Abstract

Investigating the interaction between liposomes and proteins is of paramount importance in the development of liposomal formulations with real potential for bench-to-bedside transfer. Upon entering the body, proteins are immediately adsorbed on the liposomal surface, changing the nanovehicles’ biological identity, which has a significant impact on their biodistribution and pharmacokinetics and ultimately on their therapeutic effect. Albumin is the most abundant plasma protein and thus usually adsorbs immediately on the liposomal surface. We herein report a comprehensive investigation on the adsorption of model protein bovine serum albumin (BSA) onto liposomal vesicles containing the zwitterionic lipid 1,2-dipalmitoyl-sn-glycero-3-phosphocholine (DPPC), in combination with either cholesterol (CHOL) or the cationic lipid 1,2-dioleoyl-3-trimethylammoniumpropane (DOTAP). While many studies regarding protein adsorption on the surface of liposomes with different compositions have been performed, to the best of our knowledge, the differential responses of CHOL and DOTAP upon albumin adsorption on vesicles have not yet been investigated. UV–vis spectroscopy and sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) revealed a strong influence of the phospholipid membrane composition on protein adsorption. Hence, it was found that DOTAP-containing vesicles adsorb proteins more robustly but also aggregate in the presence of BSA, as confirmed by DLS and TEM. Separation of liposome–protein complexes from unadsorbed proteins performed by means of centrifugation and size exclusion chromatography (SEC) was also investigated. Our results show that neither method can be regarded as a golden experimental setup to study the protein corona of liposomes. Yet, SEC proved to be more successful in the separation of unbound proteins, although the amount of lipid loss upon liposome elution was higher than expected. In addition, coarse-grained molecular dynamics simulations were employed to ascertain key membrane parameters, such as the membrane thickness and area per lipid. Overall, this study highlights the importance of surface charge and membrane fluidity in influencing the extent of protein adsorption. We hope that our investigation will be a valuable contribution to better understanding protein–vesicle interactions for improved nanocarrier design.
1. Introduction
As current advances in nanotechnology and materials science allow for the continuous development of complex drug carriers, often with multifunctional purposes, there has been an increasing interest in describing the exact details of nanovehicle selective responses to specific biological microenvironments.1 Lipid nanocarriers such as liposomes are currently regarded as one of the most promising transport systems for precise drug delivery.2 Yet, basic aspects regarding the interactions between liposomes and blood components, which play a major role in nanoparticle stability and kinetic properties, are often not exhaustively described.3
Upon entering a biological environment, liposomes become coated by a dynamic layer of proteins, known as protein corona, which often controls particle transport properties.4 This topic has stimulated a lot of discussions in the scientific community.5−7 The proteins attached to the liposomes upon entering the body change the biological identity of the nanovehicles, and therefore, their stability, biodistribution, and pharmacokinetics are changed as well. There are situations when liposomes, but not limited to, are quickly opsonized and cleared by macrophages, thereby limiting their circulation time.8 The composition of the protein corona is affected by a plethora of factors including vesicle size, charge, and lipid composition.9 In this context, stealth liposomes prepared by coating the vesicles with polymers and polysaccharides are a strategy used in the development of liposomal formulations for stability improvement. Yet, the use of molecules that are incorporated inside the bilayer without coating with macromolecules is equally reported.10,11 For this work, we chose to investigate the stability of the prepared liposomes by incorporating various amounts of CHOL or DOTAP followed by their behavior in the presence of albumin. Although not the subject of the current investigations, it is worth mentioning that not all adsorbed proteins have a negative impact on the stability of liposomes and their circulation in the bloodstream.6 Therefore, exploring the tumor-targeting ability of proteins coating liposomes is a strategy for improving the accumulation of drug-loaded liposomes at the tumor site. In addition, the protein shell(s) can act as additional controlling layers of drug release behavior, thus improving the ability of carrier-controlled release. Albumins are the most abundant soluble components in plasma, having many physiological functions, including binding and transport of a wide variety of compounds. Since albumin is usually one of the most frequently identified proteins in the protein coronas of nanocarriers,12 understanding the driving forces behind albumin and lipid vesicle interaction could contribute to novel nanocarrier design for improved drug delivery and reduced immune recognition. Notably, it has been recently demonstrated that nanocarrier surface chemistry can be modulated to either stabilize or denature the structure of the adsorbed albumin. This modulation will then either promote immune evasion or lead to nonspecific clearance in vivo.13 In addition, conflicting evidence exists in terms of the types of molecular forces involved in albumin stabilization at the liposomal surface. For example, Yokouchi et al. found that albumin adsorbed on uncharged DPPC and negatively charged DPPC/DPPG mixed liposomes, but not on pure negatively charged DPPG vesicles or positively charged DPPC/SA or DPPC/DCP liposomes.14 They concluded that the hydrophobic interaction is most likely the driving force behind liposome-BSA stabilization. Kristensen et al. also propose that albumin prefers to interact with liposomal hydrophobic domains, exposed through packing defects.15 Tretiakova et al., on the other hand, found that albumin adsorption on liposomes with various compositions primarily affects lipid polar headgroups by disrupting surface water hydrogen-bonding networks, leading to the formation of novel hydrogen bonds between lipid headgroups and the protein surface.16 Albumin adsorption to DMPC vesicles has also been described as being governed by electrostatic interactions by the work of Sabín et al., although hydrophobic interactions, mainly due to bilayer core penetration by the protein, are also acknowledged.17
For preparation of liposomes with marketable value, long-term stability is critical.18 One of the key factors affecting this property is the degree of unsaturation of the carbon chain. It is well acknowledged that unsaturation is prone to oxidation and thus to degradation. Therefore, liposomes made of natural phospholipids with a high unsaturation degree are expected to have a shorter life. To address this limitation, phospholipids with as low a degree of unsaturation as possible would be a more suitable choice for providing liposomes with a high level of clinical translation.19,20 Indeed, there are already commercially approved liposomes made of saturated phospholipids, such as AmBisome, Lipo-Dox, or with only one unsaturated bond in the carbon chain, such as Visudyne.19 Considering this, we selected the saturated carbon chain-containing DPPC as the main lipid in the composition of the prepared liposomes in the current study.
Cholesterol (CHOL) is the most abundant lipid species in animal cell membranes, with a typical concentration of 20–30 mol %, and up to 70 mol %, depending on cell type.21 Numerous studies have shown that the presence of CHOL in bilayer structures can induce important structural changes, which can have an impact on protein binding. This includes increased headgroup spacing and hydration,22 increased hydrophobicity in the membrane core,23 increased bilayer thickness,24 as well as increased nanocarrier stability through membrane fluidity modulation.25 The ability of CHOL to interfere with phospholipid packing has also been demonstrated to influence drug encapsulation by directly affecting membrane permeability.26 In POPC/POPS membranes, CHOL was found to modulate the binding of a retroviral Gag protein by inducing structural changes in the bilayer which promote electrostatic interactions between protein lysine residues and lipid headgroups.27 However, the authors did not find any evidence of a direct interaction between the protein and CHOL, thus excluding hydrophobic contact stabilization, and postulated that the observed enhanced protein binding is caused by CHOL’s ability to affect the electrostatic and solvation properties of the membrane while condensing the membrane area.
DOTAP is one of the most widely used cationic lipids in liposomal transfection reagents due to its ability to form stable complexes with nucleic acids (called lipoplexes) and fuse with the cell membrane to release its encapsulated cargo in the cytosol.28 In addition, many DOTAP-containing liposomes are currently under clinical investigation for cancer gene therapy.9 DOTAP incorporation into liposomes has been shown to increase membrane fluidity when compared to CHOL29 and improve nanocarrier uptake in cell cultures through direct binding to negatively charged cell membranes.30 Coating cationic lipoplexes with albumin was shown to improve transfection efficiency in a dose-dependent fashion,31 although the exact details of albumin binding to vesicles were not described. A subsequent investigation on the differential binding behavior of BSA to CHOL and DOTAP, as well as other cationic or negatively charged lipids, revealed that BSA attaches to lipids via both hydrophilic and hydrophobic interactions, with similar overall binding constants.32 The study also showed that DOTAP alone could induce BSA partial protein unfolding, while CHOL stabilized protein structure apart from controlling the stiffness of the membrane, which is another critical parameter for liposomal formulations and for carriers in general.33,34 CHOL incorporation in lipoplexes has been shown to decrease serum protein adsorption35 and enhance the resistance of lipoplexes to serum-induced aggregation.36
While many studies regarding protein adsorption on the surface of liposomes with different compositions have been performed, to the best of our knowledge, the differential behavior of CHOL and DOTAP upon albumin adsorption on vesicles has not yet been investigated. We herein report the interaction of model protein BSA with liposomal vesicles containing the zwitterionic lipid DPPC and either CHOL or the cationic lipid DOTAP in a systematic investigation that uses a combined spectroscopic and electrophoretic approach. This investigation aims to bring a significant contribution to a better understanding of the underlying forces behind the protein–vesicle interaction for improved nanocarrier design.
2. Experimental Section
2.1. Materials
Cholesterol (CHOL, >98%), 1,2-dipalmitoyl-sn-glycero-3-phosphocholine (DPPC, >99%), and 1,2-dioleoyl-3-trimethylammonium-propane (DOTAP, >99%) were purchased from Avanti Polar Lipids, Inc. (Alabaster, AL). Aqueous acrylamide and bis(acrylamide) stock solution (30%, 29:1), bovine serum albumin (BSA) fraction V (≥97%), 2-mercaptoethanol (≥99%), sodium dodecyl sulfate (SDS, ≥99%), 1,2-bis(dimethylamino)-ethane, N,N,N′,N′-tetramethylethylenediamine (TEMED, ≥97%), and tris(hydroxymethyl)aminomethane hydrochloride (Tris-HCl, >99%) were obtained from Carl Roth GmbH & Co. KG (Karlsruhe, Germany). Disodium hydrogen phosphate (Na2HPO4, ≥99%), potassium dihydrogen phosphate (KH2PO4, ≥99%), anhydrous iron chloride(III) (FeCl3, ≥98%), glycerol (≥99%), ammonium peroxodisulfate (APS, ≥98%), ammonium thiocyanate (NH4SCN, ≥98%), Tris base (>99%), methanol (99.8%), and acetic acid (99.5%) were bought from Merck KGaA (Darmstadt, Germany). Sodium chloride (NaCl, 99.97%) and potassium chloride (KCl, 100%) were purchased from Lach-Ner, s.r.o. (Neratovice, Czech Republic). Molecular weight standards for electrophoresis, bromphenol blue, Coomassie Blue, as well as the protein assay dye reagent concentrate for protein quantification were obtained from Bio-Rad Laboratories, Inc. (Hercules, CA). Sepharose CL-4B was purchased from Cytiva (Marlborough, MA) and washed with fresh buffer prior to use. All other reagents and solvents were purchased in the highest available purity and used as received. Unless otherwise stated, all aqueous solutions were prepared using water purified though a Barnstead GenPure Pro water purification system (Thermo Fisher Scientific, Waltham, MA) and filtered through 0.22 μm pore poly(ether sulfone) (PES) membranes. Buffer compositions were as follows: phosphate-buffered saline (PBS): 8.54 mM Na2HPO4, 1.46 mM KH2PO4, 2.7 mM KCl, and 137 mM NaCl; phosphate buffer (PB): 8.54 mM Na2HPO4, 1.46 mM KH2PO4. SDS-PAGE sample loading buffer: 2% SDS, 10% glycerol, 5% 2-mercaptoethanol, 0.01% bromophenol blue, and 62.5 mM Tris-HCl, pH 6.8. Nucleopore track-etched hydrophilic membranes for extrusion were purchased from Cytiva.
2.2. Liposomal Preparation
Liposomes were generated through thin-film hydration (TFH),37 followed by sonication and extrusion. In brief, lipids were dissolved in chloroform, and the solution was slowly evaporated under reduced pressure. Lipid films were kept under vacuum and the system was flushed with N2 to remove any traces of organic solvent. Dry lipid films were hydrated for 1h with 10 mM PBS (pH 7.4) or 10 mM PB (pH 7.4) at 50 °C. The final lipid concentration in aqueous suspension was 7.5 mM. Liposomal formulations were probe sonicated at 26 kHz at 3% amplitude for 15 min using an ultrasonic homogenizer (UP200Ht, 200W, Hielscher Ultrasonics, Teltow, Germany, equipped with a S26d2 sonotrode) and extruded 11 times through 200 nm pore size polycarbonate membranes using a Mini-Extruder (Avanti Polar Lipids, Inc.), at 55 °C on a heating plate. All prepared formulations were stored at 4 °C for a maximum of one month.
2.3. Vesicle Characterization
Particle size and size distribution were determined through dynamic light scattering (DLS) by using an Amerigo instrument (Cordouan Technologies, Pessac, France). Zeta (ζ) potential was measured by using the same instrument through Laser Doppler Electrophoresis (LDE). All measurements were performed at 25 °C, with appropriate dilutions for ζ-potential determinations.
TEM images were acquired by using a Hitachi HT 7000 Transmission Electron Microscope (Hitachi Ltd. Tokyo, Japan) operating at 100 kV and equipped with a high-resolution 8-Megapixel CCD camera. For analysis, 200 μL of each liposomal suspension were mixed with 1200 μL of ultrapure water and 1 μL of 4% osmium tetroxide (OsO4). Samples were incubated at 4 °C for 60 min and then centrifuged at 12,000g for 5 min. The supernatant was removed, and the liposomes were resuspended in 200 μL of ultrapure water. Five μL of this liposomal suspension was deposited on TEM grid support films of Carbon 400 mesh. After 2–5 min, the excess liquid was removed with a filter paper and the sample was allowed to dry at room temperature.
The number of lipids per liposome was approximated from the size determined through DLS, with liposomes being regarded as spherical and with a unimodal size distribution, with a mean bilayer thickness and cross-sectional area calculated from 30 ns molecular dynamics production simulations of bilayer sections of the appropriate lipid composition.
2.4. Protein Adsorption
Liposomes (7.5 mM, 500 μL) were incubated for 1 h at 37 °C and 250 rpm with BSA prepared in either PB or PBS (10 mg/mL, 100 μL), with a total incubation volume of 1 mL.
2.4.1. Separation of Protein-Coated Liposomes from Unbound Protein through Centrifugation
The liposome–protein assemblies were recovered upon centrifugation for 30 min at 29,000g at 4 °C, and the obtained pellet was resuspended and centrifuged again in fresh buffer to remove loosely bound proteins. The final volume of resuspended pellet varied between 50 and 90 μL. This procedure was repeated three times. Centrifugation protocol was adapted from elsewhere.38
Direct quantification of adsorbed protein was performed through SDS-PAGE electrophoresis using 10% resolving and 5% stacking polyacrylamide gels from the final resuspended pellet. Electrophoresis was performed at constant voltage (110 V) in a Mini-PROTEAN Tetra Cell using a PowerPac Basic Power supply (Bio-Rad Laboratories, Inc.) until the dye front reached the end of the gel. Staining was performed overnight with Coomassie Brilliant Blue R-250, while destaining was done with a mixture of 50:10:40 (v/v/v) CH3OH/CH3COOH/H2O. Imaged gels were processed using ImageJ (NIH), with band intensities being determined through densitometric analysis. SDS-PAGE protein separation protocol was adapted from elsewhere.39 Protein binding (PB) values for SDS-PAGE experiments are calculated as grams of protein per mole of lipid following phospholipid quantification from final pellets.
Indirect quantification of adsorbed protein was performed through UV–Vis spectroscopy on a Shimadzu 1900i apparatus, using a modified Bradford assay.40 In brief, aliquots from the supernatant from the first centrifugation (20 μL) were incubated with 1.8 mL of protein assay dye for at least 15 min and absorbance was read at 595 nm. Calibration curve was linear in the range of 0–800 μg/mL BSA (R2 = 0.993). Determinations were made in triplicate for at least three incubation experiments. DOTAP was found to interfere with the assay; therefore, incubation blanks, without any protein, were also performed for each liposomal formulation. Supernatants from the first centrifugation of each incubation blank were used as references. CHOL did not interfere with the assay. PB values for UV–Vis experiments are calculated as gram protein per mole lipid following phospholipid quantification from the incubated suspension.
2.4.2. Separation of Protein-Coated Liposomes from Unbound Protein through Size Exclusion Chromatography (SEC)
Eight hundred μL of liposome–protein mixtures was loaded onto a Sepharose CL-4B column (20 cm × 1.5 cm) equilibrated with either PBS or PB, depending on the formulation dispersion medium, similar to how others have done.41,42 After 5 min, 30 fractions of 1 mL were collected and assayed for protein and lipid content using the previously described Bradford assay and a modified Stewart assay, respectively. Flow rate was kept constant at a rate of 0.75 mL/min by using a peristaltic pump (Rotarus flow 50, Hirschmann, Eberstadt, Germany). Lipid-containing fractions were pooled together and concentrated to ∼200 μL by centrifugation using Amicon Ultra 0.5 mL centrifugal filters (100,000 MWCO, Merck KGaA, Darmstadt, Germany) following the manufacturer’s instructions. Liposomes were washed three times with 200 μL of PBS or PB. Concentrated liposomes were assayed for adsorbed protein content through SDS-PAGE electrophoresis, using 10% resolving and 5% stacking polyacrylamide gels, as described for the centrifugation protocol.
2.5. Phospholipid Quantification
The phospholipid content of liposomal suspensions or resuspended pellet after centrifugation and washing was determined through a modified Stewart assay.43 Briefly, aliquots of liposomal suspensions or resuspended pellets (4–20 μL) were diluted up to 0.5 mL of water, and 2 mL of aqueous 0.1 M FeCl3 and 0.4 M NH4SCN were added, in addition to 3 mL of chloroform. The mixture was vortexed for 1 min and centrifuged for 5 min at 2500 rpm, after which the lower organic phase was carefully transferred into a glass tube. Absorbance was read from the organic phase in quartz cuvettes at 469 nm. Determinations were made in triplicate. Calibration curve was composed of pure DPPC (linear range of 0–270 nmol; R2 = 0.995). No interference with CHOL, DOTAP, or BSA was observed (data not shown).
SEC fractions were quantified for lipid content in a smaller volume as follows: 250 μL of each 1 mL fraction were mixed with 0.5 mL of aqueous 0.1 M FeCl3 and 0.4 M NH4SCN and 800 μL of chloroform. The mixture was vortexed for 1 min and centrifuged for 2 min at 13,000g, after which the lower organic phase was transferred to the quartz cuvette. Calibration curve was composed of pure DPPC (linear range of 0–170 nmol; R2 = 0.9977).
2.6. Molecular Dynamics Simulations
Coarse-grained (CG) molecular dynamics were performed using the GPU-accelerated version of GROMACS 2020.644 with the Martini 2.2 force field.45 The insane.py script46 was used to build all initial configurations for the investigated formulations. As DOTAP is not a default lipid, its topology was defined in the insane.py script, using parameters described elsewhere.47
All bilayers contained a total of 224 molecules with varying ratios of DPPC:CHOL or DPPC:DOTAP. The aqueous phase contained either solely Martini water (4 molecules per bead) and neutralizing ions (if needed) or Martini water, neutralizing ions, and 137 mM NaCl. The exact composition of simulated systems can be found in Table S1.
Minimization, equilibration, and production runs were performed in several steps, using the general GROMACS inputs provided by CHARMM-GUI for membrane bilayers.48 Equilibration was performed in an isothermal–isobaric (NPT) ensemble for a total of 4.75 ns using 2–20 fs time steps with positional restraints on lipid headgroups. Unrestrained production runs were performed for 30 ns using 20 fs time steps, also in the NPT ensemble. The v-rescale thermostat and Berendsen barostat were used to keep the simulations at 310 K and 1 bar pressure in the case of equilibration simulations, while the v-rescale thermostat and Parrinello–Rahman barostat were used for production runs. Semi-isotropic pressure coupling was used for all steps. The Verlet cutoff scheme was used with a 1.1 cutoff for both the Coulombic (reaction field) and van der Waals interactions (potential shift method).
Visual Molecular Dynamics (VMD) version 1.9.3.49 was used for visual inspection of the simulated systems and image generation.
Membrane thickness, area per lipid (APL), and membrane area were calculated from production runs for each system using Voronoi tessellation, as implemented in FATSLiM.50 All plots were generated using ggplot2 v 3.3.6,51 as implemented in R (version 4.1.2), from .xvg files generated with FATSLiM and read into R using the readXVG function in the Peptides v 2.4.5 package.52
2.7. Statistical Analysis
Statistical significance was assessed using one-way ANOVA followed by pairwise comparisons between group levels using multiple testing adjustment. If data violated the normal distribution, the ladder of powers transformation was applied. P values less than 0.05 were considered to be statistically significant.
3. Results and Discussion
3.1. Characterization of Liposomes before BSA Incubation
Seventeen liposomal formulations of DPPC with varying CHOL (0 to 50 mol % in either PB or PBS) or DOTAP content (0 to 40 mol % in PBS) were generated (Table 1). The chemical structures of the lipids used to prepare the liposomes are depicted in Figure 1.
Table 1. Composition, Average Particle Size, Polydispersity Index (PDI), and Zeta (ζ) Potential of the Investigated Liposomal Formulations before BSA Incubation.
| formulation (mol:mol %)a | average particle size (nm) ± SD | PDI ± SD | dispersion medium |
|---|---|---|---|
| DPPC:CHOL 100:0 | 176 ± 2 | 0.114 ± 0.013 | PBSb, pH 7.4, 10 mM |
| DPPC:CHOL 90:10 | 166 ± 3 | 0.104 ± 0.041 | |
| DPPC:CHOL 80:20 | 190 ± 3 | 0.112 ± 0.059 | |
| DPPC:CHOL 70:30 | 184 | 0.089 ± 0.033 | |
| DPPC:CHOL 60:40 | 180 ± 2 | 0.096 ± 0.038 | |
| DPPC:CHOL 50:50 | 171 | 0.123 ± 0.026 | |
| DPPC:CHOL 100:0 | 162 ± 1 | 0.089 ± 0.020 | PBb, pH 7.4, 10 mM |
| DPPC:CHOL 90:10 | 174 ± 1 | 0.033 ± 0.010 | |
| DPPC:CHOL 80:20 | 163 ± 1 | 0.058 ± 0.007 | |
| DPPC:CHOL 70:30 | 163 ± 1 | 0.060 ± 0.024 | |
| DPPC:CHOL 60:40 | 167 ± 2 | 0.092 ± 0.015 | |
| DPPC:CHOL 50:50 | 163 ± 1 | 0.105 ± 0.020 | |
| DPPC:DOTAP 100:0 | 176 ± 2 | 0.114 ± 0.013 | PBSb, pH 7.4, 10 mM |
| DPPC:DOTAP 90:10 | 146 ± 1 | 0.069 ± 0.016 | |
| DPPC:DOTAP 80:20 | 151 ± 2 | 0.111 ± 0.009 | |
| DPPC:DOTAP 70:30 | 129 ± 7 | 0.306 ± 0.038 | |
| DPPC:DOTAP 60:40 | 214 ± 6 | 0.224 ± 0.043 |
All formulations are given as molar ratio (%).
PB—phosphate buffer (8.54 mM Na2HPO4, 1.46 mM KH2PO4), PBS—phosphate buffer saline (8.54 mM Na2HPO4, 1.46 mM KH2PO4, 2.7 mM KCl, 137 mM NaCl).
Figure 1.

Chemical structures of the lipids used in this work.
The presence of CHOL up to 50 mol % for PBS formulations did not have a significant impact on liposome size or particle size distribution after downsizing through mild sonication and extrusion, with populations having hydrodynamic diameters ranging from 166 ± 2 to 190 ± 2 nm and polydispersity indices (PDI) from 0.089 to 0.123. CHOL content was directly proportional with more negative ζ-potential values (−16.77 ± 1.52 mV for plain DPPC vesicles to −35.57 ± 2.87 mV for vesicles containing 50 mol % CHOL, Figure 2). As the isoelectric point of liposomes containing DPPC is around pH 4, the net electrophoretic mobility observed at physiological pH is caused by the adsorption of ions present in the solution on the membrane surface. It is worth mentioning that the ζ-potential does not provide information only on the surface of the particle itself, but it is a more complex property which involves the surface and its surroundings, particularly the ions at the interface between the Stern layer and bulk liquid in which the particle is suspended.53 For the liposomes prepared herein, the electrophoretic mobility was measured in phosphate buffer either enriched (PBS) or not (PB) with salts. For the DPPC and CHOL-based liposomes, all values are negative, which is in line with previous studies on liposomes made of zwitterionic phospholipids.54−57 It appears that the negatively charged phosphate groups attached to the cationic groups of the phospholipids are responsible for the negative ζ-potential. Based on rational deduction, the liposomes themselves have a positively charged surface. Moreover, when cations from the employed salts (Na+, K+) were added to the dispersion medium, the negative values of the ζ-potential were lower (less and less negative) due to ion binding to the membrane.57 What is opposite as compared with previous results published by other groups is not the negative value of the liposomes containing CHOL, but the trend. In our case, the increasing amount of CHOL generated liposomes with a more negative ζ-potential, suggesting an orientation and organization of the lipids favorable for adsorption of negatively charged species. On the other hand, the DPPC-DOTAP liposomes exhibit positive values for all concentrations of DOTAP in PBS, which are surprising knowing that measurements were performed in the presence of anions from the PBS. These results, in correlation with those already reported in the literature, emphasize that the ζ-potential is extremely sensitive not only to the composition of the liposomes but also to the orientation and the location of each of the lipid components in the membrane and the intermolecular interactions, providing consistent material for other investigative works. CHOL has been previously found to impact Na+ binding to the membrane in saline solutions, leading to overall more negative ζ-potential values with increasing CHOL content.58 The authors explained this phenomenon by the transport of Na+ from the interfacial water to the bulk water phase, resulting in a more neutral surface charge for the membrane. This observation is in line with our experimental results, showing more negative zeta potentials as the amount of CHOL increases in the membrane (Figure 2, vide infra). In addition, vesicle membrane rigidization in the presence of CHOL is known to improve colloidal stability,59 which can also be reflected by greater ζ-potential values. As such, it is generally assumed that absolute ζ-potential values above 30 mV indicate moderately stable systems.60 However, it is becoming increasingly recognized that although the ζ potential can provide indications on colloid stability, it does not entirely reflect it. Hence, it is not uncommon that stable colloids exhibit low ζ-potential or vice versa.53
Figure 2.

ζ-Potential for generated liposomes. Data are presented as mean ± SEM (n = 4–6).
In order to study the effect of ionic strength on the size and surface charge of CHOL-containing liposomes, the same lipid compositions were used to generate liposomes in PB without any added salt. These were overall slightly smaller and had a narrower size distribution, with hydrodynamic diameters ranging from 162 ± 1 to 174 ± 1 nm and PDI values between 0.058 and 0.105. The linearity between ζ-potential and CHOL content was not seen in the case of liposomes formulated in PB, although all CHOL-containing liposomal suspensions had ζ-potential values lower than pure DPPC (Figure 2).
PB also contains Na+ ions but at a much lower concentration than PBS. This suggests that the lack of an overall trend in ζ-potential values in formulations without added salt could be attributed to the stabilizing effect of CHOL alone, with only small effects from the presence of Na+ ions in the buffer. Thus, the observed ζ-potential trend with increasing CHOL in PBS formulations could be caused by a cumulative effect of vesicle stabilization and decreased Na+ binding at a high salt concentration. Nevertheless, the presence of CHOL can change the organization of the phospholipid membrane, which impacts the interaction between interfacial water and phosphocholine headgroups.61 These structural modifications influence the orientation of the membrane components as well as their interaction with the surrounding anions, and consequently, the values of the ζ-potential can suffer changes accordingly. Yet, the origin of the apparent values of the ζ-potential of phospholipidic membranes can be sometimes challenging, requiring additional and deeper experimental and theoretical investigation. Moreover, the results obtained from a particular system cannot be used to draw a universal conclusion for all phospholipid-based vesicles.
In the case of cationic formulations, incorporation of DOTAP up to 20 mol % generated smaller vesicle populations with essentially unimodal size distribution (Table 1), with highly positive ζ-potential values, as would be expected for cationic liposomes.62 The fact that these DOTAP-containing formulations are much smaller than the pores of the extrusion membrane (200 nm) could be an indicator of the unique structural rearrangement of DPPC:DOTAP mixtures during sonication and heating into smaller unilamellar vesicular structures, as other authors suggested.63 However, incorporation of 30 mol % DOTAP led to the generation of more heterogeneous vesicle populations (PDI > 0.2), although the average size was still small (129 ± 7 nm). At 40 mol % DOTAP, vesicles were larger and heterogeneous but did not show any signs of aggregation, even after one month (Figure S1).
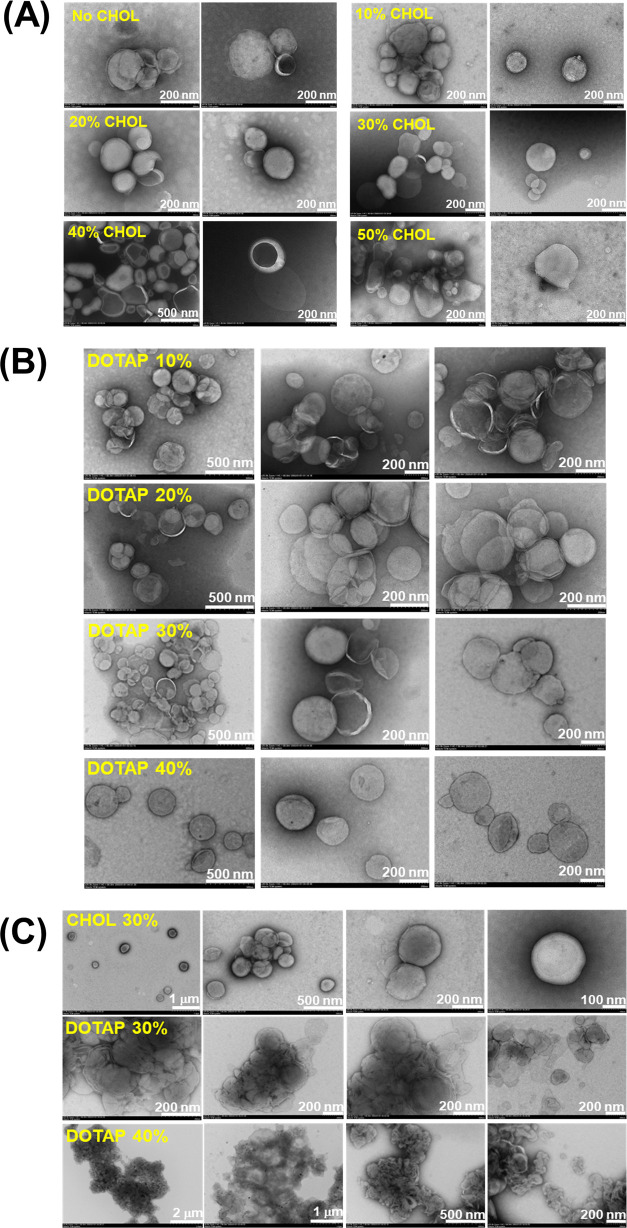
The morphology of the prepared liposomes was investigated by positive staining TEM, which implies shelling of liposomes with a material providing a high contrast. For the characterization of our samples, we employed a positive stain (OsO4) which usually interacts with the phospholipids via C=C bonds, followed by attachment to the lipid phosphate groups. However, despite its frequent use in electron microscopy to visualize structures in biological environments, the nature of the contrast mechanism is not yet known.64 For the samples investigated herein, the interaction of OsO4 with the palmitoyl-saturated chains of phospholipids did not occur due to the lack of unsaturated bonds in the carbon chain. Therefore, it is assumed that staining was achieved through the interaction with phosphate groups, particularly since staining with OsO4 in the presence of phosphate is significantly more intense than in the presence of other anions.65 In addition, the interaction is more probable due to the nearest-neighbor effect, thus creating a contrast with the empty central core of the vesicles. In this way, liposomes are stabilized without membrane damage, making assessment of their morphological properties facile. Yet, the acknowledged drawbacks of this staining approach should be considered in the morphological and architectural characterization of liposomes.66−68 Representative images collected for the samples prepared herein after extrusion, containing 0–50 mol % CHOL, 10–40 mol % DOTAP are illustrated in Figure 3. Also, liposomes after BSA adsorption (vide infra) were investigated by TEM, and illustrative images are depicted in Figure 3.
Figure 3.
Representative TEM images of aqueous suspensions of liposomes with different concentrations of CHOL in the bilayer and after extrusion. (A) CHOL-containing liposomes, (B) DOTAP-containing liposomes, and (C) (CHOL)DOTAP-containing liposomes after BSA adsorption.
Notably, the images reveal a heterogeneous population of variously sized vesicles with an estimated average of 170 nm for CHOL-based liposomes, which is in line with DLS measurements (Table 1). In most cases, unilamellar vesicles very close to each other can be seen, irrespective of bilayer CHOL concentration, indicating that CHOL content has no impact on the aggregation behavior of the vesicles. Interestingly, as the bilayer accommodates a higher amount of CHOL, the spherical morphology gradually experiences distortions or crashing. Hence, for the formulations with the highest amount of CHOL (40 and 50 mol %), undefined shapes can be observed in the TEM images. For DOTAP-containing liposomes, the expected sphere-like morphology can be observed, while size is well related with the results provided by DLS (Table 1). Of note, the sphere-like morphology of liposomes is improved as the contribution of DOTAP increases in the membrane, while the aggregation phenomenon at the same dilution is reduced. The morphology of liposomes after BSA adsorption noticeably changes due to aggregation phenomena occurring upon protein adsorption. Yet, the liposomal surface does not seem to be disrupted. However, possible artifacts caused by staining, drying, or exposure to high vacuum, which usually induce such shape alteration, are not excluded in the interpretation of the images.66−70
3.2. Behavior of Liposomes upon Centrifugation and SEC
Since the methodology used for separating protein–vesicle assemblies from unadsorbed proteins is centrifugation, it is important to describe the behavior of liposomal suspensions upon centrifugation without protein incubation. This will aid the correct interpretation of observed changes upon protein incubation when handling is performed under the same conditions. As such, upon analyzing lipid concentration, we observed that up to 57 and 38% of CHOL-containing liposomes formulated in PBS and PB, respectively, are unable to pellet, remaining in the supernatant upon centrifugation (Figure 4). In addition, perhaps relating to their overall smaller size, DOTAP-containing formulations show the most loss in the supernatant, with a lipid loss in the range of 61–88%. DLS analysis showed that vesicles with an average diameter of 116 and 126 nm are present in the supernatant after centrifugation for CHOL- and DOTAP-containing formulations, respectively (Table S2). Lipid loss and similar-sized liposomes in the supernatant following centrifugation have previously been observed even at greater centrifugal forces.71 Other studies also point out that centrifugation is suboptimal at isolating liposomal populations in adequate quantities for subsequent electrophoretic evaluations.72,73
Figure 4.

Lipid loss upon centrifugation, as determined by phospholipid content determination, relative to the expected content in the measured volume of sample. Data are presented as mean ± SEM (n = 3).
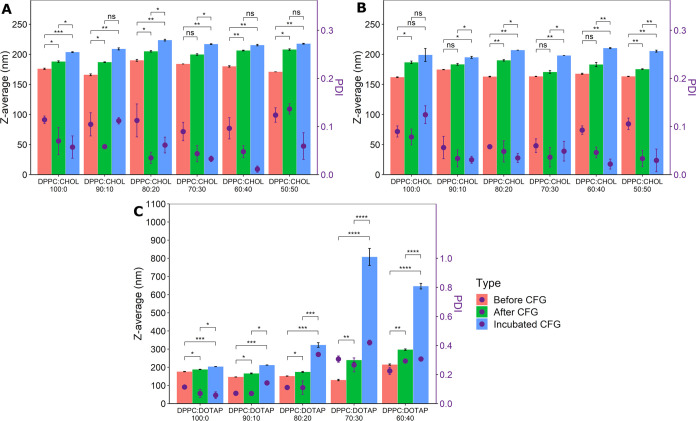
All resuspended pellets exhibited larger average hydrodynamic diameters than uncentrifuged suspensions as well as improved PDI values (Figure 5), most likely due to the smaller liposomes remaining in the supernatant upon centrifugation. A notable exception in terms of PDI improvement is the formulation containing 40 mol % DOTAP, suggesting the occurrence of aggregation or disruption phenomena due to increased shear forces resulting from high rotational speeds.74 In addition, the 30 mol % DOTAP formulation displayed almost a double average hydrodynamic diameter upon pellet resuspension and virtually no change in PDI (Figure 5) despite the high supernatant lipid loss. This suggests some aggregation of the pelleted vesicles or a dynamic restructuring of liposomes during centrifugation and/or resuspension.
Figure 5.
Liposome size and PDI before centrifugation (CFG), after centrifugation, and after incubation with BSA and centrifugation for (A) CHOL-containing liposomes in PBS, (B) CHOL-containing liposomes in PB, and (C) DOTAP-containing liposomes in PBS. Data are presented as mean ± SEM (n = 3). A one-way ANOVA followed by pairwise comparison using the Benjamini–Hochberg method for multiple testing correction was used for assessing statistical significance; “Incubated CFG” refers to the liposomes after BSA adsorption; *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001, ns—not significant.
Currently, there are no established guidelines for studying the liposomal protein corona, although many research groups have focused their attention on this subject.3,16,38,75,76 Despite some well-described caveats,77 centrifugation is currently the most used method for the separation of vesicle-protein assemblies from unbound proteins.78 Thus, the following experiments and quantitative aspects relating to liposome–protein isolation through centrifugation are described with regard to the population of liposomes that are able to pellet upon centrifugation. It is important to note that no conclusions can be drawn about the protein binding behavior of nonpelleted liposomes.77
Considering the limitation of centrifugation for separating liposome–protein complexes from unbound proteins, we additionally performed SEC for the incubated samples. However, we observed considerable lipid retention in the column, depending on the loaded formulation, as reflected by the amount of calculated lipid loss upon phospholipid determination in each fraction (Table 2) and the fact some liposomal material eluted in the washing steps between two subsequent SEC experiments (data not shown). This was surprising, as most SEC experiments performed on liposomes do not report such a considerable loss of lipids upon liposome elution.79 An interesting study highlights, however, that liposome retention in the exclusion gel is a common phenomenon which can lead to significant lipid loss, depending on bead size, especially if column presaturation with free liposomes is not performed.80 We did not perform column presaturation in our SEC setup; therefore, we attributed the observed lipid loss to the retention of liposome material in the column. Representative elution profiles for BSA-incubated liposomes with 30 mol % CHOL in PB and 40 mol % DOTAP are given in Figure 6. Upon inspecting the elution profiles, we pooled all lipid-containing fractions (usually fractions 7–18, except cases where liposomes stopped eluting earlier). Elution profiles for all other formulations are given in Figure S2, along with the elution of free BSA. Regardless of lipid loss upon column elution, the trend of more protein adsorption for liposomes with increasing DOTAP content was observed (Table 2). We could not, however, quantify the amount of lipid adsorbed through SDS-PAGE due to the very low concentration of BSA in the concentrated pooled fractions. We did observe that some smaller liposomes could penetrate the membranes used in the final concentration step in some cases (Table S5). No detectable BSA was found in the flow-through volume from the membrane ultrafiltration step (data not shown). It is possible, however, that the low amount of adsorbed BSA in the liposome-containing fractions combined with the high dilution due to fraction pooling generates only trace amounts of BSA, which are difficult to quantify.
Table 2. Average Protein Binding (PB) Values from Pooled SEC Fractions, as Determined through UV–Vis, as well as Lipid Loss during SECa.
| formulation (mol:mol %) | PB (g/mol lipid) | lipid loss (%) |
|---|---|---|
| DPPC:CHOL 100:0 (PBS) | 14.97 | 78.41 |
| DPPC:CHOL 90:10 (PBS) | 21.51 | 57.34 |
| DPPC:CHOL 80:20 (PBS) | 9.49 | 62.50 |
| DPPC:CHOL 70:30 (PBS) | 12.43 | 82.39 |
| DPPC:CHOL 60:40 (PBS) | 11.25 | 75.56 |
| DPPC:CHOL 50:50 (PBS) | 4.84 | 85.50 |
| DPPC:CHOL 100:0 (PB) | 13.68 | 57.70 |
| DPPC:CHOL 90:10 (PB) | 8.73 | 12.32 |
| DPPC:CHOL 80:20 (PB) | 15.94 | 43.05 |
| DPPC:CHOL 70:30 (PB) | 5.09 | 24.61 |
| DPPC:CHOL 60:40 (PB) | 22.02 | 68.22 |
| DPPC:CHOL 50:50 (PB) | 9.08 | 10.22 |
| DPPC:DOTAP 100:0 (PBS) | 14.97 | 78.41 |
| DPPC:DOTAP 90:10 (PBS) | 24.20 | 82.36 |
| DPPC:DOTAP 80:20 (PBS) | 25.35 | 88.48 |
| DPPC:DOTAP 70:30 (PBS) | 35.04 | 89.12 |
| DPPC:DOTAP 60:40 (PBS) | 63.11 | 65.11 |
All formulations are given as molar ratio (%). SEC was performed once per each formulation. PB values are relative to the entire quantity of liposomes loaded in the column.
Figure 6.
Size exclusion chromatography elution profiles for BSA-incubated (A) DPPC:CHOL 70:30 in PB and (B) DPPC:DOTAP 60:40 in PBS.
3.3. Physicochemical Characterization of BSA-Coated Liposomes
3.3.1. Size and ζ-Potential
Following incubation, an up to 22 nm increase in the average hydrodynamic diameter was observed for CHOL-containing vesicles formulated in PBS, but no trend relating to CHOL content was found. The same formulations in PB exhibited an up to 30 nm increase, the magnitude of which was dependent on CHOL level (Figure 5).
For DOTAP-containing formulations with more than 20 mol % cationic lipid, however, a much larger increase in size following incubation was observed, accompanied by PDI values larger than 0.2, indicating a substantial loss in size distribution homogeneity. As such, the average sizes after incubation were 212 ± 1, 322 ± 21, 807 ± 81, and 646 ± 26 nm for DOTAP-containing pellets with 10, 20, 30, and 40 mol %, respectively, following incubation (full data available in Table S3).
It is generally recognized that the formation of a protein corona manifests as an increase in the mean nanoparticle hydrodynamic diameter due to the formation of at least one protein layer on the surface.81 However, the finding that DOTAP-containing liposomes had such a significant increase in size following incubation could also indicate BSA-induced aggregation phenomena. In fact, it has previously been suggested that negatively charged proteins could act as a molecular glue between distinct DOTAP-containing vesicles,82 leading to the formation of large aggregates. Our findings seem to reinforce this hypothesis.
Following incubation, all CHOL-containing formulations exhibited an unexpected increase in ζ-potential compared to unincubated samples (Figure 7), as in principle, the net negative charge of adsorbed BSA should have resulted in more negative values for ζ-potential.83 However, in effect, this behavior indicates that adsorbed BSA lowers the electrostatic repulsion between liposomes. A similar observation has been made for polymersomes which stabilized the native protein fold upon adsorbing BSA and evaded immune recognition.13
Figure 7.
ζ-Potential before centrifugation, after centrifugation, and after incubation with BSA and centrifugation for (A) CHOL-containing liposomes in PBS, (B) CHOL-containing liposomes in PB; and (C) DOTAP-containing liposomes in PBS, “Incubated CFG” refers to the liposomes after BSA adsorption; Data are presented as mean ± SEM (n = 3).
The CHOL-dependent effect on liposome ζ-potential could be observed following both PB and PBS incubations, with increasing CHOL content leading to more negative values (Figure 7). At the used incubation temperature, which is above the gel-to-liquid phase pretransition temperature of DPPC (∼33 °C), and close to the main transition temperature (41.5 °C), the gel and liquid phases of the lipid should coexist, so packing defects are likely to occur.84 CHOL is known to have a fluidizing effect on liposomes below the gel-to-liquid phase transition temperature of the main lipid component, stabilizing a liquid ordered state and decreasing packing defects.85 Thus, more negative ζ-potential values with increasing CHOL content would be expected, in both incubated and centrifuged unincubated samples.
DOTAP-containing formulations behaved as expected in terms of ζ-potential upon BSA incubation (Figure 7), exhibiting a marked decrease, most likely due to the adsorbed BSA’s net negative charge. Similar to the case of CHOL-containing formulations, this reflects a lowered electrostatic repulsion between the BSA-coated nanocarriers. A similar effect of BSA on ζ-potential has been observed for other cationic liposomes containing DOTAP,86 although a different behavior was found upon full plasma incubation.6 However, DOTAP-induced changes in BSA secondary structure have been reported,32 and this effect was recently demonstrated to also occur in liposomes containing positively charged moieties, other than lipids, on their surface.87 CHOL, on the other hand, does not induce a change in protein secondary structure,32 while DPPC has a minimal effect.88 Thus, further investigations regarding possible changes in BSA structure upon adsorption should be performed, aimed at better understanding the biological identity of liposomes upon protein interaction.
3.3.2. BSA Adsorption Quantification
In terms of protein adsorption, SDS-PAGE experiments showed up to 5.619 ± 0.47 g/mol lipid adsorbed BSA for DOTAP-containing formulations, and more than 16-fold decreased adsorption for CHOL-containing formulations (Figure 8). Other authors also observed that cationic liposomes generally adsorb more proteins than electroneutral vesicles,89 although exceptions have also been found to occur.14 In contrast, CHOL was found to promote positively charged protein binding to negatively charged POPC/POPS vesicles.27
Figure 8.
Adsorbed BSA in g/mol lipid following SDS-PAGE from pelleted liposomes after centrifugation and washing and UV–vis determinations from the supernatant. Data are presented as mean ± SEM (n = 3). A one-way ANOVA followed by pairwise comparison using the Benjamini–Hochberg method for multiple testing correction was used for assessing statistical significance; *p < 0.05, **p < 0.01, ns–not significant.
UV–vis experiments showed about 10 times more protein adsorption than SDS-PAGE for all cases, with a maximum of 51.47 ± 2.08 g/mol lipid adsorbed BSA for DOTAP-containing formulations, which would be expected, considering the extent of lipid loss upon centrifugation. It is important to note that SDS-PAGE experiments only reflect the amounts of protein adsorbed on pelleted liposomes after subsequent washing steps, while values from UV–vis determinations also include weakly bound proteins on the surface of liposomes, which most likely detach upon pellet washing.
The formulation with 40 mol % DOTAP showed a marked decrease in protein adsorption when compared to the formulations containing 20 or 30 mol % charged lipid in SDS-PAGE experiments (Figure 8). This suggests that the observed increase in size following incubation (Figure 5) is mostly induced by aggregation and not the quantity of protein adsorbed. At the same time, UV–vis experiments showed the most protein adsorption in the case of 40 mol % DOTAP liposomes, suggesting that adsorbed BSA is only weakly bound on liposomal aggregates and is predominantly washed out during subsequent washing steps (51.470 ± 2.08 g BSA/mol lipid from UV–vis determinations vs 7.55 ± 2.41 g BSA/mol lipid from SDS-PAGE determinations, Table S4). It has previously been proposed that the increase in size due to aggregation would most likely have a larger impact on the bloodstream circulation of liposomes than the nature of adsorbed proteins, and thus should not be ignored from a clinical perspective.77 Liposomes containing more than 30 mol % cationic lipids have also been shown to aggregate in serum, which leads to their subsequent accumulation primarily in lungs and other off-target sites.90 Thus, special care needs to be taken when deciding upon lipid composition for generating formulations for intravenous administration.
The relatively low protein adsorption exhibited by the CHOL-containing formulations, as well as the one containing 10 mol % DOTAP, corroborated with their low size increase upon incubation suggest that these formulations could in principle be suitable for intravenous administration if albumin adsorption is to be avoided. This does not, however, mean that these formulations will not adsorb any proteins in plasma or serum. This aspect is currently under investigation by our group. The 30 mol % DOTAP-containing formulation, on the other hand, showed the most protein binding in SDS-PAGE, with about 38 BSA molecules per liposome being adsorbed (Table S4). However, the significant increase in size following incubation makes it unusable as a carrier, although it could be used as a model to study the molecular details of the albumin-nanocarrier interaction.
Emphasis should be placed on the significance of investigating BSA adsorption on liposomes, especially since albumin precoating has become an attractive strategy for prolonging nanocarrier plasma half-life,91,92 as well as improving the release kinetics of liposomal carriers.83 In particular, liposomes decorated with a 46-residue albumin binding domain were recently shown to specifically bind endogenous albumin, which competitively inhibited opsonin adsorption and prolonged liposomal in vivo circulation.93 At the same time, as albumin is a carrier protein that possesses ligand binding pockets for many anticancer drugs,94 it could be used for drug or prodrug complexation prior to precoating to increase drug half-life and improve targeted delivery.95 From our experiments, the 20 mol % DOTAP-containing formulation could be suitable for further albumin precoating, as it exhibits the lowest increase in size upon incubation but adsorbs a significant quantity of BSA, which could be advantageous for improving immune evasion. However, further investigations regarding the nature of adsorbed proteins from plasma are required and are of ongoing interest to our group. Finally, our results show that BSA interactions with liposomal nanocarriers are most likely governed by electrostatic and not hydrophobic interactions.
3.4. Bilayer Properties—Insights from Molecular Modeling
Because the mechanical properties of the liposomal membrane are an important aspect in the design and production of lipid-based drug delivery systems as well as in their adsorption behavior, we also investigated key membrane parameters, namely, membrane thickness, area per lipid, and membrane area as well as salt ion adsorption by coarse-grained molecular dynamics. Simulations were run for 30 ns for each experimentally investigated liposomal composition in the form of small membrane slabs with or without added salt. Molecular dynamics simulations are of particular usefulness in modeling or predicting observed experimental effects, such as lipid packing and ordering96 and protein97 or ion adsorption events.58 The membrane transverse snapshots at the final frame of production molecular dynamics simulations are illustrated in Figure 9.
Figure 9.
Membrane transverse snapshots in the final frame of production molecular dynamics simulations (30 ns). (A) DPPC:CHOL 100:0, (B) DPPC:CHOL 90:10, (C) DPPC:CHOL 80:20, (D) DPPC:CHOL 70:30, (E) DPPC:CHOL 60:40, (F) DPPC:CHOL 50:50, (G) DPPC:DOTAP 90:10, (H) DPPC:DOTAP 80:20, (I) DPPC:DOTAP 70:30, (J) DPPC:DOTAP 60:40. DPPC are depicted as cyan, CHOL are yellow and DOTAP are magenta. Na+ and Cl– are shown as blue and cyan spheres, respectively.
Our simulations indicate that CHOL exerts a condensing effect on DPPC. Specifically, membrane thickness, area per lipid, and average membrane area all decrease with increasing CHOL content (Table S6), in agreement with previously reported data.98,99 Of note, replacement of 50 mol % DPPC with CHOL reduced area per lipid and membrane area by ∼33%. We did not observe the appearance of CHOL patches in our simulations, with CHOL being distributed uniformly throughout the bilayer, regardless of concentration (Figure 9).
Systems containing DOTAP showed a slight increase in all measured parameters as a function of DOTAP content except membrane thickness (Table S6). This would be expected, since lateral expansion is likely to occur due to the electrostatic repulsion between charged lipids as the DOTAP concentration increases. This has been previously demonstrated to occur for DOPC/DOTAP systems.100 However, combined DOPC/DOTAP atomistic simulation systems have been shown to exhibit a compressive effect in terms of APL, contrary to our own findings, which use coarse-grained models for the representation of lipids.101 However, the work of Gao and Fang on the theoretical investigation of APL evolution with increasing cationic lipid content shows similar results as ours, which could be explained by the fact they used similar simulation conditions as we did.102 This also highlights the need for continuous force field optimization in order to lower the gap between atomistic and coarse-grained simulation results of the same systems. We also noticed a slight increase in the bilayer thickness in the presence of added salt for all systems. This phenomenon could be attributed to the binding of Na+ to the ester bonds of phospholipids, as other authors have shown,103 clearly showing that bilayer properties can be directly altered by the ion-lipid interactions. At the same time, from a simulation point of view, the calculated average thickness does not include the adsorbed ion diameter. Therefore, the increase in thickness is more likely a direct consequence of slightly different lipid packing at the membrane-water interface due to Na+ adsorption in the headgroup region and water rearrangement, as other authors have suggested.101 We did not perform additional measurements of average headgroup atom positions relative to the bilayer center to confirm this, but as other observations observed the same effect, we assume a similar behavior. Our simulations also demonstrate decreased Na+ binding with increased CHOL content, but also with increasing DOTAP content (Figure 10). This is in good agreement with another group’s combined experimental and computational findings, which are reflected in ζ-potential value trends for CHOL-containing systems.58 In the case of DOTAP systems, this behavior is expected, as increasing cationic lipid content naturally leads to repulsive behavior of the membrane surface toward Na+. Ions were defined as a contact if they were within the distance of 5 Å from any atom of a lipid molecule.
Figure 10.

Number of Na+: membrane contacts within 5 Å throughout the salt-containing simulations. Data are presented as mean ± SD.
4. Conclusions
In summary, we herein showed that the interaction of model protein BSA with liposomal vesicles containing the zwitterionic lipid DPPC and either CHOL (0–50%) or the cationic lipid DOTAP (0 to 40%) strongly depends on the chemical composition of the liposome. The conclusions of our study are based on a combined experimental and in silico approach, which certainly brings more value and meaning to liposome formulations as biocompatible carriers for drugs and their behavior in the presence of biologically relevant molecules, such as BSA. SDS-PAGE and UV–vis experimental results revealed protein adsorption to occur for all formulations. Yet, DOTAP-containing vesicles adsorbed proteins more tightly, but at the expense of aggregation, as demonstrated by DLS and TEM. Our results also show that while both SEC and centrifugation have limitations in the separation of liposome–protein complexes from unbound proteins, they are ultimately complementary to each other and can lead to similar conclusions. The calculated key parameters of the phospholipid membranes from molecular dynamics simulations showed a clear trend of both CHOL and DOTAP to decrease Na+ interaction with the membrane, which could also contribute to the differential behavior of liposomes relative to the adsorption of BSA, in addition to explaining the observed ζ-potential trend with increasing non-DPPC lipid. While our results suggest that protein–vesicle interactions are governed by a plethora of factors, such as size and size distribution, chemical composition, and surface charge, the exact molecular determinants of BSA adsorption are yet to be described. We expect that our study will contribute to a better understanding of protein–vesicle interactions for improved liposomal nanocarrier design, increase the fundamental knowledge in the field, and inspire other scientists in their research. As further work, we foresee extension to an in-depth investigation of the adsorption of plasma proteins on liposomes as well as gain novel insight into protein conformational changes upon adsorption.
Acknowledgments
This work was supported by two grants of the Romanian Ministry of Research, Innovation and Digitalization, CNCS-UEFISCDI, project codes PN–III-P1-1.1-PD-2021-0786, no. PD66/2022 and PN–III-P3-3.6-H2020-2020-0105, no. 35/2021, as well as a H2020 grant – ERA-Chair, no. 952390/2020.
Supporting Information Available
The Supporting Information is available free of charge. The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acsomega.3c09131.
Molecular composition of simulated systems (Table S1); average particle size and polydispersity index (PDI) from supernatants upon first centrifugation, unincubated or following BSA incubation (Table S2); average particle size, polydispersity index (PDI), and ζ-potential for resuspended pellets, unincubated or following BSA incubation (Table S3); average protein binding (PB) values from SDS-PAGE and UV–vis determinations, as well as mean number of BSA molecules per liposome for the investigated liposomal formulations (Table S4); average particle size from flow-through volumes after the final concentration step of SEC experiments for protein–liposome complexes (Table S5); average membrane thickness, area per lipid (APL) and membrane area in simulated bilayer systems from 30 ns production runs (Table S6); vesicle size as a factor of time, measured through DLS (Figure S1); and complete elution profiles for all DPPC-based liposomes in PBS and PB (Figure S2) (PDF)
Author Contributions
The manuscript was written through contributions of all authors. All authors have given approval to the final version of the manuscript.
The authors declare no competing financial interest.
Supplementary Material
References
- Moghimi S. M.; Hunter A. C.; Andresen T. L. Factors controlling nanoparticle pharmacokinetics: an integrated analysis and perspective. Annu. Rev. Pharmacol. Toxicol. 2012, 52, 481–503. 10.1146/annurev-pharmtox-010611-134623. [DOI] [PubMed] [Google Scholar]
- Tenchov R.; Bird R.; Curtze A. E.; Zhou Q. Lipid Nanoparticles horizontal line From Liposomes to mRNA Vaccine Delivery, a Landscape of Research Diversity and Advancement. ACS Nano 2021, 15 (11), 16982–17015. 10.1021/acsnano.1c04996. [DOI] [PubMed] [Google Scholar]
- Mahmoudi M.; Landry M. P.; Moore A.; Coreas R. The protein corona from nanomedicine to environmental science. Nat. Rev. Mater. 2023, 8, 422–438. 10.1038/s41578-023-00552-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundqvist M.; Cedervall T. Three Decades of Research about the Corona Around Nanoparticles: Lessons Learned and Where to Go Now. Small 2020, 16 (46), e2000892 10.1002/smll.202000892. [DOI] [PubMed] [Google Scholar]
- Ke P. C.; Lin S.; Parak W. J.; Davis T. P.; Caruso F. A Decade of the Protein Corona. ACS Nano 2017, 11 (12), 11773–11776. 10.1021/acsnano.7b08008. [DOI] [PubMed] [Google Scholar]
- Arcella A.; Palchetti S.; Digiacomo L.; Pozzi D.; Capriotti A. L.; Frati L.; Oliva M. A.; Tsaouli G.; Rota R.; Screpanti I.; et al. Brain Targeting by Liposome-Biomolecular Corona Boosts Anticancer Efficacy of Temozolomide in Glioblastoma Cells. ACS Chem. Neurosci. 2018, 9 (12), 3166–3174. 10.1021/acschemneuro.8b00339. [DOI] [PubMed] [Google Scholar]
- Clemente E.; Martinez-Moro M.; Trinh D. N.; Soliman M. G.; Spencer D. I. R.; Gardner R. A.; Kotsias M.; Sanchez Iglesias A.; Moya S.; Monopoli M. P. Probing the glycans accessibility in the nanoparticle biomolecular corona. J. Colloid Interface Sci. 2022, 613, 563–574. 10.1016/j.jcis.2021.11.140. [DOI] [PubMed] [Google Scholar]
- Yoo J. W.; Chambers E.; Mitragotri S. Factors that control the circulation time of nanoparticles in blood: challenges, solutions and future prospects. Curr. Pharm. Des. 2010, 16 (21), 2298–2307. 10.2174/138161210791920496. [DOI] [PubMed] [Google Scholar]
- Liu C.; Zhang L.; Zhu W.; Guo R.; Sun H.; Chen X.; Deng N. Barriers and Strategies of Cationic Liposomes for Cancer Gene Therapy. Mol. Ther. Methods Clin. Dev. 2020, 18, 751–764. 10.1016/j.omtm.2020.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell R. B.; Balasubramanian S. V.; Straubinger R. M. Influence of cationic lipids on the stability and membrane properties of paclitaxel-containing liposomes. J. Pharm. Sci. 2001, 90 (8), 1091–1105. 10.1002/jps.1063. [DOI] [PubMed] [Google Scholar]
- Briuglia M. L.; Rotella C.; McFarlane A.; Lamprou D. A. Influence of cholesterol on liposome stability and on in vitro drug release. Drug Delivery Transl. Res. 2015, 5 (3), 231–242. 10.1007/s13346-015-0220-8. [DOI] [PubMed] [Google Scholar]
- Nguyen V. H.; Meghani N. M.; Amin H. H.; Tran T. T. D.; Tran P. H. L.; Park C.; Lee B. J. Modulation of serum albumin protein corona for exploring cellular behaviors of fattigation-platform nanoparticles. Colloids Surf., B 2018, 170, 179–186. 10.1016/j.colsurfb.2018.05.060. [DOI] [PubMed] [Google Scholar]
- Vincent M. P.; Bobbala S.; Karabin N. B.; Frey M.; Liu Y.; Navidzadeh J. O.; Stack T.; Scott E. A. Surface chemistry-mediated modulation of adsorbed albumin folding state specifies nanocarrier clearance by distinct macrophage subsets. Nat. Commun. 2021, 12 (1), 648 10.1038/s41467-020-20886-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokouchi Y.; Tsunoda T.; Imura T.; Yamauchi H.; Yokoyama S.; Sakai H.; Abe M. Effect of adsorption of bovine serum albumin on liposomal membrane characteristics. Colloids Surf., B 2001, 20 (2), 95–103. 10.1016/S0927-7765(00)00176-4. [DOI] [PubMed] [Google Scholar]
- Kristensen K.; Urquhart A. J.; Thormann E.; Andresen T. L. Binding of human serum albumin to PEGylated liposomes: insights into binding numbers and dynamics by fluorescence correlation spectroscopy. Nanoscale 2016, 8 (47), 19726–19736. 10.1039/C6NR05455B. [DOI] [PubMed] [Google Scholar]
- Tretiakova D.; Kobanenko M.; Le-Deygen I.; Boldyrev I.; Kudryashova E.; Onishchenko N.; Vodovozova E. Spectroscopy Study of Albumin Interaction with Negatively Charged Liposome Membranes: Mutual Structural Effects of the Protein and the Bilayers. Membranes 2022, 12 (11), 1031. 10.3390/membranes12111031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabin J.; Prieto G.; Ruso J. M.; Messina P. V.; Salgado F. J.; Nogueira M.; Costas M.; Sarmiento F. Interactions between DMPC liposomes and the serum blood proteins HSA and IgG. J. Phys. Chem. B 2009, 113 (6), 1655–1661. 10.1021/jp804641e. [DOI] [PubMed] [Google Scholar]
- Sainaga Jyothi V. G. S.; Bulusu R.; Venkata Krishna Rao B.; Pranothi M.; Banda S.; Kumar Bolla P.; Kommineni N. Stability characterization for pharmaceutical liposome product development with focus on regulatory considerations: An update. Int. J. Pharm. 2022, 624, 122022 10.1016/j.ijpharm.2022.122022. [DOI] [PubMed] [Google Scholar]
- Beltrán-Gracia E.; López-Camacho A.; Higuera-Ciapara I.; Velázquez-Fernández J. B.; Vallejo-Cardona A. A. Nanomedicine review: clinical developments in liposomal applications. Cancer Nanotechnol. 2019, 10 (1), 11. 10.1186/s12645-019-0055-y. [DOI] [Google Scholar]
- Liu P.; Chen G.; Zhang J. A Review of Liposomes as a Drug Delivery System: Current Status of Approved Products, Regulatory Environments, and Future Perspectives. Molecules 2022, 27 (4), 1372. 10.3390/molecules27041372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rog T.; Pasenkiewicz-Gierula M.; Vattulainen I.; Karttunen M. Ordering effects of cholesterol and its analogues. Biochim. Biophys. Acta, Biomembr. 2009, 1788 (1), 97–121. 10.1016/j.bbamem.2008.08.022. [DOI] [PubMed] [Google Scholar]
- Simon S. A.; McIntosh T. J.; Latorre R. Influence of cholesterol on water penetration into bilayers. Science 1982, 216 (4541), 65–67. 10.1126/science.7063872. [DOI] [PubMed] [Google Scholar]
- Subczynski W. K.; Wisniewska A.; Yin J. J.; Hyde J. S.; Kusumi A. Hydrophobic barriers of lipid bilayer membranes formed by reduction of water penetration by alkyl chain unsaturation and cholesterol. Biochemistry 1994, 33 (24), 7670–7681. 10.1021/bi00190a022. [DOI] [PubMed] [Google Scholar]
- Kucerka N.; Perlmutter J. D.; Pan J.; Tristram-Nagle S.; Katsaras J.; Sachs J. N. The effect of cholesterol on short- and long-chain monounsaturated lipid bilayers as determined by molecular dynamics simulations and X-ray scattering. Biophys. J. 2008, 95 (6), 2792–2805. 10.1529/biophysj.107.122465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bozzuto G.; Molinari A. Liposomes as nanomedical devices. Int. J. Nanomed. 2015, 10, 975–999. 10.2147/IJN.S68861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramli N. A.; Ali N.; Hamzah S.; Yatim N. I. Physicochemical characteristics of liposome encapsulation of stingless bees’ propolis. Heliyon 2021, 7 (4), e06649 10.1016/j.heliyon.2021.e06649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doktorova M.; Heberle F. A.; Kingston R. L.; Khelashvili G.; Cuendet M. A.; Wen Y.; Katsaras J.; Feigenson G. W.; Vogt V. M.; Dick R. A. Cholesterol Promotes Protein Binding by Affecting Membrane Electrostatics and Solvation Properties. Biophys. J. 2017, 113 (9), 2004–2015. 10.1016/j.bpj.2017.08.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simberg D.; Weisman S.; Talmon Y.; Barenholz Y. DOTAP (and other cationic lipids): chemistry, biophysics, and transfection. Crit. Rev. Ther. Drug Carrier Syst. 2004, 21 (4), 257–317. 10.1615/CritRevTherDrugCarrierSyst.v21.i4.10. [DOI] [PubMed] [Google Scholar]
- Takechi-Haraya Y.; Sakai-Kato K.; Abe Y.; Kawanishi T.; Okuda H.; Goda Y. Atomic Force Microscopic Analysis of the Effect of Lipid Composition on Liposome Membrane Rigidity. Langmuir 2016, 32 (24), 6074–6082. 10.1021/acs.langmuir.6b00741. [DOI] [PubMed] [Google Scholar]
- Peters T.; Grunewald C.; Blaickner M.; Ziegner M.; Schutz C.; Iffland D.; Hampel G.; Nawroth T.; Langguth P. Cellular uptake and in vitro antitumor efficacy of composite liposomes for neutron capture therapy. Radiat. Oncol. 2015, 10, 52. 10.1186/s13014-015-0342-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simoes S.; Slepushkin V.; Pires P.; Gaspar R.; Pedroso de Lima M. C.; Duzgunes N. Human serum albumin enhances DNA transfection by lipoplexes and confers resistance to inhibition by serum. Biochim. Biophys. Acta, Biomembr. 2000, 1463 (2), 459–469. 10.1016/s0005-2736(99)00238-2. [DOI] [PubMed] [Google Scholar]
- Charbonneau D. M.; Tajmir-Riahi H. A. Study on the interaction of cationic lipids with bovine serum albumin. J. Phys. Chem. B 2010, 114 (2), 1148–1155. 10.1021/jp910077h. [DOI] [PubMed] [Google Scholar]
- Doole F. T.; Kumarage T.; Ashkar R.; Brown M. F. Cholesterol Stiffening of Lipid Membranes. J. Membr. Biol. 2022, 255 (4–5), 385–405. 10.1007/s00232-022-00263-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi S.; Kang B.; Yang E.; Kim K.; Kwak M. K.; Chang P. S.; Jung H. S. Precise control of liposome size using characteristic time depends on solvent type and membrane properties. Sci. Rep. 2023, 13 (1), 4728 10.1038/s41598-023-31895-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu L.; Anchordoquy T. J. Cholesterol domains in cationic lipid/DNA complexes improve transfection. Biochim. Biophys. Acta, Biomembr. 2008, 1778 (10), 2177–2181. 10.1016/j.bbamem.2008.04.009. [DOI] [PubMed] [Google Scholar]
- Betker J. L.; Gomez J.; Anchordoquy T. J. The effects of lipoplex formulation variables on the protein corona and comparisons with in vitro transfection efficiency. J. Controlled Release 2013, 171 (3), 261–268. 10.1016/j.jconrel.2013.07.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- da Costa C. A. M.; Moraes Â. M. Encapsulation of 5-fluorouracil in liposomes for topical administration. Acta Sci. Technol. 2008, 25 (1), 53–61. 10.4025/actascitechnol.v25i1.2240. [DOI] [Google Scholar]
- Caracciolo G.; Pozzi D.; Capriotti A. L.; Cavaliere C.; Foglia P.; Amenitsch H.; Lagana A. Evolution of the protein corona of lipid gene vectors as a function of plasma concentration. Langmuir 2011, 27 (24), 15048–15053. 10.1021/la202912f. [DOI] [PubMed] [Google Scholar]
- Capriotti A. L.; Caracciolo G.; Caruso G.; Cavaliere C.; Pozzi D.; Samperi R.; Lagana A. Analysis of plasma protein adsorption onto DC-Chol-DOPE cationic liposomes by HPLC-CHIP coupled to a Q-TOF mass spectrometer. Anal. Bioanal. Chem. 2010, 398 (7–8), 2895–2903. 10.1007/s00216-010-4104-y. [DOI] [PubMed] [Google Scholar]
- Langer C.; Koll-Weber M.; Holzer M.; Hantel C.; Suss R. Mitotane Nanocarriers for the Treatment of Adrenocortical Carcinoma: Evaluation of Albumin-Stabilized Nanoparticles and Liposomes in a Preclinical In Vitro Study with 3D Spheroids. Pharmaceutics 2022, 14 (9), 1891. 10.3390/pharmaceutics14091891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hadjidemetriou M.; Al-Ahmady Z.; Mazza M.; Collins R. F.; Dawson K.; Kostarelos K. In Vivo Biomolecule Corona around Blood-Circulating, Clinically Used and Antibody-Targeted Lipid Bilayer Nanoscale Vesicles. ACS Nano 2015, 9 (8), 8142–8156. 10.1021/acsnano.5b03300. [DOI] [PubMed] [Google Scholar]
- Kristensen K.; Engel T. B.; Stensballe A.; Simonsen J. B.; Andresen T. L. The hard protein corona of stealth liposomes is sparse. J. Controlled Release 2019, 307, 1–15. 10.1016/j.jconrel.2019.05.042. [DOI] [PubMed] [Google Scholar]
- Hein R.; Uzundal C. B.; Hennig A. Simple and rapid quantification of phospholipids for supramolecular membrane transport assays. Org. Biomol. Chem. 2016, 14 (7), 2182–2185. 10.1039/C5OB02480C. [DOI] [PubMed] [Google Scholar]
- Abraham M. J.; Murtola T.; Schulz R.; Páll S.; Smith J. C.; Hess B.; Lindahl E. GROMACS: High performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX 2015, 1–2, 19–25. 10.1016/j.softx.2015.06.001. [DOI] [Google Scholar]
- de Jong D. H.; Singh G.; Bennett W. F.; Arnarez C.; Wassenaar T. A.; Schafer L. V.; Periole X.; Tieleman D. P.; Marrink S. J. Improved Parameters for the Martini Coarse-Grained Protein Force Field. J. Chem. Theory Comput. 2013, 9 (1), 687–697. 10.1021/ct300646g. [DOI] [PubMed] [Google Scholar]
- Wassenaar T. A.; Ingolfsson H. I.; Bockmann R. A.; Tieleman D. P.; Marrink S. J. Computational Lipidomics with insane: A Versatile Tool for Generating Custom Membranes for Molecular Simulations. J. Chem. Theory Comput. 2015, 11 (5), 2144–2155. 10.1021/acs.jctc.5b00209. [DOI] [PubMed] [Google Scholar]
- Bruininks B. M. H.; Souza P. C. T.; Marrink S. J. A Practical View of the Martini Force Field. Methods Mol. Biol. 2019, 2022, 105–127. 10.1007/978-1-4939-9608-7_5. [DOI] [PubMed] [Google Scholar]
- Wu E. L.; Cheng X.; Jo S.; Rui H.; Song K. C.; Davila-Contreras E. M.; Qi Y.; Lee J.; Monje-Galvan V.; Venable R. M.; et al. CHARMM-GUI Membrane Builder toward realistic biological membrane simulations. J. Comput. Chem. 2014, 35 (27), 1997–2004. 10.1002/jcc.23702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humphrey W.; Dalke A.; Schulten K. VMD: visual molecular dynamics. J. Mol. Graph. 1996, 14 (1), 33–38. 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- Buchoux S. FATSLiM: a fast and robust software to analyze MD simulations of membranes. Bioinformatics 2017, 33 (1), 133–134. 10.1093/bioinformatics/btw563. [DOI] [PubMed] [Google Scholar]
- Wickham H.ggplot2: Elegant Graphics for Data Analysis; Springer-Verlag, 2016. [Google Scholar]
- Osorio D.; Rondón-Villarreal P.; Torres R. Peptides: a package for data mining of antimicrobial peptides. R J. 2015, 7 (1), 4–14. 10.32614/RJ-2015-001. [DOI] [Google Scholar]
- Bhattacharjee S. DLS and zeta potential - What they are and what they are not?. J. Controlled Release 2016, 235, 337–351. 10.1016/j.jconrel.2016.06.017. [DOI] [PubMed] [Google Scholar]
- Zimmermann R.; Kuttner D.; Renner L.; Kaufmann M.; Zitzmann J.; Muller M.; Werner C. Charging and structure of zwitterionic supported bilayer lipid membranes studied by streaming current measurements, fluorescence microscopy, and attenuated total reflection Fourier transform infrared spectroscopy. Biointerphases 2009, 4 (1), 1–6. 10.1116/1.3082042. [DOI] [PubMed] [Google Scholar]
- Chibowski E.; Szcześ A. Zeta potential and surface charge of DPPC and DOPC liposomes in the presence of PLC enzyme. Adsorption 2016, 22 (4), 755–765. 10.1007/s10450-016-9767-z. [DOI] [Google Scholar]
- Sek A.; Perczyk P.; Wydro P.; Gruszecki W. I.; Szczes A. Effect of trace amounts of ionic surfactants on the zeta potential of DPPC liposomes. Chem. Phys. Lipids 2021, 235, 105059 10.1016/j.chemphyslip.2021.105059. [DOI] [PubMed] [Google Scholar]
- Banerjee K. K.; Maity P.; Das S.; Karmakar S. Effect of cholesterol on the ion-membrane interaction: Zeta potential and dynamic light scattering study. Chem. Phys. Lipids 2023, 254, 105307 10.1016/j.chemphyslip.2023.105307. [DOI] [PubMed] [Google Scholar]
- Magarkar A.; Dhawan V.; Kallinteri P.; Viitala T.; Elmowafy M.; Rog T.; Bunker A. Cholesterol level affects surface charge of lipid membranes in saline solution. Sci. Rep. 2014, 4, 5005 10.1038/srep05005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Virden J. W.; Berg J. C. Sodium chloride-induced aggregation of dipalmitoylphoshpatidylglycerol small unilamellar vesicles with varying amounts of incorporated cholesterol. Langmuir 1992, 8 (6), 1532–1537. 10.1021/la00042a007. [DOI] [Google Scholar]
- Ostolska I.; Wisniewska M. Application of the zeta potential measurements to explanation of colloidal Cr(2)O(3) stability mechanism in the presence of the ionic polyamino acids. Colloid Polym. Sci. 2014, 292 (10), 2453–2464. 10.1007/s00396-014-3276-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dreier L. B.; Wolde-Kidan A.; Bonthuis D. J.; Netz R. R.; Backus E. H. G.; Bonn M. Unraveling the Origin of the Apparent Charge of Zwitterionic Lipid Layers. J. Phys. Chem. Lett. 2019, 10 (20), 6355–6359. 10.1021/acs.jpclett.9b02587. [DOI] [PubMed] [Google Scholar]
- Grace V. M. B.; Wilson D. D.; Guruvayoorappan C.; Danisha J. P.; Bonati L. Liposome nano-formulation with cationic polar lipid DOTAP and cholesterol as a suitable pH-responsive carrier for molecular therapeutic drug (all-trans retinoic acid) delivery to lung cancer cells. IET Nanobiotechnol. 2021, 15 (4), 380–390. 10.1049/nbt2.12028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cinelli S.; Onori G.; Zuzzi S.; Bordi F.; Cametti C.; Sennato S.; Diociaiuti M. Properties of mixed DOTAP-DPPC bilayer membranes as reported by differential scanning calorimetry and dynamic light scattering measurements. J. Phys. Chem. B 2007, 111 (33), 10032–10039. 10.1021/jp071722g. [DOI] [PubMed] [Google Scholar]
- Li R.; Wildenberg G.; Boergens K.; Yang Y.; Weber K.; Rieger J.; Arcidiacono A.; Klie R.; Kashturi N.; King S.. Contrast Mechanism of Osmium Staining in Electron Microscopy of Biological Tissues ChemRxiv 2023 10.26434/chemrxiv-2023-scskq-v2. [DOI] [PubMed]
- Wood R. L.; Luft J. H. The Influence of Buffer Systems on Fixation with Osmium Tetroxide. J. Ultrastruct. Res. 1965, 12, 22–45. 10.1016/S0022-5320(65)80004-1. [DOI] [PubMed] [Google Scholar]
- Ting-Beall H. P. Interactions of uranyl ions with lipid bilayer membranes. J. Microsc. 1980, 118 (2), 221–227. 10.1111/j.1365-2818.1980.tb00264.x. [DOI] [PubMed] [Google Scholar]
- Lindner S.; Gruhle K.; Schmidt R.; Garamus V. M.; Ramsbeck D.; Hause G.; Meister A.; Sinz A.; Drescher S. Azide-Modified Membrane Lipids: Synthesis, Properties, and Reactivity. Langmuir 2017, 33 (20), 4960–4973. 10.1021/acs.langmuir.7b00228. [DOI] [PubMed] [Google Scholar]
- Baxa U. Imaging of Liposomes by Transmission Electron Microscopy. Methods Mol. Biol. 2018, 1682, 73–88. 10.1007/978-1-4939-7352-1_8. [DOI] [PubMed] [Google Scholar]
- Bello V.; Mattei G.; Mazzoldi P.; Vivenza N.; Gasco P.; Idee J. M.; Robic C.; Borsella E. Transmission electron microscopy of lipid vesicles for drug delivery: comparison between positive and negative staining. Microsc. Microanal. 2010, 16 (4), 456–461. 10.1017/S1431927610093645. [DOI] [PubMed] [Google Scholar]
- Miko M.; Danisovic L.; Majidi A.; Varga I. Ultrastructural analysis of different human mesenchymal stem cells after in vitro expansion: a technical review. Eur. J. Histochem. 2015, 59 (4), 2528. 10.4081/ejh.2015.2528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts S. A.; Lee C.; Singh S.; Agrawal N. Versatile Encapsulation and Synthesis of Potent Liposomes by Thermal Equilibration. Membranes 2022, 12 (3), 319 10.3390/membranes12030319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pattipeiluhu R.; Crielaard S.; Klein-Schiphorst I.; Florea B. I.; Kros A.; Campbell F. Unbiased Identification of the Liposome Protein Corona using Photoaffinity-based Chemoproteomics. ACS Cent. Sci. 2020, 6 (4), 535–545. 10.1021/acscentsci.9b01222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hacene Y. C.; Loiseau A.; Maio V. D. P.; Grenier P.; Boisselier E.; Bertrand N. Isolating Nanoparticles from Complex Biological Media by Immunoprecipitation. Nano Lett. 2021, 21 (11), 4530–4538. 10.1021/acs.nanolett.0c05056. [DOI] [PubMed] [Google Scholar]
- He Y.; Grandi D. D.; Chandradoss S.; LuTheryn G.; Cidonio G.; Nunes Bastos R.; Pereno V.; Carugo D. Rapid Production of Nanoscale Liposomes Using a 3D-Printed Reactor-In-A-Centrifuge: Formulation, Characterisation, and Super-Resolution Imaging. Micromachines 2023, 14 (9), 1763 10.3390/mi14091763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foteini P.; Pippa N.; Naziris N.; Demetzos C. Physicochemical study of the protein-liposome interactions: influence of liposome composition and concentration on protein binding. J. Liposome Res. 2019, 29 (4), 313–321. 10.1080/08982104.2018.1468774. [DOI] [PubMed] [Google Scholar]
- Singh N.; Marets C.; Boudon J.; Millot N.; Saviot L.; Maurizi L. In vivo protein corona on nanoparticles: does the control of all material parameters orient the biological behavior?. Nanoscale Adv. 2021, 3 (5), 1209–1229. 10.1039/D0NA00863J. [DOI] [PMC free article] [PubMed] [Google Scholar]
- a Munter R.; Simonsen J. B. Comment on “Optimal centrifugal isolating of liposome–protein complexes from human plasma” by L. Digiacomo, F. Giulimondi, A. L. Capriotti, S. Piovesana, C. M. Montone, R. Z. Chiozzi, A. Laganá, M. Mahmoudi, D. Pozzi and G. Caracciolo, Nanoscale Adv., 2021, 3, 3824. Nanoscale Adv. 2023, 5, 290–299. 10.1039/D2NA00343K. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Digiacomo L.; Giulimondi F.; Capriotti A. L.; Piovesana S.; Montone C. M.; Chiozzi R. Z.; Lagana A.; Mahmoudi M.; Pozzi D.; Caracciolo G. Optimal centrifugal isolating of liposome-protein complexes from human plasma. Nanoscale Adv. 2021, 3 (13), 3824–3834. 10.1039/D1NA00211B. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kristensen K.; Munter R.; Kempen P. J.; Thomsen M. E.; Stensballe A.; Andresen T. L. Isolation methods commonly used to study the liposomal protein corona suffer from contamination issues. Acta Biomater. 2021, 130, 460–472. 10.1016/j.actbio.2021.06.008. [DOI] [PubMed] [Google Scholar]
- Ruysschaert T.; Marque A.; Duteyrat J. L.; Lesieur S.; Winterhalter M.; Fournier D. Liposome retention in size exclusion chromatography. BMC Biotechnol. 2005, 5, 11. 10.1186/1472-6750-5-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sangra M.; Estelrich J.; Sabate R.; Espargaro A.; Busquets M. A. Evidence of Protein Adsorption in Pegylated Liposomes: Influence of Liposomal Decoration. Nanomaterials 2017, 7 (2), 37. 10.3390/nano7020037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giulimondi F.; Digiacomo L.; Pozzi D.; Palchetti S.; Vulpis E.; Capriotti A. L.; Chiozzi R. Z.; Lagana A.; Amenitsch H.; Masuelli L.; et al. Interplay of protein corona and immune cells controls blood residency of liposomes. Nat. Commun. 2019, 10 (1), 3686 10.1038/s41467-019-11642-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei X. Q.; Ba K. Construction a Long-Circulating Delivery System of Liposomal Curcumin by Coating Albumin. ACS Omega 2020, 5 (27), 16502–16509. 10.1021/acsomega.0c00930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kneidl B.; Peller M.; Winter G.; Lindner L. H.; Hossann M. Thermosensitive liposomal drug delivery systems: state of the art review. Int. J. Nanomed. 2014, 9, 4387–4398. 10.2147/IJN.S49297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redondo-Morata L.; Giannotti M. I.; Sanz F. Influence of cholesterol on the phase transition of lipid bilayers: a temperature-controlled force spectroscopy study. Langmuir 2012, 28 (35), 12851–12860. 10.1021/la302620t. [DOI] [PubMed] [Google Scholar]
- Tsukamoto M.; Okuda T.; Okamoto H.; Higuchi Y.; Kawakami S.; Yamashita F.; Hashida M. Bovine serum albumin as a lyoprotectant for preparation of DNA dry powder formulations using the spray-freeze drying method. Biol. Pharm. Bull. 2012, 35 (7), 1178–1181. 10.1248/bpb.b110731. [DOI] [PubMed] [Google Scholar]
- Tretiakova D.; Le-Deigen I.; Onishchenko N.; Kuntsche J.; Kudryashova E.; Vodovozova E. Phosphatidylinositol Stabilizes Fluid-Phase Liposomes Loaded with a Melphalan Lipophilic Prodrug. Pharmaceutics 2021, 13 (4), 473 10.3390/pharmaceutics13040473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thakur R.; Das A.; Chakraborty A. Interaction of human serum albumin with liposomes of saturated and unsaturated lipids with different phase transition temperatures: a spectroscopic investigation by membrane probe PRODAN. RSC Adv. 2014, 4 (28), 14335–14347. 10.1039/C4RA01214C. [DOI] [Google Scholar]
- Palchetti S.; Caputo D.; Digiacomo L.; Capriotti A. L.; Coppola R.; Pozzi D.; Caracciolo G. Protein Corona Fingerprints of Liposomes: New Opportunities for Targeted Drug Delivery and Early Detection in Pancreatic Cancer. Pharmaceutics 2019, 11 (1), 31. 10.3390/pharmaceutics11010031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao W.; Zhuang S.; Qi X. R. Comparative study of the in vitro and in vivo characteristics of cationic and neutral liposomes. Int. J. Nanomed. 2011, 6, 3087–3098. 10.2147/IJN.S25399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng Q.; Zhang S.; Yang Q.; Zhang T.; Wei X. Q.; Jiang L.; Zhang C. L.; Chen Q. M.; Zhang Z. R.; Lin Y. F. Preformed albumin corona, a protective coating for nanoparticles based drug delivery system. Biomaterials 2013, 34 (33), 8521–8530. 10.1016/j.biomaterials.2013.07.102. [DOI] [PubMed] [Google Scholar]
- Kummitha C. M.; Malamas A. S.; Lu Z. R. Albumin pre-coating enhances intracellular siRNA delivery of multifunctional amphiphile/siRNA nanoparticles. Int. J. Nanomed. 2012, 7, 5205–5214. 10.2147/IJN.S34288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H.; Yin D.; Liao J.; Wang Y.; Gou R.; Tang C.; Li W.; Liu Y.; Fu J.; Shi S.; et al. Regulation of protein corona on liposomes using albumin-binding peptide for targeted tumor therapy. J. Controlled Release 2023, 355, 593–603. 10.1016/j.jconrel.2023.02.004. [DOI] [PubMed] [Google Scholar]
- Taguchi K.; Okamoto Y.; Matsumoto K.; Otagiri M.; Chuang V. T. G. When Albumin Meets Liposomes: A Feasible Drug Carrier for Biomedical Applications. Pharmaceuticals 2021, 14 (4), 296. 10.3390/ph14040296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chubarov A. S. Serum Albumin for Magnetic Nanoparticles Coating. Magnetochemistry 2022, 8 (2), 13. 10.3390/magnetochemistry8020013. [DOI] [Google Scholar]
- Benz R. W.; Castro-Roman F.; Tobias D. J.; White S. H. Experimental validation of molecular dynamics simulations of lipid bilayers: a new approach. Biophys. J. 2005, 88 (2), 805–817. 10.1529/biophysj.104.046821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latour R. A. Molecular simulation of protein-surface interactions: benefits, problems, solutions, and future directions. Biointerphases 2008, 3 (3), FC2–FC12. 10.1116/1.2965132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y.; Carter J. W.; Lervik A.; Brooks N. J.; Seddon J. M.; Bresme F. Structural organization of sterol molecules in DPPC bilayers: a coarse-grained molecular dynamics investigation. Soft Matter 2016, 12 (7), 2108–2117. 10.1039/C5SM03051J. [DOI] [PubMed] [Google Scholar]
- Edholm O.; Nagle J. F. Areas of molecules in membranes consisting of mixtures. Biophys. J. 2005, 89 (3), 1827–1832. 10.1529/biophysj.105.064329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jurkiewicz P.; Olzynska A.; Langner M.; Hof M. Headgroup hydration and mobility of DOTAP/DOPC bilayers: a fluorescence solvent relaxation study. Langmuir 2006, 22 (21), 8741–8749. 10.1021/la061597k. [DOI] [PubMed] [Google Scholar]
- Pokorna S.; Jurkiewicz P.; Cwiklik L.; Vazdar M.; Hof M. Interactions of monovalent salts with cationic lipid bilayers. Faraday Discuss. 2013, 160, 341–358. 10.1039/C2FD20098H. [DOI] [PubMed] [Google Scholar]
- Gao L.; Fang W. Communications: Self-energy and corresponding virial contribution of electrostatic interactions in dissipative particle dynamics: Simulations of cationic lipid bilayers. J. Chem. Phys. 2010, 132 (3), 031102 10.1063/1.3297889. [DOI] [PubMed] [Google Scholar]
- Saunders M.; Steele M.; Lavigne W.; Varma S.; Pandit S. A. Interaction of salt with ether- and ester-linked phospholipid bilayers. Biochim. Biophys. Acta, Biomembr. Biomembr. 2019, 1861 (5), 907–915. 10.1016/j.bbamem.2019.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.