Abstract
gp64 is the major envelope glycoprotein in the budded form of Autographa californica multicapsid nucleopolyhedrovirus (AcMNPV). gp64 is essential for AcMNPV infection, as it mediates penetration of budded virus into host cells via the endocytic pathway. In this study, we used site-directed mutagenesis to map the positions of the N-linked glycans on AcMNPV gp64, characterize their structures, and evaluate their influence on gp64 function. We found that four of the five consensus N-glycosylation sites in gp64 are used, and we mapped the positions of those sites to amino acids 198, 355, 385, and 426 in the polypeptide chain. Endoglycosidase H sensitivity assays showed that N-linked glycans located at different positions are processed to various degrees. Lectin blotting analyses showed that each N-linked glycan on gp64 contains α-linked mannose, all but one contains α-linked fucose, and none contains detectable β-linked galactose or α2,6-linked sialic acid. The amounts of infectious progeny produced by AcMNPV mutants lacking one, two, or three N-linked glycans on gp64 were about 10- to 100-fold lower than wild-type levels. This reduction did not correlate with reductions in the expression, transport, or inherent fusogenic activity of the mutant gp64s or in the gp64 content of mutant budded virus particles. However, all of the mutant viruses bound more slowly than the wild type. Therefore, elimination of one or more N-glycosylation sites in AcMNPV gp64 impairs binding of budded virus to the cell, which explains why viruses containing these mutant forms of gp64 produce less infectious progeny.
Autographa californica multicapsid nucleopolyhedrovirus (AcMNPV) is the type species of the family Baculoviridae, a large group of DNA-containing viruses that infect invertebrates (reviewed in reference 32). AcMNPV produces two distinct forms of viral progeny during infection. One form is budded virus (BV), which is produced when viral nucleocapsids migrate from the nucleus to the plasma membrane and bud from the surface of the infected cell. The other is polyhedron-derived virus (PDV), which is produced when nucleocapsids are enveloped and embedded within polyhedra, the paracrystalline structures that appear in the nucleus during the very late phase of infection. It is generally accepted that PDV and BV have different roles in the nucleopolyhedrovirus life cycle. PDV is responsible for primary infection of insects via the digestive tract, whereas BV disseminates the infection from the midgut to other tissues within an infected individual. This conclusion is supported by differences in the relative infectivities of PDV and BV for insect larvae or cultured insect cells (51). Differences in their polypeptide compositions (5) probably account for these differences in the biological roles of PDV and BV during AcMNPV infection.
One clear difference in the polypeptide compositions of PDV and BV is a major envelope glycoprotein, gp64, which is found only in BV. gp64 is concentrated on one end of the rod-shaped BV particle, where it appears to form peplomers (45). gp64 has pH-dependent fusogenic activity (4, 30, 33, 49), and a gp64-specific monoclonal antibody largely neutralizes BV infectivity without blocking viral adsorption (49, 50). Immunocytochemical studies have shown that this antibody prevents nucleocapsids from reaching the nucleus (7). Similarly, lysosomotropic agents such as ammonium chloride and chloroquine drastically reduce BV infectivity by preventing nucleocapsids from reaching the nucleus (7, 49). Finally, insertional inactivation of the AcMNPV gp64 gene precludes cell-to-cell transmission of AcMNPV infection (34). Together, these findings indicate that BV enters host cells via adsorptive endocytosis and that the function of gp64 is to mediate penetration of viral nucleocapsids into the host cell cytoplasm by promoting fusion between the BV envelope and endosomal membranes.
gp64 is synthesized during both the early and late phases of nucleopolyhedrovirus infections; the newly synthesized protein enters the cellular secretory pathway, where it is chemically modified, oligomerized, and transported to the plasma membrane (3, 6, 9, 13, 18, 20, 21, 37, 39, 42, 48, 50, 53). gp64 is incorporated into BV during the late phase of infection when progeny virions bud from the surface of the infected cell. The AcMNPV gp64 gene has been mapped, cloned, and sequenced (53). The deduced amino acid sequence includes five consensus N-glycosylation sites, consisting of the sequence Asn-X-Thr/Ser (17). Relative to the translational initiation site, which is the second in-frame ATG in the long open reading frame (20), the asparagine residues in these five potential N-glycosylation sites are located at positions 160, 198, 355, 385, and 426 (Fig. 1), and at least one site is used, as gp64 is known to be N-glycosylated in AcMNPV-infected insect cells (6, 9, 18, 20, 21, 42, 50, 53). At least one of the N-linked glycans on gp64 is processed to an endo-β-N-acetylglucosaminidase H (endo H)-resistant structure (20, 21, 46), and differential processing gives rise to two different gp64 glycoforms, both of which can reach the cell surface and assemble into progeny BV (20). However, processing of the N-linked glycans on gp64 is not required for its efficient transport to the cell surface, assembly into BV, or fusogenic activity (20, 21). By contrast, N-glycosylation is required for the efficient transport, assembly, and fusogenic activity of gp64, as all are dramatically inhibited when N-glycosylation is blocked by treating AcMNPV-infected cells with tunicamycin (6, 20, 21, 43). The inhibitory effects of tunicamycin could result directly from blocking N-glycosylation of gp64, or they might result indirectly from blocking N-glycosylation of all newly synthesized glycoproteins in the baculovirus-infected cell. In this study, we distinguished between these possibilities by using site-directed mutagenesis to selectively eliminate the consensus N-glycosylation sites in AcMNPV gp64. Analysis of the resulting gp64 mutants provided new information on the positions, structures, and functions of its N-linked glycans. Analysis of recombinant baculoviruses encoding and containing these mutant gp64s showed that elimination of one or more gp64 N-glycosylation sites reduced the rate of binding of budded virus to the cell, which resulted in a corresponding reduction in the amounts of infectious progeny production.
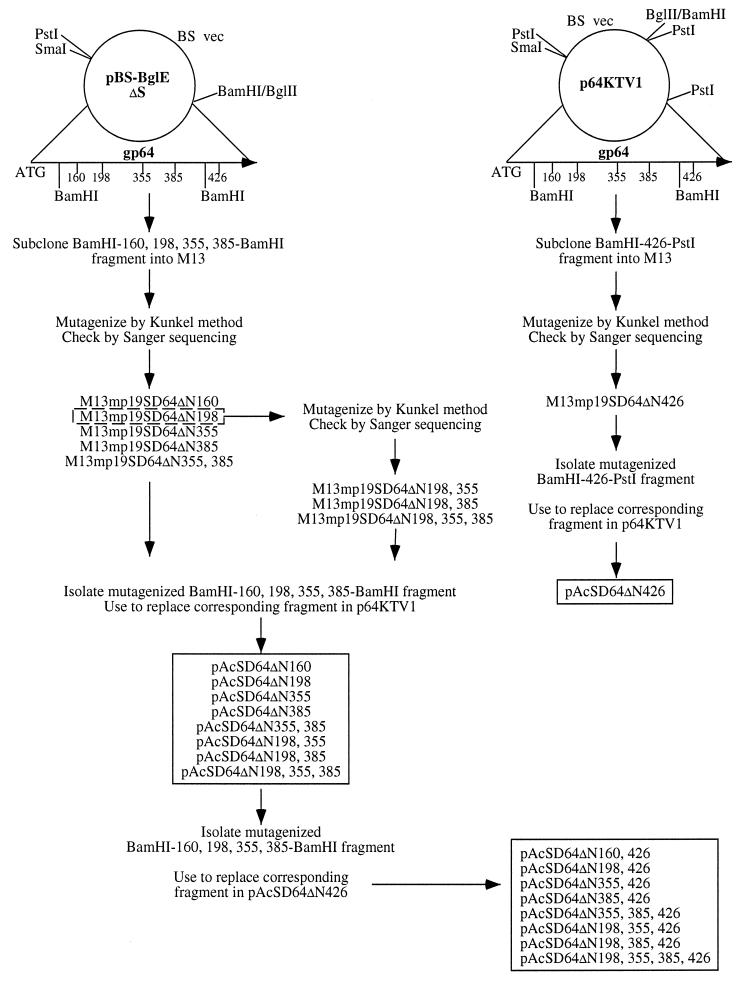
FIG. 1.
Site-directed mutagenesis of the AcMNPV gp64 gene. Two plasmids encoding full-length AcMNPV gp64 were used as the starting materials for mutagenesis of the consensus N-glycosylation sites in the gp64 protein. The BamHI fragment of pBS-BglEΔS was subcloned into M13 and used to mutagenize the consensus sites at positions 160, 198, 355, and 385, while the BamHI-PstI fragment of p64KTV1 was subcloned into M13 and used to mutagenize the consensus site at position 426. The mutagenized fragments then were excised from the M13 vectors and used to replace the corresponding wild-type fragments in p64KTV1 as described in Materials and Methods. Double and triple mutations were produced by using two oligonucleotides to simultaneously mutagenize M13 subclones, by performing a second round of mutagenesis on a single mutant in M13, or by combining different fragments from two mutants after each had been subcloned into p64KTV1. Ultimately, this strategy yielded a large set of transfer plasmids (shown in the two boxes) which was used to isolate a set of recombinant baculoviruses encoding every possible single, double, and triple N-glycosylation site mutation in AcMNPV gp64.
MATERIALS AND METHODS
Cells.
The Sf9 subclone of Spodoptera frugiperda IPLB-Sf21-AE (47) was routinely maintained as a spinner culture in TNM-FH medium (14) supplemented with fetal bovine serum, antibiotics, and pluronic F68 (35), as described previously (20). Radiolabeling medium was methionine-free Grace’s medium (10) supplemented with heat-inactivated fetal bovine serum and antibiotics as specified above. Chase medium was Grace’s medium containing 3.4 mM methionine (10 times the usual concentration), 10 μg of cycloheximide per ml, and fetal bovine serum, antibiotics, and pluronic F68 as specified above.
Construction of transfer plasmids and isolation of recombinant baculoviruses.
The strategy used to mutagenize the consensus N-glycosylation sites in AcMNPV gp64 is shown in Fig. 1. In essence, we subcloned two different AcMNPV gp64 gene fragments into M13 vectors and used oligonucleotide-directed mutagenesis (29) to change asparagine codons (AA[C/T]) in the consensus N-glycosylation sites to serine codons (TC[C/T]). The gp64 fragments were derived from either pBS-BglEΔS or p64KTV1, both of which are phagemids that include the full-length gp64 gene with the native promoter and flanking regions from the E2 strain of AcMNPV, as described previously (18). Each M13 mutant was checked by sequencing single-stranded DNA via the chain termination method (40); then the mutated gp64 fragments were excised and used to replace the corresponding wild-type fragment in p64KTV1. Double and triple N-glycosylation site mutations were produced by using two oligonucleotides to simultaneously introduce two mutations into an M13 subclone, by performing a second round of mutagenesis on a single mutant in M13, or by combining different fragments from two mutants after each had been recloned in p64KTV1, as illustrated in Fig. 1. Ultimately, these procedures yielded a set of transfer plasmids that encoded mutant forms of AcMNPV gp64 lacking various consensus N-glycosylation sites. We designated the position of each potential N-glycosylation site in AcMNPV gp64 by a number which corresponds to the Asn residue in the consensus recognition sequence (Asn-X-Thr/Ser), with the gp64 initiator methionine defined as amino acid 1 (20, 53). The numbers following ΔN in the names of the transfer plasmids indicate which sites were deleted by the mutagenesis procedure.
The transfer plasmids were used to produce recombinant baculoviruses encoding various gp64 N-glycosylation site mutants by homologous recombination in the gp64 region. A modified calcium phosphate precipitation method (44) was used to cotransfect Sf9 cells with individual transfer plasmids plus Bsu36I-linearized viral DNA from a recombinant baculovirus called AcMNPV/p34DZ5 (54). This recombinant virus has an Escherichia coli lacZ gene inserted in frame in the pp34 coding sequence, and the lacZ gene has a Bsu36I site that can be used to linearize the viral DNA prior to cotransfection. This parental viral DNA was linearized to facilitate the isolation of recombinant viruses with allelic transplacements in the gp64 region, by analogy to the strategy originally developed by Kitts and coworkers for the polyhedrin region (28). Recombinants obtained by crossing Bsu36I-digested AcMNPV/p34DZ5 viral DNA with the gp64 glycosylation site mutant transfer plasmids were tentatively identified by their white-plaque phenotypes; after two additional rounds of plaque purification, N-glycosylation site mutations were confirmed by sequencing PCR-amplified fragments of viral DNA.
Expression and electrophoretic analysis of AcMNPV gp64.
Generally, AcMNPV gp64 was expressed by transfecting Sf9 cells with plasmids or by infecting Sf9 cells with baculoviruses encoding wild-type or mutant forms of the protein. The cells were then radiolabeled and extracted, gp64 was immunoprecipitated, and the washed and disrupted immunoprecipitates were resolved by discontinuous sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). We used a modified calcium phosphate method (44) to transfect Sf9 cells for 2 h with 10 μg of a plasmid encoding AcMNPV IE1 (11) plus 10 μg of individual plasmids encoding various forms of AcMNPV gp64. After washing and feeding with TNM-FH, the transfected cells were treated as specified in the figure legends. Similarly, we infected Sf9 cells with wild-type or recombinant baculoviruses at defined multiplicities, allowed the virus to adsorb for 1 h, removed the inoculum, and treated the infected cells as detailed in the figure legends. Titers of all virus stocks were determined by plaque assays on Sf9 cells as described previously (44).
Castanospermine (Calbiochem, San Diego, Calif.) was used at a concentration of 0.2 mM to block processing of N-linked glycans in transfected or infected Sf9 cells, which accentuated differences in the relative electrophoretic mobilities of various gp64 glycoforms (20, 21). Radiolabeling was performed with 100 μCi of Trans[35S]-label (ICN Radiochemicals, Irvine, Calif.) per ml of radiolabeling medium, except for pulse-chase experiments, in which cells were treated with isotope-free radiolabeling medium, pulsed for 5 min with 500 μCi of Trans[35S]-label per ml of radiolabeling medium, and treated with chase medium for 4 h as described previously (20). gp64 was extracted by treating transfected or infected cells for 10 min on ice with cold extraction buffer (50 mM Tris-HCl [pH 8.0], 100 mM NaCl, 1% [vol/vol] Nonidet P-40 [Calbiochem] 0.2 mM leupeptin [Boehringer Mannheim Biochemicals, Indianapolis, Ind.]). The extracts were clarified by centrifugation for 10 min at 4°C in a Fisher model 235C microcentrifuge, and the clarified extracts were immunoprecipitated with AcV1 (15) and/or B12D5 (25), both of which are AcMNPV gp64-specific monoclonal antibodies. Immune complexes were absorbed with heat-inactivated, formalin-fixed Staphylococcus aureus strain Cowan I (27), washed three times with ice-cold extraction buffer supplemented with 0.1% (wt/vol) SDS and 1% (wt/vol) sodium deoxycholate, and disrupted by heating for 10 min at 65°C in Laemmli sample buffer prior to analysis by SDS-PAGE.
Endoglycosidase assays.
For endoglycosidase treatments, gp64 was extracted from transfected or infected Sf9 cells, immunoprecipitated, and recovered by resuspending the washed S. aureus pellets in 0.5% (wt/vol) SDS and 0.1 M β-mercaptoethanol, heating for 10 min at 65°C, and pelleting for 10 min in a microcentrifuge. The supernatant was harvested, adjusted to final concentrations of 0.1 M sodium phosphate (pH 6.0) and 1% (vol/vol) Nonidet P-40, and split into equal aliquots. One aliquot was incubated without any enzyme as a control, while the others were incubated with various concentrations of endo H (Boehringer Mannheim) (46). After various times of digestion at 37°C, the endo H reactions were terminated by adding an equal volume of 2× Laemmli sample buffer and heating for 10 min at 65°C, and the reaction products were analyzed by SDS-PAGE as described above.
Lectin blotting assays.
The starting material for lectin blotting assays was partially purified BV from baculovirus-infected Sf9 cells (18). Briefly, Sf9 cells were infected with wild-type or recombinant baculoviruses at a multiplicity of about 0.01 PFU per cell, and the cultures were monitored daily for the appearance of viral occlusions until at least 75% of the cells were occlusion positive. At that time, the culture media were harvested and clarified by centrifugation for 15 min at about 3,000 × g, and BV was concentrated from the supernatants by centrifugation for 30 min at 100,000 × g. The pellets were resuspended in phosphate-buffered saline (PBS; 10 mM sodium phosphate, 140 mM NaCl [pH 7.2]), layered onto 10 to 74% (wt/vol in PBS) linear sucrose gradients, and centrifuged for 1.5 h at 100,000 × g. The opaque BV bands were harvested and diluted with PBS, and the virus was reconcentrated by centrifugation for 30 min at 100,000 × g. The final BV pellets were resuspended in 0.1× TE buffer (1 mM Tris-HCl [pH 8.0], 0.1 mM EDTA), and aliquots containing equal amounts of gp64 were heated for 10 min at 65°C in Laemmli sample buffer and resolved by SDS-PAGE as described above. Proteins were transferred to Immobilon polyvinylidene difluoride membranes (Millipore Corporation, Bedford, Mass.), the membranes were cut into strips corresponding to individual lanes, and the strips were blocked by overnight incubation at 4°C in Tris-buffered saline (50 mM Tris [pH 7.5], 150 mM NaCl) containing 0.5% (wt/vol) Tween 20 (Bio-Rad Laboratories, Hercules, Calif.). After blocking, each strip was rinsed and incubated with a digoxigenylated lectin (Boehringer Mannheim) for 1 h at room temperature in the same buffer. The lectins used in this study were concanavalin A (ConA), Aleuria aurantia agglutinin (AAA), Ricinus communis agglutinin (RCA), and Sambucus nigra agglutinin (SNA). Immediately prior to use, each lectin was preincubated in buffer alone or buffer containing excess competing sugar for 2 h at room temperature to determine if lectin binding was carbohydrate specific. Competing sugars were 0.7 M α-d-methylmannopyranoside for ConA, 0.7 M l-(−)-fucose for AAA, 0.7 M D-(+)-galactose for RCA, and 0.2 M α-lactose for SNA. After the unbound lectins were washed away, secondary reactions were done with alkaline phosphatase-conjugated sheep antidigoxigenin (Boehringer Mannheim), followed by more washes and a standard color reaction (2). Some strips were probed with a gp64-specific primary antibody followed by alkaline phosphatase-conjugated secondary antibody and the same color reaction, as described previously (22).
One-step growth curves.
One-step growth curves were performed by infecting Sf9 cells at a multiplicity of 10 PFU per cell with wild-type AcMNPV or individual recombinants encoding gp64s lacking various N-glycosylation sites. The virus was allowed to adsorb for 1 h at 28°C, the inocula were removed, and the cells were washed twice with TNM-FH medium and incubated for various times at 28°C. At appropriate time points, the medium from each infected cell culture was harvested and clarified in a microcentrifuge, and infectious virus in the supernatants was quantitated by a limiting dilution assay as described previously (36). The raw data were converted to PFU per milliliter by using a Microsoft Excel spreadsheet (36) and plotted against time of infection.
Cell surface expression assays.
A lactoperoxidase-catalyzed cell surface radioiodination method (31) was used to examine the expression of various forms of gp64 on the infected cell surface. Sf9 cells were inoculated with wild-type or recombinant baculoviruses at a multiplicity of about 3 PFU per cell, the virus was allowed to adsorb for 1 h, the inocula were removed, and the cells were rinsed and fed with TNM-FH and incubated at 28°C. At 40 h postinfection, the medium was removed, the cells were washed three times with Dulbecco’s PBS, and the lactoperoxidase-catalyzed radioiodination procedure was performed. After radiolabeling, the cells were washed three more times with Dulbecco’s PBS and extracted as described above, and the extracts were immunoprecipitated with AcV1 (15) or PAb419, a monoclonal antibody specific for simian virus 40 large tumor antigen (12). Finally, the immunoprecipitates were washed, disrupted, and analyzed by SDS-PAGE as described above.
Cell fusion assays.
Sf9 cells were transfected with a plasmid encoding AcMNPV IE1 (11) plus individual plasmids encoding wild-type or mutant gp64s, as described above. At 36 h posttransfection, the cells were washed once with Grace’s medium (pH 5.0) containing 10% (vol/vol) fetal bovine serum and then treated with the same medium for 30 min at room temperature. The cells were photographed with an Olympus CK2 microscope equipped with an automatic 35-mm camera system (Olympus America Inc., Melville, N.Y.).
Binding and infectious center assays.
Sf9 cells were seeded into six-well culture plates at a density of 0.5 × 106 cells per well and incubated for 24 h at 28°C. The plates were transferred to a refrigerator and cooled to 4°C for 1 h; then the medium was removed, and triplicate wells were inoculated with 1,000 PFU of wild-type AcMNPV or recombinants encoding gp64s lacking various N-glycosylation sites. These viruses had been partially purified, concentrated, filter sterilized, and titered by plaque assay on Sf9 cells as described above. The viruses were allowed to bind to the cells for various times at 4°C; then the inocula were removed, the cells were washed three times with cold TNM-FH, and an agarose overlay was added. After 1 week at 28°C, plaques were counted and the average numbers of plaques were plotted against binding time, with error bars showing the standard deviations.
RESULTS
Number of N-linked glycans on AcMNPV gp64.
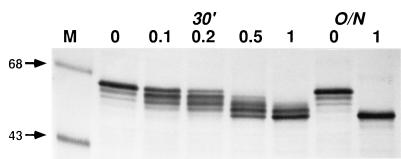
The derived amino acid sequence of AcMNPV gp64 includes five consensus N-glycosylation sites which are necessary, but not sufficient, for protein N-glycosylation in eucaryotic cells (17, 53). Previous surveys have shown that only about 30% of consensus N-glycosylation sites are actually used (52), and the rules governing site usage remain unclear. Therefore, our first goal was to determine how many of the potential glycosylation sites in AcMNPV gp64 are used during its biosynthesis. To address this question, we used endo H to produce a glycoform ladder containing various forms of gp64 with different numbers of N-linked glycans. AcMNPV-infected cells were briefly radiolabeled with Trans[35S]-label during the peak time of gp64 synthesis, the cells were extracted, and gp64 was immunoprecipitated. The immunoprecipitates were washed, disrupted, and digested with buffer alone or various concentrations of endo H, and the digestion products were analyzed by SDS-PAGE and autoradiography (Fig. 2). The position of the completely glycosylated protein is shown in the buffer control lanes, and the position of the completely deglycosylated protein is shown for a control in which gp64 was digested overnight with 1 mU of endo H. The other reactions, in which gp64 was digested for 30 min with increasing amounts of endo H, produced the glycoform ladder. This ladder included five different glycoforms, each differing from its nearest neighbors by the presence or absence of a single glycan. The smallest glycoform has no glycans, as it comigrated with the completely deglycosylated control. Each of the progressively larger glycoforms has one additional glycan, and since there were four additional glycoforms in the ladder, including one which comigrates with the completely glycosylated control, gp64 appears to have a maximum of four N-linked glycans. These results suggest that four of the five consensus N-glycosylation sites are used during biosynthesis of AcMNPV gp64.
FIG. 2.
Number of N-linked glycans on AcMNPV gp64. Sf9 cells were infected with wild-type AcMNPV at a multiplicity of 5 PFU per cell, treated with radiolabeling medium from 20 to 22 h postinfection and pulse-labeled for 5 min with 500 μCi of Tran35S-label per ml of fresh radiolabeling medium. Intracellular gp64 was extracted, immunoprecipitated, and eluted from the washed immunoprecipitates. Soluble proteins were recovered and digested overnight (O/N) with buffer alone (0) or 1.0 mU of endo H or were digested for 30 min (30′) with buffer alone or 0.1, 0.2, 0.5, or 1.0 mU of Endo H, as indicated above the lanes. The reactions were terminated by the addition of an equal volume of 2× Laemmli sample buffer, heated for 10 min at 65°C, and analyzed by SDS-PAGE and autoradiography. The numbered arrows to the left of this and subsequent figures indicate the sizes (in kilodaltons) of protein standards in lanes marked M.
Positions of N-linked glycans on AcMNPV gp64.
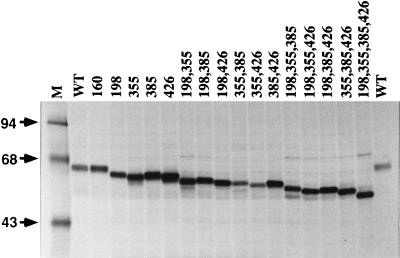
Our next goal was to confirm and extend the conclusion from the partial endoglycosidase assays by determining the positions of the N-linked glycans on AcMNPV gp64. The approach was to eliminate selected consensus N-glycosylation sites by using site-directed mutagenesis to change the asparagine codons (AA[C/T]) in those sites to serine codons (TC[C/T]). The mutagenesis and cloning strategy (described in detail in Materials and Methods and outlined in Fig. 1) yielded a large set of plasmids which encode gp64 mutants lacking one or more N-glycosylation sites. Each of these plasmids or a wild-type control was used to transfect Sf9 cells, and the cells were treated with castanospermine to block N-linked oligosaccharide processing and accentuate differences in the electrophoretic mobilities of gp64 glycoforms containing different numbers of N-linked glycans. Subsequently, the cells were radiolabeled and gp64 was extracted, immunoprecipitated, and analyzed by SDS-PAGE (Fig. 3). The results showed that all of the gp64 mutants lacking N-glycosylation sites were expressed in Sf9 cells, with no correlation between the numbers or positions of N-glycosylation site mutations and the expression levels observed by radiolabeling and immunoprecipitation. The mutant lacking one N-glycosylation site at position 160 comigrated with wild-type gp64. In contrast, mutants lacking single N-glycosylation sites at position 198, 355, 385, or 426 all migrated faster than the wild type. These results confirmed that only four of the five consensus N-glycosylation sites in AcMNPV gp64 are used and revealed that those sites are located at positions 198, 355, 385, and 426. The N-glycosylation site located at position 160 is not used during biosynthesis of AcMNPV gp64.
FIG. 3.
Transient expression of gp64 N-glycosylation site mutants. Sf9 cells were transfected with plasmids encoding wild-type (WT) or mutant gp64s lacking various consensus N-glycosylation sites, as indicated above the lanes. The cells were treated with 0.2 mM castanospermine from 16 to 20 h and labeled with 100 μCi of Tran35S-label per ml of radiolabeling medium containing 0.2 mM castanospermine from 20 to 24 h posttransfection. At the end of the labeling period, intracellular gp64 was extracted and immunoprecipitated, and the washed immunoprecipitates were analyzed by SDS-PAGE and autoradiography.
Combinatorial mutants lacking more than one N-glycosylation site migrated faster than those lacking single sites, and mutants lacking larger numbers of N-glycosylation sites migrated faster than those lacking smaller numbers of sites. Mutants lacking the same number of N-glycosylation sites did not necessarily have identical electrophoretic mobilities, but the observed differences in the electrophoretic mobilities of the gp64 mutants clearly reflected differences in N-glycosylation, as all 16 comigrated with the wild type after being completely deglycosylated with endo H (data not shown).
Processing of individual N-linked glycans on AcMNPV gp64.
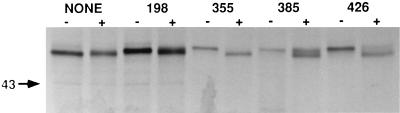
Four of the plasmids described above encoded triple-mutant gp64s with single N-linked glycans located at defined sites. Previous work had shown that at least one N-linked glycan on AcMNPV gp64 is processed to an endo H-resistant structure (20). Therefore, we used these four plasmids to examine processing of the individual N-linked glycans on gp64. Sf9 cells were transfected with the triple-mutant plasmids or a quadruple-mutant control encoding a mutant gp64 lacking all four N-glycosylation sites, which served as a marker for deglycosylated gp64. The cells were starved for methionine, pulsed briefly with Trans[35S]-label, and chased for 4 h; gp64 was extracted, immunoprecipitated, digested with endo H, and analyzed by SDS-PAGE, as described in Materials and Methods. The results showed that the glycan at position 198 was completely resistant, the glycan at position 355 was completely sensitive, and the glycans at positions 385 and 426 were about 50 and 10%, respectively, resistant to endo H (Fig. 4). These results show that only a subset of the N-linked glycans on AcMNPV gp64 are processed and reveal that processing can be highly efficient or quite inefficient, depending on the position of the glycan.
FIG. 4.
Processing of individual N-linked glycans on AcMNPV gp64. Sf9 cells were transfected with plasmids encoding mutant gp64s with no consensus N-glycosylation sites (NONE) or with single sites at position 198, 355, 385, or 426, as indicated above the lanes. The cells were treated with radiolabeling medium from 22 to 24 h, pulse-labeled for 5 min with 500 μCi of Tran35S-label per ml of fresh radiolabeling medium, and chased for 4 h as described in Materials and Methods. Intracellular gp64 was extracted, immunoprecipitated, and eluted from the washed immunoprecipitates. Soluble proteins were recovered and digested overnight with buffer alone (−) or endo H (+); then the reactions were terminated by the addition of an equal volume of 2× Laemmli sample buffer and heated for 10 min at 65°C, and the products were analyzed by SDS-PAGE and autoradiography.
Isolation and characterization of recombinant baculoviruses with mutant gp64s.
Recombinant baculoviruses encoding mutant gp64s lacking various N-linked glycans were isolated by homologous recombination between individual transfer plasmids and a parental viral DNA that had been linearized in the gp64 region as described in Materials and Methods. We used this approach to successfully isolate recombinants encoding mutant gp64s lacking every possible combination of one, two, or three N-linked glycans. Despite multiple attempts, however, we failed to isolate a recombinant encoding a quadruple-mutant nonglycosylated gp64 (data not shown). The reason for this is unclear, as we observed high-level transient expression of nonglycosylated gp64 by the quadruple mutant plasmid (Fig. 3), and we and others have shown previously that low levels of infectious progeny containing nonglycosylated gp64 can be produced by tunicamycin-treated cells (6, 20, 26, 43).
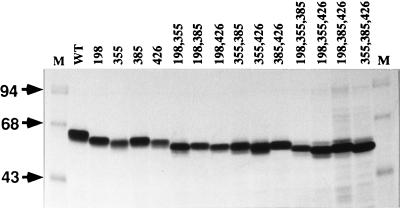
Sf9 cells were infected with each of the recombinant baculoviruses or wild-type AcMNPV, the cells were treated with castanospermine, and gp64 was radiolabeled, extracted, immunoprecipitated, and analyzed by SDS-PAGE. The results of this analysis were similar to those obtained in the transient expression experiments, as each gp64 mutant was expressed during infection and there was no correlation between expression levels and the numbers or locations of the N-glycosylation site mutations (Fig. 5). Again, the electrophoretic mobility of gp64 increased with increasing number of glycosylation site mutations, but mutants lacking the same numbers of N-linked glycans did not necessarily have the same electrophoretic mobilities.
FIG. 5.
Expression of gp64 N-glycosylation site mutants by recombinant baculoviruses. Sf9 cells were infected at a multiplicity of 2.5 PFU per cell with wild-type AcMNPV (WT) or recombinant baculoviruses encoding mutant gp64s lacking various consensus N-glycosylation sites, as indicated above the lanes. The cells were treated with 0.2 mM castanospermine from 1 to 44 h and labeled with 100 μCi of Tran35S-label per ml of radiolabeling medium containing 0.2 mM castanospermine from 44 to 48 h postinfection. At the end of the labeling period, intracellular gp64 was extracted and immunoprecipitated, and the washed immunoprecipitates were analyzed by SDS-PAGE and autoradiography.
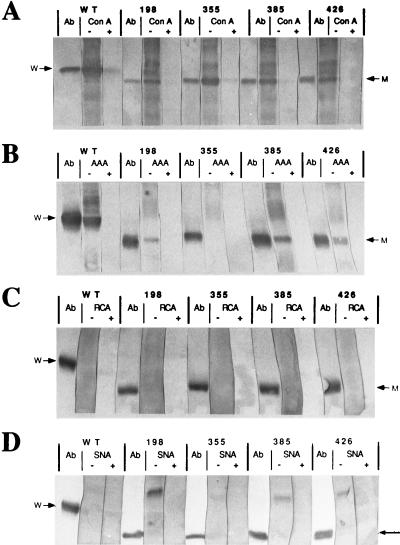
The four recombinant viruses encoding triple mutant gp64s were used to examine the carbohydrate compositions of the individual N-linked glycans on gp64. BV progeny were partially purified from Sf9 cells infected with the recombinant viruses or wild-type AcMNPV; gp64 was extracted, immunoprecipitated, and analyzed by lectin blotting as described in Materials and Methods. The lectins used for this analysis were ConA, which binds specifically to α-linked mannose, AAA, which binds specifically to α-linked fucose, RCA, which binds specifically to β-linked galactose, and SNA, which binds specifically to terminal α2,6-linked sialic acid. The results showed that ConA bound to all four triple mutant gp64s, indicating, as expected, that each N-linked glycan on AcMNPV gp64 contains α-linked mannose (Fig. 6A). The specificity of ConA binding was demonstrated by a clear reduction in binding when the lectin was preincubated with competing sugar. AAA bound to the triple-mutant gp64s with N-linked glycans at position 198, 385, or 426 but not to the one with an N-linked glycan at position 355, indicating that the former, but not the latter, contain α-linked fucose (Fig. 6B). Again, the specificity of lectin binding was demonstrated by the absence of binding when AAA was preincubated with competing sugar. Neither RCA nor SNA bound to any of the triple-mutant gp64s or to wild-type gp64, indicating that none of the N-linked glycans on gp64 contain detectable levels of β-linked galactose or α2,6-linked sialic acid. These results are consistent with the results of all of our previous lectin blotting assays on gp64 from wild-type AcMNPV (16, 18, 19).
FIG. 6.
Carbohydrate compositions of individual N-linked glycans on AcMNPV gp64. Sf9 cells were infected at a multiplicity of about 0.01 PFU per cell with wild-type AcMNPV (WT) or recombinant baculoviruses encoding mutant gp64s with single N-glycosylation sites at position 198, 355, 385, or 426, as indicated above the lanes. BV progeny were partially purified and solubilized, total virion proteins were resolved by SDS-PAGE, and the proteins were then transferred to Immobilon filters and probed with various digoxigenylated lectins, all as described in Materials and Methods. Each lectin (ConA [A], AAA [B], RCA [C], or SNA [D]) was preincubated in buffer alone (−) or buffer containing excess competing sugar (+) prior to use, and lectin binding was detected with alkaline phosphatase-conjugated sheep antidigoxigenin and a standard color reaction (2). Control strips were probed with a monoclonal antibody against gp64 (Ab) followed by alkaline phosphatase-conjugated secondary antibody and the same color reaction. Arrows at the left and right of each panel indicate the positions of wild-type (w) and mutant (m) gp64s.
In vitro growth properties of recombinant baculoviruses with mutant gp64s.
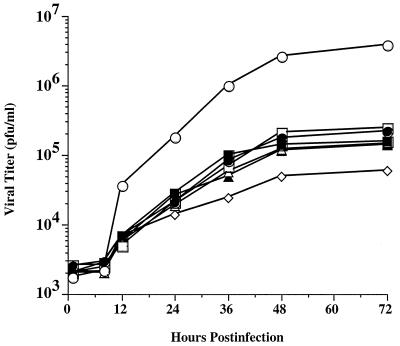
One-step growth curves were performed to compare the in vitro growth properties of wild-type AcMNPV and representative recombinants with mutant gp64s lacking various N-linked glycans as described in Materials and Methods. The results showed that the amounts of infectious progeny produced by recombinants with mutant gp64s lacking one, two, or three N-linked glycans were about 10- to 100-fold lower than the amounts produced by wild-type virus (Fig. 7). These results were confirmed and extended in an independent experiment performed nearly 2 months later, which showed that these recombinants produced about 10- to 50-fold less infectious progeny by 48 h and about 10- to 200-fold less infectious progeny by 1 week postinfection compared to the wild type (data not shown). Interestingly, the loss of a single Nglycosylation site at any position was sufficient to reduce infectious progeny production. Although one triple mutant, AcSD64KΔN355,385,426, produced slightly less progeny than all the others, there were no consistent further reductions among the mutants lacking additional N-glycosylation sites.
FIG. 7.
Replication of recombinant baculoviruses with gp64 glycosylation site mutations. Sf9 cells were infected at a multiplicity of 10 PFU per cell with wild-type AcMNPV (open circles) or recombinant baculoviruses encoding mutant gp64s lacking various consensus N-glycosylation sites (closed circles, AcSD64ΔN198; open boxes, AcSD64ΔN198,355; closed boxes, AcSD64ΔN198,355,385; open triangles, AcSD64ΔN198,355,426; closed triangles, AcSD64ΔN198,385,426; open diamonds, AcSD64ΔN355,385,426). After a 1 h adsorption period, the cells were washed and samples were harvested and clarified immediately or at various times after infection. The resulting cell-free supernatants were titered by using a limiting dilution assay as described in Materials and Methods, and the results were plotted to generate one-step growth curves for each virus.
Transport of mutant gp64s to the cell surface and incorporation into BV progeny.
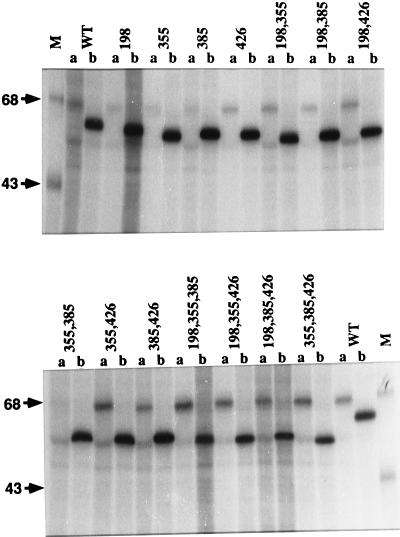
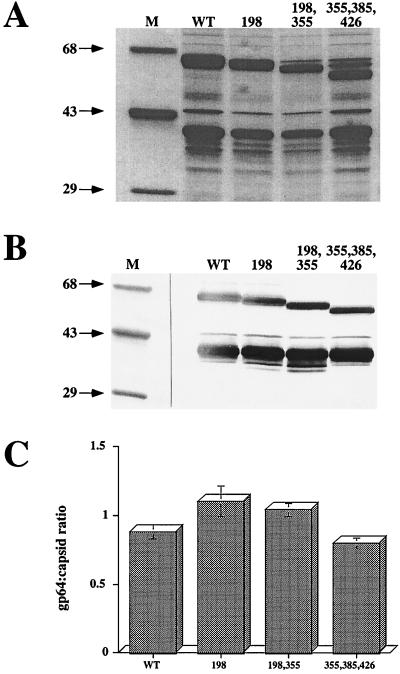
One possible explanation for the reduced levels of infectious progeny production by the recombinant viruses was that mutant gp64s lacking one or more N-linked glycans were less efficiently transported to the infected cell surface. This could reduce infectious progeny production directly if gp64 is required for budding or indirectly if BV particles require a critical mass of gp64 for optimal infectivity. To examine the expression of various gp64 mutants on the surface of cells infected with the recombinant viruses, we used a lactoperoxidase-catalyzed radioiodination method (31) that specifically radiolabels proteins exposed on the cell surface. The results showed that each mutant gp64, including all four triple mutants, reached the cell surface and each was at least as intensely radiolabeled as the wild type (Fig. 8). We recognize that it is difficult to quantify cell surface expression due to potential differences in conformation among the different forms of gp64. However, it was clear from these data that there was no correlation between the intensity of gp64 labeling and the levels of infectious progeny produced by the recombinant viruses. Thus, it was unlikely that differences in gp64 cell surface expression could account for the observed differences in infectious progeny production. To extend this observation, we evaluated the gp64 content of representative recombinant viruses by examining the ratio of gp64 to capsid protein in partially purified BV preparations. The gp64- and capsid-specific bands in the Coomassie blue-stained gel (Fig. 9A) were identified by Western blotting (Fig. 9B), and these bands in three replicate gels were quantitated by densitometry (Fig. 9C). Visual inspection of all three stained gels and the average densitometric data indicated that the gp64/capsid ratios of the mutant viruses were similar to those of the wild type. This conclusion was supported by the Western blotting results as well. Thus, there were no consistent differences in the transport of gp64 to the cell surface or its subsequent incorporation into progeny virions, which might have explained the reduced levels of infectious progeny virus produced by the recombinant viruses that contained mutant gp64s.
FIG. 8.
Cell surface expression of gp64 with various glycosylation site mutations. Sf9 cells were infected at a multiplicity of about 2.5 PFU per cell with wild-type AcMNPV (WT) or recombinant baculoviruses encoding mutant gp64s lacking various consensus N-glycosylation sites, as indicated above the lanes. The cells were incubated to 40 h postinfection, radioiodinated by the lactoperoxidase method, and extracted. The extracts were immunoprecipitated with the control antibody PAb419 (lanes a) or with AcV1 (lanes b), and the immunoprecipitates were washed, disrupted, and analyzed by SDS-PAGE and autoradiography.
FIG. 9.
Relative gp64 content of wild-type and recombinant baculovirus particles. Sf9 cells were infected at a multiplicity of about 0.01 PFU per cell with wild-type AcMNPV (WT) or recombinant baculoviruses encoding mutant gp64s lacking various consensus N-glycosylation sites, as indicated above the lanes. BV progeny were partially purified and solubilized, and total virion proteins were resolved by SDS-PAGE as described in Materials and Methods. Either the gels were stained with Coomassie blue or proteins were transferred to Immobilon filters and probed with a mixture of two different monoclonal antibodies against gp64 and one against AcMNPV capsid as described in Materials and Methods. The relevant proteins in the stained gel were quantitated by using the Gel Doc 1000 system equipped with image analysis software (Molecular Analyst version 2.1; Bio-Rad). (A) Stained gel; (B) Western blot; (C) results of image analysis shown as the gp64/capsid ratio for each type of virus.
Fusogenic activities of gp64 mutants.
AcMNPV gp64 has acid-induced fusogenic activity, which is believed to be responsible for viral penetration and cell-to-cell transmission of BV infection (4, 30, 33, 34, 49). Therefore, it was important to evaluate the inherent acid-induced fusogenic activities of the gp64 N-glycosylation site mutants. Sf9 cells were transfected with control plasmids or plasmids encoding selected gp64 mutants, and acid-induced cell fusion assays were performed at 36 h posttransfection as described in Materials and Methods. The results showed that cells transfected with pBS alone had no detectable fusogenic activity, while cells transfected with a plasmid encoding wild-type gp64 had substantial levels of fusogenic activity (Fig. 10). The levels of fusogenic activity induced by gp64 mutants lacking one or two N-glycosylation sites were indistinguishable from wild-type levels. By contrast, fusogenic activity was greatly reduced with a gp64 mutant lacking three N-glycosylation sites, and almost no activity was detected in cells transfected with a plasmid encoding nonglycosylated gp64. Cell fusion assays performed with the full set of gp64 N-glycosylation site mutants showed that all of the single and double mutants had at least wild-type levels of fusogenic activity, while all of the triple mutants were severely defective and the quadruple mutant had almost no activity (data not shown). These data showed that there was no correlation between the inherent levels of acid-induced fusogenic activity in the mutant gp64s and the lower levels of infectious progeny produced by recombinant viruses containing these mutant glycoproteins.
FIG. 10.
Fusogenic activity of gp64 with various glycosylation site mutations. Sf9 cells were transfected with a control plasmid (BS), a plasmid encoding wild-type gp64 (WT), or plasmids encoding gp64 mutants lacking various consensus N-glycosylation sites (missing sites indicated by the numbers above the panels). The cells were incubated for 36 h, then fusion assays were performed as described in Materials and Methods, and the cells were photographed at a magnification of ×20 with a phase-contrast microscope.
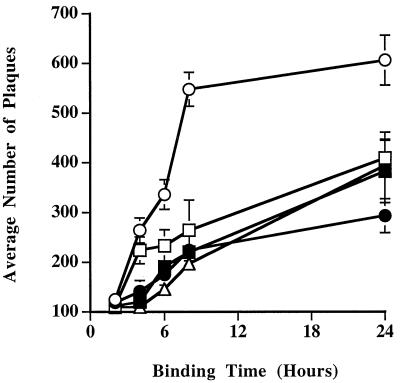
Binding kinetics of recombinant baculoviruses with mutant gp64s.
Another possible basis for the differences in the one-step growth curves of the wild-type and mutant viruses was that there might be differences in their binding properties. We addressed this possibility by comparing the binding kinetics of the wild-type and mutant viruses at 4°C. The results of these assays demonstrated that all of the mutant viruses had reduced binding kinetics relative to the wild type, irrespective of the numbers or positions of their gp64 N-glycosylation site mutations (Fig. 11). The difference in binding kinetics between the wild-type and mutant viruses correlated perfectly with the difference in their growth properties. We conclude, therefore, that elimination of one, two, or three N-glycosylation sites on AcMNPV gp64 reduces the binding kinetics of BV particles and that this reduction can account for the corresponding reduction in the levels of infectious progeny produced by these viruses.
FIG. 11.
Binding kinetics of gp64 glycosylation site mutants. Equivalent titers of wild-type AcMNPV (open circles) or recombinant baculoviruses encoding mutant gp64s lacking various consensus N-glycosylation sites (closed circles, AcSD64ΔN198; open boxes, AcSD64ΔN198,355; closed boxes, AcSD64ΔN198,355,385; open triangles, AcSD64ΔN355,385,426) were added to triplicate cultures of Sf9 cells and incubated for various times at 4°C. The viral inocula were then removed, the cells were washed three times with growth medium, and an agarose overlay was added. After 1 to 2 weeks at 28°C, plaques were counted under a dissecting microscope and the average numbers of plaques obtained with each virus were plotted against binding time.
DISCUSSION
The overall goals of this study were to further characterize the properties of AcMNPV gp64 as a glycoprotein and to examine the functional role(s) of its N-linked glycans. It had been reported previously that N-glycosylation of AcMNPV gp64 is important for its intracellular transport and fusogenic activity, as well as for BV infectivity (6, 20, 21, 43). However, those conclusions were derived from studies in which tunicamycin, a general inhibitor of protein N-glycosylation, was used to block glycosylation of gp64. Thus, it was difficult to determine whether the results of these studies reflected a direct requirement for N-glycosylation of gp64 or indirect effects of blocking N-glycosylation of all newly synthesized glycoproteins in the cell. In this study, we used site-directed mutagenesis to eliminate consensus N-glycosylation sites in AcMNPV gp64. This approach yielded a set of plasmids which encoded gp64 mutants that lacked various N-linked glycans. These plasmids were expressed transiently, and the gp64 mutants were examined biochemically to determine if the absence of N-linked glycans had any direct effects on behavior of the protein. These plasmids also were used to produce recombinant baculoviruses that encoded and contained various mutant gp64s, and the properties of these viruses were characterized to determine if the absence of N-linked glycans had any effects on the behavior and function(s) of gp64 during AcMNPV infection.
As a prelude to the site-directed mutagenesis experiments, we performed partial endo H digestions and SDS-PAGE analyses to measure the number of N-linked glycans on gp64. The results suggested that AcMNPV gp64 has a maximum of four N-linked glycans, indicating that only four of its five consensus N-glycosylation sites are used. This conclusion was confirmed and extended by examining the electrophoretic mobilities of five gp64 mutants, each lacking one N-glycosylation site at a defined location, after transient expression in Sf9 cells. Four of these mutants migrated faster than wild-type gp64, while the fifth comigrated with wild-type gp64. These results confirmed that four of the five consensus N-glycosylation sites in AcMNPV gp64 are used, identified their positions at amino acids 198, 355, 385, and 426, and revealed that the unused site is located at amino acid 160. It is widely accepted that the consensus recognition sequence Asn-X-Thr/Ser is necessary but not sufficient for protein N-glycosylation in eucaryotic cells, as some are used, some are used inefficiently, and some are not used at all (1, 8, 17). The rules governing site usage remain rather poorly defined, but some information is available. For example, it is clear that the presence of proline in the middle position or immediately following a consensus N-glycosylation site strongly reduces the likelihood of N-glycosylation (8). None of the N-glycosylation sites that are used in AcMNPV gp64 have prolines in either of these positions. In addition, consensus sites containing threonine in the third position are used more frequently than those containing serine in the third position (8, 24). The N-glycosylation site at position 160 of AcMNPV gp64, which is not used, contains serine in the third position. Two of the N-glycosylation sites that are used have threonine, and the other two have serine, in the third position. When serine is found in the third position, the identity of the second amino acid in the recognition site becomes more important and sites that have tryptophan, aspartic acid, glutamic acid, or leucine in the middle position are used inefficiently (23, 41). Both of the used N-glycosylation sites in AcMNPV gp64 that have serine in the third position have asparagine in the middle position. Thus, the rules governing N-glycosylation site usage, in general, appear to be followed during biosynthesis of gp64 in uninfected or AcMNPV-infected Sf9 cells.
Processing of each of the individual glycans on AcMNPV gp64 was assessed by analyzing the endo H sensitivities and monosaccharide compositions of the four triple mutants, each of which contained a single N-linked glycan at a defined position. The results demonstrated that only a subset of the N-linked glycans on AcMNPV gp64 are processed to endo H-resistant structures and revealed that there were differences in the efficiency of processing of individual N-linked glycans. In addition, only those glycans that acquired at least partial endo H resistance contained detectable levels of fucose, and none of the glycans on AcMNPV gp64 were processed to complex structures containing β-linked galactose or α2,6-linked sialic acid. These results indicate that processing of the N-linked glycans on AcMNPV gp64 is subject to positional effects, which is a common feature of glycoproteins, particularly membrane-bound glycoproteins, which often have both high mannose (endo H-sensitive) and processed, complex (endo H-resistant) N-linked glycans (38). The most efficiently processed glycan on AcMNPV gp64 is closest to the amino terminus of the protein, which is consistent with the previous observation that complex glycans are usually located in the N-terminal regions of glycoproteins (38). It also has been observed that complex glycans on glycoproteins which have both types of N-linked side chains are usually found on the amino-terminal side of a high-mannose glycan (38). Interestingly, the two inefficiently processed glycans on AcMNPV gp64 are located on the carboxy-terminal side of a high mannose (endo H-sensitive) glycan. Thus, based on previous observations, we might have predicted that these glycans would be inefficiently processed due to their positions with respect to the high-mannose glycan. On the other hand, if we consider their positions to be absolutely incompatible with any processing, then we would have predicted that these glycans would not have been processed, even inefficiently, based on previous observations.
AcMNPV gp64 mutants with every possible combination of N-glycosylation site mutations were expressed in transiently transfected Sf9 cells, and there was no correlation between expression levels and the numbers or positions of these mutations. As expected, mutants with more mutations migrated faster than mutants with fewer mutations. However, mutants with the same number of mutations did not necessarily comigrate. All of the mutant gp64s comigrated after being completely deglycosylated with endo H, and the differences in the electrophoretic mobilities of different mutants lacking the same number of N-glycosylation sites did not reflect differences in the sizes of their glycans because processing had been blocked at Glc3Man9GlcNAc2 by castanospermine treatment. Therefore, different N-linked glycans apparently contribute in different ways, perhaps via SDS-resistant conformational effects, to the electrophoretic migration of the mutant AcMNPV gp64s.
Recombinant baculoviruses which encoded and contained gp64 mutants lacking one, two, or three N-linked glycans were isolated, and their in vitro one-step growth curves were compared in assays using Sf9 cells as the host. The results demonstrated that the all of the mutant viruses produced lower levels of infectious BV progeny than wild-type AcMNPV. Interestingly, all of the mutants had very similar growth curves, indicating that the loss of a single N-linked glycan reduced infectious progeny production and the loss of one or two additional glycans had no additional effect. Although one triple mutant consistently produced slightly lower levels of infectious progeny than all other mutants, there was no correlation between the number or positions of N-glycosylation site mutations and the magnitude of the reduction in infectious progeny production.
Subsequent experiments were designed to determine the molecular basis for the reduction in infectious progeny production by the mutant viruses. Due to the nature of the one-step growth curve experiment, we had to consider both the relative ability of the mutants to produce BV progeny and the relative infectivity of those progeny. Under the reasonable assumption that gp64 is necessary for BV progeny production, we first examined the relationship between expression levels, cell surface expression, and the one-step growth curves of these viruses. Although not strictly quantitative, the results clearly showed that there was no correlation between either the levels of mutant gp64 expression or the levels of cell surface expression and the levels of infectious progeny production. A comparison of the relative gp64 content of wild-type and mutant BV particles demonstrated that there was little difference and no correlation between the gp64 content and growth properties of these viruses. We also found no correlation between the inherent acid-induced cell fusogenic activities of the mutant gp64s and the growth properties of recombinant viruses containing these mutant proteins. Together, these findings suggested that the differences in the one-step growth curves of the mutant viruses could not be explained by reductions in BV production or defects in the ability of the mutant gp64s to mediate viral penetration.
We have considered the possibility that the gp64 fusion assay does not accurately reflect the fusogenic activities of the wild-type and recombinant virus particles themselves. However, there is no simple way to measure the fusogenic activities of the budded virions in quantitative fashion. Furthermore, the single and double mutants both produced less infectious progeny than the wild type, and they all had wild-type levels of inherent fusogenic activity and similar gp64/capsid ratios. By contrast, the triple mutants produced the same amounts of infectious progeny as the single and double mutants but had significantly less fusogenic activity. Considering these observations, it is highly unlikely that there would be a clear correlation between the relative levels of fusogenic activity in the wild-type and mutant BV particles and their one-step growth curves. The ability of the triple gp64 mutants to produce about the same amounts of infectious progeny as the single and double mutants is significant, for it suggests that the triple-mutant gp64s, despite having much less inherent fusogenic activity than the wild type and the single or double mutants, still provide enough activity to function in viral penetration.
Subsequently, we tested the hypothesis that the growth properties of the mutant baculoviruses can be explained by differences in their binding activities. The results of binding assays demonstrated that the mutant viruses all bound to Sf9 cells more slowly than the wild type. Furthermore, there were no clear differences in binding kinetics among the three classes of mutants. The observed reduction in binding kinetics was the only property we examined that correlated perfectly with the reduction in infectious progeny production by the mutant viruses. Therefore, we conclude that elimination of one or more N-glycosylation sites in AcMNPV gp64 impairs the binding of BV particles containing the mutant protein to Sf9 cells. This explains why viruses containing these mutant forms of gp64 produce less infectious progeny than wild-type gp64. One interpretation of the findings presented in this study is that AcMNPV gp64 has a direct role in binding BV to the host cell; that is, gp64 functions as a viral attachment protein in addition to functioning in viral penetration via its recognized fusogenic activity. If this is true, the differential effect of N-glycosylation site mutations on the binding and fusogenic activities of AcMNPV gp64 would suggest that these two functions are mediated by physically independent domains of the native gp64 molecule. At this time, however, it remains equally possible that alterations in the structure of the mutant gp64s indirectly impair the binding of BV particles to the cell.
ACKNOWLEDGMENTS
We thank Loy Volkman (University of California at Berkeley) for the kind gift of anti-gp64 and anticapsid monoclonal antibodies and for a critical review of the manuscript. We also thank Peter Faulkner (Queen’s University) for providing an anti-gp64 antibody. We thank Aurelio Garcia and Eric Finn for technical assistance on various aspects of this project.
This work was supported by National Institutes of Health award GM49734.
REFERENCES
- 1.Bause E. Structural requirements of N-glycosylation of proteins. Studies with proline peptides as conformational probes. Biochem J. 1983;209:331–336. doi: 10.1042/bj2090331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Blake M S, Johnston K H, Russell-Jones G J, Gotschlich E C. A rapid, sensitive method for detection of alkaline phosphatase conjugated anti-antibody on western blot. Anal Biochem. 1984;36:175–179. doi: 10.1016/0003-2697(84)90320-8. [DOI] [PubMed] [Google Scholar]
- 3.Blissard G W, Rohrmann G F. Location, sequence, transcriptional mapping, and temporal expression of the gp64 envelope glycoprotein gene of the Orgyia pseudotsugata multicapsid nuclear polyhedrosis virus. Virology. 1989;170:537–555. doi: 10.1016/0042-6822(89)90445-5. [DOI] [PubMed] [Google Scholar]
- 4.Blissard G W, Wenz J R. Baculovirus gp64 envelope glycoprotein is sufficient to mediate pH-dependent membrane fusion. J Virol. 1992;66:6829–6835. doi: 10.1128/jvi.66.11.6829-6835.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Braunagel S C, Summers M D. Autographa californica nuclear polyhedrosis virus PDV and ECV viral envelopes and nucleocapsids: structural proteins, antigens, lipid and fatty acid profiles. Virology. 1994;202:315–328. doi: 10.1006/viro.1994.1348. [DOI] [PubMed] [Google Scholar]
- 6.Charlton C A, Volkman L E. Effect of tunicamycin on the structural proteins and infectivity of budded Autographa californica nuclear polyhedrosis virus. Virology. 1986;154:214–218. doi: 10.1016/0042-6822(86)90443-5. [DOI] [PubMed] [Google Scholar]
- 7.Charlton C A, Volkman L E. Penetration of Autographa californica nuclear polyhedrosis virus nucleocapsids into IPLB Sf 21 cells induces actin cable formation. Virology. 1993;197:245–254. doi: 10.1006/viro.1993.1585. [DOI] [PubMed] [Google Scholar]
- 8.Gavel Y, von Heijne G. Sequence differences between glycosylated and nonglycosylated Asn-X-Thr/Ser acceptor sites: implications for protein engineering. Protein Eng. 1990;3:433–442. doi: 10.1093/protein/3.5.433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Goldstein N I, McIntosh A H. Glycoproteins of nuclear polyhedrosis viruses. Arch Virol. 1980;64:119–126. doi: 10.1007/BF01318015. [DOI] [PubMed] [Google Scholar]
- 10.Grace T D C. Establishment of four strains of cells from insect tissues grown in vitro. Nature. 1962;195:788–789. doi: 10.1038/195788a0. [DOI] [PubMed] [Google Scholar]
- 11.Guarino L A, Summers M D. Functional mapping of a trans-activating gene required for expression of a baculovirus delayed-early gene. J Virol. 1986;57:563–571. doi: 10.1128/jvi.57.2.563-571.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Harlow E, Crawford L V, Pim D C, Williamson N M. Monoclonal antibodies specific for simian virus 40 tumor antigens. J Virol. 1981;39:861–869. doi: 10.1128/jvi.39.3.861-869.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hill J E, Faulkner P. Identification of the gp67 gene of a baculovirus pathogenic to the spruce budworm, Choristoneura fumiferana multinucleocapsid nuclear polyhedrosis virus. J Gen Virol. 1994;75:1811–1813. doi: 10.1099/0022-1317-75-7-1811. [DOI] [PubMed] [Google Scholar]
- 14.Hink F. Established insect cell line from the cabbage looper, Trichoplusia ni. Nature. 1970;226:466–467. doi: 10.1038/226466b0. [DOI] [PubMed] [Google Scholar]
- 15.Hohmann A W, Faulkner P. Monoclonal antibodies to baculovirus structural proteins: determination of specificities by Western blot analysis. Virology. 1983;125:432–444. doi: 10.1016/0042-6822(83)90214-3. [DOI] [PubMed] [Google Scholar]
- 16.Hollister J R, Shaper J H, Jarvis D L. Stable expression of mammalian β1,4-galactosyltransferase extends the N-glycosylation pathway in insect cells. Glycobiology. 1998;8:473–480. doi: 10.1093/glycob/8.5.473. [DOI] [PubMed] [Google Scholar]
- 17.Hunt L T, Dayhoff M O. The occurrence in proteins of the tripeptides Asn-X-Ser and Asn-X-Thr and of bound carbohydrate. Biochem Biophys Res Commun. 1970;39:757–765. doi: 10.1016/0006-291x(70)90270-6. [DOI] [PubMed] [Google Scholar]
- 18.Jarvis D L, Finn E E. Biochemical analysis of the N-glycosylation pathway in baculovirus-infected lepidopteran insect cells. Virology. 1995;212:500–511. doi: 10.1006/viro.1995.1508. [DOI] [PubMed] [Google Scholar]
- 19.Jarvis D L, Finn E E. Modifying the insect cell N-glycosylation pathway with immediate early baculovirus expression vectors. Nat Biotechnol. 1996;14:1288–1292. doi: 10.1038/nbt1096-1288. [DOI] [PubMed] [Google Scholar]
- 20.Jarvis D L, Garcia A., Jr Biosynthesis and processing of the Autographa californica nuclear polyhedrosis virus gp64 protein. Virology. 1994;205:300–313. doi: 10.1006/viro.1994.1646. [DOI] [PubMed] [Google Scholar]
- 21.Jarvis D L, Oker-Blom C, Summers M D. Role of glycosylation in the transport of recombinant glycoproteins through the secretory pathway of lepidopteran insect cells. J Cell Biochem. 1990;42:181–191. doi: 10.1002/jcb.240420402. [DOI] [PubMed] [Google Scholar]
- 22.Jarvis D L, Summers M D. Glycosylation and secretion of human tissue plasminogen activator in recombinant baculovirus-infected insect cells. Mol Cell Biol. 1989;9:214–223. doi: 10.1128/mcb.9.1.214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kasturi L, Chen H, Shakin-Eshleman S H. Regulation of N-linked core glycosylation: use of a site-directed mutagenesis approach to identify Asn-Xaa-Ser/Thr sequons that are poor oligosaccharide acceptors. Biochem J. 1997;323:415–419. doi: 10.1042/bj3230415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kasturi L, Eshleman J R, Wunner W H, Shakin-Eshleman S H. The hydroxy amino acid in an Asn-X-Ser/Thr sequon can influence N-linked core glycosylation efficiency and the level of expression of a cell surface glycoprotein. J Biol Chem. 1995;270:14756–14761. doi: 10.1074/jbc.270.24.14756. [DOI] [PubMed] [Google Scholar]
- 25.Keddie B A, Aponte G W, Volkman L E. The pathway of infection of Autographa californica nuclear polyhedrosis virus in an insect host. Science. 1989;243:1728–1730. doi: 10.1126/science.2648574. [DOI] [PubMed] [Google Scholar]
- 26.Kelly D C, Lescott T. Glycosylation of polypeptides synthesized in Trichoplusia ni nuclear polyhedrosis virus-infected cells and the effect of tunicamycin. J Gen Virol. 1983;64:1915–1926. doi: 10.1099/0022-1317-65-7-1183. [DOI] [PubMed] [Google Scholar]
- 27.Kessler S W. Rapid isolation of antigens from cells with a staphylococcal protein A-antibody adsorbent: parameters of the interaction of antibody-antigen complexes with protein A. J Immunol. 1975;115:1617–1624. [PubMed] [Google Scholar]
- 28.Kitts P A, Ayres M D, Possee R D. Linearization of baculovirus DNA enhances the recovery of recombinant virus expression vectors. Nucleic Acids Res. 1990;18:5667–5672. doi: 10.1093/nar/18.19.5667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kunkel T A. Rapid and efficient site specific mutagenesis without phenotypic selection. Proc Natl Acad Sci USA. 1985;82:488–492. doi: 10.1073/pnas.82.2.488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Leikina E, Onaran H O, Zimmerberg J. Acidic pH induces fusion of cells infected with baculovirus to form syncytia. FEBS Lett. 1992;304:221–224. doi: 10.1016/0014-5793(92)80623-O. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Marchalonis J J, Cone R E, Santer V. Enzymatic iodination: a probe for accessible surface proteins of normal and neoplastic lymphocytes. Biochem J. 1971;124:921–927. doi: 10.1042/bj1240921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Miller L K. The baculoviruses. New York, N.Y: Plenum Press; 1997. [Google Scholar]
- 33.Monsma S A, Blissard G W. Identification of a membrane fusion domain and an oligomerization domain in the baculovirus gp64 envelope fusion protein. J Virol. 1995;69:2583–2595. doi: 10.1128/jvi.69.4.2583-2595.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Monsma S A, Oomens A G P, Blissard G W. The gp64 envelope fusion protein is an essential baculovirus protein required for cell to cell transmission of infection. J Virol. 1996;70:4607–4616. doi: 10.1128/jvi.70.7.4607-4616.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Murhammer D W, Goochee C F. Scaleup of insect cell cultures: protective effects of pluronic F-68. Bio/Technology. 1988;6:1411–1418. [Google Scholar]
- 36.O’Reilly D R, Miller L K, Luckow V A. Baculovirus expression vectors. W. H. New York, N.Y: Freeman and Company; 1992. [Google Scholar]
- 37.Oomens A G, Monsma S A, Blissard G W. The baculovirus GP64 envelope fusion protein: synthesis, oligomerization, and processing. Virology. 1995;209:592–603. doi: 10.1006/viro.1995.1291. [DOI] [PubMed] [Google Scholar]
- 38.Pollack L, Atkinson P H. Correlation of glycosylation forms with position in amino acid sequence. J Cell Biol. 1983;97:293–300. doi: 10.1083/jcb.97.2.293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Roberts T E, Faulkner P. Fatty acid acylation of the 67K envelope glycoprotein of a baculovirus: Autographa californica nuclear polyhedrosis virus. Virology. 1989;172:377–381. doi: 10.1016/0042-6822(89)90145-1. [DOI] [PubMed] [Google Scholar]
- 40.Sanger F, Nicklen S, Coulson A R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shakin-Eshleman S H, Spitalnik S L, Kasturi L. The amino acid at the X position of an Asn-X-Ser sequon is an important determinant of N-linked core-glycosylation efficiency. J Biol Chem. 1996;271:6363–6366. doi: 10.1074/jbc.271.11.6363. [DOI] [PubMed] [Google Scholar]
- 42.Stiles B, Wood A H. A study of the glycoproteins of Autographa californica nuclear polyhedrosis virus (AcNPV) Virology. 1983;131:230–241. doi: 10.1016/0042-6822(83)90548-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Stiles B, Wood H A, Hughes P R. Effect of tunicamycin on the infectivity of Autographa californica nuclear polyhedrosis virus. J Invertebr Pathol. 1983;41:405–408. [Google Scholar]
- 44.Summers M D, Smith G E. A manual of methods for baculovirus vectors and insect cell culture procedures. Texas Agricultural Experiment Station Bulletin no. 1555. College Station, Tex: Texas Agricultural Experiment Station; 1987. [Google Scholar]
- 45.Summers M D, Volkman L E. Comparison of biophysical and morphological properties of occluded and extracellular nonoccluded baculovirus from in vivo and in vitro host systems. J Virol. 1976;17:962–972. doi: 10.1128/jvi.17.3.962-972.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tarentino A L, Maley F. Purification and properties of an endo-β-N-acetylglucosaminidase from Streptomyces griseus. J Biol Chem. 1974;249:811–817. [PubMed] [Google Scholar]
- 47.Vaughn J L, Goodwin R H, Thompkins G J, McCawley P. The establishment of two insect cell lines from the insect Spodoptera frugiperda (Lepidoptera:Noctuidae) In Vitro. 1977;13:213–217. doi: 10.1007/BF02615077. [DOI] [PubMed] [Google Scholar]
- 48.Volkman L E, Goldsmith P A. Budded Autographa californica NPV 64K protein: further biochemical analysis and effects of postimmunoprecipitation sample preparation conditions. Virology. 1984;139:295–302. doi: 10.1016/0042-6822(84)90375-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Volkman L E, Goldsmith P A. Mechanism of neutralization of budded Autographa californica nuclear polyhedrosis virus by a monoclonal antibody: inhibition of entry by adsorptive endocytosis. Virology. 1985;143:185–195. doi: 10.1016/0042-6822(85)90107-2. [DOI] [PubMed] [Google Scholar]
- 50.Volkman L E, Goldsmith P A, Hess R T, Faulkner P. Neutralization of budded Autographa californica NPV by a monoclonal antibody: identification of the target antigen. Virology. 1984;133:354–362. doi: 10.1016/0042-6822(84)90401-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Volkman L E, Summers M D. Autographa californica nuclear polyhedrosis virus: comparative infectivity of the occluded, alkali-liberated, and nonoccluded forms. J Invertebr Pathol. 1977;30:102–103. doi: 10.1016/0022-2011(77)90045-3. [DOI] [PubMed] [Google Scholar]
- 52.von Heijne G. Sequence analysis in molecular biology: treasure trove or trivial pursuit. San Diego, Calif: Academic Press, Inc.; 1987. [Google Scholar]
- 53.Whitford M, Stewart S, Kuzio J, Faulkner P. Identification and sequence analysis of a gene encoding gp67, an abundant envelope glycoprotein of the baculovirus Autographa californica nuclear polyhedrosis virus. J Virol. 1989;63:1393–1399. doi: 10.1128/jvi.63.3.1393-1399.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zuidema D, Klinge-Roode E C, van Lent J W, Vlak J M. Construction and analysis of an Autographa californica nuclear polyhedrosis virus mutant lacking the polyhedral envelope. Virology. 1989;173:98–108. doi: 10.1016/0042-6822(89)90225-0. [DOI] [PubMed] [Google Scholar]