Far from a random tangle, cellular DNA is packed into the nucleus with astounding precision. Indeed, there is growing appreciation for how the three-dimensional (3D) organization of the genome contributes to controlling gene expression. For instance, loops of DNA called insulated neighborhoods can protect small groups of genes from silencing or activation (1). If cancer can result from dysregulation of gene expression (2), then an enticing hypothesis is that disrupting insulated neighborhoods may lead to increased transcription of cancer genes. On page 1454 of this issue, Hnisz et al. (3) use tumor-derived sequencing data and targeted deletions in cells to show that disruption of insulated neighborhoods leads to activation of proto-oncogenes—genes with the potential to cause cancer. These findings strongly support disruption of chromatin structure as causally linked to tumorigenesis, and suggest that such disruptions may be the hidden culprit driving many tumors.
Physical separation is maintained between transcriptionally active and inactive regions of the genome (4), and recent studies have identified units of hundreds to thousands of kilobases of DNA, called topologically associating domains (TADs), where genes exhibit close spatial proximity and correlated expression (5). The boundaries of these TADs are occupied by CCCTC-binding factor (CTCF), the primary protein that mediates long-range chromatin interactions in mammals (6). Further transcriptional control can occur inside CTCF-mediated DNA loops such as insulated neighborhoods. These loops can stop the spread of silencing heterochromatin, physically separate genes from activating enhancers, or sequester genes together with powerful transcriptional activators known as superenhancers (1).
Hnisz et al. asked whether changes to this three-dimensional structure of DNA contribute to the activation of proto-on-cogenes. Proto-oncogenes are often not expressed, and in a human cell-line model of T cell acute lymphoblastic leukemia (TALL), Hnisz et al. found that many of these dormant proto-oncogenes lie in insulated neighborhoods. To determine whether opening of these DNA loops is sufficient to allow transcription of the proto-oncogenes, the authors used the DNA editing technique called CRISPR (clustered regularly-interspaced short palindromic repeats )-Cas9 to delete the CTCF sites that mark the boundaries of insulated neighborhoods containing the genes TAL1 and LMO2. TAL1 and LMO2 are transcription factors that drive hematopoiesis and can be aberrantly activated in T cell acute lymphoblastic leukemia. Remarkably, despite targeting regions thousands of base pairs away from the gene body, both deletions flanking an insulated neighborhood were sufficient to induce proto-oncogene expression in nonmalignant cells. The authors further made a key connection with patient data: Recurrent focal deletions targeting the same region were associated with increased TAL1 expression in T cell acute lymphoblastic leukemia.
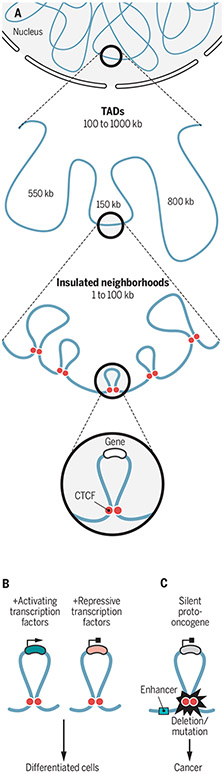
Chromatin architecture and gene expression.
(A) DNA has a hierarchical structure, with units of hundreds to thousands of kilobases organized into topologically associating domains (TADs). Smaller subdomains called insulated neighborhoods are organized by CTCF and cohesin binding at insulators. (B) Insulated neighborhoods provide protected units where genes can be co-regulated. (C) Otherwise well-behaved proto-oncogenes can become activated in cancer by disrupting the structure of insulated neighborhoods through deletions or mutations of boundary CTCF binding sites.
One model behind the increased expression is that releasing DNA loops allows physical contact between proto-oncogenes and nearby enhancers. Such commandeering of enhancers to drive expression is reminiscent of enhancer hijacking, where superenhancers are brought into proximity of proto-oncogenes through genome rearrangement events (7). Interestingly, the TAL1 locus can also undergo somatic mutations in T cell acute lymphoblastic leukemia to create a new superenhancer (8), further demonstrating the arsenal of tools with which tumors can dysregulate transcription.
CRISPR-Cas9-mediated deletions of insulated neighborhood boundaries can activate proto-oncogenes in the laboratory, but how prevalent are such disruptions in real tumors? CTCF binding sites are known to be frequently somatically mutated (9); to identify whether this is due to positive selection for chromatin disruption, Hnisz et al. separately analyzed mutations in nonboundary CTCF sites and boundary CTCF sites in different cancer types. If the target of CTCF site alterations is destruction of insulated neighborhoods, then only boundary CTCF sites should be enriched for mutations. Across a large pan-cancer cohort, the authors observed a factor of >2 enrichment for boundary CTCF site mutations. This enrichment was particularly strong in liver and esophageal carcinomas, where boundary CTCF site mutations were also significantly more likely to be found near known oncogenes. Whether this enrichment is driven primarily by activation of proto-oncogenes is not clear, and further analyses are needed to uncover the specific gene targets driving CTCF site alterations within different tumor types. Such studies may also be useful for clinical genotyping, where identification of activated oncogenes is a key step in applying the optimal targeted therapy.
Genetic events that disrupt insulated neighborhoods may be just one of many ways that cells alter their 3D chromatin structure to dysregulate gene expression. Recently, Flavahan et al. reported that disruption of TADs by DNA methylation of boundary CTCF sites allows a distant active enhancer to interact with and drive a key oncogene in brain tumors (10). Together with the findings of Hnisz et al., these pioneering studies highlight the diversity of mechanisms by which chromatin structure may be targeted and suggest that modulating 3D chromatin structure may be widespread in cancer.
By showing that disruption of insulated neighborhoods leads to activation of protooncogenes, Hnisz et al. describe a previously unrecognized mechanism by which cancers may escape transcriptional regulation. This study adds to an expanding understanding of the deep impact that alterations outside of protein-coding regions can have in driving the expression of cancer genes (11-13). Future research aimed at deciphering such noncoding alterations in cancer will need to account for perturbations to the 3D architecture of the genome, while also being alert to indications of novel methods of transcriptional dysregulation.
REFERENCES
- 1.Dowen JM et al. , Cell 159, 374 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hanahan D, Weinberg RA, Cell 144, 5 (2011). [DOI] [PubMed] [Google Scholar]
- 3.Hnisz D et al. , Science 351, 1454 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lieberman-Aiden E et al. , Science 326, 289 (2009).19815776 [Google Scholar]
- 5.Dixon JR et al. , Nature 485, 376 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Phillips-Cremins JE, Corces VG, Mol. Cell 50, 461 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Northcott PA et al. , Nature 511, 428 (2015). [Google Scholar]
- 8.Mansour MR et al. , Science 346, 1373 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Katainen R et al. , Nat. Genet 47, 818 (2015). [DOI] [PubMed] [Google Scholar]
- 10.Flavahan WA et al. , Nature 529, 110 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Davis CF et al. , Cancer Cell 26, 319 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Horn S et al. , Science 339, 959 (2013). [DOI] [PubMed] [Google Scholar]
- 13.Huang FW et al. , Science 339, 957 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]