Abstract
Background
The immune status of a patient’s tumor microenvironment (TME) may guide therapeutic interventions with cancer immunotherapy and help identify potential resistance mechanisms. Currently, patients’ immune status is mostly classified based on CD8+tumor-infiltrating lymphocytes. An unmet need exists for comparable and reliable precision immunophenotyping tools that would facilitate clinical treatment-relevant decision-making and the understanding of how to overcome resistance mechanisms.
Methods
We systematically analyzed the CD8 immunophenotype of 2023 patients from 14 phase I–III clinical trials using immunohistochemistry (IHC) and additionally profiled gene expression by RNA-sequencing (RNA-seq). CD8 immunophenotypes were classified by pathologists into CD8-desert, CD8-excluded or CD8-inflamed tumors using CD8 IHC staining in epithelial and stromal areas of the tumor. Using regularized logistic regression, we developed an RNA-seq-based classifier as a surrogate to the IHC-based spatial classification of CD8+tumor-infiltrating lymphocytes in the TME.
Results
The CD8 immunophenotype and associated gene expression patterns varied across indications as well as across primary and metastatic lesions. Melanoma and kidney cancers were among the strongest inflamed indications, while CD8-desert phenotypes were most abundant in liver metastases across all tumor types. A good correspondence between the transcriptome and the IHC-based evaluation enabled us to develop a 92-gene classifier that accurately predicted the IHC-based CD8 immunophenotype in primary and metastatic samples (area under the curve inflamed=0.846; excluded=0.712; desert=0.855). The newly developed classifier was prognostic in The Cancer Genome Atlas (TCGA) data and predictive in lung cancer: patients with predicted CD8-inflamed tumors showed prolonged overall survival (OS) versus patients with CD8-desert tumors (HR 0.88; 95% CI 0.80 to 0.97) across TCGA, and longer OS on immune checkpoint inhibitor administration (phase III OAK study) in non-small-cell lung cancer (HR 0.75; 95% CI 0.58 to 0.97).
Conclusions
We provide a new precision immunophenotyping tool based on gene expression that reflects the spatial infiltration patterns of CD8+ lymphocytes in tumors. The classifier enables multiplex analyses and is easy to apply for retrospective, reverse translation approaches as well as for prospective patient enrichment to optimize the response to cancer immunotherapy.
Keywords: CD8-Positive T-Lymphocytes, Gene Expression Profiling, Immunohistochemistry, Tumor Biomarkers, Tumor Microenvironment
WHAT IS ALREADY KNOWN ON THIS TOPIC
T-cell infiltration, most commonly classified based on CD8+T cell immunohistochemistry (IHC) staining, and various tumor microenvironment (TME)-specific resistance mechanisms, can impact response rates to cancer immunotherapy.
WHAT THIS STUDY ADDS
Our data provide new insights into the impact of tumor excision location and indication on the immune composition of the TME. We developed a transcriptome-based classifier that could accurately predict different spatial CD8+T cell infiltration patterns in the TME. We demonstrate the prognostic and predictive value of the classifier across independent patient cohorts (phase I–III trials).
HOW THIS STUDY MIGHT AFFECT RESEARCH, PRACTICE OR POLICY
Our new RNA-based tool provides a surrogate read-out for spatial IHC-based CD8 infiltration patterns, is easy to use and broadly applicable for both retrospective and prospective patient enrichment to enhance the effectiveness of cancer immunotherapy.
Introduction
Cancer immunotherapy (CIT) has been widely integrated into routine clinical practice for many tumor indications, helping to move from a disease-centric approach to a more personalized care with substantially improved outcomes.1 2 However, varied response and resistance rates can be attributed to several factors, including paucity of T-cell infiltration in the tumor microenvironment (TME) and tumor-intrinsic resistance mechanisms.3–5 Improved understanding of resistance mechanisms, along with reliable prognostic and predictive biomarkers, would help achieve the full potential of CIT.6 7 Reliable predictive biomarkers could eliminate administration of immunotherapy to unsuitable patients, minimizing the associated risk of toxicity, and reducing costs.8 9
Many biomarkers have been investigated, with various limitations, challenging their implementation into routine clinical practice.1 7 10 Among these, the immune status of a patient is commonly assessed by analyzing the TME at baseline using immunohistochemistry (IHC) and classifying the spatial infiltration of CD8+T cells in tumors as “CD8-inflamed” (a high rate of infiltration, typically showing significant immune cell diversity), “CD8-excluded” (T cells are retained in the stroma) or “CD8-desert” (very limited infiltration in intra-tumoral stroma and tumor areas).2 11 12 A significant correlation between CD8-inflamed tumors and improved clinical response to CIT has been reported,2 13 14 including for pembrolizumab treatment of metastatic melanoma (MEL)15 and oral squamous cell carcinoma.16 IHC is useful to broadly categorize tumors, but risks simplifying the complex genetic and immune picture inside the TME.6 For example, with lymphocytes, IHC often only reports CD8 positivity and may not identify other pathological pathways (eg, altered tumor metabolic pathways); more complex biomarker approaches are thus needed to further identify the dynamics of tumorigenesis. IHC also assumes all centers have equal access to reliable assays, appropriately curated samples, and equivalent means of interpreting results.6 Therefore, it may not always be the optimal approach to delineate tumor immunophenotypes and predict therapeutic response, leaving an unmet need for reliable and cohesive biomarkers to help guide patient care.
Gene expression-based models have been developed to classify tumor immunophenotypes, for example, by using RNA sequencing (RNA-seq) to analyze gene expression related to CD8+T cell activity. In line with the IHC-based assessments, the T-effector gene signature was also associated with an improved efficacy of atezolizumab versus docetaxel in patients with non-small-cell lung cancer (NSCLC).17–19 Similar signatures have been used to generate risk scores correlated with CD8+T cell infiltration,13 which could predict immunotherapy efficacy, potentially enabling early identification of non-responders.20 Importantly, gene expression profiling by RNA-seq can generate comprehensive and cost-effective datasets, with demonstrated concordance between IHC, quantitative real-time PCR and gene expression microarrays.21 An easy-to-implement gene expression-based classifier reflecting the spatial infiltration of CD8+T cells in the TME is needed to support large-scale analyses of clinical datasets, enabling retrospective and reverse translation analyses across clinical studies. However, previous attempts at identifying RNA-seq signatures of immunophenotypes and their predictive and prognostic values were limited to specific cancer types.20 22
Here, we describe the development of a transcriptome-based classifier that accurately identifies spatial infiltration patterns of CD8+T cells in the TME across indications and excision locations. This classifier is able to predict clinical outcomes in early-phase and late-phase clinical trials, which has significant implications for the use of CIT.
Methods
Patient samples
Formalin-fixed and paraffin-embedded tumor tissue collected from 11 unpublished phase I/II clinical trials (online supplemental table S1) before treatment start was retrospectively analyzed. Tumor metastases in the liver and lymph nodes were measurable and assessable as target lesions, allowing an immune score to be calculated.23
jitc-2023-008185supp001.pdf (896.7KB, pdf)
Additionally, IHC-based CD8 immunophenotypes were scored for samples extracted from three previously published open-label, phase II–III trials (NCT02008227; NCT02108652; NCT02302807), from which RNA-seq data were also available.24–26
All clinical studies were conducted in accordance with the principles of the Declaration of Helsinki and Good Clinical Practice Guidelines. Written informed consent was collected from all enrolled patients. The protocol for each clinical study was approved by the institutional review boards/ethics committees at each center (see online supplemental table S2).
Data from The Cancer Genome Atlas (TCGA) Program (National Cancer Institute; available at https://portal.gdc.cancer.gov/) were used to test the gene expression-based classifier on a large dataset, processed via the recount3 resource.27
CD8 immunophenotypes: IHC classification
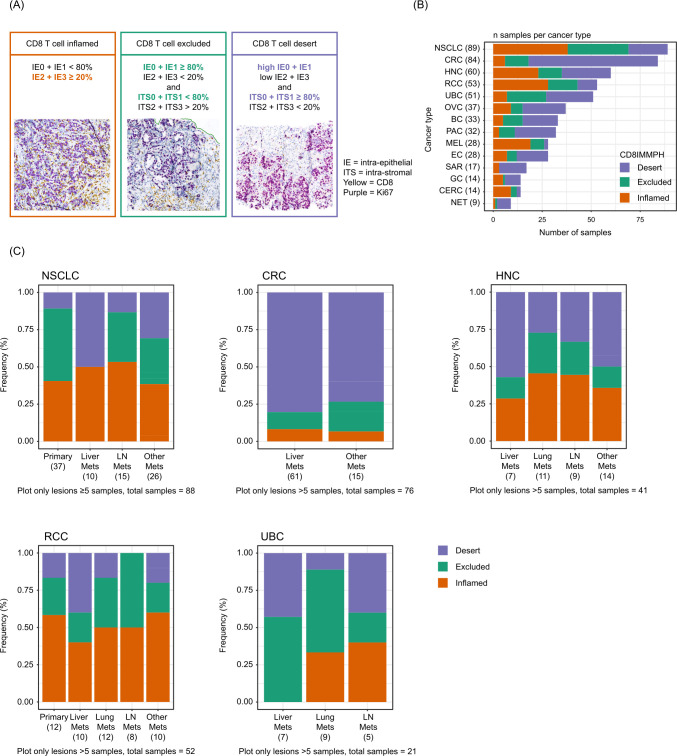
CD8/Ki67 slides were cut (2.5 µm thickness) and stained in-house (Roche Innovation Center Munich, Germany). Slides were scanned at 20× using the Ventana iScan HT and the resulting whole-slide images were sent to CellCarta (formerly known as HistoGeneX (Antwerp, Belgium)) for immunophenotyping assessment. Phenotype scoring (or density proportion scoring, adapted from Galon and Lanzi28) was based on semiquantitative CD8/Ki67 IHC staining evaluation in tumor epithelial and stroma areas by CellCarta (as performed in Mariathasan et al 29 and Powles et al 30), although only CD8 IHC staining was evaluated in our study. Briefly, CD8 IHC staining was used to identify lymphocytes and to assess their density using a four-tier scoring system: 0=no lymphocytes; 1=single dispersed lymphocyte; 2=numerous dispersed lymphocytes; 3=dense lymphocytic infiltrate. Using this scoring system, CD8 lymphocyte density was then assessed separately in tumor epithelial areas (ie, tumor cell nests) and tumor stroma (tissue areas between tumor cells/nests), resulting in the creation of four separate categories each for tumor intraepithelial (ie, IE0–IE3) and intratumoral stroma (ITS0–ITS3) lymphocyte density. Pathologists then estimated the percentage area for each category. Based on a combined density score from IE and ITS score compartments, patient samples were classified as (figure 1A): (1) CD8-inflamed, with IE2+IE3≥20%; (2) CD8-excluded, with ITS2+ITS3>20% and IE0+IE1≥80%; and (3) CD8-desert, with IE0+IE1≥80% and ITS0+ITS1≥80%.
Figure 1.
Definition and prevalence of CD8 immunophenotype categories. (A) CD8-inflamed was defined with IE2+IE3≥20%. CD8-excluded was defined with ITS2+ITS3>20% and IE0+IE1≥80%. CD8-desert was defined with IE0+IE1≥80% and ITS0+ITS1≥80%. Prevalence of immunophenotype by (B) indication and (C) tumor excision location. The “Other mets” category refers to excision sites of minor frequency (each n≤4): abdominal cavity, sinus, adrenal gland, pleura, skin and soft tissue. Tumor types with n<5 samples (25 different categories, eg, bone, fallopian tube, pelvis, thymus, testis; most of them n=1) are not included. CRC, colorectal cancer; HNC, head and neck carcinoma; IE, intraepithelial; ITS, intrastromal; LN, lymph node; mets, metastases; NSCLC, non-small-cell lung cancer; UBC, urinary bladder cancer.
RNA-seq data processing
All samples were further analyzed for genome-wide RNA expression using RNA-seq, performed on macrodissected tissue of the tumor area (ie, normal tissue was excluded). RNA-seq data were analyzed using HTSeqGenie31 in BioConductor32 as follows: first, reads with low nucleotide qualities (70% of bases with quality<23) or matches to rRNA and adapter sequences were removed. The remaining reads were aligned to the human reference genome GRCh38.p10 using GSNAP33 V.“2013-10-10-v2”, allowing maximum two mismatches per 75 base sequence (parameters: “-M 2 -n 10 -B 2 -i 1 -N 1 -w 200,000 -E 1 --pairmax-rna=200,000 --clip-overlap”). Transcript annotation was based on the Gencode database (human: GENCODE 27). To quantify gene expression levels, the number of reads mapping unambiguously to the exons of each gene was calculated. RNA-seq values were transformed using variance stabilizing transformation and DESeq2 V.1.6.3 (Bioconductor, Massachusetts, USA). Lowly expressed genes and those with vastly different expression levels between TCGA and the rest of the data were excluded to allow the classifier to be applied to TCGA. Batch correction was subsequently performed: principal component analysis transformation was used on a selected subset of lung cancer samples to identify components corresponding to the batch-effect (ie, difference in mean and variance). Finally, the effect of those components on individual genes was identified and an inverse transformation was applied to the whole TCGA dataset.
Signature scores corresponding to rank-biserial correlation values were produced using BioQC V.3.15.34 Cell-type-specific signatures were previously derived from single-cell RNA-seq data,35 while pathway-specific signatures were available from the Molecular Signature Database (MsigDB) “Hallmark” collection, release V.7.4.36 Pairwise differential expression analyses were performed to characterize gene expression differences between the three phenotypes. A linear model was fitted using the limma-voom approach defined by Law et al 37 with a threshold for log fold change of 1.5 and corrected false discovery rate (FDR) of 0.05. Biopsy location (liver, kidney, lung, urothelial, lymph node) was added as a covariate in the model. Gene set enrichment was calculated using a competitive gene set test (limma::camera) with FDR<0.05. Differential expression results are provided as online supplemental table S3 and gene set enrichment results in online supplemental table S4.
Classifier development
To ensure adequate classifier performance estimates, the data (n=2023: n=628 from phase I/II trials and n=1395 from the three additional phase II/III trials) were split into three portions: training (71%; n=1438), validation (8%; n=158), and test (21%; n=427). To build the classifier, lasso regression was implemented in the glmnet R package38 via mlr3.39 Nested cross-validation was used to identify the optimal value of lambda. Variance-stabilization transformed gene expression values were selected as an input for the classifier. TCGA RNA-seq samples were batch-corrected to match the training dataset. To explore the most important features for classification, a set of classifiers were trained that used: (1) all genes (GENCODE 27, excluding genes that were filtered out, see section RNA-seq data processing); (2) only immune-expressed genes, as derived from single-cell RNA-seq expression datasets available in besca35 40–42; (3) cell-type-specific signatures scores35; (4) hallmark pathway signature scores (MSigDB, release V.7.4)36; or (5) cancer-specific signature scores (C4 and C6, MSigDB, release V.7.4).36 A 10-fold cross-validation was repeated 100 times (on random data splits) to measure the feature importance of individual genes. For every classifier parameter, the number of times it was present in one of the models and its coefficient value were recorded. The test fold was used to assess the classifier performance. Samples from the phase III OAK study (NCT02008227; n=352) were only used in the test fold to control for classifier generalizability and translatability. Using a study for the test fold that was not included in training ensures that the prediction performance is a good approximation of classifier generalizability to completely new, unseen data. Intergene correlation between gene signatures was also calculated (see online supplemental figure S1).
jitc-2023-008185supp002.pdf (7.6MB, pdf)
Single-cell RNA-seq and spatial transcriptomics expression analysis
To explore which cell types express the top genes retained in the final classifier, previously published colorectal cancer (CRC),42 lung cancer43 and liver cancer44 datasets were reprocessed with the besca standard workflow and cell annotation workflow.35 To explore the spatial distributions of individual genes, publicly available 10×Visium data from CRC45 were reprocessed as described previously.46
Statistical analyses
The R package limma was used for the differential gene expression analysis.37 The RTCGA package V.1.26.0 was used to extract clinical information from TCGA.47 The survminer R package V.0.4.9 was used to fit Cox proportional hazard models.48 The meta V.6.1-0 package was used to produce forest plots.
Results
CD8 immunophenotypes vary across indications and tumor excision locations
Tumor samples from 628 patients enrolled into 11 phase I/II clinical trials were systematically analyzed by IHC and RNA-seq (see online supplemental table S1 for a detailed overview). Patient baseline characteristics are reported in online supplemental table S5: median age was 60 years, and 43% of patients were female. The most common diagnoses were NSCLC (n=89), CRC (n=84), and head and neck carcinoma (HNC) (n=60). A total of 79% of patients were treatment naïve.
Samples were classified based on CD8 IHC levels in tumor epithelial and stroma areas as CD8-inflamed (31%), CD8-excluded (23%), and CD8-desert (46%) (figure 1A). The distribution of immunophenotypes was highly variable across indications and tumor excision locations (figure 1B,C). As expected from previous reports, the majority of NSCLC, HNC and urinary bladder carcinoma (UBC) samples were CD8-inflamed and/or CD8-excluded. There was also a high prevalence of CD8-inflamed phenotypes in MEL and renal cell carcinoma (RCC) samples, whereas CD8-desert phenotypes were predominant in CRC. There was a higher relative fraction of CD8-desert phenotypes in liver metastases across all tumor types: approximately 50% (vs less than 25% in lymph node metastases or in the primary tumor) of liver metastases from NSCLCs had a CD8-desert phenotype. Similarly, the proportion of CD8-desert phenotype in liver metastases was 40% (vs 10% in lung metastases) for UBCs, and 80% (vs 60% in the primary tumor) for CRCs. Trends were also consistent for RCC and HNC (figure 1B,C).
CD8 immunophenotypes show distinct transcriptional profiles
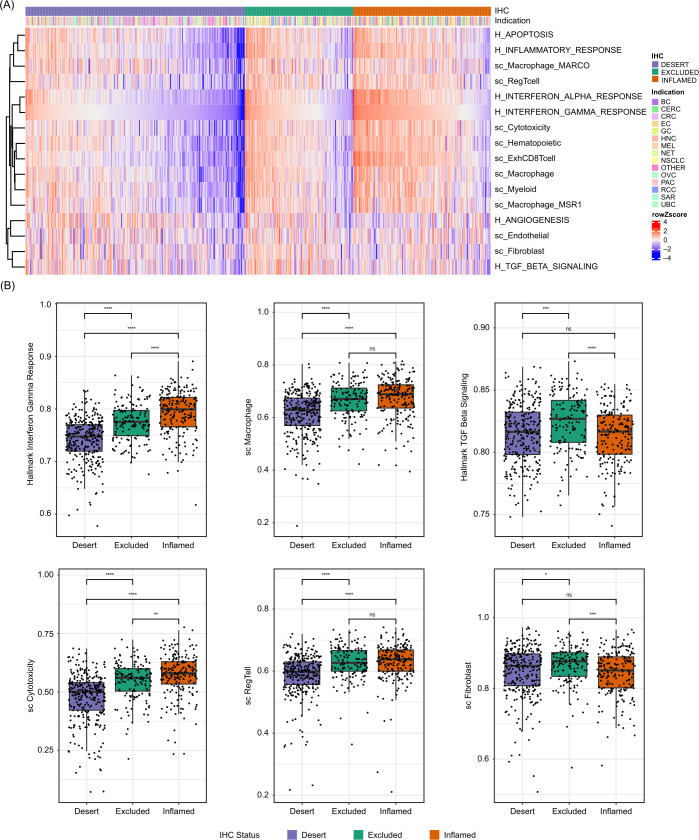
The transcriptional characteristics of the three CD8 immunophenotypes were first explored with a focus on cell-type and pathway-specific gene signatures previously positively or negatively associated with immune infiltration and/or response to CIT (figure 2A). Globally, gene expression was similar, although with magnitude differences, in the CD8-inflamed and CD8-excluded groups but showed rather distinct patterns from CD8-desert samples across indications and excision locations (figure 2A). Gene expression related to CD8+T cells, cytotoxicity and exhaustion, as well as interferon (IFN) response, was highly enriched in CD8-inflamed samples, intermediate in CD8-excluded, and low in CD8-deserts, in line with the abundance and activity of CD8+T cells in CD8-inflamed samples (figure 2A,B; online supplemental figure S2). In contrast, expression levels related to immune populations previously associated with repression of T-effector activity, such as regulatory T cells, macrophages, or myeloid cells, showed more similar expression in inflamed and excluded samples, and markedly lower expression in CD8-desert samples. Stromal-specific genes, including fibroblast and endothelial markers, as well as angiogenic and transforming growth factor-β (TGF-β) pathways, showed excluded-enriched expression (figure 2B; online supplemental figure S2).
Figure 2.
Inflamed, excluded and desert samples show distinct transcriptional profiles. (A) Heatmap showing relative signature scores per sample, sorted according to CD8 immunophenotype and “interferon gamma” signature scores; cancer types are displayed. (B) Selected signature scores across CD8 immunophenotype classes. “H_” represents pathway-specific signatures (Hallmark), while “sc_” represents cell-type specific signatures. *p≤0.05. **p≤0.01. ***p≤0.001. ****p≤0.0001. CRC, colorectal cancer; IHC, immunohistochemistry; ns, not significant; NSCLC, non-small-cell lung cancer; UBC, urinary bladder cancer.
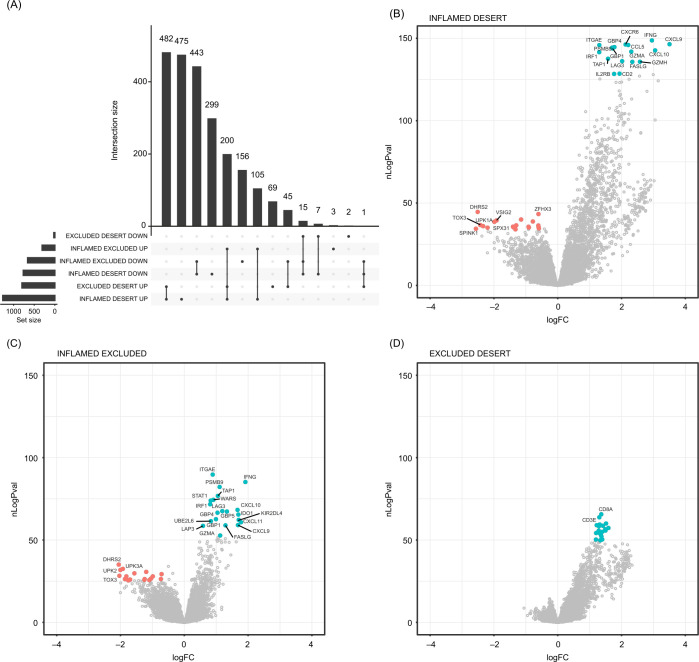
To systematically characterize gene expression differences between CD8 immunophenotypes, we next performed pairwise differential expression analyses. Given the observed heterogeneity of our phase I/II cohort, we included an additional set of 1395 samples (2023 samples in total) from three phase II–III trials to better control for the observed indication and excision location-induced variability. The greatest differences were between CD8-inflamed and CD8-desert samples, consistent with the signature-based analysis (figure 3; online supplemental table S4). Pairwise differential expression analyses also showed that T-effector and IFN-ɣ pathway-related gene expression, including CXCL9, CXCL10, IFN-ɣ, CCL5, ITGAE, LAG3, FASLG, and TAP1, was most highly enriched in CD8-inflamed samples (figure 3B–D; online supplemental figure S3A). The same genes were significantly lower expressed in CD8-excluded versus CD8-inflamed samples, but more highly expressed between CD8-excluded and CD8-desert samples; this was consistent across indications.
Figure 3.
Systematic characterization of gene expression differences across CD8 immunophenotypes. (A) Overlaps between the genes significantly differentially expressed across all pairwise comparisons. (B–D) Volcano plots highlighting the most strongly differentially expressed genes in inflamed versus desert (B), inflamed versus excluded (C), and excluded versus desert (D) tumors. Upregulated genes are in blue; downregulated genes are in red. CD8IMMPH, CD8 immunophenotype; logFC, log fold change; nLogPval=−log10 (p value).
Few (n=22) genes were more highly expressed in CD8-desert samples than in both CD8-inflamed and CD8-excluded samples (desert-enriched): such genes were also often more highly expressed in CD8-excluded versus CD8-inflamed samples (online supplemental figure S3B). Similarly, only 45 genes were more highly expressed in CD8-excluded samples versus both CD8-inflamed and CD8-desert samples. These included TGF-β signaling genes such as PLN, C7, ADH1B, OGN and SCRG1, the Wnt signaling genes SFRP4, SFRP1 and SFRP2, and genes characteristic of smooth muscle (CNN1, ACTC1, DES) and mast cells (CTSG, MS4A2). Consistently, fibroblast, endothelial, and mast cell signatures were significantly higher enriched in CD8-excluded versus both CD8-inflamed and CD8-desert samples (online supplemental figures S3C, table S4).
Given our previous observation that liver metastases showed an enrichment of desert samples, we also assessed the transcriptional differences between liver, lung, and lymph node metastases, or primary samples, respectively, among deserts (online supplemental figure S4, table S6). A large number of genes and pathways were distinctly expressed in liver metastases deserts, with a strong upregulation of liver and metabolism-related gene expression, including hepatocyte, bile acid and xenobiotic metabolism. In addition, complement and coagulation, angiogenesis, oxidative phosphorylation, and blood vessel-endothelial-specific gene expression were also increased. On the other hand, there was no consistent significant difference in immune-related expression among liver versus other deserts, including genes characteristic of classically suppressive cell types such as regulatory T cells.
IHC-based CD8 immunophenotypes can be accurately predicted from transcriptomic data
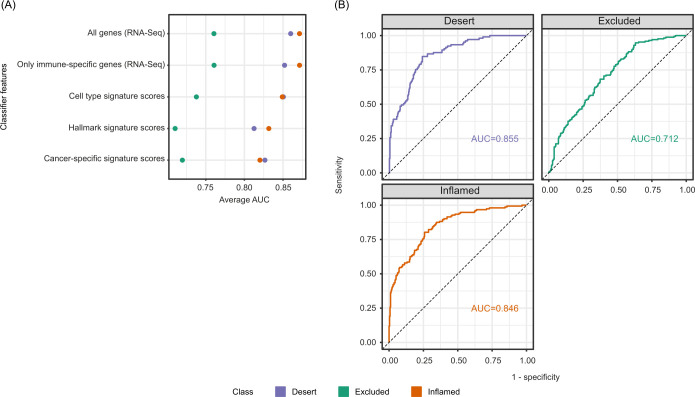
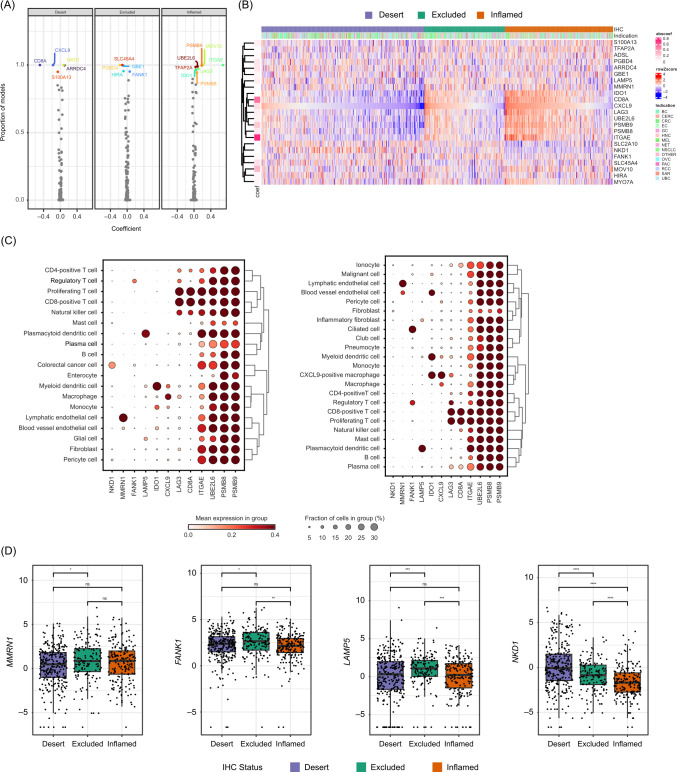
Given the large number of differentially expressed genes across the three immunophenotype classes and their high consistency across indications and excision locations, we hypothesized that the transcriptomic information may be sufficient to distinguish CD8-inflamed, CD8-excluded and CD8-desert samples, in the absence of IHC data. To test this hypothesis, as well as to better understand molecular mechanisms driving immunophenotypes by narrowing down the list of genes/pathways critical for their separation, we developed a set of classifiers using distinct input features. We used either: (1) all detected genes; (2) genes typically only expressed in immune cells; (3) a small set (97) of cell-type specific signature scores previously derived from single-cell RNA-seq data35; (4) hallmark pathway signatures (MSigDB)36; or (5) cancer-specific signatures (C4 and C6, MSigDB)36 as input features, as described in the Methods section. We developed all classifiers using the full 2023 patient sample set from 14 phase I–III trials while performing area under the curve (AUC) evaluation on 5-fold cross-validation.
All classifiers demonstrated high performance in identifying CD8-inflamed and CD8-desert samples with a mean AUC>0.8, while CD8-excluded samples were more challenging to delineate (AUC 0.70–0.76; figure 4A). The best performance was obtained when using either all genes or only immune-related genes, consistent with a large fraction of the signal being derived from the immune compartment. The classifier trained on cell-type-specific signature scores showed comparable performance, while pathway signatures and cancer-specific gene expression-trained classifiers showed the lowest accuracy (figure 4A). T-effector-related signatures were retained in this classifier: high signature values associated positively with the CD8-inflamed phenotype (eg, “Macrophage_CXCL9”, “ExhCD8Tcell”, “Interferon-alpha/gamma response”) or negatively with the CD8-desert phenotype (eg, “T cell”, “Cytotoxicity”, “Allograft rejection”) (online supplemental figure S5A). By contrast, stromal-related expression (“Fibroblast”), known immunosuppressive pathways (TGF-β signaling), or pathways related to tumor survival and aggressiveness (NOTCH signaling) were negatively associated with the CD8-inflamed phenotype. Only a few signatures were discriminatory for the CD8-excluded phenotype, most notably “Regulatory T cell”, “Exhausted B cells”, “Adipocytes”, “Adipogenesis”, “Androgen response”, and “PI3K_AKT_MTOR_Signaling”.
Figure 4.
Accurate transcriptome-based prediction of CD8 immunophenotypes. (A) Predictiveness of genes and signatures: the performance of classifiers trained on distinct features, as assessed using 5-fold cross-validation. (B) AUC curves for the final 92 gene-based classifier for CD8-desert, CD8-excluded, and CD8-inflamed phenotypes computed on the test dataset. AUC, area under curve; RNA-seq, RNA-sequencing.
As the model trained on all genes had shown the best performance overall, we trained a final classifier based on the full transcriptome. A total of 92 genes were retained (online supplemental figure S5B) in the classifier, which showed an AUC 0.855 for CD8-desert, 0.846 for CD8-inflamed, and 0.712 for CD8-excluded samples on the test dataset (figure 4B, and the Methods section). Only 17% of the retained genes were highly correlated with CD8 expression and showed the pattern of high expression in inflamed, intermediate in excluded, and low expression in desert samples described in the exploratory section above (online supplemental figure S5C).
Both CD8+ T effector-associated and non-immune-expressed genes contribute to accurate immunophenotype classification
In order to explore, in detail, the characteristics of genes critically contributing to a good separation of the immunophenotype classes, we focused on genes retained in 85% of classifiers retrained on random subsets of the training data (figure 5A,B and online supplemental table S7). We examined their expression in bulk (this study) and in publicly available colorectal, lung and liver cancer single-cell RNA-seq data.42–44 The CD8+T-effector/cytotoxicity, IFN-ɣ and antigen-processing/MHC pathway-associated genes IDO1, CD8A, CXCL9, LAG3, UBE2L6, PSMB8/9, and ITGAE formed a cohesive cluster with increasing expression in desert-to-excluded-to-inflamed samples (figure 5B and online supplemental figures S5C, S6A). Despite this strong correlation, according to single-cell RNA-seq data, not all genes were expressed by the same cell subsets. IDO1 and CXCL9 were primarily expressed in the myeloid compartment, specifically on dendritic cells and macrophages (figure 5C, online supplemental figure S6B, table S7). LAG3, CD8A, and ITGAE were CD8+T cell specific/enriched, while UBE2L6, PSMB8, and PSMB9 showed ubiquitous expression, even beyond immune cell types (figure 5C, online supplemental figures S5C, S6B).
Figure 5.
Characteristics of genes contributing highly to an accurate CD8 immunophenotype classification. (A) The frequency and magnitude of every feature in the RNA-seq classifier was measured by training and validating a classifier 100 times on random subsets of the data. The frequency of every feature and its median magnitude are displayed. Genes used in >85% of the classifiers are labeled. (B) Heatmap showing the relative expression of top classification-relevant genes across CD8 immunophenotype class cancer types as well as in (C) CRC and lung cancer single-cell RNA-seq data and (D) bulk RNA-seq data. *p≤0.05. **p≤0.01. ***p≤0.001. ****p≤0.0001. CRC, colorectal cancer; IHC, immunohistochemistry; NSCLC, non-small-cell lung cancer; LN, lymph nodes; mets, metastases; ns, not significant; RNA-seq, RNA-sequencing.
The classifier also retained genes with opposite expression patterns (enriched in desert or enriched in excluded), some of which also showed highly cell-type-specific/enriched expression (figure 5B and online supplemental figure S6A,B). For instance, the excluded-enriched LAMP5 gene was mainly expressed in plasmacytoid dendritic cells and FANK1 in regulatory T cells and malignant cells, while MMRN1 showed endothelial-restricted expression (figure 5B–D and online supplemental figure S6B). Among desert-enriched genes, we retrieved the glucose transported GLUT10 (encoded by SLC2A10) and the Wnt pathway inhibitor NKD1, which showed preferential expression in tumor cells and fibroblasts (figure 5B–D and online supplemental figure S6B). By using publicly available spatial transcriptomics data in CRC (Valdeolivas et al 46 and Wu et al 202245), we confirmed an enrichment in NKD1 expression in CD8-desert as compared with CD8-high tumor areas. In addition, we observed the strongest expression of LAMP5 in the sample showing CD8 infiltration in the stromal area only, in line with LAMP5 being excluded-enriched (online supplemental figure S6C).
In the training dataset, classifier features usually followed either desert-excluded-inflamed or inflamed-excluded-desert low-to-high expression patterns (online supplemental figure S5A, right columns of the plots). To assess relationships between immunophenotypes on the molecular level, we performed uniform manifold approximation and projection transformation on the 92 genes used by the classifier trained on the gene expression values. We observed a clear separation between inflamed and desert, whereas excluded samples were more scattered (online supplemental figure S6D). This suggests that the excluded immunophenotype is an intermediate state, rather than a distinct one. This is also consistent with lower performance of the classifiers when predicting the excluded phenotype. Multinomial model coefficient matrices show that the contribution of important transcriptional determinants to the inflamed and excluded immunophenotypes were separated in orthogonal directions, defining a separation between these tumor groups (online supplemental figure S6E).
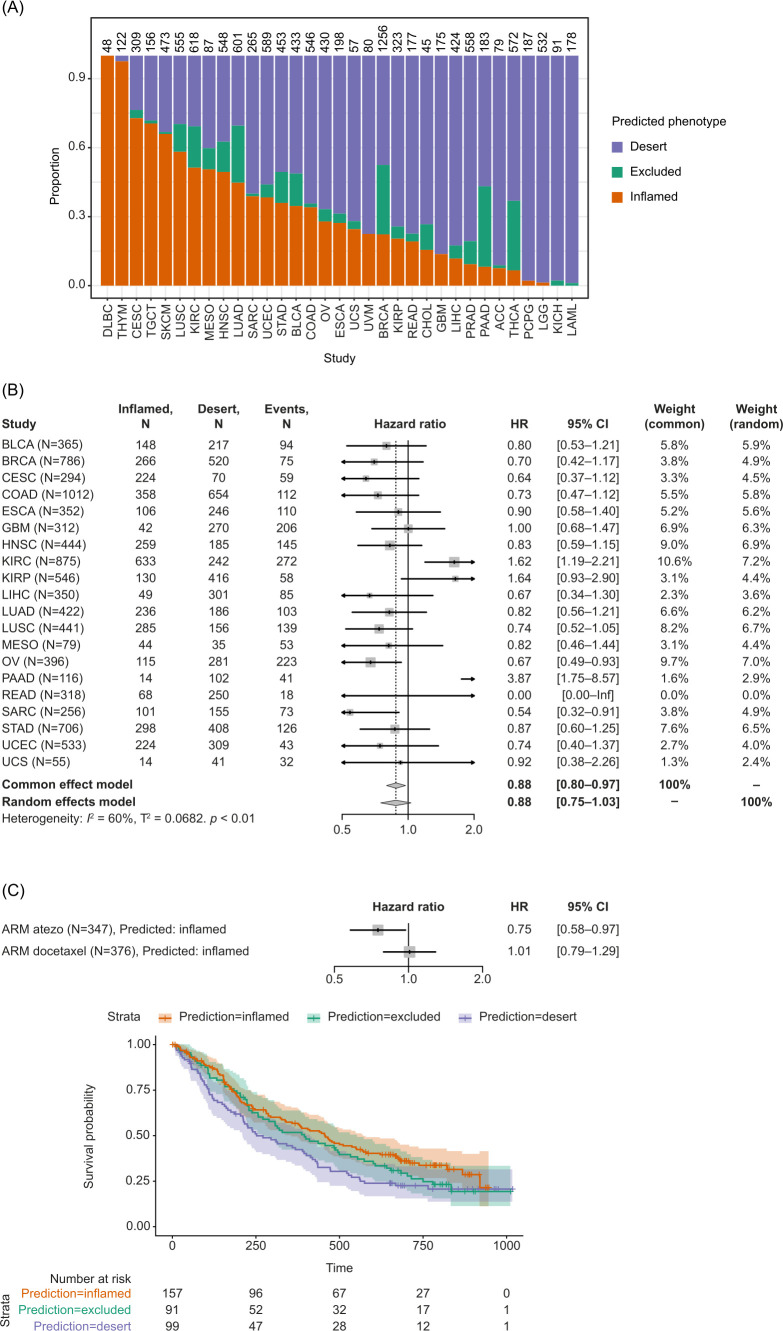
CD8 immunophenotype predictions are associated with patient survival and immunotherapy response
The developed classifier was applied to the pan-cancer TCGA dataset in order to evaluate its prognostic effect on OS. The first step toward this was to batch-correct the TCGA dataset using only lung samples as a reference. The phenotypes of hematological malignancies, diffuse large B-cell lymphoma, and thymoma were predicted as CD8-inflamed only (figure 6A). Classically highly inflamed indications including MEL, lung squamous cell carcinoma, and RCC were predicted to have ≥50% CD8-inflamed samples, compared with tumors with typically low levels of inflammation, such as pancreatic and prostate cancer, where only <10% of samples were predicted as CD8-inflamed.
Figure 6.
Application of the transcriptome-based classifier on patient data. (A) Predicted CD8 immunophenotypes across TCGA. (B) Prognostic value of predicted CD8 inflamed versus desert immunophenotype based on TCGA data. Samples predicted as excluded were removed from the analysis. (C) Predictive value of the classifier in lung cancer. CIT, cancer immunotherapy; TCGA, The Cancer Genome Atlas.
A positive association was observed between the predicted CD8 immunophenotype and OS across indications. Specifically, patients who had predicted CD8-inflamed tumors across TCGA showed prolonged OS compared with patients who had CD8-desert tumors (HR 0.88; 95% CI 0.80 to 0.97) (figure 6B). While ovarian carcinoma and sarcoma showed particularly strong survival benefits, kidney cancer was the sole indication associated with a worse prognosis in patients with inflamed phenotypes (figure 6B).
Finally, the classifier was evaluated using data from the OAK trial, including patients for whom the IHC-based CD8 immunophenotype was missing. In the atezolizumab arm, samples predicted as inflamed had an HR for mortality of 0.75 (95% CI 0.58 to 0.97), whereas in the docetaxel arm, the HR was 1.01 (95% CI 0.79 to 1.29), suggesting the CD8 immunophenotypes only had an influence on the survival of immune checkpoint inhibitor-treated patients and not on patients treated with docetaxel (figure 6C).
Finally, we investigated if newly identified immunophenotype-enriched genes showed any association with increased/decreased survival across TCGA. While no significant patterns were observed for NKD1, LAMP5-high tumors showed a higher risk of mortality compared with the overall tumor samples: the common effect model HR for mortality was 1.20 (95% CI 1.08 to 1.33) (online supplemental figures S7A–C).
Discussion
IHC analyses of CD8+T cell infiltration in tumor samples represent one widely used approach to stratify patients by immunophenotype based on the spatial location of CD8+T cells in the TME. To better understand variability in this phenotype across indications and excision locations, as well as to what extent it is reflected in transcriptional patterns, we generated and explored a large dataset (>2000 samples) from 14 phase I–III trials, in which the IHC data were scored and classified in an identical way.
Consistent with prior reports, we found that CD8 immunophenotypes showed highly distinct patterns between indications as well as between primary and metastatic sites.12 Strikingly, CD8-deserts were most abundant in liver metastases, regardless of the tumor origin. Together with the fact that the majority of biopsies in our phase I/II cohort originated from liver metastases, despite studies not normally mandating exact biopsy locations, this highlights that the patient population enrolled in CIT trials may be highly challenging to treat due to low baseline immune infiltration. In contrast, indications such as MEL, lung and bladder cancers presented a predominant CD8-inflamed and/or CD8-excluded phenotype, in line with these tumors generally being considered to be immune ‘hot’ and responsive to CIT.49 50 Our findings highlight the impact of the local microenvironment on tumor immunophenotype as well as on biomarker or pharmacodynamics analyses and treatment decisions based on biopsies from metastases.
Inflammatory and suppressive immune signatures also varied in areas of differential CD8 immunophenotypes. The strongest signal was related to T-effector and IFN-ɣ activity, which showed highest levels in inflamed, intermediate in excluded and very low levels in desert samples, in line with high consistency at protein and RNA level. Few excluded-specific signals were detected: 45 genes showed significantly higher expression in both excluded versus deserts and excluded versus inflamed comparisons, enriching for fibroblast, endothelial and mast cell-specific expression. Interestingly, mast cells can take multiple roles in shaping the TME, promote angiogenesis and support tumor invasiveness51 and have recently been shown to mediate resistance to anti-programmed death-1 therapy.52 Further, the excluded-enriched expression of stromal-specific genes and the angiogenic and TGF-β pathways is consistent with an immunosuppressive environment, underlying immune cell exclusion.2 Our findings are also in line with a recent ovarian cancer study, which found that CD8-excluded phenotypes have a higher stromal, TGF-β, and angiogenesis pathway activity.22
The good correspondence between the transcriptome and IHC-based measurements encouraged us to develop a classifier that can accurately predict IHC-derived CD8 immunophenotypes solely based on gene expression.
Our classifier yielded a high predictive performance, although better for CD8-inflamed/CD8-desert phenotypes than for the CD8-excluded phenotype. This lower performance for the excluded phenotype may be because RNA-seq provides a bulk characterization of the tumor, while the excluded phenotype is largely characterized by spatial relationships more difficult to capture at the molecular level. Notably, the described CD8 IHC immunophenotype classification relies on a pathologist’s assessment of lymphocyte location on a whole-slide tissue with regard to tumor epithelium and stroma areas, which makes the method prone to interobserver bias. Given this inherent noise, the performance of the classifier was very high.
We tested the broad applicability of the classifier by evaluating the prognostic effect on overall survival (OS) in pan-cancer TCGA and clinical trial datasets, and the predictive effect on OS in lung cancer from the OAK study. Both the content of CD8+T cells and many of the pathways and signatures identified and described in this report were previously found to be positively associated with survival and response to CIT.3 10 13 53 54 For instance, CXCL9 and CD8A (two of our top classifier genes), as well as the inflamed signature,55 which includes CD8A, LAG3 and IDO1 from our top classifier genes, were positively associated with CIT response according to a recent meta-analysis across >1000 patients.10 Consistent with these observations, patients with NSCLC in the OAK trial26 receiving atezolizumab had superior OS with classifier-predicted CD8-inflamed tumors than with CD8-desert tumors, whereas there was no OS difference between those phenotypes in those patients receiving docetaxel.
When investigating the prognostic effect of the predicted CD8 immunophenotype across the TCGA data, a positive association was found between CD8 immunophenotype and OS across indications, as expected based on previous reports.56 Patients with kidney cancer with inflamed phenotypes had a worse prognosis than those with other CD8 immunophenotypes. This atypical pattern of RCC was recently reported in a study where the proliferating CD8+T cell percentage was histologically assessed.57 Thus, our predictions are in line with previous findings but expand the range of indications for which the relation between immunophenotype and response can be evaluated.
While most of the T-effector and immune infiltration-related signals have been previously identified and described, our approach also revealed predictive features stemming from other cellular compartments. This included the desert-enriched CRC gene NKD1, which is induced to antagonize Wnt signaling and promotes cancer cell proliferation.58 Although we confirmed the tumor-associated localization of NKD1 using publicly available spatial transcriptomic data, we did not detect a significant survival association across TCGA data. However, we report here for the first time that excluded-enriched LAMP5 expression is negatively associated with OS across indications in TCGA data. Interestingly, LAMP5 depletion was recently found to significantly inhibit leukemia cell growth and to be a modulator of innate immune pathways by suppressing type I IFNs downstream of TLR9.59 60 LAMP5 was also detected on TGF-β-myofibroblastic cancer-associated fibroblasts, which are associated with an immunosuppressive environment.61 Our single-cell RNA-seq analysis confirms fibroblasts, along with plasmacytoid dendritic cells, as the main source of LAMP5 expression in tumor tissues. How LAMP5 contributes to immune cell exclusion and a survival disadvantage, and which of the two cell types is important for this association, warrants further investigation.
Potential limitations of our study include the need to further confirm our findings in a wider population and other cancer types. Notably, our RNA-seq-based classifier was solely developed to predict the CD8 immunophenotype and has thus not been optimized to predict survival benefit, in contrast to other studies highlighting CD8-related biomarkers. We solely reported here a correlation between the predicted CD8 immunophenotype and OS. Finally, the bulk RNA-seq method used in our study has previously shown limited reproducibility due to heterogeneity between tumor cells and within the microenvironment.62 More recent technologies such as single-cell RNA-seq and spatial transcriptomics could address the limitations of bulk RNA-seq and provide more accurate information such as spatial resolution.2 62
Conclusions
CD8 immunophenotyping has emerged as a powerful tool for understanding the TME and its impact on cancer. Our novel 92-gene classifier accurately predicts the spatial CD8 immunophenotype of primary and metastatic tumors. As RNA-seq provides wider information on patient samples than IHC, the new classifier could be used for retrospective and reverse translation analyses of CD8 immunophenotypes from clinical trial cohorts without the need for individual tissue section curation.
Tumor-agnostic enrichment strategies require consideration of spatial location of immune cells, immune-related patterns and lesion location,63 and the development of this RNA-based CD8 immunophenotyping classifier is a promising step in this direction, providing a reliable, cost-effective and simple tool to help optimize patient selection, response to CIT, and patient outcomes.
Acknowledgments
The authors would like to thank the patients, their families, and the participating study centers. We acknowledge Laura Jarassier for data analysis support and the Roche-wide enhanced Data and Insights Sharing (EDIS) network for providing curated and annotated clinical data. Medical writing support for the development of this manuscript, under the direction of the authors, was provided by Laura Vergoz, PhD, and Edward Neale, PhD, of Ashfield MedComms, an Inizio company, and funded by F. Hoffmann-La Roche.
Footnotes
Contributors: AR, IID, PCS, MLS-S and MAC designed the study, interpreted the data and drafted the manuscript, with additional input from all authors. IID designed and executed the machine learning experiment, including classifier training, validation and assessment of feature importance, developed and executed batch-correction methodology, and performed survival analyses. PCS performed analyses relating to signature enrichment, gene expression characteristics in bulk, and single-cell transcriptomics data. AV performed analyses related to spatial transcriptomics data. MLS-S performed analyses relating to cohort characterization. AH performed cross-clinical trial sample metadata curation. AH and CSF were responsible for CD8 IHC immunophenotype data integration and harmonization. NS performed the differential expression analyses. All authors read and approved the final manuscript.
Funding: This study was supported by F. Hoffmann-La Roche.
Competing interests: AR, IID, PCS, MLS-S, NS, AV and KK are employees of F. Hoffmann-La Roche. AH, CSF, GD and MAC are employees of Roche Diagnostics. IK was an employee of Roche Diagnostics at the time of the study. AH, AR, IID, PCS, MLS-S, NS, CSF, AV, KK and MAC are shareholders of F. Hoffmann-La Roche.
Provenance and peer review: Not commissioned; externally peer reviewed.
Supplemental material: This content has been supplied by the author(s). It has not been vetted by BMJ Publishing Group Limited (BMJ) and may not have been peer-reviewed. Any opinions or recommendations discussed are solely those of the author(s) and are not endorsed by BMJ. BMJ disclaims all liability and responsibility arising from any reliance placed on the content. Where the content includes any translated material, BMJ does not warrant the accuracy and reliability of the translations (including but not limited to local regulations, clinical guidelines, terminology, drug names and drug dosages), and is not responsible for any error and/or omissions arising from translation and adaptation or otherwise.
Data availability statement
Data are available on reasonable request.
Data availability statement
The source code used for the analysis is available via https://github.com/bedapub/CD8-immune-phenotype-paper-supplementary. The classifier is available as an R-package at https://github.com/bedapub/cd8ippred. Due to Roche company policy and patient privacy reasons, access to individual patient-level data is restricted. For up-to-date details on Roche’s Global Policy on the Sharing of Clinical Information and how to request access to related clinical study documents, see here: https://go.roche.com/data_sharing.
Ethics statements
Patient consent for publication
Not applicable.
Ethics approval
This study involves human participants and all clinical studies were conducted in accordance with the principles of the Declaration of Helsinki and Good Clinical Practice Guidelines. Written informed consent was collected from all enrolled patients. The protocol for each clinical study was approved by the institutional review boards/ethics committees at each center. A list of the ethics approval board/committee from the principal investigator of each of the 14 studies is included as supplementary table S2. Participants gave informed consent to participate in the study before taking part.
References
- 1. Sankar K, Ye JC, Li Z, et al. The role of biomarkers in personalized immunotherapy. Biomark Res 2022;10:32. 10.1186/s40364-022-00378-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Kirchhammer N, Trefny MP, Auf der Maur P, et al. Combination cancer Immunotherapies: emerging treatment strategies adapted to the tumor Microenvironment. Sci Transl Med 2022;14:eabo3605. 10.1126/scitranslmed.abo3605 [DOI] [PubMed] [Google Scholar]
- 3. Tumeh PC, Harview CL, Yearley JH, et al. PD-1 blockade induces responses by inhibiting adaptive immune resistance. Nature 2014;515:568–71. 10.1038/nature13954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Herbst RS, Soria J-C, Kowanetz M, et al. Predictive correlates of response to the anti-PD-L1 antibody Mpdl3280A in cancer patients. Nature 2014;515:563–7. 10.1038/nature14011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Cristescu R, Mogg R, Ayers M, et al. Pan-tumor genomic biomarkers for PD-1 checkpoint blockade-based immunotherapy. Science 2018;362:eaar3593. 10.1126/science.aar3593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Hegde PS, Chen DS. Top 10 challenges in cancer immunotherapy. Immunity 2020;52:17–35. 10.1016/j.immuni.2019.12.011 [DOI] [PubMed] [Google Scholar]
- 7. Havel JJ, Chowell D, Chan TA. The evolving landscape of biomarkers for checkpoint inhibitor immunotherapy. Nat Rev Cancer 2019;19:133–50. 10.1038/s41568-019-0116-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Masucci G, Ascierto PA, Villabona L. Search of predictive biomarkers in cancer immunotherapy. 2019. [Google Scholar]
- 9. Xu Y, Fu Y, Zhu B, et al. Predictive biomarkers of immune checkpoint inhibitors-related toxicities. Front Immunol 2020;11:2023. 10.3389/fimmu.2020.02023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Litchfield K, Reading JL, Puttick C, et al. Meta-analysis of tumor- and T cell-intrinsic mechanisms of sensitization to checkpoint inhibition. Cell 2021;184:596–614. 10.1016/j.cell.2021.01.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Hegde PS, Karanikas V, Evers S. The where, the when, and the how of immune monitoring for cancer immunotherapies in the era of checkpoint inhibition. Clin Cancer Res 2016;22:1865–74. 10.1158/1078-0432.CCR-15-1507 [DOI] [PubMed] [Google Scholar]
- 12. Echarti A, Hecht M, Büttner-Herold M, et al. CD8+ and regulatory T cells differentiate tumor immune phenotypes and predict survival in locally advanced head and neck cancer. Cancers (Basel) 2019;11:1398. 10.3390/cancers11091398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Yuan Y, Zhu Z, Lan Y, et al. Development and validation of a CD8+ T cell infiltration-related signature for melanoma patients. Front Immunol 2021;12:659444. 10.3389/fimmu.2021.659444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Galon J, Bruni D. Approaches to treat immune hot, altered and cold tumours with combination immunotherapies. Nat Rev Drug Discov 2019;18:197–218. 10.1038/s41573-018-0007-y [DOI] [PubMed] [Google Scholar]
- 15. Lei Y, Li X, Huang Q, et al. Progress and challenges of predictive biomarkers for immune checkpoint blockade. Front Oncol 2021;11:617335. 10.3389/fonc.2021.617335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Troiano G, Rubini C, Togni L, et al. The immune phenotype of tongue squamous cell carcinoma predicts early relapse and poor prognosis. Cancer Med 2020;9:8333–44. 10.1002/cam4.3440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Fehrenbacher L, Spira A, Ballinger M, et al. Atezolizumab versus docetaxel for patients with previously treated non-small-cell lung cancer (POPLAR): a multicentre, open-label, phase 2 randomised controlled trial. Lancet 2016;387:1837–46. 10.1016/S0140-6736(16)00587-0 [DOI] [PubMed] [Google Scholar]
- 18. Kowanetz M, Zou W, McCleland M, et al. MA 05.09 pre-existing immunity measured by teff gene expression in tumor tissue is associated with atezolizumad efficacy in NSCLC. Journal of Thoracic Oncology 2017;12:S1817–8. 10.1016/j.jtho.2017.09.485 [DOI] [Google Scholar]
- 19. Kowanetz M, Zou W, Gettinger SN, et al. Differential regulation of PD-L1 expression by immune and tumor cells in NSCLC and the response to treatment with atezolizumab (anti-PD-L1). Proc Natl Acad Sci U S A 2018;115:E10119–26. 10.1073/pnas.1802166115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hammerl D, Martens JWM, Timmermans M, et al. Spatial immunophenotypes predict response to anti-PD1 treatment and capture distinct paths of T cell evasion in triple negative breast cancer. Nat Commun 2021;12:5668. 10.1038/s41467-021-25962-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Paluch BE, Glenn ST, Conroy JM, et al. Robust detection of immune transcripts in FFPE samples using targeted RNA sequencing. Oncotarget 2017;8:3197–205. 10.18632/oncotarget.13691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Desbois M, Udyavar AR, Ryner L, et al. Integrated digital pathology and transcriptome analysis identifies molecular mediators of T-cell exclusion in ovarian cancer. Nat Commun 2020;11:5583. 10.1038/s41467-020-19408-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Eisenhauer EA, Therasse P, Bogaerts J, et al. New response evaluation criteria in solid tumours: revised RECIST guideline (version 1.1). Eur J Cancer 2009;45:228–47. 10.1016/j.ejca.2008.10.026 [DOI] [PubMed] [Google Scholar]
- 24. Balar AV, Galsky MD, Rosenberg JE, et al. Atezolizumab as first-line treatment in cisplatin-ineligible patients with locally advanced and metastatic urothelial carcinoma: a single-arm, multicentre, phase 2 trial. Lancet 2017;389:67–76. 10.1016/S0140-6736(16)32455-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Powles T, Durán I, van der Heijden MS, et al. Atezolizumab versus chemotherapy in patients with platinum-treated locally advanced or metastatic urothelial carcinoma (IMvigor211): a multicentre, open-label, phase 3 randomised controlled trial. Lancet 2018;391:748–57. 10.1016/S0140-6736(17)33297-X [DOI] [PubMed] [Google Scholar]
- 26. Rittmeyer A, Barlesi F, Waterkamp D, et al. Atezolizumab versus docetaxel in patients with previously treated non-small-cell lung cancer (OAK): a phase 3, open-label, multicentre randomised controlled trial. Lancet 2017;389:255–65. 10.1016/S0140-6736(16)32517-X [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Wilks C, Zheng SC, Chen FY, et al. Recount3: summaries and queries for large-scale RNA-seq expression and splicing. Genome Biol 2021;22:323. 10.1186/s13059-021-02533-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Galon J, Lanzi A. Immunoscore and its introduction in clinical practice. Q J Nucl Med Mol Imaging 2020;64:152–61. 10.23736/S1824-4785.20.03249-5 [DOI] [PubMed] [Google Scholar]
- 29. Mariathasan S, Turley SJ, Nickles D, et al. TGF-β attenuates tumour response to PD-L1 blockade by contributing to exclusion of T cells. Nature 2018;554:544–8. 10.1038/nature25501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Powles T, Kockx M, Rodriguez-Vida A, et al. Clinical efficacy and biomarker analysis of neoadjuvant atezolizumab in operable urothelial carcinoma in the ABACUS trial. Nat Med 2019;25:1706–14. 10.1038/s41591-019-0628-7 [DOI] [PubMed] [Google Scholar]
- 31. Htseqgenie: A NGS analysis pipeline [program]. R Package Version 2012;3. [Google Scholar]
- 32. Huber W, Carey VJ, Gentleman R, et al. Orchestrating high-throughput genomic analysis with bioconductor. Nat Methods 2015;12:115–21. 10.1038/nmeth.3252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Wu TD, Reeder J, Lawrence M, et al. GMAP and GSNAP for genomic sequence alignment: enhancements to speed, accuracy, and functionality. Methods Mol Biol 2016;1418:283–334. 10.1007/978-1-4939-3578-9_15 [DOI] [PubMed] [Google Scholar]
- 34. Zhang JD, Hatje K, Sturm G, et al. Detect tissue heterogeneity in gene expression data with BioQC. BMC Genomics 2017;18:277. 10.1186/s12864-017-3661-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Mädler SC, Julien-Laferriere A, Wyss L, et al. Besca, a single-cell transcriptomics analysis toolkit to accelerate translational research. NAR Genomics and Bioinformatics 2021;3. 10.1093/nargab/lqab102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Liberzon A, Birger C, Thorvaldsdóttir H, et al. The molecular signatures database (MSigDB) hallmark gene set collection. Cell Syst 2015;1:417–25. 10.1016/j.cels.2015.12.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Law CW, Chen Y, Shi W, et al. Voom: precision weights unlock linear model analysis tools for RNA-seq read counts. Genome Biol 2014;15:R29. 10.1186/gb-2014-15-2-r29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Friedman J, Hastie T, Tibshirani R. Regularization paths for generalized linear models via coordinate descent. J Stat Softw 2010;33:1–22. 10.18637/jss.v033.i01 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Bischel B, Lang M, Kotthoff L, et al. Mlr: machine learning in R. J Mach Learn Res 2016;17:170. [Google Scholar]
- 40. Smillie CS, Biton M, Ordovas-Montanes J, et al. Intra- and inter-cellular rewiring of the human colon during ulcerative colitis. Cell 2019;178:714–30. 10.1016/j.cell.2019.06.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Peng J, Sun B-F, Chen C-Y, et al. Single-cell RNA-seq highlights intra-tumoral heterogeneity and malignant progression in pancreatic ductal adenocarcinoma. Cell Res 2019;29:725–38. 10.1038/s41422-019-0195-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Lee H-O, Hong Y, Etlioglu HE, et al. Lineage-dependent gene expression programs influence the immune landscape of colorectal cancer. Nat Genet 2020;52:594–603. 10.1038/s41588-020-0636-z [DOI] [PubMed] [Google Scholar]
- 43. Qian J, Olbrecht S, Boeckx B, et al. A pan-cancer blueprint of the heterogeneous tumor microenvironment revealed by single-cell profiling. Cell Res 2020;30:745–62. 10.1038/s41422-020-0355-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Ma L, Hernandez MO, Zhao Y, et al. Tumor cell biodiversity drives microenvironmental reprogramming in liver cancer. Cancer Cell 2019;36:418–30. 10.1016/j.ccell.2019.08.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Wu Y, Yang S, Ma J, et al. Spatiotemporal immune landscape of colorectal cancer liver metastasis at single-cell level. Cancer Discov 2022;12:134–53. 10.1158/2159-8290.CD-21-0316 [DOI] [PubMed] [Google Scholar]
- 46. Valdeolivas A, Amberg B, Giroud N, et al. Profiling the heterogeneity of colorectal cancer consensus molecular subtypes using spatial transcriptomics. NPJ Precis Oncol 2024;8:10. 10.1038/s41698-023-00488-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. RTCGA . The Cancer Genome Atlas Data Integration [Program]. R Package Version 1.28.0 Version. 2022. [Google Scholar]
- 48. Survminer: drawing survival curves using Ggplot2 [program]. R package version 0.4.9]. 2021.
- 49. Maleki Vareki S. High and low mutational burden tumors versus immunologically hot and cold tumors and response to immune checkpoint inhibitors. J Immunother Cancer 2018;6:157. 10.1186/s40425-018-0479-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. De Guillebon E, Dardenne A, Saldmann A, et al. Beyond the concept of cold and hot tumors for the development of novel predictive biomarkers and the rational design of immunotherapy combination. Int J Cancer 2020;147:1509–18. 10.1002/ijc.32889 [DOI] [PubMed] [Google Scholar]
- 51. Komi DEA, Redegeld FA. Role of mast cells in shaping the tumor microenvironment. Clin Rev Allergy Immunol 2020;58:313–25. 10.1007/s12016-019-08753-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Somasundaram R, Connelly T, Choi R, et al. Tumor-infiltrating mast cells are associated with resistance to anti-PD-1 therapy. Nat Commun 2021;12:346. 10.1038/s41467-020-20600-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. House IG, Savas P, Lai J, et al. Macrophage-derived CXCL9 and CXCL10 are required for antitumor immune responses following immune checkpoint blockade. Clin Cancer Res 2020;26:487–504. 10.1158/1078-0432.CCR-19-1868 [DOI] [PubMed] [Google Scholar]
- 54. Saleh R, Sasidharan Nair V, Toor SM, et al. Differential gene expression of tumor-infiltrating CD8. J Immunother Cancer 2020;8:e001294. 10.1136/jitc-2020-001294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Ayers M, Lunceford J, Nebozhyn M, et al. IFN-gamma-related mRNA profile predicts clinical response to PD-1 blockade. J Clin Invest 2017;127:2930–40. 10.1172/JCI91190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Zuo S, Wei M, Wang S, et al. Pan-cancer analysis of immune cell infiltration identifies a prognostic immune-cell characteristic score (ICCS) in lung adenocarcinoma. Front Immunol 2020;11:1218. 10.3389/fimmu.2020.01218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Blessin NC, Li W, Mandelkow T, et al. Prognostic role of proliferating CD8(+) cytotoxic Tcells in human cancers. Cell Oncol (Dordr) 2021;44:793–803. 10.1007/s13402-021-00601-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Wang Y, Yang C, Li W, et al. Identification of colon tumor marker NKD1 via integrated bioinformatics analysis and experimental validation. Cancer Medicine 2021;10:7383–94. 10.1002/cam4.4224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Gracia-Maldonado G, Clark J, Burwinkel M, et al. LAMP-5 is an essential inflammatory-signaling regulator and novel immunotherapy target for mixed lineage leukemia-rearranged acute leukemia. Haematologica 2022;107:803–15. 10.3324/haematol.2020.257451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Combes A, Camosseto V, N’Guessan P, et al. BAD-LAMP controls TLR9 trafficking and signalling in human plasmacytoid dendritic cells. Nat Commun 2017;8:913. 10.1038/s41467-017-00695-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Kieffer Y, Hocine HR, Gentric G, et al. Single-cell analysis reveals fibroblast clusters linked to immunotherapy resistance in cancer. Cancer Discov 2020;10:1330–51. 10.1158/2159-8290.CD-19-1384 [DOI] [PubMed] [Google Scholar]
- 62. Li X, Wang C-Y. From bulk, single-cell to spatial RNA sequencing. Int J Oral Sci 2021;13:36. 10.1038/s41368-021-00146-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Rad HS, Rad HS, Shiravand Y, et al. The Pandora’s box of novel technologies that may revolutionize lung cancer. Lung Cancer 2021;159:34–41. 10.1016/j.lungcan.2021.06.022 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
jitc-2023-008185supp001.pdf (896.7KB, pdf)
jitc-2023-008185supp002.pdf (7.6MB, pdf)
Data Availability Statement
Data are available on reasonable request.
The source code used for the analysis is available via https://github.com/bedapub/CD8-immune-phenotype-paper-supplementary. The classifier is available as an R-package at https://github.com/bedapub/cd8ippred. Due to Roche company policy and patient privacy reasons, access to individual patient-level data is restricted. For up-to-date details on Roche’s Global Policy on the Sharing of Clinical Information and how to request access to related clinical study documents, see here: https://go.roche.com/data_sharing.