ABSTRACT
Case studies present students with an opportunity to learn and apply course content through problem solving and critical thinking. Supported by the High-throughput Discovery Science & Inquiry-based Case Studies for Today’s Students (HITS) Research Coordination Network, our interdisciplinary team designed, implemented, and assessed two case study modules entitled “You Are What You Eat.” Collectively, the case study modules present students with an opportunity to engage in experimental research design and the ethical considerations regarding microbiome research and society. In this manuscript, we provide instructors with tools for adopting or adapting the research design and/or the ethics modules. To date, the case has been implemented using two modalities (remote and in-person) in three courses (Microbiology, Physiology, and Neuroscience), engaging over 200 undergraduate students. Our assessment data demonstrate gains in content knowledge and students’ perception of learning following case study implementation. Furthermore, when reflecting on our experiences and student feedback, we identified ways in which the case study could be modified for different settings. In this way, we hope that the “You Are What You Eat” case study modules can be implemented widely by instructors to promote problem solving and critical thinking in the traditional classroom or laboratory setting when discussing next-generation sequencing and/or metagenomics research.
KEYWORDS: case study, research design, ethics, microbiome, metagenomics, next-generation sequencing, high-throughput discovery science, undergraduate life science education, active learning
INTRODUCTION
Case study-based learning is a high-impact teaching strategy in which students practice problem solving and critical thinking while engaging with a relatable narrative (1, 2). Supported by HITS (High-throughput Discovery Science and Inquiry-based Case Studies for Today’s Students) (2–7), a research coordination network sponsored by the National Science Foundation, our interdisciplinary team designed and assessed a case study entitled “You Are What You Eat” (Appendix 1). The two-part case study is an active learning tool to engage students in the experimental design process (Research Design Module) and the critical evaluation of ethical issues related to sequencing human microbiome samples (Ethics Module). By focusing on high-throughput (HT) metagenomics research, the case study also creates awareness of cutting-edge tools and techniques used to advance scientific knowledge and health practices.
In the research design module, students learn about next-generation sequencing (8) and HT metagenomics research (9), engage with scientific literature, and design a research study using a guided handout. In the ethics module, students explore ethical issues related to metagenomics research (10, 11), including data privacy, telemedicine tradeoffs, influence of food deserts, and the use of representative samples in microbiome research. Our team implemented and assessed the case study in various courses and modalities (remote and in person) at three distinct institutions. This highly adaptable and cross-disciplinary case is easily streamlined or can be offered as expandable independent modules depending on course requirements. Here, we provide details for each of our distinct implementations to illustrate how other instructors may adopt or adapt the case study for their own instructional purposes.
Intended audience
The “You Are What You Eat” case study is interdisciplinary and was implemented in three upper-level courses that cover metagenomics: Cellular & Molecular Neurotechnology, Microbiology, and Animal Physiology (Table 1). Given our cross-disciplinary implementation, a variety of STEM majors can benefit from the case, including biology and microbiology, neuroscience and psychology, biotechnology, and allied health majors, among others. The ethics module, which considers ethical issues related to microbiome research, is suitable for an even wider audience, including non-science majors and introductory survey courses.
TABLE 1.
Case study implementation metadata summary
| Institution type | Course | Course enrollment | Module(s) implemented | Classroom modality |
|---|---|---|---|---|
| Private liberal arts university in the midwestern United States | 300-level Microbiology | Two sections of 35 students | 2 | Face-to-face (F20; F21) |
| Public regional comprehensive university; Native American-serving, non-tribal institution in the southeastern United States | 400-level Physiology | Two sections from F2020- S2022 of 10–18 students | 1 & 2 | Face-to-face (S22), hybrid (F20) |
| Public research university in the southeastern United States | 400-level Neuroscience |
Four sections from F2020-F2021 of 35 students | 1 & 2 | Remote, synchronous (F20; S21), face-to-face (F21) |
Learning time
Completion of both modules of the case study, as originally designed, will require approximately 6 hours (~3 hours of class time and ~3 hours outside class time). In this study, we describe both streamlined and expanded versions of the modules, and encourage course instructors to modify the allotted learning time based on their specific course objectives and student requirements. Variations in our cross-disciplinary learning time and implementation tips are summarized in Table 2.
TABLE 2.
Cross-discipline learning time and implementation tipsa
| Module & course | Pre-class | In-class | Post-class |
|---|---|---|---|
| Neuroscience Research Design Module |
60 minutes Students explore the Lieberman Lab website, read the Lieberman Blog post, watch sequencing & microbiome videos, and answer reflection questions |
30 minutes Lesson to review pre-work material, the microbiota–gut–brain axis, neurobiological disorders associated with gut dysbiosis, NGS, study design, etc. 20 minutes Student case study research design work |
60 minutes Students continue to work on research design. The case study document is handed in as a group 2 weeks later |
| Animal Physiology Research Design Module |
60 minutes Students read the blog post, watch videos on sequencing, and answer some reflection questions. Students also prepare to speak on 1 of 3 discussion prompts |
30 minutes Lesson to review NGS, workflow, and study design, followed by group discussion 45 minutes Students are divided into teams to read an article and design an experiment |
60 minutes Students turn in a draft of their research design and then provide peer feedback on two other proposals. Students then implement the feedback to improve their design and turn in a final version |
| Neuroscience Ethics Module |
60 minutes Student groups are assigned to a topic (Data Privacy, Food Deserts, or Diverse Populations Samples). Students explore multiple articles related to their topic and answer reflection questions. They also create a summarizing slide for a jigsaw presentation in the next class period (Fig. 1) |
50 minutes Student groups alternate presentations on the assigned topics. 2–3 groups will discuss each topic and address specific questions from the assigned pre-work with their slide and verbal presentation. At the end of the group presentations for each topic, 5 minutes of small group discussions (teams of 4) take place. Students generate questions and comments to report out for a whole class discussion: ~15 minutes is devoted to the presentation of each topic; small group and whole class discussion |
60 minutes Students continue to work on their research design. The case study document is handed in at the end of the week as a group. The last day of class also has a few minutes set aside for case study group work |
| Animal Physiology Ethics Module |
60 minutes Students are assigned to one of four topic groups, and they read articles on the topic and prepare discussion questions |
75 minutes Students break into their topic groups and prepare a presentation on their topic. Each group presents to the class and leads a discussion. The slide is submitted for assessment |
15 minutes Students write up a short reflection on the module |
| Microbiology Ethics Module |
60 minutes Students are assigned reading passages and respond to the reflection prompts through the course management system |
50 minutes Debrief and classroom discussion of the assigned readings and reflection prompts |
Not applicable |
Case study materials (slides, lesson plans, etc.) are available upon request to the corresponding author. Abbreviations: NGS next-generation sequencing.
Prerequisite student knowledge
Knowledge of general biology concepts, including genetics and DNA structure, is required for the successful completion of the research design module and is helpful for the ethics module. Additionally, the ability to describe next-generation sequencing technology and chemistry would facilitate the achievement of learning objectives associated with the research design module and allow condensing of in-class lecture time. However, the pre-work for this module (Appendix 1) includes videos and resources that introduce students to HT sequencing approaches, the concept of metagenomics, the microbiome and the microbiota, and literature that explains best practices in microbiome study design (12). Students in our courses also had significant experience reading research articles, which is beneficial for both modules, especially the research design module. To train students how to engage with the research literature, we used evidence-based tools: Figure FACTS (13) and Research Analysis Worksheets (14). Minimal prerequisite knowledge is necessary for the ethics module, and it could easily be adapted for non-science majors by purging the scientific articles and focusing exclusively on the popular press science articles covering ethics topics.
Learning objectives
After completion of the modules, students should be able to do the following:
Research design module:
Summarize why next-generation sequencing approaches are HT and transformative.
Distinguish the terms microbiota, microbiome, and metagenomics.
Compare and contrast 16S and whole genome metagenomics.
Explain the underlying workflow employed to analyze HT microbiome data sets.
Describe in detail the goals, methodology, and results of a published HT microbiome study.
Create an experimental design to complete a metagenomics study.
Predict the experimental outcome of a metagenomics study.
Ethics module:
Integrate concepts of ethics and biology to construct an argument about the balance of privacy and information in data sets.
Discuss and evaluate the intersection of race, socioeconomic status, and health outcomes.
PROCEDURE
Materials
This case was offered both synchronously online and through face-to-face instruction. Some asynchronous preparation was also required for students to engage in the case. For either modality, each student should have access to an internet-capable device. The case study documentation was delivered to students using a learning management system (LMS). In-class polls were delivered using a Web-based polling system. Students accessed PubMed to practice navigating the literature for the research design module and also collaboratively presented work using Google Slides for the ethics module. The ethics module can utilize scientific essays and popular science articles (15–21) in combination with or instead of research articles. The microbiome-related research articles on data privacy (22), food deserts (23), representative samples (24), and telemedicine (25) were used for the advanced neuroscience and physiology course implementations. No other materials are needed.
Student instructions
Complete student instructions are included in Appendix 1. The full case study, including both the research design and ethics modules, is designed for two class periods, with pre- and post-class work for each module (Table 2). The case can easily be expanded to 3–4 sessions to cover topics at a greater depth or to allow more protected student work time in class (Table 2). Instructors should also consider student background knowledge, course learning objectives, and class period length when adapting student instructions for their course needs. The modules can also be spaced apart or offered in back-to-back sessions.
Metagenomics research design module
Pre-Class: Students are given a short narrative introducing the case (Appendix 1). They learn about a first-year graduate student who works in a microbiology lab. The pre-class work has three main sections. First, students explore the featured microbiology lab’s website and explore the group’s current research. Included in the pre-class work is an article from Scientific American (26) about the gut microbiome (Appendix 4). Students are directed to pick one of three discussion prompts and consider their response. Next, students watch three short videos to provide background information on HT sequencing technologies. After watching the videos, they answer a set of guided questions. Finally, students read two short sections of a paper on conducting a microbiome study and answer two questions.
In-Class: Students participate in a short lecture on HT sequencing technologies and the basics of a microbiome study. The slide deck, available upon request from the authors, includes poll questions for students to answer on their electronic devices. Then, students are divided into groups of 3–6 people. The group chooses a paper on the gut–brain axis (27) or another microbiome-related research topic tailored to the course requirements. After reading their selected paper, the group uses the provided handout to design a future experiment (Appendix 1).
Post-Class: In a larger class, students may simply submit a single case study research design module document as a group assessment. Alternatively, there is an option to add a peer review component to the case module: each student can be given handouts of two other groups to evaluate and provide peer feedback using a rubric (Appendix 1). Then, students individually revise their experimental plan and submit a one-page summary of their design.
Ethics and implications of large, complex microbiome data sets module
Pre-Class: Students are assigned to one of 4 topics, and they read 2–3 articles about the topic. The articles are a mix of scientific and popular press articles and are listed in the Materials section and Appendix 1. Before coming to class, students answer questions about the articles and can also be tasked with creating a Google slide for presentation (Fig. 1). The number and type (scientific or press) of articles can be modified to align with course goals and student background. For example, if students are not comfortable reading scientific literature, instructors may assign sections of the articles (e.g., abstract and discussion, introduction and one key figure, etc.). In addition, instructors may want to only assign popular press articles to reduce the pre-class time.
Fig 1.
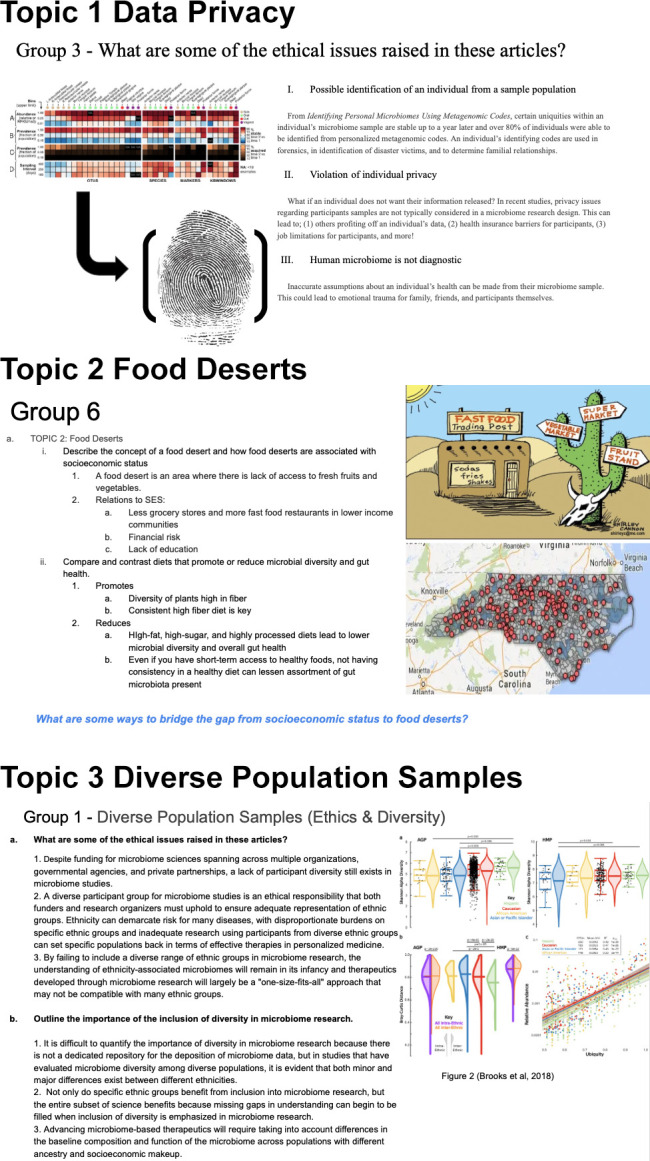
Examples of student two-minute jigsaw presentations on the ethics and implications of microbiome research. (Top image for topic 1 reproduced from reference 22; top image for topic 2 reproduced from Shirley Cannon’s original image shirleyc@me.com; bottom image for topic 2 reproduced from Rose Hoban and Steve Tell original 2014 map https://www.northcarolinahealthnews.org/interactive-food-deserts-and-farmers-markets/; topic 3 image reproduced from reference 24.)
In-Class: Students meet with other members of their assigned topic, and they decide on group roles. Together, they discuss the questions and their responses. Then, the group creates a 1–2 slide visual covering the key points of their topic. If time is limited, this may be moved to pre-class work; the variations in implementation are outlined in Table 2. The class comes back together, and each group gives a 5- to 7-minute presentation and answers questions from the class. If the class time is limited, the instructor may focus on small group discussions on the articles, leading to a large class discussion, without creating or presenting slide visuals. Instructors could also jigsaw the class, assigning each person or group one article or one figure and having groups come together to teach each other the full article.
Faculty instructions
Teaching notes and answer keys are provided in Appendix 2. Instructors should note that the pre-class assignment for the research design module is time-intensive and will take students between 60 and 75 minutes, depending on their familiarity with the topic. The first pre-class assignment asks students to pick one of three discussion questions. Instructors may wish to assign these prompts to specific students rather than having students pick for themselves. Generally, the pre-class assignment is graded on completion. Instructors may ask students to submit the answers on their LMS before coming to class to ensure adequate student preparation.
In class, the research design module starts with a slide deck. Instructors will need to set up an electronic polling system (e.g., Poll Everywhere) if they want to use technology for poll questions. Otherwise, instructors may have students write down their answers or informally discuss with the people around them. The students will then work with a group of 3–6 peers to design a new microbiome study, using an article about the gut–brain axis as a jumping-off point. The topic and articles can be altered depending on the course topic and level. For example, if the class is a physiology course, the instructor may choose a study about the link between diabetes and the gut microbiome and run the case during a unit on metabolism. Students will use the provided handout, either physical or digital (one per group), and they can submit their work in person or through the LMS. Instructors can then give each student an experimental design for peer review. Most LMS software includes a built-in peer review function.
After class, students use the rubric to grade the work of their peers. Once the peer reviews are completed, instructors can relay feedback to the original writers anonymously. Students then have a week to revise their experimental plan and turn in a one-page summary of the design. Alternatively, if the course size or time constraints do not allow for the peer evaluation step, students can simply complete the case study document (Appendix 1) in teams.
The pre-class assignment for the ethics module is less time-intensive than the research design module. However, students are required to read several articles and answer a set of reflection questions, which may take them up to an hour. Depending on student comfort with primary literature, instructors may modify the pre-class articles (by assigning only the popular press articles or only a section of the scientific articles to reduce the pre-class time). Instructors may assign students to one of the four topics (data privacy, food deserts, telemedicine, or non-representative sampling) or allow students to choose. Students may submit their work through the LMS before coming to class.
In class, instructors should divide the students into groups based on the topic (either virtually or in person) and have them discuss the questions to consider. Then, the students will need to make a visual, explaining what they learned from their articles. Each group will have approximately 15–20 minutes to work on their visual and review group roles.
Then, the students will come back together, and instructors should give each group 5 minutes to present their visuals (time may be adjusted or the visuals may be skipped entirely, depending on class size and time). Alternatively, the group creation of a slide visual can be moved to pre-class work for shorter class periods. In the end, the instructor will lead the students in a larger group discussion to assess whether they can connect what they learned to the work they have done in the class before as well as establish what they want to learn more about.
Following the ethics module, instructors may assign a short reflective writing assignment, asking the students to describe what they learned and what questions they still have. Alternatively, instructors may wish to have students submit the full case (research design and ethics modules) from either individuals or groups.
Suggestions for determining student learning
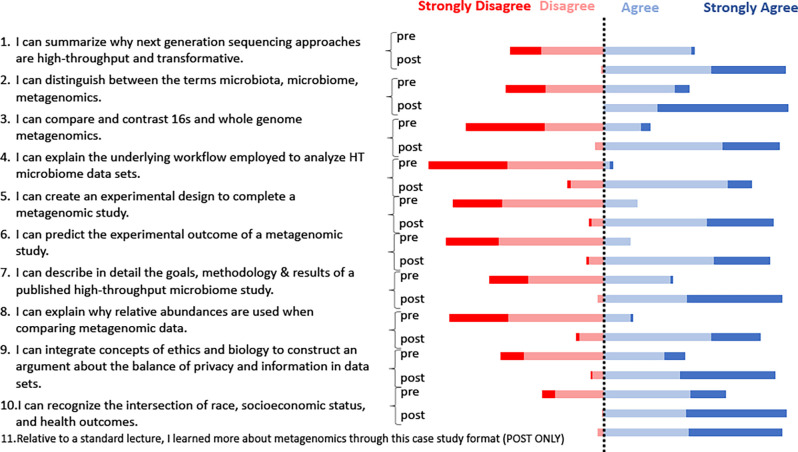
The complete assessment instrument is included in Appendix 3. Student learning was assessed with a pre- and post-knowledge-based assessment instrument. The instrument was composed of three multiple-choice questions for the research design module (Fig. 2) and two short answer questions for the ethics module. The in-class research design assignment was assessed with a rubric (Appendix 1), and the short presentation in the ethics module was assessed for completion. Alternatively, student learning can be determined by having students hand in the entire student case study document for assessment, which connects to every student learning outcome in a single comprehensive assignment (Appendix 1). Additionally, student perceptions of learning were assessed with a pre- and post-Likert scale-based survey (Fig. 3).
Fig 2.
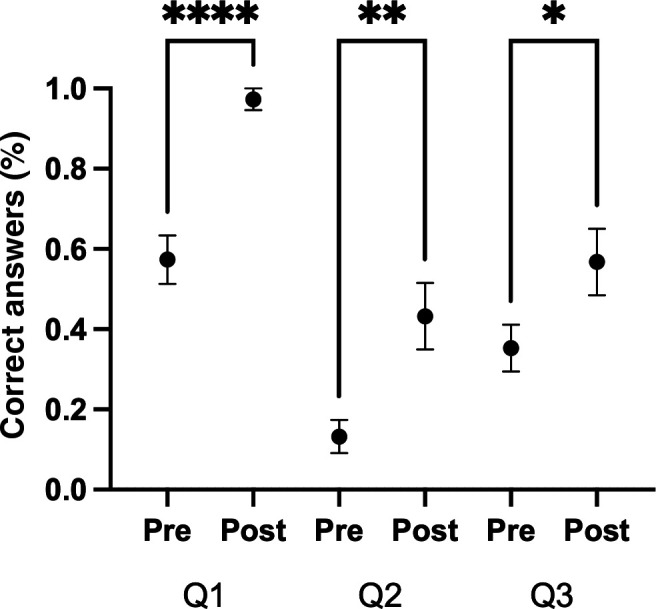
Student scores on questions linked to case study learning objectives. Student responses were collected through an optional pre -and post-course survey from the 400-level neuroscience course. The average fraction of correct answers is shown pre- and post-course (n = 71 students; One-Way ANOVA, Sidak’s multiple comparisons test, *P < 0.05; **P < 0.01; ****P < 0.0001). Error bars are ±SEM.
Fig 3.
Stacked divergent bar plot of Likert-scale data on participant perceptions of learning. Retrospective survey instrument items using a 4-point Likert scale (data represent all participating institutions and implementations, n is a range from 59 to 99 depending on the question). All retrospective question pairs exhibited significant divergence in student perception (P < 0.001, Wilcoxon signed-rank test).
Sample data
Examples of student responses from the research design module are provided in Table 3. Samples of instructor feedback comments are also included for student responses that require improvement. For the ethics module, examples of students’ slide presentations are provided for three topics (data privacy, food deserts, and diverse population samples) (Fig. 1).
TABLE 3.
Examples of student case study responses to the experimental design of metagenomics research
|
Excerpts from students’ case study writing in response to the following prompt: “Formulate a research question that can be investigated using next-generation sequencing methodology to evaluate the composition of the gut microbiota.” (See Appendix 1) *All sample data provided is from the upper level neuroscience course | |
| Meets Expectations |
How does the microbiome of individuals diagnosed with clinical depression differ from that of individuals who do not show symptoms of clinical depression? What type of gut microbiota is present within individuals with IBDs and dementia/Alzheimer’s disease, individuals with just IBDs, and individuals with dementia/ Alzheimer’s disease? In order to determine what type of gut microbiota is present within individuals with IBDs and dementia/Alzheimer’s disease, IBD, dementia/ Alzheimer’s disease, we can use 16 s ribosomal sequencing to determine this. How does the composition of one’s microbiome potentially predict their susceptibility of developing certain forms of epilepsy? As dementia rates have been shown to be influenced by various inflammatory bowel diseases (IBD), such as Crohn’s or Ulcerative Colitis, an exploratory treatment option includes exposing individuals with IBD to varying levels of an anti-nflammatory drug. This would occur as a double-blind longitudinal study to determine if rates of dementia remain high in these individuals compared to groups without the anti-inflammatory drug. With the methodology of 16 s ribosomal sequencing, varying levels of the different organisms within the microbiota of the IBD-gut could be observed throughout the timeline of the study. Therefore, negative effects from the anti-inflammatory drug on the microbiota could be monitored as well as the populations of bacteria thought to be having an association with dementia. |
|
Needs Improvement Student Response Instructor Feedback |
How does national cuisine and environment affect the microbiome, and what correlations can be seen in the microbiome of patients with a neurological disease across countries? This is a big research question exploring cuisine, environment, neurological disease and countries, making effective study not feasible. Adding specificity to have a more targeted research question is essential. We will attempt to use full shot-gun metagenomic sequencing to track which antibiotics affect which microbiota. We will be working with groups to find potential correlations between antibiotics used and bacteria populations present in the microbiome There is a disconnect here between the research question and experimental design. When looking at the fully sequenced genomes of bacteria of those diagnosed and living with major depressive disorder, are the sequenced genomes different than the average population of those living without major depressive disorder? In the study you’ve chosen, the authors conducted experiments to address this research question. What effect do various mutations of genes linked to IBD have on the susceptibility of Alzheimer’s development? This research question is not a microbiome research-related question. |
Safety issues
There are no safety issues with either module because there is no wet lab component in this case study. Furthermore, the assessment protocol received exemption status from the Institutional Review Board at UNC-Chapel Hill (IRB #20–3516).
DISCUSSION
Field testing
Implementation of the ethics module was carried out in person in a 300-level microbiology course at a private, liberal arts, primarily undergraduate institution in the midwestern United States in the fall of 2020 and 2021. Participating students were a mixture of pre-nursing and biology majors without previous exposure to concepts associated with big data and the gut microbiome. Students were assigned to read the ethics module passages and respond to the reflection prompts through the course LMS as a pre-discussion assignment. During the class session, students were randomly assigned into groups with defined roles to debrief on the assigned readings and reflection prompts for the first ethics and implications of big data and the microbiome topic. The instructor then used a random number generator to identify a group to lead the discussion for each sequential reflection prompt. This was iteratively done for each of the ethics topics during a single 50-minute session.
Both modules were implemented in a 4000-level physiology class at a public, Native American-serving, non-tribal institution in the Southeastern United States in fall 2020 and spring 2022. Students were junior and senior biology majors (general biology or biomedical), some of whom had a background in genetics. The sections were small (less than 20 students per class), and the modules were run synchronously HyFlex (fall 2020) or completely in person (spring 2022) in consecutive lab sessions. The labs run for 110 minutes each, with tables that easily facilitate group work. In the first iteration, the modules were both run in a hybrid format, with approximately 33% of the students joining the class virtually on Zoom and the other 67% of the students present in person. For pre-work, the students required approximately 1 hour to read the articles and watch the videos as background. For the research design module, the students completed pre-work that included videos on next-generation sequencing and study design, as well as an article introducing the Lieberman lab. The pre-work took approximately 1 hour to complete. In-class, the students listened to a short lecture, and they worked in established groups of 3–4 to develop a new experimental question and method (110 minutes). For the in-person students, the lab tables served as group dividers. For the virtual students, the instructor placed them in separate breakout rooms on Zoom. For the ethics module, the instructor assigned the students to 1 of 4 groups and for pre-work the students read articles about their assigned topic. In-class, the groups then came together by topic to create a visual that they presented to the larger class and turned in for assessment. The virtual students were all assigned to the same group. The long, uninterrupted class time allowed the students to work intentionally with their group and ask deep questions, especially as it relates to the sensitive topics covered in this module.
Both modules were also implemented in a 400-level cellular and molecular neurotechnology course taken predominantly by neuroscience majors at a public R1 state institution in the Southeastern United States. The case study was implemented in four distinct sections of this course beginning in fall 2022 (>120 students). The first implementations in fall 2020 and spring 2021 (two sections) were in a synchronous online environment due to the COVID-19 pandemic. In fall 2021, the case was offered in an in-person format. In the first delivery of the case two and a half 50-minute periods were dedicated. Over several rounds of iteration, the case was expanded because it falls at the end of the semester, when student workload in this course and others is high. Students required more protected collaborative class work time, and in Spring 2021, three 50-minute class periods were utilized for both modules of the case. Finally, in fall 2021, the same three class periods were utilized, but additional work time was also protected on the last day of class. Expanding the in-class, protected group work time improved the quality of case study products. The research design module occurred over two 50-minute class periods that required approximately 1 hour of student pre-work. The second ethics module was also expanded to include one-and-a-half class sessions (~75 minutes total). In the first session, student groups presented slides summarizing their topic (Fig. 1), and the whole class discussed metagenomics research-related ethical issues. Students used any remaining time to finish their group case study document that was turned in for assessment.
Evidence of student learning
To assess student perceptions of learning gains, each classroom administered a retrospective survey instrument consisting of a 4-point Likert-type item for each learning outcome. Participants self-reported significant learning gains for 11 items and indicated that they believed they learned more through the modular case study module format relative to standard lecture delivery (Fig. 3). Likert ratings indicate that both case study modules lead to significant student self-reported learning gains (P < 0.001, Wilcoxon signed-rank test).
Possible modifications
In the microbiology course, over 70% of the respondents reported particularly enjoying the engaging content on aspects of the human microbiome. Smaller subsets of students specifically cited the ethical aspects of the activity and real-world applications as items they enjoyed the most. When prompted to identify something that they did not enjoy about the activity, a majority (>55%) of the respondents could not identify an aspect that they did not enjoy and instead indicated that they enjoyed the activity. The most frequent response for activity improvement was to streamline the chosen articles assigned for reading. This cohort of students, predominantly pre-allied health, found less value in having both a popular press and a primary literature article showcasing each example compared to majors in other courses. Feedback for “ideas for resource improvement” principally followed comments suggesting to reduce two articles per topic to a single article. Allied health majors found the two articles redundant and the primary literature article a bit high in register. Additionally, a subset of students suggested allocating more class time to assist with unpacking the topics in greater detail during classroom discussion.
In the physiology course, over 75% of the students who completed the feedback section stated that they enjoyed the case study. A subset of students specifically commented on the rural medicine section in the ethics module as being interesting and relevant, especially at a rural institution. In the “ideas for resource improvement,” students asked for a reduction in the number or length of the articles they had to read for both the modules pre-work, and the addition of a video for the ethics module. A subset of students wanted class time devoted to a lecture on research design module topics and the addition of hands-on bench work to practice DNA extraction themselves or more quantitative skills in data analysis.
In the neuroscience course, 49% of students strongly agreed and 51% agreed (100% total) that, relative to a standard lecture, they learned more about metagenomics through the case study (37 total student responses). Students also reported what they enjoyed most, what they did not enjoy, and ideas they had for case improvement. Students cited a variety of case features as most enjoyable: learning new content at the intersection of neuroscience and microbiology (39%), discussions of the real-world implications of the material, for example, relevance to their own health or how microbiome data are impacted by food deserts and socioeconomic status (23%), designing their own experiment (15%), critical evaluation of microbiome research studies (10%), and the discussion of microbiome-related ethics (10%). Several students also pointed out their preference for the case study as a pedagogical approach. When asked to identify aspects of the case that they did not enjoy, the majority of students reported nothing (76%). However, some students shared recommendations to improve case formatting, how they struggled with the difficulty of the assigned reading, that too much reading was required, or that they wished the case study had been done in person. Although 52% of students did not identify an idea for improvement, several students provided suggestions we used to improve the case iteratively over time. For example, students had formatting suggestions that improved the readability of the case to clarify expectations. Students also suggested providing a more extensive list of microbiome studies to explore before the research design component, which we have incorporated into the latest version of the case and continue to add to each semester. Some students shared how they would have enjoyed learning about more than one topic for the ethics discussion. Student ideas also reveal potential modifications of the case for different courses that they may enjoy, such as expanding the case to include actual data analysis rather than just experimental design. Some students also simply requested more time. All of these potential modifications could be utilized to adapt the case and tailor it to a particular course’s needs. Before completing the manuscript, our team consulted with the author of the Scientific American blog “What’s in my poop?” (26). This article serves as an initial prompt for students and encourages them to think critically with skepticism about interpreting data from a single microbiome sample. In this regard, Dr. Lieberman shared how differences between the two samples discussed in the article may have resulted from bacterial blooming (28) during sample transport. Adding a discussion of this topic with upper-level microbiology students could be an interesting addition to the case for some courses. Regardless of the overall point of the blog, a call to remain cautious when analyzing microbiome data remains.
ACKNOWLEDGMENTS
This work was supported by the HITS Case Network (NSF-RCN grant no. 1730317).
We thank Carlos Goller, the leader of the HITS Network, undergraduate learning assistants, students in Neurotechnology (NSCI 423), students in Microbiology (BIO 314), and students in Animal Physiology (BIO 4610).
Contributor Information
Sabrina D. Robertson, Email: sabrinae@email.unc.edu.
Stanley Maloy, San Diego State University, San Diego, California, USA.
SUPPLEMENTAL MATERIAL
The following material is available online at https://doi.org/10.1128/jmbe.00074-23.
The student handout and instructions for students to engage in the case study.
Answers to questions from the student handout as well as detailed teaching notes for instructors to implement the case study in their classroom.
Assessment questions that were used to show evidence of student learning from the case study.
Tami Lieberman’s Scientific American Blog post in PDF, format included with her permission. The PDF is helpful for instructors because the figures no longer show up appropriately on Scientific American's website.
ASM does not own the copyrights to Supplemental Material that may be linked to, or accessed through, an article. The authors have granted ASM a non-exclusive, world-wide license to publish the Supplemental Material files. Please contact the corresponding author directly for reuse.
REFERENCES
- 1. Herreid CF. 1997. What is a case? J Coll Sci Teach 27:92–94. doi: 10.1177/183335839702700213 [DOI] [Google Scholar]
- 2. Bixler A, Eslinger M, Kleinschmit AJ, Gaudier-Diaz MM, Sankar U, Marsteller P, Goller CC, Robertson S. 2021. Three steps to adapt case studies for synchronous and asynchronous online learning. J Microbiol Biol Educ 22:22.1.22. doi: 10.1128/jmbe.v22i1.2337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Robertson SD, Bixler A, Eslinger MR, Gaudier-Diaz MM, Kleinschmit AJ, Marsteller P, O’Toole KK, Sankar U, Goller CC. 2021. HITS: Harnessing a collaborative training network to create case studies that integrate high-throughput, complex datasets into curricula. Front Educ 6:711512. doi: 10.3389/feduc.2021.711512 [DOI] [Google Scholar]
- 4. Goller CC, Johnson GT, Casimo K. 2022. Does organelle shape matter?: exploring patterns in cell shape and structure with high-throughput (HT) imaging. CourseSource 9. doi: 10.24918/cs.2022.3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Samsa LA, Eslinger M, Kleinschmit A, Solem A, Goller CC. 2021. Single cell insights into cancer transcriptomes: a five-part single-cell RNAseq case study lesson. CourseSource 8. doi: 10.24918/cs.2021.26 [DOI] [Google Scholar]
- 6. Miller HB, Robertson SD, Srougi MC. 2021. Seq’ Ing the cure: Neuroscience edition | NSTA. Available from: https://www.nsta.org/ncss-case-study/seq-ing-cure-neuroscience-edition
- 7. Goller CC. 2021. New tricks for old drugs: using high-throughput screening to repurpose medications. Available from: https://static.nsta.org/case_study_docs/case_studies/ht_screening.pdf
- 8. Slatko BE, Gardner AF, Ausubel FM. 2018. Overview of next‐generation sequencing technologies. Curr Protoc Mol Biol 122:e59. doi: 10.1002/cpmb.59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Weinstock GM. 2012. Genomic approaches to studying the human microbiota. Nature 489:250–256. doi: 10.1038/nature11553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Rhodes R. 2016. Ethical issues in microbiome research and medicine. BMC Med 14:156. doi: 10.1186/s12916-016-0702-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Ma Y, Chen H, Lan C, Ren J. 2018. Help, hope and hype: ethical considerations of human microbiome research and applications. Protein Cell 9:404–415. doi: 10.1007/s13238-018-0537-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Goodrich JK, Di Rienzi SC, Poole AC, Koren O, Walters WA, Caporaso JG, Knight R, Ley RE. 2014. Conducting a microbiome study. Cell 158:250–262. doi: 10.1016/j.cell.2014.06.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Round JE, Campbell AM. 2013. Figure facts: encouraging undergraduates to take a data-centered approach to reading primary literature. CBE Life Sci Educ 12:39–46. doi: 10.1187/cbe.11-07-0057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Johnson ZA, Sciolino NR, Plummer NW, Harrison PR, Jensen P, Robertson SD. 2021. Assessment of Mapping the brain, a novel research and neurotechnology based approach for the modern neuroscience classroom. JUNE 19:A226–A259. [PMC free article] [PubMed] [Google Scholar]
- 15. Paige . 2019. All people have a right to healthy gut Microbes. LIFE Apps LIVE LEARN. Available from: https://lifeapps.io/nutrition/all-people-have-a-right-to-healthy-gut-microbes
- 16. Finding diversity in the microbiome. 6. 2019. Nat Med 25:863–863. doi: 10.1038/s41591-019-0494-3 [DOI] [PubMed] [Google Scholar]
- 17. Brooks AW. 2019. How could ethnicity-associated Microbiomes contribute to personalized therapies Future Microbiol 14:451–455. doi: 10.2217/fmb-2019-0061 [DOI] [PubMed] [Google Scholar]
- 18. Ishaq SL, Rapp M, Byerly R, McClellan LS, O’Boyle MR, Nykanen A, Fuller PJ, Aas C, Stone JM, Killpatrick S, Uptegrove MM, Vischer A, Wolf H, Smallman F, Eymann H, Narode S, Stapleton E, Cioffi CC, Tavalire HF. 2019. Framing the discussion of microorganisms as a facet of social equity in human health. PLoS Biol 17:e3000536. doi: 10.1371/journal.pbio.3000536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Justin R. 2020. Tex Trib. In Rural Texas, the COVID-19 Pandemic Has Brought More Accessible Mental Health Care. Available from: https://www.texastribune.org/2020/06/09/coronavirus-texas-rural-mental-health-telehealth/
- 20. Stein R. 2013. Getting Your Microbes Analyzed Raises Big Privacy Issues. Cent Genet Soc. Available from: https://www.geneticsandsociety.org/article/getting-your-microbes-analyzed-raises-big-privacy-issues
- 21. Callaway E. 2015. Microbiome privacy risk. Nature 521:136. 10.1038/521136a [DOI] [PubMed] [Google Scholar]
- 22. Franzosa EA, Huang K, Meadow JF, Gevers D, Lemon KP, Bohannan BJM, Huttenhower C. 2015. Identifying personal microbiomes using metagenomic codes. Proc Natl Acad Sci USA 112:E2930–8. doi: 10.1073/pnas.1423854112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Sonnenburg ED, Smits SA, Tikhonov M, Higginbottom SK, Wingreen NS, Sonnenburg JL. 2016. Diet-induced extinctions in the gut microbiota compound over generations. Nature 529:212–215. doi: 10.1038/nature16504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Brooks AW, Priya S, Blekhman R, Bordenstein SR, Cadwell K. 2018. Gut microbiota diversity across ethnicities in the United States. PLoS Biol 16:e2006842. doi: 10.1371/journal.pbio.2006842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Vaudreuil R, Langston DG, Magee WL, Betts D, Kass S, Levy C. 2022. Implementing music therapy through telehealth: considerations for military populations. Disabil Rehabil Assist Technol 17:201–210. doi: 10.1080/17483107.2020.1775312 [DOI] [PubMed] [Google Scholar]
- 26. Agapakis C. 2014. Which bacteria are in my poop? It depends where you look. Sci Am Blog Netw. Available from: https://blogs.scientificamerican.com/oscillator/which-bacteria-are-in-my-poop-it-depends-where-you-look/
- 27. Morais LH, Schreiber HL, Mazmanian SK. 2021. The gut microbiota-brain axis in behaviour and brain disorders. Nat Rev Microbiol 19:241–255. doi: 10.1038/s41579-020-00460-0 [DOI] [PubMed] [Google Scholar]
- 28. Amir A, McDonald D, Navas-Molina JA, Debelius J, Morton JT, Hyde E, Robbins-Pianka A, Knight R, Arumugam M. 2017. Correcting for microbial blooms in fecal samples during room-temperature shipping. mSystems 2:e00199-16. doi: 10.1128/mSystems.00199-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The student handout and instructions for students to engage in the case study.
Answers to questions from the student handout as well as detailed teaching notes for instructors to implement the case study in their classroom.
Assessment questions that were used to show evidence of student learning from the case study.
Tami Lieberman’s Scientific American Blog post in PDF, format included with her permission. The PDF is helpful for instructors because the figures no longer show up appropriately on Scientific American's website.