Summary
Plant cell walls constitute complex polysaccharidic/proteinaceous networks whose biosynthesis and dynamics implicate several cell compartments. The synthesis and remodeling of homogalacturonan pectins involve Golgi-localized methylation/acetylation and subsequent cell wall-localized demethylation/deacetylation. So far, TRICHOME BIREFRINGENCE-LIKE (TBL) family members have been described as Golgi-localized acetyltransferases targeting diverse hemicelluloses or pectins. Using seed mucilage secretory cells (MSCs) from Arabidopsis thaliana, we demonstrate the atypical localization of TBL38 restricted to a cell wall microdomain. A tbl38 mutant displays an intriguing homogalacturonan immunological phenotype in this cell wall microdomain and in an MSC surface-enriched abrasion powder. Mass spectrometry oligosaccharide profiling of this fraction reveals an increased homogalacturonan acetylation phenotype. Finally, TBL38 displays pectin acetylesterase activity in vitro. These results indicate that TBL38 is an atypical cell wall-localized TBL that displays a homogalacturonan acetylesterase activity rather than a Golgi-localized acetyltransferase activity as observed in previously studied TBLs. TBL38 function during seed development is discussed.
Subject areas: Plant biology, Plant cytology, Plant development
Graphical abstract

Highlights
-
•
Known plant TBLs are Golgi-localized acetyltransferases for cell wall polysaccharides
-
•
TBL38 is peculiarly localized to a wall microdomain in Arabidopsis seed epidermis
-
•
TBL38 has an uncommon acetylesterase activity targeted toward homogalacturonans
-
•
TBL38 is a new actor of the complex maturation of plant cell wall polysaccharides
Plant biology; Plant cytology; Plant development;
Introduction
Plant cells specifically design, synthesize and constantly remodel their cell wall (CW) to achieve proper cell shape and function along the development to face environmental constraints. Such dynamics lead to specific composition and properties of CW polymers. Accumulating evidence of chemical group modifications on particular polysaccharides involved in key developmental processes can be found in the literature. The importance of pectin methylesterification dynamics has been largely studied, particularly for homogalacturonans (HGs). In the Golgi apparatus, HGs are methylated on the COOH of galacturonic acids (GalA) through methyl-transferase activity1 and later undergo additional remodeling once in the CW. Methylated HG can be demethylesterified by pectin methyl esterases (PMEs), whose activity can be regulated by PME inhibitors (PMEIs). The joint activity of both enzyme categories is the source of many developmental roles.2 Additionally, O-acetylation of CW polymers such as pectins and hemicelluloses in the Golgi lumen has been described in multiple studies, highlighting three groups of proteins involved in this process.3 In Arabidopsis thaliana, (i) ALTERED XYLOGLUCAN 9 (AXY9) is encoded by a unique gene likely indirectly responsible for the non-specific acetylation of xyloglucan and xylan hemicelluloses4 leading to smaller plants when mutated. (ii) The REDUCED WALL O-ACETYLATION (RWA) family has four members in A. thaliana which also participate in the O-acetylation machinery of xylans, mannans, xyloglucans, and pectins.5,6 (iii) The TRICHOME BIREFRINGENCE-LIKE (TBL) family comprises 46 members distributed in four phylogenetic clades in A. thaliana that target specific CW polysaccharides (Figure 1; Table S1). Originally named based on the observed changes in the birefringence properties of a mutant of TRICHOME BIREFRINGENCE (TBR),7 29 TBLs have been characterized for their O-acetyltransferase activity specific for the xyloglucan backbone and side chains, xylans, mannans, rhamnogalacturonan I (RG-I) or HGs (Figure 1; Table S1, reviewed in.3,8 These activities could be either highly clade-specific, as exemplified by nine TBLs belonging to a similar clade that displayed xylan-acetyltransferase activity, or more phylogenetically widespread, as exemplified by the four homogalacturonan acetyltransferases (Figure 1; Table S1). Mutations on particular TBLs led to (i) defects in CW structure and overall plant development as observed in tbr, tbl3, tbl29, and tbl37,9,10,11 and (ii) increased resistance to drought stress12 or pathogen infection13 in tbl10 and tbl44/pmr5, respectively (Table S1). Like AXY9, TBLs have the conserved GDSL domain, which is typically associated with esterase activity/lipase,14,15 and a DXXH motif also found in a rhamnogalacturonan acetylesterase of Aspergillus aculeatus.16 The TBL acetylesterase activity allows the formation of an acyl-enzyme intermediate necessary for the O-acetyltransferase activity targeting various CW polymers3,17 (Figure 1; Table S1). Out of the 23 characterized TBLs, 12 have been localized to the Golgi alongside with AXY9 and RWAs, supporting their role as O-acetyltransferases (Figure 1; Table S1). Surprisingly, TBR was localized to the CW.18 Yet, although its precise activity is still unknown, an overall decrease in pectin acetylation was observed in tbr, suggesting an acetyltransferase activity that is surprising considering that the existence of a CW-localized acetyl donor is unclear.18 In silico prediction of the targeting function of the N-terminal hydrophobic sequence sorted the 46 TBLs in two groups: 35 with a transmembrane anchoring domain, and 11 with a cleavable signal peptide. Six TBLs have been identified in CW-enriched fractions without clear relationship with the in silico targeting predictions (Table S1). Overall, experimental evidence depicts TBLs as acetylesterases forming an acyl-enzyme intermediate to further perform an O-acetyltransferase activity targeted on various CW polymers during their synthesis in the Golgi lumen.
Figure 1.
The TBL family mostly comprises Golgi-localized acetyltransferases specifically acting on various CW polymers
The 46 members show high sequence homology and two conserved GDSL and DXXH motifs associated with esterase activities. Previous work showed that the esterase activity is not targeted toward CW polymers but rather allows the formation of an acyl-enzyme intermediate. Multiple TBLs were shown to participate in the O-acetylation of a wide range of CW polymers in the Golgi with the notable exception of TBR which was positioned to the CW. Twenty nine recombinant enzymatic activities (yellow), 13 fluorescent protein (FP) localization (green), and six CW proteome occurrence https://www.polebio.lrsv.ups-tlse.fr/WallProtDB/(orange) were reported. http://aramemnon.uni-koeln.de/predicts that the N-terminus hydrophobic domain either acts as a transmembrane (TM) domain (35 non-underlined proteins) or as a cleaved signal peptide (11 underlined proteins). The AT activities are either grouped in a phylogenic cluster (e.g., xylan-ATs) or distributed within TBLs in distinct phylogenic clusters (e.g., HG-ATs). The phylogenetic tree was adapted from.9 AE, acetylesterase; AT, acetyltransferase; PAE, pectin acetyl esterase; HG, homogalacturonan; RGI, rhamnogalacturonan I; XG, xyloglucan. See Table S1 for detailed information and references. This study reports on TBL38 peculiar localization and activity.
Initially, we screened genes involved in A. thaliana seed mucilage release with particular interest in candidates that could modify the esterification status of HGs. The mucilage is a polysaccharidic hydrogel synthesized by the seed mucilage secretory cells (MSCs) in the outer most cell layer of the seed coat of the so-called myxospermous species.19,20,21 During A. thaliana seed maturation, the HG methylesterification status is modified through the PECTIN METHYLESTERASE INHIBITOR6 6 (PMEI6) activity on an unknown PME in a CW microdomain at the top of the radial primary wall of MSCs, leading to an HG partially methylesterified pattern.22,23 This pattern acts as an anchoring platform for PEROXIDASE36 (PRX36) which then induces a local weakening and thinning of the CW microdomain.22,24 After seed imbibition, the swelling mucilage breaks the pre-weakened CW microdomain and is extruded around the seed. We identified TBL38 (At1g29050) for its high co-expression level with PRX36 and PMEI6 during A. thaliana seed coat development. Based on the literature cited above and considering its co-expression with PMEI6, we first hypothesized TBL38 to be a Golgi-localized protein acting as an acetyltransferase onto CW polymers possibly including HGs. Hereafter, we examined TBL38 localization, activity, and putative role in MSCs and uncovered that TBL38 is an atypical CW-localized HG acetylesterase rather than a Golgi-localized acetyltransferase. Although TBLs are not thought to influence the methylesterification status of CW polymers, we investigated the putative indirect role of TBL38-dependent HG O-deacetylation on the HG methylation status and on PRX36 anchoring, thus adding another layer of complexity on the fine-tuning of HG remodeling during plant development.
Results
TBL38 is co-expressed with PEROXIDASE36 and PECTIN METHYLESTERASE INHIBITOR6 in mucilage secretory cells at intermediate developmental stages of seed development
Using the tissue-specific seed developmental kinetics GSE12404 dataset,25 we previously built a co-expression network centered on PRX36 in which PMEI6 came as the second hit22 (Table S2). We noticed the high co-expression of TBL38, which was ranked 13 in this network, with a strong specific seed coat expression at the linear cotyledon stage (Figures 2A and 2B; Table S2). We filtered TBL expression data, which was available for 34 out of the 46 TBLs, although only 23 were above the detection limit, and confirmed that TBL38 was likely to be the only TBL highly co-expressed with PRX36 and PMEI6 (Table S2). The seed coat transcriptomic data encompasses five cell layers: ii1 (inner integument), ii1′, ii2, oi1 (outer integument), and oi2, commonly addressed here as MSCs.19,26 Therefore, we determined MSC-specific TBL38 expression using in situ hybridization on tissue array paraffin sections, encompassing the whole developmental kinetics of wild-type seeds. TBL38 anti-sense probes displayed a clear purple/violet hybridization signal in MSCs, particularly between the early linear and early bending cotyledon developmental stages (Figure 2C). No such signal could be observed at earlier and later stages with the anti-sense probe and at all developmental stages with the sense probe used as a negative control to account for the overall non-specific background and natural pigmentation. We could associate the specific TBL38 anti-sense probe signal to MSCs, excluding the chalazal tissues, in agreement with -and refining-the transcriptomic data (Figures 2A–2C).
Figure 2.
TBL38 is co-expressed with PRX36 and PMEI6 in the seed coat and transcript the localization of TBL38 is tied to the mucilage secretory cells (outer integument 2)
(A) eFP browser pictogram displaying the similarity in the spatiotemporal expression of PRX36, PMEI6, and TBL38 in A. thaliana seed coat. Absolute tissue-specific expression level is represented in color variation from yellow (lowest) to red (highest). Adapted from http://bar.utoronto.ca/efp/cgi-bin/efpWeb.cgi.
(B) PRX36 co-expression network using the same dataset highlighting the similar profiles of PRX36, PMEI6, and TBL38 (see Table S2 for details).
(C) In situ hybridization of dig-labeled RNA probes targeting TBL38 on paraffin sections of developing wild-type seeds. Anti-sense probes showed highest purple signal in the outermost layer corresponding to MSCs between the early linear and early bending developmental stages. These signals should be subtracted from the overall background obtained on serial sections of the same seeds with the sense probe control and natural pigmentation. Bars: 200 μm.
TBL38-TagRFP is transiently localized to the PEROXIDASE36-PECTIN METHYLESTERASE INHIBITOR6 outer periclinal/radial cell wall microdomain
We generated a p35S::TBL38-TagRFP construct to transiently transform N. benthamiana leaves. The TBL38-TagRFP fluorescence did not colocalize with a Golgi-YFP marker, but rather displayed a cellular delineation (Figure 3A). Under plasmolysis conditions, we could clearly attribute the TBL38-TagRFP signal to the CW separating from the receding plasma membrane YFP marker signals (Figure 3A). We concluded that TBL38 is a CW-localized protein similarly to TBR18 and contrarily to the 12 other Golgi-localized TBLs (Figure 1; Table S1). We then moved on to assess protein localization in A. thaliana seed MSCs. As seed development advances, MSCs become mostly made of different CWs, namely the primary CW, the mucilage, and the columella.19 Within MSCs, increasing evidence have positioned CW proteins in only one or two types of CW22,27,28 or in sub-layer (microdomain) of one CW.22,29 To localize TBL38 in A. thaliana MSCs, we generated a pPRX36:TBL38-TagRFP construct which was stably transformed in tbl38 plants. We localized TBL38-TagRFP to the radial primary CW of MSCs at the onset of pPRX36 activity at early developmental stages (Figure 3B; Figure S1). Orthogonal projection allowed to refine TBL38-positonning at the top of the radial primary CW, corresponding to the CW microdomain encompassing PRX36.22 Intriguingly, while PRX36-TagRFP was stably accumulated at this location using the same promoter,22 TBL38-TagRFP appeared to be naturally delocalized to the future mucilage pocket and eventually pushed toward the inner and outer margins of the pocket when filled with mucilage after 12 DAP (Figure 3B; Figure S1).
Figure 3.
TBL38 is a cell wall protein transiently localized to the radial primary CW microdomain of MSCs
(A) Confocal microscopy visualization of both TBL38-TagRFP and a Golgi-YFP marker transiently overexpressed in N. benthamiana leaves indicates that TBL38 is not a Golgi-localized protein. Under plasmolysis, the TBL38-TagRFP signal and the plasma membrane-YFP marker signal are separated, indicating the localization of TBL38-TagRFP to the CW. First row images are extracted from a maximum projection of 17 stacks. Second row images are a representative view of a single confocal plan. Bars: 10 μm.
(B) Confocal spinning disk observations of TBL38-TagRFP in A. thaliana developing seeds. At the onset of pPRX36 activity,22 TBL38-TagRFP fluorescence was positioned to the radial primary CW of MSCs. Orthogonal projection refined this localization to the CW microdomain consisting in the top of the radial primary CW similarly to PRX36-TagRFP.22 However, contrary to the previously observed stable localization of PRX36-TagRFP along seed development, TBL38-TagRFP fluorescence was delocalized to the future mucilage pocket at 8–12 DAP and eventually to the outer and inner margins of the mucilage-filled pocket at > 12 DAP. Sum intensity projections were done using similarly sized projection of 30 Z stacks with no singular edition. rcw, radial CW microdomain; m, mucilage pocket; im and om, inner and outer margins of mucilage pocket, respectively. Bars: 25 μm. (see Figure S1 for full kinetics performed on three individual transformed lines).
(C) αTag-RFP western blot performed using total protein extracts from developing seeds harvested at stages corresponding to the three sequential patterns of fluorescence shows that the band #2 corresponding to the TBL38-TagRFP fusion protein is only seen at the early stage when TBL38-TagRFP is localized to the CW microdomain. Non-specific bands #1 and #3 are also present in Col-0 samples (see Figure S2 for full membrane visualization).
We accurately sampled developing seeds at stages corresponding to the three above mentioned fluorescence patterns and performed a αTagRFP western blot (Figure 3C; Figure S2). Excluding the non-specific bands (#1 and 3) also observed with similarly staged Col-0 seeds, a single specific band (#2) corresponding to the apparent molecular mass of TBL38-TagRFP fusion protein (70 kDa) was only observed at 6–8 DAP when TBL38-TagRFP was localized to the CW microdomain. The absence of such specific band when the fluorescence was later localized to the mucilage pocket suggests that this additional fluorescence pattern rather corresponds to the delocalized TagRFP possibly cleaved from TBL38. This hypothesis is strengthened by the appearance of a 28 kDa band that may correspond to cleaved TagRFP at 8–12 DAP that unfortunately migrated with an additional non-specific band of similar molecular mass also observed in Col-0 after 12 DAP (Figure S2).
Altogether, the expression of TBL38-TagRFP in both N. benthamiana and A. thaliana showed an unexpected CW localization, making TBL38 the second TBL localized to the CW with TBR. Because its localization to the outer radial CW microdomain was strikingly similar to that of PRX36, we investigated TBL38 involvement in the above mentioned PRX36-PMEI6 molecular module and particularly its influence on HG modification.
The mucilage release and the anchoring of PEROXIDASE36 are not significantly modified in tbl38
Mutation in either PRX36 or PMEI6 results in impaired mucilage release because the primary CW microdomain improperly brakes up upon seed imbibition.22,23,24 Because TBL38 and PRX36 gene products have similar spatiotemporal expression and positioning, we assessed whether mucilage release was affected in mutated seeds. We isolated a homozygous KO line for tbl38 (Figure S3) and used it for comparison with Col-0 wild type seeds using ruthenium red staining of adherent mucilage. We did not see any differences between WT and tbl38 seeds (Figure 4A), contrarily to impaired mucilage release previously observed in prx36 or pmei6.22,23,24 Quantification of adherent mucilage area and circularity on > 1,000 seeds confirmed the similarity of the patterns of Col-0 and tbl38 (Figure 4B). We did not see any obvious change in mucilage staining intensity as sometimes observed in mutants responsible for mucilage modification.30
Figure 4.
tbl38 seeds show no defect in mucilage release and accordingly no obvious consequence on PRX36 anchoring
(A) Example of screenshot from scan of Ruthenium red staining of released adherent mucilage shows similar patterns between Col-0 and tbl38. Note the yellow masks used in (B). tbl38 does not show the phenotypes previously observed in prx36 and pmei6.22,23,24
(B) Violin plot representation of the measurement of adherent mucilage area and circularity using the yellow masks shown in (A) confirm the absence of any obvious phenotype.
(C) αPRX36 labeling of paraffin sections positioned PRX36 at the top of radial primary cell wall from MSC in both Col-0 and tbl38 contrary to the previously demonstrated loss of labeling in prx36 and pmei6 [S1]. Bars: 25 μm.
(D) PRX36-TagRFP localization is not affected in tbl38 developing seeds. Confocal spinning disk observations of stable proPRX36::PRX36-TagRFP in A. thaliana reveal no obvious mislocalization of the signal in tbl38. Similar protein accumulation at 6 DAP was observed in both genotypes. Images are the result of a maximum projection of similar stacks (Z > 30) which were not edited. Bars: 50 μm.
Since PRX36 anchoring to a specific HG platform and subsequent CW weakening are required for proper mucilage release, we investigated PRX36 positioning in tbl38. For this, we used the αPRX36 antibody on paraffin sections of developing seeds from Col-0 or tbl38. Anti-PRX36 similarly labeled the top of the radial primary CW in Col-0 and tbl38 at all developmental stages (Figure 4C). tbl38 did not show the loss of signal previously observed in prx36 or pmei6.22 To further validate this result, we generated a pPRX36::PRX36-TagRFP construct, which we expressed in prx36 and tbl38 backgrounds. Observation of the PRX36-TagRFP signal was made on developing seeds aged from 6-to-12 DAP, a period which encompasses the onset of the PRX36 promoter activity up to the last stage of the clearly visible PRX36-TagRFP fluorescence signal.22 PRX36-TagRFP showed very similar signal localization at the top of the radial primary CW in both tbl38 and prx36 (Figure 4D). Again, no delocalization of the signal was observed in tbl38 contrary to the PRX36-TagRFP delocalization in the mucilage pocket previously observed in pmei6.22
So far, the particular PMEI6-specific HG methylation pattern that exists at the top of the radial primary CW linked proper mucilage release with PRX36 anchoring.22 Our results did not show such a relationship between TBL38, mucilage release, and PRX36 positioning, suggesting that no changes in the HG methylation pattern occur in tbl38 or that putative changes do not impact PRX36 anchoring. However, TBL38 could be involved in the acetylation of CW polymers similarly to other described TBLs. We first studied the HG methylation pattern in tbl38 for the following reasons: (i) changes in HG acetylation could lead to different HG methylesterification status, possibly through the indirect modulation of PME activity, or hinder the dimerization of pectin chains,31,32 (ii) TBL38 colocalized with partially methylesterified HG JIM7 and LM20 epitopes at the top of radial CW in MSCs.22
The partially methylated homogalacturonan JIM7 epitope corresponding to the PEROXIDASE36-PECTIN METHYLESTERASE INHIBITOR6 cell wall microdomain is maintained in tbl38, while the supposed similar LM20 epitope is surprisingly lost
The specific HG platform enabling PRX36 anchoring, is generated through the inhibition of an unknown PME by PMEI6 leading to a partially methylesterified pattern of HGs at the top of the radial primary CW in MSCs. This molecular pattern could be labeled by both JIM7 and LM20, two monoclonal antibodies routinely used to characterize partially methylesterified HGs, and these labelings were lost in pmei6.22 Thus, we incubated JIM7 and LM20 on paraffin sections of developing seeds from tbl38 and Col-0.
As a control, JIM7 signals were correctly positioned at the top of the radial primary CW in Col-0 developmental kinetics, particularly as MSC development advanced to later stages (Figure 5A). Very similar JIM7 labeling was observed in tbl38 (Figure 5A) contrary to the loss of signal previously observed in pmei6.22 A similar pattern was observed with LM20 along the developmental kinetics of the Col-0 control genotype (Figure 5B). However, the LM20 labeling was intriguingly lost in tbl38 (Figure 5B). This was related to the tbl38 mutation since this labeling was restored along the developmental kinetics of tbl38 plants complemented with a pPRX36::TBL38-TagRFP construct (Figure 5B). While both antibodies are generally used in parallel to similarly label partially methylesterified HG, their partially characterized epitopes may be somehow different.33,34 Overall, we concluded that tbl38 mutation resulted in lack of the LM20 signal, but not of that of JIM7. Yet, it was still unclear if this could be due to putative changes in the acetylation of HGs which may prevent LM20 binding through steric hindrance, or to changes in HG methylation as an indirect effect of acetylation. Regardless, we sought to investigate TBL38 biochemical function in MSCs.
Figure 5.
LM20 but not JIM7 labeling is lost in tbl38
(A) In Col-0, JIM7 labeling of partially methylesterified HG is visible as early as 6 DAP at the MSC surface and is restricted to the top of the radial primary CW microdomain of more mature MSCs (7–10 DAP). A very similar labeling pattern occurs in tbl38 as well as in two complemented lines (tbl38 transformed with pPRX36::TBL38-TagRFP).
(B) LM20 was previously thought to bind to similar epitopes as JIM7 in MSCs since both epitopes were lost in pmei6.22 However, contrary to JIM7, the LM20 labeling at the top of the radial primary CW microdomain was lost in tbl38 and restored in the two tested complemented lines. 40× scans of fluorescence and brightfield channels were merged. The same parameters were used to analyze each image which are a representative of n > 20 MSCs at each developmental stage. Additional stages positioned between the staged ranks are shown. Bars: 25μm.
TBL38 displays an atypical acetylesterase activity toward cell wall polymers
We took advantage of MSC surface accessibility to generate an MSC surface-enriched CW abrasion powder from dry seeds using home-made abrasion columns (see STAR Methods). The resulting homogeneous powder (about 1 mg per 50 mg seeds) was collected through a nylon mesh and subsequently analyzed (Figures 6A–6F). We confirmed that the abrasion process was restricted to the MSC surface, as MSC CW autofluorescence was strongly reduced but could still be barely observed in abraded seeds (Figures 6G and 6H) which released residual amount of adherent mucilage when imbibed (Figures 6I and 6J). Although the overall MSC surface morphology was affected, we could still see the base of broken columella and radial CW (Figures 6M and 6N). We successfully recovered a surface-abraded powder containing the top of the radial primary wall, considering the loss of LM20 signal in Col-0 abraded seeds (Figures 6K and 6L). We next used this MSC surface-enriched CW abrasion powder from various genotypes to study the reactivity of antibodies specific for various CW epitopes including antibodies against HGs with various levels of methylation, rhamnogalacturonan I, and xyloglucan, all these polysaccharides being known to be present in mucilage or in the same CW microdomain as PRX36 and PMEI6 (Figure S4). Only JIM7 and LM20 displayed a differential reactivity between the genotypes of interest. We validated the abrasion method since JIM7 and LM20 dot blot signal intensities in pmei6 were both about 5% as compared to Col-0 (Figures 7A and 7B), in agreement with the previously observed loss of JIM7 and LM20 immunofluorescence signals in development kinetics of pmei6 MSCs.22 Again, in agreement with the present immunofluorescence study on development kinetics (Figure 5), the JIM7 signal intensity from dry seed surface abraded fractions was very similar in all genotypes except pmei6, since tbl38 and three complemented lines all showed 91–95% of Col-0 intensity (Figure 7A). However, the LM20 signal intensity in tbl38 was only 4% of that in Col-0 control while the three complemented lines showed a strong -though partial-signal restoration with 66–79% of Col-0 intensity (Figure 7B).
Figure 6.
Characterization of the abrasion process of dry seed surface to produce a mucilage secretory cell (MSC) surface-enriched fraction
Scanning electron microscopy (SEM) view of sand paper (A), nylon mesh (B) included in the home-made abrasion column (see STAR methods), dry seed before (C) and after (D) 5 × 1 min abrasion illustrating the homogeneous etching of the dry seed surface. SEM view of the recovered MSC surface-enriched powder (E, F) illustrating the small size and relatively homogeneous particles. Note that the same scale is used for A to E to better compare the sizes. UV imaging of dry seed before (G) and after (H) abrasion illustrating the loss of autofluorescence of MSC radial walls and the decrease of autofluorescence of columella. Ruthenium red (RR) staining of dry seed before (I) and after (J) abrasion showing the remaining thin layer of adherent mucilage. LM20 immunofluorescence of dry seed before (K) and after (L) abrasion showing that the LM20 cell wall microdomain is abraded. Toluidine blue O (TBO) staining of dry seed before (M) and after (N) abrasion further showing that the etching occurred only at the surface of the MSCs. ∗, radial primary wall, c, columella, oi2, outer integument 2, ii1, inner integument 1, PE, peripheral endosperm.
Figure 7.
Dry seed surface enriched fraction shows a decreased LM20 epitope signal intensity and an increased acetic acid content in tbl38 compared to Col-0, and the wild type phenotype is restored in complemented lines
JIM7 (A) and LM20 (B) immuno dot blots of dry seed surface CW-enriched fraction from Col-0, pmei6, tbl38, and three tbl38 complemented lines expressing pPRX36:TBL38-TagRFP (PTR2.4, 3.5 and 5.2). On top of the panels are shown screen shots of individual dots from the same nitrocellulose membrane. On the bottom of the panel are shown boxplot representations of the data analysis.
(C) Boxplot representation of the acetic acid content released following acid hydrolysis of dry seed surface CW-enriched fraction from Col-0, tbl38, and three tbl38 complemented lines expressing pPRX36::TBL38-TagRFP (PTR2.4, 3.5 and 5.2). Three biological replicates each with at least two technical replicates were analyzed. Letters above the bars show the statistical distribution of the results using ANOVA and TUKEY HSD tests.
So far, all the described TBLs were either directly characterized as acetyltransferases or associated with lower acetylation content of CW polymers in tbl mutants (Table S1). Therefore, we quantified the acetylation of CW polymers using the surface-enriched CW abrasion powders. Surprisingly, the mean amount of released acetic acid in tbl38 was 199% of that in Col-0 and the level dropped back to 117–153% of that of Col-0 in the three complemented lines (Figure 7C). This suggested that the TBL38 activity was akin to an acetylesterase rather than the previously described acetyltransferase of other characterized TBLs. Because the increased acetylation level in tbl38 could be achieved through the acetylation of multiple polymers, we sought to identify TBL38 substrates in these fractions.
TBL38 is an homogalacturonan acetylesterase
Surface-enriched CW abrasion powders were enzymatically hydrolyzed with a pectin lyase (PL) or a polygalacturonase (PG), displaying specificity toward highly and lowly methylesterified HGs, respectively. The resulting oligogalacturonates (OGAs) from Col-0, tbl38 as well as pmei6 and the complemented line PTR3.5 used as additional controls, have then been analyzed by mass spectrometry, which allowed for the determination of the degree of polymerization (DP), degree of methylation (DM) and degree of acetylation (DA). Technical repeats showed the low standard deviation of the intensities of identified species validating the approach despite the low amount of biological material (Table S3A). We first used a PL with a high affinity for HG with a high DM. The intensity of the identified OGA species was summed for the identified OGA species of same DA and DP regardless the DM, or same DM and DP regardless the DA (Table S3B) and for each genotype, the results were plotted as DA vs. DP (Figures 8A–8D), or DM vs. DP (Figures 8E–8H). We observed a clear qualitative HG acetylation phenotype in tbl38 as compared to Col-0 and pmei6 (Figures 8A–8C) and a qualitative complementation of PTR3.5 (Figure 8D). This was in agreement and refined the high acetylation phenotype observed in tbl38 with the enzymatic assay (Figure 7C) since new OGA species of higher DA appeared in tbl38 as compared to the three other genotypes. This strongly suggests that TBL38 could be an HG acetylesterase. Conversely, a quantitative HG methylation phenotype was observed for pmei6 as compared to Col-0, tbl38, and PTR3.5 (Figures 8E–8H).
Figure 8.
Enzymatic profiling reveals the HG acetylation qualitative phenotype of tbl38 and the quantitative HG methylation phenotype of pmei6
MSC surface abrasion powders of Col-0, tbl38, pmei6 and tbl38 transformed with pPRX36::TBL38-TagRFP (line PTR3.5) were digested with pectin lyase (PL) specific for highly methylesterified HG (A–H; Table S3), polygalacturonase (PG) specific for low-methylesterified HG (I–N; Table S4) or rhamnogalacturonan hydrolase (RG) specific for RGI backbone (O–P; Table S5). The identified HG species (A–N) or RGI species (O–P) were sorted according to their degree of acetylation (DA), degree of methylation (DM) and degree of polymerization (DP). The mean intensities were plotted either as DA vs. DP or DM vs. DP, regardless their DM and DA, respectively. See Tables S3B, S4A and S5A for details on the data treatment. The individual scales are provided on the Figure. Note the specific HG acetylation phenotype of tbl38 with PL (B), the qualitative complementation of PTR3.5 with PL (D), the absence of RGI acetylation phenotype for tbl38 (P) and the quantitative HG methylation phenotype of pmei6 (G, N). These results argue for an HG acetylesterase role of TBL38 and are consistent with the PME inhibitory activity of PMEI6.23
The same analysis and data processing were performed with PG which has an affinity for lowly methylated HG (Figures 8I–8N; Tables S4A–S4D). The DA vs. DP profiles of Col-0, tbl38, and pmei6 were qualitatively similar though a slight increase of species of DA > 0 occurred for tbl38 (Figures 8I–8K; Tables S4A–S4D. The DM vs. DP plots showed a quantitative phenotype of pmei6 as compared to Col-0 and tbl38 in agreement with the PMEI activity of PMEI623 (Figures 8L–8N). We also noticed an about 10-fold higher intensity of OGA species of low DM identified with PG (Figures 8I–8N; Table S4A) vs. those of high DM species identified with PL (Figures 8A–8H; Table S3B). This could reflect the nature of the major HGs in MSCs (low DM and low DA) and/or be related to a possible lower activity of PL toward HGs with relatively high DA.35 To strengthen the HG high acetylation phenotype of tbl38, we additionally compared Col-0 and tbl38 rhamnogalacturonate acetylation profiles to see whether TBL38 could also target the abundant RGI present in seed mucilage (Figures 8O–8P; Table S5A). No increased acetylation phenotype occurred in tbl38 for RGI since the very strong intensity of the identified RGI species that were non-acetylated had similar profiles in both Col-0 and tbl38. This result demonstrates the specificity of the acetylation phenotype of tbl38 toward HGs rather than RGI.
Finally, we produced a recombinant TBL38 (rTBL38) in Pichia pastoris (Figure 9A) which was partially purified using affinity chromatography. rTBL38, at a predicted molecular mass of 42.7 kDa, was gradually enriched in elution fractions when using increasing imidazole concentrations (from 5 mM to 100 mM) as compared to the NaP fraction, used as a negative control. We next used the NaP, 25 mM and 100 mM fractions to assay the acetyl esterase activity of rTBL38 in vitro toward three acetylated substrates: the generic acetylated triacetin, acetylated pectins from sugarbeet and acetylated xylans from tomato. When using pectins, an increasing esterase activity was detected in the 25 and 100 mM native fractions as compared to boiled samples, which correlated with the rTBL38 protein amount inferred from the SDS-PAGE. In contrast, no activity was detected when using the NaP fraction (Figure 9B). Similar results were obtained when using triacetin as a substrate and the 25 and 100 mM fractions (Figure S5A) while no significant activity could be detected when using acetylated xylans as a substrate (Figure S5B).
Figure 9.
Pectin acetylesterase activity of the recombinant TBL38 and molecular modeling of TBL38 displaying its putative catalytic triad and substrate binding pocket
(A) SDS-PAGE showing the production of recombinant TBL38 (rTBL38) in Pichia pastoris and its partial purification using affinity chromatography and increasing concentrations of imidazole. NaP refers to the buffer used to wash the column after the loading of concentrated supernatant.
(B) In vitro enzymatic activity of rTBL38 in NaP and 25/100 mM imidazole fractions using acetylated sugar beet pectins as substrate and the K-ACET enzymatic acetic acid assay kit (n = 3). Boiled samples were used as negative control, mean values ±SD, ∗, p-value <0.05.
(C) Amino acid sequence of TBL38 (At1g29050). The predicted signal peptide (https://services.healthtech.dtu.dk/services/SignalP-6.0/) is underlined. The amino acids in italics correspond to the 15% of the protein not covered by the molecular modeling performed in (D). GDSL and DxxS tetrads that are highly conserved among the 46 TBR/TBLs including TBL389 are highlighted in red and blue, respectively. The amino acids of the putative catalytic triad S136-H360-D357 appear in bold underlined, in red, green and blue, respectively. L139 in TBL38 underlined in yellow replaces R219 in TBL29/ESK1/XOAT1 that was demonstrated to be necessary for acetyltransferase activity.17
(D) The TBL38 structural model proposed in this study is a Phyre2 prediction36 with the highest ranking score (Confidence: 100.0%; Coverage: 85%). The template was the crystallographic structure (X-ray diffraction, 1.85 Å) of A. thaliana TBL29 (At3g55990; PDB 6CCI; Phyred2 c6cciA).17
(E) Zoom on the putative catalytic triad S136-H360-D357 originally identified in Ser proteases,37 and necessary for the acyl-enzyme formation and acetyltransferase activity of TBL29.17
(F and G) Structure and zoom on the catalytic triad of TBL29 for comparison.17 Note that R219 necessary for acetyltransferase activity in TBL2917 is replaced by L139 in TBL38.
Discussion
TBL38 is atypically positioned in the PEROXIDASE36/PECTIN METHYLESTERASE INHIBITOR6 cell wall microdomain of mucilage secretory cells with a transient pattern
Originally, we searched for genes involved in the PMEI6-JIM7/LM20 HG epitopes-PRX36 molecular module22 with a particular interest in HG remodeling enzymes. Using spatiotemporal co-expression with PRX36, we selected TBL38 for its putative role in HG acetylation. Previously described TBLs are mostly anchored to the Golgi membrane (Table S1) with the exception of TBR.18 Although the authors did not refute the possibility of some TBR-GFP signal in the Golgi, they concluded that TBR was a CW protein. The Golgi experimental localization matches with the N-terminal transmembrane domain prediction for 10 TBLs (TBL03, 27, 28, 29, 30, 31, 32, 33, 34, 35) while the Golgi-localized TBL37 has a predicted cleavable signal peptide (akin to a secreted CW protein), and conversely, TBR that has been localized to the CW has a predicted N-terminal transmembrane domain (Table S1). Six TBLs including TBL38 have been identified in CW proteomes but these proteins did not match perfectly to the targeting prediction: there is a good match with cleavable signal peptide prediction for TBL9, 38, 39 and 45, but a poor match with transmembrane domain prediction for TBL15 and TBL40 (Table S1). Here, we clearly localized TBL38-TagRFP in the CW of tobacco leaves or A. thaliana MSCs (Figure 3) in agreement with TBL38 presence in a CW proteome and its predicted cleavable signal peptide (Table S1). In MSCs, we showed that this targeting is restricted to the CW microdomain harboring the PMEI6-JIM7/LM20 HG epitopes-PRX36 molecular module with an intriguing transient localization pattern (6–8 DAP). The localization to the CW microdomain occurs as soon as the fusion protein is produced just after the onset of PRX36 promoter activity. We chose this promoter considering the strong co-expression of PRX36 and TBL38 and because it worked well with stable localization in various CW microdomains in previous studies.22,24,27,28 The physiological relevance of the transient localization pattern of TBL38-TagRFP is attested by the fact that the PTR lines displayed functional complementation in our different assays discussed hereafter. Then, the early delocalization of the fusion protein could be related to the loss of the putative anchoring motif (acetylated HG) as soon as TBL38 performed its acetylesterase activity, leading to the degradation of the protein.
TBL38 is an homogalacturonans acetylesterase
So far, all described TBLs have been associated with a Golgi-localized acetyltransferase activity, either on hemicelluloses or pectins (Figure 1; Table S1). Therefore, the question arose on whether the CW-localization of TBL38 could lead to similar activity. Yet, both acetic acid quantification and mass spectrometry analysis, as well as in vitro enzymatic assays with rTBL38 indicate that TBL38 is an acetylesterase targeting acetylated HGs as opposed to other TBLs (Figures 7, 8, and 9). Interestingly, in a recent elegant study, the screening of the acetyltransferase activities of 25 TBLs including TBL38 did not show any acetyltransferase activity on pectins, mannan, xyloglucan or xylan for TBL38.38 Sequence analysis of the TBL family highlighted the conservation of two characteristic domains: the GDS(L) and DXXH motifs commonly found in esterases.14,16 Crystallography analysis of TBL29/ESK1/XOAT1 pointed to the spatial reunion of these two domains forming the conserved Ser-His-Asp catalytic site, Ser coming from GDS(L) and Asp and His coming from DXXH.17 TBL29 displayed a xylan acetyltransferase activity involving the formation of an acyl-enzyme intermediate on Ser129.17 Therefore, the acetyltransferase activity of TBL29 necessitated a generic acetylesterase activity on both the acetyl donor and the acyl-enzyme intermediate. Based on the crystallography structure of TBL29, we built an in silico model of TBL38 to determine the position of the above mentioned amino acids. The predicted catalytic site was correctly positioned in a putative polysaccharide binding groove (Figures 9D and 9E). Interestingly, in TBL29, the nearby R219 (Figures 9F and 9G) was shown to stabilize the catalysis17 as R219A site mutation led to loss of transfer of acetyl groups for the acetyltransferase activity without loss of the acetylesterase activity necessary for the acyl-enzyme formation.17 TBL multiple alignment17 shows that nine TBLs, including TBL38, do not have this particular Arg residue: TBL12 (E135), TBL30 (Y156), TBL37 (L143), TBL38 (L139) (Figures 9C–9E), TBL39 (T125), TBL40 (L123), TBL41 (L112), TBL42 (N115), TBL43 (L122). All the other TBLs have an Arg except TBL44/PMR5 which possess another polar positive residue at this position (K145) and an acetyltransferase activity.13 Among these nine TBLs with no polar positive residue at this position, four correspond to the six TBLs identified in CW proteomes (Table S1). It is therefore tempting to postulate that the absence of this residue could fit with the absence of a (Golgi-localized) acetyltransferase activity and the presence of a (CW-localized) acetylesterase activity as for TBL38. However, this cannot be the sole determinant since TBL37 and TBL30/XOAT3 both display a Golgi-localized acetyltransferase activity.11,39 The characterization of TBL12, TBL39-43 could help deciphering the importance of this residue at this specific position. Since we could not identify in TBL38 a particular motif or conformation which could explain such opposite esterase vs. transferase activity, we propose that the opposite activity of TBL38 is primarily related to its CW localization. Indeed, TBL-dependent polysaccharide acetylation is done in two consecutive steps: hydrolysis of the acetyl group from the donor as evidence by the formation of the Acyl-enzyme intermediate, followed by transfer of the O-acetyl moiety to its polysaccharide target. To our knowledge, neither acetyl-CoA nor another putative acetyl donor is present in the CW, which could prevent the CW-localized TBL38 to complete the successive steps of the transferase reaction; the enzyme could then rather function as a direct acetylesterase on acetylated HGs. It should be noted that an esterase activity targeting CW polymers was not observed for TBR, despite its CW localization. tbr was associated with lower acetylation level of HGs (the opposite phenotype of tbl38), possibly through the TBR-mediated protection of HG from pectin acetylesterases.18 Although is it unclear how such a mechanism could play out, it is difficult to explain the differences between the measured acetylation levels. Characterization of additional TBLs with regards to subcellular localization and acetylesterase activity is required for a better understanding of TBL roles.
Homogalacturonans fine-tuning of esterification is affected in tbl38
Although the control of HG acetylation and methylation is achieved by different proteins, proof of the direct and indirect interplay between both chemical groups was given. Indeed, HG acetylation could inhibit PME activity by steric hindrance.31,40 Additionally, methyl ester groups are preferentially added on non-acetylated galacturonic acid moieties, which may explain the overall low level of HG acetylation observed in our results with Col-0 and in most plants,41,42 (Figure 8). Although we did not quantify HG methylesterification, the total number of identified species was particularly low in tbl38, when using a PL that has a high affinity for highly methylesterified HGs (Figure 8). Even if we cannot exclude the possibility that the acetyl groups in tbl38 negatively impact the PL activity, this could be associated with reduced HG methylesterification in tbl38 considering the lack of LM20 labeling in tbl38. It should be noted, however, that the combined use of PG/PL might not give the full representation of HG species, since randomly methylated HG of DM around 50% are mediocre substrates for both PL and PG.43 Additionally, JIM7 antibody still labels tbl38 seeds suggesting that the lack of LM20 binding might rather be due to steric hindrance caused by HG acetylation, rather than changes in the pattern of methylesterification. Therefore, LM20 could display specificity for partially methylesterified and non-acetylated HGs, while JIM7 would be specific for partially methylesterified HGs regardless of the acetylation status. Interestingly, nanofibrils of HGs could be observed with LM20 but not with JIM7 in epidermis anticlinal CW from pavement cells.44,45 Considering the proposed different specificities of the two antibodies, one interpretation would be that the reduced steric hindrance and/or altered HG gelling properties32 related to the absence of acetyl groups would be necessary for proper assembly of HGs in nanofibrils. Evaluation of PME activity in tbl38 developing MSCs could give clues as to TBL38 indirect influence on HG methylesterification. Regardless of the DM, our results confirmed increased amounts of HG acetylation in tbl38 seed surface extracts. Contrary to methylesterification, acetylation does not influence the charge density and therefore the cation binding capacity of HGs.32,46 However, experimental evidence have consistently associated highly acetylated HGs with reduced affinity to cations binding such as Ca2+32,40 Conceptually, this could be due to the direct influence of acetylation to conformation, access or cations binding properties, but also to the indirect effect on the PME activity, as stated above.47,48
Increased homogalacturonans acetylation does not influence PEROXIDASE36 anchoring
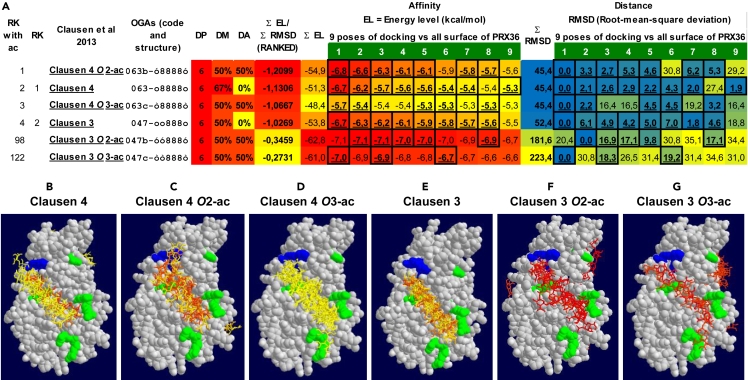
Our results have identified TBL38 as a CW localized HG acetylesterase with increased HG acetylation level in tbl38 and no obvious effect on PRX36 positioning, despite the colocalization of TBL38 and PRX36 in the same CW microdomain. Previous work has demonstrated that PRX36 binding to HG was dependent on both methyl and Ca2+ as the PRX36-TagRFP localization was severely impacted in pmei6 or using EDTA, respectively.22 To better understand the absence of PRX36 delocalization in tbl38, we investigated the implication of acetylated HGs on PRX36 docking using an in silico model based on PRX53 crystallographic data.49 The previous in silico docking study tested the five OGAs of DP6 used to define JIM7 specificity.22,33 It turned out that the three OGAs (so-called Clausen 3, 4 and to a lesser extend 5) that were the best recognized by JIM7 could be used for in silico docking on a PRX36 valley that was validated by site-directed mutagenesis.22 Here, we first randomly screened in silico PRX36 anchoring on 124 OGAs representing all the combinations of methylesterification from DP2 to DP6 (Table S6). The in silico models consisted in nine poses of decreasing affinity (increasing energy level). To rationalize the distribution of these poses on the protein surface, we systematically measured the root-mean-square deviation (RMSD) showing the distance between the best pose and the eight other poses. We integrated the sum of energy levels and sum of RMSD for each OGA as a proxy of affinity and distances (Table S6; see STAR methods). The 124 OGAs could then be sorted in a rationale -though of course imperfect manner-for their ability to bind to the PRX36 valley. This strongly reinforced the previous docking experiments made with a priori with Clausen1-5 OGAs22 since Clausen 4 and Clausen 3 ranked 1 and 2 (Figure 10; Table S6). Finally, we tested the impact on the in silico docking of the addition of 2-O or 3-O acetyl on the non-methylated galacturonic acids of Clausen 3 and 4 (Figure 10). The addition of acetyl on hit number 2 (Clausen 3) strongly impaired PRX36 docking capacity, which does not fit with the observed results (no obvious modification of PRX36 anchoring and no mucilage release phenotype in tbl38). However, the addition of acetyl on hit number 1 (Clausen 4) did not have a strong impact on the docking capacity (Figure 10). In turn, this is consistent with the immunolocalization of αPRX36 and confocal imaging of PRX36-TagRFP in tbl38 background (Figures 4C and 4D) which did not show any obvious mislocalization of PRX36, suggesting that the Clausen 4 model might have a higher affinity toward PRX36 in muro.
Figure 10.
The acetylation of the best oligogalacturonate (OGA) hit for PRX36 docking does not impair the docking prediction
(A) The two best hits obtained following the random systematic screening of molecular docking of PRX36 on 124 OGAs (DP6-DP2) covering all theoretical demethylesterification patterns correspond to two OGAs of DP6 constituting the epitope of JIM7 (Table S6). These two OGAs named Clausen 3 and Clausen 422 were further acetylated in silico in their non-methylated galacturonic acids either in O-2 or O-3 positions. The molecular docking performed on the whole PRX36 surface was compared between the non-acetylated and acetylated OGAs using autodock vina showing the 9 best poses including their individual energy level (EL) and Root-mean-square deviation (RMSD) indicating the affinity of the interaction (the lower, the better) and the distance in Angstrom of the 9 poses for a given OGA (the lower, the shorter), respectively. (i) Data mining was performed as detailed in Table S6 and STAR Methods and the OGAs were sorted according to ∑EL/∑RMSD used as a proxy to integrate the affinity and the distance (the lowest, the highest affinity and shorter distance). Red-to-yellow and blue-to-yellow heatmaps were drawn for EL and RMSD, respectively. The EL or RMSD values appearing in bold underlined frames correspond to poses fitting to the PRX36 valley previously demonstrated22 to accommodate the hexagalacturonates Clausen 3, 4 and 5 (047-oo888o, 063-o8888o and 027-8o8o8o) that are recognized by the JIM7 monoclonal antibody.33 Note that the docking specificity of the best hit (Clausen 4) is not significantly modified between the non-acetylated and the two acetylated versions whereas the docking of the second best hit (Clausen 3) is strongly impaired following acetylation.
(B–G) Visualization of the docking models for the six combinations of hexagalacturonates. Key: RK, rank; DP, degree of polymerization; DM, degree of methylation; DA, degree of acetylation; OGA, oligogalacturonate; o, demethylesterified galacturonic acid; ó, O2-acetylated demethylesterified galacturonic acid; ò, O3-acetylated demethylesterified galacturonic acid; 8, methylated galacturonic acid. The red and blue amino acids in (B–G) correspond to polar positive and hydrophobic amino acids from the OGA docking valley, purported to interact with negatively charged demethylesterified and neutral methylesterified galacturonic acids, respectively, and whose experimental mutation impaired both in silico PRX36 docking and in vivo PRX36 localization.22 The OGAs in (B–G) were color-coded following the energy level heatmap shown in (A).
In light of our results, we conclude that TBL38 acts as an HG acetylesterase in the PMEI6-JIM7/LM20 HG epitopes-PRX36 CW microdomain with no effect on PRX36 anchoring observed in tbl38 contrarily to pmei6. The uncommon acetylesterase activity could be related to the localization to the CW where acetyl-CoA is absent. We could hypothesize that TBL38 could have an acetyltransferase activity if artificially targeted in a different environment such as the Golgi considering the presence of the canonical amino acids required for acetyltransferase activity. It remains to be understood why evolution has selected a acetyltransferase-capable protein to deacetylate HGs instead of the traditionally described PAEs, though in A. thaliana, no PAE has yet been experimentally linked to HG deacetylation. Beyond this case study, this work paves the way for further understanding of the role of fine-tuning of complex CW polysaccharide structures.
Limitations of the study
This study links the remote localization of TBL38 in a CW microdomain achieved through thorough cell biology studies and a biochemical phenotype with state-of-the art mass spectrometry oligosaccharide profiling. Our original seed abrasion method allowed for the obtaining fractions enriched in the CW microdomain but do not strictly correspond to the CW microdomain observed under the microscope. The increasing development of mass spectrometry imaging technologies may reach in a near future a spatial resolution and semi-quantitative improvements that will enable observing CW microdomain phenotypes in situ. We were not yet able to associate the increased HG acetylation observed in tbl38 to any clear developmental phenotype. However, considering the localization of this process to a remote CW microdomain, additional tools will be required to search for subtle developmental phenotypes, similarly to the development of the new original abrasive method allowing the characterization of the biochemical phenotype of tbl38.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| JIM7 (Rat IgA primary antibody) specific for partially ME HG | Kerafast | Cat# ELD005 |
| LM20 (Rat IgM primary antibody) specific for partially ME HG | Kerafast | Cat# ELD003; RRID:AB_2734789 |
| JIM5 (Rat IgG primary antibody) specific for low-to-no ME HG | Plant Probes | Cat# JIM5 |
| LM19 (Rat IgM primary antibody) specific for low-to-no ME HG | Plant Probes | Cat# LM19; RRID: AB_2734788 |
| LM7 (Rat IgM primary antibody) specific for partially ME HG | Plant Probes | Cat# LM7 |
| LM18 (Rat IgG2c primary antibody) specific for partially-to-no ME HG | Plant Probes | Cat# LM18 |
| INRA-RU2 (mouse IgG primary antibody) specific for RGI backbone | Ralet et al. (2010)50 | N/A |
| LM25 (Rat IgM primary antibody) specific for xyloglucan (XXLG & XLLG) | Plant Probes | Cat# LM25 |
| αPRX36 (Rabbit polyclonal primary antibody) specific for CSLIGSMENIPSPES PRX36 peptide-KLH conjugate | Genscript private order Francoz et al. (2019)22 | αPRX36 |
| αTagRFP (Rabbit polyclonal primary antibody) | Thermo-Fisher (Invitrogen) | Cat# R10367; RRID: AB_10563941 |
| Goat αRat IgG-A488 secondary antibody | Thermo-Fisher (Invitrogen) | Cat# A-11006; RRID:AB_2534074 |
| Goat αRat IgG-AP secondary antibody | Sigma-Aldrich | Cat# A8438; RRID:AB_258391 |
| Goat αmouse IgG-AP secondary antibody | Sigma-Aldrich | Cat# A5153; RRID:AB_258225 |
| Goat αRabbit IgG-A488 secondary antibody | Thermo-Fisher (Invitrogen) | Cat# A-11034; RRID:AB_2576217 |
| Goat αRabbit IgG-AP secondary antibody | Sigma-Aldrich | Cat# A3687; RRID:AB_258103 |
| αDig Fab-AP secondary antibody | Sigma-Aldrich (Roche) | Cat# 11093274910; RRID:AB_2734716 |
| Bacterial and virus strains | ||
| Agrobacterium tumefaciens (strain GV3101:pMP90) | Koncz and Schell (1986)51 | N/A |
| Chemicals, peptides, and recombinant proteins | ||
| Ruthenium red | Schuchardt Müchen | N/A |
| Prolong Gold antifade mounting medium | Molecular probes | Cat# P36934 |
| Critical commercial assays | ||
| Enzymatic Acetic Acid Assay (ACS Manual Format) | Megazyme | K-ACET |
| Enzymatic Acetic Acid Assay (Acetate Kinase Manual Format) | Megazyme | K-ACETRM |
| Deposited data | ||
| eFP browser | http://bar.utoronto.ca/efp/cgi-bin/efpWeb.cgi?dataSource=Seed | RRID: SCR_006748 |
| Arabidopsis thaliana seed development tissue specific transcriptomics | Belmonte et al. (2013)25 | GEO: GSE12404 |
| Protein DataBank no. 1pa2, PRX53/At5g06720 | Ostergaard et al. (2000)49 | https://www.ebi.ac.uk/pdbe/entry/pdb/1pa2 |
| Protein DataBank no. 6cci, TBL29/ESK1/XOAT1/At3g55990 | Lunin et al. (2020)17 | https://www.ebi.ac.uk/pdbe/entry/pdb/6cci |
| Glyco3D: α-D-(1–4) polygalacturonic acid structural model (Braccini et al. (1999)52) | Perez et al. (2015)53 | http://glyco3d.cermav.cnrs.fr/mol.php?type=polysaccharide&molecule=2504 |
| Experimental models: Organisms/strains | ||
| Arabidopsis thaliana: Col-0 wild-type | NASC | N1093 (https://arabidopsis.info/) |
| Arabidopsis thaliana: tbl38 transfer DNA insertion mutant | This study | SAIL_34_C03 (https://arabidopsis.info/) |
| Arabidopsis thaliana: prx36 transfer DNA insertion mutant | Kunieda et al. (2013)24 | SAIL_194_G03 |
| Arabidopsis thaliana: pmei6 transfer DNA insertion mutant | Saez-Aguayo et al. (2013)23 | SM_3.19557 |
| Arabidopsis thaliana: proPRX36:TBL38-TagRFP/tbl38 | This study | Lines PTR2.4, PTR3.5, PTR5.2 |
| Arabidopsis thaliana: proPRX36:PRX36-TagRFP/tbl38 | This study | Line 4.2 |
| Arabidopsis thaliana: proPRX36:PRX36-TagRFP/prx36 | Francoz et al. (2019)22 | Line 3.8 |
| Oligonucleotides (5′-3′). | ||
|
tbl38 genotyping: CTTGGCATTGGTGGACTCACAAAG |
This study | AT1G29050_For2 (BF2) |
|
tbl38 genotyping: CATCGAACTCAACACTTTCGACACTACTCC |
This study | AT1G29050_Rev2 (BR2) |
|
tbl38 SAIL mutant genotyping: GCCTTTTCAGAAATGGATAAATAGCCTTGCTTCC |
https://abrc.osu.edu/help/genotyping | LB1 (T2) |
|
proPRX36 Golden Gate lvl0/Genotyping: GGTCTCCGGAGGGCCCATATAAGTT Primer not used for cloning: previously cloned22 The underlined bases are identical to the template DNA. |
This study | pPRX36_GGAG_F (F10) |
|
proPRX36 Golden Gate lvl0: GGTCTCGCATTTTGGACTCTCAGC Primer not used for cloning: previously cloned22 |
This study | pPRX36_CATT_R (R10) |
|
PRX36 CDS Golden Gate lvl0: GGTCTCCAATGAATACAAAAACGGT Template: PRX36 cDNA (RIKEN pda20378) |
This study | PRX36CDS_AATG_F (F11) |
|
PRX36 CDS Golden Gate lvl0) GGTCTCGTAGCAACATCATGGTTAA Template: PRX36 cDNA (RIKEN pda20378) |
This study | PRX36CDS_TAGC_R (R11) |
|
TagRFP CDS Golden Gate lvl0: GGTCTCCGCTACCGGTATGGTGAG Template: TagRFP-AS-N entry clone (Evrogen FP149) |
This study | TagRFP_GCTA_F (F12) |
|
TagRFP CDS Golden Gate lvl0: GGTCTCGAAGCAAGAAAGCTGGGT Template: TagRFP-AS-N entry clone (Evrogen FP149) |
This study | TagRFP_AAGC_R (R12) |
|
TBL38 CDS Golden Gate lvl0: GGTCTCCAATGATGGGTTTCAAACT Template: TBL38 cDNA (RIKEN pda09004) |
This study | TBL38CDS_AATG_F (F15) |
|
TBL38 CDS Golden Gate lvl0: GGTCTCCTAGCCATCGTAAGAGCTG Template: TBL38 cDNA (RIKEN pda09004) |
This study | TBL38CDS_TAGC_R (R15) |
| Golden Gate Lvl 0 sequencing: GTAAAACGACGGCCAGT Template: clones in pGEM-T easy/pCR ZeroBlunt vectors |
This study | M13_FOR (F25) |
| Golden Gate Lvl 0 sequencing: CAGGAAACAGCTATGACCA Template: clones in pGEM-T easy/pCR ZeroBlunt vectors |
This study | M13_REV (R25) |
|
Pro35S Golden Gate lvl0/Genotyping: GGTCTCCGGAGCCAGTGAATTGT Template: pEAQ-HT-DEST1; Primers not used for cloning: Genscript synthesis pUC57-p35S (Genscript) |
This study | p35S CaMV_GGAG_F (F31) |
|
Pro35S Golden Gate lvl0: GGTCTCGCATTATCGAATTTGGG Template: pEAQ-HT-DEST1; Primers not used for cloning: Genscript synthesis pUC57-p35S (Genscript) |
This study | 5′UTR CPMV_CATT_R (R31) |
| Golden Gate Lvl1 (pL1V-R2 recombinant vector) sequencing: CATCCAACAAATTAAAAGCC | This study | pPRX36_3'_F (F33∗) |
| Golden Gate Lvl1 (pL1V-R2 recombinant vector) sequencing: TACTTCTGCTTGACGAGGTATTGTT |
This study | 5′UTR CPMV_3'_F (F35∗) |
| Golden Gate Lvl1 (pL1V-R2 recombinant vector) sequencing: GTACACTTAAAGTGGTGATTATTGACAG |
This study | TagRFP_N_R (R36∗) |
| Golden Gate Lvl1 (pL1V-R2 recombinant vector) sequencing: AAGCAGACAAAGAAACTTACGTGGA |
This study | TagRFP_C_F (F37∗) |
| Golden Gate Lvl1 (pL1V-R2 recombinant vector) sequencing: ATTATATGATAATCATCGCAAGAC |
This study | T-Nos_5'_R (R40∗ |
| Golden Gate Lvl1 (pL1V-R2/pL2V-HYG/KAN) sequencing/genotyping: GCCAATATATCCTGTCAAACACTG | This study | pL1V-R2_RBshort_F (F50) |
| Golden Gate Lvl1 (pL1V-R2 recombinant vector) sequencing: TAGACAACTTAATAACACATTGCGGAC |
This study | pL1V-R2_LB_R (R51) |
| Golden Gate Lvl2 (pL2V-HYG/KAN recombinant vector) sequencing: TTGGAGTAGACCAGAGTGTCGTG | This study | pL2V-3′p35S_F (F52) |
|
TBL38 RT-PCR: AAAACCCAAAACCTTTGATATTTGTCTAA |
This study | TBL38_RT_OUT_F (F61) |
|
TBL38 RT-PCR: CACTAGAAGATTTACGAATGTCATTTACAGA |
This study | TBL38_RT_OUT_R (R61) |
|
ACT2 RT-PCR: GGTAACATTGTGCTCAGTGGTGG |
Fulton and Cobbett (2003)54 | ACT-2_RT_OUT_F (F65) |
|
ACT2 RT-PCR: CTCGGCCTTGGAGATCCAC |
Fulton and Cobbett (2003)54 | ACT-2_RT_OUT_R (R65) |
| Recombinant DNA | ||
| Golgi marker: Binary vector expressing p35S::GmMan1-YFP | Nelson et al. (2007)55 | G-yb CD3-966 |
| Plasma membrane marker: Binary vector expressing p35S::AtPIP2A-YFP | Nelson et al. (2007)55 | pm-yb CD3-1006 |
| PRX36 (At3g50990) ORF |
Seki et al. (2002)56; Seki et al. (1998)57 |
RAFL22-03-B11 (pda20378) |
| TBL38 (At1g29050) ORF |
https://www.brc.riken.jp Seki et al. (2002)56; Seki et al. (1998)57 |
RAFL09-90-M11 (pda09004) |
| TagRFP ORF (codon use optimized for Arabidopsis and Saccharomyces) | https://evrogen.com/ | Gateway TagRFP-AS-N entry clone (FP149) |
| Golden Gate Level 0 pPRX36 (can be cloned with primers F10/R10 not used here: previously cloned by gene synthesis) The promoter sequence is according to Kunieda et al. (2013)24 | Francoz et al. (2019)22 https://www.genscript.com/ |
pUC57-promPRX36 (code 1) |
| Golden Gate Level 0 p35S_edit (can be cloned with primers F31/R31 not used here: gene synthesis). This includes Promoter of CaMV and 5′UTR of CPMV from pEAQ-HT-DEST1 GQ497235 (5906–6769) (Sainsbury et al. (2009).58 The sequence was adapted for easier Golden Gate cloning by adding GGTCTCCGGAG upstream of promoter and AATGCGAGACC downstream of 5′UTR as 5′ and 3′ security margin, respectively, and by removal of two BbsI sites (6530: A to C; 6549: T to C) before synthesis. | This study https://www.genscript.com/ |
pUC57-prom35S (code 3) |
| Golden Gate Level 0 PRX36 CDS (cloned with primers F11/R11 using PRX36 (At3g50990) ORF (see above)) | This study | pCR Zero Blunt-PRX36 CDS (code 4) |
| Golden Gate Level 0 TBL38 CDS (cloned with primers F15/R15 using TBL38 (At1g29050) ORF (see above)) | This study | pCR Zero Blunt-TBL38 CDS (code 5) |
| Golden Gate Level 0 Tag-RFP (cloned with primers F12/R12 using TagRFP ORF (see above)) | This study | pCR Zero Blunt-TagRFP CDS (code 20) |
| Golden Gate Level 0 T-Nos (3′UTR, polyadenylation signal/NOS terminator (A. tumefaciens) | Engler et al. (2014)59 Gift from P.M. Delaux (LRSV, Auzeville, France) |
pICH41421 |
| Golden Gate Level 1 p35S::TBL38-TagRFP T-Nos (combination of 3-5-20-21 level 0 plasmids cloned in pICH47811 “pL1V-R2” vector (Weber et al. (2011)60) | This study | Lvl1 p35S::TBL38-TagRFP T-Nos |
| Golden Gate Level 2 p35S::TBL38-TagRFP T-Nos (Level 2 plasmid backbone was EC15027 “pL2V-HYG” with pICH471744 “pL1M-ELE-2” as a linker-containing plasmid (gift from Dr. P.M. Delaux, LRSV, Auzeville-Tolosane, France)) | This study | Lvl2 p35S::TBL38-TagRFP T-Nos |
| Golden Gate Level 1 pPRX36::TBL38-TagRFP T-Nos (combination of 1-5-20-21 level 0 plasmids cloned in pICH47811 “pL1V-R2” vector (Weber et al. (2011)60) | This study | Lvl1 pPRX36::TBL38-TagRFP T-Nos |
| Golden Gate Level 2 pPRX36::TBL38-TagRFP T-Nos (Level 2 plasmid backbone was EC15027 “pL2V-HYG” with pICH471744 “pL1M-ELE-2” as a linker-containing plasmid (gift from Dr. P.M. Delaux, LRSV, Auzeville-Tolosane, France)) | This study | Lvl2 pPRX36::TBL38-TagRFP T-Nos |
| Golden Gate Level 1 pPRX36::PRX36-TagRFP T-Nos (combination of 1-4-20-21 level 0 plasmids cloned in pICH47811 “pL1V-R2” vector (Weber et al. (2011)60) | This study | Lvl1 pPRX36::PRX36-TagRFP T-Nos |
| Golden Gate Level 2 pPRX36::PRX36-TagRFP T-Nos (Level 2 plasmid backbone was EC15027 “pL2V-HYG” with pICH471744 “pL1M-ELE-2” as a linker-containing plasmid (gift from Dr. P.M. Delaux, LRSV, Auzeville-Tolosane, France)) | This study | Lvl2 pPRX36::PRX36-TagRFP T-Nos |
| Software and algorithms | ||
| ImageJ |
https://imagej.net/ Schneider et al. (2012)61 |
RRID: SCR_003070 |
| R package | https://www.r-project.org/ | RRID: SCR_001905 |
| Microsoft Excel | https://www.microsoft.com/ | RRID: SCR_016137 |
| Corel-Photo-Paint | www.corel.com/ | RRID: SCR_016137 |
| Phyre2 |
http://www.sbg.bio.ic.ac.uk/phyre2/html/page.cgi?id=index Kelley et al. (2015)36 |
N/A |
| AutoDock Tools |
http://autodock.scripps.edu/resources/tools https://autodocksuite.scripps.edu/adt/ Morris et al. (2009)62 |
RRID: SCR_012746 |
| AutoDock Vina SCR_011958 |
http://vina.scripps.edu/ Trott and Olson (2010)63 |
RRID: SCR_011958 |
| Swiss-PdbViewer |
http://www.expasy.org/spdbv/ https://spdbv.unil.ch/ Guex and Peitsch (1997)64 |
RRID: SCR_002668 |
| Volocity 3D Image Analysis Software | http://www.perkinelmer.com/pages/020/cellularimaging/products/volocity.xhtml | RRID: SCR_002668 |
| NDP view | https://www.hamamatsu.com/ | N/A |
| Quartz software (Waters embedded software, release 5) | https://www.waters.com/ | N/A |
| Mass Lynx 4.2 (Waters) | https://www.waters.com/ | N/A |
| MZmine |
http://mzmine.github.io/ Pluskal et al. (2010)65 |
RRID: SCR_012040 |
| Other | ||
| Epson perfection V700 photo scanner | https://epson.com/ | N/A |
| Leica DM IRB/E inverted microscope equipped with a Leica MC190HD digital camera | https://www.leica-microsystems.com/ | N/A |
| Nanozoomer 2.0RS scanner | https://www.hamamatsu.com/ | N/A |
| Upright confocal laser scanning microscope (LEICA SP8) | https://www.leica-microsystems.com/ | N/A |
| Spinning disk confocal microscopy: Yokogama CSU-X1 scan head, two EmCCD Hamamatsu C9100-13 cameras |
https://www.hamamatsu.com/ | N/A |
| MED 020 modular high vacuum coating and Quanta 250 FEG FEI scanning electron microscopy | https://trigenotoul.com/en/ressources-tech/electron-microscopy/scanning-electron-microscopy/ | N/A |
| Retsch MM400 ball mill | https://www.retsch.com/products/milling/ball-mills/ | N/A |
| FastPrep-24™ Classic bead beating grinder and lysis system | https://www.mpbio.com/eu/fastprep-24-classic-instrument-1-each/ | Cat# 116004500 |
| IP-RP-UHPC-MS. Acquisitions were performed on a Select Series Cyclic IMS (Waters, Wilmslow, UK) coupled with a UHPLC system (Acquity H-Class Plus, Waters, Manchester, UK). | https://www.waters.com/ | N/A |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Vincent Burlat (vincent.burlat@univ-tlse3.fr).
Materials availability
Material generated in this study will be shared by the lead contact upon request.
Data and code availability
-
•
All data including Microscopy data reported in this paper will be shared by the lead contact upon request.
This paper mainly concerns the three following genes: TBL38 (AT1G29050), PMEI6 (AT2G47670) and PRX36 (AT3G50990) (https://www.arabidopsis.org/index.jsp). The previously published transcriptomics data25 that we used for co-expression analysis corresponds to GSE12404 dataset (https://www.ncbi.nlm.nih.gov/geo/). Homology models for PRX36 and TBL38 (UniProt accession number Q9SD46 and Q8VY22, respectively) were built using as a template, the crystallographic structure of A. thaliana PRX53 (At5g06720; Protein DataBank no.1PA2)49 and TBL29/ESK1/XOAT1 (At3g55990; 6cci pdb file),17 respectively.
-
•
Any additional information required to reanalyze the data reported in this work paper is available from the lead contact upon request.
Experimental model and study participant details
Plant material and growth conditions
A. thaliana mutants were ordered from the NASC for tbl38: SAIL_34_C03 (https://arabidopsis.info/; Figure S3), or were previously available for prx36 (SAIL_194_G03)24 and pmei6 (SM_3.19557).23 Homozygous lines for tbl38 were identified by PCR (Figure S3; key resources table). The knock out status of tbl38 was determined by RT-PCR (Figure S3; key resources table). A. thaliana culture was performed as previously described.22
Nicotiana benthamiana were cultivated in a growth chamber under a 16 h day/8 h night cycle at 23°C (Neon (86.90 μmol/m/s-1)/22°C upon 70% humidity. They were transplanted after 14 days and fertilized each week.
Method details
Transcriptomic data mining
PRX36, PMEI6 and TBL38 seed-specific expression profiles were obtained using the seed data source of eFP browser (http://bar.utoronto.ca/efp/cgi-bin/efpWeb.cgi?dataSource=Seed). Tissue-specific seed development transcriptomic data25 was used to build the PRX36 co-expression network.22 The TBL family9 was filtered out.
Bioinformatic analysis of the TBL family
Previous TBL38 phylogeny9 was used. The occurrence of TBLs in cell wall proteomes was assessed using http://www.polebio.lrsv.ups-tlse.fr/WallProtDB/. The topological prediction of the N-terminal hydrophobic sequence as being a transmembrane domain or a signal peptide was determined using http://aramemnon.uni-koeln.de/.
Ruthenium red mucilage release test, image analysis and statistical analysis
The high-throughput adherent mucilage release semi-quantitative phenotyping used the ruthenium red (key resources table) staining method previously described allowing to calculate adherent mucilage area and circularity.22 Briefly, following staining, seeds were transferred in 12-well microplates and scanned at 6400 dpi with an Epson Perfection V700 photo scanner. Images were analyzed using ImageJ 1.8 (https://imagej.nih.gov/ij/) without edition of native images with an updated.txt ImageJ script described below that facilitates cleaning of the data and allows automatic storage of the data. Seed area (without mucilage) and whole seed area (seed + mucilage) were manually edged in a semi-automatic fashion. Non-individualized seeds were automatically removed from the analysis and selected parameters were measured. A manual examination allowed to double check the absence of artifactual data. The cleaned data were extracted and pooled into a Microsoft Excel sheet for further analyses. At least two technical repeats, each with about 50–100 seeds were performed for seed batches coming from 6 individual Col-0 and tbl38 plants simultaneously grown in the same conditions. Results are presented as mean ± SD with n > 1,000 seeds. Statistical data were obtained using ANOVA tests with n ≥ 3.
ImageJ script for ruthenium red staining semi quantitative analysis
//13 may 2022, A. LE RU, FRAIB.
//quantification of seed surface with and without mucilage calibrated for 6400dpi scan images.
//reset to 0 the ROI and Tables of results.
//adjustment by the user of threshold on seed with mucilage (black + pink) and then threshold for seed area without mucilage (black only)
//(use the same thresholds for a serie of images to be compared)
//creation and numbering of seeds in the ROI manager.
//curation by the user of the data on the image.
//(move with the hand tool, deselect the hand tool (select e.g., the line tool) click on a mask and remove it with the keyboard back key)
//(make sure that for an aberrant seed, both masks (seed+ mucilage and seed only) are deleted to avoid a gap in the result Table
//after validation: reset of result Table and measurement of remaining ROIs.
//draw of Roi on the original image + backup the image.
//backup of ROI manager and results
run("Clear Results"); roiManager("Reset"); roiManager("UseNames", "true"); run("Set Measurements … ", "area mean perimeter shape display redirect = None decimal = 3"); setForegroundColor(255, 255, 0);
run("Line Width … ", "line = 1"); roiManager("UseNames", "true");
title = getTitle();
nom = File.nameWithoutExtension(); dir = File.directory;
run("Duplicate … ", " ");
title2 = getTitle();
selectWindow(title);
run("Properties … ", "channels = 1 slices = 1 frames = 1 unit = um pixel_width = 4.1344 pixel_height = 4.1344 voxel_depth = 4.1344 global");
//run("Subtract Background … ", "rolling = 100 light separate sliding");
run("Split Channels");
selectWindow(title+" (blue)");
close();
//threshold graines ALL (graine + mucillage indiv
selectWindow(title+" (green)");
run("Threshold … "); waitForUser("graines entiere","mettre en rouge les graines entieres"); setOption("BlackBackground", false); run("Convert to Mask"); run("Watershed"); run("Set Measurements … ", "area shape display redirect = None decimal = 3"); run("Analyze Particles … ", "size = 60000-Infinity circularity = 0.7–1.00 show = Masks exclude add");
//run("Analyze Particles … ", "size = 200000-Infinity show = Masks exclude add");
c = roiManager("Count");
for(i = 0; i<c; i++)
{
roiManager("Select", i); roiManager("Rename","graine ALL"+i+1);
}
//threshold graines seules et add manager = graines seules global
selectWindow(title+" (red)");
run("Threshold … "); waitForUser("center des graines","mettre en rouge le center des graines"); setOption("BlackBackground", false);
run("Create Selection"); roiManager("Add"); roiManager("Select", c); roiManager("Rename", "graines noires global");
for(i = 0; i<c; i++)
{
roiManager("Select", newArray(i,c)); roiManager("AND"); roiManager("Add"); roiManager("Select", c+i+1); roiManager("Rename", "graine"+i+1);
}
roiManager("Show All"); roiManager("Measure"); selectWindow(title2); roiManager("Show All"); waitForUser("trier les ROI","trier le ROI"); run("Clear Results"); roiManager("Show All"); roiManager("Measure"); saveAs("Results", dir+nom); roiManager("draw"); saveAs("jpeg", dir+nom+"_DRAW");
roiManager("save", dir+nom+".zip").
TagRFP reporting constructs, plant transformation and selection
The primers used for vector construction are listed in key resources table. Full length cDNA clones and custom-ordered DNA plasmids used as template DNA for further cloning are listed in key resources table. Level 0 GoldenGate generated constructs are listed in key resources table. Level 1 GoldenGate plasmids assembled in pL1V-R2 vector and finalized Level 2 GoldenGate generated constructs are detailed in key resources table. Level 1 plasmid was pICH47811 “pL1V-R2”pL1V-R2 vector.60 Level 2 plasmid was EC15027 “pL2V-HYG” with pICH471744 “pL1M-ELE-2” as a linker-containing plasmid (gift from Dr Pierre-Marc Delaux, LRSV, Auzeville-Tolosane, France). Every construct was checked by restriction analysis and sequencing prior to its transfer into Agrobacterium tumefaciens GV3101:pMP90 strain.51 The transformed bacteria were grown in the presence of 15 μg mL−1 gentamycin, 50 μg mL−1 rifampicin and 25 μg mL−1 hygromycin. A. thaliana plants were transformed by floral-dipping66 or spraying flower buds with an Agromix containing 0.05% (v/v) Silwet L-77 (De Sangosse 2000235). proPRX36::PRX36-TagRFP construct was transformed in prx36 and tbl38 plants. The proPRX36::TBL38-TagRFP construct was transformed in tbl38 plants. These last complemented lines were referred as PTR2.4, PTR3.5 and PTR5.2 in the manuscript. Seeds were selected on Murashige and Skoog (MS) medium containing 25 μg mL−1 hygromycin. Three independent homozygous transformed plant lines were studied for each construct.
The pro35S::TBL38-TagRFP construct was transiently co-transformed with either G-yb CD3-966 sialyltransferase-YFP Golgi marker or pm-yb-CD3-1006 aquaporin-YFP plasma membrane marker55 in 30-day-old N. benthamiana leaves. The final inoculum consisted in a mix of various A. tumefaciens lines: 10% (v/v) subcellular YFP-marker line, 80% (v/v) fusion protein-Tag-RFP line of interest, and 10% (v/v) P19 silencing inhibitor-containing vector line. Leaf infiltration was done on the abaxial face through the stomata with a 1 mL syringe and pieces of leaves were mounted and analyzed under confocal microscopy 48 h-post agro-infiltration.
A. thaliana silique fixation, paraffin tissue array embedding and microtomy
The protocol was as previously described,22 with minor improvement using biopsy foam enabling handling more samples. Briefly, for each genotype, flowers were marked on the developing floral stem at the petal emergence stage used as a proxy to define the pollination time. At the end of floral stem development, sequential kinetics of developing siliques of each genotype were systematically harvested, placed in order (pedicel on top) and sandwiched in Deltalab histoset 2 embedding cassettes (Dutscher 039751) between two 30,2 × 25,4 × 2 mm SimPort biopsy foams (Dutscher 040666). Selected marked siliques that were initially labeled to account for a given day after pollination (DAP) were placed with pedicels facing down for better identification. Shortly after, cassettes were submerged in FAA fixative (3.7% (v/v) formaldehyde from 37% formaldehyde solution (Sigma 1.04002); 5% (v/v) acetic acid; 50% (v/v) ethanol; 35% (v/v) Milli-Q water or RNase-free DEPC-treated water, depending on the necessity), vacuum infiltrated for 8 × 1 min and fixed at 4°C for 6–8 h. Cassettes were then washed 4 times in 50% ethanol and placed at 4°C for 72–84 h. Sample dehydration was done in an H2O/ethanol/tert butanol gradient series (50/40/10, 30/50/20, 15/50/35, 0/45/55, 0/25/75, 1 h; 0/0/100 overnight, 0/0/100 for 1 h), followed by a tert butanol/paraplast gradient series (50/50 overnight, 0/100 three times over 2 days) with final embedding in paraplast.22,67,68 The ordered siliques containing seed development kinetics were used to assemble organized tissue arrays each encompassing up to 1000 developing seeds22,67 To assemble paraffin blocs, cassettes were delicately opened on a slide warmer set to 60°C (LabScientific XH-2001) and siliques were deposited on metallic mold with respect to the initial order. The molds were covered with a fitting plastic ring (Simport M460), the paraffin tissue-array blocks were allowed to solidify at least overnight at 4°C and could be stored for months/years. Ten-to 12 μm-thick serial sections were made with a rotary microtome and spread on silane coated slides.
In situ RNA hybridization
In situ RNA hybridization experiments were performed as described.67,68 In short, a plasmid (RAFL09-90-M11, pda09004) containing TBL38 full length cDNA including 5′-UTR and 3′-UTR was ordered at RIKEN BioResource Center (https://www.brc.riken.jp)56,57 to be used as a template for riboprobe synthesis. The plasmid was first linearized with EcoRI at the 5′ end of the TBL38 ORF and with BamHI at the 3′ end to further generate anti-sense and sense probe used as a negative control, respectively. The synthesis of digoxygenin-labeled riboprobes was performed by in vitro transcription using DigRNA labeling mix (Roche 11277073910) and T3 (Promega P2083) or T7 RNA polymerase (Promega P2075), respectively. Serial sections of paraffin-embedded wild-type seed development kinetics encompassing all developmental stages were first deparaffinized with xylene and rehydrated with an inverted ethanol series up to water. Each probe was hybridized overnight on paraffin serial sections of seeds. Following stringent washing steps, detection of hybridized riboprobes was performed with the anti-digoxigenin-AP conjugates (key resources table) revealed with its chromogenic substrate (BCIP/NBT) overnight. Finally, slides were mounted in EUKITT (Dutscher 045799) and scanned with the nanozoomer HT (Hamamatsu, https://www.hamamatsu.com/). Image analysis was performed using NDPview (Hamamatsu, https://www.hamamatsu.com/) and figures were assembled using Corel Photopaint.
Immunofluorescence on tissue-array sections
We used our previously described protocol.22 Briefly, serial sections of paraffin-embedded Col-0 or t bl38mutant seed development kinetics were first deparaffinized with xylene and rehydrated with an inverted ethanol series up to water. The slides were placed in a 20-slide plastic rack and collectively blocked in 200 mL 5% TTBS-milk (5% non-fat dry milk (Regilait), 0.01 M Tris-HCl pH 7.5, 0.5 M NaCl, 0.3% Tween 20) for 30 min at RT. Slides containing Col-0 and tbl38 samples were separately incubated with 150 μL of anti-PRX36 primary antibodies (key resources table) diluted 1:10 dilution in TTBS-milk, for a 3–4 h under a coverslip in a home-made humid chamber. The coverslip was carefully removed by dipping the slide in TTBS-milk and the slides were collectively washed 6 × 5 min in 200 mL TTBS. 150 μL of goat-anti rabbit-A488 secondary antibodies (key resources table) of diluted 1:100 in TTBS-milk was incubated for 1–2 h at RT. The coverslips were carefully removed and the slides were collectively washed 3 × 5 min in 200 mL TTBS and 3 × 1 min in Milli-Q water. Slides were mounted in Prolong Gold antifade (Molecular probes P36934) or Fluoromount-G (Invitrogen 15586276). Serial section of Col-0, tbl38, PTR2.4 and PTR3.5 developmental kinetics were similarly labeled with JIM7 or LM20 primary antibodies followed by Goat anti-rat A488 secondary antibodies (key resources table). Slides were scanned using a Nanozoomer 2.0RS scanner (Hamamatsu) at 20 × or 40 ×. The FITC (excitation: 482/18 nm; dichroic mirror 488 nm; emission: 525/30 nm) filter sets and the bright field (BF) mode were sequentially used to visualize Alexa 488 fluorescence and the morphology, respectively. Scans were analyzed using NDP view (Hamamatsu, Hamamatsu City, Japan).
Confocal spinning disk microscopy of seed development kinetics of A. thaliana stable transformants (high throughput strategy)
Developing siliques taken from plants expressing the different TagRFP constructs in various genetic backgrounds were dissected and the replums containing the seeds were mounted under a coverslip in distilled water. Image were taken with a PLAN APO 20x/0.75 dry objective using the confocal spinning disk microscope from PerkinElmer driven by the Volocity 6.3.0 software and equipped with a YokogamaCSU-X1 scan head, two EmCCD Hamamatsu C9100-13 cameras (Hamamatsu) and a 580 nm beam splitter to separate dual staining on the two cameras as described.22 Images were acquired for TagRFP fluorescence with a 561 nm laser (Laser power intensity: 7% or 15%, exposure time: 200 msec; gain: 5; sensitivity: 148) and the fluorescence was selected between 580 and 612 nm. ImageJ was used to analyze the Z distribution of the fluorescence intensity profile along regions of interest (ROIs). The set display range was set to 16-bit (0–65635) for the 16-bit spinning disk images that were further calibrated using the set scale option (1 pixel = 0.66 μm). Three modes were used: (i) the maximum intensity Z projection mode was used to build a stack image of a given number of 1 μm-slices taking the maximum fluorescence value among the slices for each pixel; (ii) the sum intensity Z projection mode was used for fair comparison of relative fluorescence intensities among the analyzed lines since all the intensity values from all selected slices were summed for each pixel; (iii) the orthogonal view along an axis positioned in the XY plan was used to better visualize the Z distribution of the fluorescence along the chosen XY axis.
Confocal microscopy of leaves of N. Benthamiana transient transformants
Transiently transformed N. benthamiana leaves were observed 48 h post agroinfiltration using an upright confocal laser scanning microscope (LEICA SP8) with a 40 x apochromatic water immersion lens. TagRFP and YFP fluorescences were imaged with the following settings: excitation: 561 nm/emission: 582–622 nm; excitation 514 nm/emission: 527–530 nm, respectively. Z stacks (maximum intensity) were performed using ImageJ.
SDS-PAGE and anti-TagRFP western-blots from tbl38 complemented lines (PTR)
The TagRFP fluorescence patterns of PTR lines was controlled using spinning disk confocal microscopy along developmental kinetics of seeds from staged floral stem. The seeds that displayed the early (6–8 DAP), medium (8–12 DAP) and late (>12 DAP) fluorescence patterns were carefully extracted from the dissected siliques (3–4 siliques per fluorescence pattern) and each pooled in 2 mL tubes with a metallic grinball before freezing in liquid nitrogen. Total proteins were extracted and analyzed by western blot as previously described22 using anti-TagRFP primary and anti-rabbit-AP secondary antibodies (key resources table) with minor modifications:
For SDS-PAGE analysis, total proteins were extracted as follows: frozen weighed samples were ground using a Mixer Mill MM 400 (RETSCH) with a metallic grindball twice 30 s at 30 Hz. Eight μL of 5 mM sodium acetate pH 4.6, 0.2 M CaCl2 extemporaneously complemented with 1 μL mL−1 of β-mercaptoethanol and 1.5 μL mL−1 of plant protease inhibitor cocktail (Sigma P9599) were added per mg of seed and the tubes were plunged in liquid nitrogen. The samples were further ground in the frozen buffer 3 times 30 s at 30 Hz. The samples were spun and thoroughly vortexed. Then, they were vertically agitated at 400 rpm on an orbital shaker at 4°C for 45 min. After 10 min of centrifugation at 14,000 rpm, the supernatant was discarded (controls showed that no protein of interest was present in this fraction). The remaining pellet was mixed with 8 μL of denaturation solution containing 10% (v/v) β-mercaptoethanol, 6 M urea, 10% (v/v) glycerol, 5% (v/v) SDS69 complemented with 0.01% bromophenol blue and additional 0.2 M CaCl2 per mg of initial seeds. The samples were run for a second vertical extraction again at 400 rpm on an orbital shaker at 4°C for 45 min. After 10 min of centrifugation at 14,000 rpm, the supernatant was carefully transferred in a clean 1.5 mL tube. Twenty-five μL of each sample were analyzed by SDS-PAGE in a 13% or 8% resolving and 4% polyacrylamide denaturing stacking gels under electric current set at 120 mA and 500 V.
Protein were transferred onto nitrocellulose membranes for 45 min to 1 h under electric current set at 20 V and 800 mA. Membranes were transiently stained with Ponceau Red solution and imaged. Nitrocellulose membranes were blocked overnight in 0.02 M Tris-HCl pH 7.5, 0.5 M NaCl, 0.5 g.L−1 Tween 20 and 5% Régilait non-fat milk (TTBS-milk) under constant agitation at 4°C and 60 rpm on an orbital shaker. Membranes were incubated at RT for 1 h 30 min and then overnight at 4°C in 10 mL of anti-TagRFP primary antibody (key resources table) diluted 1:3,000 in TTBS-milk, then rinsed 6 × 5 min with TTBS. Secondary antibody incubation was performed for 2 h in 10 mL of goat anti-rat AP secondary antibody (key resources table) diluted 1:5000 in TTBS-milk at room temperature. Membranes were rinsed 3 × 5 min with TTBS and once with deionized water before revelation in 50 mL of alkaline phosphatase BCIP/NBT chromogenic substrate for 30 min. Membranes were imaged with a Canon EOS 550D digital camera.
Dry seed MSC surface wall abrasive fraction
Abrasion columns were a home-made design allowing for homogeneous dry seed MSC surface wall abrasive enrichment using plastic column and collector tube from the GeneJET Plasmid Miniprep Kit (Thermo Scientific K0502). The bottom of the column was cut with a razor blade, the silica layer of the column was removed and replaced with a 50 μm Seffar Nylon mesh (Dutscher 074010) wrapped around the bottom of the open column. Then, a 2.1 × 1.6 cm piece of P500 sand-paper (Dexter, Castorama) was gently rolled in the column and placed with the abrasive side inward. The P500 sand paper theoretically corresponded to 30.2 μm average abrasive particle size (https://www.fine-tools.com/G10019.html), i.e., about 1:10 of A. thaliana seed length) Finally, the so-called abrasive column was put back in the collector tube. The collector tube was weighted at the time of use and 50 mg dry seeds (about 2,500 seeds) were deposited inside the paper sand roll before locking the cap. Abrasion was done with a FastPrep (MP Biomedicals 116004500) for 5 to 6 cycles of 1 min at 6.5 m s−1. The abrasion was not linear and the powder usually appeared in the collection tube after the 4th or 5th cycle probably corresponding to a breaking limit. While most of the extracted surface wall powder passes through the nylon mesh during the last cycles of abrasion, a final centrifugation step for 5 min at 8,500 rpm allowed total recovery of the powder. The powder was carefully weighted in the collector tube (in the mg range per 50 mg dry seeds) and additionally ground with a metallic grindball for 3 min 30 s at 30 Hz (Mixer Mill MM 400, RETSCH). To evaluate the abrasion efficiency, Col-0 dry seeds were analyzed through four different methods for comparison before and after abrasion. (i) Ruthenium red mucilage release assay using the protocol described above: seeds are shaken at 250 rpm in Tris Buffer (0.01 M Tris-HCl pH 7.5) during 1 h at RT, rinsed with Tris Buffer, and shaked at 250 rpm in a 0.02% Ruthenium Red solution in Tris Buffer during 1 h. Following 2 Tris washing steps, seeds are transferred in a 24-well microscopy plate and imaged. (ii) UV autofluorescence of dry seed surface: Seeds were imaged by epifluorescence using an UV filter set (excitation: 387/11 nm; dichroic mirror: 405 nm; emission: 440/40 nm) using a Leica DM IRB/E inverted microscope equipped with a Leica MC190HD digital camera. (iii) Fixation for 2 h in 1.25% glutaraldehyde/2% paraformaldehyde in 0.05 M PIPES, 5 mM EGTA, 5 mM MgSO4, pH 6.9 buffer complemented with 0.1% Triton X-100 and 50% ethanol and embedding in LRW acrylic resin using automatic microwave tissue processor for electron microscopy (Leica EM AMW),22 followed by immunofluorescence on 1 μm semi-thin sections70 using JIM7 (1:10) and goat anti-rat A488 (1:100) antibodies (key resources table). (iv) Finally, abraded and non-abraded seeds, 100 μm nylon mesh and the abrasion powder were imaged by scanning electron microscopy. Samples were metallized using a MED 020 modular high vacuum coating and directly observed with no further treatment using a Quanta 250 FEG FEI electron microscope. Images were taken with a 2.0 spot size and 10.00 kV acceleration. Following these setup controls, powder from various genotypes was analyzed by immuno dotblots, enzymatic assays and mass spectrometry. > 10 seeds were observed with similar results.
Immuno dotblot and semi-quantitative analysis from dry seed MSC abrasive fractions
Powder of MSC surface CW was chemically extracted in the collector tube with extraction buffer [80 μL of 5 mM sodium acetate pH 4.6 containing 1 μL mL−1 of plant protease inhibitor cocktail (Sigma P9599)] per mg of powder. The collector tube with cap was shaken at 250 rpm during 1 h at 4°C to ensure proper extraction. After a 5 min centrifugation at 14,400 rpm, the supernatant was recovered, diluted (1 μL in 50 μL milliQ-H2O) and deposited into sample wells onto nitrocellulose membrane disposed into a 96-well Bio-Dot microfiltration apparatus (BioRad) previously soaked with in TBS (0.02 M Tris-HCl pH 7.5, 0.15 M NaCl) for 10 min. A 1 min 30 s of vacuum insured homogeneous transfer among the wells. The membrane was briefly air-dried to ensure proper adsorption and blocked in 5% milk (Regilait non fat dry milk) in TTBS (0.05% Tween 20, 0.02 M Tris-HCl pH 7.5, 0.5 M NaCl) overnight at 4°C, under a 50 rpm gentle shaking. The membranes were rolled in a hemolysis tube containing 5 mL of JIM7, LM20, JIM5, LM19, LM7, LM18, LM25 or INRA-RU2 primary antibodies (key resources table) diluted 1:100 in TTBS-milk for 3–4 h at RT. Membranes were washed 6 × 5 min each in about 50 mL TTBS before incubation for 1 h in a hemolysis tube filled with 5 mL of 1:5000 alkaline phosphatase (AP) conjugated secondary antibody adapted to the species used to raise the primary antibody (key resources table). After 3 × 5 min wash in about 50 mL TTBS and 3 × 1 min in deionized water, membranes were immersed in the alkaline phosphatase (AP) chromogenic substrate 0.3 mg mL−1 NBT 0.15 mg mL−1 BCIP in 100 mM Tris-HCl pH 9.5, 100 mM NaCl, 10 mM MgCl2 for 5 to 15 min. The membranes were air dried and imaged with a Canon EOS 550D digital camera. JIM7 and LM20 dot blots were analyzed using ImageJ 1.8 without edition of native images. Multiple ROIs of a similar selected size were used to measure the mean intensity of the signal. Two negative controls were used (i) the extraction buffer that was blotted on the membrane and (ii) for each lane, another ROI placed outside of the dots to account for possible vertically different background noises. Measurements were extracted on an Excel file ready for R statistical analyses. Corresponding Col-0 values were assigned as “100% of signal” and percentage of mutant signal values were calculated accordingly ±standard deviation (SD). Statistical data was obtained using ANOVA/Tukey HSD tests with n ≥ 3.
Acetyl group deesterification and acetic acid semi-quantitative analysis from dry seed MSC abrasive fractions
The acetylation of cell wall polymer in MSC surface wall abrasive samples was analyzed directly in the collection tube following adaptation of previously described method.12 Acetyl groups were released by ester alkaline hydrolysis with 1 M NaOH (80 μL of NaOH per mg powder). The samples were vigorously shaken horizontally for 1 h at 250 rpm at 4°C. The reaction was stopped with an equal volume of 1 M HCl which stabilizes pH at a neutral value. Acetic acid content was then determined using the K-ACET enzymatic Acetic Acid Assay Kit (Megazyme) by measuring the stoichiometric conversion of acetic acid to NADH following three enzymatic reactions. The same volume of samples (100 μL) was used following the suggested protocol with the following adaptation (1.5 mL of final reaction volume was used instead of 2.84 mL). NADH was measured at 340 nm with a ©Agilent Cary 60 UV-Vis spectrophotometer using the Eppendorf Uvette (Dutscher 033189). The linearity of the assay was checked with acetic acid standard provided with the kit and with an acetic acid standard curve (0-5μg). The final results were expressed as mean μg of acetic acid released per mg of original powder ±standard deviation (SD) with n ≥ 3. Statistical data was obtained using ANOVA/Tukey HSD tests.
Structural oligosaccharide composition of dry seed MSC abrasive fractions by IP-RP-UHPC-MS analysis
MSC surface wall powders were resuspended in 1 mL distilled water for 1 h. An aliquot (450 μL) was mixed with 50 μL of 500 mM sodium acetate buffer pH 5.0 and polysaccharides were hydrolyzed for 24 h at 40°C under 500 rpm shaking with either a pectin lyase (PL) specific for methylesterified HGs,43 a polygalacturonase (PG)71 specific for non-methylesterified HGs or a rhamnogalacturonase (RG, Swiss-Prot Q00018, provided by Novozymes, Copenhagen, Denmark) specific for the RGI backbone.50 Resulting OGA digests were filtered on 0.45 μm and analyzed by IP-RP-UHPC-MS. Acquisitions were performed on a Select Series Cyclic IMS (Waters, Wilmslow, UK) coupled with a UHPLC system (Acquity H-Class Plus, Waters, Manchester, UK). The chromatographic separations were performed on an hypersil GOLD (100 mm × 1 mm, packed with 1.9 μm porosity particles; Thermo-Fisher Scientific, Bremen, Germany). A ternary gradient was used (A: Milli-Q water, B: 100% methanol, and C: 20 mM heptylammonium formate, pH 6), from 2% to 25% of solvent B for 10 min, then up to 73% at 23.5 min and maintained at 73% for 4 min. Percentage of solvent C was kept constant at 25%. The flow rate was 0.175 mL min−1, and the column was heated to 45°C. Spectra were recorded in positive electrospray ionization (ESI) ionization mode in the m/z range 150–2000, with the TOF operating in the V-mode. The source parameters were as follows: capillary voltage 2.8 kV; cone voltage 120 V; source temperature 100°C; desolvation temperature 350°C; desolvation gas 350 L/h; and nebulization gas 6 bar. Data were recorded with the Quartz software (Waters embedded software, release 5) and processed using Mass Lynx 4.2 (Waters) and MzMine software.65 MzMine was used to produce a database of annotated structures (m/z; retention time; intensity). The identified species (charge states and anomers were merged) were annotated according to their degree of polymerization (DP), degree of methylesterification (DM) and degree of acetylation (DA). The data was analyzed using Microsoft Excel.
Production of recombinant TBL38 in Pichia pastoris and enzymatic activity assays
The coding sequence of TBL38 (Q8VY22) was codon optimized for Pichia pastoris and synthesized without signal peptide in frame with His-tag in pPCIZ-αB by ProteoGenix (Schiltigheim, France). rTBL38 was produced in transformed P. pastoris cells grown on buffered glycerol-complex medium at 30°C before the induction step, which begins with the transfer of the cells into a medium with 0.5% methanol and without glycerol. During 72 h, the medium is kept at this concentration by daily addition of methanol. Following centrifugation (1 500×g, 8 min, 4°C) supernatants are recovered and filtered with GD/X 0.45 μm PES filter (Whatman, Maidstone, United Kingdom).72 rTBL38 purification was carried out using 1 mL HisTrap excel column (GE Healthcare, Chicago, Illinois, United State). 100 mL of the culture supernatant was loaded onto the column at 1 mL/min flow rate. Column was washed with 10 column volumes of wash buffer (50 mM NaP pH 7.2, 250 mM NaCl, 5 mM imidazole). rTBL38 was purified using 10 column volumes of elution buffer (50 mM NaP pH 7.2, 250 mM NaCl, 100 mM imidazole). The elution fraction of rTBL38 was concentrated with Amicon Ultra Centrifugal filter with a 10 kDa cut-off (Merck Millipore, Burlington, Massachusetts, United States) up to a volume between ∼150 μL. The buffer was exchanged to the activity buffer (Mcilvaine’s 50 mM pH 6.5, 100 mM NaCl) using PD SpinTrap G-25 (Cytiva, Björkgatan, Uppsala, Sweden). Enzyme purity and molecular weight were estimated by 12% SDS-PAGE using mini-PROTEAN 3 system (BioRad, Hercules, California, United States). Gels were stained using PageBlue Protein Staining Solution (Thermo Fisher Scientific) according to the manufacturer’s protocol. Activity assays of rTBL38 were performed with the acetic acid assay kit (K-ACETRM, Megazyme) using the activity buffer at 40°C and with three different acetylated substrates (Triacetine 100 mM, Xylan 24% acetylation, 10 mg mL−1 sugar beet pectins 31% acetylation 10 mg mL−1). Activity of rTBL38 was expressed as nmole acetic acid.μg protein−1.min−1 and compared to that of boiled sample.
In silico model and docking simulations
Homology models for PRX36 and TBL38 (UniProt accession number Q9SD46 and Q8VY22, respectively) were built with the Phyre2 server.36 For PRX36, the template was the crystallographic structure (X-ray diffraction, 1.45 Å) of A. thaliana PRX53 (At5g06720; Protein DataBank no.1PA2).49 For TBL38, the template was the crystallographic structure (X-ray diffraction, 1.85 Å) of A. thaliana TBL29/ESK1/XOAT1.17 For comparison with the TBL38 model, the TBL29/ESK1/XOAT1 structure was drawn as well using the 6cci pdb file17 visualized and analyzed with Swiss-PdbViewer (http://www.expasy.org/spdbv/).64
For PRX36 docking experiments, α-D-(1–4) polygalacturonic acid structural model52 was retrieved from the Glyco3D portal53 http://glyco3d.cermav.cnrs.fr/mol.php?type=polysaccharide&molecule=2504) and was modified to initially build the five hexagalacturonates models used to establish JIM7 specificity.33 This selection was extended to the 124 oligogalacturonates (OGAs) from DP2 to DP6 covering all the theoretical combination of methylation (64 OGAs of DP6 + 32 OGAs of DP5 + 16 OGAs of DP4 + 8 OGAs of DP3 +4 OGAs of DP2). AutoDock Tools62 and AutoDock Vina63 were used for simulating the binding of the 124 OGAs to PRX36 within a search box encompassing the whole target protein enabling to recover the 9 best poses for each OGA. Energy level and Root-mean-square deviation (RMSD) upper bound values were obtained with AutoDock tools, representing the affinity of each pose and distances from pose 1 for a given OGA, respectively. RMSD upper bound matches each atom of a given OGA in each conformations (poses) with itself in the conformation of reference (pose 1). As an imperfect mean to sort and rank the 124 OGAs using Microsoft Excel, we used the following proxy: We summed the energy levels (negative values) of the nine poses (the lower, the higher affinity) and summed the nine RMSD for each OGA (the lower, the less dispersed). The ratio of both was used as a proxy integrating both parameters (the lower ratio, the higher affinity and higher gathering of the poses on the protein). Structural models were visualized and analyzed with Swiss-PdbViewer (http://www.expasy.org/spdbv/).64 The visualization of the models and color edition were performed using Swiss-PdbViewer. Finally, an additional docking simulation of PRX36 was made with the two best hits (OGA of DP 6, so-called Clausen 4 and Clausen 3) to which an acetyl was first added in silico at O-2 or O-3 position. The same calculation as above were made and the results for the four new acetylated OGAs (Clausen-3 O-2 ac, Clausen 3 O-3 ac, Clausen 4 O-2 ac and Clausen 4 O-3 ac) were integrated in the previous ranking.
Quantification and statistical analysis
All microscopy images: No individual treatment of images on a given Figure was performed to enable comparison between patterns/intensities.
Figure 2: The images were extracted from Nanozoomer scans of the whole tissue arrays containing hundreds of seeds in each serial section. The same exact seed on serial sections used for anti-sense and sense probe control are shown. Each image is representative of at least ten seeds for each stage.
Figures 3, S1, and S2: The confocal images in Figure 3A are representative of numerous observations (n > 10 MSCs). The result shown in Figure 3B for one genetic line is representative of three independent genetic lines as shown in Figure S1. All images in Figures 3B and S1 are representative of 2–3 observations of >10 seeds. The sum view and orthogonal projection were performed on stacks of 40 slices (Figure 3B) or 70 slices (Figure S1) for each seed. The whole membrane views of the western blot shown in Figure 3C are provided in Figure S2. The Western blots have been performed twice with similar results. Two gel acrylamide percentages were used and both results are displayed. Experiments for Figures 3B and 3C have been performed on plants grown in the same conditions and selection of silique ranks used for western blot sampling has been assisted by spinning disk observation of the fluorescence patterns observed in Figure 3B.
Figure S3: The RT-PCR experiments were performed twice with similar results.
Figure 4, panel B: At least two technical repeats, each with about 50–100 seeds were performed for seed batches coming from 6 individual Col-0 and tbl38 plants simultaneously grown in the same conditions. Results are presented as mean ± SD with n > 1,000 seeds. Statistical data were obtained using ANOVA tests with n ≥ 3 using the R package. Panel C: The images were extracted from Nanozoomer scans of the whole tissue arrays containing hundreds of seeds. Each image is representative of at least ten seeds for each stage. Panel D: The confocal images are representative of numerous observations (n > 10 MSCs). All images are representative of 2–3 observations of >10 seeds. Images are the result of a maximum projection of similar stacks (Z > 30) which were not edited.
Figure 5: The images were extracted from Nanozoomer scan of the whole tissue arrays containing hundreds of seeds. Each image is representative of at least ten seeds for each stage.
Figure 6: The characterization of the efficiency of the seed surface abrasion was performed by five different approaches all giving consistent and complementary results. The images shown are representative of at least 5 observations for each technique.
Figures 7 and S4: The initial antibody screening performed in Figure S4 was performed with two technical repeats for all samples (two biological repeats for Col-0 and one biological repeat for the mutants). The thorough repeat and quantification of results for JIM7 and LM20 antibodies are shown in Figures 7A and 7B. In Figure 7, the five analyzed genotypes including three independent complemented lines were grown in parallel in the same condition. Three biological replicates (individual abrasions from 50 mg of seeds) each with at least two technical replicates were analyzed. The results of ANOVA and TUKEY HSD tests performed with the R package are shown by letters on Figure 7.
Figure 8 and Tables S3–S5: The detailed data of the results in Figure 8 are presented in Table S3. Homogalacturonan (HG) acetylation phenotype of tbl38 illustrated by the comparative view of the mean intensity of each HG species identified following pectin lyase (PL) digestion of dry seed abrasion powder from the four genotypes, related to Figure 8, Table S4. Homogalacturonan (HG) methylation phenotype of pmei6 illustrated by the comparative view of the mean intensity of each HG species identified following the digestion of dry seed abrasion powder from the three genotypes with polygalacturonase (PG) specific for low-methylated HGs, related to Figure 8, Table S5. Absence of significant rhamnogalacturonan I (RGI) acetylation phenotype of tbl38 illustrated by the comparative view of the mean intensity of each RG species identified following the rhamnogalacturonase (RG) digestion of dry seed abrasion powder from two genotypes, related to Figure 8. Six technical repeats in Table S3A illustrated the low standard deviation of intensities for identified homogalacturonan species (2–7% for the five species of highest intensity). Tables S3B, S4A and S5A show the detailed the mean results of three biological repeat and 1–2 technical repeats used to build Figure 8. The corresponding individual intensity values as well as the mean and SD for the four genotypes studied with pectin lyase (PL), the three genotypes studied with polygalacturonase (PG) and the two genotypes studied with rhamogalacturonase (RG) are shown in Tables S3C–S3F, Tables S4B–S4E and Tables S5B and S5C, respectively.
Figures 9B and S5: The measurement of the enzymatic activity of TBL38 was performed using three different substrates on three purification fractions each either native or boiled. Results are mean ± SD (n = 3). The native samples were compared to boiled samples used as negative controls. Students t-tests were performed using Microsoft Excel. ∗p < 0.05.
Table S6 and Figure 10: For each of the 124 OGAs (Table S6) and six hexagalacturonates (Figure 10A) tested, the nine docking pose models were superimposed on PRX36 structural model and the corresponding energy level (EL) values were heat mapped in red-to-yellow. Root-mean-square deviations (RMSD) were additionally heat mapped in blue-to-yellow. The same red-to-yellow heatmaps color coding was used to label the individual hexagalacturonates in Figure 10B. In Table S6, data mining was performed by calculating the sum of the EL (ΣEL) reflecting the global affinity of each OGA for PRX36 and the sum of RMSD (ΣRMSD) reflecting the dispersion vs. concentration of the nine poses on the protein surface. (ii) Then, ΣEL/ΣRMSD was used as a proxy to sort the OGAs in order to integrate both parameters (the lowest ΣEL/ΣRMSD, the highest affinity and shorter distance).
Acknowledgments
We thank the Paul Sabatier Toulouse 3 University and the Center National de la Recherche Scientifique (CNRS) for granting this work. This work was specifically supported by the French National Research Agency project ‘‘MicroWall’’ (ANR-18-CE20-0007). It was also supported by the French Laboratory of Excellence project ‘‘TULIP’’ (ANR-10- LABX-41; ANR-11-IDEX-0002-02). We are grateful to Dr. H. North (IJPB, Versailles, France) and I.-H. Nishimura (Kyoto University, Japan) for providing seeds and to Dr. P.-M. Delaux (LRSV, Auzeville-Tolosane, France) for the gift of GoldenGate plasmids and helpful discussions about Golden Gate cloning.
Author contributions
Conceptualization, V.B.; methodology, all authors; investigation, B.G.D, D.R., M.R., P.R., C.C., S.O., J. M.-B., P.T., A.G., and V.B.; writing – original draft writing, B.G.D. and V.B.; review and editing, B.G.D., D.R., P.R., E.J., C.D., J.P., M.-C.R., and V.B. All authors read and approved the article; funding acquisition, J.P., M.-C.R., and V.B.; resources, J.P., M.-C.R., and V.B; supervision, J.P., M.-C.R., and V.B.
Declaration of interests
The authors declare no competing interests.
Published: April 5, 2024
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2024.109666.
Supplemental information
References
- 1.Driouich A., Follet-Gueye M.L., Bernard S., Kousar S., Chevalier L., Vicré-Gibouin M., Lerouxel O. Golgi-mediated synthesis and secretion of matrix polysaccharides of the primary cell wall of higher plants. Front. Plant Sci. 2012;3:79. doi: 10.3389/fpls.2012.00079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wolf S., Mouille G., Pelloux J. Homogalacturonan methyl-esterification and plant development. Mol. Plant. 2009;2:851–860. doi: 10.1093/mp/ssp066. [DOI] [PubMed] [Google Scholar]
- 3.Gille S., Pauly M. O-acetylation of plant cell wall polysaccharides. Front. Plant Sci. 2012;3:12. doi: 10.3389/fpls.2012.00012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schultink A., Naylor D., Dama M., Pauly M. The role of the plant-specific ALTERED XYLOGLUCAN9 Protein in Arabidopsis cell wall polysaccharide O-Acetylation. Plant Physiol. 2015;167:1271–1283. doi: 10.1104/pp.114.256479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Manabe Y., Nafisi M., Verhertbruggen Y., Orfila C., Gille S., Rautengarten C., Cherk C., Marcus S.E., Somerville S., Pauly M., et al. Loss-of-function mutation of REDUCED WALL ACETYLATION2 in Arabidopsis leads to reduced cell wall acetylation and increased resistance to Botrytis cinerea. Plant Physiol. 2011;155:1068–1078. doi: 10.1104/pp.110.168989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Manabe Y., Verhertbruggen Y., Gille S., Harholt J., Chong S.L., Pawar P.M.A., Mellerowicz E.J., Tenkanen M., Cheng K., Pauly M., Scheller H.V. Reduced wall acetylation proteins play vital and distinct roles in cell wall O-acetylation in Arabidopsis. Plant Physiol. 2013;163:1107–1117. doi: 10.1104/pp.113.225193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Potikha T., Delmer D.P. A mutant of Arabidopsis thaliana displaying altered patterns of cellulose deposition. Plant J. 1995;7:453–460. doi: 10.1046/j.1365-313X.1995.7030453.x. [DOI] [Google Scholar]
- 8.Pauly M., Ramírez V. New insights into wall polysaccharide O-acetylation. Front. Plant Sci. 2018;9:1210. doi: 10.3389/fpls.2018.01210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bischoff V., Nita S., Neumetzler L., Schindelasch D., Urbain A., Eshed R., Persson S., Delmer D., Scheible W.R. TRICHOME BIREFRINGENCE and Its homolog AT5G01360 encode plant-specific DUF231 proteins required for cellulose biosynthesis in Arabidopsis. Plant Physiol. 2010;153:590–602. doi: 10.1104/pp.110.153320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lefebvre V., Fortabat M.N., Ducamp A., North H.M., Maia-Grondard A., Trouverie J., Boursiac Y., Mouille G., Durand-Tardif M. ESKIMO1 disruption in Arabidopsis alters vascular tissue and impairs water transport. PLoS One. 2011;6 doi: 10.1371/journal.pone.0016645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sun A., Yu B., Zhang Q., Peng Y., Yang J., Sun Y., Qin P., Jia T., Smeekens S., Teng S. MYC2-activated TRICHOME BIREFRINGENCE-LIKE37 acetylates cell walls and enhances herbivore resistance. Plant Physiol. 2020;184:1083–1096. doi: 10.1104/pp.20.00683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stranne M., Ren Y., Fimognari L., Birdseye D., Yan J., Bardor M., Mollet J.C., Komatsu T., Kikuchi J., Scheller H.V., Sakuragi Y. TBL10 is required for O-acetylation of pectic rhamnogalacturonan-I in Arabidopsis thaliana. Plant J. 2018;96:772–785. doi: 10.1111/tpj.14067. [DOI] [PubMed] [Google Scholar]
- 13.Chiniquy D., Underwood W., Corwin J., Ryan A., Szemenyei H., Lim C.C., Stonebloom S.H., Birdseye D.S., Vogel J., Kliebenstein D., et al. PMR5, an acetylation protein at the intersection of pectin biosynthesis and defense against fungal pathogens. Plant J. 2019;100:1022–1035. doi: 10.1111/tpj.14497. [DOI] [PubMed] [Google Scholar]
- 14.Akoh C.C., Lee G.C., Liaw Y.C., Huang T.H., Shaw J.F. GDSL family of serine esterases/lipases. Prog. Lipid Res. 2004;43:534–552. doi: 10.1016/j.plipres.2004.09.002. [DOI] [PubMed] [Google Scholar]
- 15.Lai C.P., Huang L.M., Chen L.F.O., Chan M.T., Shaw J.F. Genome-wide analysis of GDSL-type esterases/lipases in Arabidopsis. Plant Mol. Biol. 2017;95:181–197. doi: 10.1007/s11103-017-0648-y. [DOI] [PubMed] [Google Scholar]
- 16.Mølgaard A., Larsen S. Crystal packing in two pH-dependent crystal forms of rhamnogalacturonan acetylesterase. Acta Crystallogr. Sect. D-Struct. Biol. 2004;60:472–478. doi: 10.1107/s0907444903029767. [DOI] [PubMed] [Google Scholar]
- 17.Lunin V.V., Wang H.T., Bharadwaj V.S., Alahuhta M., Peña M.J., Yang J.Y., Archer-Hartmann S.A., Azadi P., Himmel M.E., Moremen K.W., et al. Molecular mechanism of polysaccharide acetylation by the Arabidopsis xylan O-acetyltransferase XOAT1. Plant Cell. 2020;32:2367–2382. doi: 10.1105/tpc.20.00028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sinclair S.A., Larue C., Bonk L., Khan A., Castillo-Michel H., Stein R.J., Grolimund D., Begerow D., Neumann U., Haydon M.J., Krämer U. Etiolated seedling development requires repression of photomorphogenesis by a small cell-wall-derived dark signal. Curr. Biol. 2017;27:3403–3418.e7. doi: 10.1016/j.cub.2017.09.063. [DOI] [PubMed] [Google Scholar]
- 19.Francoz E., Ranocha P., Burlat V., Dunand C. Arabidopsis seed mucilage secretory cells: regulation and dynamics. Trends Plant Sci. 2015;20:515–524. doi: 10.1016/j.tplants.2015.04.008. [DOI] [PubMed] [Google Scholar]
- 20.Viudes S., Burlat V., Dunand C. Seed mucilage evolution: Diverse molecular mechanisms generate versatile ecological functions for particular environments. Plant Cell Environ. 2020;43:2857–2870. doi: 10.1111/pce.13827. [DOI] [PubMed] [Google Scholar]
- 21.Western T.L. The sticky tale of seed coat mucilages: production, genetics, and role in seed germination and dispersal. Seed Sci. Res. 2012;22:1–25. doi: 10.1017/s0960258511000249. [DOI] [Google Scholar]
- 22.Francoz E., Ranocha P., Le Ru A., Martinez Y., Fourquaux I., Jauneau A., Dunand C., Burlat V. Pectin demethylesterification generates platforms that anchor peroxidases to remodel plant cell wall domains. Dev. Cell. 2019;48:261–276.e8. doi: 10.1016/j.devcel.2018.11.016. [DOI] [PubMed] [Google Scholar]
- 23.Saez-Aguayo S., Ralet M.C., Berger A., Botran L., Ropartz D., Marion-Poll A., North H.M. PECTIN METHYLESTERASE INHIBITOR6 promotes Arabidopsis mucilage release by limiting methylesterification of homogalacturonan in seed coat epidermal cells. Plant Cell. 2013;25:308–323. doi: 10.1105/tpc.112.106575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kunieda T., Shimada T., Kondo M., Nishimura M., Nishitani K., Hara-Nishimura I. Spatiotemporal secretion of PEROXIDASE36 is required for seed coat mucilage extrusion in Arabidopsis. Plant Cell. 2013;25:1355–1367. doi: 10.1105/tpc.113.110072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Belmonte M.F., Kirkbride R.C., Stone S.L., Pelletier J.M., Bui A.Q., Yeung E.C., Hashimoto M., Fei J., Harada C.M., Munoz M.D., et al. Comprehensive developmental profiles of gene activity in regions and subregions of the Arabidopsis seed. Proc. Natl. Acad. Sci. USA. 2013;110:E435–E444. doi: 10.1073/pnas.1222061110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Debeaujon I., Nesi N., Perez P., Devic M., Grandjean O., Caboche M., Lepiniec L. Proanthocyanidin-accumulating cells in Arabidopsis testa: Regulation of differentiation and role in seed development. Plant Cell. 2003;15:2514–2531. doi: 10.1105/tpc.014043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McGee R., Dean G.H., Mansfield S.D., Haughn G.W. Assessing the utility of seed coat-specific promoters to engineer cell wall polysaccharide composition of mucilage. Plant Mol. Biol. 2019;101:373–387. doi: 10.1007/s11103-019-00909-8. [DOI] [PubMed] [Google Scholar]
- 28.Šola K., Gilchrist E.J., Ropartz D., Wang L., Feussner I., Mansfield S.D., Ralet M.C., Haughn G.W. RUBY, a putative galactose oxidase, influences pectin properties and promotes cell-to-cell adhesion in the seed coat epidermis of Arabidopsis. Plant Cell. 2019;31:809–831. doi: 10.1105/tpc.18.00954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dauphin B.G., Ranocha P., Dunand C., Burlat V. Cell-wall microdomain remodeling controls crucial developmental processes. Trends Plant Sci. 2022;27:1033–1048. doi: 10.1016/j.tplants.2022.05.010. [DOI] [PubMed] [Google Scholar]
- 30.Turbant A., Fournet F., Lequart M., Zabijak L., Pageau K., Bouton S., Van Wuytswinkel O. PME58 plays a role in pectin distribution during seed coat mucilage extrusion through homogalacturonan modification. J. Exp. Bot. 2016;67:2177–2190. doi: 10.1093/jxb/erw025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kohn R., Furda I. Binding of Calcium ions to acetyl derivatives of pectin. Collect. Czech Chem. Commun. 1968;33:2217–2225. doi: 10.1135/cccc19682217. [DOI] [Google Scholar]
- 32.Ralet M.C., Crépeau M.J., Buchholt H.C., Thibault J.F. Polyelectrolyte behaviour and calcium binding properties of sugar beet pectins differing in their degrees of methylation and acetylation. Biochem. Eng. J. 2003;16:191–201. doi: 10.1016/s1369-703x(03)00037-8. [DOI] [Google Scholar]
- 33.Clausen M.H., Willats W.G.T., Knox J.P. Synthetic methyl hexagalacturonate hapten inhibitors of antihomogalacturonan monoclonal antibodies LM7, JIM5 and JIM7. Carbohydr. Res. 2003;338:1797–1800. doi: 10.1016/s0008-6215(03)00272-6. [DOI] [PubMed] [Google Scholar]
- 34.Verhertbruggen Y., Marcus S.E., Haeger A., Ordaz-Ortiz J.J., Knox J.P. An extended set of monoclonal antibodies to pectic homogalacturonan. Carbohydr. Res. 2009;344:1858–1862. doi: 10.1016/j.carres.2008.11.010. [DOI] [PubMed] [Google Scholar]
- 35.Zeuner B., Thomsen T.B., Stringer M.A., Krogh K.B.R.M., Meyer A.S., Holck J. Comparative characterization of Aspergillus pectin lyases by discriminative substrate degradation profiling. Front. Bioeng. Biotechnol. 2020;8:873. doi: 10.3389/fbioe.2020.00873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kelley L.A., Mezulis S., Yates C.M., Wass M.N., Sternberg M.J.E. The Phyre2 web portal for protein modeling, prediction and analysis. Nat. Protoc. 2015;10:845–858. doi: 10.1038/nprot.2015.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dodson G., Wlodawer A. Catalytic triads and their relatives. Trends Biochem. Sci. 1998;23:347–352. doi: 10.1016/s0968-0004(98)01254-7. [DOI] [PubMed] [Google Scholar]
- 38.Zhong R., Cui D., Richardson E.A., Ye Z.H. Acetylation of homogalacturonan and rhamnogalacturonan-I is catalyzed by a suite of trichome birefringence-like proteins. Plant J. 2024;117:1084–1098. doi: 10.1111/tpj.16540. [DOI] [PubMed] [Google Scholar]
- 39.Zhong R., Cui D., Ye Z.H. Regiospecific acetylation of xylan is mediated by a group of DUF231-containing O-acetyltransferases. Plant Cell Physiol. 2017;58:2126–2138. doi: 10.1093/pcp/pcx147. [DOI] [PubMed] [Google Scholar]
- 40.Celus M., Kyomugasho C., Van Loey A.M., Grauwet T., Hendrickx M.E. Influence of pectin structural properties on interactions with divalent cations and its associated functionalities. Compr. Rev. Food Sci. Food Saf. 2018;17:1576–1594. doi: 10.1111/1541-4337.12394. [DOI] [PubMed] [Google Scholar]
- 41.Pérez S., Mazeau K., du Penhoat C.H. The three-dimensional structures of the pectic polysaccharides. Plant Physiol. Biochem. 2000;38:37–55. doi: 10.1016/s0981-9428(00)00169-8. [DOI] [Google Scholar]
- 42.Ralet M.C., Cabrera J.C., Bonnin E., Quéméner B., Hellìn P., Thibault J.F. Mapping sugar beet pectin acetylation pattern. Phytochemistry. 2005;66:1832–1843. doi: 10.1016/j.phytochem.2005.06.003. [DOI] [PubMed] [Google Scholar]
- 43.Ralet M.C., Williams M.A.K., Tanhatan-Nasseri A., Ropartz D., Quéméner B., Bonnin E. Innovative enzymatic approach to resolve homogalacturonans based on their methylesterification pattern. Biomacromolecules. 2012;13:1615–1624. doi: 10.1021/bm300329r. [DOI] [PubMed] [Google Scholar]
- 44.Haas K.T., Wightman R., Meyerowitz E.M., Peaucelle A. Pectin homogalacturonan nanofilament expansion drives morphogenesis in plant epidermal cells. Science. 2020;367:1003–1007. doi: 10.1126/science.aaz5103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Haas K.T., Wightman R., Peaucelle A., Höfte H. The role of pectin phase separation in plant cell wall assembly and growth. Cell Surf. 2021;7 doi: 10.1016/j.tcsw.2021.100054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dronnet V., Renard C.M.G.C., Axelos M.A.V., Thibault J.F. Characterisation and selectivity of divalent metal ions binding by citrus and sugar beet pectins. Carbohydr. Polym. 1996;30:253–263. doi: 10.1016/s0144-8617(96)00107-5. [DOI] [Google Scholar]
- 47.Oosterveld A., Beldman G., Searle-Van Leeuwen M.J.F., Voragen A.G.J. Effect of enzymatic deacetylation on gelation of sugar beet pectin in the presence of calcium. Carbohydr. Polym. 2000;43:249–256. doi: 10.1016/s0144-8617(00)00174-0. [DOI] [Google Scholar]
- 48.Renard C.M.G.C., Jarvis M.C. Acetylation and methylation of homogalacturonans - 2: Effect on ion-binding properties and conformations. Carbohydr. Polym. 1999;39:209–216. doi: 10.1016/s0144-8617(99)00015-6. [DOI] [Google Scholar]
- 49.Ostergaard L., Teilum K., Mirza O., Mattsson O., Petersen M., Welinder K.G., Mundy J., Gajhede M., Henriksen A. Arabidopsis ATP A2 peroxidase. Expression and high-resolution structure of a plant peroxidase with implications for lignification. Plant Mol. Biol. 2000;44:231–243. doi: 10.1023/a:1006442618860. [DOI] [PubMed] [Google Scholar]
- 50.Ralet M.C., Tranquet O., Poulain D., Moïse A., Guillon F. Monoclonal antibodies to rhamnogalacturonan I backbone. Planta. 2010;231:1373–1383. doi: 10.1007/s00425-010-1116-y. [DOI] [PubMed] [Google Scholar]
- 51.Koncz C., Schell J. The promoter of TL-DNA GENE 5 controls the tissue-specific expression of chimeric genes carried by a novel type of Agrobacterium binary vector. Mol. Gen. Genet. 1986;204:383–396. doi: 10.1007/bf00331014. [DOI] [Google Scholar]
- 52.Braccini I., Grasso R.P., Pérez S. Conformational and configurational features of acidic polysaccharides and their interactions with calcium ions: a molecular modeling investigation. Carbohydr. Res. 1999;317:119–130. doi: 10.1016/s0008-6215(99)00062-2. [DOI] [PubMed] [Google Scholar]
- 53.Pérez S., Sarkar A., Rivet A., Breton C., Imberty A. Glyco3D: a portal for structural glycosciences. Methods Mol. Biol. 2015;1273:241–258. doi: 10.1007/978-1-4939-2343-4_18. [DOI] [PubMed] [Google Scholar]
- 54.Fulton L.M., Cobbett C.S. Two alpha-L-arabinofuranosidase genes in Arabidopsis thaliana are differentially expressed during vegetative growth and flower development. J. Exp. Bot. 2003;54:2467–2477. doi: 10.1093/jxb/erg269. [DOI] [PubMed] [Google Scholar]
- 55.Nelson B.K., Cai X., Nebenführ A. A multicolored set of in vivo organelle markers for co-localization studies in Arabidopsis and other plants. Plant J. 2007;51:1126–1136. doi: 10.1111/j.1365-313X.2007.03212.x. [DOI] [PubMed] [Google Scholar]
- 56.Seki M., Narusaka M., Kamiya A., Ishida J., Satou M., Sakurai T., Nakajima M., Enju A., Akiyama K., Oono Y., et al. Functional annotation of a full-length Arabidopsis cDNA collection. Science. 2002;296:141–145. doi: 10.1126/science.1071006. [DOI] [PubMed] [Google Scholar]
- 57.Seki M., Carninci P., Nishiyama Y., Hayashizaki Y., Shinozaki K. High-efficiency cloning of Arabidopsis full-length cDNA by biotinylated CAP trapper. Plant J. 1998;15:707–720. doi: 10.1046/j.1365-313x.1998.00237.x. [DOI] [PubMed] [Google Scholar]
- 58.Sainsbury F., Thuenemann E.C., Lomonossoff G.P. pEAQ: versatile expression vectors for easy and quick transient expression of heterologous proteins in plants. Plant Biotechnol. J. 2009;7:682–693. doi: 10.1111/j.1467-7652.2009.00434.x. [DOI] [PubMed] [Google Scholar]
- 59.Engler C., Youles M., Gruetzner R., Ehnert T.M., Werner S., Jones J.D.G., Patron N.J., Marillonnet S. A Golden Gate modular cloning toolbox for plants. ACS Synth. Biol. 2014;3:839–843. doi: 10.1021/sb4001504. [DOI] [PubMed] [Google Scholar]
- 60.Weber E., Gruetzner R., Werner S., Engler C., Marillonnet S. Assembly of designer TAL effectors by Golden Gate cloning. PLoS One. 2011;6 doi: 10.1371/journal.pone.0019722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Schneider C.A., Rasband W.S., Eliceiri K.W. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods. 2012;9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Morris G.M., Huey R., Lindstrom W., Sanner M.F., Belew R.K., Goodsell D.S., Olson A.J. AutoDock4 and AutoDockTools4: Automated docking with selective receptor flexibility. J. Comput. Chem. 2009;30:2785–2791. doi: 10.1002/jcc.21256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Trott O., Olson A.J. Software news and update AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2010;31:455–461. doi: 10.1002/jcc.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Guex N., Peitsch M.C. SWISS-MODEL and the Swiss-PdbViewer: An environment for comparative protein modeling. Electrophoresis. 1997;18:2714–2723. doi: 10.1002/elps.1150181505. [DOI] [PubMed] [Google Scholar]
- 65.Pluskal T., Castillo S., Villar-Briones A., Oresic M. MZmine 2: Modular framework for processing, visualizing, and analyzing mass spectrometry-based molecular profile data. BMC Bioinf. 2010;11:395. doi: 10.1186/1471-2105-11-395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Clough S.J., Bent A.F. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998;16:735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- 67.Francoz E., Ranocha P., Dunand C., Burlat V. Medium-throughput RNA in situ hybridization of serial sections from paraffin-embedded tissue microarrays. Methods Mol. Biol. 2019;1933:99–130. doi: 10.1007/978-1-4939-9045-0_6. [DOI] [PubMed] [Google Scholar]
- 68.Francoz E., Ranocha P., Pernot C., Le Ru A., Pacquit V., Dunand C., Burlat V. Complementarity of medium-throughput in situ RNA hybridization and tissue-specific transcriptomics: case study of Arabidopsis seed development kinetics. Sci. Rep. 2016;6 doi: 10.1038/srep24644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kato N., Pontier D., Lam E. Spectral profiling for the simultaneous observation of four distinct fluorescent proteins and detection of protein-protein interaction via fluorescence resonance energy transfer in tobacco leaf nuclei. Plant Physiol. 2002;129:931–942. doi: 10.1104/pp.005496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Oudin A., Mahroug S., Courdavault V., Hervouet N., Zelwer C., Rodríguez-Concepción M., St-Pierre B., Burlat V. Spatial distribution and hormonal regulation of gene products from methyl erythritol phosphate and monoterpene-secoiridoid pathways in Catharanthus roseus. Plant Mol. Biol. 2007;65:13–30. doi: 10.1007/s11103-007-9190-7. [DOI] [PubMed] [Google Scholar]
- 71.Safran J., Tabi W., Ung V., Lemaire A., Habrylo O., Bouckaert J., Rouffle M., Voxeur A., Pongrac P., Bassard S., et al. Plant polygalacturonase structures specify enzyme dynamics and processivities to fine-tune cell wall pectins. Plant Cell. 2023;35:3073–3091. doi: 10.1093/plcell/koad134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Lemaire A., Duran Garzon C., Perrin A., Habrylo O., Trezel P., Bassard S., Lefebvre V., Van Wuytswinkel O., Guillaume A., Pau-Roblot C., Pelloux J. Three novel rhamnogalacturonan I- pectins degrading enzymes from Aspergillus aculeatinus: Biochemical characterization and application potential. Carbohydr. Polym. 2020;248 doi: 10.1016/j.carbpol.2020.116752. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
All data including Microscopy data reported in this paper will be shared by the lead contact upon request.
This paper mainly concerns the three following genes: TBL38 (AT1G29050), PMEI6 (AT2G47670) and PRX36 (AT3G50990) (https://www.arabidopsis.org/index.jsp). The previously published transcriptomics data25 that we used for co-expression analysis corresponds to GSE12404 dataset (https://www.ncbi.nlm.nih.gov/geo/). Homology models for PRX36 and TBL38 (UniProt accession number Q9SD46 and Q8VY22, respectively) were built using as a template, the crystallographic structure of A. thaliana PRX53 (At5g06720; Protein DataBank no.1PA2)49 and TBL29/ESK1/XOAT1 (At3g55990; 6cci pdb file),17 respectively.
-
•
Any additional information required to reanalyze the data reported in this work paper is available from the lead contact upon request.