Abstract
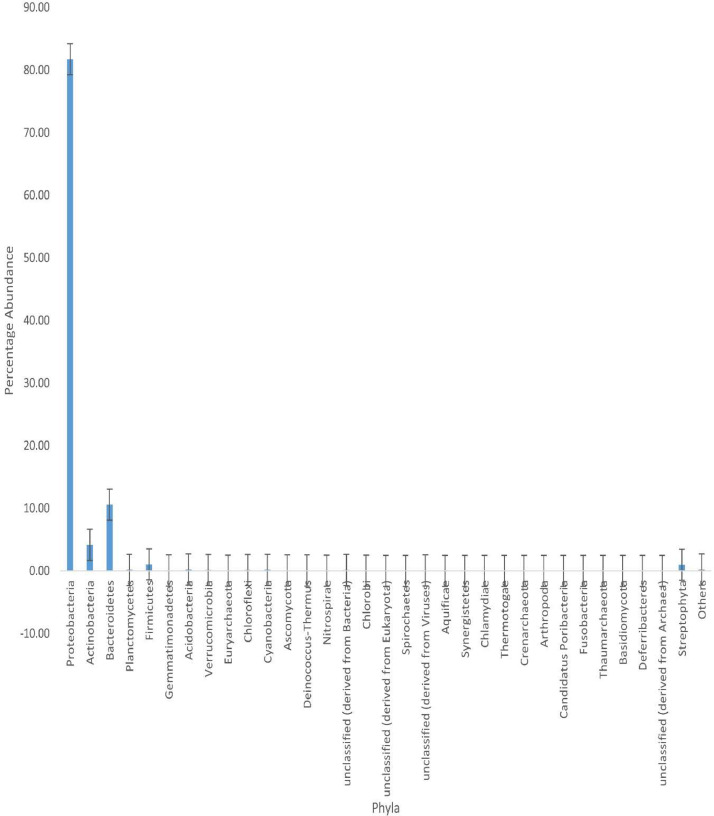
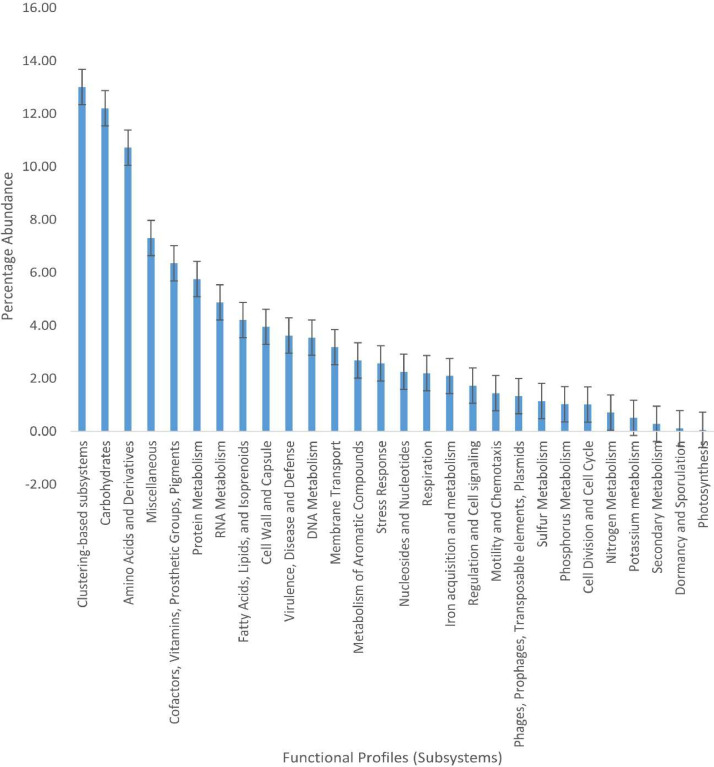
Microorganisms inhabiting caves exhibit medical or biotechnological promise, most of which have been attributed to factors such as antimicrobial activity or the induction of mineral precipitation. This dataset explored the shotgun metagenomic sequencing of the Cango cave microbial community in Oudtshoorn, South Africa. The aimed to elucidate both the structure and function of the microbial community linked to the cave. DNA sequencing was conducted using the Illumina NovaSeq platform, a next-generation sequencing. The data comprises 4,738,604 sequences, with a cumulative size of 1,180,744,252 base pairs and a GC content of 52%. Data derived from the metagenome sequences can be accessed through the bioproject number PRJNA982691 on NCBI. Using an online metagenome server, MG-RAST, the subsystem database revealed that bacteria displayed the highest taxonomical representation, constituting about 98.66%. Archaea accounted for 0.05%, Eukaryotes at 1.20%, viruses were 0.07%, while unclassified sequences had a representation of 0.02%. The most abundant phyla were Proteobacteria (81.74%), Bacteroidetes (10.57%), Actinobacteria (4.16%), Firmicutes (SK‒1.03%), Acidobacteria (0.20), and Planctomycetes (SK‒0.16%). Functional annotation using subsystem analysis revealed that clustering based on subsystems had 13.44%, while amino acids and derivatives comprised 11.41%. Carbohydrates sequences constituted 9.55%, along with other advantageous functional traits essential for growth promotion and plant management.
Keywords: Cave microbiome, Sustainable plant growth, One health, Anthropogenic interference, Unclassified microbiome, Food safety
Specification Table
| Subject | Microbiology |
| Specific subject area | Microbiome |
| Type of data | Raw metagenomics data |
| How the data were acquired | Metagenomic DNA extraction from soil samples from Cango Cave, Next generation sequencing on Illumina (NovaSeq) instrument and metagenomics classification using Ribosomal Database Project (RDP) Technology |
| Data format | Raw data (fastq.gz.file) |
| Data source location | Soil samples from cave located at Oudtshoorn (33o23’32.9886” S 22o12’51.9906” E), Western Cape Province, South Africa |
| Data accessibility | Repository name: National Centre for Biotechnology Information SRA Data Identification Number: PRJNA982691 URL: https://www.ncbi.nlm.nih.gov/bioproject/PRJNA982691 |
1. Value of the Data
-
•
The dataset provides information on the functional and community diversities of microbes associated with cave soil.
-
•
It indicates the influence and peculiarities of cave soil on selecting important microbiome.
-
•
The dataset can provide further insight into the distinctive features (harboring resistomes) of cave metagenome, especially as a means to actualize the objectives of “one Health”.
-
•
This dataset provides preliminary insights into the possibly untapped roles of the culturable and unculturable soil microbes.
-
•
The dataset provides the prospects of finding novel genes of biotechnological importance
2. Background
The advent of next-generation DNA sequencing (NGS) technology, including metagenomics analysis, has provided opportunities to deepen our understanding of the composition and functionality of microbial communities in soil. In this study, we aim to reveal the microbial diversity and functioning of Cango Cave using a shotgun metagenomics approach.
3. Data Description
The metagenomics files (ST1‒SRR24958369, ST3‒SRR24958368, ST4‒SRR24958367, STC‒SRR24958366, STC3‒SRR24958365, STC4‒SRR24958364) under the Bioproject Number PRJNA982691 at NCBI comprises of raw sequences acquired via the shotgun sequencing of soils from Cango cave (ST) and Lawn/control (STC) samples, Western Cape, South Africa. Details of the microbial community and functional structure determined using SEED subsystem were shown in Fig. 1, Fig. 2, respectively.
Fig. 1.
Phyla obtained according to the taxonomic annotation of the Cango cave soil microbiome. Each bar represents the mean ± standard error of each phyla component recovered.
Fig. 2.
Combined functional profiles of Cango cave soil microbiome dataset using SEED subsystem. Each bar represents the mean ± standard error of each functional component recovered.
4. Experimental Design, Materials and Methods
Soil samples from the cave located at Western Cape (33o23’32.9886” S 22o12’51.9906” E) were collected from three different locations inside the dark zone of the cave (ST), about 20–50 m from the cave entrance (within/core of the cave) and also lawn area (surrounding soil) of the cave to serve as a control. The gathered samples were conveyed to the laboratory in a cooler box filled with ice and were subsequently stored at ‒20°C for a duration of one week [1]. DNA extraction from 5 g of each soil sample was performed using the DNeasy PowerMax soil kit in accordance with the manufacturer's instructions. Subsequently, the libraries were prepared using the Nextera DNA Flex library preparation kit (New York, USA). To prepare the libraries, 20 to 50 ng of DNA was used. The samples underwent fragmentation, followed by the addition of adapter sequences. The final concentrations of the libraries were assessed using the Qubit double-stranded DNA (dsDNA) HS assay kit from Life Technologies, and the average DNA fragment lengths were determined using a 2100 Bioanalyzer from Agilent Technologies. Subsequently, the libraries were pooled, diluted to 0.6 nM, and subjected to paired-end sequencing for 300 cycles using the NovaSeq system from Illumina. The downstream analysis of the reads was conducted using the default settings of the Metagenomic Rapid Annotations using Subsystems Technology (MG-RAST) server v4.0.3. Within the MG-RAST server, quality control of raw reads was executed through SolexaQA to trim low-quality reads and dereplicate the metagenomic data. Assessment of sample sequencing error, based on artificial duplicate read measurements, was accomplished using duplicate read inferred sequencing error estimation (DRISEE). Additionally, the pipeline employed the Bowtie aligner to screen the reads for unwanted genomes associated with model organisms such as mice, humans, cows, and other animals [2]. Using the same pipeline, the BLAST-like alignment tool (BLAT) algorithm was applied to annotate the sequences [3] against the M5NR database [4], which offers a non-redundant compilation of various databases.
Limitations
Not Applicable.
Ethics Statement
The study follows the ethical requirements for publication in Data in Brief. It does not involve human subjects, animal experiments, or any data collected from social media platforms.
CRediT authorship contribution statement
Olubukola Oluranti Babalola: Conceptualization, Methodology, Validation, Formal analysis, Investigation, Writing – review & editing, Supervision. Afeez Adesina Adedayo: Methodology, Validation, Formal analysis, Visualization. Saheed Adekunle Akinola: Conceptualization, Methodology, Validation, Formal analysis, Investigation, Writing – original draft, Visualization.
Acknowledgment
This research was funded by the National Research Foundation (ZA) grant (UID123634 and UID132595) awarded to OOB.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.dib.2024.110381.
Appendix. Supplementary materials
Data Availability
References
- 1.Babalola O.O., Akinola S.A., Ayangbenro A.S. Shotgun metagenomic survey of maize soil rhizobiome. Microbiol. Resour. Announc. 2020;9(39) doi: 10.1128/MRA.00860-20. e00860-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Meyer F., et al. The metagenomics RAST server—a public resource for the automatic phylogenetic and functional analysis of metagenomes. BMC Bioinform. 2008;9:386. doi: 10.1186/1471-2105-9-386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kent W.J. BLAT—the BLAST-like alignment tool. J. Genome Res. 2002;12(4):656–664. doi: 10.1101/gr.229202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wilke A., et al. The M5nr: a novel non-redundant database containing protein sequences and annotations from multiple sources and associated tools. BMC Bioinform. 2012;13(1):141. doi: 10.1186/1471-2105-13-141. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.