Abstract
We have previously shown that NF-κB nuclear translocation can be observed upon human immunodeficiency virus type 1 (HIV-1) binding to cells expressing the wild-type CD4 molecule, but not in cells expressing a truncated form of CD4 that lacks the cytoplasmic domain (M. Benkirane, K.-T. Jeang, and C. Devaux, EMBO J. 13:5559–5569, 1994). This result indicated that the signaling cascade which controls HIV-1-induced NF-κB activation requires the integrity of the CD4 cytoplasmic tail and suggested the involvement of a second protein that binds to this portion of the molecule. Here we investigate the putative role of p56lck as a possible cellular intermediate in this signal transduction pathway. Using human cervical carcinoma HeLa cells stably expressing CD4, p56lck, or both molecules, we provide direct evidence that expression of CD4 and p56lck is required for HIV-1-induced NF-κB translocation. Moreover, the fact that HIV-1 stimulation did not induce nuclear translocation of NF-κB in cells expressing a mutant form of CD4 at position 420 (C420A) and the wild-type p56lck indicates the requirement for a functional CD4-p56lck complex.
The CD4 protein is an integral membrane glycoprotein of 58 kDa that contains four extracellular domains showing structural homology with immunoglobulin (Ig) Vκ regions and that is predominantly expressed at the surface of helper T lymphocytes (29, 36, 48). CD4 function as an adhesion or accessory molecule that facilitates cell-to-cell contact by interacting directly with the major histocompatibility complex (MHC) class II molecules at the surface of the antigen-presenting cells and stabilizing the T-cell receptor (TCR)–MHC-II interaction (8, 26). Furthermore, CD4 can actively participate in transmembrane signal transduction, since coaggregation of the TCR-CD3 complex and CD4 in multimeric clusters (40, 49) potentiates a variety of biochemical responses, including protein tyrosine phosphorylation, production of cytoplasmic inositol triphosphate, and release of intracellular Ca2+ (58), that ultimately regulate cell proliferation (2). During the past few years, some ligands of CD4 were shown to modulate T-cell activation in MHC-independent systems, suggesting that activation signals can be transduced directly through the CD4 molecule (3, 5, 10, 16).
Beside its crucial role in immune function, the CD4 molecule has been identified as the primary high-affinity cellular receptor for human immunodeficiency virus type 1 (HIV- 1) (19, 32). The initial step in the infection of human T lymphocytes by HIV- 1 involves binding of the viral envelope glycoprotein (gp120) to the cell surface CD4 molecule. Because it is a ligand capable of cross-linking CD4, the possibility that HIV- 1 can activate T cells has been considered, and it is now generally accepted that HIV- 1 and recombinant HIV- 1 gp120 can modulate T-cell activation, although there is some controversy as to the nature of the signals delivered to the target cells (5, 10, 15, 16, 27, 28, 31, 33). Conceivably, the noted differences derive, at least in part, from differences in experimental design, the origin of the ligand for CD4 (heat-inactivated HIV- 1, gp120– anti-gp120 immune complexes, virus-extracted gp120, recombinant gp120/gp160), and the nature of the CD4+ cells used (peripheral blood mononuclear cells [PBMCs], purified CD4+ lymphocytes, CD4+ T-cell lines, CD4-transfected cell lines). Moreover, for viral ligands, differences in the interactions between molecules (of viral or cellular origin) expressed on the virus envelope and cell surface molecules other than the virus receptors may also influence signaling.
Using CD4-transfected T-lymphoblastoid cell lines as a model, we reported direct evidence indicating that heat-inactivated HIV- 1 (iHIV- 1)-mediated oligomerization of CD4 triggers the delivery of an activation signal to T cells which can be monitored by measuring the nuclear translocation of NF-κB (5). This result was confirmed by the work from Chirmule and coworkers (15). Next, we demonstrated similar effects of iHIV- 1 on primary lymphocytes; the binding of iHIV- 1 to infected resting PBMCs promotes progression in the cell cycle, induces cell surface expression of CD25, stimulates provirus integration, induces NF-κB translocation, and commits the cell to produce virus (10). Indeed, it is well established that virus production requires cell activation and that nuclear translocation of NF-κB enhances the κB-dependent early transcription of HIV- 1. These results suggest that besides using CD4 as a receptor, HIV- 1 takes advantage of the signal-tranduction function of CD4 to modulate the intracellular virus life cycle and/or to regulate the equilibrium between viral latency, viral replication, and virus-induced apoptosis. However, the mechanism(s) by which HIV- 1 induces immune activation is still poorly understood.
To better understand the mechanism of cell signaling that results from HIV- 1 interaction with CD4, signal transduction studies have been performed which demonstrate that CD4 ligation by HIV- 1 or gp120 stimulates protein kinase C (PKC) (60), generates PKC-dependent phosphorylation of CD4 (25), induces a rise in intracellular calcium (33), and activates p56lck (27, 28), as well as phosphatidylinositol-3-kinase (PI-3K) (9), phosphatidylinositol-4-kinase (PI-4K) (50), Ras (34), Raf- 1 (43), and extracellular-regulated protein kinase (ERK) (6). Besides the identification of a panel of molecules that are activated upon engagement of CD4 with HIV- 1, the consequences of activation for the virus and the cell and the signaling pathway(s) used remain unclear.
CD4 lacks intrinsic tyrosine kinase activity but associates with p56lck, a 56-kDa cytoplasmic membrane-associated member of the Src family of nonreceptor protein tyrosine kinases expressed primarily in T lymphocytes (46, 57) through interaction of its cytoplasmic domain with two cysteine residues located at the N-terminal domain of the kinase (51, 56). Although association with Lck was demonstrated to be necessary for CD4- mediated antigen responsiveness, it has not been clearly established that the kinase activity of Lck plays a role in CD4-dependent T-cell activation. Although p56lck is the usual partner of CD4 in CD4-dependent signal transduction, a number of results suggest that p56lck also plays a major role in the transduction of signals following HIV- 1 binding to CD4. Cross-linking of CD4 with gp120, gp120-derived peptides, or anti-CD4 monoclonal antibodies (MAbs) known to be specific for the HIV- 1 gp120 binding site results in a rapid phosphorylation of p56lck on both tyrosine and serine residues and an increase in p56lck activity (9, 27, 28, 31, 42, 54). Moreover, we found that the integrity of the CD4 cytoplasmic domain is required for HIV- 1-induced nuclear translocation of NF-κB (5) and HIV-1-induced activation of ERK (6), which represents a possible downstream substrate for p56lck (23). Finally, p56lck–Raf- 1 coimmunoprecipitation after HIV- 1 binding to CD4 has been reported (43).
The objective of this study was to assess the involvement of the p56lck in the transduction of activation signal(s) induced by iHIV- 1 oligomerization of CD4. To this end, we used a series of nonlymphoid HeLa cell lines stably transfected with wild-type or mutant forms of the human CD4 molecule with or without murine wild-type p56lck. We demonstrate that the p56lck-CD4 interaction is required for triggering the NF-κB nuclear translocation that follows HIV- 1 interaction with CD4.
Main characteristics of transfected HeLa cell lines used in this study.
All cell lines included in the present study derive from the HeLa parental cell line. The HeLa CD4.2G3 cell line (37), referred to below as HeLa CD4+, expresses the human wild-type CD4. The HeLa CD4+/p56lck line was obtained by supertransfection of the HeLa CD4.2G3 cell line with both the pSM expression vector encoding p56lck and the pBabe/Hygro vector encoding the gene conferring hygromycin resistance (41). HeLa CD4+ Cyt− expresses a tailless CD4 molecule truncated at amino acid 402 (38). This truncation deletes all but seven amino acids of the cytoplasmic domain. HeLa CD4 (S408A)/p56lck and CD4 (C420S)/p56lck express mutant forms of CD4 with an alanine and a serine substitution for S408 and C420, respectively (41). Both cell lines were supertransfected with the pSM vector and the pBabe/Hygro vector and express the murine p56lck. The S408A mutation removes a critical residue in the cytoplasmic domain of CD4 which has been shown to be phosphorylated in response to phorbol esters (52), and the Ser-408 CD4 mutant molecule does not dissociate from p56lck under phorbol myristate acetate (PMA) stimulation (7, 53). The cysteine residue at position 420 of CD4 is required for binding to p56lck, and mutation at this site completely disrupts CD4-p56lck association (56). Finally, HeLa CH4 cells express a chimeric molecule consisting of the first two domains of CD4 linked to Thy1, a cell surface antigen with an external domain attached to the membrane by a glycosylphosphatidylinositol (GPI) anchor (30). An additional cell line, HeLa p56lck, was included, consisting of HeLa cells expressing p56lck but not CD4. All cell lines were grown in Dulbecco’s modified Eagle’s medium (DMEM), 10% fetal calf serum, 100 U of penicillin per ml, and 0.1 mg of streptomycin (Gibco BRL Life Technologies, Paisley, Scotland) per ml. One milligram of G418 (Gibco) per ml was added to culture medium of cells expressing the human CD4. Two hundred micrograms of hygromycin (Gibco) per ml was added to the culture medium of cells expressing p56lck. The pSM-Lck construct and HeLa CH4 cells were kindly provided by Dan Littman (Skirball Institute of Biomolecular Medicine, New York University Medical Center, New York).
CD4 and p56lck expression in HeLa-transfected cell lines.
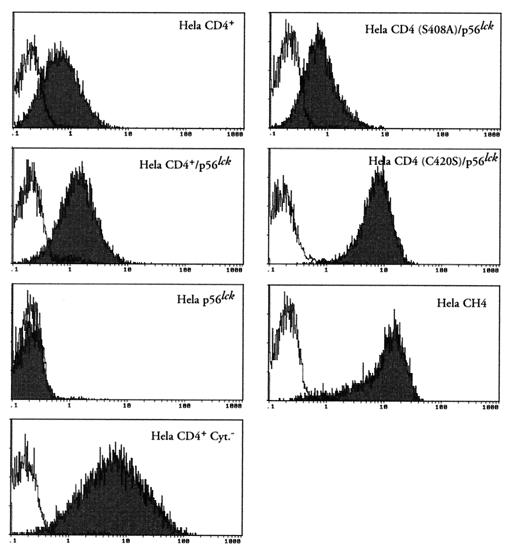
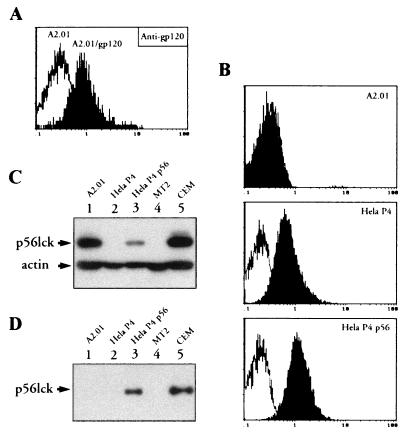
CD4 expression was assessed by indirect immunofluorescence staining and flow cytometry. Cells (5 × 105) were incubated for 30 min at 4°C with 50 μg of BL4 anti-CD4 antibody (Immunotech-Coulter Comp., Marseille, France) per ml directed against the D1-D2 region of CD4. After washing in phosphate-buffered saline–bovine serum albumin (PBS-BSA), bound MAb was revealed by addition of 50 μl of a 1/50 dilution of fluoresceinated goat anti-mouse (GAM) Ig (Immunotech-Coulter). Fluorescence intensity was recorded in the log mode on an EPICS PROFILE XL4C cytometer (Coulter, Coultronics, Margency, France). Representative cytofluorometric profiles are shown in Fig. 1. As expected, no CD4 expression was detected on HeLa p56lck, which was only transfected with the pBabe/Hygro vector and the pSM vector encoding the p56lck gene. Although variations in the expression level of CD4 were observed among the different cell lines, this antigen was expressed at the surface of HeLa cells transformed with wild-type and mutant forms of CD4 or the chimeric CH4 molecules.
FIG. 1.
Cell surface expression of CD4 molecules in HeLa cell lines. Cells were incubated with medium alone (white histograms) or 50 μg of anti-CD4 MAb BL4 per ml (black histograms). MAb binding was detected by a fluorescein isothiocyanate-labeled GAM Ig. The fluorescence intensity was recorded in the log mode.
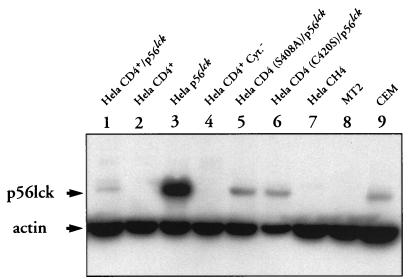
Expression of the p56lck human gene was tested by Western blotting. Cell lysates were electrophoresed onto sodium dodecyl sulfate-polyacrylamide (10%) gel electrophoresis (SDS-PAGE) gels and blotted to polyvinylidene difluoride (PVDF) membrane (Millipore, St Quentin Yvelines, France). The blot was saturated for 1 h in PBS–10% milk–0.05% Tween 20 prior to the addition of anti-p56lck (Santa-Cruz Biotechnologies, Santa Cruz, Calif.) or antiactin (Immunotech-Coulter) antibody. Antibody staining was revealed by addition of a 1:3,000 dilution of peroxidase-GAM Ig. After three washes, bound antibody was detected by incubating the membrane with the ECL (enhanced chemiluminescence) reagent (Amersham, Les Ullis, France). Western blot detection of p56lck is shown in Fig. 2. Expression of p56lck was detected in the HeLa CD4+/p56lck, HeLa p56lck, HeLa CD4 (S408A)/p56lck, and HeLa CD4 (C420S)/p56lck cell lines, with the highest expression level found in the HeLa p56lck cell line. As expected, no p56lck protein was detected in total protein lysates from HeLa CD4+, HeLA CD4+ Cyt−, and HeLa CH4 cell lines that were not transfected with the p56lck expression vector.
FIG. 2.
Detection of p56lck expression by Western blot analysis. HeLa CD4+/p56lck, HeLa CD4+, HeLa p56lck, HeLa CD4+ Cyt−, HeLa CD4 (C420S)/p56lck, HeLa CD4 (S408A)/p56lck, and HeLa CH4 extracts containing 50 μg of total cellular proteins were electrophoresed in an SDS–10% polyacrylamide gel and blotted to a PVDF membrane. The membrane was incubated with a mixture of anti-p56lck and antiactin MAbs and then reacted with GAM Ig-peroxidase conjugate. Bound MAbs were revealed by incubation of the membrane with ECL reagent and exposure to Hyperfilm-ECL. Controls consist of lysates from MT2 cells (a human T-cell leukemia virus type 1-transformed CD4+ T-cell line which lacks p56lck expression) and CEM cells (a CD4+ T-cell line which expresses p56lck).
Analysis of CD4-p56lck interaction by coimmunoprecipitation.
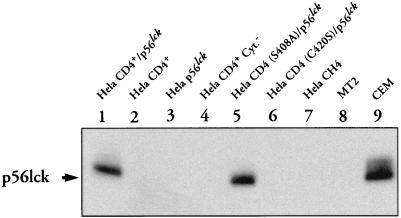
The nature of the CD4-p56lck interactions in each cell line was characterized by a coimmunoprecipitation assay. Adherent cells were washed once in Ca2+-, Mg2+-free PBS, harvested by scraping into PBS, and centrifuged at 1,500 rpm for 5 min at 4°C in a GR412 Jouan (Jouan, St. Herblain, France). Cell pellets were resuspended in lysis buffer (20 mM Tris-HCl [pH 8.0], 3% Nonidet P-40, 150 mM NaCl, 2 mM EDTA) and protease inhibitors (1 mM phenylmethylsulfonyl fluoride [PMSF] and 10 μg [each] of leupeptin, antipain, and pepstatin per ml). Detergent-insoluble material was removed by centrifugation at 4°C for 20 min at full speed in an Eppendorf microcentrifuge. The supernatants were collected, and the protein concentration in each sample was determined. Supernatants were precleared by incubation for 30 min with 50 μl of packed prewashed protein A-Sepharose (Sigma Chemical Company, Ltd.). CD4 was immunoprecipitated at 4°C by adding 4.5 μg of 13B8-2 anti-CD4 MAb (Immunotech-Coulter) for 1 h and protein A-Sepharose for an additional 1.5 h. The beads were collected by centrifugation, washed three times in lysis buffer, resuspended in an equal volume of SDS-PAGE sample buffer containing 50 mM dithiothreitol (DTT), and analyzed on SDS-PAGE (10% polyacrylamide) gels. Proteins were transferred onto PVDF membrane (Millipore), and the blot was saturated for 1 h in PBS–10% milk–0.05% Tween 20. CD4-p56lck immune complexes were revealed by addition of anti-p56lck antibody (Santa-Cruz Biotechnologies) for 1 h. After three washes, MAb staining was revealed by addition of a 1:3,000 dilution of peroxidase-GAM Ig (Immunotech-Coulter). After three washes, bound MAb was detected by incubation of the membrane with ECL reagent (Amersham).
As shown in Fig. 3, CD4-associated-p56lck was detected in the HeLa CD4+/p56lck and HeLa CD4 (S408A)/p56lck cell lines. In contrast, proteins migrating at the expected size for p56lck were not observed in HeLa CD4+, HeLa CH4, or HeLa CD4+ Cyt− cells, which only express CD4 molecules, nor in HeLa p56lck cells, which express p56lck but lack expression of the CD4 protein. Finally, CD4/p56lck coimmunoprecipitates were not detected by immunoprecipitation with anti-CD4 MAb in a HeLa CD4 (C420S)/p56lck cell line that expresses a CD4 mutant molecule and p56lck, indicating, as previously shown (56), that the p56lck kinase does not interact with the mutated form of CD4 expressed in these cells.
FIG. 3.
Analysis of CD4-p56lck interaction by coimmunoprecipitation. One milligram of total cellular protein was immunoprecipitated with 13B8-2 anti-CD4 MAb. After washing, the immunoprecipitates were electrophoresed in SDS-PAGE (10% polyacrylamide) gels, transferred to PVDF membrane, and hybridized with anti-p56lck MAbs. MAb staining was revealed by incubation of the membrane with a 1:3,000 dilution of GAM Ig. Immunoprecipitates from MT2 and CEM cellular extracts are shown as controls.
CD4-p56lck interaction is required for iHIV- 1-induced nuclear translocation of NF-κB in HeLa cell lines.
We have previously demonstrated that iHIV- 1 binding to CD4 induced NF-κB activation in T-lymphoblastoid cell lines expressing a wild-type CD4 molecule (5) and in primary lymphocytes (10). To assess whether such an activation signal requires p56lck expression, electrophoretic mobility shift assays (EMSAs) were performed to analyze the ability of iHIV- 1–CD4 interaction to stimulate NF-κB nuclear translocation in HeLa cell lines expressing either wild-type or mutated CD4 molecules and/or p56lck. To this end, 2 × 106 cells were exposed for 4 h either to 100 μl of an iHIV- 1 solution stock corresponding to 1,000 50% tissue culture infective doses (TCID50) of infectious virus per ml or to 20 ng of PMA per ml (Sigma). Briefly, cells were washed three times in PBS and lysed in buffer containing 10 mM KCl, 2 mM MgCl2, 0.1 mM EDTA, 1 mM DTT, 0.1 mM PMSF, 4 μg of leupeptin per ml, and 10 mM HEPES (pH 7.8). After 15 min on ice, a 50-μl solution of 10% Nonidet P-40 was added to the sample, and cells were microcentrifuged at 4°C for 30 s. The pellets were resuspended in 100 μl of buffer containing 50 mM KCl, 300 mM NaCl, 0.1 mM EDTA, 1 mM DTT, 0.1 mM PMSF, 4 μg of leupeptin per ml, 10% glycerol, and 50 mM HEPES (pH 7.8) and incubated for 20 min at 4°C. The supernatants were collected after centrifugation for 5 min at 4°C. The EMSAs were performed with 2 μg of protein of nuclear extract, 105 cpm of a 32P-labeled oligonucleotide corresponding to the NF-κB sequence binding site from the HIV- 1 long terminal repeat (LTR) (5), and 100 mM KCl, 1 mM DTT, 1 mM ZnSO4, 20% glycerol, 0.01% Nonidet P-40, and 50 mM HEPES (pH 7.9), supplemented with BSA, tRNA, and poly(dI-dC). After 20 min at room temperature, the mixture was run at 120 V in a 10% polyacrylamide gel.
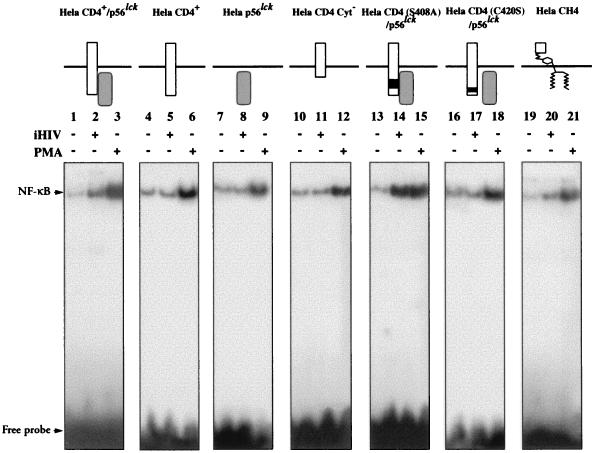
As shown in Fig. 4, a shift of labelled NF-κB oligonucleotide was observed when mixed with nuclear extracts from HeLa CD4+/p56lck cells exposed to iHIV- 1 compared with the basal activation level identified from unstimulated HeLa CD4+/p56lck cells (lanes 2 and 1, respectively). In contrast, no shift was observed when HeLa CD4+ (lane 5) or HeLa p56lck cells (lane 8), lacking either CD4 expression or p56lck expression, respectively, were exposed to iHIV- 1. As a control, a strong activation of NF-κB was observed when these two cell lines were incubated in presence of 20 ng of PMA per ml (lanes 6 and 9). These observations show that in nonlymphoid cell lines expressing wild-type CD4 and p56lck molecules, iHIV- 1 binding to CD4 induces an activation signal that leads to NF-κB nuclear translocation and is transduced via the CD4 and p56lck molecules.
FIG. 4.
Effect of iHIV treatment on NF-κB nuclear translocation in different HeLa cell lines analyzed by EMSA. Nuclear extracts prepared from HeLa CD4+/p56lck, HeLa CD4+, HeLa p56lck, HeLa CD4+ Cyt−, HeLa CD4(S408A)/p56lck, HeLa CD4(C420S)/p56lck, and HeLa CH4 cell lines cultured for 4 h in medium alone, medium containing iHIV- 1 (iHIV +) or medium supplemented with 20 ng of PMA per ml (PMA +) were reacted with radiolabeled double-stranded NF-κB oligonucleotide (HIV- 1Lai LTR sequence). The samples were electrophoresed and analyzed by autoradiography.
Our experiments provide direct evidence that the coexpression of CD4 and p56lck is required for full signal transduction after oligomerization of CD4 by the envelope glycoprotein of HIV. It is worth noting that the absence of the CD4 receptor abolishes the nuclear translocation of NF-κB that is usually observed upon iHIV- 1 stimulation, providing direct evidence that the activation events that we describe are highly specific for the interaction of the virus envelope glycoprotein gp120 with CD4. The analysis of the HeLa CD4+ cell line demonstrates the crucial role of p56lck in signal transduction induced by HIV-CD4 interaction. These data corroborate recent data from other groups; Di Somma et al. (21) reported that activation of NF-κB and AP- 1 induced by CD4 triggering with anti-CD4 MAb is strongly inhibited by a dominant-negative mutant of Lck. Furthermore, Merzouki and coworkers (39) demonstrated that HIV- 1 gp120/160-expressing cells upregulate HIV- 1 LTR-directed gene expression in CD4+ CEM cells transfected with an HIV- 1 LTR-reporter gene construct, whereas expression of the reporter gene was not induced in CD4+ U937 cells, which lack p56lck.
Mutation of CD4 at C420 and S408 influences iHIV- 1 stimulation of NF-κB translocation.
The HeLa CD4 (C420S)/ p56lck cell line expresses a mutant CD4 molecule, CD4 (C420S), in which cysteine residue 420 was replaced by a serine. This mutation disrupts a site in the cytoplasmic tail of CD4 that is required for interaction with p56lck (51, 56). The coimmunoprecipitations shown in Fig. 3 and an in vitro kinase assay performed after immunoprecipitation with an anti-CD4 antiserum (41) confirmed the lack of association between CD4 (C420S) and p56lck. When this cell line was incubated with iHIV- 1, no NF- κB shift was observed by EMSA (Fig. 4, lane 17), although a strong activation was induced with PMA (lane 18). Thus, a physical interaction of the cytoplasmic tail of CD4 with p56lck is required to induce NF-κB activation upon CD4 cross-linking.
Additional information was provided by the analysis of the HeLa CD4 (S408A)/p56lck cell line coexpressing the S408A-mutated form of CD4 and p56lck. The S408A mutation replaces a critical residue in the cytoplasmic domain of CD4 that is phosphorylated in response to phorbol esters (52) and is involved in both CD4 endocytosis and the dissociation of p56lck from CD4 (7, 53). When NF-κB activation was studied with this cell line, a very strong shift was observed upon stimulation by both iHIV- 1 and PMA (Fig. 4, lanes 14 and 15, respectively). This shift was significantly higher than that observed in HeLa CD4+/p56lck cells expressing both CD4 and p56lck wild-type molecules. The high activation observed in HeLa CD4 (S408A)/p56lck may be a consequence of high expression levels of p56lck or may be due to the lack of S408 phosphorylation, thus preventing p56lck dissociation. Altogether, these data suggest that the physical interaction of CD4 and p56lck is a prerequisite for transduction of the activation signal(s) induced by the CD4–iHIV- 1 interaction.
Finally, we have studied an additional cell line, HeLa CH4, expressing a CD4–Thy- 1 fusion protein on the cell surface which contains the HIV- 1 binding site (D1 domain of CD4) and is anchored to the membrane by a GPI tail in place of a conventional membrane-spanning domain. The CD4–Thy- 1 fusion protein can serve as an HIV- 1 receptor, and HeLa CH4 cells can therefore be infected by HIV- 1 (30). In addition, this molecule, like CD4, is downmodulated in its cell surface expression by exogenous gangliosides. Incubation of the HeLa CH4 cell line with iHIV- 1 showed a slight shift of the NF-κB transcription factor. The signal transduction observed through CH4 must involve interactions between the PI-linked molecule and a second messenger expressed in HeLa cells.
Our results indicate that signals transduced in the cell through CD4 upon iHIV- 1 stimulation involve p56lck and require cysteine 420 in the cytoplasmic tail of CD4.
Involvement of p56lck in HIV- 1 LTR activation induced after HIV- 1 gp120-CD4 interaction.
NF-κB nuclear translocation is a major start signal for HIV- 1 early gene transcription. Having determined that p56lck is required for NF-κB activation induced by iHIV- 1 binding to CD4, we next assessed the role of p56lck in HIV- 1 LTR activation generated upon HIV- 1 envelope binding to CD4. To this end, we used two HeLa CD4+ HIV- 1 LTR–β-galactosidase indicator cell lines (CD4-LTR/β-gal) expressing either the CD4 molecule alone and referred to as HeLa P4 (22), or coexpressing the CD4 and p56lck molecules and referred to as HeLa P4p56 (50a). These indicator cell lines either were exposed to medium alone, heat-inactivated virus (iHIV- 1), or infectious HIV- 1 or were cocultured with the CD4− human T-cell line A2.01 or A2.01 cells expressing HIV- 1 gp120. A2.01 cells expressing HIV- 1 gp120 (referred below to as A2.01/gp120), were constructed by transient transfections of A2.01 cells with the pV3 plasmid. (This plasmid derives from the pBRU3 vector that contains the complete HIV- 1Lai genome [provided by L. Montagnier at the Pasteur Institute, Paris, France], in which the PstI-ApaI gag sequence was deleted to construct a defective provirus. pV3 [45a] was used as an env expression vector.)
As shown in Fig. 5A, A2.01/gp120 cells were found by flow cytometry to express gp120 at the cell membrane following transfection compared to the parental A2.01 cells. Figure 5B shows that both HeLa P4 and HeLa P4p56 cells expressed cell surface CD4 and can therefore bind HIV- 1. Moreover, Fig. 5C and D indicate that HeLa P4p56 cells express p56lck and that p56lck associates with CD4. The functional assays were performed as follows. The CD4-LTR/β-gal indicator cell lines were plated in 12-well plates at 8 × 105 cells/ml in DMEM with 10% FCS. On the next day, the cells were stimulated with 500 μl of iHIV- 1 (iHIV- 1 at a concentration equivalent to 1,000 × TCID50 of infectious virus/ml) or exposed to 500 μl of infectious HIV- 1 (at 1,000 × TCID50 of infectious virus/ml). The plates were rocked every 30 to 45 min. After 2 h, the cells were washed, and 1 ml of DMEM supplemented with 10% FCS per well was added. In some experiments, stimulation were performed by coculturing the HeLa CD4+-LTR/β-gal cell lines with 4 × 104 A2.01 or A2.01/gp120 cells. After 3 days in culture, nonadherent (A2.01) cells were removed, and adherent cells were washed three times in PBS and lysed in 300 μl of buffer containing 60 mM Na2HPO4, 40 mM NaH2PO4, 10 mM KCl, 10 mM MgSO4, 2.5 mM EDTA, 50 mM 2-β-mercaptoethanol, and 0.125% Nonidet P-40 for 15 min at room temperature. Cell lysates were clarified by centrifugation for 15 min at 13,000 rpm at 4°C. For β-galactosidase activity determination, 150 μl of total cellular extract was reacted for 1 h at 37°C in 500 μl of buffer containing 80 mM Na2HPO4, 10 mM MgCl2, 1 mM 2-β-mercaptoethanol, and 6 mM O-nitrophenyl-β-d-galactopyranoside (ONPG). β-Galactosidase activity was evaluated by measuring A410.
FIG. 5.
Induction of HIV- 1 LTR transactivation by gp120-CD4 interaction. (A) Expression of gp120 on A2.01/gp120 cells (black histogram) was monitored by indirect flow cytometry. Background reactivity of anti-gp120 antibodies with gp120-negative A2.01 parental cells is shown as a control (white histogram). (B) Expression of CD4 on HeLa P4 and HeLa P4p56 (black histogram) was monitored by flow cytometry, as described in the legend to Fig. 1. The background of the probe is shown (white histogram). The CD4− A2.01 cell line was used as a control. (C) Detection of p56lck expression in HeLa P4p56 by Western blot analysis. The experiment was performed as described in the legend to Fig. 2. (D) Analysis of CD4-p56lck interaction in HeLa P4p56 cells by coimmunoprecipitation. The experiment was performed as described in the legend of Fig. 3. The CD4− p56lck-positive A2.01 cell line was used as a control.
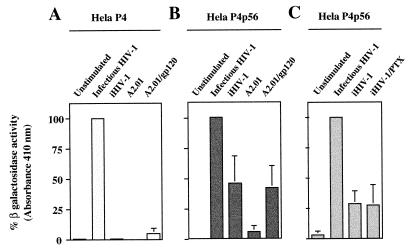
β-Galactosidase activities in HeLa P4 and HeLa P4p56 cells infected with HIV- 1 were determined. The values obtained under such experimental conditions were used to fix the 100% β-galactosidase activity for each cell line. (Note that HIV- 1 infection provides Tat transactivator to the target cell.) Subsequent values obtained in each assay were expressed as percentages of maximal β-galactosidase activity. As shown in Fig. 6A, a very weak β-galactosidase activity was detected in the HeLa P4 cell line after iHIV- 1 stimulation or after coculture with the A2.01/gp120 cells. In contrast (Fig. 6B), significant β-galactosidase activity was observed when the HeLa P4p56 cell line was incubated in the presence of either iHIV- 1 (45.6% of maximal activation) or A2.01/gp120 cells (41.5% of maximal activation). When HeLa P4p56 cells were cocultured with gp120-negative parental A2.01 cells, no significant β-galactosidase gene expression driven by the HIV- 1 LTR was detected in cell lysates. This result demonstrates that the reporter gene activity was specifically induced by gp120 expressed at the cell surface of the A2.01/gp120-stimulating cells. It is worth noting that the extent of stimulation of β-galactosidase synthesis by HIV- 1 gp120-CD4 interaction was lower than that by infectious HIV- 1. The higher β-galactosidase activity generated by infectious HIV- 1 can probably be ascribed to Tat transactivation of the HIV- 1 LTR and to virus propagation in cell cultures (the concentration of virus after 3 days of cell culture being much higher than the virus input), thereby increasing the percentage of β-galactosidase-positive cells.
FIG. 6.
β-Galactosidase activities in HeLa P4 and P4p56 cells. HeLa P4 (A) and HeLa P4p56 (B) cell lines expressing the β-galactosidase reporter gene cloned downstream of the HIV- 1 LTR promoter were cultured in the presence of medium alone (column 1) or medium supplemented with infectious HIV- 1 (column 2) or iHIV- 1 (column 3). In columns 4 and 5, the LTR–β-galactosidase indicator cell lines were cocultured with A2.01 cells or the A2.01/gp120 cells, respectively. After 3 days in culture, adherent cells were harvested and lysed, and β-galactosidase activities were determined by incubation of 150 μl of total cellular extracts with ONPG in an appropriated buffer. (C) HeLa P4p56 cells were treated for 16 h with 250 ng of PTX per ml (column 4) or with control medium (columns 1 to 3) and next cultured for 3 days in the presence of medium alone (column 1), medium supplemented with infectious HIV- 1 (column 2), or iHIV- 1 (columns 3 and 4). β-Galactosidase activities were determined as described above. In order to compare the results obtained from different experiments, all values were normalized according to the β-galactosidase activities obtained following infection of cells by HIV- 1 (100% β-galactosidase activity). Mean absorbances measured with HIV- 1-infected samples were 0.944, 0.472, and 0.452 in panels A, B, and C, respectively.
PTX-sensitive G protein signalling is not required for iHIV-1-induced HIV- 1 LTR activation.
The G protein-coupled seven transmembrane chemokine receptor CXCR4 (24), also called fusin, has been identified as a cell surface coreceptor for T-cell-tropic viruses such as HIV- 1Lai, which was used in the present study. Recently, CXCR4 was shown to transduce signals to T cells following interaction with HIV- 1 (20). This molecule is expressed at the surface of HeLa cells (24), and we previously reported that HeLa P4 cells express about two- to fourfold excess of surface CXCR4 compared to HeLa P4p56 cells (18). This observation may explain why higher β-galactosidase activities were measured in HeLa P4 cells infected by HIV- 1 compared to HIV- 1-infected HeLa P4p56 cells (mean A410 of 0.944 for HIV- 1-infected HeLa P4 cells and 0.472 for HIV- 1-infected HeLa P4p56 cells). Although we have found (see above) that iHIV- 1 binding to CD4 does not induce NF-κB translocation in HeLa CD4+, HeLa CD4+ Cyt−, HeLa p56lck, and HeLa CD4 (C420S)/p56lck cells, indicating that the interaction of HIV- 1 gp120 with CXCR4 cannot account, by itself, for signal transduction triggering NF-κB translocation, we wanted to exclude the possibility that cosignaling through CXCR4 is required to activate HIV- 1 LTR. We studied the antagonist activity of pertussis toxin (PTX), an inhibitor of protein Gi-mediated signals, on HIV- 1 LTR activation induced after HIV- 1 gp120 binding to the CD4-CXCR4 receptor complex by using a previously described protocol (18). Briefly, HeLa P4p56 cells were treated for 16 h with 250 ng of PTX per ml (a concentration of PTX that inhibits the CXCR4 natural ligand stromal cell-derived factor 1α [SDF- 1α] induction of calcium fluxes in HeLa P4 cells without affecting the cells’ viability [18]) or control medium. Next, the cells were stimulated with 500 μl of iHIV- 1 or exposed to 500 μl of infectious HIV- 1, and β-galactosidase activity was evaluated, as described above, after 3 days of cell culture. Under these experimental conditions (Fig. 6C), PTX did not significantly reduce the activation of HIV- 1 LTR triggered by iHIV- 1 gp120, indicating that CXCR4 signal transduction through Gi proteins is not required for HIV- 1 LTR activation induced in HeLa P4p56 cells after iHIV- 1 gp120 binding to the CD4-CXCR4 receptor complex. It is worth noting that we previously found that PTX did not modify the transactivation of HIV- 1 LTR in HeLa P4 and HeLa P4p56 cells infected by HIV- 1 (18).
Altogether, these data strongly support the hypothesis that NF-κB activation induced by HIV- 1 gp120-CD4 receptor interaction requires the formation of a functional CD4-p56 complex. Activation signals generated upon CD4 ligation are able to transactivate the HIV- 1 LTR and to induce early viral gene transcription. These observations are in agreement with our previous results demonstrating that uninfectious HIV- 1 and gp120–anti-gp120 immune complexes binding to CD4 on latently infected quiescent CD4+ PBMCs upregulate latent HIV- 1 and commit cells to produce virus (10).
p56lck plays a key role in transduction of signal(s) initiated upon HIV- 1 interaction with CD4.
During the past few years, many research groups have demonstrated the association of the p56lck tyrosine kinase with the cytoplasmic tail of CD4, providing a mechanism whereby CD4 could transduce signals through this kinase. However, a requirement for the association of CD4 with p56lck in transduction of signal(s) originating from CD4 is disputed. Indeed, p56lck-independent CD4-mediated enhancement and inhibition of the T-cell activation pathway have been described (4, 17, 18, 35, 59). In contrast, it has been also demonstrated that p56lck linked to CD4 is critical for CD3- and antigen-dependent T-cell activation, since cells expressing a mutant form of p56lck lacking kinase activity demonstrated a profound inhibition of tyrosine phosphorylation in response to stimulation by anti-CD3 MAb (1, 14). Similar observations were performed with interleukin- 16 (IL- 16), a natural ligand of CD4, that was described to induce T-cell migration in a p56lck-dependent fashion, whereas the motile response generated by IL- 16–CD4 interaction was independent of CD4-p56lck coupling (47). Despite the increasing number of reports indicating the ability of gp120/160 to activate CD4-associated p56lck (9, 28, 31, 42, 54), there was no clear demonstration concerning the precise role played by p56lck in CD4-mediated T-cell activation, and the possibility remained that HIV- 1-mediated signaling could be transduced following CD4 cross-linking independently of p56lck activation.
Beside activation of p56lck, the ligation of multimeric gp120 to the CD4-CXCR4 receptor complex induces a variety of effects, including mobilization of intracellular Ca2+ (33), induction of protein phosphorylation, and activation of a number of cellular proteins, including PKC (60), Shc (3), PI-3K (9, 44) and PI-4K (44, 50), p59fyn (34), Zap70 (34), p95vav (34), Ras (34), Raf- 1 (43), the Raf- 1-related 110-kDa polypeptide (45), MEK- 1 (12), ERK- 1 (12), and ERK-2 (6), which have been identified as molecules activated in response to such stimuli. Some of these molecules are likely required (or have been shown to act) as signal messengers in activation of NF-κB (5, 10, 12, 13, 15) and AP- 1 (5, 10, 12, 16) transcription factors that are specifically induced by virus-host interactions in lymphoblastoid cell lines and in primary T lymphocytes. We have previously suggested a pivotal role for p56lck in transduction of a CD4-dependent T-cell activation signal upon HIV- 1 binding to CD4. (i) CD4 oligomerization by iHIV- 1 induces p42erk activation in lymphoblastoid cell lines expressing the wild-type CD4 and a functional p56lck molecule but not in cells that express CD4 lacking the cytoplasmic domain (6). (ii) NF-κB translocation was induced by iHIV- 1 in lymphoblastoid cell lines expressing the wild-type CD4 and a functional p56lck molecule but not in cells expressing the truncated CD4 (5). (iii) iHIV induced expression of a reporter gene driven by HIV- 1 LTR in cells expressing the wild-type CD4 and a functional p56lck molecule but not in cells expressing a truncated form of CD4. Similar results have been obtained by Merzouki and coworkers (39), who reported that the activation of reporter gene driven by HIV- 1 LTR was enhanced following envelope binding to CD4 in p56lck-expressing cell lines but not in U937 cells (from the promonocytic lineage) that lack expression of this kinase. These data argued for the critical involvement of p56lck in CD4-mediated T-cell activation. We provide here the first direct demonstration that both CD4 and p56lck are required to transduce an activation signal leading to NF-κB nuclear translocation after iHIV- 1 gp120 binding to CD4. The fact that cells expressing a mutant form of CD4 at position 420 (C420A) and the wild-type p56lck did not respond to iHIV- 1 stimulation further indicates the requirement for a functional CD4-p56lck complex.
Understanding how HIV takes advantage of the cellular signaling pathways and modifies the physiology of a host cell is an important goal to improve our knowledge of the pathogenesis of AIDS (11, 55). Our results provide a molecular basis by which gp120 misappropriates the transduction function of the CD4-p56lck complex to act on CD4 T-cell signaling.
Acknowledgments
We are very grateful to Olivier Schwartz (Laboratoire Rétrovirus et Transfert Génétique, Institut Pasteur, Paris) for providing the previously unpublished HeLa P4p56 cell line and for helpful discussions.
This work was partially supported by institutional funds from the Centre National de la Recherche Scientifique (CNRS) and the Institut National de la Santé et de la Recherche Médicale (INSERM) and grants from the Agence Nationale de Recherche sur le SIDA (ANRS) and Fondation pour la Recherche Médicale (FRM)-SIDACTION program. L.B. is a fellow of the FRM-SIDACTION program.
REFERENCES
- 1.Abraham N, Miceli M C, Parnes J R, Veillette A. Enhancement of T-cell responsiveness by the lymphocyte-specific tyrosine kinase p56lck. Nature (London) 1991;350:62–66. doi: 10.1038/350062a0. [DOI] [PubMed] [Google Scholar]
- 2.Anderson P, Blue M-L, Morimoto C, Schlossman S F. Cross-linking of T3 (CD3) with T4 (CD4) enhances the proliferation of resting T lymphocytes. J Immunol. 1987;139:678–682. [PubMed] [Google Scholar]
- 3.Baldari C T, Pelicci G, Di Somma M M, Milia E, Giuli S, Pelicci P G, Telford J L. CD4 triggering results in tyrosine phosphorylation of the Shc adaptor protein. Oncogene. 1995;10:1141–1147. [PubMed] [Google Scholar]
- 4.Benkirane M, Corbeau P, Housset V, Devaux C. An antibody that binds the immunoglobulin CDR3-like region of the CD4 molecule inhibits provirus transcription in HIV-infected T cells. EMBO J. 1993;12:4909–4921. doi: 10.1002/j.1460-2075.1993.tb06185.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Benkirane M, Jeang K-T, Devaux C. The cytoplasmic domain of CD4 plays a critical role during the early stages of HIV infection in T-cells. EMBO J. 1994;13:5559–5569. doi: 10.1002/j.1460-2075.1994.tb06893.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Benkirane M, Schmid-Antomarchi H, Littman D R, Hirn M, Rossi B, Devaux C. The cytoplasmic tail of CD4 is required for inhibition of human immunodeficiency virus type 1 replication by antibodies that bind to the immunoglobulin CDR3-like region in domain 1 of CD4. J Virol. 1996;69:6904–6910. doi: 10.1128/jvi.69.11.6904-6910.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Beyers A D, Davies S J, Cantrell D A, Izquierdo M, Williams A F. Autonomous roles for the cytoplasmic domains of the CD2 and CD4 T cell surface antigens. EMBO J. 1991;10:337–385. doi: 10.1002/j.1460-2075.1991.tb07959.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Biddison W E, Rao P E, Talle M A, Golstein G, Shaw S. Possible involvement of the T4 molecule in T cells recognition of class II HLA antigens. Evidence from studies of CTL-target cell binding. J Exp Med. 1984;159:783–797. doi: 10.1084/jem.159.3.783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Briand G, Barbeau B, Tremblay M. Binding of HIV- 1 to its receptor induces tyrosine phosphorylation of several CD4-associated proteins including the phosphatidylinositol 3-kinase. Virology. 1997;228:171–179. doi: 10.1006/viro.1996.8399. [DOI] [PubMed] [Google Scholar]
- 10.Briant L, Coudronnière N, Benkirane M, Robert-Hebmann V, Devaux C. The binding of HIV- 1 virions or gp120-anti-gp120 immune complexes to HIV- 1 infected quiescent peripheral blood mononuclear cells reveals latent infection. J Immunol. 1996;156:3994–4004. [PubMed] [Google Scholar]
- 11.Briant L, Devaux C. Baseline HIV type 1 load predicts the long-term clinical outcome of infected patients: the positive feedback theory. Immunol Lett. 1997;55:123–125. doi: 10.1016/s0165-2478(96)02688-0. [DOI] [PubMed] [Google Scholar]
- 12.Briant L, Robert-Hebmann V, Sivan V, Brunet A, Pouysségur J, Devaux C. Involvement of ERK module in human immunodeficiency virus-mediated CD4 signals controlling activation of NF-κB and AP- 1 transcription factors. J Immunol. 1998;160:1875–1885. [PubMed] [Google Scholar]
- 13.Briant L, Signoret N, Gaubin M, Robert-Hebmann V, Zhang X, Murali R, Greene M I, Piatier-Tonneau D, Devaux C. Transduction of activation signal that follows HIV- 1 binding to CD4 and CD4 dimerization involves the immunoglobulin CDR3-like region in domain 1 of CD4. J Biol Chem. 1997;272:19441–19450. doi: 10.1074/jbc.272.31.19441. [DOI] [PubMed] [Google Scholar]
- 14.Caron L, Abraham N, Pawson T, Veillette A. Structural requirements for enhancement of T-cell responsiveness by the lymphocyte-specific tyrosine protein kinase p56lck. Mol Cell Biol. 1992;12:2720–2729. doi: 10.1128/mcb.12.6.2720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chirmule N, Kalyanaraman V S, Pahwa S. Signals transduced through the CD4 molecule on T lymphocytes activate NF-κB. Biochem Biophys Res Commun. 1994;203:498–505. doi: 10.1006/bbrc.1994.2210. [DOI] [PubMed] [Google Scholar]
- 16.Chirmule N, Goonewardena H, Pahwa S, Pasieka R, Kalyanaraman V S, Pahwa S. HIV- 1 envelope glycoproteins induce activation of activated protein- 1 in CD4+ T cells. J Biol Chem. 1995;270:19364–19369. doi: 10.1074/jbc.270.33.19364. [DOI] [PubMed] [Google Scholar]
- 17.Collins T, Burakoff S J. Tyrosine kinase activity of CD4-associated p56lck may not be required for CD4-dependent T-cell activation. Proc Natl Acad Sci USA. 1993;90:11885–11889. doi: 10.1073/pnas.90.24.11885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Coudronnière, N., J. Corbeil, V. Robert-Hebmann, J.-M. Mesnard, and C. Devaux. The lck protein tyrosine kinase is not involved in antibody-mediated CD4 (CDR3-loop) signal transduction that inhibits HIV- 1 transcription. Eur. J. Immunol., in press. [DOI] [PubMed]
- 19.Dalgleish A G, Beverley P C L, Clapham P R, Crawford D H, Greaves M R, Weiss R A. The CD4 (T4) antigen is an essential component of the receptor for the AIDS retrovirus. Nature (London) 1984;312:763–767. doi: 10.1038/312763a0. [DOI] [PubMed] [Google Scholar]
- 20.Davis C B, Dikic I, Unutmaz D, Hill C M, Arthos J, Siani M A, Thompson D A, Schlessinger J, Littman D R. Signal transduction due to HIV- 1 envelope interactions with chemokine receptor CXCR4 or CCR5. J Exp Med. 1997;186:1793–1798. doi: 10.1084/jem.186.10.1793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Di Somma M M, Majolini M B, Burastero S E, Telford J L, Baldari C T. Cyclosporine A sensitivity of the HIV- 1 long terminal repeat identifies distinct p56lck-dependent pathways activated by CD4 triggering. Eur J Immunol. 1996;26:2181–2188. doi: 10.1002/eji.1830260933. [DOI] [PubMed] [Google Scholar]
- 22.Dragic T, Charneau P, Clavel F, Alizon M. Complementation of murine cells for human immunodeficiency virus envelope/CD4-mediated fusion in human/murine heterokaryons. J Virol. 1992;66:4794–4802. doi: 10.1128/jvi.66.8.4794-4802.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ettehadieh E, Sanghera J S, Pelech S L, Hess-Bienz D, Watts J, Shastri N, Aebersold R. Tyrosyl-phosphorylation and activation of MAP kinases by p56lck. Science. 1992;255:853–855. doi: 10.1126/science.1311128. [DOI] [PubMed] [Google Scholar]
- 24.Feng Y, Broder C C, Kennedy P E, Berger E A. HIV- 1 entry cofactor: functional cDNA cloning of a seven-transmembrane, G protein-coupled receptor. Science. 1996;272:872–876. doi: 10.1126/science.272.5263.872. [DOI] [PubMed] [Google Scholar]
- 25.Field A P, Bednarik D P, Hers A, May W S. Human immunodeficiency virus induces phosphorylation of its cell surface receptor. Nature (London) 1988;333:278–280. doi: 10.1038/333278a0. [DOI] [PubMed] [Google Scholar]
- 26.Gay D, Maddon P, Sekaly R, Taile M A, Godfrey M, Long E, Goldstein G, Chess L, Axel R, Kappler J, Marrack P. Functional interaction between human T-cell protein CD4 and the major histocompatibility complex HLA-DR antigen. Nature (London) 1987;328:626–629. doi: 10.1038/328626a0. [DOI] [PubMed] [Google Scholar]
- 27.Goldman F, Jensen W A, Johnson G L, Heasley L, Cambier J. gp120 ligation of CD4 induces p56lck activation and TCR desensitization independent of TCR tyrosine phosphorylation. J Immunol. 1994;153:2905–2917. [PubMed] [Google Scholar]
- 28.Hivroz C, Mazerolles F, Soula M, Fagard R, Graton S, Meloche S, Sekaly R, Fischer A. Human immunodeficiency virus gp120 derived peptides activate protein tyrosine kinase p56lck in human CD4 T lymphocytes. Eur J Immunol. 1993;23:600–607. doi: 10.1002/eji.1830230303. [DOI] [PubMed] [Google Scholar]
- 29.Hwang J, Yan Y, Garett T P J, Liu J, Rodgers D W, Garlick R L, Tarr G E, Husain Y, Reinherz E L, Harrison S C. Atomic structure of a fragment of human CD4 containing two immunoglobulin-like domains. Nature (London) 1990;348:411–428. doi: 10.1038/348411a0. [DOI] [PubMed] [Google Scholar]
- 30.Jasin M, Page K A, Littman D R. Glycosylphosphatidylinositol-anchored CD4/Thy- 1 chimeric molecules serve as human immunodeficiency virus receptors in human, but not mouse, cells and are modulated by gangliosides. J Virol. 1991;65:440–444. doi: 10.1128/jvi.65.1.440-444.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Juszczak R T, Turchin H, Truneh A, Culp J, Kassis S. Effect of human immunodeficiency virus gp120 glycoprotein on the association of the protein tyrosine kinase p56lck with CD4 in human T lymphocytes. J Biol Chem. 1991;266:11176–11183. [PubMed] [Google Scholar]
- 32.Klatzmann D, Champagne E, Chamaret S, Gruest J, Guetard D, Hercend T, Gluckman J C, Montagnier L. T-lymphocyte T4 molecule behaves as the receptor for human retrovirus LAV. Nature (London) 1984;312:767–768. doi: 10.1038/312767a0. [DOI] [PubMed] [Google Scholar]
- 33.Kornfeld H, Cruikshank W W, Pyle S E, Berman J S, Center D M. Lymphocyte activation by HIV- 1 envelope glycoprotein. Nature (London) 1988;335:445–448. doi: 10.1038/335445a0. [DOI] [PubMed] [Google Scholar]
- 34.Lakshmi Tamma S M, Chirmule N, Yagura H, Oyaizu N, Kakyanaraman V, Pahwa S. CD4 cross-linking (CD4XL) induces Ras activation and tumor necrosis factor-α secretion in CD4+ T cells. Blood. 1997;90:1588–1593. [PubMed] [Google Scholar]
- 35.Lemasson I, Briant L, Hague B, Coudronnière N, Héron L, David C, Rebouissou C, Kindt T, Devaux C. An antibody that binds domains 1 of CD4 inhibits replication of HIV- 1 but not HTLV-I in a CD4positive/p56lck negative HTLV-I-transformed cell line. J Immunol. 1996;156:859–865. [PubMed] [Google Scholar]
- 36.Littman D R. The structure of the CD4 and CD8 genes. Annu Rev Immunol. 1987;5:561–584. doi: 10.1146/annurev.iy.05.040187.003021. [DOI] [PubMed] [Google Scholar]
- 37.Maddon P J, Dalgleish A G, McDougal J S, Fagard R, Marsh M. The T4 gene encodes the AIDS virus receptor and is expressed in the immune system and brain. Cell. 1986;47:333–348. doi: 10.1016/0092-8674(86)90590-8. [DOI] [PubMed] [Google Scholar]
- 38.Maddon P J, McDougal J S, Clapham P R, Dalgleish A G, Jamal S, Weiss R A, Axel R. HIV infection does not require endocytosis of its receptor CD4. Cell. 1988;54:865–874. doi: 10.1016/s0092-8674(88)91241-x. [DOI] [PubMed] [Google Scholar]
- 39.Merzouki A, Patel P, Cassol S, Ennaji M, Tailor P, Turcotte F R, O’Shaughnessy M, Arella M. HIV- 1 gp120/160 expressing cells upregulate HIV- 1 LTR directed gene expression in a cell line transfected with HIV- 1 LTR-reporter gene constructs. Cell Mol Biol. 1995;41:445–452. [PubMed] [Google Scholar]
- 40.Mittler R S, Goldman S J, Spitalny G L, Burakoff S J. T cell receptor-CD4 physical association in a murine T cell hybridoma: induction by antigen receptor ligation. Proc Natl Acad Sci USA. 1989;86:8531–8535. doi: 10.1073/pnas.86.21.8531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pelchen-Matthews A, Boulet I, Littman D R, Fagard R, Marsh M. The protein tyrosine kinase p56lck inhibits CD4 endocytosis by preventing entry of CD4 into coated pits. J Cell Biol. 1992;117:279–290. doi: 10.1083/jcb.117.2.279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Phipps D J, Stanley E R, Piovesan J P, Mills G B, Branch D R. HIV infection in vitro enhances the activity of src-family protein tyrosine kinases. AIDS. 1996;10:1191–1198. doi: 10.1097/00002030-199609000-00003. [DOI] [PubMed] [Google Scholar]
- 43.Popik W, Pitha P M. Binding of human immunodeficiency virus type 1 to CD4 induces association of Lck and Raf- 1 and activates Raf- 1 by a Ras-independent pathway. Mol Cell Biol. 1996;16:6532–6541. doi: 10.1128/mcb.16.11.6532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Prasad K V S, Kapeller R, Janssen O, Repke H, Duke-Cohan J S, Cantley L C, Rudd C E. Phosphatidylinositol (PI) 3-kinase and PI 4-kinase binding to the CD4-p56lck complex: the p56lck SH3 domain binds to PI 3-kinase but not PI 4-kinase. Mol Cell Biol. 1993;13:7708–7717. doi: 10.1128/mcb.13.12.7708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Prasad K V S, Rudd C E. A Raf- 1-related p110 polypeptide associates with the CD4-p56lck complex in T cells. Mol Cell Biol. 1992;12:5260–5267. doi: 10.1128/mcb.12.11.5260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45a.Robert-Hebmann, V. Unpublished data.
- 46.Rudd C E, Trevillyan J M, Dasgupta J D, Wong L L, Schlossman S F. The CD4 receptor is complexed in detergent lysates to a protein-tyrosine kinase from human T lymphocytes. Proc Natl Acad Sci USA. 1988;85:5190–5194. doi: 10.1073/pnas.85.14.5190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ryan T C, Cruikshank W W, Kornfeld H, Collins T L, Centers D M. The CD4-associated tyrosine kinase p56lck is required for lymphocyte chemoattractant factor-induced T lymphocyte migration. J Biol Chem. 1995;270:17081–17086. doi: 10.1074/jbc.270.29.17081. [DOI] [PubMed] [Google Scholar]
- 48.Ryu S E, Kwong P D, Truneh A, Porter T G, Arthos J, Rosenberg M, Dai X, Xuong N H, Axel R, Sweet R W, Hendrickson W A. Crystal structure of an HIV-binding recombinant fragment of human CD4. Nature (London) 1990;348:419–426. doi: 10.1038/348419a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Saizawa K, Rojo J, Janeway C A., Jr Evidence for a physical association of CD4 and CD3:αβ T cell. Nature (London) 1987;328:260–263. doi: 10.1038/328260a0. [DOI] [PubMed] [Google Scholar]
- 50.Schmid-Antomarchi H, Benkirane M, Beittmayer V, Husson H, Ticchioni M, Devaux C, Rossi B. HIV induces activation of phosphatidylinositol 4-kinase and mitogen-activated protein kinase by interacting with T cell CD4 surface molecules. Eur J Immunol. 1996;26:717–720. doi: 10.1002/eji.1830260331. [DOI] [PubMed] [Google Scholar]
- 50a.Schwartz, O. Unpublished data.
- 51.Shaw A S, Amrein K E, Hammond C, Stern D F, Sefton B M, Rose J K. The Lck tyrosine protein kinase interacts with the cytoplasmic tail of the CD4 glycoprotein through its unique amino-terminal domain. Cell. 1989;59:627–636. doi: 10.1016/0092-8674(89)90008-1. [DOI] [PubMed] [Google Scholar]
- 52.Shin J, Doyle C, Yang Z, Kappes D, Strominger J L. Structural features of the cytoplasmic region of CD4 required for internalization. EMBO J. 1990;9:425–434. doi: 10.1002/j.1460-2075.1990.tb08127.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sleckman B P, Shin J, Igras V E, Collins T L, Strominger J L, Burakoff S J. Disruption of the CD4-p56lck complex is required for rapid internalization of CD4. Proc Natl Acad Sci USA. 1992;89:7566–7570. doi: 10.1073/pnas.89.16.7566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Soula M R, Fagard R, Fischer S. Internalization of human immunodeficiency virus glycoprotein 160 with CD4 in Jurkat cells increases p56lck autophosphorylation and kinase activity. Int Immunol. 1992;4:295–299. doi: 10.1093/intimm/4.2.295. [DOI] [PubMed] [Google Scholar]
- 55.Than S, Oyaizu N, Tetali S, Romano J, Kaplan M, Pahwa S. Upregulation of human immunodeficiency virus (HIV) replication by CD4 cross-linking in peripheral blood mononuclear cells of HIV-infected adults. J Virol. 1997;71:6230–6232. doi: 10.1128/jvi.71.8.6230-6232.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Turner J M, Brodsky M M, Irving B A, Levin S D, Perlmutter R M, Littman D R. Interaction of the unique N-terminal region of tyrosine kinase p56lck with cytoplasmic domains of CD4 and CD8 is mediated by cysteine motifs. Cell. 1990;60:755–765. doi: 10.1016/0092-8674(90)90090-2. [DOI] [PubMed] [Google Scholar]
- 57.Veillette A, Bookman M A, Horak E M, Samelson L E, Bolen J B. Signal transduction through the CD4 receptor involves the activation of the internal membrane tyrosine-protein kinase p56lck. Nature (London) 1989;338:257–259. doi: 10.1038/338257a0. [DOI] [PubMed] [Google Scholar]
- 58.Weiss A, Littman D R. Signal transduction by lymphocyte antigen receptors. Cell. 1994;76:263–274. doi: 10.1016/0092-8674(94)90334-4. [DOI] [PubMed] [Google Scholar]
- 59.Zerbib A C, Reske-Kunz A B, Lock P, Sekaly R P. CD4-mediated enhancement or inhibition of T-cell activation does not require the CD4/p56lck association. J Exp Med. 1994;179:1973–1983. doi: 10.1084/jem.179.6.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zorn N E, Weill C L, Russel D H. The HIV protein gp120 activates nuclear protein kinase C in nuclei from lymphocytes and brain. Biochem Biophys Res Commun. 1990;166:1133–1139. doi: 10.1016/0006-291x(90)90984-u. [DOI] [PubMed] [Google Scholar]