Abstract
The 55-kDa E1B protein of adenovirus, which binds to and inactivates the tumor suppressor protein p53, is not expressed in the adenoviral mutant termed ONYX-015 (i.e., dl1520). It was reported that the mutant virus due to a deletion in E1B is able to replicate only in cells deficient for wild-type p53. Accordingly, dl1520 is currently being evaluated as a potential tool in the therapy of p53 deficient cancers. In contrast, we report here that dl1520 replicates independently of the p53 status in various tumor cell lines (U87, RKO, A549, H1299, and U373). In addition, the inhibition of p53-mediated transcriptional activation in wild-type p53 containing U2OS cells, by overexpression of a transdominant negative p53 mutant, did not render the cells permissive for dl1520 replication. Finally, we show that, depending on the multiplicity of infection, the deleted virus is able to replicate in and to kill primary human cells. Thus, the molecular basis for the growth differences of dl1520 within different cell types remains to be determined.
Replication-defective recombinant adenoviruses are widely used as gene therapeutic vectors in clinical protocols, including several for the treatment of cancer (25). Compared with other vector systems, adenoviruses efficiently mediate gene transfer into a large number of tissue types. Yet, the desirable objective of reaching all or most of the tumor cells within a given cancer, while leaving healthy tissue unaffected, is still far from a reality (36). Recently, a new concept has been developed based on an adenovirus mutant (ONYX-015), which reportedly replicates efficiently only in tumor cells and which appears to overcome this limitation (7). The apparently tumor-restricted adenovirus was originally constructed by Barker and Berk (5) and is called dl1520. The basis for the described tumor specificity has been postulated to be a genetic deletion of the viral E1B gene function, resulting in the loss of the expression of the viral 55-kDa protein (E1B 55K) (5). E1B 55K is known to bind to the tumor suppressor protein p53 and, as a consequence, to block p53-mediated transcriptional activation (28, 37, 38). Bischoff et al. (7) surmised that the binding of E1B 55K to p53 is necessary for efficient wild-type (wt) adenovirus replication in p53-positive cells. In support of this hypothesis, Bischoff et al. presented data indicating that replication of ONYX-015 (i.e., dl1520) was restricted to wt p53-deficient tumor cells. Since loss of function of the p53 tumor suppressor gene occurs in more than 50% of all types of human cancers (14, 18), the virus was proposed to be widely applicable in cancer therapy. Accordingly, phase I clinical trials have been initiated in head and neck cancer patients and are now being expanded to phase II. In addition, phase I trials have been initiated in patients with pancreatic cancer, ovarian cancer, and gastrointestinal cancer with liver metastasization. More recently, the same investigators reported that dl1520 is also able to replicate in some p53-positive tumor cells but not in primary cells of multiple origin (13). We also found replication of dl1520 in p53-positive tumor cells, including U87 and RKO cells, described by Bischoff et al. (7) and Heise et al. (13) to be resistant to dl1520 replication. Furthermore, we could show that p53-negative U373 cells are resistant to dl1520 replication, whereas Bischoff et al. (7) and Heise et al. (13) described U373 cells to be permissive for dl1520 replication. In addition, inactivation of transcriptionally active wt p53 in dl1520-resistant U2OS cells did not render the cells permissive for dl1520 replication. In our studies dl1520 replication seems, therefore, to be independent of the p53 status in tumor cells. Finally, we show that the deleted virus is able to replicate in and kill primary human cells in a multiplicity of infection (MOI)-dependent manner.
MATERIALS AND METHODS
Cell culture.
Dulbecco modified Eagle medium (DMEM), RPMI, cell culture supplements, and serum were obtained from Life Technologies (Gibco BRL, Eggenstein, Germany), and keratinocyte growth medium was obtained from Promocell (Heidelberg, Germany). HeLa, 911, H1299, U373, C33a, U87, A549, and U2OS cells were maintained as monolayers in DMEM; RKO cells were maintained in RPMI supplemented with 10% fetal calf serum (FCS) according to standard procedures. Normal single-donor primary human mammary epithelial cells (MEC; donor age, 24 years) were prepared and maintained as described previously (3, 34). Foreskin keratinocytes (donor 105; age, 5 years) and foreskin fibroblasts (donors 105 and 106; age, 6 years) were prepared essentially as described earlier (23). For cultivation of isolated foreskin keratinocytes, the plates were coated with a mixture of fibronectin (0.001% [wt/vol]), collagen (0.003% [wt/vol]), and bovine serum albumin (0.01% [wt/vol]). The foreskin fibroblasts were maintained in DMEM supplemented with 10% FCS, and the foreskin keratinocytes were maintained in keratinocyte growth medium (20).
Viruses.
Adenovirus wt (adenovirus type 2 [Ad2]) and dl1520 were propagated in 911 cells (9) and purified as described earlier (26). The titer of the frozen viral stocks of Ad2 (4.9 × 1010 PFU/ml) and dl1520 (3.2 × 1010 PFU/ml) was determined by plaque assay with 911 cells (9). The point mutation and deletion (5) in the dl1520 stocks (preventing the expression of the E1B 55K gene product) was confirmed by sequencing the respective genomic region (data not shown). In addition, PCR procedures were performed to exclude the possibility of wt adenovirus contamination in dl1520 stocks (data not shown). One day before infection, 3 × 105 cells were plated in duplicate onto 6-well dishes. Cells were incubated in 0.3 ml of appropriate serum-free medium containing the adenovirus wt virus (Ad2) or dl1520 at an MOI of 1 or 100 PFU per cell. After 1 h of incubation at 37°C with gentle swirling every 10 min, 3 ml of the respective growth medium was added to each dish. After 5 h the medium was replaced with 3 ml of fresh growth medium. For infection at an MOI of 100, the cells were washed twice with phosphate-buffered saline before fresh medium was added. After 72 h the cells were scraped into the culture medium and lysed by three cycles of freezing and thawing, and the supernatant was tested for virus production by plaque assay in 911 cells (9). Duplicates were titrated independently. As controls, cells were harvested immediately after fresh medium was added (5 h postinfection), and the remaining input virus was determined. For infection at an MOI of 1 PFU per cell, the titer was <5 × 103 PFU/ml, and for infection at an MOI of 100 PFU per cell, the titer was <5 × 104 PFU/ml. All viral titers, determined at 72 h postinfection, were at least 2 orders of magnitude higher, indicating that we determined the amount of newly produced virus at this time period. For cytopathic effect (CPE) assays the medium was not changed after 5 h.
Plaque assay.
At 72 h postinfection, the cells were scraped into the culture medium and lysed by three cycles of freezing and thawing, and the supernatant was tested for virus production by plaque assay in 911 cells (9). In brief, the supernatant was serially diluted in DMEM, and 0.3-ml portions of the dilutions were added to a 90% confluent 911 cell monolayer on 6-well dishes in duplicates. After 1 h of incubation at 37°C with gentle swirling every 10 min, 3 ml of 1% SeaPlaque agarose (FMC, Rockland, Maine) in DMEM supplemented with 3% FCS was added to each dish. The cells were fed with an additional overlay 4 days later. The plaques were visualized by staining with neutral red in an agarose overlay at 7 days after infection.
CPE assays.
At the indicated time points postinfection the cells were stained with crystal violet or photomicrographs were taken. For staining with crystal violet medium was removed, and the cells were fixed for 3 min in 3.7% formaldehyde at room temperature. The formaldehyde was discarded, and the cells were incubated for 3 min in 1% crystal violet. After the staining, the crystal violet solution was removed, and the cells were rinsed two times in 3 ml of water and then air dried.
Luciferase assay.
One day before transfection, 3 × 105 cells were plated in triplicate into 6-well dishes. The cells were transfected with 2 μg of luciferase reporter vector p53CON (p53 responsive) or pGUP.PA.8 (negative control) (10) from two independent plasmid preparations by Lipofectamine (Life Technologies, Eggenstein, Germany) according to the manufacturer’s instructions. Lysates were prepared 48 h after transfection, and luciferase assays were performed as reported previously (26).
U2OS cell cloning.
Logarithmically growing U2OS cells were transfected by Lipofectamine with pRc/CMV/p53-135 (substitution of cysteine at p53 codon 135 by tyrosine). G418 (Life Technologies) was added to a final concentration of 180 μg/ml at 48 h posttransfection. U2OS cells were cloned and tested by Western blot analysis with antibody p53(Ab6) (Dianova, Hamburg, Germany) for increased p53 protein expression according to standard procedures.
RESULTS
Replication of dl1520 in human tumor cell lines with different p53 status.
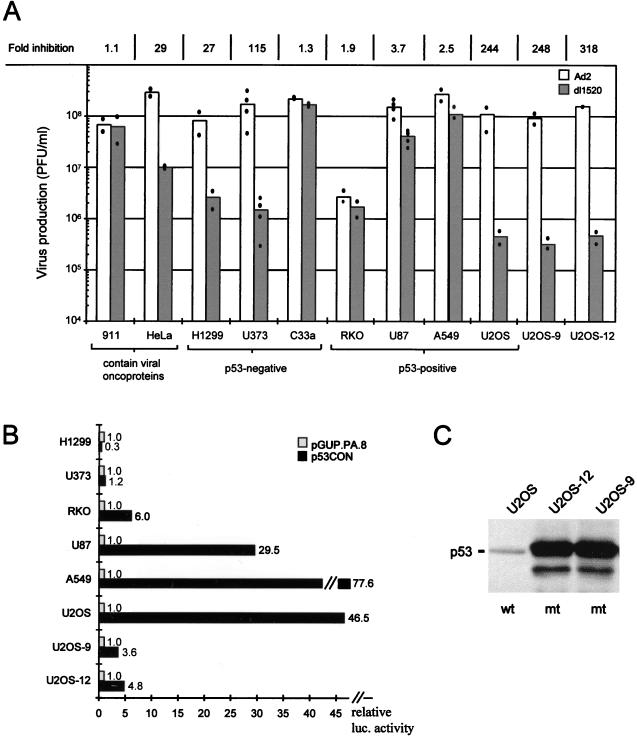
Since oncosuppressive and antiproliferative effects also have been described for adeno-associated virus infections (4, 16), we initially became interested in analyzing whether coamplification with adeno-associated virus would increase the antitumor effect of dl1520 (i.e., ONYX-015) in vivo. In initial experiments we attempted to confirm the results revealing selective growth of dl1520 in p53-negative cells (7). For this a panel of p53-positive and -negative cell lines derived from different tissues was infected with wt adenovirus or with dl1520, respectively, at an MOI of 1 (defined as plaque forming units per cell). At 72 h postinfection, the viral titers were determined by a plaque assay (9) (Fig. 1A). In agreement with the results reported previously (7), our data confirmed dl1520 replication in the p53-negative cell line C33a (cervical carcinoma, p53 codon 273 mutation) and inhibition of replication in the p53-positive cell line U2OS (osteosarcoma), in which dl1520 virus yields were reduced by more than 2 orders of magnitude (Fig. 1A). However, apart from the results for the positive control cell line 911, in which the E1B 55K is complemented by an integrated copy of the adenoviral E1 region (9), replication of dl1520 in all other cell lines was independent of the p53 status. In our studies, dl1520 titer was more than 100-fold lower in p53-negative U373 glioblastoma cells (p53 codon 273 mutation), while in the p53-positive A549 lung carcinoma, RKO colon carcinoma, and U87 glioblastoma cells, we obtained only a 1.9- to 3.7-fold inhibition compared to wt adenovirus infection. The overall low titers obtained in RKO cells can be explained by the low infectability of these cells, as determined by infection with a recombinant adenovirus expressing the β-galactosidase reporter gene (data not shown). Furthermore, in H1299 (lung carcinoma, p53 null) and HeLa cells (cervical carcinoma, p53 inactivated by E6 of human papillomavirus-18 (HPV-18 [29–32]) we found a 27- to 29-fold inhibition of dl1520 compared to wt adenovirus.
FIG. 1.
(A) Replication efficiency of dl1520 (shaded bar) and wt adenovirus (Ad2) (open bar) in various cell lines, each with a different p53 status as indicated in the figure (p53 negative; p53 null [H1299] or mutant [U373 and C33a contain p53 codon 273 mutation]). U2OS-9 and U2OS-12 cells are clones of parental U2OS cells expressing mutant p53 (p53 codon 135 mutation). All cells were infected at an MOI of 1 PFU per cell with either Ad2 or dl1520, and virus production was measured at 72 h postinfection by plaque assay. Dots represent the average values of duplicate determinations of a single infection. The bars represent the mean of 2 to 4 independent infections. At the top of the column, the fold inhibition of dl1520 replication compared to Ad2 is given as the ratio of Ad2 titer (PFU/milliliter) to dl1520 titer (PFU/milliliter). (B) Determination of transcriptionally active endogenous p53 in various cell lines by transactivation of a p53-responsive reporter plasmid. Relative luciferase activities of p53-responsive reporter plasmid p53CON (solid bar) are expressed as the fold activation above the basic vector pGUP.PA.8 (shaded bar) after transfection of the indicated cells. The results represent the average values of at least three independent transfections, each performed in triplicate. (C) Detection of mutant p53 (p53mt; codon 135 mutation) in the U2OS-derived cell clones U2OS-9 and U2OS-12 by Western blotting.
Measurement of transcriptionally active p53 in human tumor cell lines.
In transient-transfection assays with p53-responsive reporter plasmids (10), we confirmed both the absence of transcriptionally active p53 in the p53-negative cell lines H1299 and U373 and the presence of transcriptionally active p53 in the p53-positive cell lines RKO, U87, A549, and U2OS (Fig. 1B). As previously mentioned, dl1520 replication was inhibited in only one p53-positive cell line (U2OS); however, a comparable inhibition (>100 fold) also occurred in the p53-negative cell line U373. We therefore observed no correlation between the presence of transcriptionally active p53 and the ability of dl1520 to replicate in cells.
Expression of transdominant-negative mutant p53 in U2OS cell clones.
To further confirm that dl1520 replication is not dependent on the p53 status, we performed experiments to examine the permissiveness for virus replication in cells in which p53 was inactivated. To this end, U2OS cell clones expressing a dominant negative p53 allele (p53 codon 135 mutation) were established. Two of the clones (U2OS-9 and U2OS-12) expressing the highest level of mutant p53, as determined by Western blot analysis (Fig. 1C), were tested in transient transfections for p53 transactivation activity and in infections for their ability to replicate the deleted adenovirus. While the transcriptional activity of p53 in these cells was reduced to 10% of the parental U2OS cells (Fig. 1B), neither clone showed an increased permissiveness for dl1520 production (Fig. 1A). This finding is consistent with our results with the various tumor cell lines (Fig. 1A) and indicates that the p53 status in tumor cells does not correlate with dl1520 replication.
Replication of dl1520 in primary cells at different MOIs.
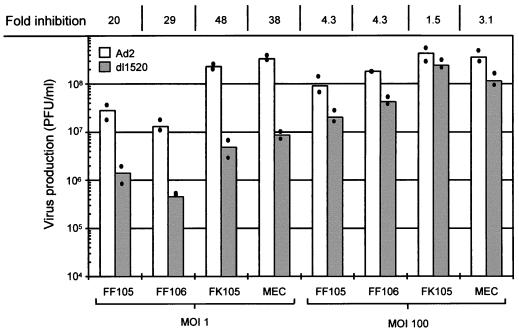
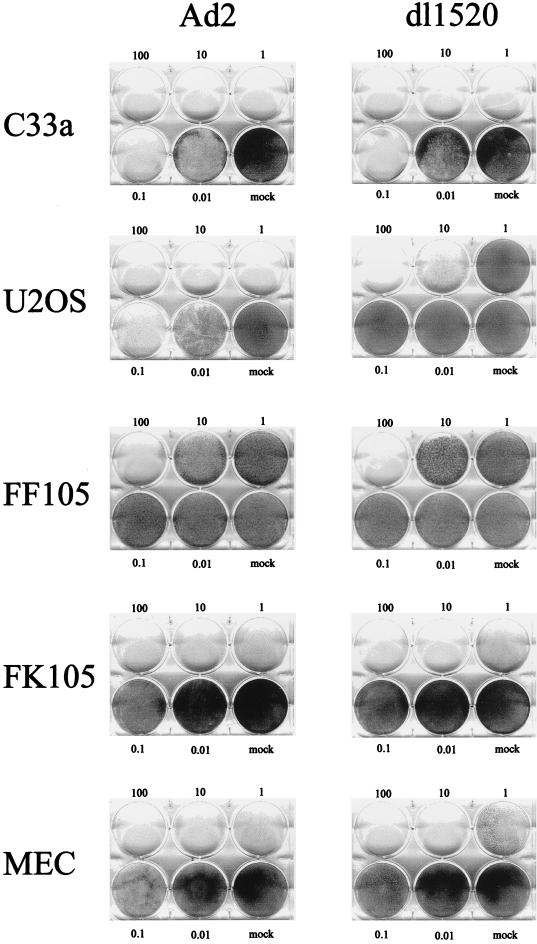
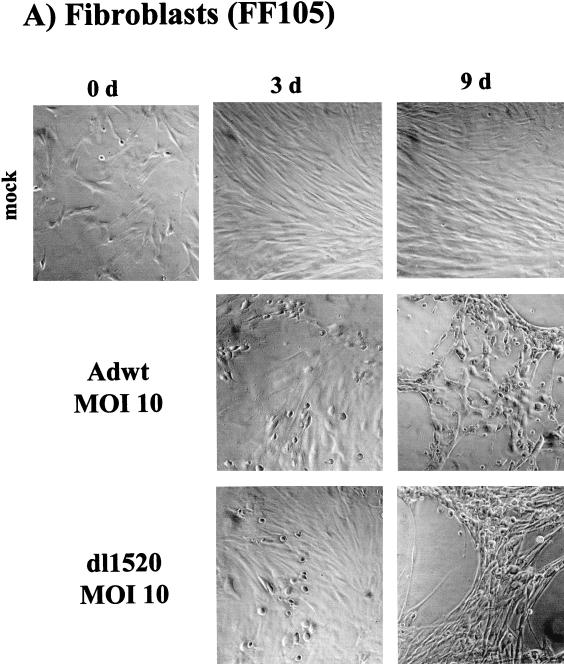
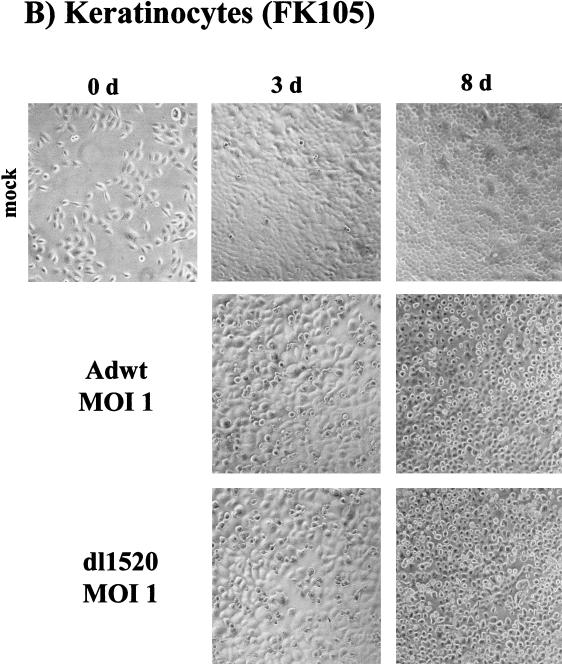
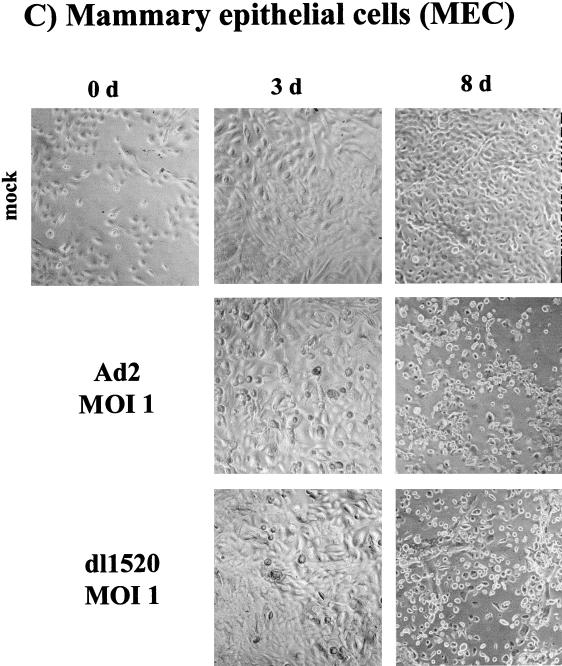
We subsequently tested the ability of dl1520 to replicate in primary cells. For this, we determined virus replication titers in adult human mammary epithelial cells (MEC), in foreskin keratinocytes (FK105), and in foreskin fibroblasts (FF105 and FF106), at 3 days postinfection at MOIs of 1 and 100 (Fig. 2). While at an MOI of 1 replication of dl1520 was reduced 20 to 48 fold, at an MOI of 100 replication approached wt adenovirus levels (1.5- to 4.3-fold reduction) (Fig. 2). The primary cells were then tested for the sensitivity to dl1520 and wt adenovirus infection at MOIs of 0.01 to 100 in CPE assays. As controls, the p53-negative cell line C33a (cervical carcinoma, p53 codon 273 mutation) and the p53-positive cell line U2OS (osteosarcoma) were infected in parallel. The CPE was monitored by staining the remaining cells on the plate with crystal violet. While U2OS cells were resistant against dl1520 infection at MOIs of 1 and less, wt adenovirus killed U2OS cells at MOIs of 0.1 to 0.01 (Fig. 3). C33a cells were killed with comparable efficiency by dl1520 and wt adenovirus at MOIs of 0.1 to 0.01 (Fig. 3). In all of the primary cell types tested, both viruses exhibited increasing CPEs with increasing MOIs. In MEC dl1520 was about 10-fold less cytopathic than wt adenovirus (see especially the MOI of 0.1; Fig. 3). In keratinocytes and fibroblasts, hardly any difference in terms of CPE between dl1520 and wt adenovirus-infected cells was apparent (Fig. 3). It should be noted that fibroblasts were found to be more resistant to adenovirus-induced CPE than were MEC and keratinocytes. Only at an MOI of 10 was destruction of the fibroblast monolayer observed at 9 days postinfection (Fig. 3). The resistance of fibroblasts to adenovirus-induced CPE correlated with the lower infectability of fibroblasts, as determined by infection with a recombinant adenovirus expressing the β-galactosidase reporter gene (data not shown). To monitor the CPE in more detail, we evaluated the dl1520 and wt adenovirus-infected cells by taking photomicrographs of the cell cultures in a time course of infection with initial MOIs of 10 for fibroblasts and 1 for keratinocytes and MEC (Fig. 4A to C). While mock-infected cells showed no CPE, adenovirus wt- and dl1520-infected primary cells showed a weak CPE at 3 days postinfection and a comparably strong CPE at 8 to 9 days postinfection (Fig. 4A to C). It should be noted that fibroblasts showed a morphological difference in the CPE upon wt adenovirus and dl1520 infection (Fig. 4A). While wt adenovirus-infected fibroblasts are rounded by 9 days after infection, dl1520 infected cells tend to become more fusiform.
FIG. 2.
Replication efficiency of dl1520 (shaded bar) and wt adenovirus (Ad2) (open bar) in primary foreskin fibroblasts (FF105 and FF106), foreskin keratinocytes (FK105), and MEC. Cells were infected at an MOI of 1 PFU per cell and at an MOI of 100 PFU per cell (as indicated) with either Ad2 or dl1520. Virus production was measured 72 h after infection by plaque assay. Dots represent the average values of duplicate determinations of a single infection. The bars represent the mean of two independent infections. At the top of the column, the fold inhibition of dl1520 replication compared to Ad2 is given as the ratio of Ad2 titer (PFU/milliliter) to dl1520 titer (PFU/milliliter).
FIG. 3.
CPE in C33a, U2OS, foreskin fibroblasts (FF105), foreskin keratinocytes (FK106), and MEC after infection with wt adenovirus (Ad2) or dl1520. Cells were infected with increasing MOIs (as indicated) and monitored for CPE by staining of the remaining cells on the plate with crystal violet (U2OS and C33a, 6 days after infection; FF105, FK106, and MEC, 9 days after infection).
FIG. 4.
Determination of CPE in foreskin fibroblasts (FF105) (A), foreskin keratinocytes (FK105) (B), and MEC (C) after infection with wt adenovirus and dl1520. Cells were infected with MOIs (as indicated) and monitored for CPE by photomicrographic examination at 3 and at 8 to 9 days after infection.
MOI dependency of dl1520 replication in tumor cells.
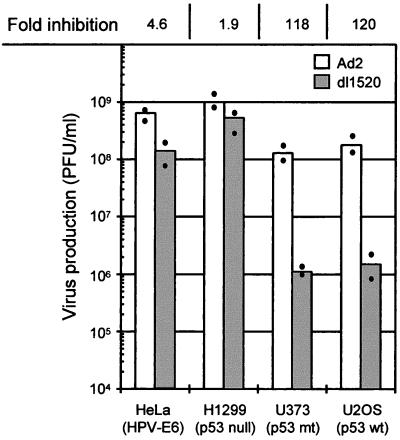
Since we observed an MOI-dependent replication of dl1520 in the primary cells, we infected the cell lines HeLa, H1299, U373, and U2OS, in which we had previously observed reduced titers at low MOIs, at an MOI of 100. Indeed, under these conditions, in H1299 (p53 null) and HeLa cells (p53 inactivated by E6 of HPV-18), dl1520 replicated to nearly the same levels as did wt adenovirus (1.9- to 4.6-fold reduction). In the p53-positive U2OS cells and the p53-negative U373 cells (p53 codon 273 mutation), however, dl1520 titers were still more than 100-fold reduced (Fig. 5). These data indicate that the p53 status of a cell alone is also not responsible for the MOI-dependent replication of dl1520.
FIG. 5.
Replication efficiency of dl1520 (shaded bar) and wt adenovirus (Ad2) (open bar) in HeLa, H1299, U373, and U2OS cells after infection at an MOI of 100 PFU per cell. The p53 status of the cell is indicated. Virus production was measured by plaque assay at 72 h after infection. Dots represent the average of duplicate determinations. The bars represent the mean of two independent infections. At the top of the column, the fold inhibition of dl1520 replication compared to that for Ad2 is given as the ratio of Ad2 titer (PFU/milliliter) to dl1520 titer (PFU/milliliter).
DISCUSSION
The report from Bischoff et al. (7) indicated a correlation between the p53 status and the susceptibility to dl1520 infection. In a CPE assay, it was shown that dl1520 killed tumor cells lacking normal p53 function but not cells (primary cells and tumor cells) with normal p53 function (7). Recently, Heise et al. (13) and Goodrum and Ornelles (11) reported exceptions to this rule, showing that some p53-positive tumor cells are susceptible to dl1520 infection. The strongest argument for a p53 dependency of dl1520 replication came from the results of Bischoff et al. (7), who reported that p53-positive RKO cells (colon carcinoma) are resistant to dl1520-induced CPE, while RKO.p53.13 cells, in which wt p53 function has been ablated by expression of a dominant-negative p53 allele, were 100-fold more sensitive to a dl1520-induced CPE. In our studies we found that dl1520 produced as much progeny virus as did wt adenovirus in RKO cells and that in p53-positive U2OS cells the inhibition of wt p53 activity by the overexpression of mutant p53 did not render the cells permissive for dl1520 replication. In addition, we observed no correlation between the p53 status in U87, A549, H1299, and U373 cells and the ability of dl1520 to replicate. While it was reported that U373 cells (p53 codon 273 mutation) were sensitive and U87 cells (p53 wild-type) were resistant to dl1520 infection in CPE assays (7, 13), our experiments indicated that dl1520 produced nearly as much progeny virus as did wt adenovirus in p53-positive U87 cells. In the p53-negative U373 cells, however, the dl1520 titer was reduced more than 100 fold. The reason for the conflicting results obtained in the CPE assays compared to the determination of viral titers remains to be determined. It should be noted, however, that results obtained in these assays may not be directly comparable since viral capsid proteins, even in the absence of viral DNA replication, may induce cytopathic changes (21, 34). Recently, Ridgway et al. (24) reported that wt adenovirus does not cause CPE in p53 mutant tumor cells, while we could clearly show that wt adenovirus produced high titers of progeny virus in the p53 mutant tumor cells C33a and U373. For these reasons, titrations of newly produced virus appears to be a better indication for permissiveness of specific cell types than the morphological evaluation of infected cells in CPE assays.
It was recently reported that, upon infection of MEC, dl1520 produced 100-fold less infectious virus compared to wt adenovirus (13). In addition, in CPE assays, MEC were found to be resistant to dl1520-induced CPE at MOIs of 0.01 to 1. In our experiments, dl1520 replication was 38-fold reduced in MEC at an MOI of 1, which is consistent with the data reported. However, we also observed clear signs of CPE by dl1520 infection at MOIs of 0.1 to 1. This difference may be explained by the use of different mammary epithelial isolates. Similarly, in human foreskin keratinocytes and foreskin fibroblasts, while dl1520 replication was clearly inhibited in viral burst experiments, the differences between dl1520 and wt adenovirus-induced CPE were even less remarkable. Thus, these primary cells are less permissive for dl1520 replication, yet they are still sensitive to the dl1520-induced killing effect.
In the present study we found an MOI-dependent dl1520 replication in the tumor cell lines H1299 (p53 null) and HeLa (p53 inactivated by E6 of HPV-18), while in the tumor cell lines U2OS (wt p53) and U373 (p53 codon 273 mutation) the replication of dl1520, even at high MOIs, was reduced more than 100 fold compared to wt adenovirus. Similarly, as previously reported, an increase in virus replication of different E1B-deleted adenoviruses correlated with an increase of the MOI in HeLa cells (1, 6, 33). Our data extend this observation, indicating that E1B is also dispensable for virulent virus replication at high MOIs of infection in adult human primary cells. In addition, we have shown that the MOI-dependent phenotype of dl1520 replication does not correlate with the p53 status of a cell. It is presently unknown whether the ability of the deleted virus to replicate in normal cells in vitro is also reflected in the in vivo situation, i.e., the patient with cancer. In a recent publication, an S-phase dependence for dl1520 replication was shown in HeLa cells (11). Since we tested tumor and primary cells under proliferating conditions in vitro, it is possible that dl1520 replication may occur preferentially in fast-proliferating tumor cells under in vivo conditions. The direct injection of dl1520 into the tumor mass, leading to a high local virus dose and high MOIs only in tumor cells, may also help to prevent undesired virus replication in normal cells. For the same reasons, systemic administration of the virus in combination with chemotherapy, as recently suggested by Heise et al. (13), does not appeal as a rational concept for the use of dl1520 in cancer therapy.
Bischoff et al. (7) could show that a U2OS cell line stably expressing the E1B 55-kDa protein complements for dl1520 replication, while we found that inactivation of transcriptionally active p53 in U2OS by overexpression of a mutant p53 does not complement for dl1520 replication. However, in both of the mutant p53-containing U2OS cell lines, the residual amount of transcriptional active p53 is comparable (ca. 10% of the parental level). It seems likely, therefore, that other functions of E1B, apart from inactivation of p53, are necessary for efficient virus replication in U2OS cells. It has been shown that during the late stage of lytic adenovirus infection, the E1B protein in complex with E4 34-kDa preferentially facilitates the transport of viral mRNA, while the export of most cellular RNAs is inhibited (2, 8, 12, 17, 19, 22, 27). As a consequence, adenoviral mutants that failed to express E1B were defective for late viral protein expression and the titers are low. It is possible that as-yet-unknown mechanisms in some tumor cells may complement for the E1B RNA transport functions and, as a result, in these cells the titers of E1B-deleted viruses are comparable to those of wt adenovirus. Furthermore, Bischoff et al. (7) reported that a stable U2OS cell line expressing an E1B deletion mutant that is unable to bind to p53 does not complement for dl1520 replication. This indicates that while the p53 status does not appear to be relevant, the epitope of E1B which is involved in binding to p53 is important. This may suggest that the interaction of E1B with p53-related proteins, such as the newly discovered p73 protein, may be crucial for adenovirus replication (15).
In this study, we could clearly demonstrate that the p53 status of a tumor cell alone does not determine the ability of dl1520 to replicate. Therefore, the p53 status does not appear to be a suitable marker for the prognosis of dl1520 replication and tumor destruction. In addition, we could show that dl1520 was able to kill and replicate in human primary cells in a manner dependent on the MOI. Moreover, while it appears that some tumor cells (C33a, A549, and U87) are more permissive for dl1520 replication than are primary cells, others (U2OS and U373) are even more resistant to the deleted virus. The molecular basis for the growth differences of dl1520 within these different cell types remains to be determined.
ACKNOWLEDGMENTS
We thank A. Berk (University of California, Los Angeles, Calif.) for providing dl1520 virus, J. M. Shay (University of Texas, Dallas, Tex.) for plasmids p53CON and P53GUP.PA.8, A. van der Eb for 911 cells, and R. Schmidt (Angewandte Tumorvirologie, Deutsches Krebsforschungszentrum, Heidelberg, Germany) for help with culturing the primary cells.
This work was supported by the Jung-Stiftung-Hamburg.
REFERENCES
- 1.Babiss L E, Ginsberg H S. Adenovirus type 5 early region 1b gene product is required for efficient shutoff of host protein synthesis. J Virol. 1984;50:202–212. doi: 10.1128/jvi.50.1.202-212.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Babiss L E, Ginsberg H S, Darnell J E., Jr Adenovirus E1B proteins are required for accumulation of late viral mRNA and for effects on cellular mRNA translation and transport. Mol Cell Biol. 1985;5:2552–2558. doi: 10.1128/mcb.5.10.2552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Band V, Sager R. Distinctive traits of normal and tumor-derived human mammary epithelial cells expressed in a medium that supports long-term growth of both cell types. Proc Natl Acad Sci USA. 1989;86:1249–1253. doi: 10.1073/pnas.86.4.1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bantel Schaal U. Adeno-associated parvoviruses inhibit growth of cells derived from malignant human tumors. Int J Cancer. 1990;45:190–194. doi: 10.1002/ijc.2910450134. [DOI] [PubMed] [Google Scholar]
- 5.Barker D D, Berk A J. Adenovirus proteins from both E1B reading frames are required for transformation of rodent cells by viral infection and DNA transfection. Virology. 1987;156:107–121. doi: 10.1016/0042-6822(87)90441-7. . (Erratum, 158:263.) [DOI] [PubMed] [Google Scholar]
- 6.Bernards R, de Leeuw M G, Houweling A, van der Eb A J. Role of the adenovirus early region 1B tumor antigens in transformation and lytic infection. Virology. 1986;150:126–139. doi: 10.1016/0042-6822(86)90272-2. [DOI] [PubMed] [Google Scholar]
- 7.Bischoff J R, Kirn D H, Williams A, Heise C, Horn S, Muna M, Ng L, Nye J A, Sampson Johannes A, Fattaey A, et al. An adenovirus mutant that replicates selectively in p53-deficient human tumor cells [see comments] Science. 1996;274:373–376. doi: 10.1126/science.274.5286.373. [DOI] [PubMed] [Google Scholar]
- 8.Dobbelstein M, Roth J, Kimberly W T, Levine A J, Shenk T. Nuclear export of the E1B 55-kDa and E4 34-kDa adenoviral oncoproteins mediated by a rev-like signal sequence. EMBO J. 1997;16:4276–4284. doi: 10.1093/emboj/16.14.4276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fallaux F J, Kranenburg O, Cramer S J, Houweling A, Van Ormondt H, Hoeben R C, van der Eb A J. Characterization of 911: a new helper cell line for the titration and propagation of early region 1-deleted adenoviral vectors. Hum Gene Ther. 1996;7:215–222. doi: 10.1089/hum.1996.7.2-215. [DOI] [PubMed] [Google Scholar]
- 10.Funk W D, Pak D T, Karas R H, Wright W E, Shay J W. A transcriptionally active DNA-binding site for human p53 protein complexes. Mol Cell Biol. 1992;12:2866–2871. doi: 10.1128/mcb.12.6.2866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Goodrum F D, Ornelles D A. The early region 1B 55-kDa oncoprotein of adenovirus relieves growth restrictions imposed on viral replication by the cell cycle. J Virol. 1997;71:548–561. doi: 10.1128/jvi.71.1.548-561.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Halbert D N, Cutt J R, Shenk T. Adenovirus early region 4 encodes functions required for efficient DNA replication, late gene expression, and host cell shutoff. J Virol. 1985;56:250–257. doi: 10.1128/jvi.56.1.250-257.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Heise C, Sampson Johannes A, Williams A, McCormick F, Von Hoff D D, Kirn D H. ONYX-015, an E1B gene-attenuated adenovirus, causes tumor-specific cytolysis and antitumoral efficacy that can be augmented by standard chemotherapeutic agents. Nat Med. 1997;3:639–645. doi: 10.1038/nm0697-639. [DOI] [PubMed] [Google Scholar]
- 14.Hollstein M, Sidransky D, Vogelstein B, Harris C C. p53 mutations in human cancers. Science. 1991;253:49–53. doi: 10.1126/science.1905840. [DOI] [PubMed] [Google Scholar]
- 15.Kaghad M, Bonnet H, Yang A, Creancier L, Biscan J C, Valent A, Minty A, Chalon P, Lelias J M, Dumont X, Ferrara P, McKeon F, Caput D. Monoallelically expressed gene related to p53 at 1p36, a region frequently deleted in neuroblastoma and other human cancers. Cell. 1997;90:809–819. doi: 10.1016/s0092-8674(00)80540-1. [DOI] [PubMed] [Google Scholar]
- 16.Katz E, Carter B J. Effect of adeno-associated virus on transformation of NIH 3T3 cells by ras gene and on tumorigenicity of an NIH 3T3 transformed cell line. Cancer Res. 1986;46:3023–3026. [PubMed] [Google Scholar]
- 17.Leppard K N, Shenk T. The adenovirus E1B 55-kd protein influences mRNA transport via an intranuclear effect on RNA metabolism. EMBO J. 1989;8:2329–2336. doi: 10.1002/j.1460-2075.1989.tb08360.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Levine A J, Momand J, Finlay C A. The p53 tumour suppressor gene. Nature. 1991;351:453–456. doi: 10.1038/351453a0. [DOI] [PubMed] [Google Scholar]
- 19.Logan J S, Shenk T. Transcriptional and translational control of adenovirus gene expression. Microbiol Rev. 1982;46:377–383. doi: 10.1128/mr.46.4.377-383.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Peehl D M, Ham R G. Growth and differentiation of human keratinocytes without a feeder layer or conditioned medium. In Vitro (Rockville) 1980;16:516–525. doi: 10.1007/BF02626465. [DOI] [PubMed] [Google Scholar]
- 21.Pereira H G. A protein factor responsible for early cytopathic effect of adenoviruses. Virology. 1958;6:601–611. doi: 10.1016/0042-6822(58)90109-0. [DOI] [PubMed] [Google Scholar]
- 22.Pilder S, Moore M, Logan J, Shenk T. The adenovirus E1B-55K transforming polypeptide modulates transport or cytoplasmic stabilization of viral and host cell mRNAs. Mol Cell Biol. 1986;6:470–476. doi: 10.1128/mcb.6.2.470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rheinwald J G, Green H. Serial cultivation of strains of human epidermal keratinocytes: the formation of keratinizing colonies from single cells. Cell. 1975;6:331–343. doi: 10.1016/s0092-8674(75)80001-8. [DOI] [PubMed] [Google Scholar]
- 24.Ridgway P J, Hall A R, Myers C J, Braithwaite A W. p53/E1b 58-kDa complex regulates adenovirus replication. Virology. 1997;237:404–413. doi: 10.1006/viro.1997.8782. [DOI] [PubMed] [Google Scholar]
- 25.Roth J A, Cristiano R J. Gene therapy for cancer: what have we done and where are we going? J Natl Cancer Inst. 1997;89:21–39. doi: 10.1093/jnci/89.1.21. [DOI] [PubMed] [Google Scholar]
- 26.Rothmann T, Katus H A, Hartong R, Perricaudet M, Franz W M. Heart muscle-specific gene expression using replication defective recombinant adenovirus. Gene Ther. 1996;3:919–926. [PubMed] [Google Scholar]
- 27.Sarnow P, Hearing P, Anderson C W, Halbert D N, Shenk T, Levine A J. Adenovirus early region 1B 58,000-dalton tumor antigen is physically associated with an early region 4 25,000-dalton protein in productively infected cells. J Virol. 1984;49:692–700. doi: 10.1128/jvi.49.3.692-700.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sarnow P, Ho Y S, Williams J, Levine A J. Adenovirus E1b-58kd tumor antigen and SV40 large tumor antigen are physically associated with the same 54 kd cellular protein in transformed cells. Cell. 1982;28:387–394. doi: 10.1016/0092-8674(82)90356-7. [DOI] [PubMed] [Google Scholar]
- 29.Scheffner M, Huibregtse J M, Vierstra R D, Howley P M. The HPV-16 E6 and E6-AP complex functions as a ubiquitin-protein ligase in the ubiquitination of p53. Cell. 1993;75:495–505. doi: 10.1016/0092-8674(93)90384-3. [DOI] [PubMed] [Google Scholar]
- 30.Scheffner M, Munger K, Byrne J C, Howley P M. The state of the p53 and retinoblastoma genes in human cervical carcinoma cell lines. Proc Natl Acad Sci USA. 1991;88:5523–5527. doi: 10.1073/pnas.88.13.5523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Scheffner M, Werness B A, Huibregtse J M, Levine A J, Howley P M. The E6 oncoprotein encoded by human papillomavirus types 16 and 18 promotes the degradation of p53. Cell. 1990;63:1129–1136. doi: 10.1016/0092-8674(90)90409-8. [DOI] [PubMed] [Google Scholar]
- 32.Schwarz E, Freese U K, Gissmann L, Mayer W, Roggenbuck B, Stremlau A, zur Hausen H. Structure and transcription of human papillomavirus sequences in cervical carcinoma cells. Nature. 1985;314:111–114. doi: 10.1038/314111a0. [DOI] [PubMed] [Google Scholar]
- 33.Shenk T, Jones N, Colby W, Fowlkes D. Functional analysis of adenovirus-5 host-range deletion mutants defective for transformation of rat embryo cells. Cold Spring Harb Symp Quant Biol. 1980;44(pt.1):367–375. doi: 10.1101/sqb.1980.044.01.041. [DOI] [PubMed] [Google Scholar]
- 34.Stampfer M R. Isolation and growth of human mammary epithelial cells. J Tissue Cult Methods. 1985;9:107. [Google Scholar]
- 35.Valentine R C, Pereira H G. Antigens and structure of the adenovirus. J Mol Biol. 1965;13:13–20. doi: 10.1002/rmv.375. [DOI] [PubMed] [Google Scholar]
- 36.Verma I M, Somia N. Gene therapy—promises, problems and prospects [news] Nature. 1997;389:239–242. doi: 10.1038/38410. [DOI] [PubMed] [Google Scholar]
- 37.Yew P R, Berk A J. Inhibition of p53 transactivation required for transformation by adenovirus early 1B protein. Nature. 1992;357:82–85. doi: 10.1038/357082a0. [DOI] [PubMed] [Google Scholar]
- 38.Yew P R, Liu X, Berk A J. Adenovirus E1B oncoprotein tethers a transcriptional repression domain to p53. Genes Dev. 1994;8:190–202. doi: 10.1101/gad.8.2.190. [DOI] [PubMed] [Google Scholar]