Abstract

Drug discovery is a lengthy and intricate process, and in its early stage, crucial steps are the selection of the therapeutic target and the identification of novel ligands. Most targets are dysregulated in pathogenic cells; typically, their activation or deactivation leads to the desired effect, while in other cases, interfering with the target-natural binder complex achieves the therapeutic results. Biophysical assays are a suitable strategy for finding new ligands or interferent agents, being able to evaluate ligand–protein interactions and assessing the effect of small molecules (SMols) on macromolecular complexes. This mini-review provides a detailed analysis of widely used biophysical methods, including fluorescence-based approaches, circular dichroism, isothermal titration calorimetry, microscale thermophoresis, and NMR spectroscopy. After a brief description of the methodologies, examples of interaction and competition experiments are described, together with an analysis of the advantages and disadvantages of each technique. This mini-review provides an overview of the most relevant biophysical technologies that can help in identifying SMols able not only to bind proteins but also to interfere with macromolecular complexes.
1. Introduction
Binding events between macromolecules, such as proteins or nucleic acids, or between a macromolecule and endogenous or exogenous ligand (SMols or peptides) are crucial for the regulation of all biological events. This process plays a central role in regulating all of the biological and cellular mechanisms vital for life, such as self-replication, metabolic pathways, and information processing. Accordingly, the modulation of macromolecule activity represents the typical strategy against several diseases. Therefore, the investigation into these interactions is essential for elucidating the underlying mechanism of biological or physiopathological regulations and serves as an invaluable resource in guiding the discovery of novel pharmacological targets and new molecules capable of engaging them for new therapeutic intervention.1
Molecular recognition is the foundation of medicinal chemistry. To explain the mechanism of drug action, Emil Fischer in 1894 proposed the model named “lock and key” that was revised by Koshland in 1958. Briefly, to be effective, the drugs must fit into specific active sites, or binding sites, of a target (enzymes or receptors), acting like a key that must fit into a lock. This model is therefore based on the principle of complementarity and has led to the development of numerous drugs.2 However, it often oversimplifies the complexity of molecular interactions in biological systems. Indeed, many diseases involve intricate networks of proteins and other macromolecules interacting with each other, which the lock and key model does not adequately address. With advancements in molecular biology and structural biochemistry, medicinal chemistry has broadened its focus on the interactions between macromolecules (such as in the interactome, which involves protein–protein or protein–nucleic acid complexes) and is developing new methods to influence these interactions. As an example, developing SMols or peptides to inhibit or stabilize protein–protein interactions (PPIs) has represented a suitable strategy for controlling these networks especially in cancer, infectious diseases, and neurodegeneration. Despite initial challenges in targeting PPIs, significant progress has been made in identifying PPI inhibitors or disruptors. This approach led to the discovery of Venetoclax, the first in-class drug of this category that specifically targets the B-cell lymphoma-2 (BCL-2) protein and exhibits efficacy in chronic leukemia treatment. Other PPI inhibitors, such as ABT-737 and idasanutlin, are under investigation in advanced clinical trials. Conversely, stabilizing PPIs, especially in unstructured proteins, is still an emerging, promising field, exemplified by molecular glue.3
Contemporary drug target research extends beyond PPIs to protein DNA/RNA interaction [PD(R)I]. Processes such as DNA replication, transcription, and repair, along with RNA functions such as splicing and translation, are driven by the dynamic interactions between nucleic acids and specific proteins. There are two distinct protein–DNA interaction events: nonspecific, like in histones, and highly specific, sequence-dependent interactions such as in transcription factors. These patterns are mirrored in the RNA–protein interactions. Both types of interactions at the DNA/RNA level are crucial for gene expression, influencing epigenetic modifications or the binding of protein factors to certain nucleotide sequences. Dysfunctions in timing and location of protein complex recruitment to DNA or RNA nucleotide-binding sites are frequently linked to the development of cancer and neurodegenerative diseases.4 Thus, the exploitation of DNA/RNA binding proteins (D/RBPs) as drug targets represents another cutting-edge frontier in therapeutic development. RBPs are involved in RNA splicing, translation control, mRNA localization and other forms of post-transcriptional gene regulation. Most RBPs bind to specific RNA sequences or secondary structures. Among RBPs, the most studied ones are LIN28, Musashi (MSI), TDP, TTP, and ELAV/Hu proteins.5
The investigation of the ability of SMols to interfere with molecular complexes is often monitored using cell-based functional assays. They are very useful to study the final biological effect of molecules; however, they do not demonstrate the direct molecular interaction with the target complex. Biophysical techniques may be useful to prove the direct interaction of SMols with the macromolecular complexes, to identify new chemical entities able to interfere with the formation, or to disrupt PP or protein nucleic acid interaction and therefore to define the structure activity relationship. Nevertheless, there are a limited number of biophysical techniques useful for studying such interactions, and this represents the major challenge in this field. The appeal of biophysical assays lies in their heightened sensitivity, which has seen considerable advancement in recent years. This enhancement has enabled the comprehensive primary screening of vast libraries, encompassing more than thousands of compounds. These compounds are not confined to a narrow range of low molecular weight fragments, offering a broader scope of investigation. Furthermore, the specificity and high fidelity of those assays have proven invaluable in acquiring detailed insights into the binding modes of new chemical entities (NCEs). These assays facilitate the measurement of kinetic constants, the determination of thermodynamic properties related to binding, and the acquisition of structural data about macromolecule–ligand interactions. Such detailed analysis is crucial for understanding the intricate dynamics at play in these molecular interactions, thereby providing a robust foundation for the development and optimization of therapeutic compounds.
This mini-review aims to provide a comprehensive overview of the foremost biophysical techniques used in studying the interactions of SMols with macromolecular complexes, with a specific focus on their efficacy in modulating the stability of these complexes. We provided information on methodologies suitable for studying the effect of SMol on macromolecular complexes, including fluorescence-based techniques, circular dichroism (CD), isothermal titration calorimetry (ITC), microscale thermophoresis (MST), and nuclear magnetic resonance (NMR) spectroscopy. As medicinal chemists, we focused on the potential of these techniques in evaluating SMols as ligands and interference compounds without delving into the physical aspects underlying them. This mini-review presents selected applications of each technique to provide an overview of their potential uses. The analysis considers the unique strengths and inherent limitations of each technique, providing a balanced perspective that is essential for a comprehensive understanding. To enhance the practical relevance of the mini-review, experimental case studies illustrating the application of these techniques in research scenarios are included.
2. Fluorescence and Luminescence-Based Approaches
2.1. Förster Resonance Energy Transfer (FRET) and Bioluminescence Resonance Energy Transfer (BRET)
Förster resonance energy transfer (FRET) involves a distance-dependent interaction between two light-sensitive molecules: an energy donor and an acceptor (Figure 2A). This nonradiative transfer of energy occurs when the donor fluorophore, in its excited state, transfers energy to a nearby acceptor fluorophore, leading to emission at a different wavelength. The efficiency of this transfer of energy decreases significatively with the sixth power of the distance between the donor and acceptor, making FRET highly sensitive to changes in molecular proximity. Typically, fluorescent probes are linked to specific sites on the proteins. When these proteins are in close proximity (typically within 1–10 nm), FRET can occur. By monitoring changes in energy transfer efficiency, it is possible to get insight on the binding or dissociation of protein complexes and how this is affected by the presence of SMols. The high sensitivity and specificity of this technique in detecting minimal distance changes between proteins is pivotal for assessing the impact of SMols on protein complexes. Its capacity for real-time monitoring provides dynamic insights into the kinetic aspects of these interactions, revealing the mechanistic details underpinning molecular associations. Additionally, FRET’s noninvasive nature allows for observations in physiological environments, including live cells. However, FRET is not without limitations. A significant challenge is the need to derivatize proteins with fluorescent probes. The choice of fluorophores is critical, as they must have overlapping emission and excitation spectra for efficient energy transfer. However, the protein derivatization can potentially alter the native structure and function of the proteins being studied. It not only introduces the risk of perturbing natural protein interactions but also demands a tailored and efficient labeling to ensure accurate FRET results.6 The technique faces challenges like fluorophore photobleaching, where prolonged light exposure can degrade the fluorophores, risking signal loss and inaccurate results. A significant constraint is the method’s reliance on the proximity of donor and acceptor fluorophores, with the efficiency of energy transfer diminishing significantly beyond 10 nm, thus limiting its application to interactions within this range. Furthermore, the interpretation of FRET data can be complex, heavily influenced by the orientation of the fluorophores and environmental factors such as pH and ionic strength, necessitating meticulous control and careful consideration in experimental design and data analysis.7
Figure 2.
Schematic representation of the (A) FRET assays for the study of modulation of the macromolecule–macromolecule binary complex by small molecules and (B) BRET assay applied to GPCR functional studies, discussed in the present mini-review.
The application of FRET for studying PPI and discovering new drugs that disrupt these interactions unveiled novel therapeutic strategies as testified by the literature of the past two decades (Figure 1). To name a few, FRET was exploited to highlight the potential of targeting the dimerization of human thymidylate synthase (hTS) using SMols, as a novel strategy in cancer therapy. Unlike conventional hTS inhibitors (i.e., 5-FU, raltiterxed) which target the enzyme’s active site and lead to resistance in cancer cells, the compounds reported in this study were designed to bind at the interface between hTS dimers, thus disrupting the homodimeric complex. This disruption shifts the equilibrium toward the hTS monomer which is degraded or, by binding with its own mRNA, reduces hTS expression. FRET was used to investigate the capability of the compounds to destabilize the hTS dimer and to provide insights into the molecular mechanisms by which they disrupt hTS dimer formation. Compound E7 proved its effectiveness in reducing cancer growth in mouse models, surpassing the performance of traditional drugs like 5-FU.8a Another successful application of the FRET-based assay is the identification of PCNA–p15 interaction inhibitors. PCNA is involved in DNA synthesis, repair, and cell cycle control. High PCNA levels in various tumor types are correlated with high rates of cell proliferation. The assay was initially conducted in 384-well plates and successfully scaled up to a 1536-well format to be applied to a high throughput screening (HTS) campaign. It was rigorously tested, including evaluations of the effects of nonfluorescent PCNA and the well-characterized inhibitor T2AA. The assay allowed an accurate determination of the dissociation constant of the PCNA–p15 interaction, aligning well with previously published binding affinity data (as per isothermal titration calorimetry). The assay was applied to an in-house database of chemical compounds for the identification of new classes of PCNA–p15 inhibitors.8b
Figure 1.
Hit compound modulators of PPIs and RNA–protein complexes identified by FRET.
Recent advancements in dye-labeled nucleic acids have significantly advanced FRET-based methodologies for analyzing protein–DNA interactions, ranging from traditional steady-state and time-resolved methods to more nuanced applications that examine distances, conformational changes, and enzymatic reactions in protein–DNA complexes. In intramolecular FRET applications, both fluorescent dyes are attached to the same biomolecule, typically the protein. This setup is particularly useful for studying DNA conformational alterations induced by protein interactions and for elucidating the structure and assembly dynamics of various nucleoprotein complexes. Conversely, in intermolecular FRET applications, the protein and DNA are separately labeled. The interaction and subsequent complex formation between these two molecules led to observable FRET events. Conversely, the FRET efficiency decreases upon complex dissociation. This technique has proven instrumental in investigating nucleic acid cleavage reactions catalyzed by DNase enzymes. Lastly, the FRET-based approach has been extended to identify a range of small bioactive molecules that interact with RNAs in protein complexes. This methodology has been effective in studying interactions such as the HIV-1 Tat-TAR and RNA repeat expansions with RNA-binding proteins.8c A notable example is the RBP Lin28 protein’s inhibition of the tumor-suppressing microRNA, let-7, by binding to its precursor RNA and impeding maturation. The Lin28/let-7 interaction is particularly significant due to let-7 miRNA’s role in downregulating multiple oncogenes, including HMGA2, c-Myc, and Ras. In this context, a GFP-tagged Lin28 and quencher-labeled let-7 FRET assay was developed to target this interaction.
Another technique based on resonance energy transfer that is also noteworthy is bioluminescence resonance energy transfer (BRET) (Figure 2B). Like FRET, it relies on the nonradiative energy transfer between a luciferase energy donor and an acceptor fluorophore that are close to each other (<10 nm). In this assay, external illumination is not required since the donor emits luminescence as a consequence of an enzymatic reaction (the oxidation of a luciferase substrate). In recent years, BRET has been employed mainly in the study of G-protein-coupled receptors (GPCRs), mainly to investigate GPCR dynamics and signaling activity and to discover new modulators of the GPCR–protein complex. In BRET assays related to GPCRs, Renilla luciferase (RLuc) is commonly employed to tag the studied receptor or the G-protein. Since BRET is typically characterized by a low sensitivity connected to the weak nature of bioluminescence,9a RLuc is frequently substituted with the smaller and brighter nanoluciferase (NanoLuc) to amplify BRET signals. Within the context of PPI modulation, a NanoBRET protocol was developed to study the allosteric modulation of GPCRs. Using a fluorescently labeled Gα peptide miming the G protein and a fluorescently tagged β2-adrenoceptor, a specific binding between the two entities was detected only in the presence of isoproterenol, an activator of β2-adrenoceptor.9b Similarly, the NanoBRET approach was applied for studying the functional activity of a series of synthetic cannabinoids and their metabolites, via ligand-induced interaction of β-arrestin2 (βarr2) with cannabinoid receptors CB1 and CB2.9c Briefly, both CB1 and CB2 and βarr2 were coupled to inactive subunits of NanoLuc, large BiT, and small BiT, respectively. When a CB1–2 agonist binds to the GPCR, it causes the association between CB1–2 receptor and βarr2. This allows large BiT and small BiT subunits to bind together, thus restoring the NanoLuc activity, which produces light when it reacts with the furimazine substrate.
In summary, FRET, and the more recently developed BRET, represent a pivotal technique in the elucidation of interactions among proteins, DNA, and RNA at the molecular level and provide valuable insights into the dynamic nature of these interactions, particularly in evaluating their modulation by SMols.
2.2. Fluorescence Polarization (FP)
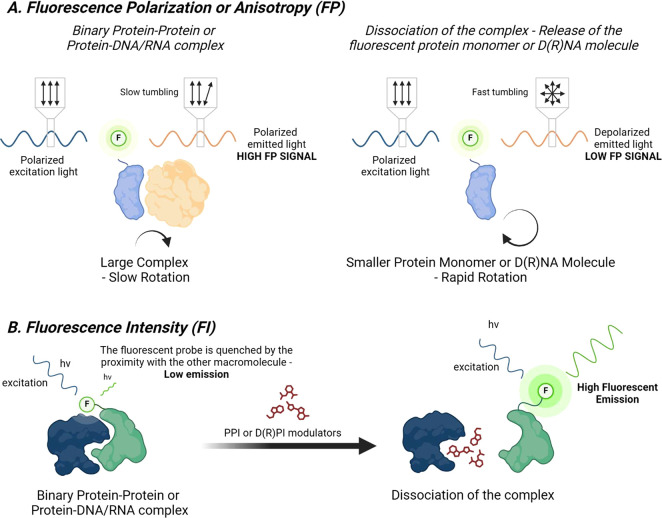
Fluorescence polarization (FP) is based on the principle of polarized light and the rotational mobility of molecules (Figure 3A). The core concept of FP involves the excitation of fluorescently labeled molecules with polarized light and the subsequent measurement of polarization of the emitted light. This polarization is influenced by the rotational motion of the fluorescent molecules. Smaller and unbound fluorescent molecules rotate rapidly. Due to their fast rotational movement, the orientation of these molecules changes significantly between the time they are excited and when they emit light. This results in the emission of light that is largely depolarized. Conversely, when the fluorescently labeled molecule binds to a macromolecule, the rotational mobility of the fluorescent molecule is significantly reduced. The slower rotation means that the orientation of the fluorescent probe does not change much between the absorption and emission. As a result, the emitted light remains more polarized. Measurement of the changes in the polarization of the emitted light serves as a reliable indicator of molecular binding events.10
Figure 3.
Schematic representation of the (A) fluorescence polarization and (B) fluorescence intensity assays for the study of modulation of a macromolecule–macromolecule binary complex by a small molecule.
FP is typically utilized as an alternative to FRET for studying macromolecular–macromolecular interactions, both PPI and PD(R)I, and the capability of SMols to affect the stability of the complex, since it offers several advantages for studying molecular interactions, particularly in HTS. One of the primary benefits is the elimination of immobilization procedures, which are often time-consuming and resource-intensive. FP enables the analysis of interactions in a homogeneous format, obviating the need for wash steps or physical separation processes. This aspect streamlines the experimental process, significantly enhancing the efficiency. Moreover, FP assays typically require small volumes of reagents and samples, such as 20–30 μL per well in a 384-well plate format, thus minimizing the total amount of compounds and materials employed in the assay. In addition, thanks to the high sensitivity of this technique, the tested compounds can be assessed at low concentrations (e.g., a maximum concentration of 10 μM). This is particularly beneficial for molecules that have solubility issues in aqueous buffers, thereby reducing potential solubility-related complications. Despite these advantages, FP requires the use of fluorescently labeled molecules, which introduces the potential for the fluorescent probe to interfere with the interaction assay, thus compromising the accuracy of the results. Moreover, FP can be challenging when investigating weak-affinity interactions and in the presence of compounds that induce light scattering, affecting the reliability of the results (Figure 4).11
Figure 4.
Representation of new modulators of PPIs and R(D)NA–protein interactions.
FP assays have been used to study a wide variety of targets including kinases, nuclear receptors, phosphatases, G-protein-coupled receptors (GPCRs), proteases, as well as PPI and D(R)BPI. Focusing on the latter two applications, FP has been extensively used to measure inhibition or enhancement of the PPI by SMols or other modulators (e.g., antibodies, peptides). The main advantage relies on the application of this assay for both primary HTS (in 96- and 384-well plates up to 1536-well plates) and dose–response assays. As an example, venetoclax and navitoclax, the two first-in-class PPI inhibitors to be approved for human use, originated from a FP-based HTS of compound repositories against Bcl-xL, followed by extensive lead-opt and development.12a Another interesting application of FP can be found in targeting the 14–3–3 proteins and the YAP–TEAD complex, which are relevant in cancer and other diseases. A highly sensitive FP-based assay was developed to monitor the interactions of 14–3–3 proteins with client proteins using a fluorescently labeled phosphopeptide from Raf-1. The assay’s specificity was validated with known 14–3–3 antagonists, like the R18 peptide, in a competitive assay. Another study reported an FP-based assay specifically optimized for identifying and evaluating inhibitors of the YAP–TEAD PPI at the YAP Ω-loop binding region of TEAD, crucial in the Hippo signaling pathway linked to cancer. This pathway’s inhibition can lead to cell transformation and tumor development. The assay’s application was validated using YAP mutant peptides and resulted in a patented small molecule with confirmed efficacy as a YAP–TEAD PPI inhibitor. The aforementioned assays and others reported in the literature revealed that the application of FP in HTS results in a high signal-to-background ratio and a Z′ factor greater than 0.7, indicating its reliability, robustness, and economic efficiency, making it suitable for hit identification and inhibitor assessment in drug discovery processes.
Although tethering remains one of the main approaches for the identification of selective PPI stabilizers, a streamlined methodology was recently proposed for this purpose, combining FP and tethering. By employing the DNA-binding site of the nuclear receptor estrogen related receptor γ (ERRγ), a specific cysteine located within the 14–3–3 PPI interface was targeted. Through this approach, multiple fragments capable of establishing a disulfide bond with the ERRγ, thereby enhancing the stability of the complex, were identified. The results, and the binding mode of the identified hits, were confirmed by crystallography.12b
Lastly, starting from broad inhibitors of 14–3–3 proteins, the same authors used molecular docking to design a selective stabilizer for the 14–3–3/ChREBP interaction. The capability of the design compounds to stabilize the complex was monitored by FP using fluorescently labeled ChREBP peptide which was titrated with an increasing concentration of 14–3–3β in the presence of fixed concentration of the ligands.12c
In a series of groundbreaking studies aimed at targeting D(R)BP–nucleic acid complexes, FP has been exploited for the identification (especially in HTS) of SMols that are able to interact with R(D)BP and/or nucleic acids and for studying their capability to destabilize their formed complexes. A worthy example is the application of FP for the identification of SMols able to interfere with the HuR–RNA complex, aimed at the development of novel anticancer therapeutic strategies. HuR is a RBP whose dysregulation is associated with immune disorders and various cancers, including breast, pancreatic, and glioblastoma.12 FP competitive experiments have been performed using an ad hoc synthesized truncated protein and TNFα fluorescently labeled mRNA. Several compounds have been identified from different research groups, thus suggesting the essential role of FP analysis for discovering new HuR–mRNA disruptors and anticancer agents.12d
The use of FP assays in HTS has been demonstrated across various studies for the purpose of identifying molecules capable of disrupting key molecular interactions. As an example, FP assays were applied to screen the National Cancer Institute’s diverse library and a separate library of approximately 6000 unique compounds, leading to the identification of agents that interfere with the HuR–mRNA complex and protein oligomerization.12e FP application was further expanded to pinpoint inhibitors of the STAT3 DNA-binding domain, a critical factor in tumor development, by utilizing a truncated version of STAT3 and a Biodipy–DNA conjugate.11,12f
These efforts collectively highlight the utility and efficiency of FP in identifying molecules that interfere with essential protein–RNA and protein–DNA interactions, contributing significantly to advancements in the development of new therapeutic drugs.
2.3. Fluorescence Intensity (FI)
Fluorescence intensity (FI) assay monitors changes in light output emitted by a fluorescent probe resulting from biochemical reactions or binding events involving the fluorescent molecule (Figure 3B). FI can be classified as either fluorogenic assays or fluorescence quench assays depending on the event being monitored. In fluorogenic assays, the reactants involved in the biochemical process are initially nonfluorescent. However, the reaction products formed are fluorescent, enabling their detection and quantification through an increase in the fluorescence intensity. This feature makes fluorogenic assays particularly useful in monitoring enzyme activities, where the reaction product exhibits fluorescence. Conversely, fluorescence quenching assays involve a substrate covalently linked to a fluorescent group. In its initial state, the fluorescence of this group is quenched or suppressed. Upon the occurrence of a specific biochemical event, such as the cleavage of the substrate, the fluorescent group is released. This release results in an observable increase in fluorescence intensity, thereby indicating the occurrence of a biochemical event. While FI assays offer several advantages, including ease of use and cost-effectiveness, one significant challenge is their sensitivity to fluorescent interference. This interference can arise from various sources, such as the color of the test compounds, the presence of organic fluorophores in assay buffers, or even the inherent fluorescence of the microplate used in the assay. These interferences can affect the accuracy and reliability of the fluorescence intensity measurements, necessitating careful consideration and control in the experimental design and interpretation. FI finds application in studying the modulation of protein–RNA interaction using SMols (Figure 5). Typically, RNA termini are modified with a fluorescent probe. A notable example is the development of a straightforward FI-based assay for detecting modulators of the oncogenic interaction between Lin28 and the let-7 microRNA. This approach was effectively adapted for HTS, leading to the discovery of pyrazolyl thiazolidinedione-type molecules as potent small-molecule inhibitors of protein–microRNA.14
Figure 5.

Hit compound identified as a modulator of RNA–protein interaction using the FI approach.
3. Circular Dichroism (CD)
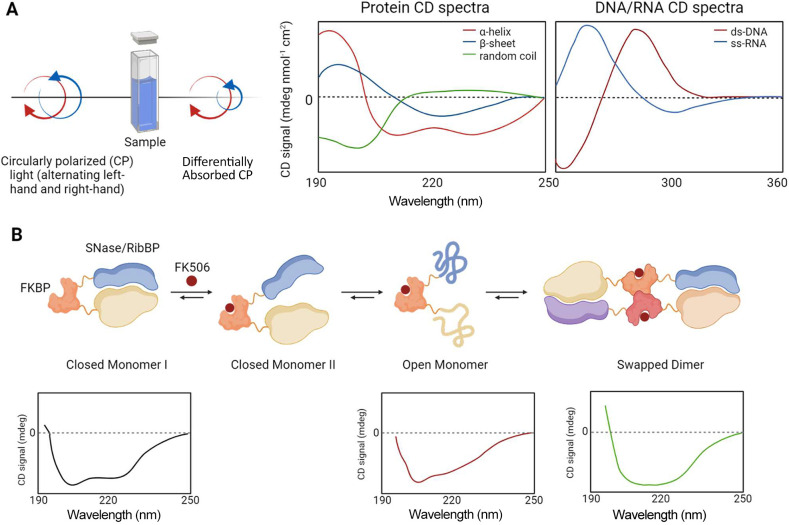
Circular Dichroism is an absorption spectroscopic technique based on the unique interaction of chiral molecules with circularly polarized light. Briefly, chiral substances, such as proteins and nucleic acids, absorb the left- and right-handed components of circularly polarized light to varying degrees due to their asymmetric structures. This disparity in absorption is measured across a range of wavelengths, yielding a CD spectrum (Figure 6A). For its ability to analyze chiral molecules, CD spectroscopy has a wide range of applications spanning chemistry to biochemistry and structural biology. Particularly, in biomolecular research, from CD spectra one can deduce the secondary structure composition of proteins, detect conformational changes, and assess protein stability and folding under varying conditions. In the near-UV region, CD provides tertiary structure insights, particularly around aromatic amino acids. For nucleic acids, CD aids in identifying distinct conformations like A-form, B-form, and Z-form DNA.15 Additionally, CD spectroscopy can yield information on the thermodynamics of molecular interactions, detect protein aggregates, and explore the kinetics of the binding processes. For these reasons, circular dichroism (CD) is particularly useful in revealing structural changes in proteins during protein–protein, protein–ligand, and protein–nucleic acid interactions since nucleic acids show strong CD signals in the near-UV range, where proteins absorb weakly. Not less important, chiral small molecules, which typically have a weak CD signal when free, may lead to significant changes in CD signals upon binding. This technique presents several advantages, including sensitivity to structural changes, minimal sample requirements, and the ability to perform analysis at various temperatures. However, interpreting CD spectra can be challenging, despite recent advances in analytical methods and dedicated databases that have significantly aided the analysis and interpretation of CD data for both proteins and nucleic acids (Figure 6B). The CD technique was successfully applied to the study of the protein conformational change that occurs in PPI during induced domain swapping (INDOS). Briefly, domain swapping occurs when two identical proteins exchange reciprocal segments to generate functional dimers. In INDOS, the domain swapping is induced by a small molecule. Thus, INDOS requires the fusion of two distinct proteins: a recognition protein and a target protein. The recognition protein is able to bind to a specific triggering molecule, leading to a domain swap with the target protein. In this pilot study, the recognition protein FKBP (able to bind to FK506 ligand) was fused with two distinct target bacterial nucleases (staphylococcal nuclease, SNase, and ribose binding protein - RibBP). Structural changes in the mutant complexes that occurred upon FK506 addition were monitored by using CD. A two-phase kinetic process involving unfolding and refolding was observed suggesting a first transition from a closed monomer to an open monomer in which the helical structure of the target protein is disrupted, followed by a second transition to the domain-swapped complex in which both the target and recognition protein are natively structured.16
Figure 6.
(A) Schematic representation of the CD principle (left) and representative Far-UV CD spectra of the characteristic secondary structure motifs detected in proteins: random coil (green), α-helix (red), β-sheet (blue), and representative Near-UV CD spectra of the secondary structure of ds-DNA (red) and ss-RNA (blue). (B) Depiction of the molecular events involved in induced domain swapping (INDOS), accompanied by representative CD spectra to illustrate the conformational changes in the proteins during this process.
Although CD spectroscopy is a valuable tool for studying macromolecule–macromolecule and macromolecule–ligand interactions, as it reveals the changes induced by these bonds, it is currently underutilized for studying the effect of SMols as modulators on the stability or conformation of macromolecule–macromolecule complexes. It could be important to explore this area further.
4. Isothermal Titration Calorimetry (ITC)
Isothermal titration calorimetry (ITC) is a biophysical technique based on measuring the heat released or absorbed during molecule−molecule interactions, thereby providing thermodynamic information about the binding process and thus allowing the thermodynamic characterization of interactive systems, such as protein–ligand interactions (Figure 8A). ITC operates using a power compensation mechanism to detect heat changes. The apparatus comprises two cells within an adiabatic chamber, both maintained at a constant temperature. One cell contains the protein solution, while the other is filled with a buffer or water. During the experiment, a ligand is incrementally introduced into the protein-containing cell via a needle, leading to heat absorption or release due to the interaction between the protein and ligand. The temperature change in this chamber is measured and compensated for by the instrument to maintain equilibrium between the two cells. The differential power applied to the protein cell, relative to the control cell, is continuously monitored over time, producing the raw ITC data, which are later processed to yield the ITC curve. ITC provides both stoichiometric and thermodynamic insights into binding events, aiding in the understanding of interaction mechanisms and supporting the validation of structural studies, even in the absence of detailed structural information.17 However, the heat signal generated from interactions can be subtle and sometimes difficult to detect, although this can be mitigated by using larger sample volumes. ITC data interpretation requires a solid understanding of thermodynamics. Moreover, due to the lengthy nature of the analysis, ITC is not ideally suited for HTS campaigns. Despite these challenges, ITC is a valuable and frequently employed tool in the study of biomolecular interactions, offering a detailed view of the thermodynamic forces driving these processes.18 ITC is a well-established technique in drug discovery for studying the binding between SMols and targets including proteins and nucleic acids. Although the ability of ITC to evaluate the influence of complex-binding modulator compounds on a variety of biomolecular complexes, including protein–protein, protein–DNA, and protein–small molecule interactions, was proved several years ago, its application in examining the influence of ligands on the stability of macromolecule–macromolecule binary complexes is underexplored.
Figure 8.
(A) Schematic drawing of an adiabatic microcalorimeter and resulting raw titration data and binding curve obtained from concentration-normalized integrated peak areas of raw data. In a typical experiment, the full binding curve was obtained by titrating the partner protein/peptide into the solution containing the counterpart protein of the binary PPI complex in the presence or absence of the small molecule or by titrating the Smol into a solution containing the PPI complex. (B) Illustrative application of ITC in studying the cooperative mechanism between PPARγ-cofactor-rosiglitazone accompanied by representative ITC curves obtained from the titration of rosiglitazone to PPARγ in the presence of various concentrations of MED1 (Adapted with permission from Pim J. de Vink et al. Chem. Sci.2022, 13, 2744–2752.19d Copyright 2022 Royal Society of Chemistry).
The first example dated in 2008 was the stabilization of the lysozyme–NAG3 complex by the LM11 compound, which highlighted the modulation of protein–small molecule interactions (Figure 7). The results indicated that NAG3′s binding affinity to lysozyme was enhanced 2-fold in the presence of the compound. Additionally, they identified a compound that targeted the interaction between porcine trypsin and turkey egg white trypsin inhibitor, as a model for protein–protein interaction.19a
Figure 7.
Hit compounds as modulators of PPIs and the DNA–protein complex identified by ITC.
Exploring the strategy of enhancing PPI through small molecule stabilizers, research focused on understanding the mechanism behind AMP’s regulation of the carbohydrate-responsive element-binding protein (ChREBP) via a direct PPI with the 14–3–3 protein. Using ITC, AMP was deciphered to act as an allosteric ligand, binding directly to the ChREBP-14–3–3 heterodimer and stabilizing it. The experiment involved titrating ChREBP 2-peptide into 14–3–3 in the absence and presence of AMP. This binding was specifically observed when the heterodimer was present, as neither (N)-ChREBP nor 14–3–3 alone exhibited binding to AMP under identical conditions. The ITC results were confirmed using FP as an orthogonal technique and further supported by crystallographic analysis which showed how AMP binds within a pocket at the interface of the complex, mildly stabilizing the protein–protein interaction.19b Similarly, the use of ITC facilitated the analysis and comparison of the thermodynamics characterizing the interaction between the ERα-peptide and 14–3–3σ proteins, specifically examining the effects of both covalent and noncovalent stabilizers on this interaction.19c
In another recent example, both FP and ITC were utilized for studying the cooperativity mechanism for small molecule modulation of nuclear receptors, influencing the interaction with coregulator proteins. Through FP titrations with varying concentrations of PPARγ and the ligand rosiglitazone, the synergistic effects on the binding of the ligand and coregulator to PPARγ were delineated. Specifically, PPARγ was titrated against a fixed concentration of an MED1 coregulator peptide across a rosiglitazone concentration gradient (0–200 mM), revealing how rosiglitazone alters the PPARγ/MED1 interaction dynamics. ITC was employed as an orthogonal method to confirm the affinities observed in the FP titrations. ITC experiments were conducted by titrating either rosiglitazone or tesaglitazar to PPARγ in the presence of MED1 (Figure 8B).19d The results of the ITC experiments provided an interesting alternative viewpoint on how the ligand and coregulator interact with the nuclear receptor.
5. Microscale Thermophoresis (MST)
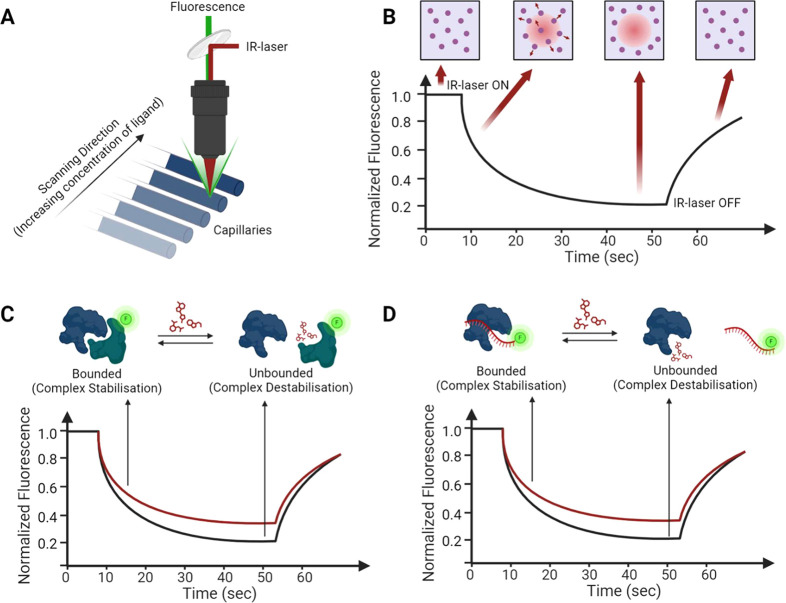
Microscale thermophoresis (MST) is a novel and robust biophysical technique to investigate the interactions between molecules. It relies on a very small temperature gradient to monitor how fast a fluorescent molecule (usually a protein) moves in a solution. The movement of the fluorescent molecule is influenced by whether it forms a complex with another molecule (usually a ligand). When the ligand binds, the fluorescent molecule slows down because its size, charge, shape, and hydration layer are altered. These changes affect how the fluorescent molecule reacts with the temperature gradient. By using a capillary and a laser, MST can measure the movement of the molecules and enables the measurement of a broad spectrum of interactions due to its high sensitivity to changes in molecular size, charge, and hydration shell (Figure 10A–D). Moreover, it is user-friendly, with low sample consumption and no limitations on molecular size or weight, and can be translated for HTS purposes. However, in most cases, a fluorescent labeling procedure is necessary if the macromolecule is not intrinsically fluorescent. One of the primary limitations of this technique is the intrinsic fluorescence of proteins rich in aromatic moieties, which can generate interfering signals. In such cases, the ligand may be labeled instead of the protein.18 However, the fluorescent moiety may occasionally interfere with protein–ligand binding, particularly if the ligand has a very low molecular weight. For these reasons, labeling the macromolecule is preferred. MST began to be used to study a wide range of protein interactions, such as protein–protein, protein–DNA, protein–RNA, protein–lipid, protein–small molecule, and protein–antibody interactions. While there are still few examples in literature, to the best of our knowledge, MST can also be used to screen for compounds that modulate PPI or D(R)PI, such as stabilizers or disruptors (Figure 9).
Figure 10.
(A) Overview of MST optics. MST measurements are conducted in capillaries holding approximately 4 μL. Fluorescence inside the capillary is both excited and detected using the same lens. A concentrated IR laser heats a specific sample volume, and the movement of fluorescent molecules through the induced temperature gradient, known as thermophoresis, is monitored. (B) Typical thermogram curve and MST experiment phases: initial uniform molecule distribution with steady fluorescence and IR laser activation, followed by thermophoretic molecule movement and fluorescence changes over 30 s, ending with molecule backdiffusion post IR laser deactivation. (C, D) Representative depiction of changes in the thermogram curve, indicative of the stabilization or destabilization of a PPI or a D(R)PI complex upon the addition of a small molecule as a modulator.
Figure 9.
Hit compound modulators of PPIs and the RNA–protein complex identified by MST.
In the context of the research for PPI modulators, a recent study exemplifies the utility of MST in fragment-based drug discovery, to investigate the ligandability of the 14–3–3/Amot-p130 interface, a key regulator of the Hippo signaling pathway, and to find SMols that can stabilize the complex and facilitate rational design. FP and other biophysical methods were combined with MST to identify and characterize SMols that modulate and stabilize the 14–3–3/Amot-p130 complex. In the FP assay, a solution containing a fixed concentration of the 14–3–3/Amot-p130 complex was titrated with fragments, in both the presence and absence of 14–3–3, to eliminate false-positive results. MST was used as an orthogonal assay, with conditions optimized for the 14–3–3η/Amot-p130 complex. MST data showed that the fragments had an affinity for the complex in the micromolar range, and the dose–response curves indicated a 2-fold stabilization effect on the 14–3–3/Amot-p130 complex.20a MST assay was employed to assess the interaction between the histone methyltransferase G9a, crucial for epigenetic regulation and cancer involvement, and the inhibitor BIX-01294. The assay consisted of preincubating G9a with its peptide substrate, β-H3, followed by titration with BIX-01294. The results demonstrated that increasing concentrations of β-H3 led to decreased apparent dissociation constant values for BIX-01294, indicating competitive binding to G9a.20b
MST showed its versatility and sensitivity in detecting and quantifying the interaction between proteins and nucleic acids. A recent study demonstrated how MST measures the affinity of epigenetic protein Df31 for nucleic acid fragments, including ssRNA, ssDNA, and dsDNA. The nucleic acid fragments were labeled with a Cy5 fluorescent dye to enable optical tracking during the assay. In the protein–nucleic acid MTS assay, this is the typical strategy adopted since the nucleic acids can be directly and easily derivatized with a fluorescent probe to the 5′ or 3′ ends. As an example, MST was employed for studying the disruption of the RNA–Fl-Rev complex by neomycin. The RNA utilized in this study was the human immunodeficiency virus (HIV) type 1 Rev response element (RRE) RNA, which is a known target of the Rev peptide. The interaction between the Rev peptide and RRE RNA is crucial for the translocation of viral mRNA into the nucleus, which is a process essential for HIV replication. The experimental procedure involved initial preincubation of RRE RNA with Gl-Rev, followed by the addition of neomycin to quantify its competitive displacement ability. In a subsequent experiment, RRE RNA and neomycin were first combined, followed by the addition of the peptide to assess competitive binding. Interestingly, MST was very recently exploited for studying the capability of SMols to interact and interfere with the RNA–RBP complex. Briefly, EXOSC3 is an RNA-binding structural cap protein that helps degrade RNA in the exosome. The binding of wtEXOSC3 and of some EXOSC3 disease-causing mutations to G-rich RNA sequences was measured using both surface plasmon resonance and MST, and the results guided a VS for the identification of potential SMols able to alter the RNA–EXOSC3 complex. Following docking, STD-NMR was used to test the ability of compounds to bind to EXOSC3, whereas ELISA and MST were exploited to assess the capability of the compounds to disrupt EXOSC3–RNA interactions. In the MST assay, EXOSC3 was incubated with fixed concentrations of the tested compounds and several concentrations of both long and short G-rich RNA. Compound ERD03 resulted in decreasing EXOSC3 binding to G-rich RNA in a dose-dependent manner.20c
6. NMR-Based Approach
Nuclear magnetic resonance (NMR) spectroscopy is a powerful and versatile technique extensively used for analyzing ligand-target binding processes in a wide affinity range, from nM to mM. To study ligand–protein complexes, NMR spectra are obtained on samples containing the ligand in molar excess, from 10:1 to 1000:1 with respect to target protein, according to the KD of the complex. NMR spectroscopy allows us to study these complexes focusing on the protein or the ligand resonances, respectively. More recently, NMR protein-based assays have been used for studying the effect of SMols on protein–protein interactions in the presence of selectively labeled proteins.
A ligand-based NMR approach (applied to both proton or fluorine nuclei) used the NOE effect or the analysis of transverse and longitudinal relaxation to identify the exchange between the bound and free state of a ligand.21 Competition ligand-based NMR experiments were developed to avoid many of the limitations associated with the ligand-based NMR experiments, such as the lack of information about the protein binding site. In these experiments, a chemical mixture or the new ligand under study can be tested against the biomolecule target in the presence of a weak- to medium-affinity ligand of known binding constant, referred to as the spy molecule or reporter. All the NMR observables of the spy molecule or of the analyzed ligand can be monitored (such as intensity of the NMR signals, intermolecular NOE, and transverse and longitudinal relaxation rates).
Saturation transfer difference (STD) NMR is a widely used ligand-based technique particularly useful to perform the interaction screening of compounds and fragments toward macromolecules and allows the identification of the so-called epitope, the moieties of the ligand involved in the binding to the target (the intensity of the signal is related to its proximity to the protein binding site).22 STD is also used for competition experiments (Figure 12) since a stronger binder (such as a natural ligand) can displace a weaker binder from the same binding site, interfering with its signals in the STD spectra. STD spectra allow us to describe in detail the ligand binding mode and to measure the KD, as demonstrated for two α-fucosylamide-based mimics of LewisX to the DC-SIGN extracellular domain. In the case of DC-SIGN, STD competition experiments in the presence of both the natural ligand LewisX(OMe) and the mimic confirmed a competition for the same binding site, evidencing a higher relative affinity of mimic in comparison to the natural ligand.23
Figure 12.
Competition NMR experiment. An example of an STD-NMR spectrum of the ligand (green peaks) complexed with the protein is reported. In the presence of a strong competitor, the reference’s signals decrease (or disappear) and the competitor’s STD signals increase (red peaks).
Other techniques can be considered as a robust complement to the STD method in the protein–ligand analysis, such as transferred-NOESY (tr-NOESY, a 2D-NOESY experiment relying on different tumbling times τc of free and bound molecules) which is useful to detect different conformations in the free and bound forms of the ligands and to establish if two ligands compete for the same binding site.13,24a In the case of artemisinin, a combined strategy of STD, tr-NOESY, and interligand NOEs for pharmacophore mapping (INPHARMA) experiments allows us to confirm that it can bind human and bovine serum albumin (HSA/BSA), competing for the binding sites of warfarin and ibuprofen (two common BSA/HSA binder drugs). On the contrary, in the case of two Cholera toxin variants, known for their ability to bind the oligosaccharide GM1 (the canonical binder present on the host membranes), STD and tr-NOESY allow the identification of a second binding site able to accommodate the histo blood group antigens (HBGA).24b
NMR spectroscopy can also be applied to investigate the effect of SMols on protein–protein complexes (Figure 11). An example is the screening of the interaction between a SMols library with a simplified system comprising FGF2 (fibroblast growth factor) and the FGFR-D2 domain (the D2 domain being a significant segment of the fibroblast growth factor receptor’s extracellular portion). This study evaluated the ability of selected members of the library to trigger the dissociation of protein–protein complexes. Observing the ligand, chemical shift, and line broadening of the SMols peaks (induced by the presence of the macromolecule) were used to confirm their specific interactions with the extracellular portion of the FGF2/FGFR system. From the protein point of view, NMR diffusion experiments demonstrated the capability of some ligands to disrupt FGF2/D2 assembly, and NMR HSQC-based experiments on 15N-labeled D2 or FGF2 proteins allowed the identification of preferential binding epitopes and allosteric sites.25a
Figure 11.
Compound modulators of PPIs identified by NMR.
To study the effect of antagonists on PPIs, the antagonist induced dissociation assay (AIDA)-NMR has been developed. This assay necessitates the use of two proteins with distinct molecular weights (a protein fragment larger than 30 kDa and a small reporter protein less than 20 kDa), with one labeled protein to serve as a molecular reporter. In the presence of a complex, the reporter exhibits increased transverse relaxation, leading to a diminished or absent NMR signal intensity. Notably, the PPI interaction spectrum undergoes significant alterations following the introduction of an interfering agent. This technique was successfully applied to monitor the influence of potential antagonists on the p53–MDM2 interaction and to study the disruption of the PD-1/PD-L1 complex (using a mutated 15N-labeled PD1), a critical target in cancer therapy. The AIDA-NMR study confirmed the fragments’ interaction with the complex, identifying SMols as potential disruptors of the PD1/PD-L1 complex.25b,25c
7. Concluding Remarks and Future Outlook
Modulation of macromolecule activity through SMols is the classical approach for finding novel therapeutic agents against several diseases. In the last 10 years, together with conventional SMols able to bind and modulate a specific target, new approaches have attracted the attention of both Academia and Industry researchers. The emerging modalities to design novel drugs include proteolysis-targeting chimera (PROTAC), molecular glues, and modulators of protein–protein and D(R)NA–protein complexes. To identify new modulators, in the earlier phases of the drug discovery process a key role is exerted by the evaluation of the molecules’ ability to bind the targets and therefore to modulate them. Several assays have been developed along the years to perform so-called binding studies, including the consolidated radioligand binding assays. The progresses in the technologies available for the study of biomolecular structure and intermolecular interactions led to the development of several biophysical techniques useful for studying the binding of SMols to selected target proteins and therefore to perform a preliminary screening.1 A literature recognition revealed that only a few of them have potential for studying ternary systems. Given the emerging chemical modalities, it is evident how important it is to have techniques that can evaluate the ability of SMols to interfere with macromolecular complexes. In this mini-review, we have highlighted the biophysical assays that can be used to screen molecules for this purpose. The goal is to provide a resource that is informative and practically beneficial for medicinal chemists working in this emerging and promising field. By providing comprehensive information about these biophysical techniques, we aim to enable researchers to choose the most suitable methodological approach for their specific research objectives. Our analysis of the literature shows that these techniques cannot be considered universal but rather should be selected based on the specific purposes of the research and on staging of the drug discovery process.
The employment of FRET, FP, and MST techniques in the early stages of the drug discovery process offers a synergistic approach that significantly enhances the efficiency and efficacy of identifying Hit compounds able to engage the target of interest. The initial phase of drug discovery requires methods that can rapidly and accurately assess the interaction between large libraries of compounds and potential targets. The fluorescence-based techniques provide researchers with powerful tools for HTS. Their adaptability to HTS formats allows for the screening of thousands of compounds against potential drug targets, facilitating the rapid identification of molecules with the desired biological activity and providing rapid feedback on the affinity and specificity of interactions. Additionally, these techniques provide quantitative data on binding affinities, kinetics, and thermodynamics, offering valuable insights on the mechanism of action and potential off-target effects, thus guiding further compound optimization. Importantly, thanks to their sensitivity and efficiency, the miniaturization of these methods makes effective use of resources, reducing the volume of reagents required and, consequently, the overall cost of screening, and enables the investigation of a wide range of targets, including challenging targets such as the herein discussed PPI and D(R)PI. However, these approaches come with inherent limitations as discussed above. Addressing these limitations through the parallel application of these techniques can compensate for individual weaknesses while taking advantage of the unique strengths of each method. Indeed, each technique provides different types of information: FP offers insights into binding affinity and specificity, MST sheds light on binding thermodynamics and kinetics, and FRET reveals proximity and conformational changes. Moreover, using these techniques in tandem allows for cross-validation of the results. For instance, a compound identified as a Hit by FP can be further examined by MST to confirm the binding interaction and elucidate the mechanism of action. This cross-validation process helps in filtering out false positives and significantly increases the Hit rate, allowing at the same time to focus on the most promising candidates.
Conversely, to gain detailed structural and binding information, beyond the identification of Hit compounds or the calculation of binding constants, techniques such as CD, ITC, and NMR spectroscopy are more informative. These techniques are particularly useful during the Hit-to-lead and lead optimization stages of the drug discovery process, as they offer in-depth insights into molecular interactions despite being slower and less high throughput. Although these advanced approaches have not been widely used to investigate macromolecular complexes and their susceptibility to SMol, they have gained increasing attention in recent years due to their high potential in medicinal chemistry and structural biology applications. Considering these factors, it is not possible to select a universal biophysical technique for studying the effect of SMol on macromolecular complexes. Each technique has its own advantages and limitations, allowing researchers to focus on specific aspects of complex molecular recognition events, particularly in the field of PPI and D(R)PI as targets. To gain a thorough understanding of biomolecular interactions, it is essential to combine various techniques. This integration enables high-throughput identification of potential modulators and detailed characterization of their binding mechanisms, structural basis, and thermodynamics. Cross-validation of results enhances the reliability of conclusions and facilitates the discovery of biologically relevant interactions.
In conclusion, the complex and dynamic nature of macromolecular complexes and how these might be affected and controlled by SMol necessitate an integrated interdisciplinary approach. By integrating various techniques, such as FRET, FP, MST, CD, ITC, and NMR, researchers can gain a complete understanding of macromolecular complexes as targets and how ligands can interact with them. This can accelerate the drug discovery process toward new therapeutic frontiers.
Acknowledgments
This work was supported by the European Union – PON R&I 2014-2020-Asse IV “Istruzione e Ricerca per il recupero–REACT-EU”, Azione IV.6 “Contratti di Ricerca su tematiche Green”.
Biographies
Martina Garbagnoli graduated in Industrial Pharmacy at the University of Pavia (IT) in 2021. She is currently a Ph.D. student in medicinal chemistry; her Ph.D. project is focused on the design and synthesis of new molecules able to interfere with the HuR–RNA complexes. She spent four months at the Helmholtz Institute for Pharmaceutical Research Saarland (HZI, DE).
Pasquale Linciano is Associate Professor at the Department of Drug Sciences of the University of Pavia (IT). He received his Ph.D. in Pharmaceutical Sciences from the University of Chieti-Pescara (IT) in 2014. He spent six years as a postdoctoral research fellow at the University of Modena (IT) where he collaborated on several European and AIRC projects. His interests mainly focus on the drug design and synthesis of compounds with anti-infective, anticancer, and neuroprotective activity.
Roberta Listro graduated in Pharmacy at the University of Palermo (IT) and obtained her Ph.D. in Chemical and Pharmaceutical Sciences in 2022 at the University of Pavia (IT). Her research activity is focused on the synthesis of new ligands and the studies of their interaction with target proteins. Her current research activity is mainly focused on the identification of anti-infective compounds.
Giacomo Rossino is a Type A Assistant Professor at the Department of Drug Sciences of the University of Pavia (IT). After graduating in Chemistry, he obtained his Ph.D. in Chemical and Pharmaceutical Sciences at the University of Pavia (IT). His research interests are in the field of medicinal chemistry, with a particular focus on multitarget ligands. His current research also aims to implement green chemistry techniques to reduce the environmental impact of the drug discovery process.
Francesca Vasile is Associate Professor at the Department of Chemistry of the University of Milan (IT) since 2020. After graduating in Pharmacy in 1998 at the University of Pisa (IT), she obtained her Ph.D. in biophysics in 2003 at the University of Genoa (IT). She was postdoc at University of Genoa and Milan (IT) for eight years, and then she worked at CISI (Interdepartmental Center of Biomolecular Study and Industrial Applications) of the University of Milan (IT). From 2017 to 2020 she was a researcher at Department of Chemistry of the University of Milan (IT). Her main research interests include the analysis of the interactions between macromolecules and small ligands applying ligand- and target-based NMR techniques. In particular, she studied the interaction of small molecules with soluble proteins, protein expressed on the membrane of living cells, and RNA fragments.
Simona Collina is Full Professor of Medicinal Chemistry at the Department of Drug Sciences of the University of Pavia (IT). She has 30 years of experience in drug design and synthesis of small molecules with therapeutic application, including the design and preparation of drug discovery libraries and the preparation and characterization of chiral compounds. During these years, she has established an interdisciplinary network of highly qualified national and international medicinal chemistry research groups with complementary expertise, including in silico design, biophysical techniques to study ligand–protein interaction, and in vitro and in vivo biological evaluation. Among the various research topics, the discovery of small molecules able to interact with ELAV protein/mRNA complexes, modulators of sigma receptors, and Quorum Sensing inhibitors are the most challenging.
The authors declare no competing financial interest.
References
- Du X.; Li Y.; Xia Y.-L.; Ai S.-M.; Liang J.; Sang P.; Ji X.-L.; Liu S.-Q. Insights into Protein-Ligand Interactions: Mechanisms, Models, and Methods. Int. J. Mol. Sci. 2016, 17 (2), 144. 10.3390/ijms17020144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gatiatulin A. K.; Ziganshin M. A.; Gorbatchuk V. V. Smart Molecular Recognition: From Key-to-Lock Principle to Memory-Based Selectivity. Front. Chem. 2020, 7, 933. 10.3389/fchem.2019.00933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ran X.; Gestwicki J. E. Inhibitors of Protein-Protein Interactions (PPIs): An Analysis of Scaffold Choices and Buried Surface Area. Curr. Opin. Chem. Biol. 2018, 44, 75–86. 10.1016/j.cbpa.2018.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cozzolino F.; Iacobucci I.; Monaco V.; Monti M. Protein-DNA/RNA Interactions: An Overview of Investigation Methods in the -Omics Era. J. Proteome Res. 2021, 20 (6), 3018–3030. 10.1021/acs.jproteome.1c00074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang D.; Lee Y.; Lee J.-S. RNA-Binding Proteins in Cancer: Functional and Therapeutic Perspectives. Cancers 2020, 12 (9), 2699. 10.3390/cancers12092699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace B.; Atzberger P. J. Förster Resonance Energy Transfer: Role of Diffusion of Fluorophore Orientation and Separation in Observed Shifts of FRET Efficiency. PloS One 2017, 12 (5), e0177122 10.1371/journal.pone.0177122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leavesley S. J.; Rich T. C. Overcoming Limitations of FRET Measurements. Cytom. Part J. Int. Soc. Anal. Cytol. 2016, 89 (4), 325–327. 10.1002/cyto.a.22851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- a Costantino L.; Ferrari S.; Santucci M.; Salo-Ahen O. M. H.; Carosati E.; Franchini S.; Lauriola A.; Pozzi C.; Trande M.; Gozzi G.; Saxena P.; Cannazza G.; Losi L.; Cardinale D.; Venturelli A.; Quotadamo A.; Linciano P.; Tagliazucchi L.; Moschella M. G.; Guerrini R.; Pacifico S.; Luciani R.; Genovese F.; Henrich S.; Alboni S.; Santarem N.; da Silva Cordeiro A.; Giovannetti E.; Peters G. J.; Pinton P.; Rimessi A.; Cruciani G.; Stroud R. M.; Wade R. C.; Mangani S.; Marverti G.; D’Arca D.; Ponterini G.; Costi M. P. Destabilizers of the Thymidylate Synthase Homodimer Accelerate Its Proteasomal Degradation and Inhibit Cancer Growth. eLife 2022, 11, e73862 10.7554/eLife.73862. [DOI] [PMC free article] [PubMed] [Google Scholar]; b Hardebeck S.; Schreiber S.; Adick A.; Langer K.; Jose J. A FRET-Based Assay for the Identification of PCNA Inhibitors. Int. J. Mol. Sci. 2023, 24 (14), 11858. 10.3390/ijms241411858. [DOI] [PMC free article] [PubMed] [Google Scholar]; c Cao H.; Tamilarasu N.; Rana T. M. Orientation and Affinity of HIV-1 Tat Fragments in Tat-TAR Complex Determined by Fluorescence Resonance Energy Transfer. Bioconjugate Chem. 2006, 17 (2), 352–358. 10.1021/bc050277u. [DOI] [PubMed] [Google Scholar]
- a Dale N. C.; Johnstone E. K. M.; White C. W.; Pfleger K. D. G. NanoBRET: The Bright Future of Proximity-Based Assays. Front Bioeng Biotechnol 2019, 7, 56. 10.3389/fbioe.2019.00056. [DOI] [PMC free article] [PubMed] [Google Scholar]; b Farmer J. P.; Mistry S. N.; Laughton C. A.; Holliday N. D. Development of Fluorescent Peptide G Protein-Coupled Receptor Activation Biosensors for NanoBRET Characterization of Intracellular Allosteric Modulators. FASEB J. 2022, 36 (11), e22576 10.1096/fj.202201024R. [DOI] [PubMed] [Google Scholar]; c Cannaert A.; Storme J.; Franz F.; Auwärter V.; Stove C. P. Detection and Activity Profiling of Synthetic Cannabinoids and Their Metabolites with a Newly Developed Bioassay. Anal. Chem. 2016, 88 (23), 11476–11485. 10.1021/acs.analchem.6b02600. [DOI] [PubMed] [Google Scholar]
- Du Y. Fluorescence Polarization Assay to Quantify Protein-Protein Interactions in an HTS Format. Methods Mol. Biol. Clifton NJ. 2015, 1278, 529–544. 10.1007/978-1-4939-2425-7_35. [DOI] [PubMed] [Google Scholar]
- Hall M. D.; Yasgar A.; Peryea T.; Braisted J. C.; Jadhav A.; Simeonov A.; Coussens N. P. Fluorescence Polarization Assays in High-Throughput Screening and Drug Discovery: A Review. Methods Appl. Fluoresc. 2016, 4 (2), 022001 10.1088/2050-6120/4/2/022001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- a Wendt M. D. Discovery of ABT-263, a Bcl-Family Protein Inhibitor: Observations on Targeting a Large Protein-Protein Interaction. Expert Opin. Drug Discovery 2008, 3 (9), 1123–1143. 10.1517/17460441.3.9.1123. [DOI] [PubMed] [Google Scholar]; b Sijbesma E.; Somsen B. A.; Miley G. P.; Leijten-van de Gevel I. A.; Brunsveld L.; Arkin M. R.; Ottmann C. Fluorescence Anisotropy-Based Tethering for Discovery of Protein-Protein Interaction Stabilizers. ACS Chem. Biol. 2020, 15 (12), 3143–3148. 10.1021/acschembio.0c00646. [DOI] [PMC free article] [PubMed] [Google Scholar]; c Sijbesma E.; Visser E.; Plitzko K.; Thiel P.; Milroy L.-G.; Kaiser M.; Brunsveld L.; Ottmann C. Structure-Based Evolution of a Promiscuous Inhibitor to a Selective Stabilizer of Protein-Protein Interactions. Nat. Commun. 2020, 11 (1), 3954. 10.1038/s41467-020-17741-0. [DOI] [PMC free article] [PubMed] [Google Scholar]; d Della Volpe S.; Linciano P.; Listro R.; Tumminelli E.; Amadio M.; Bonomo I.; Elgaher W. a. M.; Adam S.; Hirsch A. K. H.; Boeckler F. M.; Vasile F.; Rossi D.; Collina S. Identification of N,N-Arylalkyl-Picolinamide Derivatives Targeting the RNA-Binding Protein HuR, by Combining Biophysical Fragment-Screening and Molecular Hybridization. Bioorganic Chem. 2021, 116, 105305 10.1016/j.bioorg.2021.105305. [DOI] [PubMed] [Google Scholar]; e Wu X.; Lan L.; Wilson D. M.; Marquez R. T.; Tsao W.-C.; Gao P.; Roy A.; Turner B. A.; McDonald P.; Tunge J. A.; Rogers S. A.; Dixon D. A.; Aubé J.; Xu L. Identification and Validation of Novel Small Molecule Disruptors of HuR-mRNA Interaction. ACS Chem. Biol. 2015, 10 (6), 1476–1484. 10.1021/cb500851u. [DOI] [PMC free article] [PubMed] [Google Scholar]; f Shih P.-C.; Yang Y.; Parkinson G. N.; Wilderspin A.; Wells G. A High-Throughput Fluorescence Polarization Assay for Discovering Inhibitors Targeting the DNA-Binding Domain of Signal Transducer and Activator of Transcription 3 (STAT3). Oncotarget 2018, 9 (66), 32690–32701. 10.18632/oncotarget.26013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasile F.; Rossi D.; Collina S.; Potenza D. Diffusion-Ordered Spectroscopy and Saturation Transfer Difference NMR Spectroscopy Studies of Selective Interactions between ELAV Protein Fragments and an mRNA Target. Eur. J. Org. Chem. 2014, 2014 (29), 6399–6404. 10.1002/ejoc.201403014. [DOI] [Google Scholar]
- Byun W. G.; Lim D.; Park S. B. Discovery of Small-Molecule Modulators of Protein-RNA Interactions by Fluorescence Intensity-Based Binding Assay. Chembiochem Eur. J. Chem. Biol. 2020, 21 (6), 818–824. 10.1002/cbic.201900467. [DOI] [PubMed] [Google Scholar]
- Martin S. R.; Schilstra M. J. Circular Dichroism and Its Application to the Study of Biomolecules. Methods Cell Biol. 2008, 84, 263–293. 10.1016/S0091-679X(07)84010-6. [DOI] [PubMed] [Google Scholar]
- Wallace B. A.; Janes R. W. Synchrotron Radiation Circular Dichroism (SRCD) Spectroscopy: An Enhanced Method for Examining Protein Conformations and Protein Interactions. Biochem. Soc. Trans. 2010, 38 (4), 861–873. 10.1042/BST0380861. [DOI] [PubMed] [Google Scholar]
- Saponaro A. Isothermal Titration Calorimetry: A Biophysical Method to Characterize the Interaction between Label-Free Biomolecules in Solution. Bio-Protoc. 2018, 8 (15), e2957 10.21769/BioProtoc.2957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jerabek-Willemsen M.; Wienken C. J.; Braun D.; Baaske P.; Duhr S. Molecular Interaction Studies Using Microscale Thermophoresis. Assay Drug Dev. Technol. 2011, 9 (4), 342–353. 10.1089/adt.2011.0380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- a Cai Z.; Greene M. I.; Berezov A. Modulation of Biomolecular Interactions with Complex-Binding Small Molecules. Methods San Diego Calif 2008, 46 (1), 39–46. 10.1016/j.ymeth.2008.05.008. [DOI] [PubMed] [Google Scholar]; b Sato S.; Jung H.; Nakagawa T.; Pawlosky R.; Takeshima T.; Lee W.-R.; Sakiyama H.; Laxman S.; Wynn R. M.; Tu B. P.; MacMillan J. B.; De Brabander J. K.; Veech R. L.; Uyeda K. Metabolite Regulation of Nuclear Localization of Carbohydrate-Response Element-Binding Protein (ChREBP): ROLE OF AMP AS AN ALLOSTERIC INHIBITOR. J. Biol. Chem. 2016, 291 (20), 10515–10527. 10.1074/jbc.M115.708982. [DOI] [PMC free article] [PubMed] [Google Scholar]; c Konstantinidou M.; Visser E. J.; Vandenboorn E.; Chen S.; Jaishankar P.; Overmans M.; Dutta S.; Neitz R. J.; Renslo A. R.; Ottmann C.; Brunsveld L.; Arkin M. R. Structure-Based Optimization of Covalent, Small-Molecule Stabilizers of the 14–3-3σ/ERα Protein-Protein Interaction from Nonselective Fragments. J. Am. Chem. Soc. 2023, 145 (37), 20328–20343. 10.1021/jacs.3c05161. [DOI] [PMC free article] [PubMed] [Google Scholar]; d De Vink P. J.; Koops A. A.; D’Arrigo G.; Cruciani G.; Spyrakis F.; Brunsveld L. Cooperativity as Quantification and Optimization Paradigm for Nuclear Receptor Modulators. Chem. Sci. 2022, 13 (9), 2744–2752. 10.1039/D1SC06426F. [DOI] [PMC free article] [PubMed] [Google Scholar]
- a Centorrino F.; Andlovic B.; Cossar P.; Brunsveld L.; Ottmann C. Fragment-Based Exploration of the 14–3-3/Amot-P130 Interface. Curr. Res. Struct. Biol. 2022, 4, 21–28. 10.1016/j.crstbi.2021.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]; b Seidel S. A. I.; Dijkman P. M.; Lea W. A.; van den Bogaart G.; Jerabek-Willemsen M.; Lazic A.; Joseph J. S.; Srinivasan P.; Baaske P.; Simeonov A.; Katritch I.; Melo F. A.; Ladbury J. E.; Schreiber G.; Watts A.; Braun D.; Duhr S. Microscale Thermophoresis Quantifies Biomolecular Interactions under Previously Challenging Conditions. Methods San Diego Calif 2013, 59 (3), 301–315. 10.1016/j.ymeth.2012.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]; c François-Moutal L.; Jahanbakhsh S.; Nelson A. D. L.; Ray D.; Scott D. D.; Hennefarth M. R.; Moutal A.; Perez-Miller S.; Ambrose A. J.; Al-Shamari A.; Coursodon P.; Meechoovet B.; Reiman R.; Lyons E.; Beilstein M.; Chapman E.; Morris Q. D.; Van Keuren-Jensen K.; Hughes T. R.; Khanna R.; Koehler C.; Jen J.; Gokhale V.; Khanna M. A Chemical Biology Approach to Model Pontocerebellar Hypoplasia Type 1B (PCH1B). ACS Chem. Biol. 2018, 13 (10), 3000–3010. 10.1021/acschembio.8b00745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quintana J. I.; Atxabal U.; Unione L.; Ardá A.; Jiménez-Barbero J. Exploring Multivalent Carbohydrate-Protein Interactions by NMR. Chem. Soc. Rev. 2023, 52 (5), 1591–1613. 10.1039/D2CS00983H. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monaco S.; Ramírez-Cárdenas J.; Carmona A. T.; Robina I.; Angulo J. Inter-Ligand STD NMR: An Efficient 1D NMR Approach to Probe Relative Orientation of Ligands in a Multi-Subsite Protein Binding Pocket. Pharm. Basel Switz. 2022, 15 (8), 1030. 10.3390/ph15081030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guzzi C.; Angulo J.; Doro F.; Reina J. J.; Thépaut M.; Fieschi F.; Bernardi A.; Rojo J.; Nieto P. M. Insights into Molecular Recognition of Lewis(X) Mimics by DC-SIGN Using NMR and Molecular Modelling. Org. Biomol. Chem. 2011, 9 (22), 7705–7712. 10.1039/c1ob05938f. [DOI] [PubMed] [Google Scholar]
- a Vasile F.; Reina J. J.; Potenza D.; Heggelund J. E.; Mackenzie A.; Krengel U.; Bernardi A. Comprehensive Analysis of Blood Group Antigen Binding to Classical and El Tor Cholera Toxin B-Pentamers by NMR. Glycobiology 2014, 24 (8), 766–778. 10.1093/glycob/cwu040. [DOI] [PubMed] [Google Scholar]; b Vasile F.; Panigada M.; Siccardi A.; Potenza D.; Tiana G. A Combined NMR Computational Study of the Interaction between Influenza Virus Hemagglutinin and Sialic Derivatives from Human and Avian Receptors on the Surface of Transfected Cells. Int. J. Mol. Sci. 2018, 19 (5), 1267. 10.3390/ijms19051267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- a Pagano K.; Listro R.; Linciano P.; Rossi D.; Longhi E.; Taraboletti G.; Molinari H.; Collina S.; Ragona L. Identification of a Novel Extracellular Inhibitor of FGF2/FGFR Signaling Axis by Combined Virtual Screening and NMR Spectroscopy Approach. Bioorganic Chem. 2023, 136, 106529 10.1016/j.bioorg.2023.106529. [DOI] [PubMed] [Google Scholar]; b Krajewski M.; Rothweiler U.; D’Silva L.; Majumdar S.; Klein C.; Holak T. A. An NMR-Based Antagonist Induced Dissociation Assay for Targeting the Ligand-Protein and Protein-Protein Interactions in Competition Binding Experiments. J. Med. Chem. 2007, 50 (18), 4382–4387. 10.1021/jm070365v. [DOI] [PubMed] [Google Scholar]; c Musielak B.; Janczyk W.; Rodriguez I.; Plewka J.; Sala D.; Magiera-Mularz K.; Holak T. Competition NMR for Detection of Hit/Lead Inhibitors of Protein-Protein Interactions. Mol. Basel Switz. 2020, 25 (13), 3017. 10.3390/molecules25133017. [DOI] [PMC free article] [PubMed] [Google Scholar]