Abstract
Consumption of certain probiotic strains may be beneficial for reducing the risk of acute upper respiratory tract infections (URTIs), however, underlying immunological mechanisms are elusive. Bifidobacterium lactis Bl-04™ has been reported in humans to significantly reduce the risk of URTIs, affect the innate immunity in the nasal mucosa, and reduce nasal lavage virus titer after a rhinovirus (RV) challenge. To study the immunological mechanisms, we investigated the effect of Bl-04 on cytokine production and transcriptomes of human monocyte-derived macrophages (Mfs) and dendritic cells (DCs), and further on RV replication and cytokine production in MRC-5 fibroblasts. The results showed that Bl-04 modulates antiviral immune responses and potentiates cytokine production during viral challenge mimic in immune cells. However, effect of Bl-04 on RV replication and cytokine production in fibroblasts was negligible. Overall, the findings suggest that Bl-04 mildly stimulates antiviral immunity in Mfs and DCs, and potentially influences viral replication in fibroblasts that however warrants further investigations.
1. Introduction
Viruses are the major causative agents of acute upper respiratory tract infections (URTIs) or common colds that are usually short and transient but can also lead to more severe diseases including lower respiratory tract illnesses such as pneumonia, COVID-19, and bronchiolitis. Over 200 virus types can infect the respiratory tract, of which the most predominant are rhinoviruses (RV) (>50 % of cases), followed by coronaviruses, influenza, and other viruses [1,2]. Viral URTIs are one of the most common diseases in humans in all age groups and account for a significant part of visits to general practitioners and absence from day care, school, or work, causing significant costs for the society. Despite the incidence of common cold, no preventative treatments are currently available. However, the symptoms can be alleviated with nasal decongestants and anti-inflammatory drugs.
The effect of probiotic consumption on URTI episodes has been investigated in meta-analyses that indicate reduction in the incidence and severity of acute URTI in comparison to placebo [[3], [4], [5]]. However, the meta-analyses also show that in the clinical studies the efficacy ranges from neutral to favoring probiotic over placebo. These differences derive, at least partly, from strain specific effects of probiotics on immunity [6] and demonstrate the need to investigate these effects and acute URTI episodes for further development of specific probiotic solutions.
The effect of the probiotic Bifidobacterium animalis subspecies lactis Bl-04 (Bl-04) has been clinically studied on acute URTIs in adults and more specifically on a RV challenge model. The clinical studies showed significant reduction in the risk of acute URTI episodes by 27 % over 5-month winter season in healthy active adults supplemented with Bl-04 [7]. However, in follow up studies no significant differences were found in immunological analysis between Bl-04 and placebo groups [8,9]. In another clinical study the effect of Bl-04 was shown on preventing and mitigating URTIs as a yogurt supplement in haze areas [10]. In a RV challenge study, ingestion of Bl-04 for one month prior to infection resulted in higher CXCL8 levels and reduced virus titer in nasal lavage of Bl-04 group compared to placebo [11]. The result suggested an effect on the baseline state of innate immunity in the nasal mucosa influencing the subsequent response of the host to RV challenge. In a follow-up study, no effect on viral titer, blood transcriptome or CXCL8 was observed, however, IL-1β levels were higher prior to RV infection in the Bl-04 group [12].
Bl-04 has been scarcely studied in pre-clinical models, however, one study assessing the immunomodulatory in vitro and in vivo correlations of lactic acid bacteria showed that Bl-04 induced high IL-10/IL-12p70 ratio in human peripheral blood mononuclear cells (PBMC). This resulted in an efficient protection from colitis in a following in vivo mouse study by possibly slowing down early Th1 response [13]. However, specific studies on the effect of Bl-04 on antiviral immunity are lacking. In general, in vitro studies conducted predominantly with monocyte-derived cells and in vivo studies in mouse influenza challenge models suggest that stimulation of type 1 immunity and interferon stimulated genes by probiotics could be critical in improving antiviral defense [14]. However, pre-clinical and clinical data correlations remain inconclusive for specific probiotic strains.
The pathogenesis of respiratory viruses varies from strain-to-strain, as the viruses have different tropism and entry receptors, yet the host antiviral responses to the various pathogens tend to be similar and conserved across cell types. Macrophages (Mfs), dendritic cells (DCs), and fibroblasts, among other cell types, respond to viruses that can infect cells and replicate inside of them. Invading virus is recognized through pattern recognition receptors (PRRs) of the host cell, for example Toll-like receptors (TLR)-3 and TLR-7/8 that recognize viral nucleotides. PRRs recognize pathogen associated molecular patterns that are conserved in micro-organisms and not expressed in the host [15]. This interaction leads to activation of transcription factors such as Nuclear Factor-kappa B (NF-κB) and members of interferon regulatory factor (IRF) family [16] and a subsequent production of cytokines and chemokines. Mfs and DCs express a variety of different PRRs. These innate immune cells are key players in maintaining the homeostasis in immune responses and participating in host-viral interactions. Upon viral or bacterial infection, Mfs are responsible for the early pathogen recognition, initiation of inflammation and possible tissue repair. DCs act as messengers between innate and adaptive immune system and determine the type of immune responses after a viral challenge. DCs mediate the T-cell polarization into Th1, Th2, Th17 or regulatory T cells thus contributing to specific responses against a variety of different pathogens [17].
Airway fibroblasts located in the subepithelial layer are also important players in the innate immune responses, and may e.g. maintain the communication between epithelium and mesenchyme [18]. Respiratory viruses, such as RV, have been detected to infect human primary fibroblasts and contribute to inflammatory response by producing pro-inflammatory cytokines and chemokines, such as IL-6 and CXCL8 [19,20]. RV infection in fibroblasts may contribute to neutrophil chemoattraction leading to increased vascular permeability mediated by e.g. CXCL8 and ENA-78. In addition to these, vascular endothelial growth factor (VEGF) can induce vascular permeability with subsequent inflammatory reaction and enable the spread of the infection [21,22].
Understanding the effect of probiotics on the host innate immunity in response to viral infections is of interest for developing new probiotic-based solutions for the common cold and other viral URTIs. In this study, the responses of Mfs and DCs to Bl-04 and synthetic TLR-ligands, mimicking viral infection, were evaluated by immunological and transcriptomics methods to study immunomodulatory effects of Bl-04 on viral stimulation in vitro. For this purpose, a combination of polyinosinic:polycytidylic acid (pIC) and Resiquimod (R848), agonists for TLR3 and TLR7/8 receptors, respectively, were used. Priming the immune cells with these receptor ligands is a well-established model for RNA virus induced innate immune response and have been shown to induce similar responses in vitro [[23], [24], [25], [26]] and in vivo [27]. Furthermore, the effect of Bl-04 on RV replication and immune marker responses in human lung fibroblasts was evaluated in vitro. This study provides data on the immunomodulatory mechanisms of Bl-04 that help to unveil the mode of action in clinical studies.
2. Materials and methods
2.1. Bacteria and viruses
To prepare Bl-04™ bacteria for immune cell treatment, Bifidobacterium animalis subspecies lactis Bl-04 (Danisco Global Culture collection DGCC2908 Niebüll, Germany and American Type Culture Collections ATCC SD5219, Manassas, VA, USA) was cultured anaerobically in N2 atmosphere at 37 °C in Bifidobacterium medium 58 (DSMZ, Braunschweig, Germany). Bacteria were grown to logarithmic growth phase, collected by centrifugation, washed once with cell culture grade PBS (Life Technologies, Paisley, UK), and suspended in RPMI w/o supplements using optical density (OD) 600 to correspond to a 10 bacteria/cell treatment concentration. Previously the relationship between absolute bacterial counts and the OD600 value was determined by flow cytometry (BD FACSCalibur, Franklin Lakes, NJ, USA) using SYTO™24 Green Fluorescent Nucleic Acid Stain (Invitrogen, Paisley, UK). Briefly, bacteria were fixed in 4 % formaldehyde and stored at 4 °C, until appropriately diluted and stained with 0.1 μM SYTO™24 for 15 min in dark at room temperature. The total number of microbes was analyzed by flow cytometry, and the trendline between the number of bacteria and OD600 was calculated.
For MRC-5 fibroblast experiments, lyophilized powder of Bl-04, provided from IFF Health & Biosciences (Madison, WI, USA), was suspended in cell culture medium EMEM without antibiotics for 30 min to a final concentration of 2.5 × 106 CFU/ml. The bacterial suspension used in experiments was at a concentration corresponding to 25 bacteria/cell.
The virus used in this study was RV type 39 (RV39), originally purchased from ATCC (Manassas, VA, USA) and expanded by serial passage in MRC-5 cells in the laboratory. The virus pool was diluted to target concentrations of 104.5 (high titer, multiplicity of infection (MOI) 0.3), 102.5 (medium titer, MOI 0.003), or 101.5 (low titer, MOI 0.0003) TCID50/mL in cell culture medium for use in the experiments.
2.2. Monocyte purification and differentiation to macrophages and dendritic cells
Monocytes were purified from recently collected buffy coats, rich in leukocytes, that were obtained from healthy and anonymous volunteers (Finnish Red Cross Blood Service, Helsinki, Finland). The use of human blood was approved by the Ethics Committee of the Hospital District of Helsinki and Uusimaa (HUS), Finland (HUS-2563-2019-3). PBMCs were isolated by Ficoll-Paque (GE Healthcare, Chicago, IL, USA) density gradient centrifugation using SepMate tubes (Stemcell technologies, Grenoble, France) followed by purification of monocytes by MACS CD14+ magnetic beads according to manufacturer's instructions (Miltenyi Biotech GmbH, Bergisch Gladbach, Germany). Monocytes were further differentiated into Mfs on 24 well plates at a density of 3 × 105 cells/well (Falcon, Corning, NY, USA) and cultured 7 d in Macrophage-SFM (Gibco, Life Technologies, Grand Island, NY, USA) with recombinant human GM-CSF (Miltenyi Biotech) 200 ng/ml and 1 % Antibiotic-Antimycotic. To yield immature DCs, monocytes were plated on 12 well plates at a density of 5 × 105 cells/well and cultured for 7 d in RPMI-1640 (Sigma-Aldrich, Saint Louis, MI, USA). Media was supplemented with 1 % Antibiotic-Antimycotic, 10 % foetal bovine serum (FBS) (both from Life Technologies, Grand Island, NY, USA), IL-4 80 ng/ml, and GM-CSF 200 ng/ml (both from Miltenyi Biotech). Fresh medium was changed (Mf) or added (DC) every second day. Typical Mf and DC morphology was verified by microscopy. Cell differentiation was confirmed by flow cytometry (Fig. S1), and they expressed characteristic monocyte-derived Mf and DC markers.
2.3. Immune cell stimulation experiments
Cells from six blood donors without technical replicates (except for TLR agonist mix treatments) were stimulated with probiotic bacteria alone (10 bacteria/cell) or in combination with TLR agonist mix, synthetic dsRNA pIC 15 μg/ml + synthetic ssRNA analog R848 5 μM (both from Sigma-Aldrich, St. Louis, MO, USA). Mfs or DCs without stimulation were used as a control, whereas dimethyl sulfoxide (DMSO) (Sigma-Aldrich), a carrier for the TLR-ligands, was used as a control for pIC + R848 containing treatments. Cells were stimulated for 24 h and 48 h, for Mfs and DCs respectively, afterwards cell culture supernatants for ELISA analyses and RNA for transcriptomics were collected and stored at −80 °C until analyzed.
2.4. Fibroblast stimulation experiments
The effect of the probiotic Bl-04 on virus replication and cytokine production in vitro was assessed in human embryonic lung fibroblast cells (MRC-5, Quidel, Athens, OH, USA). MRC-5 cells were grown to confluence in 24-well plates (EMEM with 2 % FCS). The growth medium was removed from the wells and the cells were washed three times with growth medium without antibiotics. One ml of medium containing 2.5 × 106 CFU of Bl-04; a bacterium to cell ratio of 25:1, was added to 12 of the wells and medium without bacteria was added to the other 12 wells as a control. The plates were then incubated at 37 °C in 5 % CO2 for 24 h. After incubation with Bl-04, the media from both the control wells and the Bl-04 exposed wells were removed and replaced with 1 ml of medium containing the desired concentration of RV39. For adsorption step, the virus was incubated with the cells for 2 h at 33 °C in 5 % CO2. All wells were then washed three times with cell culture media and the last wash was collected from three control wells and three Bl-04 exposed wells for a baseline viral titer analysis. The plate was then incubated at 33 °C in 5 % CO2 and the supernatant medium was collected from triplicate control wells and triplicate Bl-04 exposed wells at 24, 48 and 72 h after virus exposure. All media collected for virus titer and cytokine analysis was stored at −80 °C until assayed. Three separate experiments were done with the low titer virus challenge, three with the medium titer virus challenge and one with the high titer challenge. For cytokine analyses the supernatants for one medium titer experiment were lost due to technical reasons.
2.5. Viral titers from fibroblast cultures
The initial viral dose used in the experiments was defined by TCID50 titration and the same stock was used for all experiments. The target RV dose for low titer experiments was 101.5 TCID50/ml, for medium titer 102.5 TCID50/ml and for high titer 104.5 TCID50/ml ( ± 0.5 log). Also the quantification of the viral titer from the supernatants was done using this method. Serial 10-fold dilutions (10°–104) of each specimen for virus quantification were inoculated in quadruplicate onto MRC-5 monolayers in 96-well plates. The plates were incubated at 33 °C in 5 % CO2 for 7 days and then read by microscopy for typical RV cytopathic effect. Viral titers were calculated and expressed as TCID50/ml.
2.6. ELISA analyses
Immune cell supernatants were analyzed for IFN-γ, IL-1β, IL-6, IL-10, IL-12p70 and TNF-α cytokines by Aushon Human 6-Plex Ciraplex Array and IL-23 and TGF-β by 1-plex Assays (Aushon BioSystems, Inc., Billerica, MA, USA) according to manufacturer's instructions. Results were analyzed with CiraSoft Software (Aushon Biosystems).
Cell culture supernatants from fibroblast experiments were analyzed for IL-1β, IL-6, CXCL8 and TNF-α with CorPlex 4 plex array and for CXCL10 and VEGF with 1 plex arrays (Quanterix, Billerica, MA, USA) according to manufacturer's instructions. The plates were detected with Quanterix SP-X and results were analyzed with Quanterix SP-X Analysis Software.
2.7. Immune cell RNA extraction and transcriptomics
Immune cells were lysed in lysis/binding solution (Invitrogen, Thermo Fisher Scientific, Watlham, MA, USA) upon collection and stored at −80 °C. Total RNA was extracted from the immune cells using automated KingFisher Flex purification system with deep well magnet head (Thermo Fisher Scientific) and MagMAX™-96 Total RNA Isolation Kit (AM1830, Thermo Fisher Scientific) according to manufacturer's instructions. Briefly, lysates were thawed on ice and suspended by pipetting. A total volume of 140 μl of the cell lysate, 30 μl of lysis/binding buffer, and 20 μl of bead mix were pipetted into a sterile Microtiter Deep well 96 plate and the protocol AM1830 was used for the purification process. RNA was eluted in 40 μl of elution buffer and stored at −80 °C. RNA quantity and quality were determined on Nanodrop 1000 Spectrophotometer (Thermo Fisher, Scientific) and/or 4200 TapeStation System (Agilent, Santa Clara, CA, USA).
RNA-seq was conducted with targeted sequencing technology using Templated Oligo detection chemistry (TempO-Seq) from BioSpyder Technologies (BioSpyder, Carlsbad, CA, USA). Human Whole Transcriptome Assay containing detector oligonucleotides designed to target all human genes (∼23,000) was used. Library preparation was conducted according to BioSpyder kit user manual. In total 48 samples (24 for each cell type) were pooled, purified, quantitated, and stored frozen prior to sequencing. The library pool was sent to Roy J. Carver Biotechnology Center at the University of Illinois at Urbana-Champaign for sequencing using the Illumina NovaSeq platform (1 × 50 nt). The targeted read-depth was 5 million reads per sample with the minimum of 1 million reads per sample. The reads were first quality filtered, trimmed and demultiplexed and further aligned to the BioSpyder Human Whole Transcriptome v2.0 annotated reference using Salmon (v1.1.0) [28].
DESeq2 (v1.26.1) [29] was used for differential expression analysis in R (v 3.6.2) where Mfs and DCs were analyzed separately due to distinct expression profiles of the cell types. Genes were considered differentially expressed if the adjusted p-value was <0.05. Principal component analysis (PCA) was performed to determine overall relatedness of the samples under the various experimental conditions and visualized using ggplot2 (v 3.3.3) [30]. The correlation of two genes in a particular cell type/treatment combination were calculated with the normalized counts output from the DESeq2 method (normalized for library size/total reads per sample) using the package DGCA (v 1.0.3) [31]. The normalized counts were then used to calculate the Pearson correlation, and the resulting correlation coefficients are transformed into Z-scores and tested against a normal distribution to generate a p-value. Resulting p-values are corrected for multiple testing using the Benjamini-Hochberg method. The correlation values and the adjusted p-values were visualized in bubble plots, with the X and Y being the gene names, and the X and Y genes being ordered by function.
Pathway analysis was performed using ROntoTools (v 2.14) [32] with a KEGG pathway [[33], [34], [35]] considered significantly changed with a combined false discovery rate of <0.05.
2.8. Statistics
The data from immune cell (Mf and DC) ELISA analyses (Fig. 1) were log2-transformed prior to statistical analysis. The data were analyzed using a linear model that included the main effects of treatment and donor (2-way ANOVA). Separate models were fitted for each cytokine. Pairwise-comparisons between the treatments were performed using contrasts of estimated marginal means. The fit of the models was checked by inspecting the normality of the model residuals. All p-values from all fitted models were collected together and corrected for family-wise error rate using the Holm-Bonferroni method. One extremely low IL-6 measurement was removed from pIC + R848 high group in the analysis to meet model assumptions.
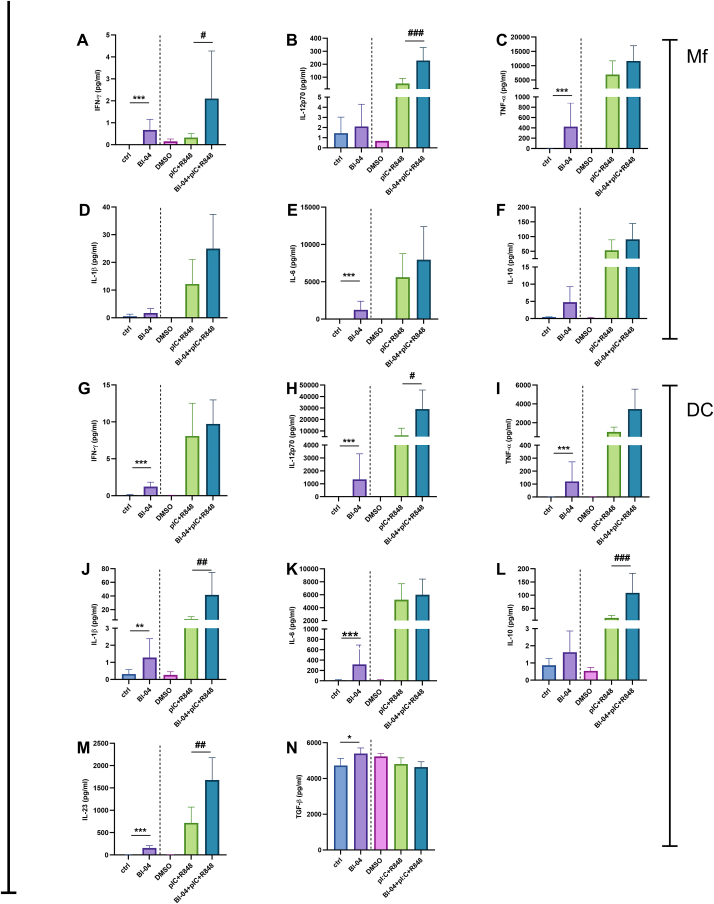
Fig. 1.
The effect of Bifidobacterium animalis subsp. lactis Bl-04 and pIC + R848 on human macrophage and dendritic cell cytokine responses. Cells were stimulated or left unstimulated with Bl-04 (10 bacteria/cell) and/or pIC (15 μg/ml) + R848 (5 μM) for 24 h (Mf) or 48 h (DC). Cytokine concentrations in the supernatants were measured by ELISA (Mf: A-F; DC: G-N) and the mean of six donors (pg/ml) with standard deviation is shown. DMSO, dimethyl sulfoxide was used as a vehicle control for pIC + R848. The data were analyzed with a linear model using change from baseline as response (Ctrl vs Bl-04; or pIC + R848 vs Bl-04 + pIC + R848). Different comparisons are separated with a dashed line. N = 6, for pIC + R848 technical replicates were used. *: 0.01 ≤ p﹤0.05, **: 0.001 ≤ p﹤0.01, ***: p < 0.001 vs ctrl and: # 0.01 ≤ p﹤0.05, ##: 0.001 ≤ p﹤0.01, ##: p < 0.001 vs pIC + R848.
Both RV replication and fibroblast cytokine data were analyzed separately at each timepoint. At each timepoint, two-factor fixed effects model with terms for experiment/concentration group, treatment, and their interaction was fitted to data. Cytokine data were log transformed prior to analysis to obtain proper model fit to data. IL-1β was measured in only one experiment and hence analyzed using a simple model with a treatment term. The treatments were compared using model contrasts, and the obtained p-values were adjusted using mvt (multivariate t distribution based) adjustment. All statistical analyses were performed using statistical software R, version 4.2.2 [36] and the model contrasts were computed using emmeans package [37] therein.
The Mf and DC ELISA data statistical analyses were done by Vincit Ltd using R version 4.1.3 (2022-03-10) and graphs were generated with GraphPad Prism. The rest of the statistical analyses were done at Inoi Ltd.
3. Results
3.1. Bl-04 enhances cytokine production from pIC + R848 stimulated Mfs and DCs
To study how probiotic Bl-04 consumption could influence human immunity and viral infections, human PBMC-derived Mfs and DCs from six blood donors were stimulated with probiotic Bl-04 alone and in combination with a viral mimic pIC + R848. Mf and DC cytokine levels were measured by ELISA from cell culture supernatants after 24 h (Mf) or 48 h (DC) stimulation (Fig. 1). Direct Bl-04 stimulation of Mfs induced higher pro-inflammatory cytokine IFN-γ, TNF-α and IL-6 (Fig. 1A–C, E) concentrations compared to control. Anti-inflammatory IL-10 (Fig. 1F) concentration change did not reach statistical significance. To study if Bl-04 could modulate viral infection-like responses, cells were treated simultaneously with Bl-04 and pIC + R848 and the secreted cytokine levels were compared to those induced by pIC + R848 alone. pIC + R848 alone strongly induced pro-inflammatory cytokines IL-12p70, TNF-α, IL-1β and IL-6 (Fig. 1B–E) in addition to the anti-inflammatory IL-10 (Fig. 1F). Modulation of pIC + R848 -induced responses by Bl-04 resulted in an enhanced production of IFN-γ and IL-12p70 in Mfs, indicating that Bl-04 could potentiate type 1 immune response during TLR3 and TLR7/8 stimulation. In DCs Bl-04 stimulation significantly induced IFN-γ, IL-12p70, TNF-α, IL-1β, IL-6, IL-23 and TGF-β (Fig. 1G–K, M, N), while IL-10 (Fig. 1L) was not significantly induced compared to control, indicating polarization of DCs towards inducing T helper cell types 1 and 17. In response to pIC + R848 in combination with Bl-04, IL-12p70, IL-1β and IL-23, as well as immunoregulatory IL-10 levels were significantly higher compared to pIC + R848 responses alone, suggesting that DC responses to TLR3 and TLR7/8 stimulation can be further enhanced by Bl-04.
3.2. Bl-04 and pIC + R848 induce broadly overlapping transcriptomes in Mfs and DCs
To evaluate the effect of Bl-04 on immune cell activation in detail and contrast that to pIC + R848 response, transcriptomic analyses were conducted. RNA was extracted for sequencing from the same experiments as described above for cytokine analysis. PCA was performed to determine overall similarity of the Mf and DC transcriptomic response to the treatments. The PCA showed clear separation of Bl-04 and pIC + R848 treatments from the controls (ctrl and DMSO) in Mfs (Fig. 2A) and DCs (Fig. 2B), however, Bl-04 treated cells were clustering closer to the controls than pIC + R848 treated in both cell types (Fig. 2). The separation of treatments from controls was clearer for DCs (PC1 49 %) than for Mfs (PC1 30 %), suggesting a larger treatment effect on the transcriptomes of DCs.
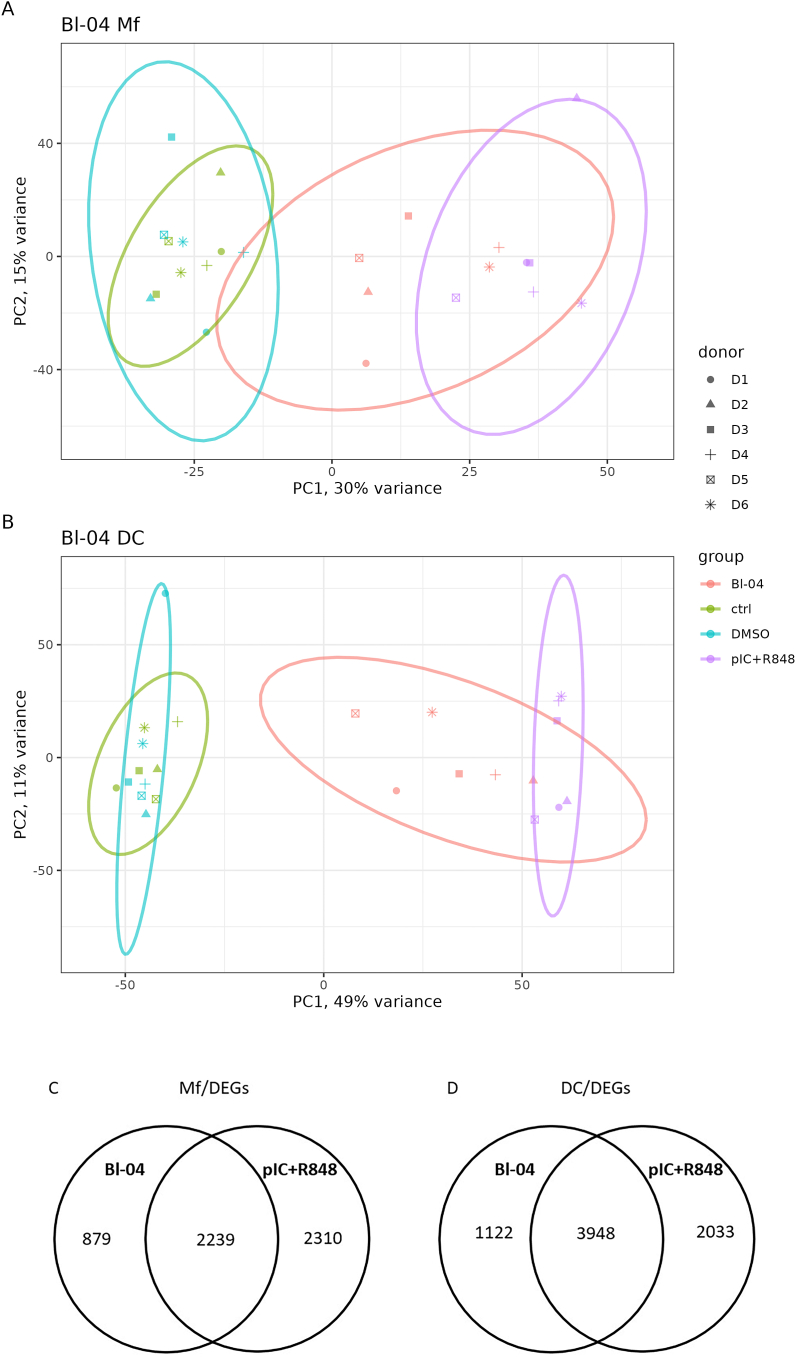
Fig. 2.
Principal component analysis of transcriptomes and differentially expressed genes of all samples tested for Mfs and DCs. PCA of all samples (A) Mfs and (B) DCs was performed. Color of the individual points and collective ellipses denotes treatment, shape of the point denotes the human donor of the cells. DEG analysis was done for each treatment. Bl-04 was compared to unstimulated controls and pIC + R848 to DMSO and the number of DEGs for Mfs (C) and DC (D) are shown as Venn diagrams. PC, Principal Component. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
In differential gene expression (DEG) analysis Bl-04 was compared to unstimulated controls and pIC + R848 to DMSO. Bl-04 and pIC + R848 treatment of Mfs resulted in 3118 and 4549 DEGs, respectively (Table S1), of which 879 were unique to Bl-04 and 2310 for pIC + R848 treatments. The number of shared DEGs between Bl-04 and pIC + R848 groups was 2239 (Fig. 2C). Treatment of DCs with Bl-04 and pIC + R848 resulted in 5070 and 5981 DEGs (Table S2), respectively, of which 1122 were unique to Bl-04 and 2033 for pIC + R848 treatments. The number of shared DEGs between Bl-04 and pIC + R848 groups was 3948 (Fig. 2D).
The overlap in the number of DEGs and PCA show broad similarity in gene activation between the Bl-04 and pIC + R848 treatments.
3.3. Bl-04 and pIC + R848 activate immune-related pathways in Mfs and DCs
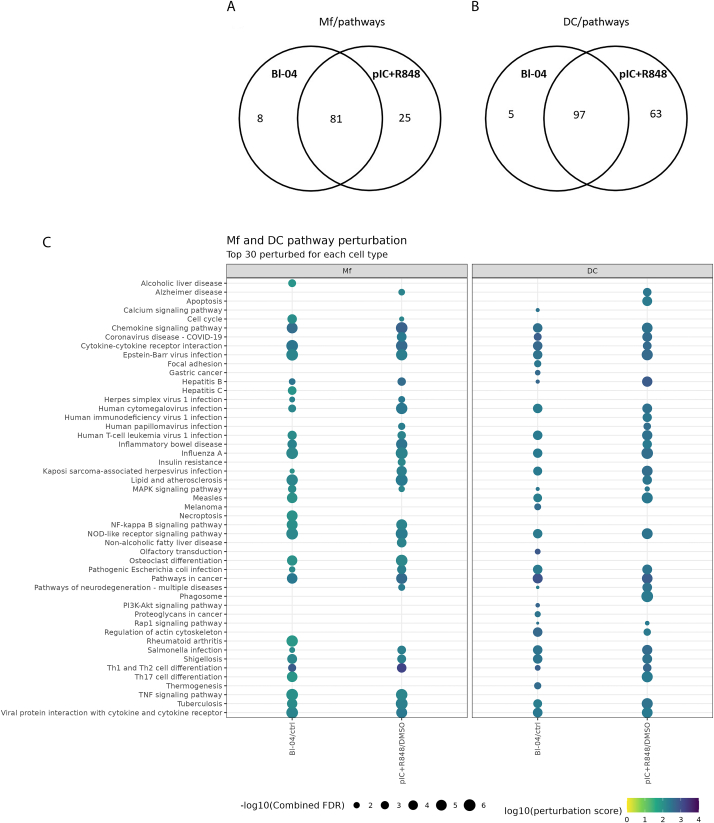
To analyze the effect of the DEGs on pathway level, we conducted pathway analysis mapping the DEGs onto KEGG pathways and calculating the probability of the pathway perturbation. A list of all perturbed pathways is provided as supplemental material (Table S3). In Mfs, there were more mutual pathways (n = 81; FDR <0.05) perturbed by both Bl-04 and pIC + R848 than by either of them alone (Bl-04 n = 8 and pIC + R848 n = 25; FDR <0.05) (Fig. 3A), suggesting that Bl-04 can activate similar pathways as the viral mimic in Mfs. Also, in DCs most (n = 97; FDR <0.05) of the perturbed pathways were mutually affected by both treatments (Fig. 3B). Of the perturbed pathways five (FDR <0.05) were unique to Bl-04 and 63 (FDR <0.05) to pIC + R848 treatment. The 30 pathways that were most affected for each cell type and treatment are presented in Fig. 3C. In both immune cell types, multiple pathways related to immune system activation were perturbed (Fig. 3C). In Mfs both Bl-04 and pIC + R848 treatments affected pathways related to chemokine and cytokine signaling, Th1 and Th2 cell as well as NF-κB activation. Additionally, NOD-like receptor signaling pathway having key roles in innate and adaptive immune responses was perturbed. Of note, in relation to antiviral gene regulation, was the perturbation of viral protein interaction with cytokine and cytokine receptor and Influenza A pathways by Bl-04. Other pathways affected among the top 30 perturbed pathways in Mfs were mostly related to different disease conditions most likely due to the activation of common inflammatory pathways during different diseases. Especially pathways related to virus infections were perturbed, such as hepatitis, measles, herpes, Epstein-Barr virus and cytomegalovirus.
Fig. 3.
Pathway perturbation in immune cells. Differentially expressed gene results were used as input to ROntoTools to determine pathways that were affected by the treatments. The number of perturbed pathways for each treatment are shown for Mfs (A) and DCs (B). The top 30 most perturbed pathways are presented, for Mfs and DCs (C). The perturbation score is shown as the color (dark blue = higher values), and the size of the balloon denotes the –log10 of the combined false discovery rate (FDR). Only pathways that have FDR <0.05 (-log10 > 5) are shown. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
In DCs, Bl-04 treatment perturbed similar pathways than in Mfs but the overall effect was detected with lower FDR rate reflecting a lower statistical significance of the perturbation. Chemokine signaling, cytokine-cytokine receptor interaction and viral protein interaction with cytokine and cytokine receptor pathways were affected with both treatments as well as Th1 and Th2 cell differentiation and NOD-like receptor signaling pathways. In line with the antiviral DEG results, Bl-04 and pIC + R848 had also significant effects on respiratory tract infection virus influenza A and COVID-19 pathways, but also more broadly on viral disease related pathways, like Epstein-Barr virus, measles and cytomegalovirus. In DCs, the top 30 pathways that were unique to Bl-04 were related to intracellular signaling and cancer pathways, but not directly linked to immune system activation.
3.4. Bl-04 stimulates anti-viral defense genes in macrophages
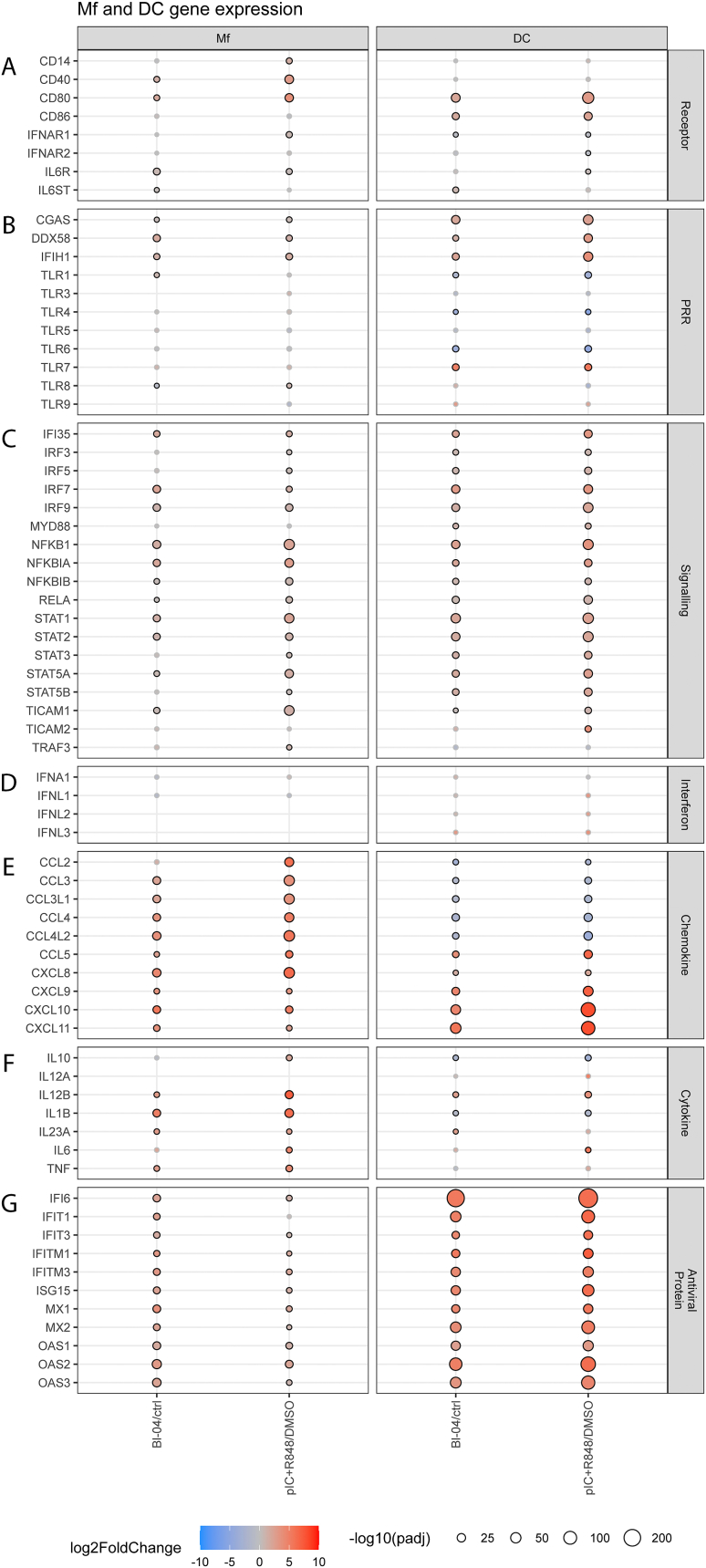
To further evaluate the effect of Bl-04 on viral defense of immune cells, genes related to host antiviral responses were more closely examined after treatment with Bl-04 and pIC + R848 (Fig. 4). Both treatments upregulated gene expression of costimulatory molecules CD40 and CD80 (Fig. 4A), indicating macrophage activation. Additionally, IL-6R gene was induced by both treatments and IL6ST by Bl-04. Interestingly, PRR genes (Fig. 4B) DDX58 (RIG-I) and IFIH1 (MDA5), recognizing viral nucleic acids, were upregulated by both treatments. Also, CGAS involved in several processes, including cellular response to exogenous dsRNA, was upregulated by both treatments. No significant differential expression of TLR3 or TLR7 after probiotic or agonist treatments was seen in Mfs at the 24 h time point. TLR8 that recognizes ssRNA was downregulated by Bl-04 and upregulated by pIC + R848. Overall, there were only minor effects on the differential expression of TLR genes in Mfs at the time point examined.
Fig. 4.
Differential expression of genes involved in antiviral responses in Mfs and DCs after Bifidobacterium animalis subsp. lactis Bl-04 or pIC + R848 treatment. Cells were stimulated with Bl-04 (10 bacteria/cell) or pIC (15 μg/ml) + R848 (5 μM) for 24 h (Mf) or 48 h (DC). RNA was collected and transcriptomic analyses were performed. Selected genes of interest were grouped to (A) Receptor, (B) PRR, (C) Signaling, (D) Interferon, (E) Chemokine, (F) Cytokine and (G) Antiviral protein categories. Color of the balloon denotes expression level (log2 fold change) with blue having reduced expression and red having increased expression compared with the control. Size of the balloon denotes significance (-log10 adjusted p-value, black outline at adjusted p-value <0.05) with the larger size having more significance. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
The upregulation of receptor genes was also reflected in signaling gene expression (Fig. 4C) as genes involved in multiple pro-inflammatory signaling pathways were upregulated. Although significant differential expression of MYD88, the major adaptor protein linking TLRs except TLR3 to downstream signaling molecules, was not seen in Mfs at the 24 h time point, NFKB1 activating downstream of MyD88 was upregulated. NFKB1 encodes p105, a DNA binding subunit of the NF-κB protein complex. Also, other genes related to NF-κB pathway, RELA, NF-κB inhibitor A (NFKB1A) and NFKB1B were upregulated by Bl-04 and pIC + R848. The NF-κB pathway is associated with the activation and controlling of pro-inflammatory and IFN responses. NFKB1A encoded protein interacts with REL dimers to inhibit NF-κB/REL complexes involved in inflammatory responses. Furthermore, genes related to IFN signaling including IFI35, IRF7, IRF9, STAT1 and STAT2 were upregulated by pIC + R848 and interestingly also by Bl-04. TLR adaptor molecule 1 gene (TICAM1), used by TLR3/4/5 to mediate NF-κB and IRF activation, was upregulated by both pIC + R848 and Bl-04. Additionally, upregulation of pro-inflammatory cytokine genes (Fig. 4 F) including IL12B, IL-1B, IL23A and TNFA suggest the activation of MyD88 and MyD88-dependent pathways by both treatments, although no statistically significant change in MyD88 gene expression was seen in Mfs.
Upregulation of IRF3 and IRF7 transcription factor genes encoding proteins that bind to type I and type III IFN promoters indicate activation of IFN genes by pIC + R848. The upregulation of STAT1 and STAT2 which are both components of the ISGF3 complex, suggests activation of type I and type III interferon pathways by both treatments. This leads to the expression of IFN-stimulated genes (ISGs) such as anti-viral protein genes. Bl-04 induced similar expression of anti-viral protein genes as pIC + R848 (Fig. 4G), including genes from IFIT, ISG, MX and OAS families. IFN genes (Fig. 4D) were not differentially expressed which may relate to the late time point of sample collection (24 h), however, subsequent activation of interferon-induced genes (Fig. 4G) can still be detected.
Chemokine genes were broadly activated in Mfs in response to Bl-04 and pIC + R848 (Fig. 4E). Chemokines belonging to CCL and CXCL classes, e.g., CCL3, CCL4, CCL5, CXCL8, CXCL9, CXCL10 and CXCL11 were most broadly activated. These molecules activate mainly macrophages, NK cells and neutrophils. CXCL8 and CXCL10 genes were strongly induced and are known to be involved in antiviral responses. Specific gene induced only in pIC + R848 treated cells was CCL2.
Compared to control, Bl-04 treatment upregulated the expression of pro-inflammatory cytokine genes (Fig. 4F) including IL1B, IL12B, IL23A and TNFA. The same cytokine genes were upregulated by pIC + R848 with higher fold changes compared to Bl-04 treatment. In addition, IL6 and IL10 were upregulated by pIC + R848 and not significantly affected by Bl-04. Stronger gene expression of cytokines by pIC + R848 is in line with secreted cytokine levels (Fig. 1).
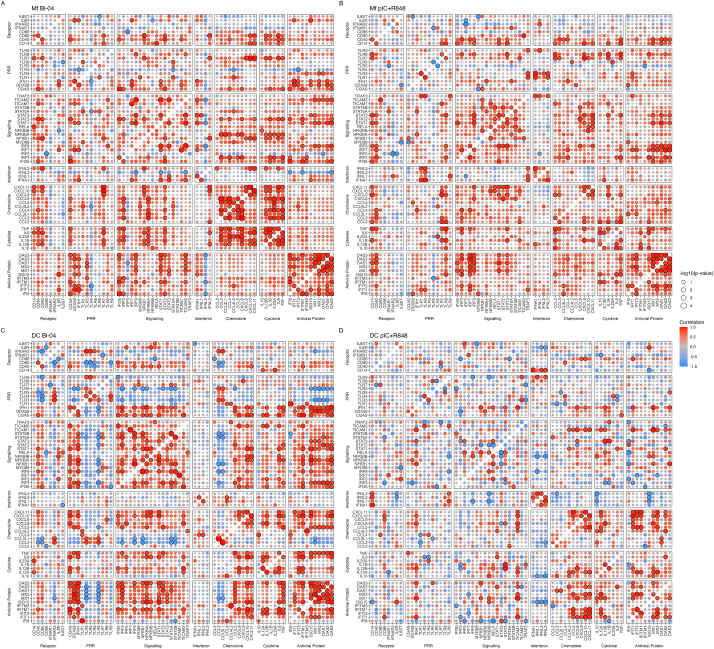
To examine the co-expression of DEGs in Mfs treated either with Bl-04 or pIC + R848, Pearson correlation was calculated and the results were functionally categorized (Fig. 5). IFN-inducible genes such as the OAS, ISG and MX gene family members showed positive correlation with the other antiviral protein genes in both Bl-04 and pIC + R848 treated cells (Fig. 5A and B). Also, the expression of STAT1, STAT2 and IRF9 that form a heterotrimeric complex promoting expression of ISGs, correlated positively with expression of IFN regulated genes in both treatments. Genes related to NF-κB pathway (NFKB1 and NFKB1A) showed a positive correlation with cytokines and chemokines when Mfs were treated with Bl-04 (Fig. 5A), however, this was not detected with pIC + R848 treatment (Fig. 5B). In addition, the costimulatory molecules CD40 and CD80 correlated positively with the pro-inflammatory cytokine DEGs with Bl-04 (Fig. 5A) but not pIC + R848 treatments (Fig. 5B), indicating a different type of activation of Mfs.
Fig. 5.
Correlation analysis on the differentially expressed genes. Mfs treated with Bl-04 (A), or pIC + R848 (B), DCs treated with Bl-04 (C), or pIC + R848 (D). The correlation values and the adjusted p-values are visualized in the bubble plots, with the X and Y axes indicating the gene names that are grouped to Receptor, pattern-recognition receptor (PRR), Signaling, Interferon, Chemokine, Cytokine and Antiviral protein categories. The blue and red colors of the balloon indicate a negative and a positive Pearson correlation, respectively. Values with an adjusted p-value <0.05 were circled with a black ring to show significance, and the size of the balloon denotes the -log10 (adjusted p-value). (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
3.5. Bl-04 and pIC + R848 induce similar antiviral gene expression in DCs
The effect of Bl-04 and pIC + R848 on genes related to host antiviral responses in DCs were more closely examined (Fig. 4). Both treatments upregulated the expression of costimulatory molecule genes including CD80 and CD86 (Fig. 4A), indicating DC maturation. Of the PRR genes that are involved in the recognition of viral nucleic acids, CGAS, DDX58, IFIH1 and TLR7 were significantly upregulated (Fig. 4B). On the other hand, transcription of some of the TLR genes, TLR1, TLR4 and TLR6, targeting bacterial PAMPs were downregulated. No significant change in TLR3 or TLR8 expression was seen with either of the treatments. DC samples were collected at 48 h after treatment, and this may reflect in the results.
Downstream of PRRs, genes involved in multiple pro-inflammatory signaling pathways were upregulated. Genes related to NF-κB pathway, NFKB1, RELA, NFKB1A and NFKB1B were upregulated with PRR adaptor MYD88 by Bl-04 and pIC + R848. Interestingly, Bl-04 upregulated genes related to IFN signaling similar to pIC + R848, including IFI35, IRF3, IRF5, IRF7, IRF9, STAT1 and STAT2. TICAM1, a gene encoding TLR adaptor molecule involved in NF-κB and IRF activation was upregulated by both pIC + R848 and Bl-04. TICAM2 that facilitates downstream signaling leading to type I IFN induction was upregulated only by pIC + R848.
Type I and III interferon genes were not significantly upregulated by Bl-04 or pIC + R848 in DCs. However, like in Mfs the activation of IFN genes may have occurred at earlier time point as multiple genes involved in IFN signaling including IRF3, IRF5, IRF7, IRF9 and STAT genes were upregulated, as well as, a wide range of antiviral protein genes including genes from IFI, IFIT, ISG, MX and OAS families.
Chemokine genes belonging to CXCL class e.g. CXCL8, CXCL9, CXCL10 and CXCL11 were most broadly activated in response to Bl-04 and pIC + R848. CCL2, CCL3 and CCL4 chemokines were downregulated while CCL5 was upregulated by both Bl-04 and pIC + R848. Compared to control, Bl-04 treatment significantly upregulated the expression of pro-inflammatory cytokine genes IL12B and IL23A in DCs. pIC + R848 significantly upregulated IL12B and IL6 gene expression. In both Bl-04 and pIC + R848 groups IL1B and IL10 were downregulated in DCs.
Correlation matrices of DEGs were calculated for DC data (Fig. 5). Positive correlation between the IFN-inducible genes in the antiviral protein category was strong in Bl-04 (Fig. 5C) but not in pIC + R848 (Fig. 5D) treated cells. Also, PRR genes CGAS, DDX58, IFIH1 correlated significantly with the antiviral protein genes in Bl-04 treated DCs (Fig. 5C) whereas only IFIH1 correlated significantly with IFIT1, OAS3, and MX1 in pIC + R848 treated cells (Fig. 5D). Negative correlation between TLR3 and TLR6 and the antiviral protein genes (MX, OAS, ISG gene families) was evident with Bl-04 (Fig. 5C) but not with pIC + R848 (Fig. 5D) treatment. pIC + R848 treated cells had a positive correlation among the interferon genes (IFNL1-3 and IFNA1) as well as CD14 receptor gene (Fig. 5D). Several genes involved in signaling (STAT1, STAT2, STAT3, NFKB1A and IRF3) correlated positively with antiviral protein genes in DCs treated with Bl-04 (Fig. 5C). There was also a positive correlation between Th1 type CXCL class chemokines (CXCL8, CXCL9, CXCL10, CXCL10) in both Bl-04 and pIC + R848 treated cells (Fig. 5C and D). Overall, Bl-04 treated DCs had more significant and positively correlated genes compared to pIC + R848 treated cells.
3.6. Effect of Bl-04 on RV replication in human fibroblasts
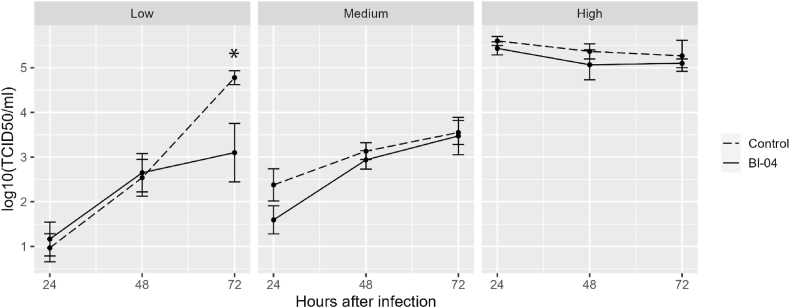
To evaluate the effect of Bl-04 on modulating antiviral responses and viral replication, we used RV-infected fibroblast culture and measured virus replication (Fig. 6). Confluent MRC-5 fibroblasts in 24-well plates were pre-treated (Bl-04) or left untreated (control) with Bl-04 for 24 h before RV challenge. Three independent experiments were performed, low virus titer with target RV dose of 101.5 TCID50/ml, medium titer with a target of 102.5 TCID50/ml and high titer with a target of 104.5 TCID50/ml. The results were analyzed separately for the low, medium, and high challenge experiments (Fig. 6). In the low titer challenge, virus replication was significantly reduced at 72 h in the Bl-04-treated wells, but otherwise significant effects were not found. The effect of Bl-04 on virus replication in fibroblasts cannot be concluded based on these findings.
Fig. 6.
The effect of Bifidobacterium animalis subsp. lactis Bl-04 on RV replication in fibroblasts. Mean values (±SEM) of viral replication from low and medium titer (three replicate experiments, target titer 101.5 TCID50/ml and 102.5 TCID50/ml) respectively or high titer (target 104.5 TCID50/ml) experiments are shown. TCID50, Median Tissue Culture Infectious Dose. *p = 0.01.
3.7. Effect of Bl-04 on RV induced cytokine responses in fibroblasts
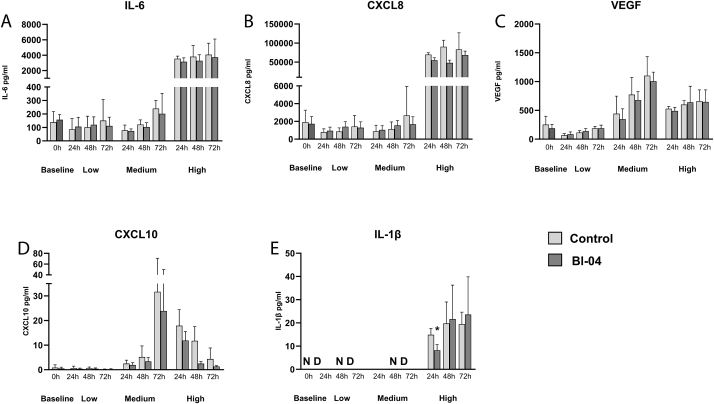
The effect of Bl-04 to modulate the cytokine response after RV infection was assessed in the same experiments as above. Fibroblasts were first treated with Bl-04 or with plain cell culture media (control) for 24 h after which the baseline (0 h) samples were collected. Wells were then washed and infected with RV. Pro-inflammatory cytokines and chemokines (IL-6, CXCL8, VEGF, CXCL10 and IL-1β, Fig. 7A–E) were measured from three replicate cell culture wells in each experiment.
Fig. 7.
Cytokine and chemokine production of MRC-5 fibroblast before and after RV infection. (A) IL-6, (B) CXCL8, (C) VEGF, (D) CXCL10, (E) IL-1β. MRC-5 fibroblast cells were treated with Bifidobacterium animalis subsp. lactis Bl-04 (25 bacteria/cell) for 24 h and baseline samples (0 h) were collected. Remaining wells were infected with low (3 experiments), medium (2 experiments) or high titer (1 experiment) of RV for 2 h. Bl-04 treated and control samples were collected after 24, 48 and 72 h of incubation and cytokines were analyzed by ELISA. Error bars represent the SD of three replicate wells. Statistically significant result of Bl-04 versus control is marked with an asterisk (IL-1β high titer 24 h p = 0.037). ND = not detected.
The baseline cytokine production of Bl-04 and control was similar in fibroblasts (0 h, Baseline; Fig. 7). It is noteworthy that the non-RV infected samples at 24 h had similar immune marker concentrations to low and medium titer samples even with 48- and 72 h time points, suggesting low immune activation with lower RV titers. IL-6 (Fig. 7A) and CXCL8 (Fig. 7B) production were highly induced with high titer infection reflecting a strong pro-inflammatory response already at 24 h. The response remained constant at 48 h and 72 h time points; however, the results were not statistically significant between Bl-04 and control.
VEGF cytokine expression was increased after the medium titer RV infection compared to the non-RV treated controls, however, Bl-04 treatment did not affect the VEGF cytokine production statistically significantly compared to control (Fig. 7C). CXCL10 was not produced with low titer infection and there was no basal production of the cytokine (Fig. 7D). With high titer the cytokine production also decreased over the time course. IL-1β was only produced with high titer infection and Bl-04 treatment decreased the cytokine production statistically significantly at 24 h compared to the control (Fig. 7E). TNF-α expression was below the limit of detection in all the experiments (data not shown).
4. Discussion
Meta-analyses suggest small but beneficial effects of probiotics on reducing the risk of acute URTIs [3,38]. However, the efficacy is considered strain specific [6], and thus requires clinical studies on specific strains, but also mechanistic studies to better understand the impact of the strain on antiviral immunity. In this study, the effect of a clinically studied probiotic strain Bifidobacterium animalis ssp. lactis Bl-04™, on modulation of immune responses in vitro was evaluated. We found that in human monocyte-derived Mf and DC models Bl-04 stimulated pro-inflammatory, Th1 and Th17 type cytokine production and activated multiple intracellular signaling pathways related to antiviral responses, suggesting that the strain could have potential to modulate the immune system function before viral infections. However, in lung fibroblasts, Bl-04 was able to restrict RV replication only at low viral infective dose at 72 h timepoint, warranting further studies on the effect of the probiotic on viral replication.
4.1. Bl-04 drives antiviral responses in Mfs and DCs similar to pIC + R848
In this study we showed that stimulation of Mfs and DCs by Bl-04 induces production of cytokines driving inflammatory response and macrophage activation, as well as polarization of DCs towards Th1 and Th17 type immunity (Fig. 1). Co-stimulation of Mfs and DCs with pIC + R848 and Bl-04 resulted in increased production of cytokines compared to pIC + R848 alone (Fig. 1), suggesting additive effects on the immune response in vitro. Previous studies have shown that the probiotic effects with different stimuli may be additive or inhibitory [39,40], highlighting the strain specificity of the immunomodulation, as well as the relevance of the comparator when studying the probiotic effects on immune system.
To further understand the effect of Bl-04 on immune and antiviral response in Mfs and DCs, we compared selected DEGs induced by Bl-04 and pIC + R848. The pIC + R848 treatment that we used as a positive control to induce antiviral like response induced expected DEGs in both cell types (Fig. 4). Many PRR genes including CGAS, DDX58 and IFIH1 involved in viral recognition were significantly upregulated by both treatments in Mfs and DCs. However, the effect of the treatments on TLR gene expression in general was modest and the expression of virus recognizing TLR3, TLR7 and TLR8 varied between cell types. More significant changes were seen in DCs which may reflect different functions of the cell types in innate immune response as well as the timepoints examined. Some of the TLR genes including TLR1, TLR4, TLR6, and TLR8 are shown to be constitutively expressed [41], which may reflect the modest changes in gene expression after Bl-04 or pIC + R848 treatments. Additionally, differential expression of MYD88 that links most TLRs to downstream signaling molecules was not detected in Mfs at the 24 h time point. Recruitment of MyD88 leads to NF-kB activation which is required for the expression of many pro-inflammatory cytokine genes [42]. We did observe upregulation of signaling molecules activated downstream of MyD88 such as NFKB1 and upregulation of pro-inflammatory cytokine genes (Fig. 4 F) including IL12B, IL-1B, IL23A and TNFA indicating the activation of MyD88 and MyD88-dependent pathways.
As expected, genes related to IFN signaling were upregulated by pIC + R848 (Fig. 4). It is noteworthy that in our results upregulation of IFN genes was not observed (Fig. 4). In the experiments, the time points of sample collection (Mfs 24 h and DCs 48 h) were not optimal for detecting IFN gene expression, as they are induced at 4–6 h and peaking at 12–16 h after stimulus [43]. Although, IFN genes were not differentially expressed in the transcriptomics analyses, upregulation of a large set of ISGs suggest that type I and III IFNs were upregulated at an earlier time point by Bl-04 and pIC + R848 treatments.
The Pearson correlations (Fig. 5) showed that with Bl-04 stimulation, more genes correlated with each other compared to the pIC + R848 stimulation, especially in the chemokines and cytokines gene groups in both Mf and DCs, and in the signaling genes in DCs. It may indicate that the bacterial stimulation induces more complex signaling via several PPRs compared to the ligand stimulation. Another possibility is a kinetic difference in signaling, where a ligand stimulation induces a faster response compared to a bacterial stimulation, since the difference between the stimulations was more distinct in the DCs which were studied in 48 h time point, than in Mfs studied in 24 h time point. Interestingly, in pIC + R848 stimulated DCs the TLR3 adaptor gene TICAM1 correlated positively with chemokine, cytokine and antiviral genes, whereas in Bl-04 stimulated DCs TICAM2 that associated with TLR4 correlated with the aforementioned gene groups.
In Mfs Bl-04 induced pro but not anti-inflammatory cytokine gene expression (Fig. 4), in line with the cytokine response (Fig. 1), indicating the ability of Bl-04 to induce macrophage activation and modulate immune responses most likely towards type 1 immune responses. In support, the chemokine DEG pattern indicates attraction of NK-cells, Th1 cells, and neutrophils by Bl-04. These results suggest that Bl-04 induces innate responses that are activated also during viral infections [44]. Interestingly, in a clinical trial IL-1β and CXCL8 levels were found to be higher in the nasal washes of subjects taking Bl-04 prior to RV infection [11,12]. Bl-04 also induced expression of the genes related to antiviral response in a similar pattern with pIC + R848 (Fig. 4), indicating activation of antiviral state in Mfs and signaling of the presence of viral infection. The above is interesting in the context of clinical findings, as in one [11], but not the other [12] human RV challenge clinical study, RV was found in lower quantity and less frequently in the nasal washes of the subjects supplemented with Bl-04, suggesting potential impact on the viral life cycle. The exact effect of Bl-04 on antiviral response in nasal mucosa of humans needs further investigations. The in vitro results on Bl-04 align with previous studies showing the ability of probiotic bacteria to induce cytokine production from human monocyte-derived Mf in a strain specific manner [13,45]. In contrast to our results on non-significant induction of IL-10 (Fig. 1), in another in vitro study Bl-04 was shown to induce relatively high IL-10 concentration from PBMCs compared to other probiotics [13], which could also reflect differences in in vitro cell systems. Although we did not study viral replication in our immune cell model, it is interesting to note that an in vitro study showed reduction in Influenza A virus replication in Mfs by Lacticaseibacillus strains that correlated with their ability to activate type I IFN-dependent antiviral genes [46].
DC response to Bl-04 induced Th1 (IL12B and STAT1) and Th17 (IL23A and STAT3) polarizing genes and increased Th1 (IFN-γ, IL-12p70) and Th17 (IL-6, IL-23, and TGF-β) cytokine production without increasing IL-10 and downregulating IL10 (Fig. 1, Fig. 4). With pIC + R848, Bl-04 induced higher IL-10, IL-12p70, and IL-23 cytokine expression (Fig. 1) showing modulation of pIC + R848 induced DC polarization. The above results suggest that Bl-04 induces mixed Th1 and Th17 polarization in DCs and can augment pIC + R848 response. Previously published study that screened the effect of 16 bifidobacterial strains on mouse bone marrow derived DCs, showed relatively constant induction of IL-6, but variable effect on IL-10, IL-12, and IL-23 production [47], suggesting that the effect of bifidobacterial strains vary on T regulatory and Th responses. Similar results were found when PBMCs were stimulated with various bifidobacterial strains [48], highlighting the need for strain specific studies on immune system effects.
In the context of the above discussed effects of Bl-04 on Mf antiviral response and on acute URTIs and RV load in human clinical studies, it is noteworthy that also in DCs Bl-04 induces transcription of several important genes for antiviral response (Fig. 4). Previously published pre-clinical study showed similar effect in the same Mf and DC model, but with the blend of five probiotics, including Bl-04 [49]. In a subsequent SARS-CoV-2 challenge ferret study, the blend stimulated gut-lung axis immunity and reduced SARS-CoV-2 load.
4.2. Bl-04 has a mild effect on RV replication and immune response in fibroblasts
In addition to immune cells, we also used human embryonic lung fibroblast cell line (MRC-5) to study the effect of Bl-04 on cytokine production and RV replication upon low, medium or high dose of RV infection. We could observe inhibition of RV replication by Bl-04 with the low titer infection at 72 h, but not with the medium or high titer infection, indicating mild viral inhibition in the in vitro conditions. This result may be reflective of the two clinical studies where one study showed reduction in nasal wash viral load by Bl-04 compared to placebo [11], but one did not [12] Likewise, Bl-04 reduced lung influenza virus load in mice compared to placebo at one time point only [50]. These results highlight that clinical and in vivo translation with small effect sizes may be difficult, when multiple factors provide variance in the studies.
To our knowledge, this is the first study where the effect of a bifidobacterium on RV infection has been studied in in vitro setting. A recent study analyzed the effect of Lactobacillus rhamnosus on RV-A16 virus in well-differentiated human nasal epithelial cells [51]. They could not detect a decrease in RV TCID50 titers at 24 h time point when the cells were pretreated with L. rhamnosus, but a decrease in cytokine and chemokine secretion was seen.
Most in vitro studies on viral replication have been conducted with lung (primary) epithelial cells. Fibroblasts, however, have been proposed to act as sentinel cells having a dual role i.e. functioning as immunoregulatory cells in addition to their structural functions [52]. Consequently, fibroblasts also take part in early innate responses responding to bacterial and viral challenges by producing a variety of cytokines and chemokines. On the other hand, excessive production of these molecules may lead to exacerbation of flu symptoms or already existing conditions such as allergy or asthma [53].
Several publications have shown that RV infection leads to increased production of proinflammatory cytokines in airway epithelial cells and fibroblasts [19,52,53]. We could also confirm the proinflammatory cytokine production in the MRC-5 cell line after RV infection, especially with the high titer infection. However, priming of the cells with Bl-04 significantly attenuated the cytokine or chemokine production compared with the un-primed control at only one test condition (IL-1β at 24 h with high titer). These results contradict with the recent publication [51] but might be explained by differences between the cell line and bacterial strain.
Overall, the results suggest that RV can induce pro-inflammatory cytokine expression in MRC-5 cells when medium or high virus titer is used, however; the effect of Bl-04 was very limited in this in vitro setting.
5. Conclusions
We showed that probiotic Bl-04 induces overlapping but also distinct activation of human Mfs and DCs in comparison to viral nucleotide analogues pIC + R848. The activation of antiviral gene expression and pathways, and Th1 and Th17 type cytokine production are suggestive of the mechanism of action of Bl-04 on the immune system. Bl-04 had negligible effect on RV replication and cytokine expression in vitro in fibroblasts. Overall, our results suggest that Bl-04 could modulate the immune system by pre-stimulating antiviral defense of immune cells before infection rather than acting directly on the stromal cells.
5.1. Limitations of the study
The caveat of the in vitro monocyte derived immune cell model is that it does not recapitulate the complexity of the gut where the phenotypically diverse tissue resident macrophages and dendritic cells likely are in contact with the orally consumed probiotic in the context of microbiota. Anaerobic, but aerotolerant Bl-04 is possibly producing only a limited number of metabolites and in a non-replicative state due to the normoxic atmosphere and antibiotics used in the cell media. Thus, some effects of the bacterial metabolome or other secreted molecules to the immune cells may be missing. In the future research, comparison between Bl-04 and other probiotics would be beneficial for understanding how broad or narrow strain specific differences are on immune function.
The cytokine and gene expression responses vary due to host genetics, age, and underlying conditions of the donors, which were not standardized in this study, however, the donor effect was taken into account in the statistical models. Having access to well-defined blood donors, ideally divided into high or low responders would remove some of the bias caused by the donor variation. The ELISA results show the cumulative cytokine production by the cells whereas the transcriptomics analyzed give a snapshot of the gene expression at the given time point. A more complete picture could be drawn from also studying the kinetics of the transcriptome and cytokine production with this model.
Fibroblast cells were grown as monocultures lacking the crosstalk with epithelial cells and immune cells that are normally present in the airways and presumably the first line of defense when host is challenges with a virus. The above-mentioned limitations for using Bl-04 in the cell culture model and performing ELISA analysis are valid also for the fibroblast in vitro experiments.
Ethics statement
The use of human blood was approved by the Ethics Committee of the Hospital District of Helsinki and Uusimaa (HUS), Finland (HUS-2563-2019-3). All blood donors provided informed consent that their blood can be used for research purposes.
Data availability statement
The transcriptomics datasets generated during the study are available in the NCBI BioSample repository under BioProject PRJNA950561.
CRediT authorship contribution statement
Sinikka Latvala: Writing – review & editing, Writing – original draft, Validation, Resources, Methodology, Investigation, Conceptualization. Markus J. Lehtinen: Writing – review & editing, Writing – original draft, Visualization, Supervision, Resources, Project administration, Methodology, Investigation, Conceptualization. Sanna M. Mäkelä: Writing – review & editing, Writing – original draft, Visualization, Methodology. Derek Nedveck: Writing – review & editing, Writing – original draft, Visualization, Software, Methodology, Investigation, Formal analysis, Data curation. Bryan Zabel: Writing – review & editing, Visualization, Software, Methodology, Investigation, Formal analysis, Data curation. Ilmari Ahonen: Writing – review & editing, Visualization, Software, Methodology, Formal analysis, Data curation. Liisa Lehtoranta: Writing – review & editing, Validation, Methodology, Conceptualization. Ronald B. Turner: Writing – review & editing, Writing – original draft, Validation, Supervision, Resources, Project administration, Methodology, Investigation, Conceptualization. Jenni Liljavirta: Writing – review & editing, Writing – original draft, Visualization, Validation, Resources, Methodology, Investigation, Conceptualization.
Declaration of competing interest
The authors declare the following financial interests/personal relationships which may be considered as potential competing interests:Markus Lehtinen, Ronald B. Turner has patent #IMMUNOMODULATORY COMPOSITION COMPRISING BIFIDOBACTERIA, PCT/E P2016/051695 issued to DUPONT NUTRITION BIOSCIENCES APS. SL, SMM, MJL, DN and JL are employees of IFF Health and Biosciences that manufactures and sells probiotics used in this study. LL and BZ are former employees of IFF Health and Biosciences. If there are other authors, they declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
Katri Holappa, Henri Ahokoski, Jaana Larsson-Leskelä and Anna Lappalainen are thanked for excellent technical assistance.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.heliyon.2024.e29588.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Heikkinen T., Järvinen A. The common cold. Lancet. 2003;361:51–59. doi: 10.1016/s0140-6736(03)12162-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jain S. Epidemiology of viral pneumonia. Clin. Chest Med. 2017;38:1–9. doi: 10.1016/j.ccm.2016.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hao Q., Dong B.R., Wu T. Probiotics for preventing acute upper respiratory tract infections. Cochrane Database Syst. Rev. 2015 doi: 10.1002/14651858.CD006895.pub3. [DOI] [PubMed] [Google Scholar]
- 4.Wang Y., Li X., Ge T., Xiao Y., Liao Y., Cui Y., Zhang Y., Ho W., Yu G., Zhang T. Probiotics for prevention and treatment of respiratory tract infections in children: a systematic review and meta-analysis of randomized controlled trials. Medicine (Baltim.) 2016;95 doi: 10.1097/md.0000000000004509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li L., Hong K., Sun Q., Xiao H., Lai L., Ming M., Li C. Probiotics for preventing upper respiratory tract infections in adults: a systematic review and meta-analysis of randomized controlled trials. Evid Based Complement Alternat Med. 2020;2020 doi: 10.1155/2020/8734140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hill C., Guarner F., Reid G., Gibson G.R., Merenstein D.J., Pot B., Morelli L., Canani R.B., Flint H.J., Salminen S., et al. Expert consensus document. The International Scientific Association for Probiotics and Prebiotics consensus statement on the scope and appropriate use of the term probiotic. Nat. Rev. Gastroenterol. Hepatol. 2014;11:506–514. doi: 10.1038/nrgastro.2014.66. [DOI] [PubMed] [Google Scholar]
- 7.West N.P., Horn P.L., Pyne D.B., Gebski V.J., Lahtinen S.J., Fricker P.A., Cripps A.W. Probiotic supplementation for respiratory and gastrointestinal illness symptoms in healthy physically active individuals. Clin. Nutr. 2014;33:581–587. doi: 10.1016/j.clnu.2013.10.002. [DOI] [PubMed] [Google Scholar]
- 8.West N.P., Horn P.L., Pyne D.B., Warren H.S., Asad S., Cox A.J., Lahtinen S.J., Lehtinen M.J., Fricker P.A., Cripps A.W., Fazekas de St Groth B. Probiotic supplementation has little effect on peripheral blood regulatory T cells. J. Allergy Clin. Immunol. 2016;138:1749–1752.e1747. doi: 10.1016/j.jaci.2016.06.055. [DOI] [PubMed] [Google Scholar]
- 9.West N.P., Horn P.L., Barrett S., Warren H.S., Lehtinen M.J., Koerbin G., Brun M., Pyne D.B., Lahtinen S.J., Fricker P.A., Cripps A.W. Supplementation with a single and double strain probiotic on the innate immune system for respiratory illness. e-SPEN Journal. 2014;9:e178–e184. doi: 10.1016/j.clnme.2014.06.003. [DOI] [Google Scholar]
- 10.Zhang H., Miao J., Su M., Liu B.Y., Liu Z. Effect of fermented milk on upper respiratory tract infection in adults who lived in the haze area of Northern China: a randomized clinical trial. Pharm. Biol. 2021;59:647–652. doi: 10.1080/13880209.2021.1929344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Turner R.B., Woodfolk J.A., Borish L., Steinke J.W., Patrie J.T., Muehling L.M., Lahtinen S., Lehtinen M.J. Effect of probiotic on innate inflammatory response and viral shedding in experimental rhinovirus infection - a randomised controlled trial. Benef. Microbes. 2017;8:207–215. doi: 10.3920/bm2016.0160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Turner R.B., Lehtoranta L., Hibberd A., Männikkö S., Zabel B., Yeung N., Huttunen T., Burns F.R., Lehtinen M.J. Effect of bifidobacterium animalis spp. lactis Bl-04 on rhinovirus-induced colds: a randomized, placebo-controlled, single-Center, phase II trial in healthy volunteers. EClinicalMedicine. 2022;43 doi: 10.1016/j.eclinm.2021.101224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Foligne B., Nutten S., Grangette C., Dennin V., Goudercourt D., Poiret S., Dewulf J., Brassart D., Mercenier A., Pot B. Correlation between in vitro and in vivo immunomodulatory properties of lactic acid bacteria. World J. Gastroenterol. 2007;13:236–243. doi: 10.3748/wjg.v13.i2.236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lehtoranta L., Latvala S., Lehtinen M.J. Role of probiotics in stimulating the immune system in viral respiratory tract infections: a narrative review. Nutrients. 2020;12 doi: 10.3390/nu12103163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Janeway C.A., Jr., Medzhitov R. Innate immune recognition. Annu. Rev. Immunol. 2002;20:197–216. doi: 10.1146/annurev.immunol.20.083001.084359. [DOI] [PubMed] [Google Scholar]
- 16.Taniguchi T., Ogasawara K., Takaoka A., Tanaka N. IRF family of transcription factors as regulators of host defense. Annu. Rev. Immunol. 2001;19:623–655. doi: 10.1146/annurev.immunol.19.1.623. [DOI] [PubMed] [Google Scholar]
- 17.de Jong E.C., Smits H.H., Kapsenberg M.L. Dendritic cell-mediated T cell polarization. Springer Semin. Immunopathol. 2005;26:289–307. doi: 10.1007/s00281-004-0167-1. [DOI] [PubMed] [Google Scholar]
- 18.Sirianni F.E., Chu F.S., Walker D.C. Human alveolar wall fibroblasts directly link epithelial type 2 cells to capillary endothelium. Am. J. Respir. Crit. Care Med. 2003;168:1532–1537. doi: 10.1164/rccm.200303-371OC. [DOI] [PubMed] [Google Scholar]
- 19.Zhu Z., Tang W., Gwaltney J.M., Jr., Wu Y., Elias J.A. Rhinovirus stimulation of interleukin-8 in vivo and in vitro: role of NF-kappaB. Am. J. Physiol. 1997;273:L814–L824. doi: 10.1152/ajplung.1997.273.4.L814. [DOI] [PubMed] [Google Scholar]
- 20.Bedke N., Haitchi H.M., Xatzipsalti M., Holgate S.T., Davies D.E. Contribution of bronchial fibroblasts to the antiviral response in asthma. J. Immunol. 2009;182:3660–3667. doi: 10.4049/jimmunol.0802471. [DOI] [PubMed] [Google Scholar]
- 21.De Silva D., Dagher H., Ghildyal R., Lindsay M., Li X., Freezer N.J., Wilson J.W., Bardin P.G. Vascular endothelial growth factor induction by rhinovirus infection. J. Med. Virol. 2006;78:666–672. doi: 10.1002/jmv.20591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ghildyal R., Dagher H., Donninger H., de Silva D., Li X., Freezer N.J., Wilson J.W., Bardin P.G. Rhinovirus infects primary human airway fibroblasts and induces a neutrophil chemokine and a permeability factor. J. Med. Virol. 2005;75:608–615. doi: 10.1002/jmv.20315. [DOI] [PubMed] [Google Scholar]
- 23.Napolitani G., Rinaldi A., Bertoni F., Sallusto F., Lanzavecchia A. Selected Toll-like receptor agonist combinations synergistically trigger a T helper type 1-polarizing program in dendritic cells. Nat. Immunol. 2005;6:769–776. doi: 10.1038/ni1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Huang C.C., Duffy K.E., San Mateo L.R., Amegadzie B.Y., Sarisky R.T., Mbow M.L. A pathway analysis of poly(I:C)-induced global gene expression change in human peripheral blood mononuclear cells. Physiol. Genom. 2006;26:125–133. doi: 10.1152/physiolgenomics.00002.2006. [DOI] [PubMed] [Google Scholar]
- 25.Vareille M., Kieninger E., Edwards M.R., Regamey N. The airway epithelium: soldier in the fight against respiratory viruses. Clin. Microbiol. Rev. 2011;24:210–229. doi: 10.1128/CMR.00014-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mäkelä S.M., Osterlund P., Julkunen I. TLR ligands induce synergistic interferon-β and interferon-λ1 gene expression in human monocyte-derived dendritic cells. Mol. Immunol. 2011;48:505–515. doi: 10.1016/j.molimm.2010.10.005. [DOI] [PubMed] [Google Scholar]
- 27.Aeffner F., Traylor Z.P., Yu E.N., Davis I.C. Double-stranded RNA induces similar pulmonary dysfunction to respiratory syncytial virus in BALB/c mice. Am. J. Physiol. Lung Cell Mol. Physiol. 2011;301:L99–L109. doi: 10.1152/ajplung.00398.2010. [DOI] [PubMed] [Google Scholar]
- 28.Patro R., Duggal G., Love M.I., Irizarry R.A., Kingsford C. Salmon provides fast and bias-aware quantification of transcript expression. Nat. Methods. 2017;14:417–419. doi: 10.1038/nmeth.4197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Love M.I., Huber W., Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wickham H. Applied Spatial Data Analysis R; 2016. Elegant Graphics for Data Analysis (Ggplot2) [Google Scholar]
- 31.McKenzie A.T., Katsyv I., Song W.M., Wang M., Zhang B. DGCA: A comprehensive R package for differential gene correlation analysis. BMC Syst. Biol. 2016;10:106. doi: 10.1186/s12918-016-0349-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Voichita C., Ansari S., Draghici S. ROntoTools: the R Onto-Tools suite. R package version. 2019;1 [Google Scholar]
- 33.Kanehisa M. Toward understanding the origin and evolution of cellular organisms. Protein Sci. 2019;28:1947–1951. doi: 10.1002/pro.3715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kanehisa M., Furumichi M., Sato Y., Ishiguro-Watanabe M., Tanabe M. KEGG: integrating viruses and cellular organisms. Nucleic Acids Res. 2021;49:D545–d551. doi: 10.1093/nar/gkaa970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kanehisa M., Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.R Core Team . R Foundation for Statistical Computing; Vienna, Austria: 2022. R: A Language and Environment for Statistical Computing.https://www.R-project.org/ [Google Scholar]
- 37.Lenth R.V. Emmeans: estimated marginal means, aka least-squares means. R package version 1.8.2. 2022 https://CRAN.R-project.org/package=emmeans [Google Scholar]
- 38.Coleman J.L., Hatch-McChesney A., Small S.D., Allen J.T., Sullo E., Agans R.T., Fagnant H.S., Bukhari A.S., Karl J.P. Orally ingested probiotics, prebiotics, and synbiotics as countermeasures for respiratory tract infections in nonelderly adults: a systematic review and meta-analysis. Adv. Nutr. 2022;13:2277–2295. doi: 10.1093/advances/nmac086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yamazaki T., Ohshio K., Sugamata M., Morita Y. Lactic acid bacterium, Lactobacillus paracasei KW3110, suppresses inflammatory stress-induced caspase-1 activation by promoting interleukin-10 production in mouse and human immune cells. PLoS One. 2020;15 doi: 10.1371/journal.pone.0237754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kaji R., Kiyoshima-Shibata J., Tsujibe S., Nanno M., Shida K. Short communication: probiotic induction of interleukin-10 and interleukin-12 production by macrophages is modulated by co-stimulation with microbial components. J. Dairy Sci. 2018;101:2838–2841. doi: 10.3168/jds.2017-13868. [DOI] [PubMed] [Google Scholar]
- 41.Miettinen M., Veckman V., Latvala S., Sareneva T., Matikainen S., Julkunen I. Live Lactobacillus rhamnosus and Streptococcus pyogenes differentially regulate Toll-like receptor (TLR) gene expression in human primary macrophages. J. Leukoc. Biol. 2008;84:1092–1100. doi: 10.1189/jlb.1206737. [DOI] [PubMed] [Google Scholar]
- 42.Duan T., Du Y., Xing C., Wang H.Y., Wang R.F. Toll-like receptor signaling and its role in cell-mediated immunity. Front. Immunol. 2022;13 doi: 10.3389/fimmu.2022.812774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pietilä T.E., Latvala S., Osterlund P., Julkunen I. Inhibition of dynamin-dependent endocytosis interferes with type III IFN expression in bacteria-infected human monocyte-derived DCs. J. Leukoc. Biol. 2010;88:665–674. doi: 10.1189/jlb.1009651. [DOI] [PubMed] [Google Scholar]
- 44.Kanauchi O., Andoh A., AbuBakar S., Yamamoto N. Probiotics and paraprobiotics in viral infection: clinical application and effects on the innate and acquired immune systems. Curr Pharm Des. 2018;24:710–717. doi: 10.2174/1381612824666180116163411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Latvala S., Miettinen M., Kekkonen R.A., Korpela R., Julkunen I. Lactobacillus rhamnosus GG and Streptococcus thermophilus induce suppressor of cytokine signalling 3 (SOCS3) gene expression directly and indirectly via interleukin-10 in human primary macrophages. Clin. Exp. Immunol. 2011;165:94–103. doi: 10.1111/j.1365-2249.2011.04408.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Miettinen M., Pietila T.E., Kekkonen R.A., Kankainen M., Latvala S., Pirhonen J., Osterlund P., Korpela R., Julkunen I. Nonpathogenic Lactobacillus rhamnosus activates the inflammasome and antiviral responses in human macrophages. Gut Microb. 2012;3:510–522. doi: 10.4161/gmic.21736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Weiss G., Christensen H.R., Zeuthen L.H., Vogensen F.K., Jakobsen M., Frøkiær H. Lactobacilli and bifidobacteria induce differential interferon-β profiles in dendritic cells. Cytokine. 2011;56:520–530. doi: 10.1016/j.cyto.2011.07.024. [DOI] [PubMed] [Google Scholar]
- 48.López P., González-Rodríguez I., Gueimonde M., Margolles A., Suárez A. Immune response to Bifidobacterium bifidum strains support Treg/Th17 plasticity. PLoS One. 2011;6 doi: 10.1371/journal.pone.0024776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lehtinen M.J., Kumar R., Zabel B., Mäkelä S.M., Nedveck D., Tang P., Latvala S., Guery S., Budinoff C.R. The effect of the probiotic consortia on SARS-CoV-2 infection in ferrets and on human immune cell response in vitro. iScience. 2022;25 doi: 10.1016/j.isci.2022.104445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zabel B., Mäkelä S.M., Nedveck D., Hibberd A.A., Yeung N., Latvala S., Lehtoranta L., Junnila J., Walters K.B., Morovic W., Lehtinen M.J. The effect of bifidobacterium animalis subsp. lactis bl-04 on influenza A virus infection in mice. Microorganisms. 2023;11 doi: 10.3390/microorganisms11102582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yarlagadda T., Zhu Y., Snape N., Carey A., Bryan E., Maresco-Pennisi D., Coleman A., Cervin A., Spann K. Lactobacillus rhamnosus dampens cytokine and chemokine secretion from primary human nasal epithelial cells infected with rhinovirus. J. Appl. Microbiol. 2024;135 doi: 10.1093/jambio/lxae018. [DOI] [PubMed] [Google Scholar]
- 52.Smith R.S., Smith T.J., Blieden T.M., Phipps R.P. Fibroblasts as sentinel cells. Synthesis of chemokines and regulation of inflammation. Am. J. Pathol. 1997;151:317–322. [PMC free article] [PubMed] [Google Scholar]
- 53.Yoshikawa M., Wada K., Yoshimura T., Asaka D., Okada N., Matsumoto K., Moriyama H. Increased CXCL10 expression in nasal fibroblasts from patients with refractory chronic rhinosinusitis and asthma. Allergol. Int. 2013;62:495–502. doi: 10.2332/allergolint.13-OA-0572. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The transcriptomics datasets generated during the study are available in the NCBI BioSample repository under BioProject PRJNA950561.