers of Madin-Darby bovine kidney (MDBK) or SW-13 cells, a human adrenocortical carcinoma cell line, in 35-mm wells were infected, respectively, with BVDV NADL at a multiplicity of infection of 0.1 in 400 μl of phosphate-buffered saline–2% horse serum or with YF 17D at a multiplicity of infection of 5 in 200 μl of Earle’s minimal essential medium (MEM)–2% fetal bovine serum (FBS) for 1 h at 37°C. After the initial infection period, Dulbecco’s MEM–1 mM sodium pyruvate–10% horse serum or Earle’s MEM–2% FBS was added to the BVDV-infected MDBK cells or the YF-infected SW-13 cells, respectively, and incubation was continued for 20 h at 37°C. The cells were then labeled for 4 h at 37°C with MEM containing 2% of the normal methionine concentration, 3% dialyzed FBS, and 100 μCi of Expre35S35S (NEN) or with MEM lacking phosphate supplemented with 3% dialyzed FBS and 400 to 500 μCi of [32P]orthophosphate (ICN) per ml, as indicated. Cell lysates were prepared, and BVDV NS5A or YF NS5 was immunoprecipitated with region-specific antisera (3, 7) and protein A-agarose as previously described (36), except that BVDV NS5A samples were subjected to two successive rounds of IP to reduce the nonspecific background. IPs were analyzed by sodium dodecyl sulfate (SDS)–8% polyacrylamide gel electrophoresis (PAGE), followed by autoradiography. Samples from mock-infected cells are shown in lanes 1, 3, 5, and 7. The sizes of molecular size marker proteins (in kilodaltons) are indicated to the left of each panel.
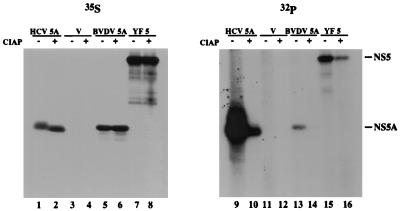
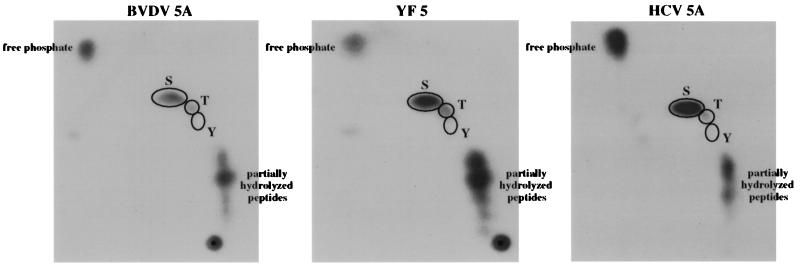
Once phosphorylation of BVDV NS5A and YF NS5 had been demonstrated in infected cells, phosphorylation of these proteins, along with that of HCV NS5A, was similarly examined in baby hamster kidney (BHK-21) cells with the vaccinia virus-T7 hybrid system (10). This system was selected for subsequent experiments because such heterologous expression systems are currently the only suitable method available for molecular analysis of HCV proteins and because the higher levels of protein expression provided by this system facilitated further analyses of the phosphorylation of all three viral proteins. Incorporation of [32P]orthophosphate into all three viral proteins was observed by using this system. The relative levels were as follows: HCV NS5A≫YF NS5>BVDV NS5A (Fig. 3, lanes 9, 13, and 15). Reduction of these signals as a result of phosphatase treatment (Fig. 3, lanes 10, 14, and 16) confirmed that the observed 32P labeling of these proteins was due to phosphorylation. The incomplete removal of 32P may be indicative of phosphorylation on threonines, which are poor substrates for calf intestinal alkaline phosphatase (CIAP), or on serines located in a phosphatase-resistant conformation. However, the incorporation of some 32P into alternative phosphorus-containing moieties, such as glycosylphosphatidyl inositol, cannot be excluded based on these data. Phosphoamino acid analyses of heterologously expressed BVDV NS5A, YF NS5, and HCV NS5A demonstrated that phosphorylation occurred preferentially on serine, although a low level of threonine phosphorylation was also observed (Fig. 4). Several lines of evidence suggest that the phosphorylation of HCV NS5A, and probably of BVDV NS5A and YF NS5 also, is mediated by cellular serine/threonine kinases: (i) none of the NS5 or NS5A proteins contains motifs characteristic of known kinases; (ii) phosphorylation is able to occur in the absence of other viral proteins, as shown in Fig. 3; (iii) phosphorylation has been observed in the absence of vaccinia virus-encoded kinases (Fig. 2 and reference 36); and (iv) phosphorylation of HCV NS5A expressed in Escherichia coli is dependent on the addition of cellular extracts (19, 36).
FIG. 3.
Phosphorylation of HCV NS5A, BVDV NS5A, and YF NS5 transiently expressed in BHK-21 cells. BHK-21 cells were infected with vTF7-3 (10), transfected with pTM3 (32) (lanes 3, 4, 11, and 12), pTM3/HCV 5A (36) (lanes 1, 2, 9, and 10), pTM3/BVDV 5A (lanes 5, 6, 13, and 14), or pBRTM/YF 5 (lanes 7, 8, 15, and 16), labeled with 80 μCi of Expre35S35S (NEN) or 100 μCi of [32P]orthophosphate (ICN) per ml, and harvested; NS5A and NS5 were then immunoprecipitated with region-specific antisera (3, 7, 14) by using Pansorbin cells (Calbiochem) and analyzed by SDS–8% PAGE and autoradiography (36). IPs in the even-numbered lanes were treated with 20 U of CIAP in 100 μl of phosphatase buffer (50 mM Tris-Cl [pH 7.5], 1 mM MgCl2, 0.1 mM ZnCl2, 1 mM spermidine) for 1 h at 37°C prior to SDS-PAGE; mock phosphatase treatments were performed on samples shown in the odd-numbered lanes. pTM3/BVDV 5A was constructed by inserting the NcoI-XhoI fragment of a PCR product amplified from pTM3/BVDV 2398-3988 (50a) with primers corresponding to the N and C termini of BVDV NS5A (BRL 367 [5′-AACCATGGCGTCCGGAAATTACATT-3′] and BRL 368 [5′-AACTCGAGCTATAGCTTCATGGCATA-3′]) into the NcoI-XhoI site of pTM3 (32). pBRTM/YF 5 was constructed by inserting the NcoI-EcoRI and EcoRI-PstI fragments of pBS.YF.NS5 (6a) into pBRTM/HCV 827-3011 (14) that had been digested with NcoI and PstI to remove the HCV sequences.
FIG. 4.
Phosphoamino acid analyses of BVDV NS5A, YF NS5, and HCV NS5A expressed transiently in BHK-21 cells. BHK-21 cells were infected with vTF7-3 and transfected with pTM3/BVDV 5A, pBRTM/YF 5, or pTM3/HCV 5A, labeled with [32P]orthophosphate (ICN), and harvested; the NS5A and NS5 proteins were then isolated and subjected to phosphoamino acid analysis as previously described (36). The positions of comigrating unlabeled phosphoamino acid standards are indicated by the outlined ovals.
Previous studies have further demonstrated that HCV NS5A is phosphorylated by an associated cellular serine/threonine kinase activity in vitro (19, 36). The observation that BVDV NS5A and YF NS5 were phosphorylated mostly on serine, like HCV NS5A, is consistent with the possibility that phosphorylation of all three proteins is catalyzed by the same cellular serine/threonine kinase. To investigate this possibility, phosphorylation of all three proteins was analyzed in an in vitro kinase assay, which, at least in the case of HCV, was shown to resemble intracellular NS5A phosphorylation (36).
Standard conditions for this assay have been described in detail elsewhere (36), but the main features were as follows: BVDV NS5A, YF NS5, and HCV NS5A were fused to the C terminus of a 26-kDa fragment of the glutathione S-transferase (GST) protein from Schistosoma japonicum (42) and transiently expressed in BHK-21 cells with the vaccinia virus-T7 hybrid system; cells expressing the fusion protein were lysed in a buffer containing nonionic detergent; the clarified lysate was incubated with glutathione-agarose to capture the GST fusion protein and associated cellular proteins, and the resulting complexes were washed with lysis buffer to remove nonspecifically bound proteins. Kinase reactions were then performed by incubating the purified complexes in buffer containing MnCl2 and [γ-32P]ATP to allow phosphorylation of the fusion protein by associated kinases, terminated by heating the reaction mixtures in protein sample buffer, and analyzed by SDS-PAGE followed by autoradiography.
As shown in Fig. 5, in vitro phosphorylation of GST-BVDV NS5A, GST-YF NS5, and GST-HCV NS5A was observed in this assay, with the GST-HCV NS5A protein exhibiting the highest level of phosphorylation. A number of additional 32P-labeled species were observed in the GST-YF NS5 in vitro phosphorylation reaction. Most of these appeared to be phosphoproteins associated specifically with YF NS5 rather than degradation products of the GST-YF NS5 fusion protein, since they disappeared after IP under denaturing conditions with NS5-specific antiserum (data not shown). Since this removal of associated phosphoproteins facilitated quantitation of GST-YF NS5 phosphorylation, subsequent in vitro assays of GST-YF NS5 phosphorylation were immunoprecipitated prior to SDS-PAGE (Fig. 6 and 7).
FIG. 5.
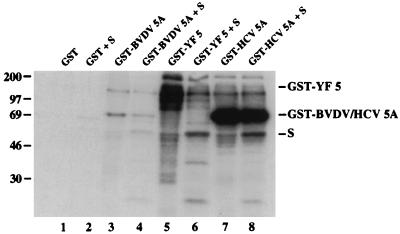
In vitro phosphorylation of fusion proteins consisting of GST and BVDV NS5A, YF NS5, or HCV NS5A by a kinase activity associated with the NS5A/NS5 region. pTM3/GST (36), pTM3/GST-BVDV 5A, pBRTM/GST-YF 5, or pTM3/GST-HCV 5A (36) was expressed in BHK-21 cells, and the respective fusion proteins were purified and assayed for associated kinase activity as previously described (36). A GST-truncated HCV NS5A substrate (S) produced in E. coli was added to the kinase reaction mixtures for which results are shown in lanes 2, 4, 6, and 8. The sizes of marker proteins (in kilodaltons) are on the left. pTM3/GST/BVDV 5A was constructed by inserting the BamHI-XhoI fragment of pGEX-3x/BVDV 5A into the BamHI-XhoI site of pTM3/GST. To construct pGEX-3x/BVDV 5A, pTM3/BVDV 5A was digested with BspEI and StuI, the 5′ overhang produced by BspEI was filled in with T4 DNA polymerase, and the resulting blunt-ended fragment was inserted into pGEX-3x (Pharmacia) that had been linearized with EcoRI and treated with T4 DNA polymerase. pBRTM/GST-YF 5 was subcloned by ligation of the XbaI-BsiCI fragment of pTM3/GST and the SfuI-SstII fragment of pGEX-3x/YF 5∗ into the XbaI-SstII site of pBRTM/YF 5. pGEX-3x/YF 5∗ was constructed by digesting pBRTM/YF 5 with NcoI and HincII, treating it with T4 DNA polymerase, and ligating the resulting fragment to the blunt ends of pGEX-3x digested with AvaI and treated with T4 DNA polymerase. The GST-truncated HCV NS5A substrate was purified as described elsewhere (21) from 10-ml cultures of E. coli TOPP2 (Stratagene), transformed with pGEX-3x/HCV 2179-2420 (36), and grown for 24 h at room temperature after induction with 0.1 mM isopropyl-β-d-thiogalactopyranoside (IPTG).
FIG. 6.
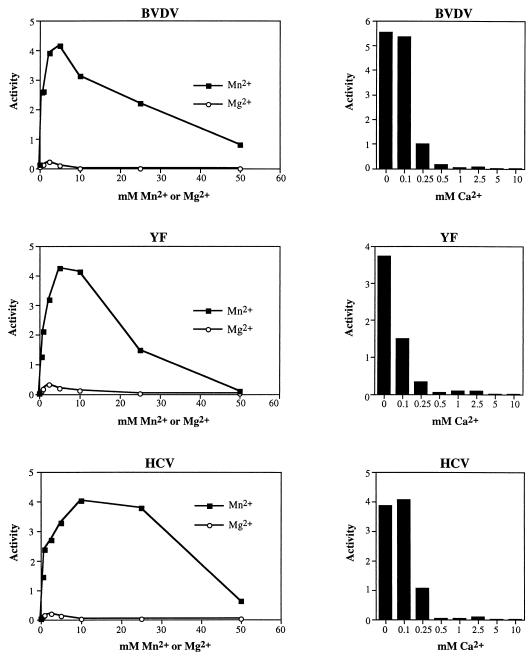
Comparison of the effects of divalent cation concentration on the in vitro phosphorylation of GST-BVDV NS5A, GST-YF NS5, and GST-HCV NS5A. pTM3/GST-BVDV 5A, pBRTM/GST-YF 5, or pTM3/GST-HCV 5A was expressed in BHK-21 cells, and the respective fusion proteins were purified and assayed for associated kinase activity as previously described (36). Clarified lysates were pooled and divided into equal aliquots prior to isolation on glutathione-agarose to ensure that all in vitro kinase reactions contained equal amounts of protein. Standard kinase wash buffers and kinase reaction buffers were used in these experiments with the following exceptions: (i) 5 mM MnCl2 was replaced with the indicated concentrations of MnCl2 in reaction mixtures for which activities are shown in panels on the left, and (ii) the indicated concentrations of CaCl2 were included, along with 5 mM MnCl2, in reaction mixtures for which activities are shown in the panels on the right. The level of phosphorylation in each reaction was determined by phosphorimager quantitation (Bio-Rad) of SDS-8% polyacrylamide gels.
FIG. 7.
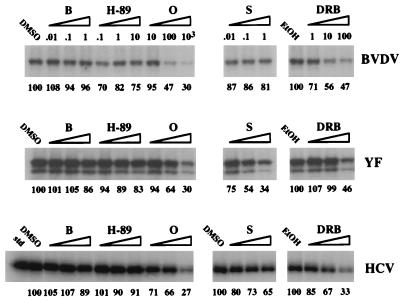
Comparison of the effects of protein kinase inhibitors on GST-BVDV NS5A, GST-YF NS5, and GST-HCV NS5A phosphorylation in vitro. Assays were performed as previously described (36), except that the kinase reaction buffers contained various concentrations of protein kinase inhibitors, as indicated in micromolar units. Provided below each lane are the percentages of NS5A or NS5 phosphorylation, as determined by phosphorimager quantitation, relative to the appropriate solvent controls (dimethyl sulfoxide [DMSO] for bisindolylmaleimide I-HCl [B], H-89, olomoucine [O], and staurosporine [S] and ethanol [EtOH] for DRB). An additional dimethyl sulfoxide control is shown for GST-HCV NS5A phosphorylation assays containing staurosporine because these samples were analyzed on a separate SDS-8% polyacrylamide gel.
GST was not a substrate for phosphorylation in this in vitro assay (Fig. 5, lane 1). To provide additional evidence that the kinase activity(-ies) responsible for in vitro phosphorylation of GST-BVDV NS5A, GST-YF NS5, and GST-HCV NS5A associated specifically with the NS5A or NS5 region and not the common GST moiety, purified GST and GST-viral fusion protein complexes were analyzed for their ability to phosphorylate an HCV NS5A substrate that was produced in E. coli and added to the kinase reaction mixture. This substrate was also expressed as a GST fusion protein, but the NS5A N terminus was truncated by 206 amino acids to distinguish its mobility from that of full-length GST-BVDV NS5A or GST-HCV NS5A expressed in BHK-21 cells. The ability of this substrate to undergo mammalian kinase-dependent phosphorylation in vitro has previously been demonstrated (36).
Phosphorylation of the truncated, E. coli-expressed substrate was observed by one or more kinases captured on glutathione-agarose bound to GST-BVDV NS5A (Fig. 5, lane 4), GST-YF NS5 (lane 6), and GST-HCV NS5A (lane 8), but not GST (lane 2). The level of phosphorylation of the truncated, E. coli-expressed substrate in the GST-BVDV NS5A reaction was low, but the level of GST-BVDV NS5A phosphorylation was also low in comparison to that of GST-YF NS5 or full-length GST-HCV NS5A expressed in BHK-21 cells. This suggests that BVDV NS5A is not as good a substrate for the kinase(s) as HCV NS5A or YF NS5 and that there may be less of the kinase(s) associated with it and available for phosphorylation of the E. coli-expressed substrate. However, the kinases responsible for phosphorylation of the E. coli-expressed and BHK-21-expressed proteins in each reaction were likely to be the same, since both types of phosphorylation occurred under the same reaction conditions and since the E. coli-expressed substrate seemed to compete for phosphorylation with the BHK-21-expressed proteins, particularly GST-BVDV NS5A and GST-YF NS5. Phosphorylation of the E. coli-expressed, GST-truncated HCV NS5A fusion protein by kinases associated specifically with BVDV NS5A and YF NS5, as well as HCV NS5A, further suggested that the same kinase(s) may be responsible for the in vitro phosphorylation of all three viral proteins.
Consistent with this hypothesis, the kinases responsible for GST-BVDV NS5A, GST-YF NS5, and GST-HCV NS5A phosphorylation in vitro exhibited strikingly similar divalent cation requirements (Fig. 6). Activity in all three cases was much higher in the presence of Mn2+ than in the presence of Mg2+, with peak activity in reaction mixtures containing 5 to 10 mM MnCl2. The phosphorylation of all three fusion proteins was also strongly inhibited by the inclusion of ≥0.25 mM CaCl2.
Additional support for the hypothesis that in vitro phosphorylation of GST-BVDV NS5A, GST-YF NS5, and GST-HCV NS5A was catalyzed by the same or closely related kinase activities was obtained from their inhibitor responses. Inhibitors selected for this analysis were bisindolylmaleimide I-HCl, a protein kinase C (PKC)-specific inhibitor; N-[2-(p-bromocinnamylamino)ethyl]-5-isoquinolinesulfonamide dihydrochloride (H-89), which preferentially inhibits cyclic nucleotide-dependent kinases; olomoucine, which inhibits several proline-directed kinases; the broad kinase inhibitor staurosporine; and 5,6-dichloro-1-β-d-ribofuranosyl benzimidazole (DRB), often described as a specific inhibitor of casein kinase II (CKII), but also reported to inhibit cyclin-dependent kinase (CDK)-activating kinase (CAK) and CKI (Table 1). The effects of these inhibitors on the in vitro phosphorylation of GST-BVDV NS5A, GST-YF NS5, and GST-HCV NS5A were quite similar (Fig. 7). Bisindolylmaleimide I-HCl and H-89 had little or no effect on phosphorylation of GST-BVDV NS5A, GST-HCV NS5A, or GST-YF NS5 in vitro at concentrations well above the bisindolylmaleimide I-HCl 50% inhibitory concentrations (IC50s) for PKC and phosphorylase kinase (PK) and the H-89 IC50s for cyclic AMP-dependent protein kinase (PKA) and cyclic GMP-dependent protein kinase (PKG). However, all three reactions were inhibited more than 50% by 1 mM olomoucine and 100 μM DRB, effects which are close to the reported IC50s of olomoucine and DRB for CDK6 and CAK, respectively. Both of these inhibitors target protein kinases in the CMGC kinase group (15) (an acronym based on the names of its four best-studied members, CDK, mitogen-activated protein kinase [MAPK], glycogen synthase kinase 3, and CKII), suggesting that one or more members of this group may be responsible for in vitro phosphorylation of GST-BVDV NS5A, GST-YF NS5, and GST-HCV NS5A. The most significant difference among the three activities was in their responses to staurosporine. In vitro phosphorylation of GST-YF NS5 was sensitive to this inhibitor, with 66% of its phosphorylation inhibited at 1 μM staurosporine, whereas GST-BVDV NS5A phosphorylation seemed to be fairly insensitive to staurosporine, with <20% inhibition observed at the same concentration. GST-HCV NS5A phosphorylation displayed an intermediate phenotype, with 35% inhibition at 1 μM staurosporine. Since staurosporine is thought to inhibit many cellular kinases besides those for which IC50s have been determined, this result raises the possibility that YF NS5 may be phosphorylated by an additional staurosporine-sensitive kinase. Alternatively, BVDV NS5A and HCV NS5A, but not YF NS5, may be phosphorylated by one or more staurosporine-resistant kinases distinct from the kinase(s) responsible for the similar characteristics of NS5A/NS5 phosphorylation in vitro.
TABLE 1.
Activity of selected kinase inhibitors
| Kinasea | IC50 or Kib (μM) of the following kinase inhibitorc:
|
||||
|---|---|---|---|---|---|
| B | H-89 | O | S | DRB | |
| CamKII | 30 | >1,000 | 0.02 | ||
| CKI | 38 | 14 | |||
| CKII | 137 | >2,000 | ∼6 | ||
| CAK | 10–50 | ||||
| CDK1 | 7–50 | ∼0.005 | |||
| CDK2 | 7 | ||||
| CDK4 | >1,000 | ||||
| CDK5 | 3 | ||||
| CDK6 | >250 | ||||
| p44 MAPK | 30 | ||||
| MLCK | 28 | >1,000 | ∼0.005 | ||
| PK | 0.7 | 0.003 | |||
| PKA | 2 | 0.048 | >2,000 | 0.008 | >30 |
| PKC | 0.01–0.02 | 32 | 800 | 0.003 | >30 |
| PKG | 0.48 | >2,000 | ∼0.009 | ||
| S6K | >1,000 | 0.005 | |||
HCV and pestiviruses are more closely related to one another than to flaviviruses (30), and, not surprisingly, certain features of the NS5 region of the flavivirus polyprotein differ significantly from those of the HCV and pestivirus NS5 regions. For instance, as previously mentioned, a single protein is produced from the flavivirus NS5 region, while the HCV and pestivirus NS5 regions are cleaved into the NS5A protein and the polymerase protein NS5B. The flavivirus NS5 protein is also thought to contain a methyltransferase activity (26) that appears to be lacking in the NS5A and NS5B proteins of HCV and pestiviruses. This activity is probably necessary for capping the flavivirus genome, which is thought to be translated by a cap-dependent mechanism (6, 50), whereas HCV (48) and pestivirus (35, 38) genomes can be translated by using internal ribosome entry sites. Structural similarity among the NS5/NS5A proteins of flaviviruses, HCV, pestiviruses, and the GB agents was further assessed by generating amino acid sequence alignments of these proteins by using the CLUSTAL V (16) CLUSTAL W (46), and/or MACAW (41) programs, followed by comparison of their secondary structures as predicted by the PHD program (39) (data not shown). Significant similarity was observed among the amino acid sequences and predicted secondary structures of the N-terminal halves of the HCV, pestivirus, and GBV NS5A proteins. In contrast, no significant similarity was detected between the amino acid sequence of any of these NS5A proteins and the N-terminal region of various flavivirus NS5 proteins. However, some common patterns were recognized among the predicted NS5/NS5A secondary structures: an α helix at the extreme N terminus and a downstream domain containing a number of beta strands, which may be folded similarly in NS5A and NS5. The possible conservation of these structural elements may reflect their involvement in some function common to the NS5 and NS5A proteins of the Flaviviridae which has been conserved throughout their evolution from a common ancestor. Whether this function includes phosphorylation and/or interaction of NS5/NS5A with cellular kinases has yet to be determined. However, given the rapid evolutionary rate of RNA viruses, phenotypes such as NS5/NS5A phosphorylation and interaction with cellular kinases might also be conserved through mechanisms that cannot be discerned from the amino acid alignments or secondary-structure comparisons.
A recent report that baculovirus-expressed HCV NS5B is weakly phosphorylated (18) also raised the possibility that phosphorylation of YF NS5 reflects conservation of polymerase protein phosphorylation rather than conservation of phosphorylation among BVDV NS5A, HCV NS5A, and a homologous or functionally analogous region of YF NS5. Determination of the location of phosphorylation sites in YF NS5 may help to settle this question, although the possibility that phosphorylation sites in the N-terminal region of NS5 influence polymerase activity or, conversely, that C-terminal phosphorylation sites affect the function(s) of N-terminal domains of NS5 cannot be excluded without further investigation. Moreover, phosphorylation of the pestivirus NS5B protein needs to be examined to determine whether phosphorylation of the polymerase protein is a trait conserved throughout the Flaviviridae. Correlations between YF NS5 phosphorylation and the phosphorylation of NS5A or NS5B may also be nonexclusive possibilities.
Although the significance of BVDV NS5A, YF NS5, and HCV NS5A phosphorylation and potential functional similarities among these phosphorylation events has yet to be determined, evidence presented here indicates that these three proteins are associated with kinases which exhibit similar activities in vitro. Furthermore, phosphopeptide mapping experiments have shown that the pattern of HCV NS5A phosphorylation in vitro closely resembles the pattern of intracellular HCV NS5A phosphorylation (36), suggesting that the same or closely related kinases may catalyze this phosphorylation in vitro and in vivo, at least in the case of HCV, and perhaps also for BVDV and YF. The functional significance of NS5/NS5A phosphorylation or of kinase interactions with these three proteins is not known, but NS5/NS5A phosphorylation and/or interaction of NS5 and NS5A with their kinases may regulate viral replication, cellular physiology related to viral pathogenesis, or some other aspect of the viral life cycle. Evidence for the first and/or second possibilities has been provided by analysis of DEN-2 NS5 phosphorylation. As alluded to previously, hyperphosphorylated forms of DEN-2 were found to localize preferentially to the nucleus, while hypophosphorylated forms tended to remain in the cytoplasm and associate with NS3 (23). Both NS3 and NS5 are presumed members of the flaviviral replication complex, since they are thought to contain, respectively, helicase (13) and polymerase (44) activities required for viral replication. Clearly, regulation of the subcellular localization or interaction of these proteins could have a dramatic effect on replication. In addition to a possible effect on viral replication, nuclear transport of NS5 could result in altered host gene expression. HCV NS5A has not been detected inside the nucleus, although amino acids 2326 to 2334 can function as a nuclear localization signal when fused to the N terminus of β-galactosidase (20). However, HCV NS5A is likely to be a member of the viral replication complex, since it has been localized in transfected cells to cytoplasmic membranes surrounding the nucleus (20, 28, 45), coincident with the proposed site of viral replication in flavivirus-infected cells (reviewed in reference 37). The subcellular localization of BVDV NS5A has yet to be examined.
In addition to its putative role in viral replication, HCV NS5A has been proposed to modulate the host interferon (IFN)-stimulated antiviral response, based on observations that it interacts with the IFN-stimulated, double-stranded RNA-dependent protein kinase PKR (12) and that variations in amino acids 2209 to 2248 of HCV NS5A correlate with the sensitivity of some, but not all, HCV strains to IFN treatment (2, 4, 8, 9, 17, 24, 25, 27, 54). This region has therefore been termed the IFN sensitivity-determining region (ISDR). Interaction of HCV NS5A and PKR through the ISDR appears to inhibit phosphorylation of the PKR substrate eIF-2α (12), a translation initiation factor subunit required in unphosphorylated form for the continuation of cellular translation. However, attempts to demonstrate phosphorylation of HCV NS5A by PKR have been unsuccessful (12, 36), suggesting that another cellular serine/threonine kinase is responsible for HCV NS5A phosphorylation. Interaction of BVDV NS5A or YF NS5 with PKR has not been reported, and although all three viruses are likely to interfere with cellular defense pathways such as the host IFN response, this interference may or may not occur through similar mechanisms.
The demonstration that BVDV NS5A, YF NS5 and HCV NS5A are phosphorylated by associated serine/threonine kinases with nearly identical in vitro properties suggests that phosphorylation of these proteins and/or their interaction with the same or closely related kinases is important for successful virus propagation. The process(es) influenced by these associated kinase activities is not known but may include viral replication and/or pathogenesis. Further analysis of NS5/NS5A phosphorylation may lead to greater understanding of NS5/NS5A function, RNA replication, and virus-host interactions among the Flaviviridae.
Acknowledgments
We are grateful to many colleagues for their help during the course of this work, especially Ernesto Mendez and Carol Read, and to Sean Amberg, Alexander Kolykhalov, and Brett Lindenbach for critical reading of the manuscript.
This work was supported by Public Health Service grant CA57973. K.E.R. was supported by a predoctoral fellowship from the National Science Foundation. A.E.G. was supported by the Netherlands Organization for Scientific Research and the Russian Fund for Basic Research (grant 96-04-49562).
REFERENCES
- 1.Asabe S-I, Tanji Y, Satoh S, Kaneko T, Kimura K, Shimotohno K. The N-terminal region of hepatitis C virus-encoded NS5A is important for NS4A-dependent phosphorylation. J Virol. 1997;71:790–796. doi: 10.1128/jvi.71.1.790-796.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Casato M, Agnello V, Pucillo L P, Knight G B, Leoni M, Del Vecchio S, Mazzilli C, Antonelli G, Bonomo L. Predictors of long-term response to high-dose interferon therapy in type II cryoglobulinemia associated with hepatitis C virus infection. Blood. 1997;90:3865–3873. [PubMed] [Google Scholar]
- 3.Chambers T J, McCourt D W, Rice C M. Production of yellow fever virus proteins in infected cells: identification of discrete polyprotein species and analysis of cleavage kinetics using region-specific polyclonal antisera. Virology. 1990;177:159–174. doi: 10.1016/0042-6822(90)90470-c. [DOI] [PubMed] [Google Scholar]
- 4.Chayama K, Tsubota A, Kobayashi M, Okamoto K, Hashimoto M, Miyano Y, Koike H, Kobayashi M, Koida I, Arase Y, Saitoh S, Suzuki Y, Murashima N, Ikeda K, Kumada H. Pretreatment virus load and multiple amino acid substitutions in the interferon sensitivity-determining region predict the outcome of interferon treatment in patients with chronic genotype 1b hepatitis C virus infection. Hepatology. 1997;25:745–749. doi: 10.1002/hep.510250342. [DOI] [PubMed] [Google Scholar]
- 5.Chijiwa T, Mishima A, Hagiwara M, Sano M, Hayashi K, Inoue T, Naito K, Toshioka T, Hidaka H. Inhibition of forskolin-induced neurite outgrowth and protein phosphorylation by a newly synthesized selective inhibitor of cyclic AMP-dependent protein kinase, N-[2-(p-bromocinnamylamino)ethyl]-5-isoquinolinesulfonamide (H-89), of PC12D pheochromocytoma cells. J Biol Chem. 1990;265:5267–5272. [PubMed] [Google Scholar]
- 6.Cleaves G R, Dubin D T. Methylation status of intracellular dengue type 2 40 S RNA. Virology. 1979;96:159–165. doi: 10.1016/0042-6822(79)90181-8. [DOI] [PubMed] [Google Scholar]
- 6a.Collett, M. Unpublished data.
- 7.Collett M S, Larson R, Belzer S K, Retzel E. Proteins encoded by bovine viral diarrhea virus: the genomic organization of a pestivirus. Virology. 1988;165:200–208. doi: 10.1016/0042-6822(88)90673-3. [DOI] [PubMed] [Google Scholar]
- 8.Enomoto N, Sakuma I, Asahina Y, Kurosaki M, Murakami T, Yamamoto C, Izumi N, Marumo F, Sato C. Comparison of full-length sequences of interferon-sensitive and resistant hepatitis C virus 1b. J Clin Invest. 1995;96:224–230. doi: 10.1172/JCI118025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Enomoto N, Sakuma I, Asahina Y, Kurosaki M, Murakami T, Yamamoto C, Ogura Y, Izumi N, Marumo F, Sato C. Mutations in the nonstructural protein 5A gene and response to interferon in patients with chronic hepatitis C virus 1b infection. N Engl J Med. 1996;334:77–81. doi: 10.1056/NEJM199601113340203. [DOI] [PubMed] [Google Scholar]
- 10.Fuerst T R, Niles E G, Studier F W, Moss B. Eukaryotic transient-expression system based on recombinant vaccinia virus that synthesizes bacteriophage T7 RNA polymerase. Proc Natl Acad Sci USA. 1986;83:8122–8126. doi: 10.1073/pnas.83.21.8122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gadbois D M, Hamaguchi J R, Swank R A, Bradbury E M. Staurosporine is a potent inhibitor of p34cdc2 and p34cdc2-like kinases. Biochem Biophys Res Commun. 1992;184:80–85. doi: 10.1016/0006-291x(92)91160-r. [DOI] [PubMed] [Google Scholar]
- 12.Gale M J, Jr, Korth M J, Tang N M, Tan S-L, Hopkins D A, Dever T E, Polyak S J, Gretch D R, Katze M G. Evidence that hepatitis C virus resistance to interferon is mediated through repression of the PKR protein kinase by the nonstructural 5A protein. Virology. 1997;230:217–227. doi: 10.1006/viro.1997.8493. [DOI] [PubMed] [Google Scholar]
- 13.Gorbalenya A E, Koonin E V, Donchenko A P, Blinov V M. Two related superfamilies of putative helicases involved in replication, recombination, repair and expression of DNA and RNA genomes. Nucleic Acids Res. 1989;17:4713–4729. doi: 10.1093/nar/17.12.4713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Grakoui A, Wychowski C, Lin C, Feinstone S M, Rice C M. Expression and identification of hepatitis C virus polyprotein cleavage products. J Virol. 1993;67:1385–1395. doi: 10.1128/jvi.67.3.1385-1395.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hardie G, Hanks S, editors. The protein kinase facts book: protein-serine kinases. I. San Diego, Calif: Academic Press, Inc.; 1995. [Google Scholar]
- 16.Higgins D G, Bleasby A J, Fuchs R. CLUSTAL V: improved software for multiple sequence alignment. Comput Appl Biosci. 1992;8:189–191. doi: 10.1093/bioinformatics/8.2.189. [DOI] [PubMed] [Google Scholar]
- 17.Hofgartner W T, Polyak S J, Sullivan D G, Carithers R L, Jr, Gretch D R. Mutations in the NS5A gene of hepatitis C virus in North American patients infected with HCV genotype 1a or 1b. J Med Virol. 1997;53:118–126. doi: 10.1002/(sici)1096-9071(199710)53:2<118::aid-jmv3>3.0.co;2-c. [DOI] [PubMed] [Google Scholar]
- 18.Hwang S B, Park K-J, Kim Y-S, Sung Y C, Lai M M C. Hepatitis C virus NS5B protein is a membrane-associated phosphoprotein with a predominantly perinuclear localization. Virology. 1997;227:439–446. doi: 10.1006/viro.1996.8357. [DOI] [PubMed] [Google Scholar]
- 19.Ide Y, Tanimoto A, Sasaguri Y, Padmanabhan R. Hepatitis C virus NS5A protein is phosphorylated in vitro by a stably bound protein kinase from HeLa cells and by cAMP-dependent protein kinase A-α catalytic subunit. Gene. 1997;201:151–158. doi: 10.1016/s0378-1119(97)00440-x. [DOI] [PubMed] [Google Scholar]
- 20.Ide Y, Zhang L, Chen M, Inchauspe G, Bahl C, Sasaguri Y, Padmanabhan R. Characterization of the nuclear localization signal and subcellular distribution of hepatitis C virus nonstructural protein NS5A. Gene. 1996;182:203–211. doi: 10.1016/s0378-1119(96)00555-0. [DOI] [PubMed] [Google Scholar]
- 21.Kaelin W G, Jr, Pallas D C, DeCaprio J A, Kaye F J, Livingston D M. Identification of cellular proteins that can interact specifically with the T/E1A-binding region of the retinoblastoma gene product. Cell. 1991;64:521–532. doi: 10.1016/0092-8674(91)90236-r. [DOI] [PubMed] [Google Scholar]
- 22.Kaneko T, Tanji Y, Satoh S, Hijikata M, Asabe S, Kimura K, Shimotohno K. Production of two phosphoproteins from the NS5A region of the hepatitis C viral genome. Biochem Biophys Res Commun. 1994;205:320–326. doi: 10.1006/bbrc.1994.2667. [DOI] [PubMed] [Google Scholar]
- 23.Kapoor M, Zhang L, Ramachandra M, Kusukawa J, Ebner K E, Padmanabhan R. Association between NS3 and NS5 proteins of dengue virus type 2 in the putative RNA replicase is linked to differential phosphorylation of NS5. J Biol Chem. 1995;270:19100–19106. doi: 10.1074/jbc.270.32.19100. [DOI] [PubMed] [Google Scholar]
- 24.Khorsi H, Castelain S, Wyseur A, Izopet J, Canva V, Rombout A, Capron D, Capron J-P, Lunel F, Stuyver L, Duverlie G. Mutations of hepatitis C virus 1b NS5A 2209-2248 amino acid sequence do not predict the response to recombinant interferon-alfa therapy in French patients. J Hepatol. 1997;27:72–77. doi: 10.1016/s0168-8278(97)80282-6. [DOI] [PubMed] [Google Scholar]
- 25.Komatsu H, Fujisawa T, Inui A, Miyagawa Y, Onoue M. Mutations in the nonstructural protein 5A gene and response to interferon therapy in young patients with chronic hepatitis C virus 1b infection. J Med Virol. 1997;53:361–365. [PubMed] [Google Scholar]
- 26.Koonin E V. Computer-assisted identification of a putative methyltransferase domain in NS5 protein of flaviviruses and lambda 2 protein of reovirus. J Gen Virol. 1993;74:733–740. doi: 10.1099/0022-1317-74-4-733. [DOI] [PubMed] [Google Scholar]
- 27.Kurosaki M, Enomoto N, Murakami T, Sakuma I, Asahina Y, Yamamoto C, Ikeda T, Tozuka S, Izumi N, Marumo F, Sato C. Analysis of genotypes and amino acid residues 2209 to 2248 of the NS5A region of hepatitis C virus in relation to response to interferon-β therapy. Hepatology. 1997;25:750–753. doi: 10.1002/hep.510250343. [DOI] [PubMed] [Google Scholar]
- 28.Manabe S, Fuke I, Tanishita O, Kaji C, Gomi Y, Yoshida S, Mori C, Takamizawa A, Yoshida I, Okayama H. Production of nonstructural proteins of hepatitis C virus requires a putative viral protease encoded by NS3. Virology. 1994;198:636–644. doi: 10.1006/viro.1994.1075. [DOI] [PubMed] [Google Scholar]
- 29.Meyer T, Regenass U, Fabbro D, Alteri E, Rösel J, Müller J, Caravatti G, Matter A. A derivative of staurosporine (CGP 41 251) shows selectivity for protein kinase C inhibition and in vitro anti-proliferative as well as in vivo anti-tumor activity. Int J Cancer. 1989;43:851–856. doi: 10.1002/ijc.2910430519. [DOI] [PubMed] [Google Scholar]
- 30.Miller R H, Purcell R H. Hepatitis C virus shares amino acid sequence similarity with pestiviruses and flaviviruses as well as members of two plant virus supergroups. Proc Natl Acad Sci USA. 1990;87:2057–2061. doi: 10.1073/pnas.87.6.2057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Morozova O V, Tsekhanovskaya N A, Maksimova T G, Bachvalova V N, Matveeva V A, Kit Y Y. Phosphorylation of tick-borne encephalitis virus NS5 protein. Virus Res. 1997;49:9–15. doi: 10.1016/s0168-1702(96)01433-5. [DOI] [PubMed] [Google Scholar]
- 32.Moss B, Elroy-Stein O, Mizukami T, Alexander W A, Fuerst T R. New mammalian expression vectors. Nature (London) 1990;348:91–92. doi: 10.1038/348091a0. [DOI] [PubMed] [Google Scholar]
- 33.Nakano H, Kobayashi E, Takahashi I, Tamaoki T, Kuzuu Y, Iba H. Staurosporine inhibits tyrosine-specific protein kinase activity of Rous sarcoma virus transforming protein p60. J Antibiot. 1987;40:706–708. doi: 10.7164/antibiotics.40.706. [DOI] [PubMed] [Google Scholar]
- 34.Niggli V, Keller H. On the role of protein kinases in regulating neutrophil actin association with the cytoskeleton. J Biol Chem. 1991;266:7927–7932. [PubMed] [Google Scholar]
- 35.Poole T L, Wang C-Y, Popp R A, Potgieter L N D, Siddiqui A, Collett M S. Pestivirus translation initiation occurs by internal ribosome entry. Virology. 1995;206:750–754. doi: 10.1016/s0042-6822(95)80003-4. [DOI] [PubMed] [Google Scholar]
- 36.Reed K E, Xu J, Rice C M. Phosphorylation of the hepatitis C virus NS5A protein in vitro and in vivo: properties of the NS5A-associated kinase. J Virol. 1997;71:7187–7197. doi: 10.1128/jvi.71.10.7187-7197.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rice C M. Flaviviridae: the viruses and their replication. In: Fields B N, Knipe D M, Howley P M, editors. Fields virology. 3rd ed. Vol. 1. Philadelphia, Pa: Lippincott-Raven Publishers; 1996. pp. 931–960. [Google Scholar]
- 38.Rijnbrand R, van der Straaten T, van Rijn P A, Spaan W J M, Bredenbeek P J. Internal entry of ribosomes is directed by the 5′ noncoding region of classical swine fever virus and is dependent on the presence of an RNA pseudoknot upstream of the initiation codon. J Virol. 1997;71:451–457. doi: 10.1128/jvi.71.1.451-457.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rost B, Sander C, Schneider R. PHD—an automatic mail server for protein secondary structure prediction. Comput Appl Biosci. 1994;10:53–60. doi: 10.1093/bioinformatics/10.1.53. [DOI] [PubMed] [Google Scholar]
- 40.Sakurada K, Ikuhara T, Seto M, Sasaki Y. An antibody for phosphorylated myosin light chain of smooth muscle: application to a biochemical study. J Biochem. 1994;115:18–21. doi: 10.1093/oxfordjournals.jbchem.a124297. [DOI] [PubMed] [Google Scholar]
- 41.Schuler G D, Altschul S F, Lipman D J. A workbench for multiple alignment construction and analysis. Proteins. 1991;9:180–190. doi: 10.1002/prot.340090304. [DOI] [PubMed] [Google Scholar]
- 42.Smith D B, Johnson K S. Single-step purification of polypeptides expressed in Escherichia coli as fusions with glutathione S-transferase. Gene. 1988;67:31–40. doi: 10.1016/0378-1119(88)90005-4. [DOI] [PubMed] [Google Scholar]
- 43.Tamaoki T, Nomoto H, Takahashi I, Kato Y, Morimoto M, Tomita F. Staurosporine, a potent inhibitor of phospholipid/Ca++ dependent protein kinase. Biochem Biophys Res Commun. 1986;135:397–402. doi: 10.1016/0006-291x(86)90008-2. [DOI] [PubMed] [Google Scholar]
- 44.Tan B-T, Fu J, Sugrue R J, Yap E-H, Chan Y-C, Tan Y H. Recombinant dengue type 1 virus NS5 protein expressed in Escherichia coli exhibits RNA-dependent RNA polymerase activity. Virology. 1996;216:317–325. doi: 10.1006/viro.1996.0067. [DOI] [PubMed] [Google Scholar]
- 45.Tanji Y, Kaneko T, Satoh S, Shimotohno K. Phosphorylation of hepatitis C virus-encoded nonstructural protein NS5A. J Virol. 1995;69:3980–3986. doi: 10.1128/jvi.69.7.3980-3986.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Thompson J D, Higgins D G, Gibson T J. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Toullec D, Pianetti P, Coste H, Bellevergue P, Grand-Perret T, Ajakane M, Baudet V, Boissin P, Boursier E, Loriolle F, Duhamel L, Charon D, Kirilovsky J. The bisindolylmaleimide GF 109203X is a potent and selective inhibitor of protein kinase C. J Biol Chem. 1991;266:15771–15781. [PubMed] [Google Scholar]
- 48.Tsukiyama-Kohara K, Iizuka N, Kohara M, Nomoto A. Internal ribosome entry site within hepatitis C virus RNA. J Virol. 1992;66:1476–1483. doi: 10.1128/jvi.66.3.1476-1483.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Vesely J, Havlicek L, Strnad M, Blow J J, Donella-Deana A, Pinna L, Letham D S, Kato J-Y, Detivaud L, Leclerc S, Meijer L. Inhibition of cyclin-dependent kinases by purine analogues. Eur J Biochem. 1994;224:771–786. doi: 10.1111/j.1432-1033.1994.00771.x. [DOI] [PubMed] [Google Scholar]
- 50.Wengler G, Wengler G, Gross H J. Studies on virus-specific nucleic acids synthesized in vertebrate and mosquito cells infected with flaviviruses. Virology. 1978;89:423–437. doi: 10.1016/0042-6822(78)90185-x. [DOI] [PubMed] [Google Scholar]
- 50a.Xu, J., and C. M. Rice. Unpublished data.
- 51.Yanagihara N, Tachikawa E, Izumi F, Yasugawa S, Yamamoto H, Miyamoto E. Staurosporine: an effective inhibitor for Ca2+/calmodulin-dependent protein kinase II. J Neurochem. 1991;56:294–298. doi: 10.1111/j.1471-4159.1991.tb02595.x. [DOI] [PubMed] [Google Scholar]
- 52.Yankulov K, Yamashita K, Roy R, Egly J-M, Bentley D L. The transcriptional elongation inhibitor 5,6-dichloro-1-β-d-ribofuranosylbenzimidazole inhibits transcription factor IIH-associated protein kinase. J Biol Chem. 1995;270:23922–23925. doi: 10.1074/jbc.270.41.23922. [DOI] [PubMed] [Google Scholar]
- 53.Zandomeni R, Zandomeni M C, Shugar D, Weinmann R. Casein kinase type II is involved in the inhibition by 5,6-dichloro-1-β-d-ribofuranosylbenzimidazole of specific RNA polymerase II transcription. J Biol Chem. 1986;261:3414–3419. [PubMed] [Google Scholar]
- 54.Zeuzem S, Lee J-H, Roth W K. Mutations in the nonstructural 5A gene of European hepatitis C virus isolates and response to interferon alfa. Hepatology. 1997;25:740–744. doi: 10.1002/hep.510250341. [DOI] [PubMed] [Google Scholar]