Abstract
Mycobacterium tuberculosis strains of the Beijing lineage are globally distributed and are associated with the massive spread of multidrug-resistant (MDR) tuberculosis in Eurasia. Here we reconstructed the biogeographical structure and evolutionary history of this lineage by genetic analysis of 4,987 isolates from 99 countries and whole-genome sequencing of 110 representative isolates. We show that this lineage initially originated in the Far East, from where it radiated worldwide in several waves. We detected successive increases in population size for this pathogen over the last 200 years, practically coinciding with the Industrial Revolution, the First World War and HIV epidemics. Two MDR clones of this lineage started to spread throughout central Asia and Russia concomitantly with the collapse of the public health system in the former Soviet Union. Mutations identified in genes putatively under positive selection and associated with virulence might have favored the expansion of the most successful branches of the lineage.
M. tuberculosis and the other members of the M. tuberculosis complex (MTBC) remain the leading bacterial killers worldwide and still account for 1.3 million deaths annually1. Of major concern is the uncontrolled spread of MDR tuberculosis (defined by resistance to at least the 2 major first-line drugs isoniazid and rifampicin) in regions such as southern Africa2 and across large Eurasian territories encompassing the Baltic countries, Russia and the 11 other current or former members and participating states of the Commonwealth of Independent States. These countries are all ranked among the 27 countries with a high MDR tuberculosis burden1.
The massive spread of MDR tuberculosis in Eurasia is predominantly driven by M. tuberculosis clones of the Beijing/East Asian lineage3,4,5,6. Strains from the Beijing lineage have also been associated with large MDR tuberculosis outbreaks elsewhere7 and appear to be rapidly expanding in population size in settings with contrasting tuberculosis incidence levels8,9. Strains of this lineage have been proposed to possess selective advantages in comparison to strains from other MTBC lineages, comprising an increased capacity to acquire drug resistance, linked to hypermutability10 or the presence of compensatory mutations mitigating the fitness cost of resistance-conferring mutations11,12, increased transmissibility, hypervirulence and/or more rapid progression to disease after infection13,14,15,16,17. However, the association of Beijing strain infection with MDR tuberculosis and/or with specific pathobiological or epidemiological manifestations is not systematic18. This heterogeneity suggests the existence of substantial intralineage biogeographical diversity, affecting pathobiological properties.
To investigate this hypothesis, we analyzed the global biogeographical structure and origin of the Beijing branch of the MTBC by standard genotyping of 4,987 clinical isolates from 99 countries linked to drug resistance. In addition, we analyzed the genome sequences of 110 isolates representing the main clonal complexes (CCs) identified to further explore the evolutionary history of this important lineage.
Results
Global biogeographical structure
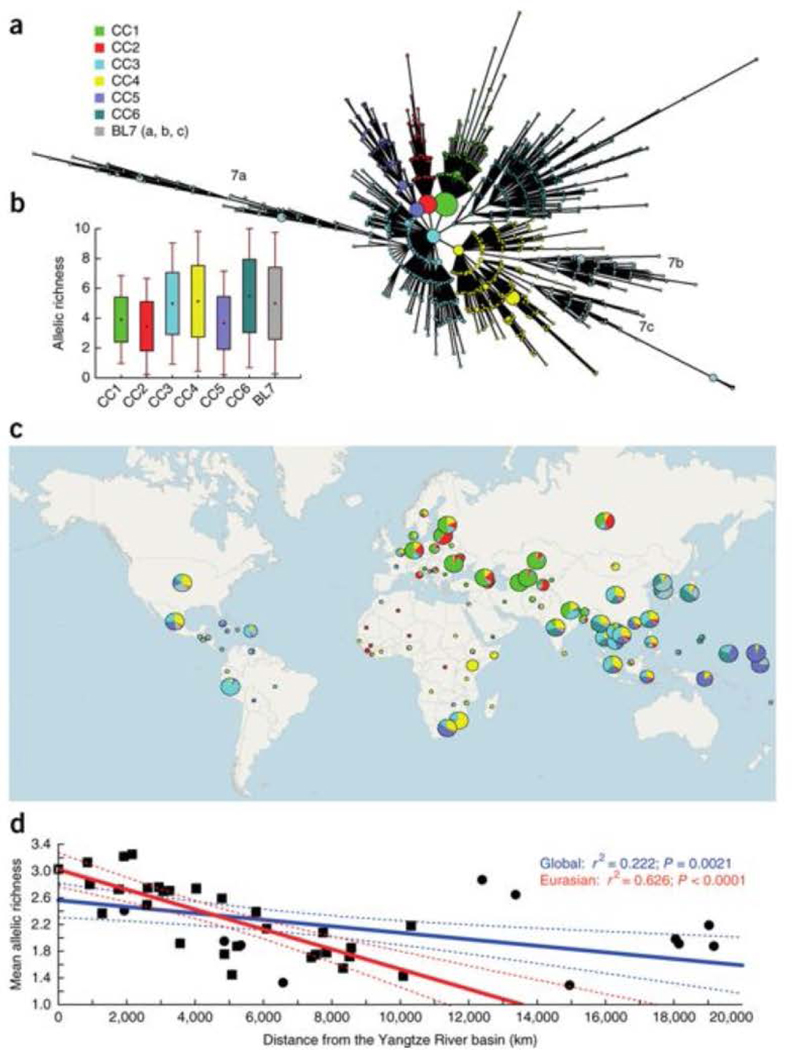
Our collection of 4,987 isolates from 99 countries (Supplementary Fig. 1) represents by far the largest Beijing lineage data set ever analyzed in terms of sample size and geographical coverage. Among these, 4,024 isolates (81%) originated from population-based or cross-sectional studies. Beijing strains were detected by screening for typical spoligotypes and best matching of 24-locus mycobacterial interspersed repetitive unit–variable-number tandem repeat (MIRU-VNTR) genotypes obtained from all the isolates15,19 (Supplementary Tables 1 and 2). To gain insight into the global population structure, we constructed a minimum-spanning tree (MSTREE) on the basis of the 24-locus MIRU-VNTR data that minimized the weights of the edges between genotypes. We initially classified the 4,987 isolates into 6 major CCs and 3 distant branches (a, b and c) collectively designated as basal sublineage 7 (BL7) (Fig. 1a). PCR analysis of the NTF region20 of 337 selected isolates showed that CC1–CC5 comprised typical/modern Beijing strains, whereas CC6 and BL7 comprised atypical ancestral Beijing variants (Supplementary Fig. 2 and Supplementary Table 3). We further analyzed the global distributions of CC1, CC2 and CC5, as the corresponding groupings were largely supported (with few to no outliers) by genome sequencing results. Likewise, the ancestral classification of CC6 and BL7 was confirmed by their deep branching in the genome-based trees.
Figure 1: Biogeographical structure of the M. tuberculosis Beijing lineage.
(a) MStree based on 24 MIRU-VNTR markers delineating the clonal complexes (CCs) gathered from a worldwide collection (n = 4,987). Major nodes and associated multi-locus variants were grouped into six CCs and a basal sublineage (BL). (b) Genetic variability in the different Beijing lineage CCs and the BL calculated using a rarefaction procedure (each CC included a subsample of 457 strains drawn randomly from its source population). Dots correspond to the mean allelic richness, boxes correspond to mean values ± s.e.m. and error bars correspond to mean values ± s.d. (c) Worldwide distribution of the Beijing CCs and BL. Each circle corresponds to a country, and circle sizes are proportional to the number of strains. Note that the results for CC3 and CC4, less supported by whole genome-based analysis, are only given as an indication. (d) Genetic erosion out of China. Mean allelic richness within geographical populations is plotted against geographical distance from the Yangtze River basin. Filled squares denote the Eurasian samples used for the regression; filled circles correspond to the global collection. Confidence intervals are represented by dashed lines.
In the MSTREE (Fig. 1a), CC1 and CC2, and to a lesser extent CC5, displayed a star-like shape typical of expanding populations, with high-frequency central genotypes surrounded by diffusing layers of variants. Such patterns were much less visible for CC6 and BL7, again suggesting more ancient populations and/or milder expansions. In accordance with this hypothesis, the mean allelic richness (number of alleles), calculated after correcting for sample size effects21 and taken as a surrogate indication of diversification time, was higher for CC6 and BL7 than for CC1, CC2 and CC5 (P < 0.05; Fig. 1b).
The spatial distribution on a worldwide scale of these CCs clearly shows a biogeographical structure and population clines in the Beijing lineage (Fig. 1c). Strikingly, the CC distribution was the most diverse in the East Asia and Far East region, suggesting that this region indeed represents the origin from which Beijing strains subsequently radiated. The gradient observed in CC5 proportions toward the Pacific Ocean suggests an eastward spread of this clone, followed by successive bottlenecks increasing its frequency in Micronesia and Polynesia. Likewise, we observed westward clines for CC1 and CC2, with these groups becoming highly dominant in central Asia and around the Black Sea (CC1) and in Russia and Eastern Europe (CC2) (Supplementary Figs. 3 and 4). In contrast, CC6 and BL7 were more confined to eastern Asia. The only other region where we retrieved substantial proportions of CC6 and BL7 was North America/Mexico, where the CC frequencies resembled those for Chinese samples (Supplementary Fig. 3), likely reflecting the effect of recent Chinese immigration.
East Asian origin, multiple epidemic waves and timing
The East Asian origin of the Beijing lineage was further supported by plotting MIRU-VNTR allelic diversity per geographical population against geographical distance from the Yangtze River basin (Fig. 1d). Allelic diversity decreased with increasing geographical distance, and 22% of the variance could be explained by geography alone when considering the full data set. Interestingly, this percentage increased to 63% when focusing solely on the Eurasian samples. This difference reflects the excess of allelic richness in the samples collected, especially in North America and in South Africa, likely resulting from substantial recent immigration from China.
We then attempted to date past expansions and to generate estimates for the time to the most recent common ancestor (TMRCA). Because it is not possible to simultaneously estimate N (the effective population size) and μ (the mutation rate), we implemented mutation rate priors and intervals covering previously reported μ values22,23,24 (Supplementary Fig. 5). By applying these rates and a generation time of 1 d for M. tuberculosis, we estimated a mean TMRCA of 6,600 years for the Beijing lineage (Table 1). According to coalescent analyses, CC6 and BL7 are the two oldest sublineages, with TMRCAs of ∼6,000 and 5,000 years, respectively, and CC5 is the youngest, with a TMRCA of ∼1,500 years.
Table 1:
MIRU-based demographic and dating estimates of the CCs and lineages detected in the Beijing clade
| Clonal complex | N 0 a | N 1 a | r = N0/N1 | t a b | TMRCAb |
|---|---|---|---|---|---|
| CC1 | 13.106 (7.996–24.713) | 0.743 (0.502–1.012) | 17.639 | 263 (190–398) | 4,415 (2,569–7,509) |
| CC2 | 8.633 (3.954–27.534) | 0.529 (0.337–0.856) | 16.319 | 216 (138–350) | 1,797 (958–3,690) |
| CC3 | 47.204 (32.510–72.062) | 1.247 (0.947–1.745) | 37.854 | 559 (445–719) | 3,151 (1,750–5,801) |
| CC4 | 32.830 (24.125–46.892) | 1.683 (1.247–2.362) | 19.507 | 699 (523–928) | 4,084 (2,616–6,764) |
| CC5 | 8.465 (4.905–18.015) | 0.581 (0.394–0.882) | 14.570 | 240 (164–360) | 1,492 (872–2,898) |
| CC6 | 66.439 (47.548–97.318) | 2.609 (2.004–3.559) | 25.465 | 1,226 (967–1,552) | 6,161 (3,419–10,725) |
| BL7 | 22.864 (17.939–29.514) | 3.030 (2.349–4.147) | 7.546 | 1,398 (1,056–1,834) | 5,212 (3,613–8,962) |
| Global lineagec | 67.098 (54.007–86.504) | 3.053 (2.331–4.160) | 21.978 | 1,275 (1,007–1,613) | 6,604 (4,270–12,514) |
N0 current effective population size; N1 ancestral population size prior expansion; ta, time elapsed since the last expansion began; TMRCA, time to the most recent common ancestor.
Effective population sizes are expressed in millions.
Datings are expressed in years. Estimates correspond to the median values, and numbers in parentheses correspond to the 95% highest posterior density (HPD) intervals generated during the Bayesian analysis.
Estimates based on 10 reiterations following a subsampling procedure of 500 strains from the full data set (±s.d.).
Genetic data can also be used to unravel recent demographic changes. By using Bayesian-based coalescent tools available for VNTR markers, we tested whether a recent decline or increase in bacterial population size occurred and calculated ta, reflecting the time elapsed since a last expansion began. All CCs displayed strong expansion signatures (Table 1). The expansion ratio r = N0/N1 (where N0 is the current effective population size and N1 is the effective population size before expansion began) ranged from 8 (BL7) to 25 (CC6). For CC1, CC2 and CC5, the expansion onset, provided by median ta values, dated back some 200–250 years. These findings clearly contrast with the much older expansions detected for CC6 and BL7 dating back to the medieval period (Table 1).
Whole genome–based phylogeny and recent population dynamics
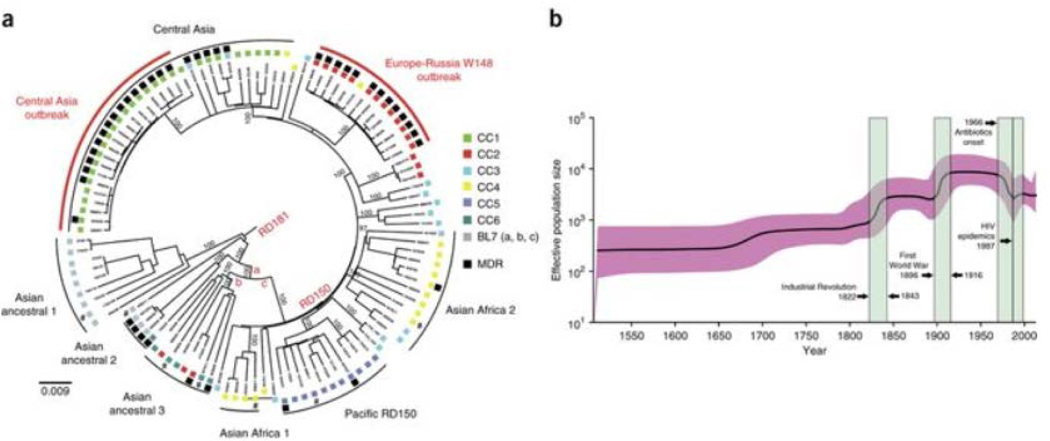
Because MIRU-VNTR loci may be affected by homoplasy25, we sequenced the genomes of 110 strains (Supplementary Fig. 6 and Supplementary Table 4) representing the 7 sublineages initially identified by genotyping to obtain a robust tree topology and confirm the ancestral clades. After removing genes associated with drug resistance, repetitive and mobile elements, and artifactual SNPs linked to indels26, we detected 6,001 polymorphic sites (SNPs). Likelihood mapping analyses27 indicated a robust phylogenetic signal (>81%), albeit with minor occurrence of star-likeness, signaling that the tree was well resolved in certain parts only (Supplementary Fig. 7). In contrast to a recent report suggesting some degree of horizontal gene transfer (HGT) in the MTBC28, analysis of neighbor nets and densitrees identified no major splits suggestive of HGT (Supplementary Figs. 8 and 9a), and the pairwise homoplasy index (PHI) test did not find evidence (P = 0.7668) for recombination that might blur phylogenetic reconstruction. The genome-based tree topology was fully consistent with Beijing linage–specific regions of difference (RD181 and RD150). Numerous subgroup-specific polymorphisms also clearly distinguished three ancestral and five modern Beijing phylogenetic clades (Fig. 2a, Supplementary Fig. 9b and Supplementary Table 5), fairly congruent with the MIRU-VNTR groupings except for CC3 and CC4 (Fig. 2a). Strains associated with BL7 clearly corresponded to the most ancestral population, followed by CC6 strains. As such, we refer to both groups as Asian ancestral subgroups 1–3. Strains from the modern CCs diverged more recently and displayed shorter branches. Central Asian, European-Russian and Pacific branches also largely confirmed the CC1, CC2 and CC5 classifications, respectively. However, CC4 strains could be clearly differentiated into two genome-based subgroups (Asian Africa 1 and 2), whereas the distribution of CC3 isolates was much more scattered on the tree. This partial incongruence in the MIRU-VNTR–based tree likely reflects homoplastic effects and/or hard polytomies in the context of recent expansions.
Figure 2: Phylogenetic reconstruction of the MTBC Beijing lineage and change in population size through time.
(a) Midpoint-rooted maximum-likelihood tree based on 110 genomes and a total of 6,001 concatenated SNPs. Characteristic mutations differentiating modern and ancestral Beijing strain types are mapped on the tree—mutT4 encoding p.Arg48Gly (branch a), ogt encoding p.Arg37Leu (branch b) and mutT2 encoding p.Gly58Arg (branch c)—as is the absence of the RD181 and RD150 regions of difference. Black squares correspond to strains with an MDR or extremely multidrug-resistant (XDR) phenotype, and a number sign indicates strains lacking drug susceptibility test information. Numbers on branches correspond to bootstrap values. The tree topology remains the same when H37Rv is used as an outgroup. (b) Bayesian skyline plot indicating changes in the Beijing lineage over time with a relaxed molecular clock set at 1 × 10−7 mutations per nucleotide per year. The shaded area represents the 95% confidence intervals, and the green colored boxes represent major socioeconomic events that might have affected the demography of M. tuberculosis.
Genome-wide SNP information provides the potential for more sensitive detection of one or even several population changes. However, such analysis requires calibration of the genome evolution rate, which is not trivial. Confident, closely matching estimates of short-term genome mutation rates, ranging between 1.0 × 10−7 and 1.3 × 10−7 substitutions per nucleotide site per year, have been independently obtained from the study of different contemporary epidemics26,29 and a macaque infection model30. However, such estimates are supposed to differ by one or two orders of magnitude from the long-term fixation rate, as less fit mutations are purged from the genomic pools31. Consequently, if the mutation rate changes through time, any mutation rate used will imply information distortion at some point. Therefore, we decided to use the previously estimated short-term rate of 1 × 10−7 substitutions per nucleotide site per year (95% confidence interval of 0.6 × 10−7 to 1.5 × 10−7)29 to depict a more likely demographic scenario over the last few hundred years.
We generated a Bayesian skyline plot that estimated changes in the pathogen’s effective population size over time (Fig. 2b) on the basis of the 110 genomes. We detected a stepping stone–like increase in population size with two sharp population growth phases, one occurring during the Industrial Revolution and the second approximately matching the period of the First World War. It was also striking that the only decrease in population observed on the skyline plot coincided with the onset of large-scale antituberculosis drug use. Although a cumulative effect cannot be excluded (also due to changes, for example, in living conditions), we do not favor a major influence from BCG vaccination, whose widespread use started earlier (in the late 1940s) and had a relatively moderate protective effect against tuberculosis32. Finally, a temporary reversal of this downward trend was noticeable. Interestingly, this late, mild bacterial expansion matched with the beginning of the HIV epidemics and the first large MDR tuberculosis outbreaks in the former Soviet Union33 and the United States34 in the 1990s.
Specific antibiotic resistance
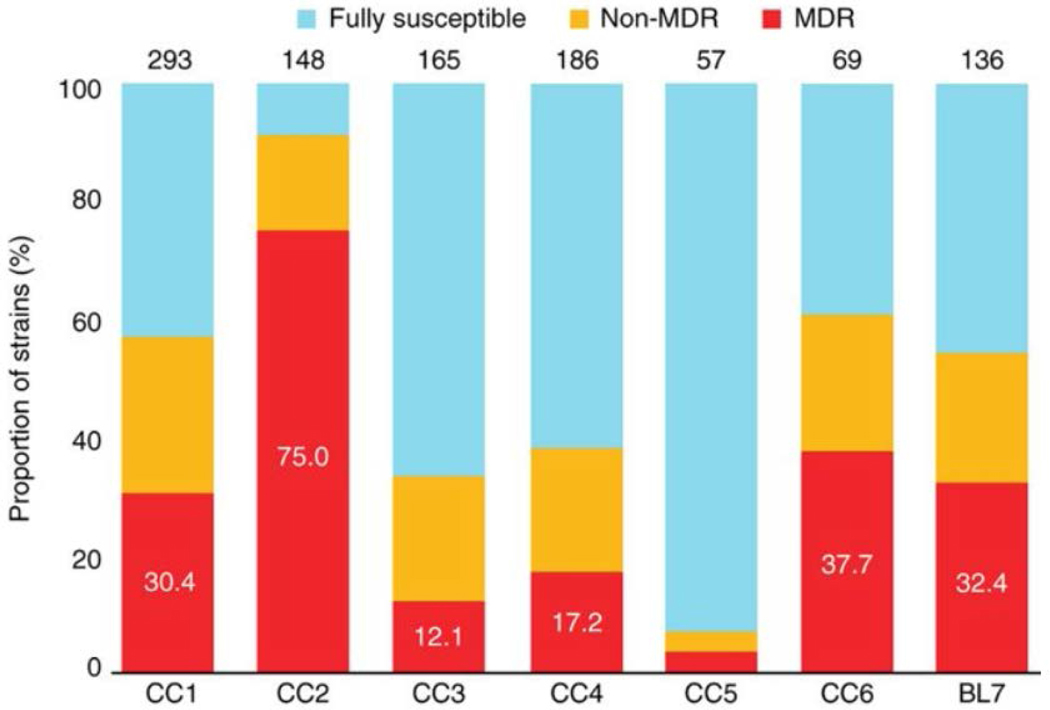
To investigate a possible association between antibiotic resistance and the identified CCs, we examined a subset of 1,054 clinical isolates with known drug resistance profiles from our global strain collection (Supplementary Table 6). Of these, 91% (965/1,054) originated from 12 different study settings that were population based or cross-sectional. We avoided including local MDR tuberculosis cohorts. The analysis showed that CC2 had the highest proportion of MDR strains (75.0%, 111/148; P < 0.0001) (Fig. 3). CC5 as well as the more heterogeneous CC3 and CC4 exhibited the lowest percentages of resistance (P < 0.01), with 3.5% (2/57), 12.1% (20/165) and 17.2% (32/186), respectively. Although the proportions of MDR isolates were similar for the modern CC1 (30.4%, 89/293) and the ancestral CC6 (37.7%, 26/69) and BL7 (32.4%, 44/136), the clustering rates (defined as the proportions of isolates with an identical MIRU-VNTR haplotype) of the MDR strains differed significantly (P < 0.0001) (Supplementary Table 7). In CC1, 94.4% (84/89) of all MDR isolates were associated with a shared MIRU-VNTR haplotype, in comparison to only 42.3% (11/26) and 56.8% (25/44) in CC6 and BL7, respectively (Supplementary Table 7). Overall, CC1 and CC2 had the highest clustering rates for MDR strains (P < 0.01), indicating population expansion amplified by the recent transmission of MDR strains, especially associated with MIRU-VNTR haplotypes termed 94–32 (CC1) and 100–32 (CC2) according to a standard nomenclature19.
Figure 3:
Proportions of MDR tuberculosis strains among the six CCs and BL of the Beijing lineage.
Note that CC2 comprises significantly (P < 0.001) more MDR strains than the other complexes. The total number of strains with available drug susceptibility test information in each group is given above the corresponding column.
This MDR outbreak hypothesis was strongly supported by the analysis of mean pairwise genetic distances among strain genomes. Strains from the central Asian outbreak (associated with CC1) and from the European-Russian W148 branch (associated with CC2 and defined as a Russian successful clone6) exhibited a lower pairwise distance than all other subgroups (P < 0.05), with respective means of only 17 and 23 SNPs differentiating pairs of isolates (Supplementary Fig. 10). These strains were all resistant to at least isoniazid and streptomycin (with resistance conferred by mutations to katG (encoding p.Ser315Thr) and rpsL (encoding p.Lys43Arg), respectively; data not shown). These data thus indicate a specific recent expansion of two MDR clones in Russia and central Asia.
Traces of positive selection
To identify genetic targets potentially linked with the expansion of modern Beijing subgroups some 200–700 years ago (Table 1), we first examined 81 polymorphisms characteristic for all modern strains (Supplementary Table 8). Among these, we found four and three SNPs in the mce (mammalian cell entry) and vapBC (virulence-associated protein) gene families, respectively. Moreover, SNPs in the coding regions of the same two gene families were fixed in the genomes of both modern and ancestral Beijing subgroups (Supplementary Table 5), possibly suggesting positive selection acting on these genes. To test this hypothesis, we calculated the mutation rates per base pair and the dN/dS ratios (global ratios of nonsynonymous to synonymous SNPs) of the concatenated gene sequences for these two gene families and compared them with corresponding values for essential and non-essential genes, genes encoding polymerases, ribosomal proteins, T cell antigens and lipoproteins, and the fad gene family (Supplementary Table 9). Within the limits imposed by the low levels of total variation, we found a two-to threefold increase in mutation rate, as well as higher dN/dS values in modern subgroups (0.91–1.83) for mce and vapBC genes than for any control set (0.26–1.07) (Supplementary Table 9). There were also more (P < 0.05) amino acid substitutions among the mce and vapBC gene products than in the control sets in modern subgroups, a situation not encountered in the ancestral subgroups (Supplementary Table 10).
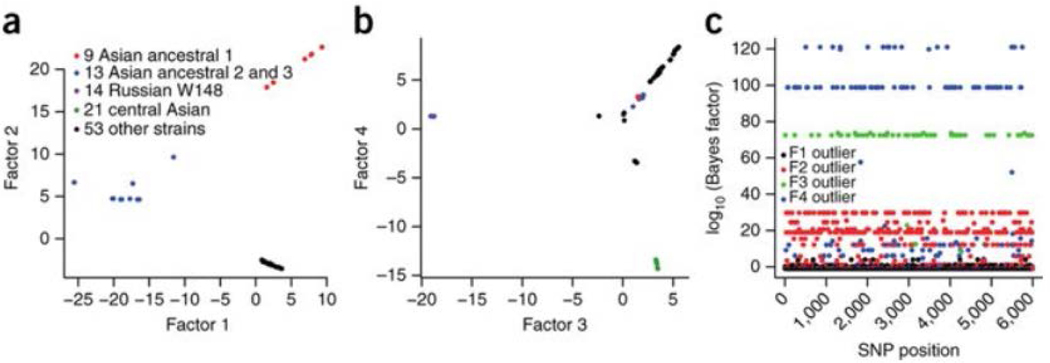
Furthermore, we searched for branch-specific SNPs and small deletions that were potential candidates for specific adaptation. Noteworthy among these was a frameshift mutation in kdpD (c.2541_2542delCA) specific to all European-Russian W148 MDR outbreak strains, predicted to result in an altered C terminus of the sensor and its fusion to the cognate regulator of the two-component system encoded by the kdpDE operon (Supplementary Table 5). A partial deletion of the kdpDE operon in M. tuberculosis has already been associated with greater virulence35. We refined the search by performing a genome scan analysis, using a Bayesian model36 that detects the structure and clustering of individuals in a population, with latent variables called factors. We thereby both inferred population structure and identified 200 ‘outlier’ SNPs, defined as those most related to the detected structure, which were distinguished from noise-containing SNPs. Inspection of the factors (Fig. 4) indicated that factor 2 distinguishes the ancestral strains from the derived ones, whereas factors 3 and 4 differentiate, respectively, the European-Russian and central Asian lineages from the other strains. Remarkably, the SNPs with the largest Bayes factors within a factor were found to be concentrated among highly plausible gene targets under positive selection (encoding drug resistance, virulence and surface-exposed proteins) (Supplementary Table 11). For factor 2, nonsynonymous SNPs affecting the bulk of the modern strains were found, for instance, in lysX (a gene required for resistance to cationic antimicrobial peptides37), fadD28 (encoding a virulence factor38) and mutT2 (a putative mutator gene39) (Supplementary Table 11). For factor 3, nonsynonymous mutations were found in pks5 (involved in the biosynthesis of surface-exposed polyketides40), mce3B (encoding an invasin-adhesin–like protein41) and Rv1877 (involved in efflux pump–mediated drug resistance in Mycobacterium smegmatis42). Finally, outlier SNPs for factor 4 included nonsynonymous mutations in fas (an essential gene involved in lipid metabolism with a potential role in antigenic recognition43) and rpoC (putatively associated with fitness cost compensation in rifampicin-resistant strains44).
Figure 4:
SNP-based Bayesian factor model analysis for detecting genes involved in positive selection in the Beijing lineage.
(a,b) Latent factors of the 6,001 SNPs and 110 strains with the first 2 factors (a) and the 2 consecutive ones (b). (c) Manhattan plot representing the selection scan and the outliers that are related to the different latent factors.
We also sought to detect SNPs undergoing convergent evolution to identify possible beneficial mutations. We scrutinized an extended data set of 6,696 polymorphic sites with loosened thresholds of variant frequency and coverage to prevent the exclusion of positions below the thresholds for some genomes. Nevertheless, candidate SNPs still had to be covered by at least ten reads to be considered. Beyond known compensatory mutations in rpoC45 and mutations in the promoter regions of drug resistance–associated genes (eis, inhA and embA), we identified 15 additional targets possibly under positive selection (Supplementary Table 12). Among these were nonsynonymous SNPs in mmpL11 (putatively involved in fatty acid transport), folC (an essential gene involved in respiration) and Rv2670c (of unknown function), exclusively found in drug-resistant isolates in different monophyletic subgroups.
Discussion
Using the largest data set of a single M. tuberculosis lineage ever investigated, we identified the population structure and reconstructed the evolutionary history of the Beijing lineage on a worldwide scale. The spatial distributions of strain haplotypes and allelic diversities, as well as the localization of ancestral CCs and branches, show that, in agreement with its historical designation, this lineage originated in the geographical zone centered on northeastern China, Korea and Japan. The time of its emergence, estimated at 6,600 years ago, is consistent with other recent data based on a much smaller strain collection46 and is compatible with the onset of agriculture in that region47. Our data lead us to conclude that the worldwide spread of Beijing sublineages from this original focus occurred in several waves and was accompanied by important changes in the pathogen’s population size, especially in the recent historical period, starting with industrialization and urbanization in the nineteenth century. The latest steps of this evolution include the specific epidemic expansion of two MDR clones throughout central Asia and Russia.
Our results suggest that, whereas the Asian sublineages (CC6 and BL7) arose during the late Neolithic, the two most recent clades appeared later (during the early medieval period) and gave rise to the European-Russian (CC2) and Pacific (CC5) branches. Interestingly, these two sublineages, as well as the central Asian (CC1) lineage, are also the ones showing the most recent traces of expansion, around 200 years ago, according to genotyping data. These recent expansions remarkably match known episodes of Chinese immigration. Major waves of Chinese settlement occurred on the Pacific Islands in the 1850s, along the navigation and trade routes across the Pacific Ocean and in North and South America, which might have promoted the expansion of CC5 in this region48. Likewise, several waves of Chinese refugees migrated to the Russian empire, especially Kyrgyzstan, Kazakhstan and Uzbekistan, as a consequence of a series of national uprisings from 1861 to 1877, which might have driven the expansion of the CC1 and CC2 strains in these regions49. These recent western expansions are probably superimposed on a more historical, continuous flux of the different Beijing sublineages westward along the Silk Road.
Consistently, a sharp increase in the population size of the Beijing lineage as a whole was also detected concomitantly with the Industrial Revolution, around 200 years before the present, by Bayesian skyline analysis of genome-wide SNPs. The amount of SNP information available allowed us to detect even more recent changes in population size. The second abrupt surge in bacterial population size, detected around the period of the First World War, is fully in line with the documented peak in the tuberculosis death rate all over the globe due to the deprivations and comortality induced by the influenza pandemics at that time50. Our data additionally disclose the probable (but not necessarily exclusive) impact on the bacterial population of the large-scale onset of antibiotic use in the 1960s, which resulted in the first drop ever observed on the skyline plot, and of the HIV epidemics and/or increase in MDR populations, interrupting this fall.
Notably, our data indicate that the expansion of the two sublineages more frequently associated with MDR genotypes (central Asian (CC1) and European-Russian (CC2)) predated the era of antibiotics. This finding indicates that the prevalence of drug resistance in these CCs is not the primary cause of expansion but rather a consequence of public health–related and clinical weaknesses superimposed on a growing bacterial population. This hypothesis is supported by the specific, extremely low mean pairwise genetic distance of around 20 SNPs among the genomes of the MDR strain subsets of CC1 and CC2 (Supplementary Fig. 10). Assuming a mutation rate of 0.3–0.5 SNPs per genome per year26,29,51, this finding indicates that the two corresponding original MDR clones started to spread epidemically only some 20–30 years ago across Eurasia, coinciding with the collapse of the public health system of the former Soviet Union. Of note, we found that a large clade (termed clade B), with limited genome-wide diversity among MDR strains from a local southwestern Russian population3, is part of the same European-Russian W148 (CC2) outbreak defined in our global study (data not shown), which thus further demonstrates the epidemic spread of this clone.
Evidence for the higher virulence of modern Beijing strains in comparison with ancestral sublineage strains has been reported52,53. This difference might have contributed to the differential historical spread observed for these two sublineage groups (with, for example, the geographical restriction of CC6 and BL7).
We also detected an ensemble of gene variants potentially associated with the expansion of the modern Beijing strains by performing whole-genome scans for candidate SNPs and genes under positive selection unrelated to known targets of drug resistance. Among these, members of the mce and vapBC multigene families, associated with mycobacterial virulence54 and the modulation of host immune response55 and with growth control56, respectively, appear as prominent candidates. Interestingly, we also identified sites within Rv0176 (encoding an MCE1-associated protein) as being under diversifying selection by analyzing 73 genomes representing 6 of the 7 main MTBC lineages57. We also defined a list of other plausible gene targets under positive selection, associated with antibiotic resistance, fitness compensation, virulence and surface-exposed proteins, using a new Bayesian model-based SNP detection method. Of special interest is the identification of a frameshift mutation in the kdpDE operon, encoding a signal transduction system, which is a hallmark of the European-Russian W148 (CC2) sublineage. As a partial deletion of kdpDE in M. tuberculosis H37Rv has been shown to result in increased virulence in a mouse infection model35, this frameshift mutation, putatively leading to a fusion protein of altered functionality, might well have contributed to the success of this clade. Hence, dismissing such phylogenetically informative SNPs as resulting from drift alone and not from selection might be misleading. This assumption is corroborated by an over-representation of genes associated with critical cell wall biosynthetic pathways among the functional families found for the other sublineage-specific SNPs detected by our Bayesian approach in comparison to those found for random SNPs (P < 0.01).
Finally, by screening for nucleotide positions under possible convergent evolution among drug-resistant isolates, we identified a set of new potential targets of drug resistance or fitness compensation mechanisms, including mmpL11 and Rv2670c, in addition to expected genes such as rpoC, embA, inhA and eis. Our screen also captured folC, recently reported as a resistance-associated target by genome sequencing of 161 isolates from China58. Interestingly, polymorphisms in these genes are especially enriched in the European-Russian W148 (CC2) sublineage and are strongly associated with the MDR tuberculosis strains of MIRU-VNTR haplotype 100–32. Intriguingly, apart from expected genes and folC, we found virtually no overlap among the targets of positive selection associated with drug resistance identified in our study and in two other recent reports based on other strain samples and patient populations58,59. This lack of overlap suggests a potential influence from differences in strain genetic backgrounds, antituberculosis drug regimens and patient-dependent pharmacokinetics on the course and targets of selection.
In conclusion, our results show for the first time, to our knowledge, the important dynamic changes that have occurred in the worldwide population of a major M. tuberculosis lineage. Although the exact timing remains dependent upon uncertainties in mutation rates, especially over the long term, the conjunction of the most recent changes in the bacterial population with specific chief events in human history, as detected by using a molecular clock calibrated according to several convergent studies, is intriguing. Among other results, our analysis of European-Russian and central Asian sublineage demography illustrates how the effect of recent human interventions (the introduction of antibiotics followed by the development of multidrug resistance) has to be differentiated from pre-existing bacterial population changes to explain the regional prevalence of particular strains. Similar approaches could therefore be envisaged to better monitor and quantify the effects of future public health interventions (for example, new drugs and/or vaccines) on the pathogen population. From a more fundamental perspective, we propose that the expansion of modern Beijing strains has been favored by mutations in a number of gene targets under positive selection. The data obtained here thus suggest further experiments to investigate which of these candidate genes were involved. Such work may ultimately contribute to the detection of new targets for combating tuberculosis.
Methods
Sampling and data collection.
The study is based on a global collection of clinical isolates of M. tuberculosis Beijing (Supplementary Tables 1 and 2). The data set contains the 24-locus MIRU-VNTR genotypes for 4,987 strains from 99 countries (Supplementary Fig. 1). M. tuberculosis isolates were genotyped by multiplex PCR amplification as described previously60,61. Amplicons were subjected to electrophoretic analysis using ABI 3100 and 3730 automated sequencers. Sizing of the PCR fragments and assignment of VNTR alleles at the 24 loci were performed using genemapper software (PE Applied Biosystems).
MIRU data analyses.
Genetic diversity estimation and population structure. The number of alleles (allelic richness) in each M. tuberculosis clonal group or biologically relevant population was estimated, and sample sizes were corrected by the rarefaction procedure using hp-rare62. Comparison tests as well as P values were estimated using the statistica v.6.1 package.
The population structure based on 24-locus MIRU-VNTR data for 4,987 clinical M. tuberculosis Beijing isolates was inferred with the minimum spanning tree (MSTREE) algorithm implemented in BioNumerics software package v6.7 (Applied Maths). Strains with an identical MIRU-VNTR haplotype were pooled in a single node in the MSTREE and thereby represent a cluster. The rate of clustered strains was considered as an indicator for the extent of recent transmission among the Beijing sublineages.
Biogeography. We evaluated the allelic richness of each geographical sample where 24-locus MIRU-VNTR haplotypes were available for at least 23 isolates using the software hp-rare21. This software computes a rarefaction to avoid the bias created by differences in sampling size. We then computed the geographical distances (shortest walking distance according to classic human migration routes) between all 40 corresponding geographical locations and the Yangtze delta region using tools implemented in Google Earth. This step was followed by the calculation of a linear correlation (r2) between the allelic richness of an area and the geographical distance from the source. For the geographical mapping of Beijing clonal complexes on the world map, we used MLVA Compare v1.03 (Ridom) and the implemented geocoding option.
Coalescence, TMRCA and demography. In a first step, we used a Bayesian-based coalescent approach63 on MIRU-VNTR data. It assumes a stepwise mutation model of MIRU evolution, and it estimates the posterior probability distributions of the genealogical and demographic parameters of a sample using Markov chain Monte Carlo (MCMC) simulations. This method permits inferences of important biological parameters such as the TMRCA of a given sample, the past and present effective population sizes, and the latest demographic changes (decline, constant population size or expansion). Given the absence of recombination (as confirmed by whole-genome sequence analysis), we inferred demographic parameters from the 24-locus data under a Bayesian-based coalescent approach implemented in the software batwing for linked tandem repeat loci64,65. The coalescent prior used for the distribution of topology and branch lengths of the gene genealogy was a three-parameter model: a constant ancestral population size experiencing an exponential population expansion at some time in the past64. The likelihood of the gene genealogy was computed under the stepwise mutation model66. The posterior probabilities of the gene genealogy, population genetics parameters and MIRU-VNTR mutation rates were approximated through the Metropolis-Hastings algorithm67,68. Time of population expansion and modern effective population size were scaled relative to the ancestral population size under the model implemented in batwing. To test for prior sensitivity and to check for convergence, we ran all the analyses using two different priors for the ancestral population size: we tested both a normal distribution N (5 × 107; 1 × 107) and a uniform distribution U (1 × 103; 1 × 108). The results were consistent between the runs, and we present only the results obtained under the normal prior. We therefore placed an independent uniform prior for the mutation rate of each MIRU-VNTR locus, bounded between 9 × 10−8 and 9 × 10−7 per locus per generation, according to previous studies69. All the analyses were run for 1 billion MCMC generations, using a thinning interval of 100,000. The MCMC output was analyzed using the library coda available under the R environment (R Development Core Team 2011) to obtain the posterior distribution and the effective sample size (ESS) of all parameters (which were all above 150).
Genome data analyses.
Strain selection for whole-genome sequencing analysis. For whole-genome sequencing and phylogenetic reconstruction, strains were selected according to two priority rules: first, to represent major nodes in our MIRU-VNTR–based minimum spanning tree and their main adjacent multi-locus variants and, second, to cover different countries of origin and/or different studies. As a result, the proportions of isolates from the 7 identified complexes were in a similar range, i.e., 29/907 (3.2%) for CC1, 19/457 (4.2%) for CC2, 14/972 (1.4%) for CC3, 18/1,027 (1.8%) for CC4, 11/542 (2.0%) for CC5, 7/475 (1.5%) for CC6 and 12/607 (2.0%) for BL7. CC1 and CC2 were somewhat over-represented to better confirm the outbreak-related nature of isolates subsequently termed as central Asian and European-Russian W148 outbreaks, respectively, and with a comparable very low mean pairwise genetic distance of around 20 SNPs. Again, this selection was carried out by considering isolates from different regions and studies (Supplementary Table 3).
Variant detection. Whole-genome sequencing was performed with Illumina technology (MiSeq) using Nextera XT library preparation kits as instructed by the manufacturer (Illumina). Raw data (fastq files) were submitted to the EMBL-EBI ENA SRA under the study accession PRJEB7281. Resulting reads were mapped to the M. tuberculosis H37Rv genome sequence (GenBank, NC_000962.3) using the exact alignment program saruman70. High-quality SNPs with a minimum of 10× coverage and 75% variant frequency were extracted and combined for all analyzed isolates (n = 110) using customized Perl scripts. We used only genome positions with high-quality variant calls for every isolate (that met the thresholds for coverage and variant frequency) for a concatenated sequence alignment. For phylogenetic inference, we excluded drug resistance–associated genes, repetitive regions and artifactual variant calls resulting from indels in single strains.
Likelihood mapping. The phylogenetic signal of the data set was investigated with the likelihood mapping method implemented in tree-puzzle71 by analyzing 10,000 random quartets. This method proceeds by evaluating, using maximum-likelihood, groups of four randomly chosen sequences (quartets). The three possible unrooted tree topologies for each quartet are weighted, and the posterior weights are then plotted using triangular coordinates, such that each corner represents a fully resolved tree topology. Therefore, the resulting distribution of the points shows whether the data are suitable for a phylogenetic reconstruction.
Recombination detection. Most of the analyses developed in our analytical framework (phylogenetics and Bayesian inference) are based on the assumption that M. tuberculosis evolution is mostly clonal and that recombination can be neglected. Therefore, in a preliminary step, we tested the presence of mosaic genomes or possible HGT with the algorithm SPLITSTREE. Each data set was analyzed for the presence of recombinant sequences using the PHI test with α = 0.001.
Phylogenetic inferences. Phylogenetic relationships were reconstructed using the maximum-likelihood approach implemented in phyml 3.412 (ref. 72). The robustness of the maximum-likelihood tree topology was assessed with bootstrapping analyses of 1,000 pseudoreplicated data sets. A transversion substitution model (TVM) was selected on the basis of Akaike’s information criterion using jmodeltest73. Phylogenies were rooted with the midpoint rooting option using FigTree software v1.4 and with the reference M. tuberculosis strain H37Rv, both resulting in the same topology.
Coalescent-based analyses. Evolutionary rates and tree topologies were analyzed using the general time-reversible (GTR) and Hasegawa-Kishino-Yano74 (HKY) substitution models with gamma distributed among-site rate variation with four rate categories (Γ4). We tested both a strict molecular clock (which assumes the same evolutionary rates for all branches in the tree) and a relaxed clock that allows different rates among branches. Constant-sized, logistic, exponentially growing coalescent models were used. We also considered the Bayesian skyline plot model75, based on a general, non-parametric prior that enforces no particular demographic history. We used a piecewise linear skyline model with ten groups, and we then compared the marginal likelihood for each model using Bayes factors estimated in tracer 1.5. Bayes factors represent the ratio of the marginal likelihood of the models being compared. Approximate marginal likelihoods for each coalescent model were calculated via importance sampling (1,000 bootstraps) using the harmonic mean of the sampled likelihoods. A ratio between 3 and 10 indicates moderate support that one model better fits the data than another, whereas values greater than 10 indicate strong support. For each analysis, 2 independent runs of 100 million steps were performed, and the chain was sampled every 10,000 generations. Examination of the MCMC samples with tracer 1.5 indicated convergence and adequate mixing of the Markov chains, with ESSs for each parameter in the hundreds or thousands. The first 10% of each chain were discarded as burn-in. We found the maximum clade credibility topology using treeannotator 1.7.5 (ref. 76), and we reconstructed the Bayesian skyline plot using tracer 1.5. The relaxed clock models provided better fit to the data (Bayes factor > 12); under the different models tested, the Bayesian skyline model provided the better fit overall (marginally better).
Analysis of genes under positive selection. We selected all genes that are associated with the four M. tuberculosis mce operons77 and known T cell antigens45. Polymerases and ribosomal proteins as well as vapBC and fad genes were selected from a free text search under http://tuberculist.epfl.ch/ using the terms “polymerase,” “vap” and “fad”. All genes with an annotated function of “lipoprotein” from the H37Rv reference genome (GenBank, NC_000962.3) were selected. Furthermore, we selected 300 essential and 300 non-essential genes by assigning random numbers with the MS Excel function = RAND () to all H37Rv genes and choosing the top 300 largest numbers in the respective category.
For the dN/dS analysis, we prepared concatenated gene sequences for all MRCAs of modern Beijing subgroups and compared them pairwise with the MRCA of the entire Beijing lineage. Ancestral states were inferred from the maximum likelihood–based phylogeny. The dN/dS ratio was calculated using the software KaKs calculator (v1.2)78 with the Nei-Gojobori method. It was not possible to calculate a dN/dS ratio for all subgroups because of the absence of either nonsynonymous or synonymous SNPs. Furthermore, we counted the number of nonsynonymous and synonymous SNPs unique to all modern or ancestral subgroups and used a χ2 test with Yates correction to compare amino acid changes affecting analyzed gene families to the random selection of essential and non-essential genes, respectively. Pairwise tests were carried out with modern Beijing subgroup SNPs and with ancestral Beijing subgroup SNPs separately. Homoplastic and convergent SNPs were identified via visual examination of the distribution of an extended set of 6,696 concatenated polymorphic sites across different Beijing subgroups.
Additionally, to capture SNPs under positive selection, we applied the software pcadapt to perform a genome scan based on a Bayesian factor model36. We chose K = 4 factors because the fifth and the sixth factors did not correspond to population structure and distinguished individuals within the same clades. The factor analysis was performed on the centered genotype matrix that was not scaled. The MCMC algorithm was initialized using singular value decomposition, and the total number of steps was equal to 400 with a burn-in of 200 steps.
Supplementary Material
Acknowledgements
We gratefully acknowledge L. Cowan and J. Posey (US Centers for Disease Control and Prevention) for providing us with significant amounts of genotyping data for M. tuberculosis Beijing isolates. We thank T. Ubben, I. Radzio, T. Struwe-Sonnenschein and J. Zallet (Research Center Borstel) for excellent technical assistance. We acknowledge J. Peh for her assistance and support in the study and I. Comas for statistical advice. Parts of this work have been supported by grants from the European Community’s Seventh Framework Programme (FP7/2007-2013) under grant agreement 278864 in the framework of the European Union PathoNGenTrace project and grant agreement 223681 in the framework of the TB-PAN-NET project. We also thank Action Transversale du Muséum National d’Histoire Naturelle ‘Les Microorganismes, Acteurs Clés dans les Ecosystèmes’ for financial support. The funders had no role in study design, data collection and analysis, decision to publish or preparation of the manuscript.
Footnotes
Competing interests
P. Supply is a consultant for Genoscreen. C.A.-B. and C.W. were or are employees of the same company. The other authors declare no competing financial interests.
Accession codes.
Sequencing reads have been submitted to the EMBL-EBI European Nucleotide Archive (ENA) Sequence Read Archive (SRA) under the study accession PRJEB7281.
Change history
10 February 2015In the version of this article initially published online, affiliation 3 was incomplete, and the middle initials of author Michael Blum were inadvertently omitted. These errors have been corrected for the print, PDF and HTML versions of this article.
Accessions
PRIMARY ACCESSIONS
European Nucleotide Archive
PRJEB7281
REFERENCED ACCESSIONS
NCBI Reference Sequence
Contributor Information
Matthias Merker, Laboratoire Biologie Intégrative des Population, Evolution Moléculaire, Ecole Pratique des Hautes Etudes, Paris, France..
Camille Blin, Institut de Systématique, Evolution, Biodiversité, UMR-CNRS 7205, Muséum National d’Histoire Naturelle, Université Pierre et Marie Curie, Ecole Pratique des Hautes Etudes, Sorbonne Universités, Paris, France.; Université Joseph Fourier, Centre National de la Recherche Scientifique, Laboratoire Techniques de l’Ingénierie Médicale et de la Complexité–Informatique, Mathématiques et Applications, Grenoble, France.
Stefano Mona, Institut de Systématique, Evolution, Biodiversité, UMR-CNRS 7205, Muséum National d’Histoire Naturelle, Université Pierre et Marie Curie, Ecole Pratique des Hautes Etudes, Sorbonne Universités, Paris, France.; Université Joseph Fourier, Centre National de la Recherche Scientifique, Laboratoire Techniques de l’Ingénierie Médicale et de la Complexité–Informatique, Mathématiques et Applications, Grenoble, France.
Nicolas Duforet-Frebourg, INSERM U1019, Center for Infection and Immunity of Lille, Lille, France..
Sophie Lecher, Centre National de la Recherche Scientifique, UMR 8204, Lille, France.; Université Lille Nord, Center for Infection and Immunity of Lille, Lille, France. Institut Pasteur de Lille, Center for Infection and Immunity of Lille, Lille, France. National Reference Center for Mycobacteria, Research Center Borstel, Borstel, Germany.
Eve Willery, Centre National de la Recherche Scientifique, UMR 8204, Lille, France.; Université Lille Nord, Center for Infection and Immunity of Lille, Lille, France. Institut Pasteur de Lille, Center for Infection and Immunity of Lille, Lille, France. National Reference Center for Mycobacteria, Research Center Borstel, Borstel, Germany.
Michael G B Blum, INSERM U1019, Center for Infection and Immunity of Lille, Lille, France..
Sabine Rüsch-Gerdes, Laboratory of Molecular Microbiology, St. Petersburg Pasteur Institute, St. Petersburg, Russia..
Igor Mokrousov, Centre for Biomedical Research, Burnet Institute, Melbourne, Victoria, Australia..
Eman Aleksic, Genoscreen, Lille, France..
Caroline Allix-Béguec, Medical Department, Médecins sans Frontières Switzerland, Geneva, Switzerland..
Annick Antierens, Department of Microbiology, National Tuberculosis and Lung Diseases Research Institute, Warsaw, Poland..
Ewa Augustynowicz-Kopeć, Institute of Social and Preventive Medicine, University of Bern, Bern, Switzerland..
Marie Ballif, Instituto de Medicina Tropical Alexander von Humboldt, Molecular Epidemiology Unit–Tuberculosis, Universidad Peruana Cayetano Heredia, Lima, Peru..
Francesca Barletta, Department of Medical Parasitology and Infection Biology, Swiss Tropical and Public Health Institute, Basel, Switzerland..
Hans Peter Beck, Tuberculosis Research Section, National Institute of Allergy and Infectious Diseases, US National Institutes of Health, Bethesda, Maryland, USA..
Clifton E Barry, III, Clinical Research Department, Epicentre, Paris, France..
Maryline Bonnet, Emerging Bacterial Pathogens Unit, San Raffaele Scientific Institute, Milan, Italy..
Emanuele Borroni, Department of Microbiology, Hospital Universitario de Gran Canaria Dr. Negrín, Las Palmas de Gran Canaria, Spain..
Isolina Campos-Herrero, Division of Medical Microbiology, University of Cape Town, Cape Town, South Africa..
Daniela Cirillo, Department of Microbiology, Hospital Universitario de Gran Canaria Dr. Negrín, Las Palmas de Gran Canaria, Spain..
Helen Cox, Department of Infectious Diseases, Alfred Hospital, Melbourne, Victoria, Australia..
Suzanne Crowe, Genoscreen, Lille, France.; Central Clinical School, Monash University, Melbourne, Victoria, Australia. National Tuberculosis Reference Laboratory, Phthysiopneumology Institute, Chisinau, Republic of Moldova.
Valeriu Crudu, Institute for Epidemiology, Schleswig-Holstein University Hospital, Kiel, Germany..
Roland Diel, Public Health England National Mycobacterial Reference Laboratory and Clinical Tuberculosis and Human Immunodeficiency Virus Group, Queen Mary’s School of Medicine and Dentistry, London, UK..
Francis Drobniewski, Department of Infectious Diseases, Imperial College, London, UK.; Tuberculosis and Mycobacteria, Scientific Institute of Public Health, Brussels, Belgium.
Maryse Fauville-Dufaux, Department of Microbiology, Public Health Agency of Sweden, Solna, Sweden..
Sébastien Gagneux, Tuberculosis Research Section, National Institute of Allergy and Infectious Diseases, US National Institutes of Health, Bethesda, Maryland, USA..
Solomon Ghebremichael, Department of Science and Technology/National Research Foundation, Centre of Excellence for Biomedical Tuberculosis Research/Medical Research Council, Centre for Tuberculosis Research, Division of Molecular Biology and Human Genetics, Faculty of Medicine and Health Sciences, Stellenbosch University, Cape Town, South Africa..
Madeleine Hanekom, Department of Diagnostics and Vaccinology, Swedish Institute for Communicable Disease Control, Solna, Sweden..
Sven Hoffner, Key Laboratory of Major Diseases in Children and National Key Discipline of Pediatrics (Capital Medical University), Ministry of Education, Beijing Pediatric Research Institute, Beijing Children’s Hospital, Capital Medical University, Beijing, China..
Wei-wei Jiao, US Agency for International Development Quality Health Care Project, Bishkek, Kyrgyzstan..
Stobdan Kalon, Samara Oblast Tuberculosis Service, Samara, Russia..
Thomas A Kohl, Laboratoire Biologie Intégrative des Population, Evolution Moléculaire, Ecole Pratique des Hautes Etudes, Paris, France..
Irina Kontsevaya, Statens Serum Institute, International Reference Laboratory of Mycobacteriology, Copenhagen, Denmark..
Troels Lillebæk, Research Institute of Tuberculosis, Japan Anti-Tuberculosis Association, Tokyo, Japan..
Shinji Maeda, World Health Organization Supranational Tuberculosis Reference Laboratory, Institut Pasteur de la Guadeloupe, Abymes, France..
Vladyslav Nikolayevskyy, Department of Infectious Diseases, Imperial College, London, UK.; Tuberculosis and Mycobacteria, Scientific Institute of Public Health, Brussels, Belgium.
Michael Rasmussen, Research Institute of Tuberculosis, Japan Anti-Tuberculosis Association, Tokyo, Japan..
Nalin Rastogi, Instituto de Investigación Sanitaria Aragón, Hospital Universitario Miguel Servet, Zaragoza, Spain..
Sofia Samper, Institute of Microbiology and Immunology, Faculty of Medicine, University of Belgrade, Belgrade, Serbia..
Elisabeth Sanchez-Padilla, Emerging Bacterial Pathogens Unit, San Raffaele Scientific Institute, Milan, Italy..
Branislava Savic, Central Tuberculosis Laboratory, Department of Pathology, Singapore General Hospital, Singapore..
Isdore Chola Shamputa, Clinical Research Department, Epicentre, Paris, France..
Adong Shen, US Agency for International Development Quality Health Care Project, Bishkek, Kyrgyzstan..
Li-Hwei Sng, Department of Immunology and Cell Biology, Institute of Biotechnology, Vilnius University, Vilnius, Lithuania..
Petras Stakenas, Tartu University Hospital United Laboratories, Mycobacteriology, Tartu, Estonia..
Kadri Toit, Medical Department, Médecins sans Frontières, Paris, France..
Francis Varaine, German Center for Infection Research, Borstel Site, Borstel, Germany..
Dragana Vukovic, Central Tuberculosis Laboratory, Department of Pathology, Singapore General Hospital, Singapore..
Céline Wahl, Medical Department, Médecins sans Frontières Switzerland, Geneva, Switzerland..
Robin Warren, Department of Diagnostics and Vaccinology, Swedish Institute for Communicable Disease Control, Solna, Sweden..
Philip Supply, Centre National de la Recherche Scientifique, UMR 8204, Lille, France.; Université Lille Nord, Center for Infection and Immunity of Lille, Lille, France. Institut Pasteur de Lille, Center for Infection and Immunity of Lille, Lille, France. National Reference Center for Mycobacteria, Research Center Borstel, Borstel, Germany.
Stefan Niemann, Laboratoire Biologie Intégrative des Population, Evolution Moléculaire, Ecole Pratique des Hautes Etudes, Paris, France..
Thierry Wirth, Institut de Systématique, Evolution, Biodiversité, UMR-CNRS 7205, Muséum National d’Histoire Naturelle, Université Pierre et Marie Curie, Ecole Pratique des Hautes Etudes, Sorbonne Universités, Paris, France.; Université Joseph Fourier, Centre National de la Recherche Scientifique, Laboratoire Techniques de l’Ingénierie Médicale et de la Complexité–Informatique, Mathématiques et Applications, Grenoble, France.
References
- 1.Global Tuberculosis Report. (World Health Organization, Geneva, 2013). [Google Scholar]
- 2.Klopper M. et al. Emergence and spread of extensively and totally drug-resistant tuberculosis, South Africa. Emerg. Infect. Dis. 19, 449–455 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Casali N. et al. Evolution and transmission of drug-resistant tuberculosis in a Russian population. Nat. Genet. 46, 279–286 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Stoffels K. et al. From multidrug-to extensively drug-resistant tuberculosis: upward trends as seen from a 15-year nationwide study. PLoS ONE 8, e63128 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Niemann S. et al. Mycobacterium tuberculosis Beijing lineage favors the spread of multidrug-resistant tuberculosis in the Republic of Georgia. J. Clin. Microbiol. 48, 3544–3550 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mokrousov I. Insights into the origin, emergence, and current spread of a successful Russian clone of Mycobacterium tuberculosis. Clin. Microbiol. Rev. 26, 342–360 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Munsiff SS et al. Persistence of a highly resistant strain of tuberculosis in New York City during 1990–1999. J. Infect. Dis. 188, 356–363 (2003). [DOI] [PubMed] [Google Scholar]
- 8.Cowley D. et al. Recent and rapid emergence of W-Beijing strains of Mycobacterium tuberculosis in Cape Town, South Africa. Clin. Infect. Dis. 47, 1252–1259 (2008). [DOI] [PubMed] [Google Scholar]
- 9.Caminero JA et al. Epidemiological evidence of the spread of a Mycobacterium tuberculosis strain of the Beijing genotype on Gran Canaria Island. Am. J. Respir. Crit. Care Med. 164, 1165–1170 (2001). [DOI] [PubMed] [Google Scholar]
- 10.Ford CB et al. Mycobacterium tuberculosis mutation rate estimates from different lineages predict substantial differences in the emergence of drug-resistant tuberculosis. Nat. Genet. 45, 784–790 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Comas I. & Gagneux S. A role for systems epidemiology in tuberculosis research. Trends Microbiol. 19, 492–500 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Casali N. et al. Microevolution of extensively drug-resistant tuberculosis in Russia. Genome Res. 22, 735–745 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Glynn JR, Whiteley J, Bifani PJ, Kremer K. & van Soolingen D. Worldwide occurrence of Beijing/W strains of Mycobacterium tuberculosis: a systematic review. Emerg. Infect. Dis. 8, 843–849 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bifani PJ, Mathema B, Kurepina NE & Kreiswirth BN Global dissemination of the Mycobacterium tuberculosis W-Beijing family strains. Trends Microbiol. 10, 45–52 (2002). [DOI] [PubMed] [Google Scholar]
- 15.Parwati I, van Crevel R. & van Soolingen D. Possible underlying mechanisms for successful emergence of the Mycobacterium tuberculosis Beijing genotype strains. Lancet Infect. Dis. 10, 103–111 (2010). [DOI] [PubMed] [Google Scholar]
- 16.Hanekom M. et al. Mycobacterium tuberculosis Beijing genotype: a template for success. Tuberculosis (Edinb.) 91, 510–523 (2011). [DOI] [PubMed] [Google Scholar]
- 17.de Jong BC et al. Progression to active tuberculosis, but not transmission, varies by Mycobacterium tuberculosis lineage in The Gambia. J. Infect. Dis. 198, 1037–1043 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kato-Maeda M. et al. Beijing sublineages of Mycobacterium tuberculosis differ in pathogenicity in the guinea pig. Clin. Vaccine Immunol. 19, 1227–1237 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Weniger T, Krawczyk J, Supply P, Niemann S. & Harmsen D. MIRU-VNTRplus: a web tool for polyphasic genotyping of Mycobacterium tuberculosis complex bacteria. Nucleic Acids Res. 38, W326–W331 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Plikaytis BB et al. Multiplex PCR assay specific for the multidrug-resistant strain W of Mycobacterium tuberculosis. J. Clin. Microbiol. 32, 1542–1546 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kalinowski ST Counting alleles with rarefaction: private alleles and hierarchicalsampling design. Conserv. Genet. 5, 539–543 (2004). [Google Scholar]
- 22.Supply P, Niemann S. & Wirth T. On the mutation rates of spoligotypes and variable numbers of tandem repeat loci of Mycobacterium tuberculosis. Infect. Genet. Evol. 11, 251–252 (2011). [DOI] [PubMed] [Google Scholar]
- 23.Reyes JF & Tanaka MM Mutation rates of spoligotypes and variable numbers of tandem repeat loci in Mycobacterium tuberculosis. Infect. Genet. Evol. 10, 1046–1051 (2010). [DOI] [PubMed] [Google Scholar]
- 24.Ragheb MN et al. The mutation rate of mycobacterial repetitive unit loci in strains of M. tuberculosis from cynomolgus macaque infection. BMC Genomics 14, 145 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Comas I, Homolka S, Niemann S. & Gagneux S. Genotyping of genetically monomorphic bacteria: DNA sequencing in mycobacterium tuberculosis highlights the limitations of current methodologies. PLoS ONE 4, e7815 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Walker TM et al. Whole-genome sequencing to delineate Mycobacterium tuberculosis outbreaks: a retrospective observational study. Lancet Infect. Dis. 13, 137–146 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nieselt-Struwe K. & von Haeseler A. Quartet-mapping, a generalization of the likelihood-mapping procedure. Mol. Biol. Evol. 18, 1204–1219 (2001). [DOI] [PubMed] [Google Scholar]
- 28.Namouchi A, Didelot X, Schock U, Gicquel B. & Rocha EP After the bottleneck: genome-wide diversification of the Mycobacterium tuberculosis complex by mutation, recombination, and natural selection. Genome Res. 22, 721–734 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Roetzer A. et al. Whole genome sequencing versus traditional genotyping for investigation of a Mycobacterium tuberculosis outbreak: a longitudinal molecular epidemiological study. PLoS Med. 10, e1001387 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ford CB et al. Use of whole genome sequencing to estimate the mutation rate of Mycobacterium tuberculosis during latent infection. Nat. Genet. 43, 482–486 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Comas I. & Gagneux S. The past and future of tuberculosis research. PLoS Pathog. 5, e1000600 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Colditz GA et al. Efficacy of BCG vaccine in the prevention of tuberculosis. Meta-analysis of the published literature. J. Am. Med. Assoc. 271, 698–702 (1994). [PubMed] [Google Scholar]
- 33.Holden C. Stalking a killer in Russia’s prisons. Science 286, 1670 (1999). [DOI] [PubMed] [Google Scholar]
- 34.Bifani PJ et al. Origin and interstate spread of a New York City multidrug-resistant Mycobacterium tuberculosis clone family. J. Am. Med. Assoc. 275, 452–457 (1996). [PubMed] [Google Scholar]
- 35.Parish T. et al. Deletion of two-component regulatory systems increases the virulence of Mycobacterium tuberculosis. Infect. Immun. 71, 1134–1140 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Duforet-Frebourg N, Bazin E. & Blum MG Genome scans for detecting footprints of local adaptation using a Bayesian factor model. Mol. Biol. Evol. 31, 2483–2495 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Maloney E. et al. The two-domain LysX protein of Mycobacterium tuberculosis is required for production of lysinylated phosphatidylglycerol and resistance to cationic antimicrobial peptides. PLoS Pathog. 5, e1000534 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sirakova TD, Fitzmaurice AM & Kolattukudy P. Regulation of expression of mas and fadD28, two genes involved in production of dimycocerosyl phthiocerol, a virulence factor of Mycobacterium tuberculosis. J. Bacteriol. 184, 6796–6802 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ebrahimi-Rad M. et al. Mutations in putative mutator genes of Mycobacterium tuberculosis strains of the W-Beijing family. Emerg. Infect. Dis. 9, 838–845 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Etienne G. et al. Identification of the polyketide synthase involved in the biosynthesis of the surface-exposed lipooligosaccharides in mycobacteria. J. Bacteriol. 191, 2613–2621 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ahmad S, El-Shazly S, Mustafa AS & Al-Attiyah R. Mammalian cell-entry proteins encoded by the mce3 operon of Mycobacterium tuberculosis are expressed during natural infection in humans. Scand. J. Immunol. 60, 382–391 (2004). [DOI] [PubMed] [Google Scholar]
- 42.Li XZ, Zhang L. & Nikaido H. Efflux pump–mediated intrinsic drug resistance in Mycobacterium smegmatis. Antimicrob. Agents Chemother. 48, 2415–2423 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Meikle V. et al. Identification of novel Mycobacterium bovis antigens by dissection of crude protein fractions. Clin. Vaccine Immunol. 16, 1352–1359 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.de Vos M. et al. Putative compensatory mutations in the rpoC gene of rifampin-resistant Mycobacterium tuberculosis are associated with ongoing transmission. Antimicrob. Agents Chemother. 57, 827–832 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Comas I. et al. Whole-genome sequencing of rifampicin-resistant Mycobacterium tuberculosis strains identifies compensatory mutations in RNA polymerase genes. Nat. Genet. 44, 106–110 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Comas I. et al. Out-of-Africa migration and Neolithic coexpansion of Mycobacterium tuberculosis with modern humans. Nat. Genet. 45, 1176–1182 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fuller DQ et al. The domestication process and domestication rate in rice: spikelet bases from the Lower Yangtze. Science 323, 1607–1610 (2009). [DOI] [PubMed] [Google Scholar]
- 48.Fang J. Atlas for Sustainability in Polynesian Island Cultures and Ecosystems (Sea Education Association, 2013). [Google Scholar]
- 49.Laruelle M. & Peyrouse S. Cross-border minorities as cultural and economic mediators between China and Central Asia. China and Eurasia Forum Quarterly 7, 93–119 (2009). [Google Scholar]
- 50.Drolet GJ World War I and tuberculosis. A statistical summary and review. Am. J. Public Health Nations Health 35, 689–697 (1945). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bryant JM et al. Inferring patient to patient transmission of Mycobacterium tuberculosis from whole genome sequencing data. BMC Infect. Dis. 13, 110 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Aguilar D. et al. Mycobacterium tuberculosis strains with the Beijing genotype demonstrate variability in virulence associated with transmission. Tuberculosis (Edinb.) 90, 319–325 (2010). [DOI] [PubMed] [Google Scholar]
- 53.Ribeiro SC et al. Mycobacterium tuberculosis strains of the modern sublineage of the Beijing family are more likely to display increased virulence than strains of the ancient sublineage. J. Clin. Microbiol. 52, 2615–2624 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gioffré A. et al. Mutation in mce operons attenuates Mycobacterium tuberculosis virulence. Microbes Infect. 7, 325–334 (2005). [DOI] [PubMed] [Google Scholar]
- 55.Stavrum R. et al. Modulation of transcriptional and inflammatory responses in murine macrophages by the Mycobacterium tuberculosis mammalian cell entry (Mce) 1 complex. PLoS ONE 6, e26295 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ahidjo BA et al. VapC toxins from Mycobacterium tuberculosis are ribonucleases that differentially inhibit growth and are neutralized by cognate VapB antitoxins. PLoS ONE 6, e21738 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Osório NS et al. Evidence for diversifying selection in a set of Mycobacterium tuberculosis genes in response to antibiotic-and nonantibiotic-related pressure. Mol. Biol. Evol. 30, 1326–1336 (2013). [DOI] [PubMed] [Google Scholar]
- 58.Zhang H. et al. Genome sequencing of 161 Mycobacterium tuberculosis isolates from China identifies genes and intergenic regions associated with drug resistance. Nat. Genet. 45, 1255–1260 (2013). [DOI] [PubMed] [Google Scholar]
- 59.Farhat MR et al. Genomic analysis identifies targets of convergent positive selection in drug-resistant Mycobacterium tuberculosis. Nat. Genet. 45, 1183–1189 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Supply P. et al. Automated high-throughput genotyping for study of global epidemiology of Mycobacterium tuberculosis based on mycobacterial interspersed repetitive units. J. Clin. Microbiol. 39, 3563–3571 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Supply P. et al. Proposal for standardization of optimized mycobacterial interspersed repetitive unit–variable number tandem repeat typing of Mycobacterium tuberculosis. J. Clin. Microbiol. 44, 4498–4510 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kalinowski ST HP-rare: a computer program for performing rarefaction on measures of allelic diversity. Mol. Ecol. Notes 5, 187–189 (2005). [Google Scholar]
- 63.Beaumont MA Detecting population expansion and decline using microsatellites. Genetics 153, 2013–2029 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wilson IJ, Weale ME & Balding DJ Inferences from DNA data: population histories, evolutionary processes and forensic match probabilities. J. R. Stat. Soc. Ser. A Stat. Soc. 166, 155–188 (2003). [Google Scholar]
- 65.Wilson IJ & Balding DJ Genealogical inference from microsatellite data. Genetics 150, 499–510 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ohta T. & Kimura M. A model of mutation appropriate to estimate the number of electrophoretically detectable alleles in a finite population. Genet. Res. 22, 201–204 (1973). [DOI] [PubMed] [Google Scholar]
- 67.Metropolis N, Rosenbluth AW, Rosenbluth MN, Teller AH & Teller E. Equations of state calculations by fast computing machine. J. Chem. Phys. 21, 1087–1091 (1953). [Google Scholar]
- 68.Hastings WK Monte Carlo sampling methods using Markov chains and their applications. Biometrika 57, 97–109 (1970). [Google Scholar]
- 69.Wirth T. et al. Origin, spread and demography of the Mycobacterium tuberculosis complex. PLoS Pathog. 4, e1000160 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Blom J. et al. Exact and complete short-read alignment to microbial genomes using Graphics Processing Unit programming. Bioinformatics 27, 1351–1358 (2011). [DOI] [PubMed] [Google Scholar]
- 71.Schmidt HA, Strimmer K, Vingron M. & von Haeseler A. TREE-PUZZLE: maximum likelihood phylogenetic analysis using quartets and parallel computing. Bioinformatics 18, 502–504 (2002). [DOI] [PubMed] [Google Scholar]
- 72.Guindon S. & Gascuel O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst. Biol. 52, 696–704 (2003). [DOI] [PubMed] [Google Scholar]
- 73.Posada D. & Crandall KA MODELTEST: testing the model of DNA substitution. Bioinformatics 14, 817–818 (1998). [DOI] [PubMed] [Google Scholar]
- 74.Hasegawa M, Kishino H. & Yano T. Dating of the human-ape splitting by a molecular clock of mitochondrial DNA. J. Mol. Evol. 22, 160–174 (1985). [DOI] [PubMed] [Google Scholar]
- 75.Drummond AJ, Rambaut A, Shapiro B. & Pybus OG Bayesian coalescent inference of past population dynamics from molecular sequences. Mol. Biol. Evol. 22, 1185–1192 (2005). [DOI] [PubMed] [Google Scholar]
- 76.Drummond AJ & Rambaut A. BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol. Biol. 7, 214 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Casali N. & Riley LW A phylogenomic analysis of the Actinomycetales mce operons. BMC Genomics 8, 60 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Zhang Z. et al. KaKs_Calculator: calculating Ka and Ks through model selection and model averaging. Genomics Proteomics Bioinformatics 4, 259–263 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.