Summary
Biological functions of glycans are intimately linked to fine details in branches and linkages, which make structural identification extremely challenging. Here, we present a protocol for automated N-glycan sequencing using multi-stage mass spectrometry (MSn). We describe steps for release/purification and derivation of glycans and procedures for MSn scanning. We then detail “glycan intelligent precursor selection” to computationally guide MSn experiments. The protocol can be used for both discrete individual glycans and isomeric glycan mixtures.
For complete details on the use and execution of this protocol, please refer to Sun et al.,1 Huang et al.,2 and Huang et al.3
Subject areas: Bioinformatics, Protein Biochemistry, Biotechnology and bioengineering, Chemistry, Computer sciences
Graphical abstract

Highlights
-
•
Protocol for identification of discrete N-glycan and isomeric glycan mixtures
-
•
Glycan identification is based on mass spectrometry (MS)
-
•
Intelligent fragmentation is proposed to instruct MS scanning
-
•
Sophisticated computer program with integrated algorithms for glycan identification
Publisher’s note: Undertaking any experimental protocol requires adherence to local institutional guidelines for laboratory safety and ethics.
Biological functions of glycans are intimately linked to fine details in branches and linkages, which make structural identification extremely challenging. Here, we present a protocol for automated N-glycan sequencing using multi-stage mass spectrometry (MSn). We describe steps for release/purification and derivation of glycans and procedures for MSn scanning. We then detail “glycan intelligent precursor selection” to computationally guide MSn experiments. The protocol can be used for both discrete individual glycans and isomeric glycan mixtures.
Before you begin
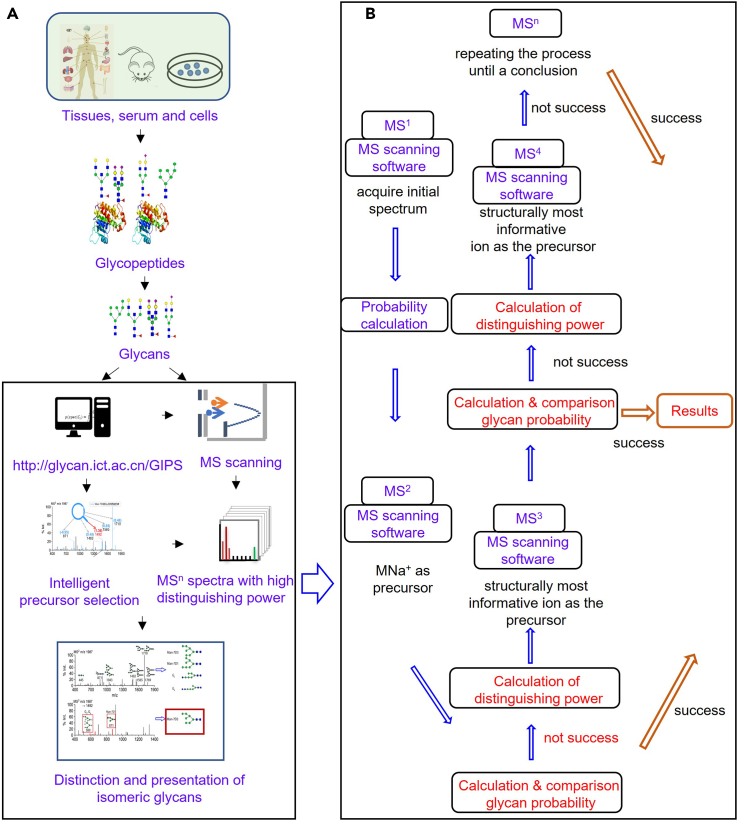
The main processes of the method involve release/purification of N-glycans/oligosaccharides and automated identification of purified individual glycans or glycan mixture containing isomeric glycans (Figure 1). The protocol below describes the specific steps for releasing N-glycans from glycoproteins, and also presents the steps for purifying carbohydrates including N-glycans released from glycoproteins/ glycolipids and discrete oligosaccharides in biological samples. This protocol presents the process of automated identification of glycan structures using GIPS strategy and sophisticated computer program.1,2,3 Before identification of glycans, buffers for enzymatic reaction, reduction solution, solution for equilibrating cartridge, and eluting solution for purification need to be prepared.
Figure 1.
Workflow of the automated glycan sequencing using mass spectrometry and computer-assisted intelligent fragmentation
(A), steps for glycan release from biological samples and mass spectrometry analysis, and (B), operations using glycan intelligent precursor selection (GIPS) strategy.
Preparation of enzymatic buffer
Timing: 30 min (Steps 1 and 2)
-
1.Preparation of Tris-HAc buffer.
-
a.Weigh 7.26 g Tris and put it into a glass bottle, and dissolve Tris in 99 mL ultra-pure water.
-
b.Add 1 mL acetic acid to Tris solution, and test the pH.
-
c.Adjust to pH 8.5 with several drops of acetic acid (usually <5 drops using 1 mL pipette).
-
d.Degas the Tris-HAc buffer by passing a slow stream of deoxygenated nitrogen through the solution with a glass dropper for 20 min to avoid the absorption of carbon dioxide in the atmosphere.
-
a.
-
2.Preparation of NH4HCO3-NH3·H2O buffer.
-
a.Weigh 0.3953 g ammonium bicarbonate (NH4HCO3), and dissolve it in the 100 mL ultra-pure water.
-
b.Add 2–3 drops of ammonia using 1 mL pipette to adjust to pH 8.5.
-
c.Prepare fresh buffer for one-time use.
-
a.
CRITICAL: Measure the pH value at the room temperature or the value is not accurate.
Preparation of reagents for solid phase extraction
Timing: 30 min (Steps 3 and 4)
-
3.Preparation of 5% acetic acid solution.
-
a.Add 10 mL acetic acid to a glass bottle.
-
b.Add 190 mL ultra-pure water to the glass bottle.
-
c.Seal the bottle to avoid evaporation of the acetic acid, and store it at room temperature (22°C–26°C).
-
a.
-
4.Preparation of 20% and 40% n-propanol.
-
a.Add 20 mL n-propanol to a clean glass bottle.
-
b.Add 80 mL 5% acetic acid to the glass bottle.
-
c.Add 40 mL n-propanol to a separate clean glass bottle.
-
d.Add 60 mL 5% acetic acid to the separate glass bottle.
-
a.
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Biological samples | ||
| Human serum | Institute of Biophysics | – |
| Animal tissues | Institute of Biophysics | – |
| Human IgG | Sigma-Aldrich | Cat#I4506 |
| Bovine serum albumin (BSA) | Sigma-Aldrich | Cat#A1933 |
| Chemicals, peptides, and recombinant proteins | ||
| DL-dithiothreitol (DTT) | Sigma-Aldrich | Cat#43815 |
| Iodoacetamide (IAM) | Sigma-Aldrich | Cat#1149 |
| 2,5-Dihydroxybenzoic acid (DHB) | Sigma-Aldrich | Cat#39319 |
| Ammonium bicarbonate | Sigma-Aldrich | Cat#09830 |
| Sodium hydroxide | Sigma-Aldrich | Cat#S8045 |
| Trypsin | Promega | Cat#V5113 |
| PNGase F | New England Biolabs | Cat#P704L |
| Dimethyl sulfoxide (DMSO) | Sigma-Aldrich | Cat#D2650 |
| Methyl iodide | Energy Chemical | Cat#W610705 |
| Tris(hydroxymethyl)aminomethane | Sigma-Aldrich | Cat#252859 |
| Man-7D3 | Dextra | Cat#MC0932 |
| Acetic acid (HAc) | Sigma-Aldrich | Cat#695092 |
| n-Propanol | Sigma-Aldrich | Cat#1.00997 |
| Ammonia | Sigma-Aldrich | Cat#5438300250 |
| Pierce RIPA buffer | Thermo Fisher Scientific | Cat#89900 |
| Chloroform | Sigma-Aldrich | Cat#372978 |
| Trifluoroacetic acid (TFA) | Sigma-Aldrich | Cat#302031 |
| Pierce BCA Protein Assay Kits | Thermo Fisher Scientific | Cat#23227 |
| Software and algorithms | ||
| GlycoWorkbench | Ceroni et al., 2008 | http://www.eurocarbdb.org/ms-tools |
| GIPS | Sun et al., 2018 | http://glycan.ict.ac.cn/GIPS/ |
| IntelliJ IDEA | – | https://www.jetbrains.com.cn/idea/ |
| Java SE Development Kit 8u391 | – | https://www.oracle.com/java/technologies/javase/javase-jdk8-downloads.html |
| Shimadzu Biotech Launchpad | – | https://shimadzu-biotech-launchpad.software.informer.com/2.8/ |
| Other | ||
| Sep-Pack C18 96-well | Waters | Cat#186003966 |
| Oasis HLB 96-well plate | Waters | Cat# WAT058951 |
| μfocus MALDI plate | Hudson Surface | Cat#PL-PC-000050-P |
| 3K ultrafiltration tube | Millipore | Cat#UFC500396 |
| Oil-less pump | AutoBo Elec. Technology | Cat#AP-550C/V |
| 96-well adaptor | Waters | Elaborated designed upon request |
Materials and equipment
-
•
20% n-Propanol in 5% acetic acid.
| Reagent | Final concentration | Amount |
|---|---|---|
| n-Propanol | 20% | 20 mL |
| Acetic acid | 4% | 4 mL |
| Deionized water | N/A | 76 mL |
| Total | N/A | 100 mL |
Note on storage conditions: room temperature (22°C–26°C), freshly prepared for use.
CRITICAL: n-Propanol is flammable. Store the prepared solution in an explosion proof freezer that can safely store flammable materials. Keep away from heat, sparks, and open flames. Steam of n-propanol can cause damage to nervous, eyes, nose and throat. Long-term exposure to n-propanol can cause dryness and chapped skin. n-Propanol is toxic if swallowed. To prevent exposure, wear protective gloves, protective clothing, and eye protection, and use in the fume hood.
-
•
40% n-Propanol in 5% acetic acid.
| Reagent | Final concentration | Amount |
|---|---|---|
| n-Propanol | 40% | 40 mL |
| Acetic acid | 3% | 3 mL |
| Deionized water | N/A | 57 mL |
| Total | N/A | 100 mL |
Note on storage conditions: room temperature (22°C–26°C), freshly prepared for use.
CRITICAL: n-Propanol is flammable. Store the prepared solution in an explosion proof freezer that can safely store flammable materials. Keep away from heat, sparks, and open flames. Steam of n-Propanol can cause damage to nervous, eyes, nose and throat. Long-term exposure to n-propanol can cause dryness and chapped skin. n-Propanol is toxic if swallowed. To prevent exposure, wear protective gloves, protective clothing, and eye protection, and use in the fume hood.
-
•
5% Acetic acid solution: add 5 mL acetic acid in 95 mL deionized water.
Note on storage conditions: room temperature (22°C–26°C), freshly prepared for use.
-
•
15% Acetonitrile (ACN) solution: add 15 mL ACN in 85 mL deionized water.
Note on storage conditions: room temperature (22°C–26°C), freshly prepared for use.
CRITICAL: Acetonitrile is highly flammable. Store the prepared solution in an explosion proof freezer that can safely store flammable materials. Acetonitrile can cause irritation to eyes, and is harmful by inhalation, in contact with skin and if swallowed. To prevent exposure, wear protective gloves, protective clothing, and eye protection, and use in the fume hood.
-
•
50% Acetonitrile (ACN) solution: add 50 mL ACN in 50 mL deionized water.
Note on storage conditions: room temperature (22°C–26°C), freshly prepared for use.
CRITICAL: Acetonitrile is highly flammable. Store the prepared solution in an explosion proof freezer that can safely store flammable materials. Acetonitrile can cause irritation to eyes, and is harmful by inhalation, in contact with skin and if swallowed. To prevent exposure, wear protective gloves, protective clothing, and eye protection, and use in the fume hood.
-
•
75% Acetonitrile (ACN) solution: add 75 mL ACN in 15 mL deionized water.
Note on storage conditions: room temperature (22°C–26°C), freshly prepared for use.
CRITICAL: Acetonitrile is highly flammable. Store the prepared solution in an explosion proof freezer that can safely store flammable materials. Acetonitrile can cause irritation to eyes, and is harmful by inhalation, in contact with skin and if swallowed. To prevent exposure, wear protective gloves, protective clothing, and eye protection, and use in the fume hood.
-
•
50% Methanol solution: add 50 mL methanol in 50 mL deionized water.
Note on storage conditions: room temperature (22°C–26°C), freshly prepared for use.
CRITICAL: Methanol is flammable. Store the prepared solution in an explosion proof freezer that can safely store flammable materials. Keep away from heat, sparks, and open flames. Methanol is toxic if swallowed. Methanol can cause eye damage, skin irritation, and breathing difficulties if inhaled. To prevent exposure, wear protective gloves, protective clothing, and eye protection and use in the fume hood.
Step-by-step method details
Extraction of proteins from different kinds of biological samples
Timing: 3 h (Steps 1–3)
Steps for extracting proteins from biological samples include treatment of the biological samples, the following centrifugation of treated biological samples and evaluation of the amounts of extracted proteins.
-
1.Treatment of biological samples.
-
a.Treatment of biological samples, tissues.
-
i.Cut the fresh/frozen tissues into pieces with a scalpel.
-
ii.Grind the crushed tissues in liquid nitrogen with a mortar and pestle until the tissues become mucilage.
-
iii.Add 1.0 mL RIPA buffer to the tissue to form a homogeneous solution.
-
i.
-
b.Treatment of biological samples, cells.
-
i.Scrap cells with a minimum number of 106 from the cell culture dishes.
-
ii.Wash cells with deionized water several times to remove any residues from the culture medium.
-
iii.Cleave cells into small debris in an ultrasonic well for 20 min.
-
i.
-
c.Treatment of biological samples, serum.
-
i.Extract 10 μL pooled serum from a Corning cryogenic vial using pipette.
-
ii.Lyophilize 10 μL serum.
-
iii.Redissolve the serum in 200 μL Tris-HAc buffer.
-
i.
-
a.
-
2.Centrifugation of treated biological samples to extract proteins.
-
a.Tissues.
-
i.Centrifuge the homogeneity of prepared tissues at 12000 g.
-
ii.Transfer the aqueous solution into a clean vial on ice.
-
i.
-
b.Cells.
-
i.Centrifuge the cell suspension for 20 min at 12000 g.
-
ii.Transfer the upper layer of aqueous solution into a clean vial on ice.
-
iii.Ultrasonic the cell pellets with addition of 500 μL water twice.
-
iv.Combine the upper layers with the aforementioned aqueous solution in step p.
-
i.
-
a.
-
3.
Evaluation of the amounts of extracted proteins.
Concentrations of extracted proteins from biological samples are determined using the Pierce BCA Protein Assay Kit which is a high-precision, detergent-compatible protein assay. We performed the concentration assay following manufacturers’ instructions, and provided detailed operations as follows.-
a.Preparation of working solution.
-
i.Mix A and B solution in the ratio of 50:1.
-
ii.Prepare BSA standards with the concentration of 0.00125 μg/μL, 0.25 μg/μL, 0.5 μg/μL, 0.75 μg/μL, 1 μg/μL, 1.5 μg/μL and 2 μg/μL.
-
i.
-
b.Reaction and incubation.
-
i.Add 95 μL working solution to 8 holes of the 96-well plate.
-
ii.Add 5 μL standard BSA solutions with 8 concentrations to the aforementioned 8 holes containing working solution.
-
iii.Triplicated BSA and working solution were added.
-
iv.Dilute the BSA and working solution with equal deionized water.
-
v.Add 5 μL samples to the diluted working solution and BSA.
-
vi.Add triplicated samples to working solution and BSA.
-
vii.Incubate the reacting solution at 37°C with continuous shaking.
-
viii.Detect the absorbance at 562 nm in all the reacted solution using Microplate reader.
-
i.
-
a.
Reduction and carboxymethylation of glycoproteins
Timing: 3 h (Steps 4–6)
This step involves the reduction of disulfide bonds in glycoproteins, the subsequent carboxymethylation of sulfydryl groups to avoid regeneration of disulfide bonds, and elimination of redundant chemicals. Reduction and carboxymethylation of glycoproteins can improve the efficiency of tryptic digestion.
-
4.Reduction of disulfide bonds in glycoproteins.
-
a.Weight out 10 mg of DTT.
-
b.Dissolve DTT in 1 mL Tris-HAc buffer to prepare 10 mg/mL reducing solution.
-
c.Add 200 μL reducing solution to 10 μg proteins extracted from biological samples.
-
d.Incubate the samples for 60 min at 37°C.
-
a.
-
5.Carboxymethylation of glycoproteins.
-
a.Weight out 10 mg of IAM.
-
b.Dissolve IAM in 1 mL Tris-HAc buffer to prepare 10 mg/mL carboxymethylation solution.
-
c.Add 200 μL IAM to the reduced glycoproteins with DTT.
-
d.Incubate the samples with wrapped foil in the dark at room temperature (22°C–26°C).
-
a.
-
6.Elimination of redundant chemicals.
-
a.Transfer the sample incubated with DTT and IAM to a 3K ultrafiltration tube.
-
b.Centrifuge the sample for 10 min at 12000 g.
-
c.Discard the solution passing through the membranes after filtration.
-
d.Add 200 μL Ambic buffer to the 3K ultrafiltration tube to change the solvent system, and mix with the sample reserved in the tube.
-
e.Centrifuge the sample for 10 min at 12000 g.
-
f.Discard the solution passing through the membranes after filtration.
-
g.Repeat the buffer transference and centrifugation steps for additional 3 times.
-
h.Transfer the reserved sample into a clean vial by inverting the 3K ultrafiltration tube during centrifugation.
-
a.
Cleavage of glycoproteins into peptides
Timing: 16–24 h (Steps 7–10)
Cleavage of glycopeptides into relatively small peptides/glycopeptides prior to digestion by glycosidases allows thoroughly release of N-glycans without the use of detergents which might influence the subsequent MS scanning.
-
7.
Add trypsin to the sample with the protein/trypsin ratio of 1:50 (w/w).
-
8.
Incubate at 37°C for 16 h.
-
9.
Terminate the tryptic digestion by heating the samples at 100°C for 2 min.
-
10.
Add 10 μL acetic acid to the sample to eliminate the activity of trypsin. Troubleshooting 1.
Extraction and purification of peptides/glycopeptides
Timing: 12 h (Steps 11–21)
Purification of peptides is performed using 96-well based solid phase extraction (SPE, Waters).4 A negative pressure extraction system is elaborately designed. The negative pressure extraction system is equipped with an Oil-less pump, a 96-well adaptor (Waters, MA) and a cut-off valve to adjust pressure. Equilibration of the 96-well plate cartridges, sample loading and sample elution are performed under negative pressure.
-
11.
Add 1 mL methanol to the 96-well cartridge.
-
12.
Add 1 mL 5% acetic acid to the 96-well cartridge.
-
13.
Add 1 mL 100% n-propanol to the 96-well cartridge.
-
14.
Add 1 mL 5% acetic acid to the 96-well cartridge.
-
15.
Repeat the addition of 1 mL 5% acetic acid twice.
-
16.
Load the sample onto the 96-well cartridge. Troubleshooting 2.
-
17.
Repeat the sample-loading step twice.
-
18.
Add 1 mL 5% acetic acid to the cartridge, and discard this fraction to remove any hexose contamination after tryptic digestion.
-
19.
Elute the peptides on the cartridge with 500 mL 20% n-propanol, and collect this fraction.
-
20.
Elute the peptides on the cartridge with 500 mL 40% n-propanol, and collect this fraction.
-
21.
Combine 20% n-propanol and 40% n-propanol fractions, and lyophilize the samples.
Release of N-glycans from glycopeptides
Timing: 24–36 h (Steps 22–25)
N-glycans are released from glycopeptides using N-glycosidase F digestion.
-
22.
Dissolve the propanol fractions after lyophilization in 200 μL ammonium bicarbonate buffer.
-
23.
Add PNGase F to the sample with the ratio 1U:10 μg proteins.
-
24.
Incubate the mixture of sample and glycosidases at 37°C for 24 h.
-
25.
Centrifuge and lyophilize the samples.
Separation and purification of N-glycans
Timing: 8–10 h (Steps 26–37)
N-glycans released from glycoproteins are separated from peptides and O-glycopeptides using 96-well based Sep-Pack C18 plates.
-
26.
Condition the Sep-Pack C18 cartridges in 96-well plate with 1 mL methanol.
-
27.
Condition the Sep-Pack C18 cartridges in 96-well plate with 1 mL 5% acetic acid.
-
28.
Condition the Sep-Pack C18 cartridges in 96-well plate with 1 mL 100% n-propanol.
-
29.
Condition the Sep-Pack C18 cartridges in 96-well plate with 1 mL 5% acetic acid in triplicated.
-
30.
Dissolve the sample in 500 μL acetic acid.
-
31.
Load the samples onto the cartridges for two times.
-
32.
Elute samples with 1 mL 5% acetic acid, and collect this fraction.
-
33.
Elute the samples with 1 mL 20% n-propanol.
-
34.
Elute the samples with 1 mL 40% n-propanol.
-
35.
Elute the samples with 1 mL 100% n-propanol.
-
36.
Collect these propanol fractions containing peptides and O-glycopeptides.
-
37.
Lyophilize the collected 5% acetic acid fractions and propanol fractions separately in clean glass tubes.
Permethylation of purified N-glycans
Timing: 2–3 h (Steps 38–56)
Permethylated N-glycans can be easily ionized during MS scanning, and the loss of acidic residues will be minimized which is extremely important for glycan identification.5
-
38.
Weigh 4 g NaOH particles, and grind the NaOH particles with a pestle gently for 3 min.
-
39.
Add 5 mL DMSO to mix with the NaOH powders.
-
40.
Grind the mixture of NaOH and DMSO with a pestle gently for 1 min to form slurry.
CRITICAL: The grinding of NaOH and mixture with DMSO should be completed fairly fast to avoid the absorption of moisture from the atmosphere. Wear protective clothing to avoid corrosion of the NaOH slurry to skin.
-
41.
Add 0.5 mL of the slurry to the lyophilized 5% acetic acid fraction with a glass pipette.
-
42.
Add 0.5 mL methyl iodide to the sample.
-
43.
Add 0.8 mL DMSO to the sample, and vortex slightly.
-
44.
Agitate the reaction mixture on an automatic shaker machine at room temperature (22°C–26°C) for 20 min until the sample becomes a white homogeneous solution as milk. Troubleshooting 3.
-
45.
Quench the reaction by dropwise addition of the ultrapure water with constant shaking to lessen the effects of the highly exothermic reaction.
-
46.
Repeat the addition of water until fizzing stops.
-
47.
Add 1 mL chloroform to the reacted sample, and mix thoroughly with the sample.
-
48.
Centrifuge the mixture at room temperature (22°C–26°C) for 5 min to allow the mixture to settle into two layers.
-
49.
Transfer the bottom layer solution to a clean tube.
-
50.
Repeat the extraction steps by adding another 1 mL chloroform to the aqueous solution.
-
51.
Combine the chloroform layers extracted from the samples.
-
52.
Wash the lower chloroform layer by adding equivalent ultrapure water, and mix thoroughly with the chloroform layers.
-
53.
Centrifuge the mixture, and discard the upper aqueous layer.
-
54.
Repeat the washing steps for additional five times.
-
55.
Dry down the chloroform layer under a gentle stream of nitrogen.
-
56.
Purify the mixture by 96-well Sep-Pack C18 purification.
Purification of permethylated N-glycans by 96-well Sep-Pack C18 cartridge
Timing: 7–8 h (Steps 57–62)
-
57.
Condition the 96-well Sep-Pack C18 cartridge by adding 1 mL methanol, 1 mL ultrapure water, 1 mL acetonitrile (ACN) and 1 mL ultrapure water in triplicated successively.
-
58.
Dissolve the sample in 200 μL 1:1 methanol: ultrapure water.
-
59.
Load the sample onto the 96-well Sep-Pack C18 cartridge twice.
-
60.
Wash the cartridge with 1 mL ultrapure water.
-
61.
Elute the sample stepwise by 1 mL 15% ACN, 50% ACN and 75% ACN, and collect these ACN fractions.
-
62.
Lyophilize the collected fractions in clean glass tubes.
Mass spectrometry analysis
Timing: 5–10 min (Steps 63–72)
-
63.
Weigh 15–20 mg 2,5-Dihydroxybenzoic (DHB), and add 50% ACN and 0.1% Trifluoroacetic acid to prepare 20 mg/mL DHB matrix solution.
-
64.
Vortex the mixture thoroughly until all the chemicals dissolved completely.
-
65.
Dissolve the lyophilized glycans by adding 10–50 μL methanol.
-
66.
Mix 1 μL glycans with 1 μL freshly prepared DHB matrix to form a mixture of sample and matrix.
-
67.
Spot 1 μL mixture of sample and matrix to the μfocus MALDI plate target, and allow it to dry at room temperature (22°C–26°C). Troubleshooting 4.
-
68.
Set the MALDI instrument working in positive mode, and perform the calibration steps using standard peptides at the mass range of 200–6000 Da to achieve mass accuracy within 10 mDa.
-
69.
Use appropriate laser power (e.g., 80–120 for Shimadzu Resonance instrument) for primary mass spectrometry scanning, and the mass range is from 200 to 6000 Da.
-
70.
Select the standard mass resolution (e.g., FWHM 250 for Shimadzu Resonance instrument) and Collision-Induced Dissociation (CID) energy (100–400 mV for Shimadzu Resonance instrument) for MSn scanning.6,7,8,9,10
Note: The mass range for MSn (n ≥ 2) scanning is dependent on the mass of permethylated precursor ions (perMNa+). The mass range is set to be from 200 Da to (perMNa++200) Da.
-
71.
Open the instrument gate through the Shimadzu Biotech MALDI-MS software, and introduce the sample plate into the gate.
-
72.
Perform the primary MS scanning, and store the mass spectra as mzXML files.
GIPS operations for identification of glycan structures
Timing: 10 s to 5 min (Steps 73–101)
-
73.
Go to the GIPS website (http://glycan.ict.ac.cn/GIPS).
Note: Researchers can go to the GIPS website or download the GIPS software for identification. The GIPS software is available upon request, and detailed operating procedure is presented in Methods video S1.
-
74.
GIPS searches glycans in CarbBank11 database with the same molecular mass as that in the primary MS spectrum, and list these glycans as candidates.
-
75.
GIPS enumerates all the possible combinations of all the candidates, and lists these combinations as candidate groups.
Note: Researchers can skip this step if a single glycan is known to be present in the sample.
-
76.
Acquire the product-ion spectrum (MS2 spectra) of the precursor ions in the primary mass spectrum, and store the mass spectra as mzXML files.
-
77.
Click the central blank area to select the file or drag the file you want to upload, and click the Upload File button after selecting the target file.
-
78.
GIPS begins to analyze the MS2 spectrum file after uploading the file, and calculate the probability of each candidate or candidate group.
-
79.
GIPS presents all candidate structures or candidate groups by ranking probability.
-
80.
If the probability of one candidate or candidate group is above 0.70, MS scanning and calculation terminate.
-
81.
Consider the candidate structure or candidate group with probability above 0.70 as the final result.
Note: In this step, GIPS ranks all candidate glycans or candidate groups according to probability. If MSn is not required to perform, from the list of ranked possible structures, researchers can determine one or several structures as the final result according to the rank of probability.
-
82.
If the probability of each candidate or candidate group is below 0.70, GIPS calculates the distinguishing power (DP) of peaks in the MS2 spectrum.4,5,6
-
83.
Click the Select Peaks button, and GIPS then starts to calculate their DPs using its functions and algorithm.
-
84.
GIPS marks the fragment ion peak with the highest DP in red color, and this fragment ion peak is selected to use as the precursor.
-
85.
Users utilize fragment ion peak in red as precursor for the next-stage MS scanning through the mass spectrometer software.
-
86.
Perform MS3 scanning, and store the product-ion spectrum as mzXML file. Troubleshooting 5.
-
87.
Click on the text area of the GIPS website to open the uploading window, and then upload the newly generated MS3 spectrum from the selected precursor ions.
-
88.
GIPS begins to analyze the file after uploading the file, and recalculates and updates the probability.
-
89.
GIPS presents all candidate structures or candidate groups by ranking probabilities in the website page.
-
90.
If the probability of one candidate or candidate group is above 0.70, MS scanning and calculation terminate.
-
91.
If the probability of each candidate or candidate group is below 0.70, GIPS calculates the DPs of all ion peaks in the MS2 and MS3 spectra.
-
92.
Click the Select Peaks button again in the GIPS website, and DP values will then be labeled on the peaks.
-
93.
GIPS differentiates peaks in different colors without detailed DP values, and click the Zoom in button to see detailed DP values.
CRITICAL: There is more than one chart on this page.
-
94.
Select the peak with the highest DP value as the precursor for next-stage MS scanning using the mass spectrometer software.
-
95.
Perform the next-stage MS scanning, and store the product-ion spectrum as the mzXML file. Troubleshooting 5.
-
96.
Upload the newly generated product-ion spectrum in the GIPS website.
-
97.
GIPS begins to analyze the file after uploading, and recalculates the probability of each candidate or candidate group.
-
98.
Repeat the peak selection and MS scanning steps until the probability of one candidate or candidate group is above 0.70.
-
99.
The candidate structure with probability above 0.70 is defined as the actual glycan in the sample. For isomeric glycans in the mixture, glycan structures in the candidate group with probability above 0.70 are determined as the glycans simultaneously present in the sample.
-
100.
Click the Export menu to export results in the website page, and store the result file in right places.
-
101.
Click Restart menu to start a new analysis project.
Expected outcomes
The primary MS and MS2 spectra, and the produced MS3 and MS4 spectra guided by GIPS instructions (the left panel), and the corresponding probabilities after addition of new MS spectra (right panel).
The protocol and strategy presented here was adapted and optimized from previously published protocols for glycan identification.1,2,3 Using the high mannose N-glycan Man-7D3 as an example, we describe the main anticipated outcomes and results of the protocol.
MS1 of Man-7D3 gave a MNa+ at m/z 1988.0 (Figure 2), indicating a glycan with a molecular mass of 1965.0 Da. A total of 9 candidate glycans were extracted from the database and listed as G1–G97. The probability of each candidate glycan was initially calculated as equal at 0.11. As this is below the termination threshold 0.70, the GIPS program instructed the instrument to carry out MS2 using the only ion MNa+ at m/z 1988.0 as the precursor (Figure 2).
Figure 2.
Identification process of Man-7D3 using mass spectrometry and computer-assisted intelligent fragmentation
When the MS2 spectrum was obtained, the probability of the 9 candidate glycans were re-calculated and updated. The underlying reason for recalculation is that the fragment ions produced in MS2 could favor certain candidates and discriminate against others, and thus the probabilities are no longer equal. In the present case, the updated probabilities for three candidate glycans increased from 0.11 to 0.32, while others decreased to nearly 0.00 (Figure 2). As none of the probabilities exceeded 0.70, further round of product-ion scanning (MS3) is required. Unlike MS2, which uses the only molecular ion MNa+ in the MS1 spectrum as the precursor, multiple fragment ions were present in the spectrum of MS2 and could be used as the precursor for MS3. The distinguishing power of the fragment ion peaks was calculated, and fragment ions with m/z 1492.2 had the highest DP value compared with other ions which were selected as the precursor for next-stage fragmentation. Based on the newly generated spectrum of MS3 (Figure 2), together with that of MS2, the probability of each candidate glycan was re-calculated. Among the updated probabilities, G1 had the highest DP value (0.57), and G2 and G3 had a moderate (0.21), while all others were close to 0 (Figure 2). Again, as none of the probabilities exceeded 0.70, further round of product-ion scanning is required. Fragment ions at m/z 825.2 were selected as the precursor ions for further MS scanning due to the highest DP value. Based on the newly generated spectrum of MS4 (Figure 2), together with those of MS2 and MS3, the probability of each candidate glycan was re-calculated. Among the updated probabilities, G1 had the highest (0.90), while all others were close to 0 (Figure 2). Thus, the identification process stopped with G1 reported as the actual glycan, as its probability exceeded the termination threshold 0.70.
The protocol and strategy presented here for mixed isomeric glycans was adapted and optimized from previously published protocols for glycan mixture identification.1,2,3 Using the analysis of neutral human milk oligosaccharide (HMO) fractions, DP4 to DP9 obtained from HPLC as an example, we describe the main anticipated outcomes and results of the protocol in analysis of natural biological samples.
In fraction DP4, MNa+ at m/z 926.6 was detected, and the oligosaccharide with a composition of Hex3HexNAc1 (labeled as H3N1) was assigned based on the primary MS spectrum. Using the present GIPS protocol, a single linear branching pattern (Figure 3) was identified in this fraction.12 In fraction DP5, a composition of H3N1F1 was deduced based on MS1. Using GIPS protocol, two different branching patterns both with MNa+ at m/z 1100.7 were identified to be present. These two branching patterns included a linear blood group H-containing and a branched Lea/x-containing sequence, and were in agreement with previous reports.12,13,14
Figure 3.
Identification of mixed isomeric oligosaccharides in fractions extracted from human milk
The compositions of oligosaccharide were provided in blue, and specific branching patterns of isomeric structures identified using the present protocol were presented in red square frame. H refers to Hexose, and N refers to HexNAc, and F refers to fucose.
Glycans with two molecular masses at m/z 1274.8 and m/z 1375.9 were identified in fraction DP6, and compositions were assigned as H3N1F2 and H4N2 based on MS1 spectrum. Isomeric branching structures were identified to be present for these two compositions. Specifically, two different branching patterns were identified for the difucosylated hexasaccharide at m/z 1274.8. One of the Fuc residues is at the GlcNAc while the other is at either the non-reducing terminal Gal forming a Leb/y or the reducing terminal Glc forming a Lea/x epitope (Figure 3).12,14 The non-fucosylated hexasaccharide at m/z 1375.9 was identified as a branched hexasaccharide backbone sequence (Figure 3).12,14,15
In DP7 fraction, three different structures with MNa+ at m/z 1550.0 were identified using GIPS protocol, and these branching patterns included monofucosylated linear and branched hexasaccharide backbones (Figure 3).12,15,16 The Fuc residue is either on the terminal Gal or an internal GlcNAc, forming a blood group H and Lea/x antigen for the two branched sequences respectively.
With the composition of H4N2F2 in MS1, one linear and two branched backbone sequences were identified to be mixed in fraction DP8 at m/z 1724.1 (Figure 3). For the linear structure, both Fuc residues were at the GlcNAc residues comprising a terminal and an internal Lea/x epitope. As for the two branched sequences, the two Fuc residues were either on the same branch to constitute a Leb/y or at different branches forming a H or Lea/x antigens.12,17,18
A linear and a branched backbone sequences at m/z 1898.2 were determined in the fraction of DP9 (Figure 3). For the linear backbone sequence, the three Fuc residues constituted a Leb/y and an internal Lea/x epitope while for branched sequence the Leb/y and Lea/x epitopes are on the different branches as the reported TFLNH and TFpLNH structures.12,19,20
Limitations
Glycosidic cleavages are mainly considered in current protocol, and linkage sites between two residues are not identified, which requires plenty of fragment ions (e.g., A and X ions) produced from cross-ring cleavages. One possible resolution is producing cross-ring fragments under the negative mode of the MS spectrometer. In addition, the identification strategy is based on the database-search process, so that abnormal glycans those are not documented in the database cannot be identified using current protocol. The protocol is applicable to mass spectrometer with Electrospray ionization (ESI) which is frequently coupled to High Performance Liquid Chromatography (HPLC) with adjustment of parameters such as charge state and doped cations besides sodium ions. The expansion to ESI-MS for GIPS will be added and updated in the following version.
Troubleshooting
Problem 1
After the termination of tryptic digestion by heating and adding acetic acid, precipitates appeared in the sample.
Potential solution
-
•
Glycopeptides may not be completely digested by the tryptic enzyme. After being heated and exposed in the acetic solution, the remaining proteins denatured and congregated to precipitates. In this case, the precipitated proteins need to be collected through centrifugation, and further to be combined with the glycopeptide fractions. Then, the combined proteins and glycopeptides are digested by N-glycosidase F. So that, the glycomic information of precipitated proteins is not missing (related to Step 10).
Problem 2
When extracting glycans from biological samples, N-glycans with large molecular mass may not detected in the MS spectrum (related to Step 16).
Potential solution
-
•
Glycopeptides may not be adsorbed onto the cartridge because of the capacity of the cartridge, and some glycopeptides were discarded. As a result, partial N-glycans especially N-glycans with large molecular mass which were in low abundance for certain glycoproteins were not detected. In order to avoid the missing information, quantification of peptides/ glycopeptides is required before loading the sample onto the cartridge. Only appropriate amounts of peptides/glycopeptides within the capacity of the cartridge are allowed. Otherwise, researchers need to repeat the extraction and purification steps with separating equivalent samples.
Problem 3
After reacting at room temperature (22°C–26°C) for 20 min, the sample does not become a white homogeneous solution as milk. The sample becomes suspension or the sample becomes dried solid. Consequently, incomplete permethylated glycan peaks are present in the mass spectrum (related to Step 44).
Potential solution
-
•
One reason for incomplete permethylation is that NaOH and DMSO slurry is not prepared in right ratio. The potential solution is grinding NaOH particles and DMSO strictly according to the protocol, and add the lower-layer slurry to the sample. Another reason for incomplete permethylation is the variance of room temperature between different days or different seasons. The possible solution is extending the reaction time to 25 min in cold days. Alternatively, laboratory space with constant temperature (25°C) is required for reaction.
Problem 4
The sample spotted in the MALDI plate is not dried to form a solid layer at room temperature (22°C–26°C) even after several hours (related to Step 67).
Potential solution
-
•
DMSO in the sample is not discarded thoroughly after permethylation, and therefore the sample cannot be dried. One possible solution is drying the sample in vacuum for 3 min. Alternatively, 1 μL ethanol can be added to the spotted sample for recrystallization. The most important operation for drying in the completely removement of DMSO.
Problem 5
When producing next-stage MS spectrum using the selected precursor ions by GIPS, very few/no fragment ions were detected in the product-ion spectrum (related to Steps 86 and 97). As a result, the probability of each candidate did not change, and this precursor selection step was unhelpful.
Potential solution
-
•
Although the DPs of certain fragment ions were the highest, the product-ion spectra from this precursor did not give a definite solution probably due to the low intensity of the precursor ions and the unuseful fragmentation. In this case, fragment ions with the next highest DPs can be selected as the precursor for dissociation.
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Yan Li (yanli@ibp.ac.cn).
Technical contact
Further questions and instructions on the application of this protocol will be addressed by the technical contact, Chuncui Huang (huangchuncui@ibp.ac.cn).
Materials availability
This study did not generate any unique materials.
Data and code availability
The software generated in this study is available at the website (http://glycan.ict.ac.cn/GIPS/). The GIPS software is also available upon request to authors.
Acknowledgments
This work was supported by the National Key Research and Development of China (2022YFC3400801), Biological Resources Programme from the Chinese Academy of Sciences (KFJ-BRP-017-76), and the project of ‘‘Dengfeng Plan’’ from Foshan Hospital of Traditional Chinese Medicine. This work was also supported, in part, by the National Natural Science Foundation of Chongqing, China (CSTB2023NSCQ-MSX0131), the Strategic Priority Research Program of Chinese Academy of Sciences (XDB38010200), the National Natural Science Foundation of China (62072435), the March of Dimes Prematurity Research Center grant (22-FY18-821), and the Wellcome Trust Biomedical Resource grant (218304/Z/19/Z).
Author contributions
Y. Li, C.H., S.S., and W.C. conceived the study and established the protocol. C.H. and Y. Li designed the glycan extraction and mass spectrometry analytical methodologies. W.C. provided critical supervision on the mass spectrometry analysis. S.S. and D.B. proposed the concept of intelligent precursor selection and designed the algorithm of GIPS strategy. Y.W., H.W., M.H., and J. Zhang designed and wrote the code of GIPS strategy. J. Zhou, K.Z., Y. Liu, X.M., and J.Y. performed experiments for the establishment of protocol. Y.R. and Y.H. developed the website and software for glycan analysis. C.H., Y. Li, and W.C. wrote the manuscript of the protocol. All authors discussed the detailed steps of the protocol and commented on the manuscript.
Declaration of interests
The authors declare no competing interests.
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.xpro.2024.102976.
Contributor Information
Wengang Chai, Email: w.chai@imperial.ac.uk.
Shiwei Sun, Email: dwsun@ict.ac.cn.
Yan Li, Email: yanli@ibp.ac.cn.
References
- 1.Sun S., Huang C., Wang Y., Liu Y., Zhang J., Zhou J., Gao F., Yang F., Chen R., Mulloy B., et al. Toward automated identification of glycan branching patterns using multistage mass spectrometry with intelligent precursor selection. Anal. Chem. 2018;90:14412–14422. doi: 10.1021/acs.analchem.8b03967. [DOI] [PubMed] [Google Scholar]
- 2.Huang C., Wang H., Bu D., Zhou J., Dong J., Zhang J., Gao H., Wang Y., Chai W., Sun S., Li Y. Multistage mass spectrometry with intelligent precursor selection for N-glycan branching pattern analysis. Carbohydr. Polym. 2020;237 doi: 10.1016/j.carbpol.2020.116122. [DOI] [PubMed] [Google Scholar]
- 3.Huang C., Hou M., Yan J., Wang H., Wang Y., Cao C., Wang Y., Gao H., Ma X., Zheng Y., et al. GIPS-mix for accurate identification of isomeric components in glycan mixtures using intelligent group-opting strategy. Anal. Chem. 2023;95:811–819. doi: 10.1021/acs.analchem.2c02978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Robinson R.C., Colet E., Tian T., Poulsen N.A., Barile D. An improved method for the puriffcation of milk oligosaccharides by graphitized carbon-solid phase extraction. Int. Dairy J. 2018;80:62–68. doi: 10.1016/j.idairyj.2017.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dell A., Reason A.J., Khoo K.H., Panico M., McDowell R.A., Morris H.R. Mass spectrometry of carbohydrate-containing biopolymers. Method Enzymol. 1994;230:108–132. doi: 10.1016/0076-6879(94)30010-0. [DOI] [PubMed] [Google Scholar]
- 6.Ashline D., Singh S., Hanneman A., Reinhold V. Congruent strategies for carbohydrate sequencing. 1. mining structural details by MSn. Anal. Chem. 2005;77:6250–6262. doi: 10.1021/ac050724z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ashline D.J., Lapadula A.J., Liu Y.H., Lin M., Grace M., Pramanik B., Reinhold V.N. Carbohydrate structural isomers analyzed by sequential mass spectrometry. Anal. Chem. 2007;79:3830–3842. doi: 10.1021/ac062383a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Reinhold V., Zhang H., Hanneman A., Ashline D. Toward a platform for comprehensive glycan sequencing. Mol. Cell. Proteomics. 2013;12:866–873. doi: 10.1074/mcp.R112.026823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zaia J. Mass spectrometry and the emerging field of glycomics. Chem. Biol. 2008;15:881–892. doi: 10.1016/j.chembiol.2008.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ashline D.-J., Zhang H., Reinhold V.-N. Isomeric complexity of glycosylation documented by MSn. Anal. Bioanal. Chem. 2017;409:439–451. doi: 10.1007/s00216-016-0018-7. [DOI] [PubMed] [Google Scholar]
- 11.Doubet S., Albersheim P. Letter to the Glyco-Forum: CarbBank. Glycobiology. 1992;2:505–507. doi: 10.1093/glycob/2.6.505. [DOI] [PubMed] [Google Scholar]
- 12.Kobata A. Structures and application of oligosaccharides in human milk. Proc. Jpn. Acad. Ser. B Phys. Biol. Sci. 2010;86:731–747. doi: 10.2183/pjab.86.731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Huang C., Sun S., Yan J., Wang H., Zhou J., Gao H., Xie W., Li Y., Chai W. Identification of carbohydrate peripheral epitopes important for recognition by positive-ion MALDI multistage mass spectrometry. Carbohydr. Polym. 2020;229 doi: 10.1016/j.carbpol.2019.115528. [DOI] [PubMed] [Google Scholar]
- 14.Kunz C., Rudloff S., Baier W., Klein N., Strobel S. Oligosaccharides in human milk: structural, fnctional and metabolic aspects. Annu. Rev. Nutr. 2000;20:699–722. doi: 10.1146/annurev.nutr.20.1.699. [DOI] [PubMed] [Google Scholar]
- 15.Yamashita K., Tachibana Y., Kobata A. Oligosaccharides of human milk: structures of three lacto-N-hexaose derivatives with H-haptenic structure. Arch. Biochem. Biophys. 1977;182:546–555. doi: 10.1016/0003-9861(77)90536-7. [DOI] [PubMed] [Google Scholar]
- 16.Dua V.K., Goso K., Dube V.E., Bush C.A. Characterization of lacto-N-hexaose and two fucosylated derivatives from human milk by high-performance liquid chromatography and proton NMR spectroscopy. J. Chromatogr. 1985;328:259–269. doi: 10.1016/s0021-9673(01)87396-9. [DOI] [PubMed] [Google Scholar]
- 17.Yamashita K., Tachibana Y., Kobata A. Oligosaccharides of human milk: structural studies of new octasaccharides, difucosyl derivatives of para-lacto-N-hexaose and para-lacto-N-neohexaose. J. Biol. Chem. 1977;252:5408–5411. [PubMed] [Google Scholar]
- 18.Strecker G., Fièvre S., Wieruszeski J.M., Michalski J.C., Montreuil J. Primary structure of four human milk octa-nona-and undeca-saccharides established by 1H-and 13C-nuclear magnetic resonance spectroscopy. Carbohydr. Res. 1992;226:1–14. doi: 10.1016/0008-6215(92)84050-3. [DOI] [PubMed] [Google Scholar]
- 19.Strecker G., Wieruszeski J.M., Michalski J.C., Montreuil J. Primary structure of human milk nona- and deca-saccharides determined by a combination of fast atom bombardment mass spectrometry and 1H-/13C- nuclear magnetic resonance spectroscopy. Evidence for a new core structure, iso-lacto-N-octaose. Glycoconj. J. 1989;6:169–182. doi: 10.1007/BF01050646. [DOI] [PubMed] [Google Scholar]
- 20.Strecker G., Wieruszeski J.M., Michalski J.C., Montreuil J. Structure of a new nonasaccharide isolated from human milk: VI2Fuc,V4Fuc,III3Fuc-p-lacto-N-hexaose. Glycoconjugate J. 1988;5:385–396. doi: 10.1007/BF01050646. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The software generated in this study is available at the website (http://glycan.ict.ac.cn/GIPS/). The GIPS software is also available upon request to authors.