Abstract
The ability of the adenovirus type 5 E1B 55-kDa mutants dl1520 and dl338 to replicate efficiently and independently of the cell cycle, to synthesis viral DNA, and to lyse infected cells did not correlate with the status of p53 in seven cell lines examined. Rather, cell cycle-independent replication and virus-induced cell killing correlated with permissivity to viral replication. This correlation extended to S-phase HeLa cells, which were more susceptible to virus-induced cell killing by the E1B 55-kDa mutant virus than HeLa cells infected during G1. Wild-type p53 had only a modest effect on E1B mutant virus yields in H1299 cells expressing a temperature-sensitive p53 allele. The defect in E1B 55-kDa mutant virus replication resulting from reduced temperature was as much as 10-fold greater than the defect due to p53 function. At 39°C, the E1B 55-kDa mutant viruses produced wild-type yields of virus and replicated independently of the cell cycle. In addition, the E1B 55-kDa mutant viruses directed the synthesis of late viral proteins to levels equivalent to the wild-type virus level at 39°C. We have previously shown that the defect in mutant virus replication can also be overcome by infecting HeLa cells during S phase. Taken together, these results indicate that the capacity of the E1B 55-kDa mutant virus to replicate independently of the cell cycle does not correlate with the status of p53 but is determined by yet unidentified mechanisms. The cold-sensitive nature of the defect of the E1B 55-kDa mutant virus in both late gene expression and cell cycle-independent replication leads us to speculate that these functions of the E1B 55-kDa protein may be linked.
The early region 1B (E1B) of adenovirus type 5 (Ad5) encodes overlapping functions essential for virus mediated cellular transformation in cooperation with E1A by suppressing p53-mediated apoptosis or a G1 growth arrest (52, 62). The E1A proteins are potent transactivators that relieve cellular growth suppression and induce quiescent cells to enter S phase by binding members of the retinoblastoma protein family and transcription factors such as p300 (reviewed in reference 17). This action of E1A results in the accumulation of p53. p53 is a cellular growth suppressor that acts as a G1 checkpoint control (reviewed in references 37 and 74). In response to viral challenge, p53 may induce a G1 growth arrest by inducing genes such as the cyclin-dependent kinase inhibitor p21/WAF1/Cip1 gene (16, 69) or apoptosis by inducing genes such as bax1 (43). Either response by p53 is expected to severely hinder Ad replication and transformation (38).
The E1B 55-kDa and 19-kDa proteins are required to fully transform cells (4, 19, 23, 56, 64). The E1B 19-kDa protein is a functional homologue of the proto-oncogene-encoded Bcl-2 and prevents apoptosis by similar mechanisms (12, 52). The E1B 55-kDa proteins of Ad5 and Ad12 have been hypothesized to permit E1A-induced DNA synthesis and transactivation by preventing a p53-mediated G1 growth arrest (51, 60, 61). The 55-kDa protein complexes with the amino-terminal end of p53 and inhibits its activity as a transcription factor (33, 72, 73). This inhibition of p53-mediated transactivation by the large E1B protein is required for transformation by both the weakly oncogenic group C and highly oncogenic group A Ads (33, 72, 75, 76). E1B 55-kDa protein-mediated inactivation of p53 has been hypothesized to be required for viral replication in the lytic infection (7); however, this has not been demonstrated.
The Ad E4orf6 protein was recently shown to bind p53 and block transcriptional activation mediated by p53 (14, 46). Subsequently, the E4orf6 protein was shown to cooperate with the E1A and E1B proteins to transform baby rat kidney cells (46), to convert the nontumorigenic 293 human cell line (24) into a tumorigenic cell line in nude mice, and to block p53-dependent apoptosis (44). In both transformed and productively infected cells, coexpression of the E1B 55-kDa and the E4orf6 proteins decreased the stability of p53 (44, 46, 50). These findings suggest that the E4orf6 protein may encode some overlapping and redundant functions with the E1B 55-kDa protein with regard to transformation.
At late times in the lytic infection, the E1B 55-kDa protein facilitates the transport of viral late mRNA while inhibiting the transport of most cellular mRNA in association with the E4orf6 protein (3, 8, 26, 36, 49). Ad mutants that fail to express the E1B 55-kDa protein are defective for expression of late viral proteins and replicate poorly. The functional interaction between the E1B 55-kDa and E4orf6 proteins may be mediated by primate-specific cellular factors (22, 47). The transport of several cellular messages, including the heat shock protein 70, β-tubulin, and interferon-inducible Mx-A and 6-16 mRNAs, requires the E1B 55-kDa protein late in Ad infection. This effect correlates with activation of their transcription during the late phase (45, 71). In addition to selectively blocking transport of most cellular mRNA, the E1B 55-kDa protein further modulates host cell shutoff by inhibiting host protein synthesis by mechanisms unrelated to the inhibition of mRNA transport (2). We have recently demonstrated that the E1B 55-kDa protein functions in promoting Ad replication independently of the cell cycle. E1B 55-kDa mutant Ads produce virus most efficiently when cells are infected during S phase and are restricted from replication in cells infected during G1 (20). These findings suggest that the E1B 55-kDa protein plays a role in deregulating the cell cycle to the advantage of the lytic infection. Perhaps this function represents a link between the functions of the E1B 55-kDa protein in the lytic infection and transformation.
Because the E1B 55-kDa protein inhibits the function of p53, it has been hypothesized that an E1B 55-kDa mutant Ad can replicate only in cells lacking a functional p53. Evidence supporting this hypothesis includes the finding that the E1B 55-kDa mutant Ad dl1520 failed to lyse U2OS cells which contain wild-type p53. Furthermore, the E1B 55-kDa mutant virus induced more severe cytopathic effect in the RKO human colon cancer cell line transfected with a dominant negative p53 gene than in the parental cell line containing a wild-type p53 gene (7). However, p53 null tumor cells were not more susceptible to lysis by the E1B 55-kDa mutant virus than some cancer cells containing wild-type p53 (28). Furthermore, the relative ability of the E1B 55-kDa mutant virus to suppress tumor growth compared to the wild-type Ad was unaffected by the status of p53 (7). By contrast, Ridgway et al. (54) suggest that the interaction between p53 and the E1B 55-kDa protein is necessary for efficient Ad replication and that neither the wild-type nor E1B 55-kDa mutant virus replicated in p53-mutant cell lines. These authors reported that the wild-type Ad failed to efficiently shut off α-actin synthesis, synthesize viral DNA, and induce cytopathic effect in the p53 mutant cell lines T98G and 143B. They suggest that p53 may mediate the interaction between the E1B 55-kDa and E4orf6 proteins in the lytic infection (54). However, Rubenwolf et al. (55) demonstrated that p53 is not required for coimmunoprecipitation of the E1B 55-kDa and E4orf6 proteins although the two proteins have been shown to bind p53 independently (14, 55, 57).
The work presented here demonstrates that the inability of the E1B mutant virus to replicate efficiently and produce virus in all infected cells is not strictly due to the failure to abrogate p53 function. Among a variety of cell lines analyzed, the capacity of the E1B 55-kDa mutant virus to replicate, synthesize viral DNA, and produce virus in all infected cells did not correlate with the status of p53. However, the ability of the E1B 55-kDa mutant virus to replicate differed among the cell types studied. Furthermore, the ability of the E1B 55-kDa mutant virus to induce cell killing correlated with permissivity to virus growth and not the status of p53. In a cell line expressing a temperature-sensitive p53 allele, active p53 only moderately affected the replication of the E1B 55-kDa mutant virus. The defect in E1B 55-kDa mutant virus replication due to reduced temperature was as much as 10-fold greater than the defect due to p53 function. Indeed, the E1B 55-kDa mutant virus produced equivalent yields of virus and synthesized equivalent levels of late viral proteins compared to the wild-type virus in cells maintained at 39°C but not at 32°C. The cell cycle restriction of the E1B 55-kDa mutant virus was also partially overcome at 39°C, and progeny virus were produced in a larger fraction of infected cells. By contrast, the apparent cell cycle restriction was exacerbated at 32°C. Since both the cell cycle restriction and the defect in late gene expression exhibited a cold-sensitive phenotype, we speculate that the functions of the E1B 55-kDa protein in promoting mRNA transport and cell cycle-independent viral replication may be linked.
MATERIALS AND METHODS
Cell culture.
Cell culture media, cell culture supplements, and serum were obtained from Life Technologies (Gibco/BRL, Gaithersburg, Md.) through the Tissue Culture Core Laboratory of the Comprehensive Cancer Center of Wake Forest University. HeLa (ATCC CCL 2; American Type Culture Collection, Rockville, Md.), A549 (ATCC CCL 185), and 293 (ATCC CRL 1573) cells were maintained as monolayers in Dulbecco’s modified Eagle’s minimal essential medium (DMEM) supplemented with 10% newborn calf serum (CS), 100 U of penicillin per ml, and 100 μg of streptomycin per ml. Saos-2 cells were a generous gift of Jerry Zambetti (St. Jude Children’s Research Hospital, Memphis, Tenn.) and were maintained as monolayers in DMEM supplemented with 10% fetal bovine serum (FBS), 100 U of penicillin per ml, and 100 μg of streptomycin per ml. NCI-H460 (ATCC HTB 117) and NCI-H358 (ATCC CTL 5807) cells were maintained in antibiotic-free RPMI 1640 supplemented with 10% FBS and essential amino acids. U2OS cells (ATCC HTB 96) were maintained in McCoy’s 5A medium supplemented with 10% FBS, 100 U of penicillin per ml, and 100 μg of streptomycin per ml. H1299 cells and H1299-p53 cells were a generous gift of Nancy Raab-Traub (University of North Carolina, Chapel Hill) and were maintained in DMEM supplemented with 10% FBS, 100 U of penicillin per ml, and 100 μg of streptomycin per ml. The H1299-p53 cells were maintained at 39°C with 0.5 mg of Geneticin (Life Science Technologies, Gibco/BRL, Gaithersburg, Md.) per ml. p53 in H1299-p53 cells is in the wild-type conformation when cells are shifted to 32°C. Cells were maintained in subconfluent adherent cultures in a 5% CO2 atmosphere at the appropriate temperature by passage twice weekly. Cells were preserved in liquid nitrogen in 93% FBS–7% dimethyl sulfoxide.
Synchronization of the HeLa cell cycle was achieved by a combination of mitotic detachment and hydroxyurea block as described previously (20). HeLa cells were passaged 1:5 into 75-cm2 flasks for synchronization 12 to 16 h prior to the mitotic detachment. All but 5 ml of growth medium was removed, and the flasks were tapped sharply six times on each side. The detached cells were resuspended in DMEM supplemented with 10% CS and 2 mM hydroxyurea (Sigma, St. Louis, Mo.), replated at 2 × 105 cells per ml, and incubated at 37°C in a 5% CO2 atmosphere. After 1 h, the medium and nonadherent cells were replaced with fresh DMEM supplemented with 10% CS and 2 mM hydroxyurea and incubated for 12 h at 37°C in a 5% CO2 atmosphere. At the completion of the incubation with hydroxyurea, the cells were washed once with warm phosphate-buffered saline (PBS; 137 mM NaCl, 3 mM KCl, 1.76 mM KH2PO4, 10 mM Na2HPO4), and the last wash was replaced with normal growth medium to release the G1/S block.
Viruses.
The phenotypically wild-type Ad5 parent virus, dl309, used in these studies lacks a portion of the region encoding the E3 region that has been shown to be dispensable for growth in tissue culture (30). The E1B mutant virus dl338 contains a 524-bp deletion in the 55-kDa protein-coding region (49). Mutant virus dl1520D contains an 827-bp deletion in the region encoding the 55-kDa protein in combination with a stop codon to ensure that a truncated 55-kDa product cannot be expressed (4). The E1B 19-kDa mutant virus, dl337, contains a 146-bp deletion in the E1B 19-kDa protein-coding region between nucleotide sequence positions 1770 and 1916 (48).
The propagation of these viruses has been described elsewhere (30). In brief, virus stocks were prepared by infecting 293 cells at a multiplicity of infection (MOI) of 1. Virus was harvested 4 days postinfection from a concentrated freeze-thaw lysate by sequential centrifugation in discontinuous and equilibrium cesium chloride gradients (31). The gradient-purified virus was supplemented with 5 volumes of 12 mM HEPES (pH 7.4)–120 mM NaCl–0.1 mg of bovine serum albumin (fraction V; Life Technologies Inc.) per ml–50% glycerol (Fisher Scientific, Pittsburgh, Pa.) and stored at −20°C. The titer of each virus stock was determined by plaque assays on 293 cells (31).
For infection with Ad, cells were passaged 16 to 24 h prior to infection to a density of 2 × 105 cells per ml. Cells were washed once with PBS, and the final wash was replaced with virus (3 to 20 PFU per cell) in Ad infection medium (PBS supplemented with 0.2 mM CaCl2, 0.2 mM MgCl2, 2% CS, 100 U of penicillin per ml, and 100 μg of streptomycin per ml). The virus was added at one-fourth the normal culture volume, and the cells were gently rocked for 60 to 90 min at 37°C. The virus suspension was then replaced with normal growth medium, and the infected cells were returned to 37°C. For temperature-sensitive and cold-sensitive experiments, cells were infected at 39 or 32°C.
Infectivity.
The infectivities of HeLa, U2OS, Saos-2, C33A, and A549 cells were determined by infecting each cell line with dl309, dl338, or dl1520 at MOIs of 1, 10, and 30 PFU per cell. The virus titers were determined by plaque assays on 293 cells (24). At 14 h postinfection, infected monolayers of cells were harvested with trypsin to achieve a single-cell suspension. Cells were fixed in 4% formaldehyde for 20 min, permeabilized with 0.1% Triton X-100 for 4 min, and secondarily fixed in cold methanol for 5 min. The Ad E2A 72-kDa DNA binding protein was stained with the E2A 72-kDa protein-specific monoclonal antibody (clone B6-8) (53) used as hybridoma cell medium diluted 1:2 in Tris-buffered saline (137 mM NaCl, 3 mM KCl, 25 mM Tris-Cl [pH 8.0], 1.5 mM MgCl2, 0.5% bovine serum albumin, 0.1% glycine, 0.05% Tween 20, 0.02% sodium azide). The primary antibody was detected with fluorescein isothiocyanate (FITC)-conjugated goat antibodies specific for mouse immunoglobulin G (Jackson ImmunoResearch, West Grove, Pa.). Cells were resuspended in PBS, filtered through a nylon mesh, and passed through a 27.5-gauge needle to achieve a single-cell suspension. Cells stained with the E2A 72-kDa protein-specific antibody were counted by fluorescence-activated cell sorting (FACS) using a Coulter Epics XL flow cytometer (Coulter Corp., Miami, Fla.) with an argon laser as the excitement source. In each case, 40,000 events were measured. H1299, H460, and H358 cell infectivity was determined similarly except that cells were fixed and stained as a monolayer. Cells staining with the E2A 72-kDa protein-specific antibody were identified by epifluorescence with a Leitz Dialux 20 EB microscope. Approximately 200 cells were evaluated for each experiment.
Electron microscopy.
Cells were fixed for transmission electron microscopy with 2.5% glutaraldehyde (Polysciences, Warrington, Pa.) in PBS–1.5 mM MgCl2 or in 0.1 M sodium cacodylate at 20 to 30 h postinfection. The fixed cell pellet was postfixed with osmium tetroxide in cacodylate buffer and dehydrated in a graded series of alcohol. Specimens were infiltrated with Spurr’s resin-propylene oxide and cut into approximately 100-nm-thick sections with a diamond knife. Sections were collected on copper grids, stained with uranyl acetate and lead citrate, and analyzed at 80 keV with a Philips 400 transmission electron microscope. Specimens were embedded and sectioned by MicroMed, the Electron Microscopy Core Laboratory of the Comprehensive Cancer Center of Wake Forest University.
Flow cytometry.
HeLa cells were harvested by treatment with trypsin and fixed in 70% ethanol for 1 h to overnight. The ethanol was removed, and the cells were resuspended to approximately 106 cells per ml in propidium iodide buffer (100 mM NaCl, 36 mM sodium citrate, 50 μg of propidium iodide per ml, 0.6% Nonidet P-40) supplemented with 0.04 mg of RNase (Sigma) per ml. The cells were filtered through nylon mesh and passed through a 27.5-gauge needle to achieve a single-cell suspension. The DNA content of individual cells was measured by FACS using a Coulter Epics XL flow cytometer (Coulter Corp.) with an argon laser as the excitement source (488 nm). The emitted light was analyzed for forward and 90°C scatter, pulse width (to discriminate doublets), and red fluorescence (>630 nm) of propidium iodide to determine the DNA content per nucleus; 40,000 events were measured in each analysis. The resulting data were acquired in list mode for discriminatory analysis such as the use of standard gating procedures to define distinct populations of cells, doublets, and debris. All flow cytometric analyses were conducted by the Steroid Receptor Laboratory in cooperation with the Hematology Flow Cytometry Laboratory of North Carolina Baptist Hospital.
Plaque assays for viral yields.
Detailed methods for Ad plaque assays have been described elsewhere (31). In brief, virus was harvested from cells in culture medium 48 to 72 h postinfection by multiple cycles of freezing and thawing. The cell lysates were clarified by centrifugation and serially diluted in infection medium for infection of 293 cells for plaque assays. After incubation with dilute virus for 1 h, the infected cells were overlaid with 0.7% SeaKem ME agarose (FMC, Rockland, Maine) in DMEM containing 0.75% sodium bicarbonate and 4% CS. The cells were fed with additional agar overlays every third day for 7 days. The plaques were visualized by staining with 0.01% neutral red in an agarose overlay on the seventh day.
DNA slot blotting.
The DNA slot blotting procedure has been described in detail previously (1, 32). Briefly, total cellular DNA was isolated from infected HeLa, A549, U2OS, and C33A cells. Cells were collected, pelleted, and resuspended in 10 mM Tris (pH 8.0). An equal volume of lysis buffer (400 mM Tris [pH 8.0], 100 mM EDTA [pH 8.0], 1% sodium dodecyl sulfate [SDS], 200 μg of proteinase K per ml) was added, and the cells were kept at 50°C for 1 h. DNA was extracted with phenol-chloroform, precipitated, and quantified by spectrophotometry (A260). Equivalent amounts of total cellular DNA was blotted onto Nytran nylon membranes (Schleicher & Schuell, Keene, N.H.) by using a manifold device (Life Technologies) and vacuum. The immobilized DNA was denatured, neutralized, and cross-linked to the matrix with UV light (Stratalinker; Stratagene, La Jolla, Calif.). The DNA was then hybridized with an excess of [α-32P]dATP-labeled (ICN, Costa Mesa, Calif.) DNA probe generated by random-primed synthesis of wild-type Ad DNA (1). Hybridized probe was quantified with the use of a Molecular Dynamics (Sunnyvale, Calif.) PhosphorImager and ImageQuant analysis software.
Late viral protein synthesis.
Late viral protein synthesis was analyzed by pulse-labeling infected H1299 cells for 1 h with 0.1 mCi of 35S-labeled amino acids (Trans35S label; ICN Biochemicals) per ml in cysteine- and methionine-free DMEM supplemented with 2% FBS at 32 or 64 h postinfection. Cells were then scraped and resuspended and lysed in 2× protein sample buffer (2% SDS, 125 mM Tris [pH 6.8], 20% glycerol, 5% β-mercaptoethanol, 100 mM dithiothreitol, 0.01% bromophenol blue). Protein from 1 × 105 to 2 × 105 cell equivalents was separated by SDS-polyacrylamide gel electrophoresis (PAGE) on an 8% polyacrylamide gel (36:1 acrylamide/N,N′-methylenebisacrylamide ratio; Polysciences). Polyacrylamide gels were fixed in 15% glacial acetic acid (Fisher Scientific)–7.5% methanol. Proteins were quantified with the use of a Molecular Dynamics PhosphorImager and ImageQuant analysis software. Ad late proteins were identified by reference to virion standards that were synthesized in the presence of 14C-labeled mixed amino acids (Amersham, Arlington Heights, Ill.) and gradient purified from 293 cells infected with the wild-type Ad dl309 as described elsewhere (31).
Cell killing assays.
Virus-induced cell killing was measured in a time course fashion by using calcein uptake and ethidium exclusion (LIVE/DEAD assay; Molecular Probes, Eugene, Oreg.). Synchronized HeLa cells or asynchronous population of A549, C33A, U2OS, or Saos-2 cells were plated in six-well dishes at 5 × 104 cells per ml. Cells were mock infected or infected with dl309 or dl1520 at 3 and 10 PFU per cell. At 24, 48, 72, 96, 120, 168, 192, and 240 h after infection, cells were trypsinized and collected with the nonadherent cells from the media. Cells were washed once with PBS, exposed as living cells to 0.1 ml of 4 μM ethidium homodimer and 2 μM calcein AM in PBS for 40 min, and then diluted with 0.4 ml of PBS, Calcein AM is converted from a nonfluorescent cell-permeant to an intensely fluorescent nonpermeant compound by ubiquitous intracellular esterase activity in live cells. Ethidium homodimer enters cells with damaged membranes, binds nucleic acids, and fluoresces bright red in dead cells but is excluded from live cells. The fraction of live and dead cells was measured by FACS using a Coulter Epics XL flow cytometer (Coulter Corp.) with an argon laser as the excitement source (488 nm) and standard gating procedures. All flow cytometric analyses were conducted by the Hematology Flow Cytometry Laboratory of North Carolina Baptist Hospital.
Apoptosis assay.
HeLa cells synchronized to S phase or G1 were infected with dl309, dl1520, or dl337 at 10 PFU per cell or were mock infected. At 24 h postinfection, phosphatidylserine exposed on the surface of cells was visualized with annexin V-FITC conjugate (3 μg/ml) in binding buffer (10 mM HEPES [pH 7.4], 150 mM NaCl, 5 mM KCl, 1 mM MgCl2, 1.8 mM CaCl2; Trevigen, Gaithersburg, Md.). The annexin V-FITC reagent was generously provided by Greg Kucera (Wake Forest University School of Medicine, Winston-Salem, N.C.). Cells were washed in binding buffer and fixed in 4% formaldehyde with 1.8 mM CaCl2. Cell nuclei were visualized by staining with 4,6-diamidino-2-phenylindole (DAPI; Sigma), a nonspecific DNA binding stain. Cells were examined by epifluorescence with a Leitz Dialux 20 EB microscope.
Computer-assisted graphics.
Radioactive samples were visualized with the use of a Molecular Dynamics PhosphorImager and ImageQuant analysis software. Radioactive images were obtained as 16-bit gray scale images and then converted to 8 bit gray scale images. The chemiluminescent images were recorded on film, which was scanned at 300 dots/in. with a Hewlett-Packard scanner fitted with a transparency adapter into an 8-bit gray scale image. The density ranges were adjusted for printing without further contrast or image enhancement. The digitized images were imported at 300 dots/in. into the graphic software Canvas (Deneba Software, Miami, Fla.) operating on a Macintosh microcomputer to create the final figures.
RESULTS
Replication of the E1B 55-kDa mutant virus is not correlated with the status of p53.
Levels of replication of the phenotypically wild-type Ad dl309 and two E1B 55-kDa mutant viruses, dl338 and dl1520, were compared among cell lines of known p53 status. Both mutant viruses contain large deletions within the gene encoding the E1B 55-kDa protein. However, dl1520 contains a termination codon at the second position to preclude expression of any E1B 55-kDa-related protein (4), whereas the deletion present in the dl338 genome could allow expression of a truncated (17-kDa) protein. The eight cell lines analyzed included HeLa cells, three p53 wild-type cell lines, and four p53 null or mutant cell lines. HeLa cells have a wild-type p53 sequence but are functionally mutant due to the presence of the human papillomavirus E6 protein (59, 66). However, during an Ad infection, active p53 accumulates in infected HeLa cells (10, 25). Therefore, in Ad-infected HeLa cells, p53 might be expected to inhibit virus replication in the absence of the E1B 55-kDa protein (10). Table 1 summarizes the status of p53 in each of the other cell lines tested, which include three p53 wild-type cell lines, A549 (35), H460 (9, 63), and U2OS (13), and four p53-deficient cell lines, C33A (11, 58), H358 (9, 63), H1299 (42), and Saos-2 (39).
TABLE 1.
p53 status in human cell lines
| Name | Cell origin | p53 status | Reference |
|---|---|---|---|
| A549 | Lung carcinoma | Wild type | 35 |
| H460 | Large-cell lung carcinoma | Wild type | 9, 63 |
| U2OS | Osteogenic sarcoma | Wild type | 13 |
| C33A | Non-HPV cervical carcinoma | A→C substitution at codon 273 | 11, 58 |
| H358 | Non-small-cell lung carcinoma | Homozygous deletion | 9, 63 |
| H1299 | Non-small-cell lung carcinoma | Homozygous deletion | 42 |
| Saos-2 | Osteogenic sarcoma | Homozygous deletion | 39 |
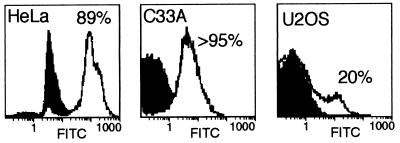
The infectivity of Ad varied significantly among the cell lines analyzed, as illustrated by the results shown in Fig. 1. For this experiment, HeLa, C33A, or U2OS cells were infected with approximately 1 PFU of dl1520 per cell (the virus titer was determined on 293 cells). Cells were fixed and labeled by indirect immunofluorescence for a representative early gene product, the E2A 72-kDa protein. Cells expressing the E2A 72-kDa protein were quantified by flow cytometry using mock-infected cells as the negative control. Typically, infectivity in HeLa cells reflected the titer measured with 293 cells. In this experiment, we measured 89% infected HeLa cells. The C33A cells appeared to infect more readily, and more than 95% of these cells were infected by 1 PFU per cell. By contrast, only 20% of the U2OS cells were infected by the same amount of virus. We measured similar results by indirect immunofluorescence microscopy of infected U2OS cells even after allowing the infection to continue for 3 days.
FIG. 1.
Infectivity determined after infection with a constant amount of virus varies significantly between different cell lines. The cell lines indicated in each panel were infected with approximately 1 PFU of dl1520 (as measured by titer on 293 cells) per cell. At 15 h postinfection, cells were stained for the E2A 72-kDa protein by indirect immunofluorescence, and the fraction of infected cells was determined by FACS. The abscissa indicates the relative number of cells counted at the fluorescence intensity indicated on the ordinate. The filled curves were derived from uninfected cells; the nonfilled curves were obtained from infected cells. The percentage of positive cells indicated in each panel was determined by fitting the data to two Gaussian curves.
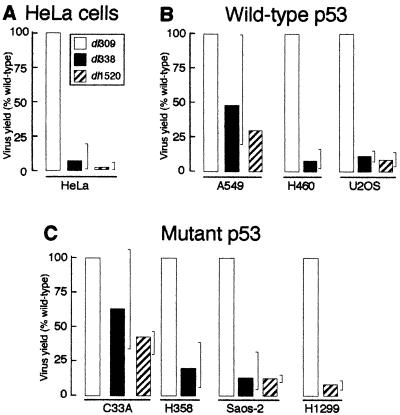
The amount of virus required to infected 80 to 90% of the cells was determined for each cell line by the method illustrated in Fig. 1. As defined by titer on 293 cells, HeLa, A549, and C33A cells required 1 to 3 PFU per cell to infect 80 to 90% of the cells. H1299, H358, and H460 cells required 3-fold more virus whereas Saos-2 and U2OS cells required 10-fold more virus to infect an equivalent number of cells. Each cell line was infected at an MOI sufficient to infect at least 80% of the cells. At 48 to 72 h postinfection, the cells were harvested and virus yields were measured by titer on 293 cells. The results of these measurements, summarized in Fig. 2, suggest that the status of p53 does not predict the ability of the cell line to support growth of the E1B 55-kDa mutant virus.
FIG. 2.
The inability of the E1B 55-kDa mutant virus to replicate is not mediated by p53. Monolayer cultures of HeLa cells, p53 wild-type cell lines, or p53 mutant cell lines were infected with the wild-type virus dl309 or the E1B 55-kDa mutant virus dl338 or dl1520 at an MOI sufficient to infect >80% of the cells (3 to 30 PFU/cell). Cells were lysed at 48 h postinfection, and virus yields were measured by plaque assay on 293 cells. The yields shown, expressed as percentages of virus produced relative to the wild-type virus, are averages of at least three independent experiments; the range of values is indicated by the brackets.
As we and others have reported previously, HeLa cells did not permit efficient replication of the E1B 55-kDa mutant viruses (20). The E1B 55-kDa mutant viruses dl338 and dl1520 grew to averages of 7 and 2%, respectively, of the wild-type virus yield in HeLa cells (Fig. 2A). Two of the three wild-type p53 cell lines restricted growth of the mutant viruses. In H460 and U2OS cells, the mutant virus produced approximately 8 and 10% of the virus obtained from an infection with wild-type virus (Fig. 2B). However, the E1B 55-kDa mutant virus grew to an average of 30 to 50% of the wild-type yield in A549 cells (Fig. 2B). In some experiments, the yield of the mutant virus was the same as the yield of the wild-type virus from A549 cells. Some of the p53 mutant cell lines also restricted growth of the E1B 55-kDa mutant viruses (Fig. 2C). Saos-2 and H1299 cells restricted growth of the mutant viruses to an averages of 14 and 8% of the level produced by the wild-type virus, respectively. H358 cells exhibited lesser restriction, producing an average of 20% of the wild-type virus yield, with a maximum value of 38% recorded in one of eight independent experiments. Finally, the p53 mutant C33A cell line was least restrictive to the E1B 55-kDa mutant viruses, producing between 40 and 65% of the yield obtained from a wild-type virus infection. In summary, the E1B 55-kDa mutant viruses grew to the highest titer in cell lines containing a mutant p53 (C33A) and a wild-type p53 (A549) gene. It should be noted that human embryonic kidney cells, which would have wild-type p53, have also been reported to be permissive for growth of E1B 55-kDa mutant Ads (6). These results suggest that although levels of replication of the E1B 55-kDa mutant viruses varied among cell types, these differences may not be mediated by p53.
E1B 55-kDa mutant Ads synthesize viral DNA to levels equivalent to those of the wild-type virus in a permissive and a restrictive cell line irrespective of the status of p53 in the infected cell.
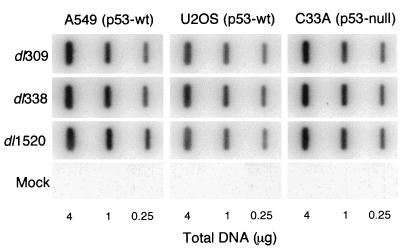
Based on the known function of p53, it is reasonable to expect that p53 could inhibit the replication of Ad in the absence of the E1B 55-kDa protein by inducing a G1 growth arrest and inhibiting viral DNA synthesis. This effect may not be readily apparent in the virus yields measured in Fig. 2; therefore, viral DNA synthesis was measured in two p53-wild-type cell lines, A549 and U2OS, and in one p53 mutant cell line, C33A. Of the two wild-type p53 cell lines chosen, A549 cells were permissive for E1B 55-kDa mutant virus replication and U2OS cells were restrictive for E1B 55-kDa mutant virus replication (Fig. 2). Hybridization analysis of total DNA isolated from cells infected with wild-type or E1B 55-kDa mutant Ad demonstrated that cells infected with either E1B 55-kDa mutant virus (dl338 or dl1520) synthesized viral DNA to levels equivalent to that of the same cells infected with the wild-type virus (Fig. 3). We previously reported that HeLa cells infected with the E1B 55-kDa mutant virus dl338 also synthesized wild-type levels of viral DNA (20). Among these four cell lines, neither the permissivity toward replication of the E1B 55-kDa mutant virus nor p53 status affected viral DNA synthesis. These results are consistent with the suggestion that the crucial role for the E1B 55-kDa protein in viral replication occurs after the onset of viral DNA synthesis during the late phase of virus replication.
FIG. 3.
E1B 55-kDa mutant virus-infected p53 wild-type (p53-wt) and p53 mutant cells synthesize viral DNA to the same level as the wild-type virus-infected cells. A549, U2OS, and C33A cell lines were mock infected or infected with the wild-type virus dl309 or the E1B 55-kDa mutant virus dl338 or dl1520 at an MOI sufficient to infect >80% of the cells (10 to 30 PFU/cell). Total DNA was isolated from equal numbers of Ad-infected cells at 20 h postinfection. Total DNA in the amount indicated below each lane was transferred to a nylon membrane, denatured, and hybridized with radioactive Ad-specific DNA probes generated by random-primed synthesis. Hybridized probe was quantified with a PhosphorImager, and mock-infected background was subtracted. Identical amounts of viral DNA were measured in each cell line infected with the E1B 55-kDa mutant viruses and the wild-type virus.
Cells that are permissive for the growth of the E1B 55-kDa mutant virus produce progeny in a greater fraction of infected cells.
The cell cycle-restricted phenotype of the E1B 55-kDa mutant virus was first identified in HeLa cells by using electron microscopy (20). In infected populations of HeLa cells, only 20% of the E1B 55-kDa mutant Ad-infected cells produced progeny virus, whereas nearly all cells infected with the wild-type Ad produced progeny virus. The p53 wild-type and p53 null or mutant cell lines infected with the wild-type or E1B 55-kDa mutant virus were examined by electron microscopy at 32 h postinfection to determine the fraction of infected cells producing E1B 55-kDa mutant virus progeny. The MOI was adjusted for each cell line to infect greater than 80% of the cells. Because the mutant virus-infected cells synthesized wild-type levels of viral DNA, the infected cells were readily identified by the presence of viral inclusions in the cell nucleus. The results of these experiments are summarized in Table 2.
TABLE 2.
Virus production in cell lines with either wild-type or mutant p53
| Cell line | Cells producing virus (%)a
|
|
|---|---|---|
| dl309 | dl1520 | |
| Wild-type p53 | ||
| A549 | 95 | 70 |
| U2OS | 90 | <10 |
| Mutant p53 | ||
| C33A | 95 | 90 |
| H1299 | 92 | 28 |
| Saos-2 | 90 | 22 |
Average of two independent experiments from each of which at least 200 cells were counted. Cells that were clearly infected were scored for the presence or absence of virus particles. Cells were infected with each virus at an MOI sufficient to infect at least 80% of the cells and processed 24 to 32 h postinfection for transmission electron microscopy.
In all five cell lines analyzed, the wild-type virus produced progeny virus in at least 90% of the infected cells. By contrast, cell lines that restricted replication of the E1B 55-kDa mutant virus, H1299, Saos-2, and U2OS, contained progeny mutant virus in 28% or less of the infected cells. This value is comparable to the value of 22% reported for asynchronously infected HeLa cells (20). The two cell lines that best supported replication of the E1B 55-kDa mutant virus, A549 and C33A, contained progeny virus in a greater fraction of cells compared to the restrictive cell lines. An average of 70% of the A549 and 90% of C33A cells infected with dl1520 contained progeny virus. These results suggest that replication of the E1B 55-kDa mutant virus is not subjected to a cell cycle growth restriction in A549 and C33A cells. Furthermore, because the E1B 55-kDa mutant virus produced progeny in 70% or more of the infected p53 wild-type cells (A549) as well as the p53 mutant cell line (C33A), it seems likely the cell cycle growth restriction is not strictly mediated by p53.
Cell killing by the E1B 55-kDa mutant virus correlates with permissivity to mutant virus replication and not the status of p53 in the infected cell.
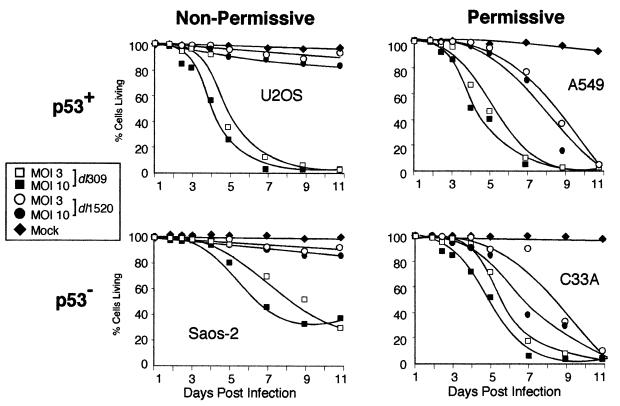
It was suggested that dl1520 (ONYX-015) selectively kills cells that lack normal p53 function (7) although dl1520 was found to also kill some tumor cells of wild-type p53 status (28). Therefore, the relationship between p53, replication of an E1B 55-kDa mutant virus, and virus-induced cell killing was analyzed in two p53 wild-type cell lines (A549 and U2OS) and two p53 mutant cell lines (C33A and Saos-2). As demonstrated by the results in Fig. 2, A549 and C33A cells were permissive for growth of an E1B 55-kDa mutant virus whereas U2OS and Saos-2 cells were restrictive for the growth of the E1B 55-kDa mutant virus. For these experiments, replicate cultures of cells were mock infected or infected with the wild-type virus dl309 or the E1B 55-kDa mutant virus dl1520 at 3 and 10 PFU per cell for A549 and C33A cells and at 9 and 30 PFU per cell for U2OS and Saos-2 cells. These multiplicities correspond to 3 and 10 infectious units per cell for the respective cell lines. For 11 days after infection, cells were harvested at the times indicated and the percentage of live cells and dead cells was determined by a flow cytometric assay. The results of these experiments suggest that cells that permitted growth of the E1B 55-kDa mutant Ad were susceptible to killing by the E1B mutant virus, irrespective of the status of p53.
The wild-type Ad dl309 killed more than 50% of the cells by day 6 or 7 in all cell lines examined (Fig. 4). By 9 days postinfection, 95% of the cells were killed by the wild-type virus for each cell line except Saos-2 cells. By contrast, the E1B 55-kDa mutant virus completely lysed only A549 and C33A cell cultures. The onset of E1B 55-kDa mutant-induced cell killing was slightly delayed in these cell lines compared to the wild-type virus; the E1B 55-kDa mutant virus induced complete lysis by 11 days postinfection, in contrast to the wild-type virus, which required 9 days. U2OS and Saos-2 cells were resistant to lysis by the E1B 55-kDa mutant virus. By 11 days postinfection, fewer than 20% of the U2OS and Saos-2 cells in the E1B mutant-infected population had been killed. Cell killing did not demonstrate a striking multiplicity effect over the range examined since cells infected at the lower multiplicities (3 or 9 PFU per cell) were lysed nearly as efficiently as cells infected at the higher multiplicities (10 or 30 PFU per cell). The cells most susceptible to killing by the E1B 55-kDa mutant virus were those that produced the highest yields of virus as measured in Fig. 2.
FIG. 4.
Cell killing induced by the E1B 55-kDa mutant virus correlates with permissivity to viral replication, not the status of p53. U2OS, Saos-2, A549, and C33A cells cultured in six-well plates were mock infected or infected with either the wild-type virus dl309 or the E1B 55-kDa mutant virus dl1520 at 3 and 10 infectious units per cell. During a time course after infection, replicate dishes of cells were harvested and the percentage of live cells was determined by flow cytometry using the LIVE/DEAD assay. Live cells with active esterases converted the nonfluorescent calcein AM to a brightly fluorescing molecule (green). Dead cells were stained with ethidium homodimer (red), which was excluded from living cells. Typically 40,000 cells were analyzed. The data shown represent a single experiment performed in duplicate. The top two graphs represent data from wild-type p53 (p53+) cell lines (U2OS and A549); the bottom two graphs represent data from a p53-mutant (p53−) cell lines (Saos-2 and C33A); the two graphs on the left represent data from infection of relatively non permissive cell lines (U2OS and Saos-2); the two graphs on the right represent data from infection of relatively permissive cell lines (A549 and C33A).
S-phase cells are more susceptible than G1 cells to cell killing by the E1B 55-kDa mutant virus.
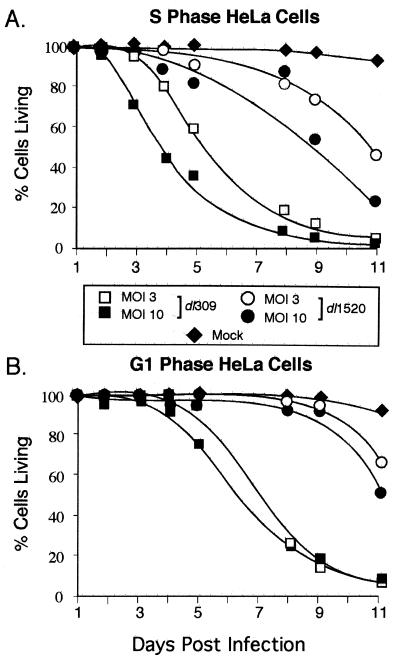
We previously showed that HeLa cells infected during S phase were permissive for E1B 55-kDa mutant virus replication whereas HeLa cells infected during G1 phase restricted E1B mutant virus replication. S-phase cells infected with the E1B 55-kDa mutant virus also exhibited more severe cytopathic effect than G1-infected cells (20). Therefore, the possibility that HeLa cells infected during S phase were more susceptible to virus-induced cell killing than HeLa cells infected during G1 phase was tested. Replicate cultures of synchronized HeLa cells were infected during S phase or G1 with the wild-type (dl309) or E1B 55-kDa mutant virus (dl1520) at 3 and 10 PFU per cell or were mock infected. As before, cells were harvested at the indicated times, and the fraction of living cells was determined. The wild-type virus killed both cells infected at the onset of S phase and during G1 with almost equal efficiencies (Fig. 5). The wild-type virus induced complete cell lysis by 11 days postinfection in cells infected during S phase or G1; however, killing was slightly delayed in cells infected during G1 phase of the cell cycle. Unlike the wild-type virus, the E1B 55-kDa mutant virus failed to kill cells infected in early G1 until day 11, at which time a maximum of 50% of cell death was measured (Fig. 5B). By contrast, S-phase-infected cells were killed by the mutant virus as soon as 4 days postinfection. By 11 days postinfection, only 20% of the cells infected during S phase with the E1B 55-kDa mutant virus were still living (Fig. 5A). These data further support the idea that cells permissive for growth of the mutant virus are most susceptible to killing by the virus.
FIG. 5.
HeLa cells infected during S phase are more susceptible than cells infected during G1 to cell killing by the E1B 55-kDa mutant virus. HeLa cells were synchronized and cultured in six-well plates and then mock infected or infected with the wild-type virus dl309 or the E1B 55-kDa mutant virus dl1520 at MOIs of 3 and 10 at the onset of S phase or G1. During a time course after infection, replicate dishes of cells were harvested and the percentage of live cells was determined by flow cytometry using the LIVE/DEAD assay as described in Materials and Methods. The data shown are from a single experiment and are representative of two independent experiments each performed in duplicate.
To determine if the mechanism of cell killing differed between cells infected with the wild-type virus and the E1B 55-kDa mutant virus, synchronized and infected cells were labeled with annexin V 24 h postinfection (Table 3). Annexin V is a calcium-dependent phospholipid-binding protein with high affinity for phosphatidylserine. Phosphatidylserine is normally restricted to the inner surface of the plasma membrane. However, early in the apoptotic pathway, the asymmetry of the plasma membrane is lost and phosphatidylserine is externalized (reviewed in reference 65). The E1B 19-kDa mutant virus dl337 induces apoptosis in infected cells and was used as a positive control (12, 48, 52). Only 2 to 3% of mock-infected S-phase or G1 HeLa cells stained with annexin V. By contrast, 71% of the HeLa cells infected during S phase and 90% of the HeLa cells infected during G1 with dl337 underwent apoptosis. The reason for increased apoptosis in G1-phase cultures infected with dl337 is not known. The fraction of apoptotic cells in cultures infected with dl309 or dl1520 was similar to or only slightly greater than that in mock-infected cultures. Furthermore, S-phase HeLa cells infected with the E1B 55-kDa mutant virus were not more likely to undergo apoptosis than cells infected during G1. These results suggest that although S-phase HeLa cells are more susceptible to killing by the E1B 55-kDa mutant virus, the increased cell killing is not due to increased apoptosis.
TABLE 3.
Fraction of apoptotic HeLa cells in cultures infected during S phase or G1a
| Virus | Cells labeled with annexin V (mean % ± SD)
|
|
|---|---|---|
| S | G1 | |
| dl309 | 3.1 ± 1.0 | 5.1 ± 1.2 |
| dl1520 | 2.4 ± 0.9 | 3.4 ± 1.0 |
| dl337 | 71.1 ± 4.0 | 90.2 ± 5.0 |
| Mock | 2.3 ± 0.8 | 2.9 ± 0.9 |
Cells were infected at the stage of the cell cycle indicated with 10 PFU of virus per cell. At 24 h postinfection, cells were labeled with FITC-conjugated annexin V, and at least 300 cells were evaluated to determine the percentage of apoptotic cells.
p53 has a modest effect on growth of the E1B 55-kDa mutant viruses in a cell line expressing a temperature-sensitive p53 allele.
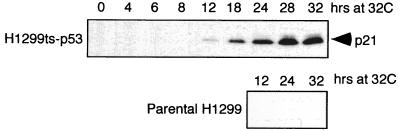
A limitation to using cell lines of defined p53 status to test the role of p53 in virus growth is that other cell-specific changes that may affect virus growth are not controlled between the cell lines. To circumvent this limitation, growth of the E1B 55-kDa mutant viruses was analyzed in H1299 cells that express a temperature-sensitive p53 allele (18). The parental H1299 human lung carcinoma cell line contains a homozygous deletion of the p53 gene (42) and therefore allows analysis of ectopic p53 function without interference from endogenous p53 protein. H1299 cells were stably transfected with the temperature-sensitive mouse p53 allele tsVal135 to create the H1299-p53 cell line (18). The p53 protein expressed in H1299-p53 cells is largely mutant at 39°C but is found predominantly in the wild-type tetramer conformation at 32°C. Accumulation of active p53 leads to the induction of the cell cycle inhibitor p21/WAF-1. p21 inhibits cyclin-dependent kinase complex formation and can therefore induce a G1 growth arrest (15, 16, 27, 69, 70). To determine the point at which p53 became functional after the shift to 32°C, the induction of p21 was analyzed in a time course fashion following the shift from 39 to 32°C by Western blotting (Fig. 6).
FIG. 6.
p21 expression is induced in H1299-p53 cells by 12 h after the shift to the permissive temperature. H1299-p53 cells and parental H1299 cells were shifted from 39°C (p53 mutant) to 32°C (p53 wild-type). Equivalent numbers of cells were lysed at the time indicated above each lane. Total cellular protein from 2 × 105 cells was separated by SDS-PAGE and then electrophoretically transferred to nitrocellulose. p21 expression was analyzed by standard Western blotting methods. The p21/WAF-1 protein present in cellular lysates was visualized with the monoclonal antibody clone EA10 (Santa Cruz Biotechnology, Inc., Santa Cruz, Calif.).
Expression of p21 was readily apparent within 12 h after the shift of H1299-p53 cells to 32°C (Fig. 6). p21 levels continued to increase through 32 h following the shift to the lower temperature. As expected, p21 was not induced in the parental H1299 cell line at 32°C. Cells expressing the temperature-sensitive p53 allele stopped dividing after being shifted to 32°C for 24 h (18, 41). H1299-p53 cells shifted to 32°C were also examined for changes in cell cycle distribution (i.e., G1 arrest) by FACS. Growth of the parental H1299 cell line at 32°C did not appreciably alter the cell cycle distribution. After 12 h at 32°C, the cell cycle distribution of the H1299-p53 cells was the same as for cells maintained at 39°C. However, by 32 h after the shift to the lower temperature, the percentage of cells in S phase had decreased from 45 to 13%, with a concomitant increase in the fraction of cells in G2/M phase (data not shown). This alteration in cell cycle distribution due to prolonged expression of p53 at 32°C is consistent with the findings of Michalovitz et al. (41). These data indicate that p53 adopts the wild-type conformation within 12 h of the shift to the lower temperature. However, p53 function has not appreciably altered the fraction of cells in S phase after 12 h at 32°C. A decrease in the fraction of cells in S phase would be expected to affect the growth of the E1B 55-kDa mutant (20).
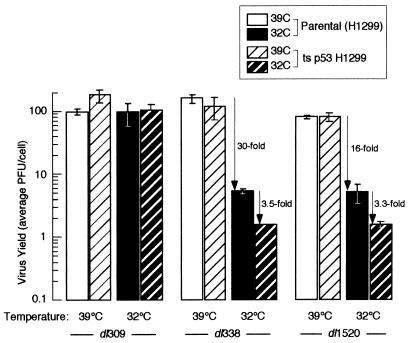
Portions of both H1299 and H1299-p53 cells were shifted from 39 to 32°C for 12 h and then infected with the wild-type virus (dl309) or an E1B 55-kDa mutant virus (dl338 or dl1520) at an MOI of 9 PFU per cell (Fig. 7). Virus yields were determined by titer on 293 cells at 48 h postinfection for cells at 39°C and 96 h postinfection for cells at 32°C. The wild-type virus produced equivalent yields of virus in both cell lines irrespective of the status of p53. Strikingly, the E1B 55-kDa mutant viruses produced wild-type virus yields in both the parental and H1299-p53 cell lines when cells were maintained at 39°C. However, growth of the E1B 55-kDa mutant Ads was restricted in the parental H1299 cells at 32°C. Approximately 30-fold (dl338) and 16-fold (dl1520) less virus was produced in cells maintained at 32°C than in those maintained at 39°C. Wild-type p53 modestly diminished the growth of the E1B 55-kDa mutant virus in H1299-p53 cells at 32°C, reducing virus yields 3.5-fold (dl338) and 3.3-fold (dl1520) beyond that of reduced temperature alone. Thus, p53 played a significant, albeit minimal, role in inhibiting growth of the E1B 55-kDa mutant Ad in H1299-p53 cells. By contrast, the growth defect of the E1B 55-kDa mutant viruses in these experiments was primarily due to reduced temperature and was no longer evident at the elevated temperature.
FIG. 7.
p53 only modestly affects replication of the E1B 55-kDa mutant virus in H1299-p53 cells. Portions of H1299-p53 and parental H1299 cells were shifted to 32°C or maintained at 39°C. Twelve hours later, cells maintained at 39°C or shifted to 32°C were infected with either the wild-type virus dl309 or the E1B 55-kDa mutant viruses dl338 and dl1520 at an MOI of 9 PFU per cell. Virus yields were measured by plaque assay on 293 cells, using lysates harvested at 48 h postinfection for cells infected at 39°C and 4 days postinfection for cells infected at 32°C. Virus yields are expressed as average PFU per cell derived from two independent experiments performed in duplicate; error bars indicate standard errors of the means.
The cell cycle restriction of the E1B 55-kDa mutant virus may be overcome at the elevated temperature.
The ability of the E1B 55-kDa mutant virus to replicate to near wild-type virus yields in H1299 cells at 39°C may reflect the absence of a cell cycle restriction at the elevated temperature. This possibility was examined by using electron microscopy to measure the fraction of virus-producing cells at 39 and 32°C. H1299 cells were maintained at 39 or 32°C and infected with the wild-type virus dl309 or the E1B 55-kDa mutant virus dl1520. Cells were fixed and processed for transmission electron microscopy at 24 h postinfection for cells maintained at 39°C and 48 h postinfection for cells maintained at 32°C. Over 200 infected cells were scored for the presence of progeny virus in each of two independent experiments, and the results are summarized below.
Virus production in H1299 cells infected with the wild-type virus was not affected by temperature. At both temperatures, 92% of the infected cells contained progeny wild-type virus. The apparent cell cycle restriction to E1B 55-kDa mutant virus replication in H1299 cells may have been partially overcome at 39°C. The E1B 55-kDa mutant virus produced progeny in 73% ± 7% of the infected cells that were maintained and infected at 39°C. Similar results were obtained for HeLa cells grown and infected at elevated temperature. When HeLa cells were maintained and infected at 39°C, the E1B 55-kDa mutant virus produced progeny in approximately 60% of the infected cells, suggesting that the cell cycle restriction to E1B 55-kDa mutant virus replication in HeLa cells can also be partially overcome by elevated temperature.
In contrast to H1299 cells infected at the elevated temperature, H1299 cells maintained and infected at 32°C with the E1B 55-kDa mutant virus contained progeny virus in only 12% ± 3% of the infected cells. Because 28% of the H1299 cells maintained and infected at 37°C contained E1B 55-kDa mutant virus progeny (Table 2), it seems likely that the cell cycle restriction is more severe at the lower temperature.
The cold-sensitive growth phenotype of the E1B 55-kDa mutant Ad in H1299 cells appears to be linked to the defect in late gene expression.
The E1B 55-kDa mutant viruses hr6 and hr13 were reported to be cold-sensitive for growth (29). Yields of hr6 and hr13 were reduced 213- and 75-fold, respectively, in HeLa cells maintained at 32.5°C compared to 38.5°C. Furthermore, the virus-mediated mRNA transport defect of the E1B 55-kDa mutant virus was shown to be more severe at 32°C than at 37 or 39°C (36, 68). Thus, it seems likely that the cold-sensitive phenotype of E1B 55-kDa mutant Ad growth in H1299 cells is related to the defects in mRNA transport or late gene expression.
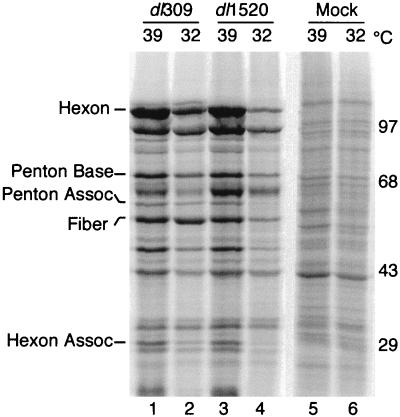
Late protein synthesis was analyzed in H1299 cells maintained at 32 or 39°C for 24 h and infected with the wild-type (dl309) or the E1B 55-kDa mutant (dl1520) Ad. Cells were pulse-labeled with 35S-amino acids at 32 h postinfection for cells maintained at 39°C and 64 h postinfection for cells maintained at 32°C. Cells maintained and infected at 39°C with the E1B 55-kDa mutant virus dl1520 synthesized late viral proteins to levels equivalent to or exceeding that of the wild-type virus (Fig. 8; compare lanes 1 and 3). However, the E1B 55-kDa mutant Ad was defective for late viral gene expression compared to the wild-type virus when cells were maintained and infected at 32°C (compare lanes 2 and 4). This defect in late gene expression in H1299 cells at 32°C is similar to that observed in HeLa cells at 37°C (21, 68). H1299 cells grown at 32°C and infected with either the wild-type or E1B 55-kDa mutant virus synthesized reduced levels of late viral proteins compared to cells grown at 39°C. The reason for this effect of reduced temperature is not known, although it may reflect lower rates of protein synthesis at 32°C.
FIG. 8.
H1299 cells infected with the E1B 55-kDa mutant virus synthesize wild-type levels of late viral proteins when infected and maintained at 39°C. Parental H1299 cells were shifted from 37 to 39 or 32°C for 24 h and then mock infected or infected with either the wild-type virus dl309 or the E1B 55-kDa mutant virus dl1520 at an MOI of 30. Cells were labeled with 35S-labeled amino acids for 1 h at 32 h postinfection for cells maintained at 39°C or at 64 h postinfection for cells maintained at 32°C. Proteins from 105 cells (per lane) were separated by SDS-PAGE. Five viral late proteins were visualized and quantified with a PhosphorImager. The positions of migration and masses (in kilodaltons) of molecular weight standards are indicated on the right. The positions of five Ad late proteins were determined by using Ad virion standards labeled with 14C-amino acids; these proteins are identified on the left. Assoc, associated.
The amount of radioactivity in the five late viral proteins identified in Fig. 8 was measured with a PhosphorImager in two independent experiments. The amount of radioactivity present in each of these proteins synthesized in the E1B 55-kDa mutant virus-infected cells is expressed as a percentage of the corresponding protein synthesized in the wild-type virus-infected cell. These results, summarized in Table 4, confirm that the E1B 55-kDa mutant virus directed the synthesis of wild-type levels of protein at 39°C. By contrast, dl1520-infected cells synthesized late viral proteins to an average of 44% of the level synthesized in wild-type virus-infected cells at 32°C. This findings also reveals that virus production is not accurately reflected by the amount of late viral protein synthesized; the 2-fold average reduction in late viral protein synthesis measured in the mutant virus-infected cells is accompanied by a 16- to 30-fold reduction in virus yield.
TABLE 4.
Relative levels of late viral proteins synthesized in dl1520-infected H1299 cells at 32 and 39°C
| Late protein | % of wild-type proteina
|
|
|---|---|---|
| 39°C | 32°C | |
| Hexon | 101 | 31 |
| Penton base | 127 | 60 |
| Penton associated | 131 | 52 |
| Fiber | 120 | 23 |
| Hexon associated | 103 | 57 |
| Avg | 116 | 44 |
H1299 cells were infected with dl309 or dl1520 at 10 PFU per cell. At 32 h postinfection for cells maintained at 39°C or 64 h postinfection for cells maintained at 32°C, cells were pulse-labeled with 35S-amino acids for 1 h and then lysed. Proteins were separated on an SDS–7.5% polyacrylamide gel and quantified with a PhosphorImager. Late proteins synthesized in cells infected with dl309 at 39 or 32°C were normalized to 100 to calculate the relative percentages of protein synthesized in cells infected with dl1520 at 39 or 32°C. Each value represents average of two independent experiments. All measurements were within ±5%.
DISCUSSION
In this work, we demonstrated that the inability of E1B 55-kDa mutant Ads to replicate efficiently is not due to a failure to abrogate the function of p53. Replication of the E1B 55-kDa mutant viruses (dl338 and dl1520) was not correlated with the status of p53 in the seven cell lines examined in this study (Fig. 2). Furthermore, viral DNA synthesis (Fig. 3), cell cycle-independent replication (Table 3), and virus-induced cell killing (Fig. 4) in cells infected with the E1B 55-kDa mutant virus did not correlate with the status of p53. Rather, cell killing by the E1B 55-kDa mutant virus correlated with permissivity to viral replication (Fig. 4 and 5). Yields of the E1B 55-kDa mutant viruses were reduced only modestly in an H1299-p53 cell line expressing a temperature-sensitive allele of p53 in the wild-type conformation (Fig. 7). The defect in E1B 55-kDa mutant virus replication resulting from reduced temperature was as much as 10-fold greater than that due to p53. Thus, E1B 55-kDa mutant virus replication is restricted by mechanisms independent of p53. In addition to replication, the defect in late viral gene expression (Fig. 8 and Table 4) and the cell cycle restriction (see Results) of the E1B 55-kDa mutant virus exhibited a cold-sensitive phenotype in H1299 cells. At elevated temperatures, the E1B 55-kDa mutant virus synthesized wild-type levels of viral late proteins and replicated independently of the cell cycle. Taken together, these results suggest that the ability of the E1B 55-kDa protein to promote late gene expression may contribute to cell cycle-independent replication of Ad.
The E1B 55-kDa protein has been hypothesized to contribute to both lytic infection and transformation by preventing p53-mediated G1 growth arrest or apoptosis. This response by p53 to viral challenge would be expected to severely hinder the ability of the virus to transform cells, synthesize viral DNA, and establish a lytic infection in the absence of the E1B 55-kDa protein (12, 38, 67). Furthermore, p53 could potentially inhibit cell cycle-independent Ad replication in the absence of the E1B 55-kDa protein. Recently, Bischoff et al. (7) have suggested that the E1B 55-kDa mutant virus (dl1520) selectively replicates in and kills p53-deficient cells, suggesting that the interaction between p53 and the E1B 55-kDa protein is essential to the lytic infection. In accordance with the results of Fig. 2 and 4, they found that U2OS cells (wild-type p53) were resistant to E1B mutant virus-induced cell killing and defective for E1B mutant virus replication whereas C33A cells (p53 mutant) were susceptible to E1B mutant-induced cell killing and permissive for replication of the E1B mutant virus. However, contrary to the hypothesis offered by Bischoff et al. (7), A549 cells which express wild-type p53 were permissive for E1B 55-kDa mutant virus replication and cell killing (Fig. 2 and 4). In addition, Bernards et al. (6) reported that human embryonic kidney cells, which contain wild-type p53, produced near-wild-type yields of the E1B 55-kDa mutant virus. Furthermore, a wild-type p53 status did not affect viral DNA synthesis in E1B 55-kDa mutant-infected cells (Fig. 3). However, Bischoff et al. (7) reported that the RKO human colon cancer cell line transfected to express a dominant negative p53 gene exhibited more severe cytopathic effects when infected with ONYX-015 compared to the parental RKO cell line with normal p53 function. Alternatively, Ridgway et al. (54) reported that wild-type Ads failed to replicate efficiently in p53 mutant cells lines because of a requirement of the E1B 55-kDa protein-p53 complex for viral growth and the shutoff of host protein synthesis. The results of these investigators are inconsistent with the work presented here in that we do not find a correlation between viral replication and the status of p53. The E1B 55-kDa mutant virus most efficiently replicated in and lysed p53 mutant C33A cells (Fig. 2 and 4). Replication of the E1B 55-kDa mutant viruses relative to the wild-type virus differed among various cell types. These differences could not be attributed to the status of p53.
The E1B 55-kDa mutant virus ONYX-015 (dl1520) has been shown to selectively kill cancerous cell lines while sparing normal cells and to induce regression of human tumor xenografts in nude mice (28). The application of oncolytic Ad vectors for cancer therapy through the ability of Ad to induce tumor regression by lytic replication also has been demonstrated by others (77). However, clinical applications of replication-competent virus vectors require selectivity for their efficacy. Bischoff et al. (7) analyzed the ability of the wild-type and E1B 55-kDa mutant viruses to induce regression of p53 null human tumor xenografts transplanted into nude mice. In both p53 null and p53 wild-type tumors, the wild-type virus was better able to retard tumor growth than the E1B 55-kDa mutant virus. The ability of the E1B 55-kDa mutant virus to suppress tumor growth compared to that of the wild-type virus was unaffected by the status of p53 (7). Furthermore, cancer cell lines that were mutant for p53 did not exhibit significantly greater susceptibility to killing by the E1B 55-kDa mutant virus compared to cells with a functional p53 (28). Therefore, the failure of the E1B 55-kDa mutant virus to replicate and lyse infected cells is not strictly dictated by the status of p53 in the infected cell.
We found that cell killing induced by the E1B 55-kDa mutant virus correlated with permissivity to viral replication and not the status of p53 (Fig. 4). In the absence of the E1B 55-kDa protein, our work has demonstrated that HeLa cells infected during S phase are more permissive than HeLa cells infected during G1 for replication of E1B 55-kDa mutant viruses (20). We report here that S-phase HeLa cells are more susceptible than G1 HeLa cells to killing by the E1B 55-kDa mutant virus (Fig. 5). Perhaps this host range restriction of the E1B 55-kDa mutant virus could be used to effectively target those tumors that contain a significant fraction of cells in S phase (5, 34, 40) for virus-induced killing. Because selective killing of S-phase cells did not depend on increased apoptosis (Table 3), E1B 55-kDa mutant viruses may be appropriate oncolytic vectors for the treatment of tumor cells that have lost the capacity to initiate an apoptotic pathway. We propose that selectivity to replication of an E1B 55-kDa mutant virus is linked to the cell cycle. The molecular basis for selective replication of cell cycle-restricted Ads continues to be investigated.
The p53 null cell lines (H358, Saos-2, U2OS, and H1299) were defective for E1B 55-kDa mutant virus replication (Fig. 2) and produced virus in only a fraction (≤28%) of the infected cells (Table 2). This defect in replication resembles the cell cycle-restricted phenotype previously described for HeLa cells (20). By contrast, A549 (p53 wild-type) and C33A (p53 mutant) cells produced virus in 70 and 90%, respectively, of the E1B 55-kDa mutant-infected cells, as determined by electron microscopy. Therefore, the cell cycle-mediated growth restriction is diminished or absent in these permissive cells. Perhaps these cell lines can be used to elucidate the nature of the cell cycle restriction.
The effect of p53 on E1B 55-kDa mutant virus replication was examined in H1299 cells stably expressing a temperature-sensitive p53 allele (18). This approach was chosen to minimize unknown differences between cell lines other than the status of p53 that may affect replication of an E1B 55-kDa mutant virus. Growth of the E1B 55-kDa mutant virus was substantially (16- to 30-fold) less than wild-type virus growth at the lower temperature in the parental H1299 (p53 null) cell line (Fig. 7). By contrast, the presence of wild-type p53 at the lower temperature modestly (3.3- to 3.5-fold) reduced replication of the E1B 55-kDa mutant beyond the effect of temperature alone. These results demonstrate that p53 affects E1B 55-kDa mutant virus replication. However, the defect in replication of E1B 55-kDa mutant virus resulting from reduced temperature was as much as 10-fold greater than that from p53 alone. Therefore, the E1B 55-kDa protein may interact with cellular regulatory factors in addition to p53 to permit virus replication in the wild-type Ad infection.
Interestingly, the E1B 55-kDa mutant virus produced near-wild-type yields of virus in H1299 or H1299-p53 cells maintained and infected at 39°C, as previously reported for HeLa cells (29). At the higher temperature, the defect in late gene expression (Fig. 8 and Table 4) and the cell cycle restriction for replication (see Results) of the E1B 55-kDa mutant virus were no longer evident. H1299 cells maintained and infected at 39°C synthesized wild-type levels of viral late proteins and produced virus in 73% of the infected cells. These results resemble those obtained after infection of HeLa cells during S phase (20). HeLa cells infected during S phase produced greater virus yields and produced virus in up to 75% of the infected cells, compared to 25% of randomly cycling cells. By contrast, the cell cycle restriction was exacerbated when H1299 cells were maintained and infected at 32°C (see Results) or when HeLa cells were infected during G1 phase (20). Under each of these conditions, E1B 55-kDa mutant virus was produced in no more than 12% of the infected cells. Furthermore, H1299 cells infected with the E1B 55-kDa mutant virus produced lower virus yields (Fig. 7) and synthesized reduced levels of late viral proteins at 32°C (Fig. 8 and Table 4). The defect in mRNA transport and, therefore, late viral gene expression by the E1B 55-kDa mutant virus was previously shown to be exacerbated at reduced temperatures in HeLa cells (36, 68).
Both the cell cycle restriction and the defect in late viral gene expression of the E1B 55-kDa mutant virus were exacerbated by reduced temperatures and ameliorated by elevated temperatures. Therefore, it is conceivable that a cellular activity at 39°C that compensates for the loss of the E1B 55-kDa protein is the same activity in S phase that permits efficient replication of the E1B 55-kDa mutant virus. For example, a cellular factor that promotes virus replication may be made available or cellular factors that hinder virus replication may be absent when cells are maintained and infected at 39°C or infected during S phase. Such factors may participate in virus-mediated mRNA transport and cell cycle-independent viral replication. We would speculate that the proposed cellular factor(s) interacts with the E1B 55-kDa–E4orf6 protein complex in Ad infection (22, 47) or, alternatively, depends on the E1B-E4 complex for efficient expression late in Ad infection.
ACKNOWLEDGMENTS
This work was supported in part by Public Health Service grant AI35589 from the National Institute of Allergy and Infectious Disease to D.A.O. and grant CA12197 from the National Cancer Institute to the Comprehensive Cancer Center of Wake Forest University. Tissue culture reagents and services were provided by the Tissue Culture Core Laboratory of the Comprehensive Cancer Center of Wake Forest University, supported in part by NIH grant CA12197.
We gratefully acknowledge Tom Shenk (Princeton University) for the dl309, dl337, and dl338 viruses, Arnie Berk (UCLA) for the dl1520D virus, and Arnie Levine (Princeton University) for the B6-8 hybridoma cell line. We also thank Nancy Raab-Traub and Katherine Fries (University of North Carolina, Chapel Hill) for the H1299 and H1299-p53 cells and Gerry Zambetti (St. Jude Children’s Research Hospital) for the Saos-2 cells. The annexin V-FITC reagent was the generous gift of Greg Kucera (Wake Forest University School of Medicine, Winston-Salem, N.C.). We also thank Natalie Walker for assistance with the FACS analysis.
REFERENCES
- 1.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K S, editors. Current protocols in molecular biology. Vol. 2. New York, N.Y: Greene Publishing Associates and John Wiley and Sons, Inc.; 1993. [Google Scholar]
- 2.Babich A, Feldman L T, Nevins J R, Darnell J E, Weinberger C. Effect of adenovirus on metabolism of specific host mRNAs: transport control and specific translational discrimination. Mol Cell Biol. 1983;3:1212–1221. doi: 10.1128/mcb.3.7.1212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Babiss L E, Ginsberg H S, Darnell J E. Adenovirus E1B proteins are required for accumulation of late viral mRNA and for effects on cellular mRNA translation and transport. Mol Cell Biol. 1985;5:2552–2558. doi: 10.1128/mcb.5.10.2552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barker D D, Berk A J. Adenovirus proteins from both E1B reading frames are required for transformation of rodent cells by viral infection and DNA transfection. Virology. 1987;156:107–121. doi: 10.1016/0042-6822(87)90441-7. [DOI] [PubMed] [Google Scholar]
- 5.Barlogie B, Raber M N, Schumann J, Johnson T S, Drewinko B, Swatzendruber D F, Gohde W, Andreeff M, Freireich E J. Flow cytometry in clinical cancer research. Cancer Res. 1983;43:3982–3997. [PubMed] [Google Scholar]
- 6.Bernards R, de Leeuw M G W, Houweling A, van der Eb A J. Role of the adenovirus early region 1 B tumor antigens in transformation and lytic infection. Virology. 1986;150:126–139. doi: 10.1016/0042-6822(86)90272-2. [DOI] [PubMed] [Google Scholar]
- 7.Bischoff J R, Kim D H, Williams A, Heise C, Horn S, Muna M, Ng L, Nye J A, Sampson-Johannes A, Fattaey A, McCormick F. An adenovirus mutant that replicates selectively in p53-deficient human tumor cells. Science. 1996;274:373–376. doi: 10.1126/science.274.5286.373. [DOI] [PubMed] [Google Scholar]
- 8.Bridge E, Ketner G. Interaction of adenoviral E4 and E1B products in late gene expression. Virology. 1990;174:345–353. doi: 10.1016/0042-6822(90)90088-9. [DOI] [PubMed] [Google Scholar]
- 9.Brower M, Carney D N, Oie H K, Gazdar A F, Minna J D. Growth of cell lines and clinical specimens of human non-small cell lung cancer in a serum-free defined medium. Cancer Res. 1986;46:798–806. [PubMed] [Google Scholar]
- 10.Chiou S-K, Tseng C-C, Rao L, White E. Functional complementation of the adenovirus E1B 19-kilodalton protein with Bcl-2 in the inhibition of apoptosis in infected cells. J Virol. 1994;68:6553–6566. doi: 10.1128/jvi.68.10.6553-6566.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Crook T, Wrede D, Vousden K H. p53 point mutations in HPV negative human cervical carcinoma cell lines. Oncogene. 1991;6:873–875. [PubMed] [Google Scholar]
- 12.Debbas M, White E. Wild-type p53 mediates apoptosis by E1A, which is inhibited by E1B. Genes Dev. 1993;7:546–554. doi: 10.1101/gad.7.4.546. [DOI] [PubMed] [Google Scholar]
- 13.Diller L, Kassel J, Nelson C E, Gryka M A, Litwak G, Gehardt M, Bressac B, Ozturk M, Baker S J, Vogelstein B, Friend S H. p53 functions as a cell cycle control protein in osteosarcomas. Mol Cell Biol. 1990;10:5772–5781. doi: 10.1128/mcb.10.11.5772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dobner T, Horikoshi N, Rubenwolf S, Shenk T. Blockage by adenovirus E4orf6 of transcriptional activation by the p53 tumor suppressor. Science. 1996;272:1470–1473. doi: 10.1126/science.272.5267.1470. [DOI] [PubMed] [Google Scholar]
- 15.El-Deiry W S, Harper J W, O’Conner P M, Velculescu V E, Canman C E, Jackman J, Pietenpol J A, Burrell M, Hill D E, Wang Y, Wiman K G, Mercer W E, Kastan M B, Kohn K W, Elledge S J, Kinzler K W, Vogelstein B. WAF1/CIP1 is induced in p53-mediated G1 arrest and apoptosis. Cancer Res. 1994;54:1169–1174. [PubMed] [Google Scholar]
- 16.El-Deiry W S, Tokino T, Velculescu V E, Levy D B, Parsons R, Trent J M, Lin K, Mercer W E, Kinzler K W, Vogelstein B. WAF-1, a potential mediator of p53 tumor suppression. Cell. 1993;75:817–825. doi: 10.1016/0092-8674(93)90500-p. [DOI] [PubMed] [Google Scholar]
- 17.Flint J, Shenk T. Viral transactivating proteins. Annu Rev Genet. 1997;31:177–212. doi: 10.1146/annurev.genet.31.1.177. [DOI] [PubMed] [Google Scholar]
- 18.Fries K L, Miller W E, Raab-Traub N. Epstein-Barr virus latent membrane protein 1 blocks p53-mediated apoptosis through the induction of the A20 gene. J Virol. 1996;70:8653–8659. doi: 10.1128/jvi.70.12.8653-8659.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gallimore P H, Sharp P A, Sambrook J. Viral DNA in transformed cells. II. A study of the sequences of adenovirus 2 DNA in nine lines of transformed rat cells using specific fragments of the viral genome. J Mol Biol. 1974;89:49–72. doi: 10.1016/0022-2836(74)90162-4. [DOI] [PubMed] [Google Scholar]
- 20.Goodrum F D, Ornelles D A. The early region 1B 55-kilodalton oncoprotein of adenovirus relieves growth restrictions imposed on viral replication by the cell cycle. J Virol. 1997;71:548–561. doi: 10.1128/jvi.71.1.548-561.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Goodrum, F. D., and D. A. Ornelles. Unpublished results.
- 22.Goodrum F D, Shenk T, Ornelles D A. Adenovirus early region 4 34-kilodalton protein directs the nuclear localization of the early region 1B 55-kilodalton protein in primate cells. J Virol. 1996;70:6323–6335. doi: 10.1128/jvi.70.9.6323-6335.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Graham F L, Abrahams P J, Mulder C, Heijneker H L, Warnaar S O, de Vries A J, Fiers W, van der Eb A J. Studies on in vitro transformation by DNA and DNA fragments of human adenoviruses and simian virus 40. Cold Spring Harbor Symp Quant Biol. 1974;39:637–650. doi: 10.1101/sqb.1974.039.01.077. [DOI] [PubMed] [Google Scholar]
- 24.Graham F L, Smiley J, Russell W C, Nairn R. Characteristics of a human cell line transformed by DNA from human adenovirus type 5. J Gen Virol. 1977;36:59–72. doi: 10.1099/0022-1317-36-1-59. [DOI] [PubMed] [Google Scholar]
- 25.Grand R J A, Grant M L, Gallimore P H. Enhanced expression of p53 in human cells infected with mutant adenoviruses. Virology. 1994;203:229–240. doi: 10.1006/viro.1994.1480. [DOI] [PubMed] [Google Scholar]
- 26.Halbert D N, Cutt J R, Shenk T. Adenovirus early region 4 encodes functions required for efficient DNA replication, late gene expression, and host cell shutoff. J Virol. 1985;56:250–257. doi: 10.1128/jvi.56.1.250-257.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Harper J W, Adami G R, Wei N, Keyomarsi K, Elledge S J. The p21 Cdk-interacting protein Cip1 is a potent inhibitor of G1 cyclin-dependent kinases. Cell. 1993;75:805–816. doi: 10.1016/0092-8674(93)90499-g. [DOI] [PubMed] [Google Scholar]
- 28.Heise C, Sampson-Johannes A, Williams A, McCormick F, von Hoff D D, Kirn D H. ONYX-015, an E1B gene-attenuated adenovirus, causes tumor-specific cytolysis and antitumoral efficacy that can be augmented by standard chemotherapeutic agents. Nat Med. 1997;3:639–644. doi: 10.1038/nm0697-639. [DOI] [PubMed] [Google Scholar]
- 29.Ho Y-S, Galos R, Williams J. Isolation of type 5 adenovirus mutants with a cold-sensitive host range phenotype: genetic evidence of an adenovirus transformation maintenance function. Virology. 1982;122:109–124. doi: 10.1016/0042-6822(82)90381-6. [DOI] [PubMed] [Google Scholar]
- 30.Jones N, Shenk T. Isolation of Ad5 host range deletion mutants defective in transformation of rat embryo cells. Cell. 1979;17:683–689. doi: 10.1016/0092-8674(79)90275-7. [DOI] [PubMed] [Google Scholar]
- 31.Jones N, Shenk T. Isolation of deletion and substitution mutants of adenovirus type 5. Cell. 1978;13:181–186. doi: 10.1016/0092-8674(78)90148-4. [DOI] [PubMed] [Google Scholar]
- 32.Kafotos F C, Jones C W, Efstratiadis A. Determination of nucleic acid sequence homologies and relative concentrations by a dot hybridization procedure. Nucleic Acids Res. 1979;7:1541–1552. doi: 10.1093/nar/7.6.1541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kao C C, Yew P R, Berk A J. Domains required for in vitro association between the cellular p53 and the adenovirus 2 E1B 55kD proteins. Virology. 1990;179:806–814. doi: 10.1016/0042-6822(90)90148-k. [DOI] [PubMed] [Google Scholar]
- 34.Kute T E, Muss H B, Anderson D, Crumb K, Miller B, Burns D, Dube L. Relationship of steroid receptor, cell kinetics, and clinical status in patients with breast cancer. Cancer Res. 1981;41:3524–3529. [PubMed] [Google Scholar]
- 35.Lehman T A, Bennett W P, Metcalf R A, Welsh J A, Ecker J, Modali R V, Ullrich S, Romano J W, Appella E, Testa J R, Gerwin B I, Harris C C. p53 mutations, ras mutations, and p53-heat shock protein complexes in human lung carcinoma cell lines. Cancer Res. 1991;51:4090–4096. [PubMed] [Google Scholar]
- 36.Leppard K N, Shenk T. The adenovirus E1B-55 kD protein influences mRNA transport via an intranuclear effect on RNA metabolism. EMBO J. 1989;8:2329–2336. doi: 10.1002/j.1460-2075.1989.tb08360.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lin J, Wu X, Chen J, Chang A, Levine A J. Functions of p53 protein in growth regulation and tumor suppression. Cold Spring Harbor Symp Quant Biol. 1994;59:215–223. doi: 10.1101/sqb.1994.059.01.026. [DOI] [PubMed] [Google Scholar]
- 38.Lowe S W, Ruley H E. Stabilization of the p53 tumor suppressor is induced by adenovirus 5 E1A and accompanies apoptosis. Genes Dev. 1993;7:535–545. doi: 10.1101/gad.7.4.535. [DOI] [PubMed] [Google Scholar]
- 39.Masuda H, Miller C, Koeffler H P, Battifora H, Cline M J. Rearrangement of the p53 gene in human osteogenic sarcomas. Proc Natl Acad Sci USA. 1987;84:7716–7719. doi: 10.1073/pnas.84.21.7716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.McDivitt R W, Stone K R, Craig R B, Palmer J O, Meyer J S, Bauer W C. A proposed classification of breast cancer based on kinetic information. Cancer. 1986;57:269–276. doi: 10.1002/1097-0142(19860115)57:2<269::aid-cncr2820570214>3.0.co;2-d. [DOI] [PubMed] [Google Scholar]
- 41.Michalovitz D, Halevy O, Oren M. Conditional inhibition of transformation and of cell proliferation by a temperature-sensitive mutant of p53. Cell. 1990;62:671–680. doi: 10.1016/0092-8674(90)90113-s. [DOI] [PubMed] [Google Scholar]
- 42.Mitsudomi T, Steinberg S M, Nau M M, Carbone D, D’Amico D, Bodner S, Oie H K, Linnoila R I, Mulshine J L, Minna J D. p53 gene mutations in non-small-cell lung cancer lines and their correlation with the presence of ras mutations and clinical features. Oncogene. 1992;7:171–180. [PubMed] [Google Scholar]
- 43.Miyashita T, Reed J C. Tumor suppressor p53 is a direct transcriptional activator of the human bax gene. Cell. 1995;80:293–299. doi: 10.1016/0092-8674(95)90412-3. [DOI] [PubMed] [Google Scholar]
- 44.Moore M, Horikoshi N, Shenk T. Oncogenic potential of the adenovirus E4orf6 protein. Proc Natl Acad Sci USA. 1996;93:11295–11301. doi: 10.1073/pnas.93.21.11295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Moore M, Schaack J, Baim S B, Morimoto R I, Shenk T. Induced heat shock mRNAs escape the nucleocytoplasmic transport block in adenovirus-infected HeLa cells. Mol Cell Biol. 1987;7:4505–4512. doi: 10.1128/mcb.7.12.4505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nevels M, Rubenwolf S, Spruss T, Wolf H, Dobner T. The adenovirus E4orf6 protein can promote E1A/E1B-induced focus formation by interfering with p53 tumor suppressor function. Proc Natl Acad Sci USA. 1997;94:1206–1211. doi: 10.1073/pnas.94.4.1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ornelles D A, Shenk T. Localization of the adenovirus early region 1B 55-kilodalton protein during lytic infection: association with nuclear viral inclusions requires the early region 4 34-kilodalton protein. J Virol. 1991;65:424–439. doi: 10.1128/jvi.65.1.424-429.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pilder S, Logan J, Shenk T. Deletion of the gene encoding the adenovirus 5 early region 1B 21,000-molecular-weight polypeptide leads to degradation of viral and host cell DNA. J Virol. 1984;52:664–671. doi: 10.1128/jvi.52.2.664-671.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pilder S, Moore M, Logan J, Shenk T. The adenovirus E1B-55K transforming polypeptide modulates transport or cytoplasmic stabilization of viral and host cell mRNAs. Mol Cell Biol. 1986;6:470–476. doi: 10.1128/mcb.6.2.470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Querido E, Marcellus R C, Lai A, Charbonneau R, Teodoro J G, Ketner G, Branton P E. Regulation of p53 levels by the E1B 55-kilodalton protein and E4orf6 in adenovirus-infected cells. J Virol. 1997;71:3788–3798. doi: 10.1128/jvi.71.5.3788-3798.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Quinlan M P. Enhanced proliferation, growth factor induction and immortilization by adenovirus E1A 12S in the absence of E1B. Oncogene. 1994;9:2639–2647. [PubMed] [Google Scholar]
- 52.Rao L, Debbas M, Sabbatini D, Hockenberry D, Korsmeyer S, White E. The adenovirus E1A proteins induce apoptosis, which is inhibited by the E1B 19-kDa and Bcl-2 proteins. Proc Natl Acad Sci USA. 1992;89:7742–7746. doi: 10.1073/pnas.89.16.7742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Reich N C, Sarnow P, Duprey E, Levine A J. Monoclonal antibodies which recognize native and denatured forms of the adenovirus DNA-binding protein. Virology. 1983;128:480–484. doi: 10.1016/0042-6822(83)90274-x. [DOI] [PubMed] [Google Scholar]
- 54.Ridgway P, Hall A R, Myers C J, Braithwaite A W. p53/E1b58kDa complex regulates adenovirus replication. Virology. 1997;237:404–413. doi: 10.1006/viro.1997.8782. [DOI] [PubMed] [Google Scholar]
- 55.Rubenwolf S, Scutt H, Nevels M, Wolf H, Dobner T. Structural analysis of the adenovirus type 5 E1B 55-kilodalton–E4orf6 protein complex. J Virol. 1997;71:1115–1123. doi: 10.1128/jvi.71.2.1115-1123.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ruley H E. Adenovirus early region 1A enables viral and cellular transforming genes to transform primary cells in culture. Nature. 1983;304:602–606. doi: 10.1038/304602a0. [DOI] [PubMed] [Google Scholar]
- 57.Sarnow P, Ho Y S, Williams J, Levine A J. Adenovirus E1b-58 kd tumor antigen and SV40 large tumor antigen are physically associated with the same 54kd cellular protein in transformed cells. Cell. 1982;28:387–394. doi: 10.1016/0092-8674(82)90356-7. [DOI] [PubMed] [Google Scholar]
- 58.Scheffner M, Munger K, Byrne J C, Howley P M. The state of the p53 and retinoblastoma genes in human cervical carcinoma cell lines. Proc Natl Acad Sci USA. 1991;88:5523–5527. doi: 10.1073/pnas.88.13.5523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Scheffner M, Werness B A, Huibregtse J M, Levine A J, Howley P M. The E6 oncoprotein encoded by human papillomavirus types 16 and 18 promotes the degradation of p53. Cell. 1990;63:1129–1136. doi: 10.1016/0092-8674(90)90409-8. [DOI] [PubMed] [Google Scholar]
- 60.Shepherd S E, Howe J A, Mymryk J S, Bayley S T. Induction of the cell cycle in baby rat kidney cells by adenovirus type 5 E1A in the absence of E1B and a possible influence of p53. J Virol. 1993;67:2944–2949. doi: 10.1128/jvi.67.5.2944-2949.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Steegenga W T, van Laar T, Shvarts A, Terleth C, van der Eb A J, Jochemsen A G. Distinct modulation of p53 activity in transcription and cell cycle regulation by the large (54 kDa) and small (21 kDa) adenovirus E1B proteins. Virology. 1995;212:543–554. doi: 10.1006/viro.1995.1512. [DOI] [PubMed] [Google Scholar]
- 62.Stillman B. Functions of the adenovirus E1B tumor antigens. Cancer Surv. 1986;5:389–404. [PubMed] [Google Scholar]
- 63.Takahashi T, Nau M M, Chiba I, Birrer M J, Rosenberg R K, Vinocour M, Levitt M, Pass H, Gazdar A F, Minna J D. p53: a frequent target for genetic abnormalities in lung cancer. Science. 1989;246:491–494. doi: 10.1126/science.2554494. [DOI] [PubMed] [Google Scholar]
- 64.van den Elsen P J, Houweling A, van der Eb A J. Morphological transformation of human adenoviruses is determined to a large extent by gene products of region E1A. Virology. 1983;131:242–246. doi: 10.1016/0042-6822(83)90549-4. [DOI] [PubMed] [Google Scholar]
- 65.van Engeland M, Nieland L J W, Ramaekers F C S, Schutte B, Reutelingsperger C P M. Annexin V-affinity assay: a review on an apoptosis detection system based on phosphatidylserine exposure. Cytometry. 1998;31:1–9. doi: 10.1002/(sici)1097-0320(19980101)31:1<1::aid-cyto1>3.0.co;2-r. [DOI] [PubMed] [Google Scholar]
- 66.Werness B A, Levine A J, Howely P M. Association of human papilloma virus types 16 and 18 E6 proteins p53. Science. 1990;248:76–79. doi: 10.1126/science.2157286. [DOI] [PubMed] [Google Scholar]
- 67.White E. Function of the adenovirus E1B oncogene in infected and transformed cells. Semn Virol. 1994;5:341–348. [Google Scholar]
- 68.Williams J, Karger B D, Ho Y S, Castiglia C L, Mann T, Flint S J. The adenovirus E1B 495R protein plays a role in regulating the transport and stability of the viral late messages. Cancer Cells. 1986;4:275–284. [Google Scholar]
- 69.Xiong Y, Hannon G J, Zhang H, Casso D, Kobayashi R, Beach D. p21 is a universal inhibitor of cyclin kinases. Nature. 1993;366:701–704. doi: 10.1038/366701a0. [DOI] [PubMed] [Google Scholar]
- 70.Xiong Y, Zhang H, Beach D. Subunit rearrangement of the cyclin-dependent kinases is associated with cellular transformation. Genes Dev. 1993;7:1572–1583. doi: 10.1101/gad.7.8.1572. [DOI] [PubMed] [Google Scholar]
- 71.Yang U-C, Huang W, Flint S J. mRNA export correlates with activation of transcription in human subgroup C adenovirus-infected cells. J Virol. 1996;70:4071–4080. doi: 10.1128/jvi.70.6.4071-4080.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Yew P R, Berk A J. Inhibition of p53 transactivation required for transformation by adenovirus early 1B protein. Nature. 1992;357:82–82. doi: 10.1038/357082a0. [DOI] [PubMed] [Google Scholar]
- 73.Yew R P, Lui X, Berk A J. Adenovirus E1B oncoprotein tethers a transcriptional repression domain to p53. Genes Dev. 1994;8:190–202. doi: 10.1101/gad.8.2.190. [DOI] [PubMed] [Google Scholar]
- 74.Zambetti G P, Levine A J. A comparison of the biological activities of wild-type and mutant p53. FASEB J. 1993;7:855–865. doi: 10.1096/fasebj.7.10.8344485. [DOI] [PubMed] [Google Scholar]
- 75.Zantema A, Fransen J A M, Davis-Oliver A, Ramaekers F C S, Vooijs G P, DeLeys B, van der Eb A J. Localization of the E1B proteins of adenovirus type 5 in transformed cells as revealed by interaction with monoclonal antibodies. Virology. 1985;142:44–58. doi: 10.1016/0042-6822(85)90421-0. [DOI] [PubMed] [Google Scholar]
- 76.Zantema A, Schrier P I, Davis-Oliver A, van Laar T, Vaessen R T M J, van der Eb A J. Adenovirus serotype determines association and localization of the large E1B tumor antigen and cellular tumor antigen p53 in transformed cells. Mol Cell Biol. 1985;5:3084–3091. doi: 10.1128/mcb.5.11.3084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Zhang JF, Hu C, Geng Y, Selm J, Klien S B, Orazi A, Taylor M W. Treatment of a human breast cancer xenograft with an adenovirus vector containing an interferon gene results in rapid regression due to viral oncolysis and gene therapy. Proc Natl Acad Sci USA. 1996;93:4513–4518. doi: 10.1073/pnas.93.9.4513. [DOI] [PMC free article] [PubMed] [Google Scholar]