Abstract
The Epstein-Barr virus (EBV) Zta and Mta regulatory proteins were previously found to be required for efficient replication of oriLyt in cotransfection-replication assays, but the contribution of Mta to the replication process was unknown. We now demonstrate that Mta regulates replication gene expression. Using the polymerase processivity factor BMRF1 as an example, we found that in transfected cells, total BMRF1 mRNA levels were unaffected by Mta but that the amounts of cytoplasmic BMRF1 RNA and protein were greatly increased in the presence of Mta. Mta also increased cytoplasmic accumulation of the BALF2, BALF5, BSLF1, and BBLF4 replication gene mRNAs but did not affect cytoplasmic levels of BBLF2/3 mRNA. Thus, five of the six core replication genes require Mta for efficient accumulation of cytoplasmic RNA. The contribution of Mta to posttranscriptional RNA processing was examined. Examination of Mta localization in transfected cells by indirect immunofluorescence revealed that Mta colocalized with the splicing factor SC35. We also found that Mta has RNA binding activity. Glutathione S-transferase–Mta bound to BMRF1 and BMLF1 transcripts but not to a control cellular gene RNA. Mta contains a consensus leucine-rich nuclear export signal. Such signal sequences are characteristic of proteins that undergo nuclear export. Examination of Mta localization in a heterokaryon assay provided evidence that Mta shuttles between the nucleus and the cytoplasm. Our experiments indicate that Mta functions in RNA processing and transport and mediates cytoplasmic accumulation of a number of EBV early mRNAs.
Epstein-Barr virus (EBV) encodes three transactivators, Zta (BZLF1, ZEBRA), Rta (BRLF1), and Mta (BMLF1), that together regulate EBV lytic cycle gene expression. Zta is a bZIP family transcriptional activator that is represented only in the gamma 1 class of herpesviruses (4). The Rta transcriptional activator has homologs encoded within both gamma 1 and gamma 2 herpesvirus genomes (52, 66), while homologs of the Mta protein are encoded by alpha-, beta-, and gammaherpesviruses (13, 32, 50, 53, 57, 62, 79, 82, 90, 91, 93, 94). The most extensively studied Mta homolog is herpes simplex virus (HSV) IE63 (ICP27). HSV mutants that lack a functional IE63 gene overexpress immediate early and early viral genes and are deficient in late gene expression (42, 45, 63–65, 80, 81). Further analysis has shown that IE63 represses expression of genes containing introns by inhibiting cellular pre-mRNA splicing. IE63 associates with and reorganizes proteins associated with small nuclear ribonucleoprotein particles (snRNPs). This activity contributes to, but is not sufficient for, splicing inhibition (27, 59, 61, 69, 70). HSV IE63 also has a posttranscriptional stimulatory activity (71). IE63 expression leads to enhanced binding of cleavage stimulation factor (CstF) to the polyadenylation signal of HSV genes (43, 44), and IE63 has recently been shown to shuttle between the nucleus and cytoplasm, indicating a role in facilitating RNA transport (60, 68, 83).
EBV Mta is a phosphoprotein (12, 92) that migrates in denaturing polyacrylamide gels as a series of polypeptides with a major species of 60 kDa (11, 92). Mta has been less well characterized than HSV IE63 but is also recognized to have a posttranscriptional mechanism of action. In transient expression assays, Mta was initially recognized to stimulate reporter gene expression from a variety of heterologous promoters (40, 54). Activity in these assays was subsequently shown to be reporter gene dependent, indicating a posttranscriptional mechanism (8, 12, 37). Mta has also been implicated as a contributor to replication via the lytic origin of replication, oriLyt. In an initial evaluation of the requirements for oriLyt replication in a cotransfection replication assay, both Zta and Mta were required to obtain replication in transfected Vero cells in addition to the six core replication genes, BMRF1 (polymerase-associated factor), BALF2 (single-stranded DNA binding protein), BALF5 (DNA polymerase), BSLF1 (primase), BBLF4 (helicase), and BBLF2/3 (primase accessory protein) (19). In this replication assay, HSV IE63 could partially substitute for Mta, suggesting that Mta may have been contributing to oriLyt replication indirectly by augmenting replication gene expression as has been described for IE63 (87). However, IE63 may also contribute to HSV DNA replication in other ways. IE63 has been found to locate within replication compartments in infected cells (95). We have now examined the effects of Mta on expression of EBV replication genes and show that five of the six core replication genes require Mta for cytoplasmic accumulation of their mRNA transcripts. Further, an examination of the mechanism of action of Mta revealed that Mta associates with splicing factors, binds specific RNAs, and shuttles between the nucleus and cytoplasm. Thus, Mta functions analogously to HSV IE63 in facilitating RNA processing and export of viral transcripts.
MATERIALS AND METHODS
Plasmids.
Expression plasmids in which the six core replication genes (BMRF1, BSLF1, BBLF4, BBLF2/3, BALF2, and BALF5) were expressed from heterologous promoters but contain natural 3′ untranslated sequences have been described elsewhere (20), as have the set in which the replication gene open reading frames (ORFs) were inserted into an SG5-based vector (72). The cDNA version of BBLF2/3, pEF76A, has been described elsewhere (19). To generate SG5-Flag-Mta vector pDH304, PCR amplification of the genomic region containing the BSLF2 and BMLF1 ORFs was performed with pTS6 as the template (11) and the primers 5′ 2195 (GTCAAGATCTATGGTTCCTTCTCAGAGA) and 3′ 1438 (TCAGAGATCTTTATTGATTTAATCCAGG). The PCR product was cleaved with BglII and ligated into the BglII-cut SG5-Flag vector pJH253. Glutathione S-transferase (GST)–Mta was generated by using the template pTS6 and the PCR primers 5′ 1439 (TCAGGGATCCGAGAGCCACATTCTGGAA) and 3′ 1438. The PCR product was cleaved with BamHI and BglII and ligated into the BglII site of pGH254 such that the BMLF1 ORF was expressed as a fusion with GST. pMBP-Tax was constructed by inserting sequences corresponding to the complete Tax cDNA into the BamHI site of pMAL-cR1 (New England Biolabs). SG5-CBF1(pJH156) contains the CBF1 coding sequence ligated as a BglII/BamHI fragment into the BglII site of SG5 (Stratagene, La Jolla, Calif.).
Cotransfection-replication assay.
The assay was performed in Vero cells as previously described (20), using the oriLyt plasmid pSL77 and SG5-based expression vectors for the six core replication genes, Mta, Rta, and Zta (72).
Western blotting.
293T cells were seeded at 106 in 100-mm-diameter dishes the day before transfection. Cells were transfected with 20 μg of DNA, using calcium phosphate-BES [N,N′-bis(2-hydroxyethyl)-2-aminoethanesulfonic acid]-buffered saline (10). Cells were harvested 48 h after transfection, washed, and lysed in sample buffer (25 mM Tris, 2% sodium dodecyl sulfate [SDS], 10% glycerol, 5% β-mercaptoethanol, 0.02% bromophenol blue). Cell lysates were electrophoresed through an SDS–10% polyacrylamide gel, and the separated proteins transferred to nitrocellulose (Bio-Rad, Hercules, Calif.). After blocking in TBS-T (100 mM Tris [pH 7.5], 100 mM NaCl, 1% Tween 20) plus 5% nonfat dry milk for 1 h, the filter was incubated with anti-Flag antibody (1:1,000; Eastman Kodak Co., New Haven, Conn.) to detect Flag-Mta.
BMRF1 protein expression was examined in Vero cells grown in six-well cluster dishes and transfected with a total of 2 μg of plasmid DNA per well. BMRF1 was detected with anti-BMRF1 monoclonal antibody (MAb; 1:2,000; Advanced Biotechnologies Inc., Columbia, Md.). The actin protein control was detected with antiactin MAb (1:3,000; Sigma, St. Louis, Mo.). Bacterially expressed GST fusion proteins were detected with anti-GST MAb (1:500; Santa Cruz Biotechnology Inc., Santa Cruz, Calif.). Protein bands were visualized using chemiluminescence (Amersham Life Sciences, Arlington Heights, Ill.).
Immunofluorescence.
Expression and colocalization assays were performed with Vero cells grown on glass tissue culture chamber slides (Nunc) and transfected with 2 μg of DNA by the calcium phosphate-BES procedure. Fixing and staining were performed as previously described (88). Anti-Flag antibody (Eastman Kodak) was diluted 1:1,500, and anti-SC35 culture supernatant (a gift from X. D. Fu and T. Maniatis) (23) was diluted 1:50.
The heterokaryon assay used HeLa cells grown on glass coverslips and transfected with Flag-Mta by the calcium phosphate method. The cells were seeded at approximately 30% confluent. After removal of the calcium-DNA precipitates by washing, the cells were allowed to grow for 24 h. Subsequently, Cos-TdRev cells were seeded onto the same dishes when approximately 50% confluent. This cell line stably expresses RevM10, a mutant of human immunodeficiency virus (HIV) Rev which is unable to export from the nucleus. The strong nucleolar staining of RevM10 was used to identify recipient cells. The combined cell populations were allowed to grow for 8 h to resume normal cell shape. Protein synthesis was inhibited by addition of cycloheximide (100 mg/ml) 30 min prior to heterokaryon formation to allow for clearance of residual cytoplasmic protein. Culture medium was removed, and fusion was mediated by incubation in 50% polyethylene glycol in phosphate-buffered saline (PBS) for 90 s. The polyethylene glycol was removed with four washes of PBS, and the fused cells were incubated for an additional hour in complete medium containing cycloheximide. The cells were then washed three times with PBS and fixed by incubation in 4% paraformaldehyde at room temperature for 12 min. The fixed cells were then permeabilized with a 2-min incubation in 100% methanol at room temperature. Flag-tagged Mta was identified with anti-Flag MAb, and anti-Rev rabbit polyclonal antibody was used to recognize RevM10. The secondary antibodies were tetramethyl rhodamine isothiocyanate (TRITC)-conjugated goat anti-mouse and fluorescein isothiocyanate (FITC)-conjugated anti-rabbit. The immunostained coverslips were attached to glass slides with VectaShield (Vector Laboratories, Inc., Burlingame, Calif.), and the samples were analyzed on a Nikon microscope.
RNA analyses.
Total RNA was isolated by the guanidinium thiocyanate-phenol method. Approximately 107 HeLa cells were lysed by addition of 1 ml of denaturing solution (4 M guanidinium thiocyanate, 25 mM sodium acetate [pH 7.0], 100 mM 2-mercaptoethanol, 0.5% Sarkosyl) directly to the culture dish following removal of the medium. The homogenate was transferred to a 5-ml polypropylene tube, 0.1 ml of 2 M sodium acetate (pH 3.9) was added and mixed, and then 1 ml of water-saturated phenol and 0.2 ml of chloroform-isoamyl alcohol (49:1) were added. After 15 min on ice, the phases were separated by centrifugation at 10,000 × g. The upper aqueous phase containing the RNA was subjected to repeated precipitation (three times) in isopropanol. The RNA was then washed in ethanol and stored at −20°C.
For preparation of cytoplasmic RNA, culture medium was removed, and the cells were scraped off in 1 ml of ice-cold PBS and collected by centrifugation (1,000 rpm for 5 min). The cell pellet was resuspended in 375 μl of ice-cold lysis buffer (50 mM Tris-Cl [pH 8.0], 100 mM NaCl, 5 mM MgCl2, 0.5% Nonidet P-40, 1,000 U of RNAs per ml, 1 mM dithiothreitol) and incubated on ice for 5 min. Nuclei and cell debris were removed by centrifugation (15,000 rpm) in a microcentrifuge for 2 min at 4°C. The supernatant was removed to a clean microcentrifuge tube containing 4 μl of 20% SDS and mixed. Proteinase K was added at 200 μg/ml and allowed to incubate at 37°C for 15 min. The supernatant was extracted twice with 400 μl of phenol-chloroform-isoamyl alcohol (25:24:1) and once with chloroform-isoamyl alcohol (24:1). RNA was precipitated from the extracted aqueous phase by addition of 40 μl of 3 M sodium acetate (pH 5.2)–1 ml of ethanol and centrifugation at 15,000 rpm for 15 min at 4°C. The RNA pellet was rinsed with ethanol, air dried, and resuspended in water.
Total BMRF1 mRNA was detected by primer extension analysis. The BMRF1-specific probe, CCTTGGTCTAAAGCGGAG (positions 79937 to 79918 on the EBV genome [4]) was designed to yield a 128-base extension product. The oligonucleotide was end-labeled by standard methods (3). Isolated RNA (20 μg) was mixed with 105 cpm of labeled oligonucleotide and precipitated. The precipitate was dissolved in 30 μl of hybridization buffer [80% formamide, 40 mM piperazine-N,N′-bis(2-ethanesulfonic acid) (PIPES; pH 6.4), 400 mM NaCl, 1 mM EDTA (pH 8.0)] and incubated at 30°C overnight. RNA and oligonucleotide were precipitated by addition of 170 μl of 0.3 M sodium acetate and 500 μl of ethanol. The pellet was washed once with ethanol and air dried. Reverse transcription was done with avian myeloblastosis virus reverse transcriptase at 42°C for 90 min. The reaction was halted by addition of 1 μl of 0.5 M EDTA and digested with 1 μg of RNase A at 37°C for 30 min. The mixture was extracted with phenol-chloroform-isoamyl alcohol (25:24:1) and precipitated by addition of 100 μl of 2.5 M sodium acetate and 300 μl of ethanol. The pellet was washed once with ethanol and air dried. The extended products were analyzed on an 8% acrylamide-urea gel.
Total and cytoplasmic RNAs were also detected by Northern blotting. RNA (30 μg) was electrophoresed through a 1% formaldehyde-agarose gel and transferred onto a nylon membrane (Schleicher & Schuell Inc., Keene, N.H.). The membrane was incubated at 65°C with individual 32P-labeled DNA probes which were generated by using a Random Primed DNA labeling kit (Boehringer Mannheim). The DNAs used as probes were as follows: BBLF2/3, 0.8-kb BglI fragment from pRTS25; BALF2, 1-kb XbaI/HindIII/BglII fragment from pRTS12; BMRF1, 0.8-kb EcoRI/HindIII fragment from pMH2; BALF5, 3.0-kb Xba/HindIII fragment from pRTS13; BSLF1, 2.6-kb Xba/HindIII fragment from pRTS11; and BBLF4, 2.4-kb Xba/HindIII fragment from pRTS28.
RNA-protein interactions.
RNA probes for Northwestern analysis were synthesized by in vitro transcription in the presence of [32P]UTP. The BMRF1 probes were transcribed from pGEM74 linearized with EcoRI, resulting in an 847-base RNA product corresponding to BMRF1/2 message from positions 82081 to 82920 plus vector sequences. The BMLF1 probe was transcribed from pGEM74 linearized with HindIII, resulting in a 853-base RNA product corresponding to BMLF1 message from 82920 to 82081 plus vector sequences. All other RNA probes were synthesized by in vitro transcription of PCR-amplified genomic sequences containing a complete T7 site in the 5′ oligomer primer. The A1 probe primers were GGATCCTAATACGACTCACTATAGGGAGGTCTCTCTCCGGGCACT (82800) and TACAGTGAGGTTACACAGGTG (82662), resulting in a 138-base RNA product. The A2 probe primers were GGATCCTAATACGACTCACTATAGGGA (82924) and ATGGCCCTGACAAGTCGGCTG (82804), resulting in a 132-base RNA probe which also contained some vector sequences. The A2a probe primers were GGATCCTAATACGACTCACTATAGGGA (82924) and AAAAGGGAGCTTAGCGTG (82870), resulting in a 63-base RNA product of which 10 nucleotides were vector sequences. The A2b probe primers were GGATCCTAATACGACTCACTATAGGGAGGGACAGAGGCCGTGGAG (82870) and ATGGCCCTGACAAGTCGGCTG(82801), resulting in a 69-base RNA product. All RNA probes were purified by spin-gel exclusion chromatography to >98% purity as determined by percent incorporated radioactivity. The overall integrity of each probe was examined by 6% acrylamide-urea gel electrophoresis.
Glutathione-agarose purified GST-Mta was separated by polyacrylamide gel electrophoresis (PAGE) on an SDS–12% polyacrylamide gel. The proteins were transferred onto Immobilon-P (Millipore) by semidry transfer using an Immunoblot Transfer Cell (Bio-Rad). The filter-bound proteins were denatured by immersion in 6 M guanidine-HCl in binding buffer (50 mM Tris-HCl [pH 7.5], 50 mM NaCl, 1 mM EDTA) for 10 min at room temperature. The denatured protein was then renatured by stepwise dilution of the guanidine-HCl with binding buffer. Dilutions (50%) were separated by 10-min incubations, with the fifth change being binding buffer. Nonspecific binding was reduced by incubation in prehybridization buffer (2.5% nonfat dry milk–1 mM dithiothreitol in binding buffer) for 60 min at room temperature. The filters were then washed twice in binding buffer for 5 min at room temperature. Labeled RNA probes were added at 106 cpm/ml/filter in a total volume of 5 ml of hybridization buffer [10 mg each of calf thymus DNA and poly(I)-poly(C) per ml in binding buffer]. The protein and probes were incubated at room temperature for 1 h. Unbound probe was removed with four 10-min washes at room temperature. The filters were allowed to air dry, then wrapped in plastic wrap, and visualized by autoradiography.
RESULTS
The requirement for Mta for oriLyt replication in transfected cells is vector dependent.
In the original oriLyt cotransfection replication assays, the six core EBV replication proteins were expressed from vectors that provided a strong heterologous promoter but no other regulatory sequences and the individual genes retained their natural 3′ untranslated sequences (19). In this setting, oriLyt replication required Zta and Mta in addition to the core replication proteins, while the Rta transactivator was not essential. Zta acts as an oriLyt origin binding protein (2, 19, 72–74) and facilitates tethering of the core replication complex (24). The role of Mta in this replication assay was not defined.
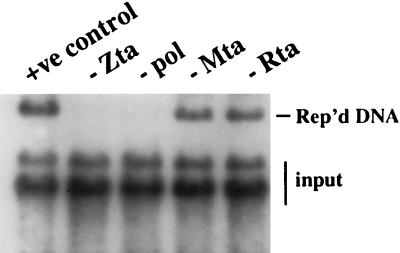
The ORFs for the six core replication genes and the Zta, Rta, and Mta transactivators were recloned into the SG5 vector such that all 5′ and 3′ regulatory sequences would be vector derived. SG5 provides the rabbit β-globin intron that facilitates splicing of expressed transcripts along with an efficient polyadenylation signal. A cotransfection replication assay using the recloned replication genes was performed with Vero cells (Fig. 1). The recloned genes produced a more robust replication signal than was obtained in assays using the original expression constructions. As previously observed, oriLyt replication did not require the presence of Rta and was dependent on Zta and on the viral DNA polymerase. However, the use of the recloned replication genes rendered oriLyt replication Mta independent.
FIG. 1.
oriLyt replication can occur in transfected cells in the absence of Mta. A cotransfection-replication assay was performed with Vero cells which were transfected with recloned expression plasmids for the six core EBV replication genes plus the three lytic transactivators Zta, Rta, and Mta. When strong splicing and polyadenylation signal sequences for replication gene expression are provided by the vector, oriLyt replication is independent of Mta. oriLyt replication remains dependent on Zta and the EBV DNA polymerase. +ve, positive; Rep’d, replicated.
Mta increases expression of BMRF1 protein.
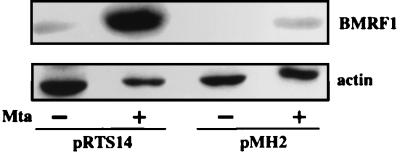
Mta is a posttranscriptional transactivator, and it seemed likely that the original requirement for Mta lay at the level of replication gene expression. To assess whether this is the case, we determined the level of protein expression of the BMRF1 protein (polymerase accessory factor) in Vero cells transfected with the original BMRF1 expression vector, pMH2, or the recloned SG5-BMRF1 vector, pRTS14. The BMRF1 protein was undetectable in cells transfected with pMH2 but became detectable upon cotransfection of pMH2 with Mta (Fig. 2). Cells transfected with the SG5-based vector pRTS14 expressed detectable BMRF1 in the absence of Mta, and the amount increased upon cotransfection with Mta (Fig. 2). Thus, a requirement for Mta in the cotransfection replication assays correlates with the need for Mta to obtain detectable levels of protein expression.
FIG. 2.
Mta increases BMRF1 protein expression. Western analysis was used to determine BMRF1 protein levels in Vero cells transfected with BMRF1 expression vectors that provide a heterologous strong promoter and use either natural 3′ downstream sequences (pMH2) or vector-provided splicing and 3′ processing signals (pRTS14). BMRF1 protein was detected with anti-BMRF1 MAb, and the actin control was detected on a duplicate membrane with antiactin MAb. In the presence of Mta (+), the amount of BMRF1 protein was increased in cells transfected with either vector. However, expression from pMH2 was dependent on Mta, and no BMRF1 protein was detected in the absence of Mta (−).
Mta increases cytoplasmic transport of replication gene mRNA.
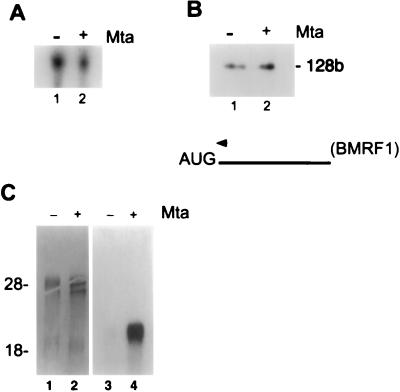
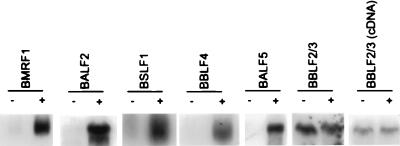
To determine whether the increased accumulation of BMRF1 protein in the presence of Mta correlated with changes in accumulation of BMRF1 transcripts, we determined the levels of BMRF1 mRNA expressed from pMH2 in the presence and absence of Mta. Cotransfection of Mta had no effect on the amount of total BMRF1 RNA in transfected cells, as measured by either Northern or S1 analysis (Fig. 3A and B). In contrast, the level of cytoplasmic BMRF1 mRNA dramatically increased upon cotransfection of Mta (Fig. 3C). Expression of the other five core replication genes was evaluated similarly in cells transfected with the original replication gene constructions (19) in which expression is directed from a strong heterologous promoter but the genes retain their natural 3′ untranslated sequences. These assays revealed that Mta was also required for efficient cytoplasmic accumulation of the transcripts for BALF2 (single-stranded DNA binding protein), BALF5 (DNA polymerase), BSLF1 (primase), and BBLF4 (helicase). However, Mta had no effect on the levels of cytoplasmic BBLF2/3 mRNA (Fig. 4). BBLF2/3 (primase-associated factor) is encoded by two ORFs, BBLF2 and BBLF3, and the mRNA, unlike that of the other five core replication genes, is spliced (19). To determine whether the BBLF2/3 intron was a necessary component of Mta independence, a cDNA version of BBLF2/3 was tested. Cytoplasmic accumulation of transcripts of the BBLF2/3 cDNA lacking the internal intron remained Mta independent (Fig. 4), suggesting that signals in addition to the presence of the intron itself influence the requirement for Mta.
FIG. 3.
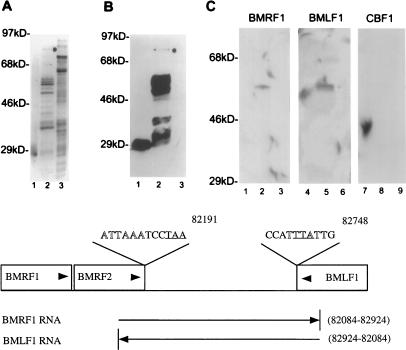
Mta increases cytoplasmic accumulation of BMRF1 mRNA. HeLa cells were transfected with the BMRF1 expression plasmid (pMH2) and cotransfected with either the Mta expression plasmid (pRTS16) or vector control. (A) Total RNA was isolated from both control (lane 1) and Mta-expressing (lane 2) cells and subjected to Northern analysis for BMRF1 message expression. As shown, there was no discernible difference in total BMRF1 message between cells cotransfected with Mta or control vector DNA. (B) Total RNA was isolated from both control (lane 1) and Mta-expressing (lane 2) cells and subjected to primer extension analysis for BMRF1 message expression. Again, there was no discernible difference in total BMRF1 message between cells cotransfected with Mta or control DNA. The band migrating at 128 bases (128b) is marked. (C) Cytoplasmic RNA was isolated from control (lanes 1 and 3) and Mta-expressing (lanes 2 and 4) cells and subjected to Northern analysis for BMRF1 message expression. Lanes 1 and 2, methylene blue stain for rRNA; lanes 3 and 4, autoradiogram of cytoplasmic RNA reactive with the BMRF1-specific probe. BMRF1 message was undetectable in the absence of Mta (lane 3) but was abundant in Mta-expressing cells (lane 4).
FIG. 4.
Mta expression increased cytoplasmic accumulation of five EBV replication gene mRNAs. HeLa cells were transfected with the original expression vector for either BMRF1, BALF2, BSLF1, BBLF4, BALF5, or BBLF2/3 as indicated and cotransfected with either control (−) or Mta-expressing (+) plasmid. Cytoplasmic RNA was isolated and subjected to Northern analysis for expression of the indicated message. Both genomic (BBLF2/3) and cDNA (BBLF2/3 cDNA) versions of BBLF2/3 were tested. Mta had no effect on BBLF2/3 message, which was found to be constitutively expressed in the cytoplasm in the absence of Mta. In contrast, the other replication mRNAs were undetectable in the cytoplasmic fraction in the absence of Mta (−) and accumulated in the presence of Mta (+).
Mta binds directly to RNA.
The HSV IE63 homolog of Mta has been shown to bind to RNA (7, 33, 46). An RNA binding assay was performed with in vitro-transcribed, 32P-labeled mRNAs transcripts and GST-Mta along with control GST and maltose binding protein (MBP)-Tax proteins (Fig. 5). The GST-Mta vector expressed the BMLF1 ORF fused to GST. Only a small amount of the intact fusion protein was detected in silver-stained gels. Most of the protein migrated as a triple band of degradation products of 55 to 60 kDa. These three protein bands contained the amino terminus of the fusion protein, as demonstrated by their interaction with anti-GST antibody (Fig. 5B) and on the basis of size, would represent the amino-terminal half of the BMLF1 polypeptide.
FIG. 5.
Mta binds target mRNAs. Purified GST-Mta, control GST, and crude control MBP-Tax were separated by SDS-PAGE and transferred to Immobilon-P filters. The membrane-bound proteins were denatured and renatured as described in the text and incubated with the indicated RNA probes. (A) Silver stain of purified GST (lane 1), purified GST-Mta (lane 2), and crude MBP-Tax (lane 3). The full-length GST-Mta is indicated by a dot. The gel shows numerous background proteins that are available for nonspecific probe binding. (B) A membrane blot containing the proteins shown in panel A was immunoprobed with anti-GST MAb and visualized by chemiluminescence. The purified GST-Mta was partially degraded. Note that the degradation was largely progressive from the carboxyl terminus since most degradation products tested positive for the amino-terminal GST fusion partner (lane 2). (C) A membrane blot of the proteins shown in panel A was incubated with BMRF1 and BMLF1 RNA probes and a control probe transcribed from the cellular CBF1 cDNA. GST-Mta, but not GST or MBP-Tax, bound to the BMRF1 and BMLF1 RNA probes (compare lanes 2 and 5 to lanes 1, 3, 4, and 6). The control CBF1 RNA did not bind to GST-Mta (lane 8). (D) Map locations of the BMRF1 and BMLF1 probes relative to the BMRF1, BMRF2, and BMLF1 ORFs. The map coordinates are those for the B95-8 genome (4). Arrowheads indicate the direction of transcription of the genes and probe RNAs. The polyadenylation signals for these genes are shown in outlined type, and the translation termination codons for BMRF2 and BMLF1 are underlined.
The BMRF1 mRNA is believed to be coterminal with that for BMRF2 and to use the polyadenylation signal located right at the 3′ end of the BMRF2 ORF (58). We generated an 847-base RNA probe that crossed the 3′ 100 bases of the BMRF2 ORF and extended across the region between the BMRF2 and BMLF1 ORFs to include sequences that might potentially be present in a primary unprocessed transcript for BMRF1 and BMRF2 (Fig. 5C). This RNA bound to the GST-Mta fusion protein. The adjacent convergent mRNA is that for BSLF2/BMLF1 (Mta). The polyadenylation site for this transcript has been mapped to the sequence coincident with the terminus of the BMLF1 ORF (67). An equivalent 853-base RNA that covered the 3′ 174 bases of the BMLF1 ORF and the region between the BMLF1 and BMRF2 ORFs was also generated to represent a primary Mta transcript. This RNA also bound to GST-Mta (Fig. 5C). There was specificity to the RNA binding in that a similarly sized RNA generated across the ORF for the cellular DNA binding protein CBF1/RBPJk (28) did not interact with GST-Mta. The BMRF1 and BMLF1 RNAs also did not interact with control GST protein or MBP-Tax (Fig. 5).
The progressive degradation of the GST-Mta fusion protein provided information on the approximate location of the RNA-binding domain within Mta. All degradation products greater than 55 kDa (GST plus 25 kDa of Mta polypeptide) bound specifically to RNA. However, degradation products containing less than 25 kDa of Mta had no RNA binding activity. An arginine-rich region of Mta that is reminiscent of domains in characterized RNA binding proteins is located between amino acids (aa) 125 and 204. This domain would be present in the RNA binding GST-Mta polypeptides.
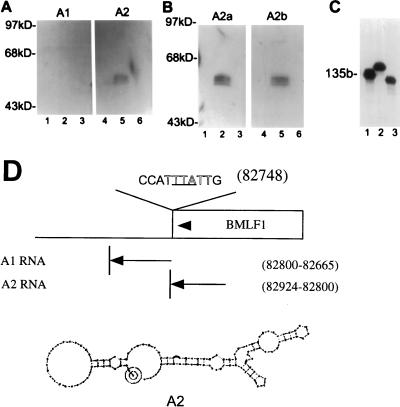
In an effort to further define the RNA region required for interaction with Mta, two smaller nonoverlapping segments of the original BMLF1 test RNA were synthesized as probes. The A1 RNA (137 bases) covered sequences that might be present in a primary BMLF1 transcript but would not be present in the mature polyadenylated mRNA. This RNA did not bind to GST-Mta (Fig. 6A). The A2 RNA, which did bind to GST-Mta, was 132 bases long and covered sequences within the BMLF1 ORF, beginning at a position 54 bp upstream of the translational termination signal. The difference in binding ability of the A1 and A2 probes was not a reflection of differences in size or RNA integrity, as illustrated in Fig. 6C. When the A2 region was again subdivided into RNAs covering 51 and 69 nucleotides of the Mta gene, both RNAs bound to GST-Mta (Fig. 6B). It is possible that there are multiple contacts between Mta and target RNAs, but this point needs further evaluation. The predicted secondary structure of the A2 probe is presented in Fig. 6D along with its relative location within the BMLF1 ORF.
FIG. 6.
Mta binds a 135-base sequence within BMLF1, as determined by Northwestern analysis of the binding of nonoverlapping BMLF1-specific RNA probes to GST-Mta. (A) Protein blots identical to those in Fig. 5A were probed with different RNAs corresponding to sequences within the BMLF1 3′ region. The A1 probe encompasses the first base of the naturally occurring poly(A) signal and continues in the 3′ direction for 139 bases. The A2 probe comprises 130 bases upstream of the start of A1 and is entirely within the BMLF1 ORF. A2 retains the ability to bind Mta, whereas A1 shows no interaction. (B) Subdivision of the A2 probe failed to identify a single interactive region. The A2a probe is the 5′ half of A2, and the A2b probe is the 3′ half of A2; both probes bound GST-Mta. (C) All probes were analyzed on 8% urea-polyacrylamide gels for integrity. Full-length probes of comparable specific activity were generated. A known 135-base (135b) control RNA (lane 1) was used as a size marker for the A1 (lane 2) and A2 (lane 3) probes. (D) Schematic showing the probes, their relative genomic positions, and the predicted secondary structures of the Mta binding A2 RNA. The polyadenylation signal (outlined type), translational termination codon (underlined), and direction of transcription (arrowheads) are indicated.
Intranuclear localization of Mta.
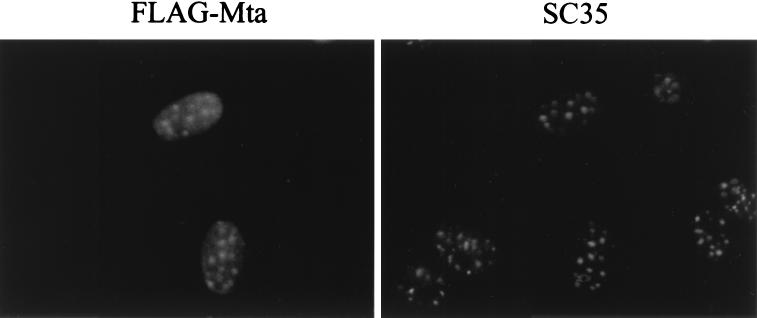
RNA binding proteins, including the HSV IE63 protein, have been found to be present in splicing bodies within the nucleus (70). The ability of Mta to colocalize with splicing factors was examined in an immunofluorescence assay. Cells transfected with Flag-Mta were stained with anti-Flag antibody and FITC-conjugated secondary antibody to detect Mta and antibody recognizing the SC35 splicing factor plus TRITC-conjugated secondary antibody to detect endogenous SC35. In transfected cells, Flag-Mta gave a diffuse nuclear staining with underlying strongly staining nuclear speckles. These speckles were found to colocalize with SC35-containing bodies (Fig. 7).
FIG. 7.
Mta colocalizes with the SC35 splicing factor. Flag-tagged Mta was transfected into Vero cells. The Mta-expressing cells were fixed and immunostained with anti-Flag (rabbit polyclonal) and anti-SG35 (mouse monoclonal) primary antibodies. Secondary antibodies were FITC-conjugated anti-rabbit and TRITC-conjugated anti-mouse. Mta expression was diffusely nuclear, with a concentration in subnuclear regions which overlap with SC35 speckles.
Mta shuttles between the nucleus and the cytoplasm.
HIV type 1 Rev serves as the mechanistic prototype for proteins that regulate cytoplasmic transport of viral transcripts. Rev recognizes a specific RNA sequence, the Rev-responsive element, via an arginine-rich region and mediates export of unspliced and incompletely spliced viral RNA (17, 18). The ability to shuttle between the nucleus and cytoplasm is believed to be an essential component of Rev function. Rev nucleocytoplasmic shuttling is facilitated by a signal for nuclear entry (nuclear localization signal) and a signal for nuclear export (nuclear export signal [NES]). It is the NES which appears to distinguish shuttling from nonshuttling nuclear proteins, and a NES has been found in several other shuttling proteins, including HSV IE63 (49, 51, 56, 68, 75). Inspection of the Mta amino acid sequence revealed the presence of a motif between aa 226 and 237 that can be aligned with the NES sequences of Rev, Rev-like proteins, and HSV IE63 (Fig. 8).
FIG. 8.
A potential NES within EBV Mta. A sequence within Mta was compared with known functional NES sequences (5, 31, 37, 49, 68, 89).
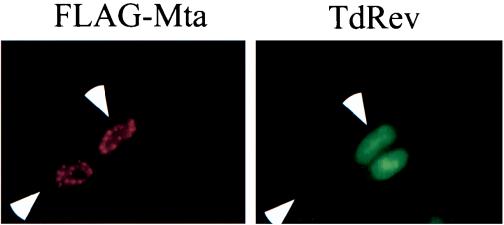
To determine whether Mta is capable of nucleocytoplasmic shuttling, we performed a heterokaryon analysis. HeLa cells transfected with Flag-Mta were fused to Cos-TdRev cells, which stably express HIV RevM10, a Rev mutant that is unable to export from the nucleus. The fused cells were incubated in medium containing cycloheximide to block de novo protein synthesis and then fixed and stained for Mta and for RevM10. In heterokaryons, the donor cell nucleus expressing transfected Flag-Mta and the recipient nucleus expressing RevM10 are located within a single cytoplasm. Proteins that are capable of shuttling will move from the nucleus of the transfected cell into the cytoplasm. From the cytoplasm, the shuttling protein can move back into either of the two available nuclei, resulting in both donor and recipient nuclei staining for the transfected protein. In this assay, Mta demonstrated the ability to shuttle from the originally transfected cells into the RevM10-staining nuclei of the recipient cells (Fig. 9).
FIG. 9.
Mta shuttles between the nucleus and the cytoplasm. In the heterokaryon assay, Flag-Mta was transfected into HeLa (the heterokaryon donor) cells. The Cos cell line TdRev, which is a stable producer of a Rev mutant incapable of nucleocytoplasmic shuttling, was used as the heterokaryon recipient. After heterokaryon formation, the cells were fixed and immunostained with anti-Flag (mouse monoclonal) and anti-Rev (rabbit polyclonal) primary antibodies. The secondary antibodies were TRITC-conjugated anti-mouse and FITC-conjugated anti-rabbit. The arrowheads point to two Mta-expressing cells in each panel. Note the even distribution of Mta between both donor (bottom arrow) and recipient (top arrow) cells.
DISCUSSION
Induction of the EBV lytic cycle by activators such as anti-immunoglobulin antibody initiates transcription of the BZLF1 and BRLF1 genes, which encode the Zta and Rta transcriptional activators (21, 47, 78, 85). Transcription across the Mta-encoding BSLF2/BMLF1 gene follows, with the Mta promoter responding to both the Zta and Rta activators (8, 36). Mta is recognized to stimulate reporter gene expression in a posttranscriptional manner, but beyond that little is known about the contribution of Mta to the regulation of EBV gene expression. We have shown that Mta is necessary for efficient cytoplasmic accumulation of mRNA for five of the EBV replication genes, BMRF1, BALF2, BALF5, BSLF1, and BBLF4. Thus, Mta contributes to EBV early gene expression. However, not all early genes are Mta dependent. Abundant cytoplasmic mRNA was detected for the BBLF2/3 replication gene in the absence of Mta, and the level was not affected by the addition of Mta, suggesting that some of the EBV early gene transcripts can be processed efficiently by cellular factors. The BBLF2/3 transcripts, unlike the other five core replication genes, is spliced, but removal of the intron in BBLF2/3 did not alter its behavior. It is possible that the partial 5′ and 3′ splicing signals bounding the intron continue to be recognized by cellular factors. However, the precise requirements for Mta independence and Mta dependence remain to be determined.
In an indirect immunofluorescence assay, Mta showed a mixture of diffuse nuclear staining and strongly staining nuclear speckles. These speckles colocalized with the nuclear speckles characteristic of the splicing factor SC35, indicating that Mta is present at sites adjacent to nascent mRNA synthesis and processing. HIV Rev and HSV IE63 also show an association with SC35 in the nucleus (35, 70). The association with SC35 together with the observation that using a vector such as SG5 that introduces the β-globin intron into expressed transcripts alleviates the dependence on Mta for BMRF1 protein expression suggests that Mta may also function in part through communication with splicing machinery. Although the SG5-BMRF1 vector, pRTS16, was not dependent on Mta for expression of BMRF1, protein expression was still increased upon cotransfection with Mta. This result would also be compatible with Mta functioning to facilitate RNA processing. The IE63 (ICP27) protein that is the HSV homolog of Mta has been demonstrated to cause a redistribution of the snRNPs that are components of the splicing complex and to colocalize with the redistributed snRNPs (59). This redistribution is believed to contribute to the repression of host cell splicing that occurs in HSV-infected cells. However, experiments performed with temperature-sensitive mutants in IE63 indicated that the repression of host cell splicing is more complex and redistribution of snRNPs alone is insufficient to bring about splicing inhibition (70). There is no evidence that Mta exerts a comparable negative effect on cellular splicing, although the question has not been rigorously examined. In our experiments, we saw no repression of expression of the BBLF2/3 replication gene upon cotransfection of Mta. Perhaps the presence of spliced lytic cycle genes in EBV is not compatible with a global splicing repression activity. The EBV lytic cycle immediate-early Zta and Rta RNAs are spliced, as are several early and late genes, including Mta itself, BHRF1, encoding the bcl-2 homolog, and BLLF1, encoding the membrane glycoprotein gp350/220.
Mta was shown to bind to RNAs from the BMRF1 and BMLF1 ORFs. The BMLF1 transcripts that showed specific binding were derived entirely from sequences within the BMLF1 ORF and included the 119 nucleotides of EBV sequence from positions 82920 to 82801 in the EBV genome. The leftward-transcribed BMLF1 ORF terminates at position 82745. The cis-acting response elements in RNA bound by the HIV Rev and human T-cell leukemia virus type 1 (HTLV-1) Rex proteins have been extensively characterized. These elements are relatively large, 233 and 254 nucleotides, respectively, and form stable stem-loop secondary structures. Deletions that result in loss of structure also result in loss of function. Mutational studies also provided evidence for the presence of additional sequence specific subregions that may be required for protein-RNA recognition (1). The HTLV-1 Rex-responsive element is located immediately downstream of the polyadenylation signal for the env gene, and the HIV Rev-responsive element is located within the env intron (41). The simpler retroviruses that do not encode Rev-like proteins are apparently able to utilize the cellular machinery for posttranscriptional processing of their RNAs. This function is mediated via specific sequence and structural elements within the transcripts. A constitutive RNA transport element that is required for cytoplasmic transport of Mason-Pfizer monkey virus intron-containing RNAs has been described elsewhere (6). This element consists of 153 nucleotides located in the 3′ untranslated region of the env RNA, and the constitutive transport element can substitute for the Rev–Rev-responsive element combination (15). Based on computer modeling, this element also forms a stable stem-loop structure (16). Computer modeling predicts that the BMLF1 RNA that bound GST-Mta is capable of stem-loop structure formation, but the predicted structure has not been validated experimentally. The location of an RNA binding element within the BMLF1 ORF may be related to the unusual placement of the BMLF1 polyadenylation signal, which is located immediately adjacent to the translational termination signal for the BMLF1 ORF. Curiously, several other EBV genes also have this unusually compact spacing between the end of the ORF and the polyadenylation signal. Examples include BORF2 (which encodes the large subunit of ribonucleotide reductase), BLRF2 (a late gene), BLLF1 (the gp350/220 membrane antigen gene), BBLF4 (the helicase gene), and BMRF2 (a late gene).
RNA binding by the Rev-like proteins and HSV IE63 is mediated by an arginine-rich region that also serves as the nuclear localization signal (29, 39, 68). The GST-Mta protein used in our binding experiments yielded specific breakdown products that based on size and interaction with anti-GST antibody represented N-terminal polypeptides comprising approximately half of the full-length 480-aa protein. An arginine-rich region is present in this N-terminal region, between aa 125 and 204 of Mta, and it is likely that this domain mediates RNA binding. The BMLF1 ORF together with BSLF2 encodes Mta. The binding of Mta to its own message implies that Mta regulates its own synthesis. The same observation has been made for the HSV IE63 (ICP27) protein (68). BMRF1 and BMRF2 have coterminal RNA transcripts, and the region of the BMRF1 message that bound to Mta would also be present in the BMRF2 transcript. It therefore seems likely that BMRF2, a late gene, is also regulated by Mta and hence that Mta can regulate both early and late classes of EBV mRNAs.
Mammalian viruses differ from the host cell by utilizing intronless transcripts to encode many of their gene products. Transcripts that are unspliced or incompletely spliced are typically retained in the nucleus. Thus, the unspliced viral messages need a mechanism to avoid nuclear retention. Viruses circumvent this problem by encoding proteins that bind to specific RNA sequences and transport unspliced and incompletely spliced viral RNA into the cytoplasm (1, 9, 25, 26, 30, 41, 68, 77). These proteins contain a leucine-rich NES that enables rapid nuclear export (5, 17, 34, 38, 48). The NES has recently been shown to interact with a protein, CRM1 or exportin 1, which is related to the karyopherin β family of nuclear import proteins (22, 55, 76, 84). Exportin 1 interacts with the NES-containing protein, and RanGTP to form a complex that is transported through the nuclear pore (86). The combined presence of a NES and a nuclear localization signal allows RNA transport proteins to shuttle between the nucleus and the cytoplasm. NES elements, in addition to the retrovirus Rev-like proteins, have been identified in the adenovirus E4 34-kDa protein (14) and most recently in the HSV IE63 (ICP27) protein (68). Mta has a leucine-rich region located between aa 215 and 250 that contains a consensus NES. The heterokaryon assay has been a valuable tool for establishing that export proteins shuttle between the nucleus and cytoplasm, and this assay was exploited to determine whether Mta functioned in this manner. Mta was readily detected in both the donor and the acceptor nuclei of heterokaryons, indicating that Mta has the ability to move from the nucleus of the donor cell into the cytoplasm and from the cytoplasm into the nucleus of the recipient cell. Thus, our experiments establish that Mta has the properties of an RNA export protein. Mta binds to specific EBV RNAs, shuttles between the nucleus and the cytoplasm, and is required for efficient cytoplasmic accumulation of EBV replication gene RNAs. EBV joins the growing list of viruses that circumvent the problem of nuclear retention of RNAs by encoding proteins that facilitate RNA processing and mediate efficient cytoplasmic accumulation of their own transcripts.
ACKNOWLEDGMENTS
We thank M.-L. Hammarskjold for the TdRev cell line, X. D. Fu and T. Maniatis for the SC35 antibody, Jon Finan for technical assistance, and Feng Chang for manuscript preparation.
This work was funded by National Institutes of Health grant RO1 CA30356. R.T.S. was supported by training grant T32 CA09243. S.D.H. is the recipient of American Cancer Society award FRA429.
REFERENCES
- 1.Ahmed Y F, Hanly S M, Malim M H, Cullen B R, Greene W C. Structure-function analyses of the HTLV-I Rex and HIV-1 Rev RNA response elements: insights into the mechanism of Rex and Rev action. Genes Dev. 1990;4:1014–1022. doi: 10.1101/gad.4.6.1014. [DOI] [PubMed] [Google Scholar]
- 2.Askovic S, Baumann R. Activation domain requirements for disruption of Epstein-Barr virus latency by ZEBRA. J Virol. 1997;71:6547–6554. doi: 10.1128/jvi.71.9.6547-6554.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ausubel F M, Brent R, Kingston R E, David D M, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. Vol. 1. New York, N.Y: John Wiley & Sons, Inc.; 1994. [Google Scholar]
- 4.Baer R, Bankier A T, Biggin M D, Deininger P L, Farrell P J, Gibson T J, Hatfull G, Hudson G S, Satchwell S C, Seguin C, Tuffnell P S, Barrell B G. Organization of the B95-8 Epstein-Barr virus genome. Nature. 1984;310:207–211. doi: 10.1038/310207a0. [DOI] [PubMed] [Google Scholar]
- 5.Bogerd A M, Fridell R A, Benson R E, Hua J, Cullen B R. Protein sequence requirements for function of the human T-cell leukemia virus type 1 Rex nuclear export signal delineated by a novel in vivo randomization-selection assay. Mol Cell Biol. 1996;16:4207–4214. doi: 10.1128/mcb.16.8.4207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bray M, Prasad S, Dubay J W, Hunter E, Jeang K-T, Rekosh D, Hammarskjold M-L. A small element from the Mason-Pfizer monkey virus genome makes human immunodeficiency virus type 1 expression and replication Rev-independent. Proc Natl Acad Sci USA. 1994;91:1256–1260. doi: 10.1073/pnas.91.4.1256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brown C R, Nakamura M S, Mosca J D, Hayward G S, Straus S E, Perera L P. Herpes simplex virus trans-regulatory protein ICP27 stabilizes and binds to 3′ ends of labile mRNA. J Virol. 1995;69:7187–7195. doi: 10.1128/jvi.69.11.7187-7195.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Buisson M, Manet E, Trescol-Biemont M-C, Gruffat H, Durand B, Sergeant A. The Epstein-Barr Virus (EBV) early protein EB2 is a posttranscriptional activator expressed under the control of EBV transcription factors EB1 and R. J Virol. 1989;63:5276–5284. doi: 10.1128/jvi.63.12.5276-5284.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chang D D, Sharp P A. Regulation by HIV Rev depends upon recognition of splice sites. Cell. 1989;59:789–795. doi: 10.1016/0092-8674(89)90602-8. [DOI] [PubMed] [Google Scholar]
- 10.Chen C, Okayama H. High-efficiency transformation of mammalian cells by plasmid DNA. Mol Cell Biol. 1987;7:2745–2752. doi: 10.1128/mcb.7.8.2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cho M-S, Jeang K-T, Hayward S D. Localization of the coding region for an Epstein-Barr virus early antigen and inducible expression of this 60-kilodalton nuclear protein in transfected fibroblasts cell lines. J Virol. 1985;56:852–859. doi: 10.1128/jvi.56.3.852-859.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cook I D, Shanahan F, Farrell P J. Epstein-Barr virus SM protein. Virology. 1994;205:217–227. doi: 10.1006/viro.1994.1637. [DOI] [PubMed] [Google Scholar]
- 13.Defechereux P, Debrus S, Baudoux L, Rentier B, Piette J. Varicella-zoster virus open reading frame 4 encodes an immediate-early protein with posttranscriptional regulatory properties. J Virol. 1997;71:7073–7079. doi: 10.1128/jvi.71.9.7073-7079.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dobblestein M, Roth J, Kimberly W T, Levine A J, Shenk T. Nuclear export of the E1B 55-kDa and E4 34-kDa adenoviral oncoproteins mediated by a rev-like signal sequence. EMBO J. 1997;16:4276–4284. doi: 10.1093/emboj/16.14.4276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ernst R K, Bray M, Rekosh D, Hammarskjold M-L. A structured retroviral RNA element that mediates nucleocytoplasmic export of intron-containing RNA. Mol Cell Biol. 1997;17:135–144. doi: 10.1128/mcb.17.1.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ernst R K, Bray M, Rekosh D, Hammarskjold M L. Secondary structure and mutational analysis of the Mason-Pfizer monkey virus RNA constitutive transport element. RNA. 1997;3:210–222. [PMC free article] [PubMed] [Google Scholar]
- 17.Fischer U, Huber J, Boelens W C, Mattaj I W, Luhrmann R. The HIV-1 Rev activation domain is a nuclear export signal that accesses an export pathway used by specific cellular RNAs. Cell. 1995;82:475–483. doi: 10.1016/0092-8674(95)90436-0. [DOI] [PubMed] [Google Scholar]
- 18.Fischer U, Meyer S, Teufel M, Heckel C, Luhrmann R, Rautmann G. Evidence that HIV-1 Rev directly promotes the nuclear export of unspliced RNA. EMBO J. 1994;13:4105–4112. doi: 10.1002/j.1460-2075.1994.tb06728.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fixman E D, Hayward G S, Hayward S D. Replication of Epstein-Barr virus oriLyt: lack of a dedicated virally encoded origin-binding protein and dependence on Zta in cotransfection assays. J Virol. 1995;69:2998–3006. doi: 10.1128/jvi.69.5.2998-3006.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fixman E D, Hayward G S, Hayward S D. trans-acting requirements for replication of Epstein-Barr virus ori-Lyt. J Virol. 1992;66:5030–5039. doi: 10.1128/jvi.66.8.5030-5039.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Flemington E K, Goldfeld A E, Speck S H. Efficient transcription of the Epstein-Barr virus immediate-early BZLF1 and BRLF1 genes requires protein synthesis. J Virol. 1991;65:7073–7077. doi: 10.1128/jvi.65.12.7073-7077.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fornerod M, Ohno M, Yoshida M, Mattaj I W. CRM1 is an export receptor for leucine-rich nuclear export signals. Cell. 1997;90:1051–1060. doi: 10.1016/s0092-8674(00)80371-2. [DOI] [PubMed] [Google Scholar]
- 23.Fu X D, Maniatis T. Factor required for mammalian spliceosome assembly is localized to discrete regions in the nucleus. Nature. 1990;343:437–441. doi: 10.1038/343437a0. [DOI] [PubMed] [Google Scholar]
- 24.Gao Z, Krithivas A, Finan J E, Semmes O J, Zhou S, Wang Y, Hayward S D. The Epstein-Barr virus proteins lytic transactivator Zta interacts with the helicase-primase replication. J Virol. 1998;72:8559–8567. doi: 10.1128/jvi.72.11.8559-8567.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hadzopoulou-Cladaras M. The Rev (Trs/Art) protein of human immunodeficiency virus type 1 affects viral mRNA and protein expression via a cis-acting sequence in the env region. J Virol. 1989;63:1265–1274. doi: 10.1128/jvi.63.3.1265-1274.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hammarskjold M-L, Heimer J, Hammarskjold B, Sangwan I, Albert L, Rekosh D. Regulation of human immunodeficiency virus env expression by the rev gene product. J Virol. 1989;63:1959–1966. doi: 10.1128/jvi.63.5.1959-1966.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hardy W R, Sandri-Goldin R M. Herpes simplex virus inhibits host cell splicing, and regulatory protein ICP27 is required for this effect. J Virol. 1994;68:7790–7799. doi: 10.1128/jvi.68.12.7790-7799.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Henkel T, Ling P D, Hayward S D, Peterson M G. Mediation of Epstein-Barr virus EBNA2 transactivation by recombination signal-binding protein Jk. Science. 1994;265:92–95. doi: 10.1126/science.8016657. [DOI] [PubMed] [Google Scholar]
- 29.Hibbard M K, Sandri-Goldin R M. Arginine-rich regions succeeding the nuclear localization region of the herpes simplex virus type 1 regulatory protein ICP27 are required for efficient nuclear localization and late gene expression. J Virol. 1995;69:4656–4667. doi: 10.1128/jvi.69.8.4656-4667.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hidaka M, Inoue J, Yoshida M, Seiki M. Post-transcriptional regulator (rex) of HTLV-1 initiates expression of viral structural proteins but suppresses expression of regulatory proteins. EMBO J. 1998;7:519–523. doi: 10.1002/j.1460-2075.1988.tb02840.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hope T J. Viral RNA export. Chem Biol. 1997;4:335–344. doi: 10.1016/s1074-5521(97)90124-1. [DOI] [PubMed] [Google Scholar]
- 32.Inchauspe G, Ostrove J M. Differential regulation by varicella-zoster virus (VZV) and herpes simplex virus type-1 trans-activating genes. Virology. 1989;173:710–714. doi: 10.1016/0042-6822(89)90584-9. [DOI] [PubMed] [Google Scholar]
- 33.Ingram A, Phelan A, Dunlop J, Clements J B. Immediate early protein IE63 of herpes simplex virus type 1 binds RNA directly. J Gen Virol. 1996;77:1847–1851. doi: 10.1099/0022-1317-77-8-1847. [DOI] [PubMed] [Google Scholar]
- 34.Kalland K H, Szilvay A M, Brokstad K A, Saetrevik W, Haukenes G. The human immunodeficiency virus type 1 Rev protein shuttles between the cytoplasm and nuclear compartments. Mol Cell Biol. 1994;14:7436–7444. doi: 10.1128/mcb.14.11.7436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kalland K H, Szilvay A M, Langhohff E, Haukenes G. Subcellular distribution of human immunodeficiency virus type 1 Rev and colocalization of Rev with RNA splicing factors in a speckled pattern in the nucleoplasm. J Virol. 1994;68:1475–1485. doi: 10.1128/jvi.68.3.1475-1485.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kenney S, Holley-Guthrie E, Mar E C, Smith M. The Epstein-Barr virus BMLF1 promoter contains an enhancer element that is responsive to the BZLF1 and BRLF1 transactivators. J Virol. 1989;63:3878–3883. doi: 10.1128/jvi.63.9.3878-3883.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kenney S, Kamine J, Holley-Guthrie E, Mar E C, Lin J C, Markovitz D, Pagano J. The Epstein-Barr virus immediate-early gene product, BMLF1, acts in trans by posttranscriptional mechanism which is reporter gene dependent. J Virol. 1989;63:3870–3877. doi: 10.1128/jvi.63.9.3870-3877.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kim F J, Beeche A A, Hunter J J, Chin D J, Hope T J. Characterization of the nuclear export signal of human T-cell lymphotropic virus type 1 Rex reveals that nuclear export is mediated by position-variable hydrophobic interactions. Mol Cell Biol. 1996;16:5147–5155. doi: 10.1128/mcb.16.9.5147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kjems J, Brown M, Chang D D, Sharp P A. Structural analysis of the interaction between the human immunodeficiency virus Rev protein and the Rev response element. Proc Natl Acad Sci USA. 1991;88:683–687. doi: 10.1073/pnas.88.3.683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lieberman P M, O’Hare P, Hayward G S, Hayward S D. Promiscuous trans-activation of gene expression by an Epstein-Barr virus-encoded early nuclear protein. J Virol. 1986;60:140–148. doi: 10.1128/jvi.60.1.140-148.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Malim M H, Hauber J, Le S Y, Maizel J V, Cullen B R. The HIV-1 rev trans-activator acts through a structured target sequence to activate nuclear export of unspliced viral mRNA. Nature. 1989;338:254–257. doi: 10.1038/338254a0. [DOI] [PubMed] [Google Scholar]
- 42.McCarthy A M, McMahan L, Schaffer P A. Herpes simplex virus type 1 ICP27 deletion mutants exhibit altered patterns of transcription and are DNA deficient. J Virol. 1989;63:18–27. doi: 10.1128/jvi.63.1.18-27.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.McGregor F, Phelan A, Dunlop J, Clements J B. Regulation of herpes simplex virus poly(A) site usage and the action of immediate-early protein IE63 in the early-late switch. J Virol. 1996;70:1931–1940. doi: 10.1128/jvi.70.3.1931-1940.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.McLauchlan J, Phelan A, Loney C, Sandri-Goldin R M, Clements J B. Herpes simplex virus IE63 acts at the posttranscriptional level to stimulate viral mRNA 3′ processing. J Virol. 1992;66:6939–6945. doi: 10.1128/jvi.66.12.6939-6945.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.McMahan L, Schaffer P A. The repressing and enhancing functions of the herpes simplex virus regulatory protein ICP27 map to C-terminal regions and are required to modulate viral gene expression very early in infection. J Virol. 1990;64:3471–3485. doi: 10.1128/jvi.64.7.3471-3485.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mears W E, Rice S A. The RGG box motif of the herpes simplex virus ICP27 protein mediates an RNA-binding activity and determines in vivo methylation. J Virol. 1996;70:7445–7453. doi: 10.1128/jvi.70.11.7445-7453.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mellinghoff I, Daibata M, Humphreys R E, Mulder C, Takada K, Sairenji T. Early events in Epstein-Barr virus genome expression after activation: regulation by second messengers of B cell activation. Virology. 1991;185:922–928. doi: 10.1016/0042-6822(91)90574-u. [DOI] [PubMed] [Google Scholar]
- 48.Meyer B E, Malim M H. The HIV-1 Rev trans-activator shuttles between the nucleus and the cytoplasm. Genes Dev. 1994;8:1538–1547. doi: 10.1101/gad.8.13.1538. [DOI] [PubMed] [Google Scholar]
- 49.Meyer B E, Meinkoth J L, Malim M H. Nuclear transport of human immunodeficiency virus type 1, visna virus, and equine infectious anemia virus Rev proteins: identification of a family of transferable nuclear export signals. J Virol. 1996;70:2350–2359. doi: 10.1128/jvi.70.4.2350-2359.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Moriuchi H, Moriuchi M, Smith H A, Cohen J I. Varicella-zoster virus open reading frame 4 protein is functionally distinct from and does not complement its herpes simplex virus type 1 homolog, ICP27. J Virol. 1994;68:1987–1992. doi: 10.1128/jvi.68.3.1987-1992.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Murphy R, Wente S R. An RNA-export mediator with an essential nuclear export signal. Nature. 1996;383:357–360. doi: 10.1038/383357a0. [DOI] [PubMed] [Google Scholar]
- 52.Nicholas J, Cameron K R, Coleman H, Newman C, Honess R W. Analysis of nucleotide sequence of the rightmost 43 kbp of herpesvirus saimiri (HSV) L-DNA: general conservation of genetic organization between HVS and Epstein-Barr virus. Virology. 1992;188:296–310. doi: 10.1016/0042-6822(92)90759-i. [DOI] [PubMed] [Google Scholar]
- 53.Nicholas J, Gompels U A, Craxton M A, Honess R W. Conservation of sequence and function between the product of the 52-kilodalton immediate-early gene of herpesvirus saimiri and the BMLF1-encoded transcriptional effector (EB2) of Epstein-Barr virus. J Virol. 1988;62:3250–3257. doi: 10.1128/jvi.62.9.3250-3257.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Oguro M O, Shimizu N, Ono Y, Takada K. Both the rightward and the leftward open reading frames within the BamHI M DNA fragment of Epstein-Barr virus act as trans-activators of gene expression. J Virol. 1987;61:3310–3313. doi: 10.1128/jvi.61.10.3310-3313.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ossareh-Nazari B, Bachelerie F, Dargemont C. Evidence for a role of CRM1 in signal-mediated nuclear protein export. Science. 1997;278:141–144. doi: 10.1126/science.278.5335.141. [DOI] [PubMed] [Google Scholar]
- 56.Pasquinelli A E, Powers M A, Lund E, Forbes D, Dahlberg J E. Inhibition of mRNA export in vertebrate cells by nuclear export signal conjugates. Proc Natl Acad Sci USA. 1997;94:14393–14399. doi: 10.1073/pnas.94.26.14394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Perera L P, Kaushal S, Kinchington P R, Mosca J D, Hayward G S, Strauss S E. Varicella-zoster virus open reading frame 4 encodes a transcriptional activator that is functionally distinct from that of herpes simplex virus homology ICP27. J Virol. 1994;68:2468–2477. doi: 10.1128/jvi.68.4.2468-2477.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Pfitzner A J, Strominger J L, Speck S H. Characterization of a cDNA clone corresponding to a transcript from the Epstein-Barr virus BamHI M fragment: evidence for overlapping mRNAs. J Virol. 1987;61:2943–2946. doi: 10.1128/jvi.61.9.2943-2946.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Phelan A, Carmo-Fonscca M, McLauchlaw J, Lamond A I, Clements J B. A herpes simplex virus type 1 immediate-early gene product, IE63, regulates small nuclear ribonucleoprotein distribution. Proc Natl Acad Sci USA. 1993;90:9056–9060. doi: 10.1073/pnas.90.19.9056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Phelan A, Clements J B. Herpes simplex virus type 1 immediate early protein IE63 shuttles between nuclear compartments and the cytoplasm. J Gen Virol. 1997;78:3327–3331. doi: 10.1099/0022-1317-78-12-3327. [DOI] [PubMed] [Google Scholar]
- 61.Phelan A, Dunlop J, Clements J B. Herpes simplex virus type 1 protein IE63 affects the nuclear export of virus intron-containing transcripts. J Virol. 1996;70:5255–5265. doi: 10.1128/jvi.70.8.5255-5265.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ren D, Lee L F, Coussens P M. Identification and characterization of Marek’s disease virus genes homologus to ICP27 and glycoprotein K of herpes simplex virus-1. Virology. 1994;204:242–250. doi: 10.1006/viro.1994.1528. [DOI] [PubMed] [Google Scholar]
- 63.Rice S A, Knipe D M. Genetic evidence for two distinct transactivation functions of the herpes simplex virus alpha protein ICP27. J Virol. 1990;64:1704–1715. doi: 10.1128/jvi.64.4.1704-1715.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Rice S A, Lam V. Amino acid substitution mutations in the herpes simplex virus ICP27 protein define an essential gene regulation function. J Virol. 1994;68:823–833. doi: 10.1128/jvi.68.2.823-833.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Rice S A, Lam V, Knipe D M. The acidic amino-terminal region of herpes simplex virus type 1 alpha protein ICP27 is required for an essential lytic function. J Virol. 1993;67:1778–1787. doi: 10.1128/jvi.67.4.1778-1787.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Russo J J, Bohenzky R A, Chein M-C, Chen J, Yan M, Maddalena D, Parry J P, Peruzzi D, Edelman I S, Chang Y, Moore P S. Nucleotide sequence of the Kaposi sarcoma-associated herpesvirus (HHV8) Proc Natl Acad Sci USA. 1996;93:14862–14867. doi: 10.1073/pnas.93.25.14862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Sample J, Lancz G, Nonoyama M. Mapping of genes in BamHI fragment M of Epstein-Barr virus DNA that may determine the fate of viral infection. J Virol. 1986;57:145–154. doi: 10.1128/jvi.57.1.145-154.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Sandri-Goldin R M. ICP27 mediates HSV RNA export by shuttling through a leucine-rich nuclear export signal and binding viral intronless RNAs through an RGG motif. Genes Dev. 1998;12:868–879. doi: 10.1101/gad.12.6.868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Sandri-Goldin R M, Hibbard M K. The herpes simplex virus type 1 regulatory protein ICP27 coimmunoprecipitates with anti-Sm antiserum, and the C terminus appears to be required for this interaction. J Virol. 1996;70:108–118. doi: 10.1128/jvi.70.1.108-118.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Sandri-Goldin R M, Hibbard M K, Hardwicke M A. The C-terminal repressor region of herpes simplex virus type 1 ICP27 is required for the redistribution of small nuclear ribonucleoprotein particles and splicing factor SC35; however, these alterations are not sufficient to inhibit host cell splicing. J Virol. 1995;69:6063–6076. doi: 10.1128/jvi.69.10.6063-6076.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Sandri-Goldin R M, Mendoza G E. A herpesvirus regulatory protein appears to act post-transcriptionally by affecting mRNA processing. Genes Dev. 1992;6:848–863. doi: 10.1101/gad.6.5.848. [DOI] [PubMed] [Google Scholar]
- 72.Sarisky R T, Gao Z, Lieberman P M, Fixman E D, Hayward G S, Hayward S D. A replication function associated with the activation domain of the Epstein-Barr virus Zta transactivator. J Virol. 1996;70:8340–8347. doi: 10.1128/jvi.70.12.8340-8347.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Schepers A, Pich D, Hammerschmidt W. Activation of oriLyt, the lytic origin of DNA replication of Espstein-Barr virus, BZLF1. Virology. 1996;220:367–376. doi: 10.1006/viro.1996.0325. [DOI] [PubMed] [Google Scholar]
- 74.Schepers A, Pich D, Hammerschmidt W. A transcription factor with homology to the AP-1 family links RNA transcription and DNA replication in the lytic cycle of Epstein-Barr virus. EMBO J. 1993;12:3921–3929. doi: 10.1002/j.1460-2075.1993.tb06070.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Schoborg R V, Clements J E. Definition of the RRE binding and activation domains of the caprine arthritis encephalitis virus Rev protein. Virology. 1996;226:113–121. doi: 10.1006/viro.1996.0633. [DOI] [PubMed] [Google Scholar]
- 76.Seedorf M, Silver P A. Importin/karyopherin protein family members required for mRNA export from the nucleus. Proc Natl Acad Sci USA. 1997;94:8590–8595. doi: 10.1073/pnas.94.16.8590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Seiki M, Inoue J, Hidaka M, Yoshida M. Two cis-acting elements responsible for posttranscriptional trans-regulation of gene expression of human T-cell leukemia virus type 1. Proc Natl Acad Sci USA. 1988;85:7124–7128. doi: 10.1073/pnas.85.19.7124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Sinclair A J, Brimmel M, Shanahan F, Farrell P J. Pathways of activation of the Epstein-Barr virus productive cycle. J Virol. 1991;65:2237–2244. doi: 10.1128/jvi.65.5.2237-2244.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Singh M, Fraefel C, Bello L J, Lawrence W C, Schwyzer M. Identification and characterization of BICP27, an early protein of bovine herpesvirus 1 which may stimulate mRNA 3′ processing. J Gen Virol. 1996;77:615–625. doi: 10.1099/0022-1317-77-4-615. [DOI] [PubMed] [Google Scholar]
- 80.Smith I L, Hardwicke M A, Sandri-Goldin R M. Evidence that the herpes simplex virus immediate early protein ICP27 acts post-transcriptionally during infection to regulate gene expression. Virology. 1992;186:74–86. doi: 10.1016/0042-6822(92)90062-t. [DOI] [PubMed] [Google Scholar]
- 81.Smith I L, Sekulovich R E, Hardwicke M A, Sandri-Goldin R M. Mutations in the activation region of herpes simplex virus regulatory protein ICP27 can be trans dominant. J Virol. 1991;65:3656–3666. doi: 10.1128/jvi.65.7.3656-3666.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Smith R H, Zhao Y, O’Callaghan D J. The equine herpesvirus 1 (EHV-1) UL3 gene, an ICP27 homolog, is necessary for full activation of gene expression directed by an EHV-1 late promoter. J Virol. 1993;67:1105–1109. doi: 10.1128/jvi.67.2.1105-1109.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Soliman T M, Sandri-Goldin R M, Silverstein S J. Shuttling of the herpes simplex virus type 1 regulatory protein ICP27 between the nucleus and cytoplasm mediates the expression of late proteins. J Virol. 1997;71:9188–9197. doi: 10.1128/jvi.71.12.9188-9197.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Stade K, Ford C S, Guthrie C, Weis K. Exportin 1 (Crm1p) is an essential nuclear export factor. Cell. 1997;90:1041–1050. doi: 10.1016/s0092-8674(00)80370-0. [DOI] [PubMed] [Google Scholar]
- 85.Takada K, Ono Y. Synchronous and sequential activation of latently infected Epstein-Barr virus genomes. J Virol. 1989;63:445–449. doi: 10.1128/jvi.63.1.445-449.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Ullman K S, Powers M A, Forbes D J. Nuclear export receptors: from importin to exportin. Cell. 1997;90:967–970. doi: 10.1016/s0092-8674(00)80361-x. . (Comment.) [DOI] [PubMed] [Google Scholar]
- 87.Uprichard S L, Knipe D M. Herpes simplex ICP27 mutant viruses exhibit reduced expression of specific DNA replication genes. J Virol. 1996;70:1969–1980. doi: 10.1128/jvi.70.3.1969-1980.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Wang Y, Finan J E, Middeldorp J M, Hayward S D. P32/TAP, a cellular protein that interacts with EBNA-1 of Epstein-Barr virus. Virology. 1997;236:18–29. doi: 10.1006/viro.1997.8739. [DOI] [PubMed] [Google Scholar]
- 89.Wen W, Meinkoth J L, Tsien R Y, Taylor S S. Identification of a signal for rapid export of proteins from the nucleus. Cell. 1995;82:463–473. doi: 10.1016/0092-8674(95)90435-2. [DOI] [PubMed] [Google Scholar]
- 90.Whitehouse A, Cooper M, Meredith D M. The immediate-early gene product encoded by open reading frame 57 of herpesvirus saimiri modulates gene expression at a posttranscriptional level. J Virol. 1998;72:857–861. doi: 10.1128/jvi.72.1.857-861.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Winkler M, Rice S A, Stamminger T. UL69 of human cytomegalovirus, an open reading frame with homology to ICP27 of herpes simplex virus, encodes a transactivator of gene expression. J Virol. 1994;68:3943–3954. doi: 10.1128/jvi.68.6.3943-3954.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Wong K M, Levine A J. Characterization of proteins encoded by the Epstein-Barr virus transactivator gene BMLF1. Virology. 1989;168:101–111. doi: 10.1016/0042-6822(89)90408-x. [DOI] [PubMed] [Google Scholar]
- 93.Zhao Y, Holden V R, Harty R N, O’Callaghan D J. Identification and transcriptional analyses of the UL3 and UL4 genes of equine herpesvirus 1, homologs of the ICP27 and glycoprotein K genes of herpes simplex virus. J Virol. 1992;66:5363–5372. doi: 10.1128/jvi.66.9.5363-5372.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Zhao Y, Holden V R, Smith R H, O’Callaghan D J. Regulatory function of the equine herpesvirus 1 ICP27 gene product. J Virol. 1995;69:2786–2793. doi: 10.1128/jvi.69.5.2786-2793.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Zhong L, Hayward G S. Assembly of complete, functionally active herpes simplex virus DNA replication compartments and recruitment of associated viral and cellular proteins in transient cotransfection assays. J Virol. 1997;3146:3160. doi: 10.1128/jvi.71.4.3146-3160.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]