Abstract
Fragments of intercellular adhesion molecule 1 (ICAM- 1) containing only the two most N terminal of its five immunoglobulin SF domains bind to rhinovirus 3 with the same affinity and kinetics as a fragment with the entire extracellular domain. The fully active two-domain fragments contain 5 or 14 more residues than a previously described fragment that is only partially active. Comparison of X-ray crystal structures show differences at the bottom of domain 2. Four different glycoforms of ICAM- 1 bind with identical kinetics.
Intercellular adhesion molecule 1 (ICAM- 1, CD54) is a cytokine-inducible cell surface receptor that binds to leukocyte integrins LFA-1 and Mac-1 (21). ICAM- 1 is also the receptor for the major group of human rhinoviruses (9, 25, 26) and Plasmodium falciparum-infected erythrocytes (2, 18). ICAM- 1 contains five immunoglobulin superfamily (IgSF) domains and is heavily glycosylated. The binding site for rhinovirus is at the tip of domain 1 in the flexible BC and FG loops and extends about halfway down domain 1 (1, 7, 17, 20, 23). The binding site for LFA-1 is on the edge of domain 1, centered on the C and D strands (1, 7, 8, 24). Soluble ICAM- 1 (sICAM) inhibits infection by rhinovirus, both by acting as a competitive inhibitor and by irreversible disruption of the capsid with release of the viral RNA (5, 10, 14, 15). However, ICAM- 1 truncated after what was predicted (24) to be the last residue of domain 2, F185, showed markedly reduced inhibition by both mechanisms compared to ICAM- 1 containing all five IgSF domains (10, 15). ICAM- 1 is unusually heavily glycosylated, and glycosylation is important for solubility (16). There are conflicting reports on the importance of the presence or absence of glycosylation for binding to rhinovirus (13, 16, 17); the influence of different classes of N-glycans, i.e., glycoforms, has not been explored. The kinetics, equilibria, and thermodynamics of IC1-5D binding to rhinovirus have been measured by using surface plasmon resonance (4, 6). The on-rate constant (kass) for binding of IC1-5D to rhinovirus is much slower than for binding to an antibody. This is consistent with either binding to a relatively inaccessible site in the rhinovirus canyon (19) or a requirement for a conformational change in the virus to permit binding. The kass is biphasic, and two putative classes of binding sites have Kds of about 0.7 and 10 μM (6). Here, we determined the kinetics with truncation and glycosylation variants of ICAM- 1.
ICAM- 1 truncated after residue 185 (IC1-2D/185), 190 (IC1- 2D/190), 199 (IC1-2D/199), 268 (IC1-3D), or 452 (IC1-5D) was expressed in insect SF9 cells (IC1-5D/SF9, IC1-3D, and IC1-2D/185), wild-type CHO-K1 cells (IC1-5D/wt), or lectin-resistant CHO Lec 3.2.8.1 cells (IC1-5D/Lec, IC1-2D/199, and IC1-2D/190) (4, 6, 7, 14). Proteins were purified with monoclonal antibody (MAb) R6.5-Sepharose and size exclusion chromatography (6). ICAM- 1 containing only one carbohydrate residue per N-linked site (IC1-5D/D) was from the flowthrough from a concanavalin A-Sepharose column that was loaded with IC1-5D/CHOLec treated with 4 mU of endoglycosidase H (endo H)/μg for 4 h at 37°C in 0.1 M phosphate buffer (pH 5.5). Purified human rhinovirus 3 (HRV3) (4) or rabbit anti-mouse Fc IgG (Pharmacia) was linked via amino groups to the dextran surface of BIAcore sensor chips to a density of about 10,000 or 8,000 resonance units, respectively (4). Sensorgrams were subjected to linear transformation to obtain kinetic constants (11). Binding of [3H]leucine-labeled HRV3 to HeLa cells and assay of HRV3 disruption by sucrose gradient centrifugation were done as previously described (15).
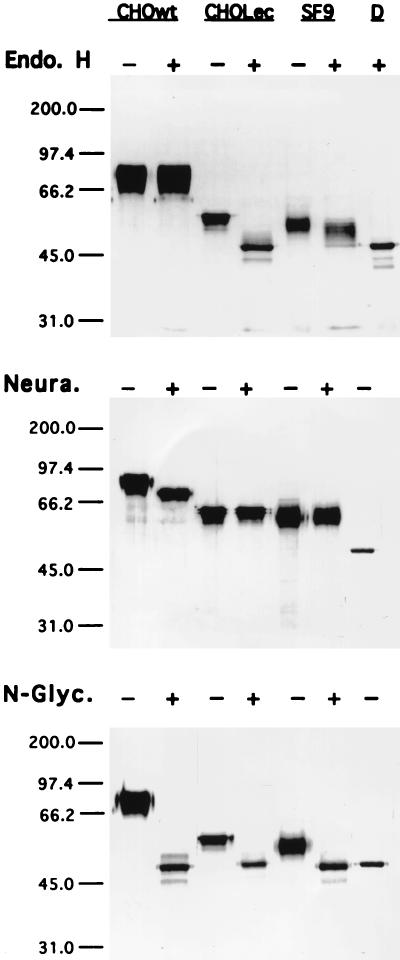
IC1-5D/wt was resistant to endo H and sensitive to neuraminidase, whereas IC1-5D/CHOLec and IC1-5D/SF9 were fully susceptible or only partially susceptible to endo H, respectively (Fig. 1). Thus, IC1-5D/wt and IC1-5D/Lec have complex-type and high-mannose N-linked glycans, respectively. Partial resistance of IC1-5D/SF9 to endo H may reflect fucosylation (3), which is absent in CHO Lec 3.2.8.1 cell N glycans (22). IC1-5D/D has only one N-acetylglycosamine per N-linked site.
FIG. 1.
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis of sICAM- 1 glycosylation variants treated with endoglycosidases. IC1-5D obtained from wild-type CHO-K1 (CHOwt), lectin-resistant CHO (CHOLec), or SF9 insect cells were treated in the presence or absence of endo H, neuraminidase (Neura.), or N-glycanase (N-Glyc.), whereas the preparation of endo H-deglycosylated material, IC1-5D/D (D), was run without further treatment. Positions of molecular weight markers are shown on the left (molecular weights are in thousands).
The four different IC1-5D glycoforms exhibited no significant differences in the kinetics of binding to HRV3 (Table 1). Association was biphasic, possibly representing two different classes of binding sites (6). A single dissociation rate constant (kdiss) was seen (6).
TABLE 1.
Kinetic and dissociation constants for binding of sICAM- 1 glycosylation variants to HRV3a
| Protein | kass1 (M−1 s−1) | kass2 (M−1 s−1) | kdiss (103) (s−1) | Kd1 (μM) | Kd2 (μM) |
|---|---|---|---|---|---|
| IC1-5D/wt | 1,910 ± 322 | 115.0 ± 1.3 | 1.84 ± 0.08 | 0.96 ± 0.17 | 16.0 ± 0.7 |
| IC1-5D/SF9 | 2,670 ± 201 | 130.0 ± 6.2 | 1.67 ± 0.16 | 0.62 ± 0.08 | 12.9 ± 1.4 |
| IC1-5D/Lec | 2,410 ± 268 | 166.0 ± 22.3 | 1.70 ± 0.19 | 0.70 ± 0.11 | 10.2 ± 1.8 |
| IC1-5D/D | 2,500 ± 203 | 221.0 ± 65.7 | 1.87 ± 0.30 | 0.75 ± 0.13 | 8.4 ± 2.8 |
Dissociation constants (Kd) were obtained from the kinetic constants as follows: Kd1 = kdiss/kass1 and Kd2 = kdiss/kass2. sICAM- 1 concentrations ranged between 1 and 10 μM. All experiments were performed at 20°C with a flow rate of 2 to 3 μl/min. The correlation coefficients obtained in the determination of the kinetic constants by the linear transformation method were about 0.99. The averages and ranges of two or standard deviations of three experiments are shown. The standard deviations of Kd1 and Kd2 were calculated as the square root of the sums of the variances of kass and kdiss.
Compared to IC1-5D and IC1-3D, three different batches of IC1-2D/185 associated with HRV3 with the same kinetics but reproducibly dissociated five- to sixfold faster (Table 2). This agrees with lower activity in inhibition of binding and in inactivation of rhinovirus (10, 15). By contrast, IC1-2D/199 reproducibly reacted with HRV3 with kinetics indistinguishable from those of IC1-5D and IC1-3D (Table 2). At a later stage in this work, we also tested the IC1-2D/190 fragment. Although less data are available, it clearly was at least as active as IC1-5D, IC1-3D, and IC1-2D/199 (Table 2).
TABLE 2.
Kinetic and dissociation constants for binding of sICAM- 1 length variants to HRV3a
| Protein | kass1 (M−1 s−1) | kass2 (M−1 s−1) | kdiss (103) (s−1) | Kd1 (μM) | Kd2 (μM) |
|---|---|---|---|---|---|
| IC1-5D | 2,240 ± 439 | 162 ± 25 | 1.65 ± 0.06 | 0.74 ± 0.15 | 10.2 ± 1.6 |
| IC1-3D | 2,060 ± 332 | 157 ± 49 | 1.80 ± 0.16 | 0.87 ± 0.16 | 11.5 ± 3.7 |
| IC1-2D/199 | 1,880 ± 361 | 182 ± 40 | 1.91 ± 0.12 | 1.02 ± 0.21 | 10.5 ± 2.4 |
| IC1-2D/190 | 2,500 | 319 | 1.11 | 0.44 | 3.5 |
| IC1-2D/185 | 2,060 ± 430 | 130 ± 21 | 9.27 ± 1.26 | 4.50 ± 1.12 | 71.6 ± 15.1 |
The concentrations of ICAM- 1 injected in BIAcore ranged from 1 to 10 μM for IC1-5D, 3D, 2D/199, and 2D/190 and from 4 to 50 μM for 2D/185. Three different preparations of IC1-2D/V185 were used; all gave similar results, which are included in the averages. IC1-3D and IC1-2D/185 were from SF9 cells, and IC1-2D/199 and 2D/190 were from CHO Lec cells; thus, all had similar glycosylation. Values for IC1-5D are from tests run in parallel with those on other variants and include data from both CHO Lec and SF9 cell preparations. The averages and standard deviations shown are from at least three experiments. For other details, see Table 1, footnote a.
The association kinetic constant (kass) for binding of ICAM- 1 fragments to MAbs was 10- to 100-fold faster than that for binding to HRV3 (Table 3). The kass increased as the number of Ig-like domains in sICAM- 1 decreased; this is likely to be related to faster diffusion of the shorter molecules, particularly in the dextran matrix to which the MAbs and virus are linked in BIAcore. The lack of such an effect on kass for HRV3 is consistent with the finding that this kass is much slower and, thus, not diffusion limited. Three MAbs to domain 1 of ICAM- 1 (12), LB2 (Table 3), 7F7, and MAY.029 (not shown), and one MAb to domain 2, R6.5 (Table 3), dissociated from ICAM- 1 fragments with kinetics that were unaffected by the truncation position. By contrast, with MAb RR1/1 to domain 1 of ICAM- 1, the IC1-2D/185 fragment dissociated 11.0-fold ± 0.8-fold more rapidly from IC1-2D/185 than from IC1-2D/190, IC1-2D/199, IC1-3D, or IC1-5D. These results confirm the differences between IC1-2D/185 on the one hand and IC1-2D/190 and IC1-2D/199 on the other.
TABLE 3.
HRV-inhibitory MAb binding to length variants of ICAM- 1a
| MAb and sICAM- 1 | kass (M−1 s−1) | kdiss (103) (s−1) | Kd (nM) |
|---|---|---|---|
| R6.5 (D2) | |||
| 5D/SF9 | 23,700 | 1.83 ± 0.24 | 77.1 |
| 5D/Lec | 34,900 | 1.82 ± 0.14 | 52.0 |
| 2D/185 | 84,800 | 1.74 ± 0.15 | 20.5 |
| 2D/199 | 123,000 | 2.02 ± 0.20 | 16.0 |
| 2D/190 | 72,000 | 1.42 ± 0.01 | 19.7 |
| LB2 (D1) | |||
| 5D/SF9 | 17,100 ± 1,230 | 1.79 ± 0.09 | 105.0 |
| 5D/Lec | 19,900 ± 710 | 2.06 ± 0.08 | 103.0 |
| 3D | 30,100 ± 2,620 | 1.82 ± 0.18 | 61.0 |
| 2D/185 | 40,400 ± 5,090 | 1.95 ± 0.26 | 48.0 |
| 2D/199 | 47,500 ± 1,300 | 2.33 ± 0.12 | 49.0 |
| RR1/1 (D1) | |||
| 5D | 47,800 | 1.80 ± 0.07 | 38.0 |
| 5D/Lec | 60,100 | 1.90 ± 0.01 | 32.0 |
| 3D | 85,200 | 1.74 ± 0.18 | 20.0 |
| 2D/185 | 197,000 | 20.40 ± 2.97 | 103.0 |
| 2D/199 | 196,000 | 1.78 ± 0.08 | 9.0 |
| 2D/190 | 214,000 | 2.10 ± 0.02 | 9.8 |
The kass and kdiss determined with BIAcore are for sICAM- 1 fragments with captured anti-ICAM- 1 MAbs. The kdiss was determined from three independent injections of 300, 500, and 750 nM sICAM- 1 at 4, 8, and 30 μl/min, respectively. The average and standard deviation for these three injections are shown. No increase in kdiss with flow rate was obtained in these experiments. The average and range (kass) or standard deviation (kdiss) of two experiments are shown for the LB2 MAb.
The crystal structures have recently been determined for two independent molecules of IC1-2D/190 (7) and one molecule of mutIC1-2D/185 that is identical to IC1-2D/185 except that it contains three Asn→Gln mutations that eliminate N-linked glycosylation sites (1). The three structures are quite similar overall, but the position of truncation has a marked effect on the structure of the bottom of domain 2. The backbone hydrogen bonds between F185 in the G strand and residues 98 and 100 in the A′ strand are lost in the 185-residue fragment, and there are significant shifts in the Cα positions of residues 183 and 184 in the G strand, 95 to 97 in the A′ strand and in the connector between the A and A′ strands on the edge of domain 2, 150 to 153 in the EF loop, and 101 and 102 in the A′B loop. Most significantly, Val186 appears to form an important component of the bottom of domain 2, with its hydrophobic side chain packing onto the side chain of Val100 and the hydrophobic portion of the Arg150 side chain. In the structure of the 185-residue fragment, Val186 is missing, the side chain of Phe185 is rotated 180°C, and the bottom of domain 2 is less compact overall. The A′G beta-sheet ladders at the bottom of domains 1 and 2 are structurally homologous in IC1-2D/190, and the Tyr83 and Trp84 side chains occupy orientations similar to those of Phe185 and Val186. Thus, the structure of Phe185 and Val186 in IC1-2D/190 is appropriate for a domain 2-3 connection structurally homologous to the domain 1-2 connection.
Our data show that structural changes at the bottom and side of domain 2 can alter the affinity of rhinovirus interaction with the top portion of domain 1 and with MAb RR1/1 to domain 1. Although this may seem surprising, the conformation of domain 1 has previously been shown to be influenced by mutagenesis of domain 2, including a conservative Ala178→ Gly mutation in the bulge of strand G near the bottom of domain 2 (23). In the three structures captured in crystals, there appear to be no differences in domain 1 that are significant for rhinovirus binding; however, the structure in solution may differ from that in crystals. We suggest that in solution, IC1-2D/185 exists in an equilibrium between two conformations, i.e., (i) an ordered conformation that can bind rhinovirus and corresponds to the conformation that crystallizes and (ii) a conformation in which a portion of domains 1 and 2 is disordered and in which the molecule cannot bind or binds markedly less well to rhinovirus and MAb RR1/1. The IC1-2D/190 fragment has a different conformation at the bottom of domain 2 and appears to be present in an ordered conformation in solution a much higher proportion of the time than the 185-residue fragment. Equilibration between the ordered and disordered forms of the IC1-2D/185 fragment that is fast compared to the rates of binding to and dissociation from HRV3 and antibodies would be consistent with our kinetic data. Our findings emphasize the intricacy and delicacy of protein structure and have important implications for viral receptors and the design of viral antagonists. Furthermore, our results show that domains 1 and 2 of ICAM- 1 are sufficient for high-affinity binding to HRV3.
Acknowledgments
This work was supported by NIH grant AI31921.
REFERENCES
- 1.Bella J, Kolatkar P R, Marlor C, Greve J M, Rossmann M G. The structure of the two amino-terminal domains of human ICAM- 1 suggests how it functions as a rhinovirus receptor and as an LFA-1 integrin ligand. Proc Natl Acad Sci USA. 1998;95:4140–4145. doi: 10.1073/pnas.95.8.4140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Berendt A R, McDowall A, Craig A G, Bates P A, Sternberg M J E, Marsh K, Newbold C I, Hogg K. The binding site on ICAM- 1 for Plasmodium falciparum-infected erythrocytes overlaps, but is distinct from, the LFA-1-binding site. Cell. 1992;68:71–81. doi: 10.1016/0092-8674(92)90207-s. [DOI] [PubMed] [Google Scholar]
- 3.Butters T D, Jones I, Clarke V A, Jacob G S. Structural characterization of the N-linked oligosaccharides derived from HIVgp120 expressed in lepidopteran cells. Glycoconj J. 1998;15:83–88. doi: 10.1023/a:1006999718552. [DOI] [PubMed] [Google Scholar]
- 4.Casanovas J M, Reed R R, Springer T A. Kinetics of receptor and virus interaction and receptor-induced virus disruption: methods for study with surface plasmon resonance. Methods (Orlando) 1994;6:157–167. [Google Scholar]
- 5.Casasnovas J M, Springer T A. The pathway of rhinovirus disruption by soluble intercellular adhesion molecule 1 (ICAM- 1): an intermediate in which ICAM- 1 is bound and RNA is released. J Virol. 1994;68:5882–5889. doi: 10.1128/jvi.68.9.5882-5889.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Casasnovas J M, Springer T A. Kinetics and thermodynamics of virus binding to receptor: studies with rhinovirus, intercellular adhesion molecule-1 (ICAM- 1), and surface plasmon resonance. J Biol Chem. 1995;270:13216–13224. doi: 10.1074/jbc.270.22.13216. [DOI] [PubMed] [Google Scholar]
- 7.Casasnovas J M, Stehle T, Liu J-H, Wang J-H, Springer T A. A dimeric crystal structure for the N-terminal two domains of ICAM- 1. Proc Natl Acad Sci USA. 1998;95:4134–4139. doi: 10.1073/pnas.95.8.4134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fisher K L, Lu J, Riddle L, Kim K J, Presta L G, Bodary S C. Identification of the binding site in intercellular adhesion molecule 1 for its receptor, leukocyte function-associated antigen 1. Mol Biol Cell. 1997;8:501–515. doi: 10.1091/mbc.8.3.501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Greve J M, Davis G, Meyer A M, Forte C P, Yost S C, Marlor C W, Kamarck M E, McClelland A. The major human rhinovirus receptor is ICAM- 1. Cell. 1989;56:839–847. doi: 10.1016/0092-8674(89)90688-0. [DOI] [PubMed] [Google Scholar]
- 10.Greve J M, Forte C P, Marlor C W, Meyer A M, Hoover-Litty H, Wunderlich D, McClelland A. Mechanisms of receptor-mediated rhinovirus neutralization defined by two soluble forms of ICAM- 1. J Virol. 1991;65:6015–6023. doi: 10.1128/jvi.65.11.6015-6023.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Karlsson R, Michaelsson A, Mattson L. Kinetic analysis of monoclonal antibody-antigen interactions with a new biosensor based analytical system. J Immunol Methods. 1991;145:229–240. doi: 10.1016/0022-1759(91)90331-9. [DOI] [PubMed] [Google Scholar]
- 12.Klickstein L B, Springer T A. CD54 (ICAM- 1) cluster report. In: Schlossman S F, Boumsell L, Gilks W, Harlan J, Kishimoto T, Morimoto T, Ritz J, Shaw S, Silverstein R, Springer T, Tedder T, Todd R, editors. Leucocyte typing. V. White cell differentiation antigens. New York, N.Y: Oxford University Press; 1995. pp. 1548–1550. [Google Scholar]
- 13.Lineberger D W, Graham D J, Tomassini J E, Colonno R J. Antibodies that block rhinovirus attachment map to domain 1 of the major group receptor. J Virol. 1990;64:2582–2587. doi: 10.1128/jvi.64.6.2582-2587.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Marlin S D, Staunton D E, Springer T A, Stratowa C, Sommergruber W, Merluzzi V. A soluble form of intercellular adhesion molecule-1 inhibits rhinovirus infection. Nature. 1990;344:70–72. doi: 10.1038/344070a0. [DOI] [PubMed] [Google Scholar]
- 15.Martin S, Casasnovas J M, Staunton D E, Springer T A. Efficient neutralization and disruption of rhinovirus by chimeric ICAM- 1/immunoglobulin molecules. J Virol. 1993;67:3561–3568. doi: 10.1128/jvi.67.6.3561-3568.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Martin S, Martin A, Staunton D E, Springer T A. Functional studies of truncated soluble ICAM- 1 expressed in Escherichia coli. Antimicrob Agents Chemother. 1993;37:1278–1284. doi: 10.1128/aac.37.6.1278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McClelland A, DeBear J, Yost S C, Meyer A M, Marlor C W, Greve J M. Identification of monoclonal antibody epitopes and critical residues for rhinovirus binding in domain 1 of intercellular adhesion molecule 1. Proc Natl Acad Sci USA. 1991;88:7993–7997. doi: 10.1073/pnas.88.18.7993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ockenhouse C F, Betageri R, Springer T A, Staunton D E. Plasmodium falciparum-infected erythrocytes bind ICAM- 1 at a site distinct from LFA-1, Mac-1, and human rhinovirus. Cell. 1992;68:63–69. doi: 10.1016/0092-8674(92)90206-r. [DOI] [PubMed] [Google Scholar]
- 19.Olson N H, Kolatkar P R, Oliveira M A, Cheng R H, Greve J M, McClelland A, Baker T S, Rossmann M G. Structure of human rhinovirus complexed with its receptor molecule. Proc Natl Acad Sci USA. 1993;90:507–511. doi: 10.1073/pnas.90.2.507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Register R B, Uncapher C R, Naylor A M, Lineberger D W, Colonno R J. Human-murine chimeras of ICAM- 1 identify amino acid residues critical for rhinovirus and antibody binding. J Virol. 1991;65:6589–6596. doi: 10.1128/jvi.65.12.6589-6596.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Springer T A. Adhesion receptors of the immune system. Nature. 1990;346:425–433. doi: 10.1038/346425a0. [DOI] [PubMed] [Google Scholar]
- 22.Stanley P. Chinese hamster ovary cell mutants with multiple glycosylation defects for production of glycoproteins with minimal carbohydrate heterogeneity. Mol Cell Biol. 1989;9:377–383. doi: 10.1128/mcb.9.2.377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Staunton D E, Dustin M L, Erickson H P, Springer T A. The arrangement of the immunoglobulin-like domains of ICAM- 1 and the binding sites for LFA-1 and rhinovirus. Cell. 1990;61:243–254. doi: 10.1016/0092-8674(90)90805-o. [DOI] [PubMed] [Google Scholar]
- 24.Staunton D E, Marlin S D, Stratowa C, Dustin M L, Springer T A. Primary structure of intercellular adhesion molecule 1 (ICAM- 1) demonstrates interaction between members of the immunoglobulin and integrin supergene families. Cell. 1988;52:925–933. doi: 10.1016/0092-8674(88)90434-5. [DOI] [PubMed] [Google Scholar]
- 25.Staunton D E, Merluzzi V J, Rothlein R, Barton R, Marlin S D, Springer T A. A cell adhesion molecule, ICAM- 1, is the major surface receptor for rhinoviruses. Cell. 1989;56:849–853. doi: 10.1016/0092-8674(89)90689-2. [DOI] [PubMed] [Google Scholar]
- 26.Tomassini J E, Graham D, DeWitt C M, Lineberger D W, Rodkey J A, Colonno R J. cDNA cloning reveals that the major group rhinovirus receptor on HeLa cells is intercellular adhesion molecule 1. Proc Natl Acad Sci USA. 1989;86:4907–4911. doi: 10.1073/pnas.86.13.4907. [DOI] [PMC free article] [PubMed] [Google Scholar]