Abstract
Different tombusviruses were able to support the replication of either homologous or heterologous defective interfering (DI) RNAs, and those infected plants usually developed typical attenuated symptoms. However, in some helper virus-DI RNA combinations the inoculated plants were necrotized, although they contained a high level of DI RNA, suggesting that the accumulation of DI RNA and the resulting suppression of genomic RNA replication were not directly responsible for the symptom attenuation. Moreover, the 19-kDa protein product of ORF 5, which is known to play a crucial role in necrotic symptom development, accumulated at the same level in the infected plants in the presence of protective homologous DI RNA and in the presence of nonprotective heterologous DI RNA. It was also demonstrated, by chimeric helper viruses, that the ability of heterologous DI RNA to protect the virus-infected plants against systemic necrosis is determined by the 5′-proximal region of the helper virus genome. The results presented suggest that DI RNA-mediated protection did not operate via the specific inhibition of 19-kDa protein expression but, more likely, DI RNAs in protective DI-helper virus combinations specifically interacted with viral products, preventing the induction of necrotic symptoms.
Defective interfering (DI) RNAs are shortened forms of viral genomes that have generally lost all essential viral genes for movement, replication, and encapsidation. DI RNAs require the presence of a helper virus to provide trans-acting factors necessary for replication and often multiply and accumulate at the expense of the helper virus from which they originated. Interference with the helper virus frequently results in remarkable symptom attenuation (16, 18). The most extensively studied plant virus DI RNA systems are those found in association with tombusviruses and carmoviruses (18, 25).
The genome of a tombusvirus is a linear, single-stranded monopartite RNA molecule of positive polarity, about 4,700 nucleotides long, that contains five open reading frames (ORFs) coding for proteins with approximate molecular masses of 33, 92, 22, and 19 kDa and for the coat protein (41 kDa) (18). The genomic RNA (ca. 4,700 nucleotides) acts as mRNA for the production of a 33-kDa protein (p33; ORF 1), and by readthrough of an amber termination codon, a 92-kDa protein (p92; ORF 2) is synthesized. It was demonstrated elsewhere that both p33 and p92 are required for viral replication (6, 12, 23). The 41- kDa coat protein (ORF 3) is translated from the subgenomic RNA 1 (sg1 RNA) (18). The two nested ORFs (4 and 5) are located at the 3′ terminus of the virus genome, encoding a 22- kDa (p22) protein and a 19-kDa (p19) protein, respectively. Both p22 and p19 are translated from sg2 RNA (18). p22 is required for cell-to-cell movement (6, 15, 20) and is also involved in symptom determination (21). Although the precise function of p19 has not been elucidated, it has an important role in necrotic symptom development (6, 15, 21). It is also suggested that p19 participates in virus spread in a host-specific manner (22).
Several reports from different laboratories demonstrated the presence and generation of DI RNAs in infection with tombusviruses: tomato bushy stunt virus cherry strain (TBSV- Ch [9]), Cymbidium ringspot virus (CymRSV [2, 3]), cucumber necrosis virus (7, 14), carnation Italian ringspot virus (CIRV [17]), and tomato bushy stunt virus pepper isolate (TBSV- P [this paper]). Sequence analyses revealed that tombusvirus DI RNAs possess a common structural arrangement containing conserved sequence blocks derived from the 5′-proximal terminus, an internal region of the replicase gene, and the 3′-proximal terminus of the viral genome (18, 26). The presence of DI RNAs in virus-infected plants suppresses the virus accumulation and attenuates the lethal necrotic symptoms regardless of whether DI RNA was in the inoculum or was provided by transgenic Nicotiana benthamiana plants expressing CymRSV DI-13 RNA (11). The implications of tombusvirus DI RNAs in symptom attenuation were considered on two levels. A general competition for viral replicase between the genomic RNA and the more competitive DI RNAs probably occurs (10), and symptom attenuation appears via a selective inhibition of expression of p19 and p22, which are important symptom determinants (24).
The aim of the present study was to analyze the mechanism of DI RNA-mediated symptom attenuation in tombusvirus-infected plants containing either homologous or heterologous DI RNAs. For this purpose, we used the previously described biologically active cDNA clones of genomic RNAs of CymRSV (6) and CIRV (3) and the corresponding DI RNAs (DI-13 RNA of CymRSV [11] and DI-7 RNA of CIRV [17]). In addition, a new tombusvirus (TBSV- P) was isolated from pepper plants (Capsicum annum L.) in a Hungarian greenhouse. The virus genome was cloned and sequenced, and a full-length TBSV- P cDNA clone was prepared (4), from which highly infectious in vitro RNA transcripts could be transcribed with T7 RNA polymerase (6). DI RNAs, related to TBSV- P, were generated by inoculation of Nicotiana clevelandii with synthetic genomic RNA followed by serial subinoculation. The predominant DI-like RNA was purified, cloned, and sequenced as previously described (17). The entire sequence of TBSV- P DI RNA (DI-5) was 550 nucleotides long, completely derived from the genomic RNA displaying a typical tombusvirus DI RNA composition (18). The full-length cDNA clone of TBSV- P DI-5 RNA was prepared (11) in order to transcribe DI-5 RNA in vitro.
Symptom development and DI RNA accumulation in DI RNA-expressing transgenic plants challenge inoculated with different tombusviruses.
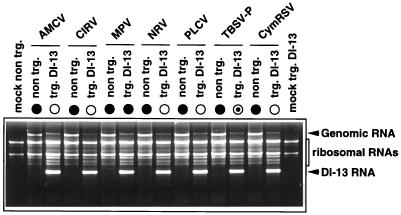
Previously prepared CymRSV DI-13 RNA-expressing transgenic N. benthamiana plants (11) were used to analyze the ability of different members of the genus Tombusvirus to support the replication and accumulation of heterologous DI RNA. Typically, three leaves of N. benthamiana transgenic plants were inoculated with 1 μg of the following per leaf: sucrose gradient-purified artichoke mottled crinkle virus, Moroccan pepper virus (MPV) (13), Neckar river virus (13), or pelargonium leaf curl virus (13) or in vitro-transcribed genomic RNA of CymRSV, CIRV, and TBSV- P. A group of six transgenic plants were inoculated with each of the above-listed viruses in two repeated experiments. All viruses used for inoculation were able to support the replication of the expressed CymRSV DI-13 RNA with similar efficiencies, and no DI RNA accumulation was observed in control nontransgenic plants (Fig. 1). Surprisingly, the infected transgenic plants displayed different symptoms. The typical DI RNA-mediated attenuated symptoms developed in plants inoculated with CymRSV, artichoke mottled crinkle virus, CIRV, pelargonium leaf curl virus, and Neckar river virus, while the nontransgenic control plants were all necrotized (data not shown). In contrast, each of the MPV-infected transgenic plants showed apical necrosis and died 2 to 3 weeks postinoculation (data not shown), although they contained a high level of DI RNA (Fig. 1). Furthermore, the TBSV- P-infected DI-13 transgenic plants exhibited only partial protection against challenge virus. Approximately 20% of the inoculated plants were necrotized and died, and the 80% of plants surviving were also strongly stunted and partially necrotized. The DI RNA accumulation in all of these plants was similar, regardless of whether they were necrotized and died or were protected (Fig. 1).
FIG. 1.
Accumulation of DI RNA in CymRSV DI-13 RNA-expressing transgenic plants challenge inoculated with different tombusviruses. The ethidium bromide-stained 1.2% agarose gel shows total RNA samples extracted 7 days after inoculation from systemically infected transgenic (trg) or nontransgenic N. benthamiana plants challenge inoculated with different tombusviruses as indicated above the lanes. Symbols • and ○ indicate whether the necrotic symptoms appeared or not, respectively, on the systemically infected leaves. Mock indicates the mock-inoculated transgenic and nontransgenic plants. AMCV, artichoke mottled crinkle virus; NRV, Neckar river virus; PLCV, pelargonium leaf curl virus.
Symptom modulation of CIRV, CymRSV, and TBSV- P DI RNAs coinoculated with homologous or heterologous helper viruses.
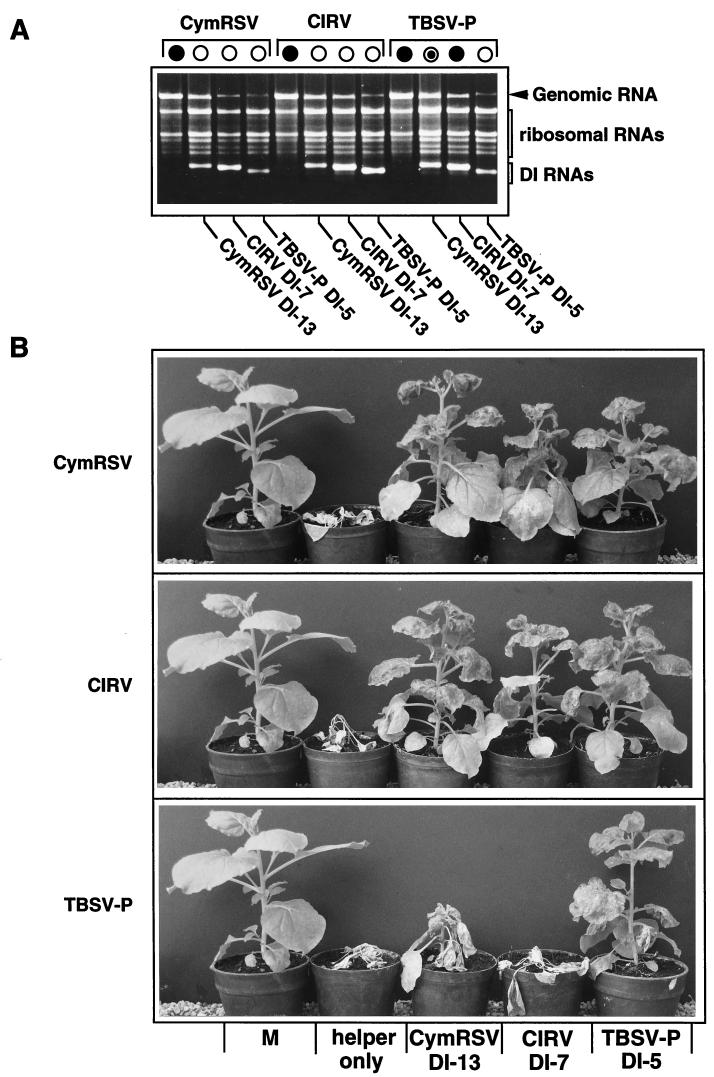
Further studies were undertaken to test the replication and symptom modulation of related tombusvirus DI RNAs in the presence of homologous and heterologous helper virus genomes. In vitro RNA transcripts of CymRSV, CIRV, and TBSV- P (1 μg of each) were coinoculated into N. benthamiana (six plants) with in vitro-synthesized CymRSV DI-13 RNA (3), CIRV DI-7 RNA (17), and TBSV- P DI-5 RNA (250 ng of each), respectively. As was expected, all of the helper virus genomes supported the replication of both homologous and heterologous DI RNAs; however, some variation in DI RNA amplification occurred (Fig. 2A). The symptoms of these plants inoculated with different helper virus-DI combinations varied from complete protection to complete necrosis. CymRSV- and CIRV-infected plants showed typical attenuated symptoms, regardless of whether the inoculum contained homologous or heterologous DI RNAs (Fig. 2B). In contrast, plants inoculated with TBSV- P and the DI-7 RNA of CIRV were necrotized and died 5 to 7 days later than the control plants inoculated with helper virus alone, although the DI-7 RNA replicated very efficiently (Fig. 2). The TBSV- P– CymRSV DI-13 RNA combination exhibited only slight protection (Fig. 2B), similarly to the TBSV- P-infected DI-13 transgenic plants. The accumulation of CymRSV DI-13 RNA in these plants was as high as that of homologous DI-5 RNA. All control plants inoculated only with different helper viruses died (Fig. 2B). We should emphasize that there was no variation in the developed symptoms (necrosis or protection) among the plants which were inoculated with the same inoculum. The only exception was the TBSV- P–CymRSV DI-13 RNA inoculum, with the plants displaying intermediate and variable symptoms (Fig. 2B).
FIG. 2.
Symptom development and the accumulation of DI and genomic RNAs in plants inoculated with in vitro RNA transcripts of genomic and DI RNAs of CymRSV, CIRV, and TBSV- P. (A) Ethidium bromide-stained 1.2% agarose gel showing total RNA samples extracted 7 days after inoculation from N. benthamiana plants inoculated with homologous or heterologous helper virus-DI combinations. Different helper viruses are indicated at the top and DI RNAs are indicated at the bottom of the gel. Symbols are defined in the legend to Fig. 1. (B) Photo presentation of symptom development in the plants inoculated with CymRSV, CIRV, and TBSV- P (indicated at the left) in the presence of heterologous and homologous DI RNAs (indicated at the bottom). The photos were taken 3 weeks after inoculation. M, mock infection.
Effect of homologous and heterologous DI RNA accumulation on the replication of TBSV- P in N. benthamiana plants.
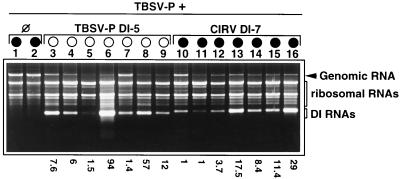
A group of seven N. benthamiana plants was inoculated with TBSV- P and either TBSV- P DI-5 (protective) or CIRV DI-7 (nonprotective) RNA. As was expected, all plants inoculated with TBSV- P and TBSV- P DI-5 RNA showed typical DI RNA attenuated symptoms, while coinoculation of TBSV- P with CIRV DI-7 RNA resulted in complete necrosis (Fig. 2B). In previous experiments, total RNA extracts from different systemically infected leaves of the same plants showed considerable variation in the level of genomic and DI RNAs (data not shown), indicating that the virus did not invade infected plant tissues homogeneously. To reduce sampling mistakes and estimate the helper virus/DI RNA ratio characteristic for the whole plant, total RNA extracts were made from samples taken from three different systemically infected leaves from the same plant and homogenized together. RNA was extracted from these mixed samples and analyzed (6). The presence of DI RNA in the TBSV- P inoculum usually, but not always, resulted in a restricted genomic RNA accumulation, regardless of whether the DI RNAs were protective (TBSV- P DI-5 RNA [Fig. 3, lanes 3 to 9]) or nonprotective (CIRV DI-7 RNA [Fig. 3, lanes 10 to 16]). The DI/genomic RNA ratio in inoculated plants varied between 1:1.4 and 1:94 for TBSV- P and TBSV- P DI-5 RNA and between 1:1 and 1:29 for TBSV- P and CIRV DI-7 RNA (Fig. 3). The observed high variation in the DI/genomic ratio did not result in variability in symptom development. For example, a high level of genomic RNA was detected in the presence of TBSV- P DI-5 RNA, but the plants were protected from necrosis (Fig. 3, lane 7), while in contrast, the replication of genomic RNA was highly reduced in the presence of various amounts of CIRV DI-7 RNA, but the plants were necrotized (Fig. 3, lanes 11 to 16). This experiment was repeated five times, and the results were always the same: considerable variation in the level of DI and genomic RNAs, regardless of which DI-helper virus combination was used. However, this variation was not reflected in the symptoms. Plants inoculated with TBSV- P and DI-5 RNAs always showed attenuated symptoms. In contrast, inoculation of plants with TBSV- P and DI-7 RNAs resulted in complete necrosis. These results clearly demonstrate that the general restriction of helper virus genomic RNA accumulation does not play a crucial role in DI RNA-mediated symptom attenuation and a relatively low level of protective DI RNA is sufficient for protection.
FIG. 3.
The accumulation of TBSV- P genomic RNA in the presence of TBSV- P DI-5 RNA (homologous) and CIRV DI-7 RNA (heterologous). The ethidium bromide-stained 1.2% agarose gel shows total RNA samples extracted 7 days after inoculation from N. benthamiana plants inoculated with homologous or heterologous helper virus-DI combinations as indicated at the top of the gel. Symbols are defined in the legend to Fig. 1. The numbers underneath indicate the DI genomic/RNA ratios, which were measured on the photograph by Analysis 2.0 (Soft-Imaging Software GmbH). A series of viral RNA samples containing 50, 75, 100, 150, 200, 300, 500, and 1,000 ng of RNA were used to create a standard curve to determine the amount of genomic and DI RNAs in the total nucleic acid extracts.
Accumulation of p19 and subgenomic RNA in the presence of protective and nonprotective DI RNAs.
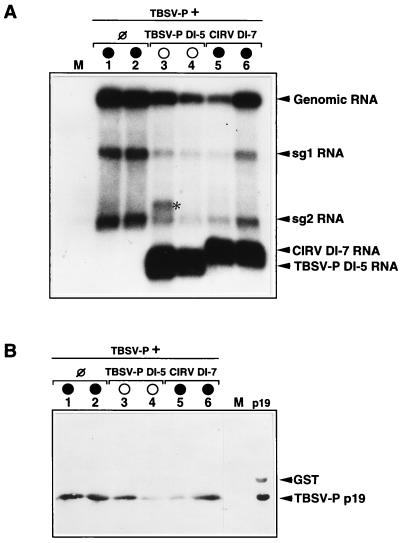
The absence of p19 or the analogous product of ORF 5 of tombusviruses resulted in symptom attenuation, and the infected plants were protected from apical necrosis and death (6, 15, 21). Therefore, we analyzed p19 and its sg2 RNA accumulation in TBSV- P-infected plants in the presence of TBSV- P DI-5 (protective) and CIRV DI-7 (nonprotective) RNAs. Northern and Western blot analyses of the same samples indicated that the accumulation of sg2 RNA and p19 correlates with the reduced accumulation of the genomic RNA regardless of the protective (TBSV- P DI-5) (Fig. 4, lanes 3 and 4) or the nonprotective (CIRV DI-7) (Fig. 4, lanes 6 and 7) nature of coreplicating DI RNAs. However, no correlation was found between the level of expressed p19 and necrotic syndrome. For instance, a low level of p19 was detected in the plant containing nonprotective CIRV DI-7 RNA just prior to the development of generalized necrosis (Fig. 4B, lane 5), and in contrast, an almost-wild-type level of p19 was detected in the TBSV- P DI-5-protected plant (Fig. 4B, lane 3). It was found again that symptom attenuation is exclusively dependent on the protective or nonprotective nature of DI RNA used for inoculation. The experiment shown in Fig. 4 was repeated three times, and the results demonstrated that the accumulation of homologous and heterologous DI RNAs in TBSV- P-infected plants did not specifically inhibit the expression of sg2 RNA or that of p19, but that inhibition occurred via a general restriction of virus replication. Furthermore, the level of p19, just above detection, was sufficient for the development of necrotic syndrome, which is very characteristic of tombusvirus infection.
FIG. 4.
Accumulation of viral RNAs and p19 in the plants inoculated with TBSV- P plus TBSV- P DI-5 or CIRV DI-7 RNA. Samples were taken from three different leaves of one plant and were first homogenized and then divided into halves which were used for Northern (A) and Western (B) blot analyses (19). Lane numbers correspond to the sample number from which both RNA and protein were extracted. A 32P-labeled probe specific to the 3′ terminus of TBSV- P was used for hybridization. The polyclonal antibody used was raised against CymRSV p19 expressed in bacterial strain BL21 and then purified with a glutathione-Sepharose 4B column according to the manufacturer’s protocol (Pharmacia Biotech). GST indicates the cleaved fusion component of the expressed recombinant protein. M and p19 indicate mock-inoculated plants and purified p19, respectively. The asterisk indicates the position of dimer form TBSV- P DI-5. Other symbols are defined in the legend to Fig. 1.
The 5′-terminal region of the helper virus genome determined the protective nature of the DI RNA.
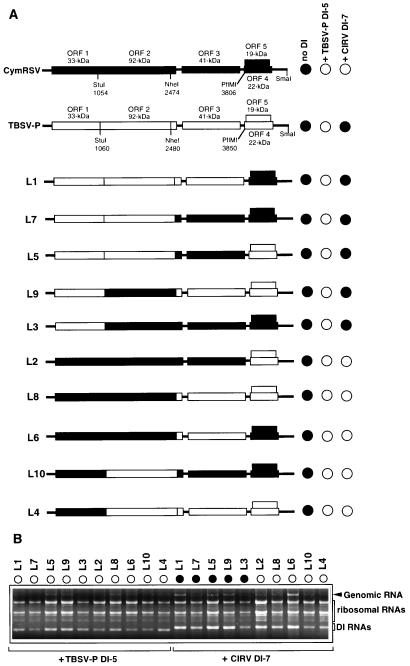
The DI-7 RNA of CIRV was able to protect the CymRSV- but not the TBSV- P-infected plants against the virus-induced lethal necrosis (Fig. 2B). This striking difference in the interference of CIRV DI-7 RNA with the two different helper viruses prompted us to attempt to identify which part of the viral genome interacts with DI RNA. For this purpose, CymRSV–TBSV- P chimeras were constructed (Fig. 5A), by exchanging viral genes and sequences with the common restriction sites (4). RNA transcripts from all CymRSV–TBSV- P chimeras were infectious and were able to elicit necrotic symptoms on the systemically infected N. benthamiana plants (data not shown). Then, plants were coinoculated with the different chimeras and TBSV- P DI-5 or CIRV DI-7 RNAs, and the appearance of necrotic symptoms on the systemically infected leaves was monitored. Coinoculation experiments demonstrated that all of the chimeras were able to support the replication both of TBSV- P DI-5 and of CIRV DI-7 RNAs efficiently (Fig. 5B). As was expected from previously reported experiments, TBSV- P DI-5 RNA protected the infected plants against all of the chimeras. However, the symptom-modulating ability of CIRV DI-7 RNA depended on the helper virus construction. Plants inoculated with TBSV- P or chimeras L1, L3, L5, L7, and L9, containing the 5′ noncoding sequence and ORF 1 of TBSV- P in the presence of CIRV DI-7 RNA, showed necrotic symptoms on the systemically infected leaves 10 to 14 days postinoculation (Fig. 5). In contrast, those plants inoculated with CymRSV or chimeras L2, L4, L6, L8, and L10, containing the 5′ noncoding sequence and ORF 1 of CymRSV, were protected in the presence of CIRV DI-7 RNA (Fig. 5). It is worth noting that the comparison of ORF 1 of CymRSV and that of TBSV- P shows only 83% amino acid sequence identity, whereas the readthrough product (ORF 2) shows 90% amino acid sequence identity. These observations indicated that the 5′-proximal region of the viral genomes, including the 5′ noncoding sequence and ORF 1, determines the nature of interference between DI RNA and helper virus.
FIG. 5.
Viral symptoms and RNA accumulation in N. benthamiana plants inoculated with TBSV- P DI-5 or CIRV DI-7 RNA in the presence of CymRSV, TBSV- P, and their chimeras. (A) Schematic representation and induced symptoms of the CymRSV–TBSV- P chimeras in the presence of the indicated DI RNAs. The organization of CymRSV and TBSV- P genomic RNAs is shown above with the ORFs and the approximate molecular masses of the encoded proteins. The common restriction endonuclease sites used for constructing chimeras are indicated. The symptom phenotypes indicated at the right represent the typical symptoms characteristic for the indicated inocula. Six plants were inoculated with each inoculum, and the experiment was repeated three times. (B) Accumulation of helper virus genomic RNA in the presence of TBSV- P DI-5 RNA and CIRV DI-7 RNA. The helper viruses are indicated on the top, and DI RNAs are indicated below. Symbols are defined in the legend to Fig. 1.
The replication ability of different DI RNAs in different tombusvirus-infected plants suggests that the cis-acting elements required for DI RNA replication (5, 8) are probably common to all tombusviruses. Although heterologous DI RNAs accumulated as efficiently as homologous DI RNAs, in some heterologous DI RNA-helper virus combinations they were not able to attenuate the helper virus-induced necrotic symptoms. This happened when CymRSV DI-13 RNA-expressing transgenic plants were challenged with MPV or TBSV- P or when nontransgenic plants were coinoculated with CymRSV DI-13 or CIRV DI-7 RNAs in the presence of TBSV- P transcripts. The same level of DI RNA accumulation in plants inoculated with TBSV- P and in plants inoculated with protective (TBSV- P DI-5) or nonprotective (CIRV DI-7) DI RNAs strongly suggests that the symptom attenuation effect of DI RNA is not a simple consequence of the competition of DI and helper virus genomic RNAs for the factors required for viral replication. More likely, DI RNAs in a protective helper virus-DI combination specifically interfere with a viral protein(s) which is able to elicit the necrotic plant response, while in the nonprotective DI-helper virus combination this interference does not occur properly. It is noteworthy that this is the first report about efficiently replicating DI RNA which is not able to protect infected plants against helper virus-induced apical necrosis in any tombusvirus system.
It has been demonstrated elsewhere that p19 plays an important role in the necrotic symptom development caused by tombusviruses (6, 15) and p19 is suggested to be solely responsible for necrotic syndrome in tombusvirus infection (21). In addition, previous study of TBSV- Ch suggested that the protective effect of DI RNA occurs due to the selective reduction in the abundance of sg2 RNA, from which p19 and p22 are expressed (24). Our analysis of the accumulation of TBSV- P genomic and sg RNAs as well as that of the virus-encoded p19 in the presence of homologous (protective) TBSV- P DI-5 RNA and heterologous (nonprotective) CIRV DI-7 RNA did not support this hypothesis. The results obtained showed that the replication of DI RNA in virus-infected plants, regardless of its protective or nonprotective nature, frequently (but not always) causes a remarkable reduction in the replication of the helper virus genome, but the accumulation of either sg RNA was not disproportionally affected by DI RNA replication. The same results were observed with tobacco plant protoplasts infected with TBSV- Ch and DI RNA (10). The overall reduction of the viral replication may directly account for the reduced level of p19 in infected plants. The inhibitory effect of DI RNA on helper virus replication is likely the consequence of direct competition for factors necessary for replication. The reduced accumulation of helper viral RNAs is clearly not responsible for symptom attenuation, which was observed at equal levels in the presence of protective and nonprotective DI RNAs. It is more likely that the inhibition of viral replication resulted in symptom delay. In addition, we could not find any direct correlation between the amount of p19 and necrotic symptom development in our system. A very low amount of p19 was also sufficient to induce necrotic symptoms. The construction of chimeric viruses containing sequences derived from CymRSV and TBSV- P has allowed us to determine viral sequences required for symptom attenuation in the presence of DI RNA. We found that chimeras which contained the 5′ leader sequence and ORF 1 of CymRSV caused attenuated symptoms, while those which contained the 5′ leader sequence and ORF 1 of TBSV- P developed necrotic symptoms in the presence of CIRV DI-7 RNA. In contrast, all of the chimeras caused attenuated symptoms in the presence of TBSV- P DI-5 RNA. These findings strongly suggest a crucial role for the 5′-proximal region of the viral genome in the mechanism of DI RNA-mediated symptom attenuation.
The mechanism of the interference between the helper virus genome and DI RNA in tombusvirus-infected plants is unknown. However, based on the presented data and our recent observation, with chimeras between CymRSV and CIRV, that a specific interaction between p33 (ORF 1) and p19 (ORF 5) is required to induce necrotic syndrome in tombusvirus-infected plants (1), a model can be proposed for the mechanism of DI RNA-mediated symptom attenuation. We suggest that the DI RNA-mediated symptom attenuation depends on the ability of DI RNA to prevent a direct or indirect interaction between p19 and p33. Maybe the protective DI RNA can specifically bind to the p33, thus inhibiting the suggested interaction between p33 and p19. This model coincides with the presented observation that the protective nature depends on the 5′-terminal region of helper virus, including the coding region of p33. However, at the moment we do not have direct evidence for the binding of DI RNA to p33; therefore, further analysis is needed for understanding the molecular mechanism of DI RNA-mediated symptom attenuation. Our unique system, with protective and nonprotective DI-helper virus combinations, could be a powerful tool for this purpose.
Nucleotide sequence accession number.
The nucleotide sequences of TBSV- P genomic and DI-5 RNAs are available in the EMBL and GenBank nucleotide databases under accession no. U80935 (genomic RNA) and AF038044 (DI-5).
Acknowledgments
This research was supported by a grant from the Hungarian OTKA (T022766). Z.H. was supported by the Ph.D. education program headed by Mihály Sajgó of the Agricultural University of Gödöllö.
We thank László Szabó for quantitation of RNA by Analysis 2.0 (Soft-Imaging Software GmbH).
REFERENCES
- 1.Burgyán J. Abstracts of EMBO Workshop on the Molecular Mechanisms in the Replicative Cycle of Viruses in Plants, Las Navas del Marques, Spain, 15 to 19 June 1997. 1997. Contribution of tombusvirus genes to necrotic symptom phenotype, abstr. 47. [Google Scholar]
- 2.Burgyán J, Grieco F, Russo M. A defective interfering RNA in cymbidium ringspot virus infections. J Gen Virol. 1989;70:235–239. [Google Scholar]
- 3.Burgyán J, Rubino L, Russo M. De novo generation of cymbidium ringspot virus defective interfering RNA. J Gen Virol. 1991;72:505–509. doi: 10.1099/0022-1317-72-3-505. [DOI] [PubMed] [Google Scholar]
- 4.Burgyán J, Rubino M, Russo M. The 5′-terminal region of a tombusvirus genome determines the origin of multivesicular bodies. J Gen Virol. 1996;77:1967–1974. doi: 10.1099/0022-1317-77-8-1967. [DOI] [PubMed] [Google Scholar]
- 5.Chang Y C, Borja M, Scholthof H B, Jackson A O, Morris T J. Host effects and sequences essential for accumulation of defective interfering RNAs of cucumber necrosis and tomato bushy stunt tombusviruses. Virology. 1995;210:41–53. doi: 10.1006/viro.1995.1315. [DOI] [PubMed] [Google Scholar]
- 6.Dalmay T, Rubino L, Burgyán J, Kollár Á, Russo M. Functional analysis of cymbidium ringspot virus genome. Virology. 1993;194:697–704. doi: 10.1006/viro.1993.1310. [DOI] [PubMed] [Google Scholar]
- 7.Finnen R L, Rochon D M. Sequence and structure of defective interfering RNAs associated with cucumber necrosis virus infections. J Gen Virol. 1993;74:1715–1720. doi: 10.1099/0022-1317-74-8-1715. [DOI] [PubMed] [Google Scholar]
- 8.Havelda Z, Dalmay T, Burgyán J. Localization of cis-acting sequences essential for tombusvirus defective interfering RNA replication. J Gen Virol. 1995;76:2311–2316. doi: 10.1099/0022-1317-76-9-2311. [DOI] [PubMed] [Google Scholar]
- 9.Hillman B I, Carrington J C, Morris T J. A defective interfering RNA that contains a mosaic of plant virus genome. Cell. 1987;51:427–433. doi: 10.1016/0092-8674(87)90638-6. [DOI] [PubMed] [Google Scholar]
- 10.Jones R W, Jackson A O, Morris T J. Defective-interfering RNAs and elevated temperatures inhibit replication of tomato bushy stunt virus in inoculated protoplasts. Virology. 1990;176:539–545. doi: 10.1016/0042-6822(90)90024-l. [DOI] [PubMed] [Google Scholar]
- 11.Kollár Á, Dalmay T, Burgyán J. Defective interfering RNA-mediated resistance against cymbidium ringspot tombusvirus in transgenic plants. Virology. 1993;193:313–318. doi: 10.1006/viro.1993.1127. [DOI] [PubMed] [Google Scholar]
- 12.Kollár Á, Burgyán J. Evidence that ORF 1 and 2 are the only virus encoded replicase gene of cymbidium ringspot tombusvirus. Virology. 1994;201:169–172. doi: 10.1006/viro.1994.1280. [DOI] [PubMed] [Google Scholar]
- 13.Martelli G P, Gallitelli D, Russo M. Tombusviruses. In: Koenig R, editor. The plant viruses. New York, N.Y: Plenum Press; 1988. pp. 13–65. [Google Scholar]
- 14.Rochon D M. Rapid de novo generation of defective interfering RNA by cucumber necrosis virus mutants that do not express the 20- kDa non-structural protein. Proc Natl Acad Sci USA. 1991;88:11153–11157. doi: 10.1073/pnas.88.24.11153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rochon D M, Johnston J C. Infectious transcripts from cloned cucumber necrosis virus cDNA. Evidence for a bifunctional subgenomic mRNA. Virology. 1991;181:656–665. doi: 10.1016/0042-6822(91)90899-m. [DOI] [PubMed] [Google Scholar]
- 16.Roux L, Simon A E, Holland J J. Effects of defective interfering viruses on virus replication and pathogenesis in vitro and in vivo. Adv Virus Res. 1991;40:181–211. doi: 10.1016/S0065-3527(08)60279-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rubino L, Burgyán J, Russo M. Molecular cloning and complete nucleotide sequence of carnation Italian ringspot tombusvirus genomic and defective interfering RNAs. Arch Virol. 1995;140:2027–2039. doi: 10.1007/BF01322690. [DOI] [PubMed] [Google Scholar]
- 18.Russo M, Burgyán J, Martelli P G. The molecular biology of Tombusviridae. Adv Virus Res. 1994;44:382–424. doi: 10.1016/s0065-3527(08)60334-6. [DOI] [PubMed] [Google Scholar]
- 19.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 20.Scholthof H B, Morris T J, Jackson A O. The capsid protein gene of tomato bushy stunt virus is dispensable for systemic movement and can be replaced for localized expression of foreign genes. Mol Plant-Microbe Interact. 1993;6:309–322. [Google Scholar]
- 21.Scholthof H B, Scholthof K-B G, Jackson A O. Identification of tomato bushy stunt virus host-specific symptom determinants by expression of individual genes from a potato virus X vector. Plant Cell. 1995;7:1157–1172. doi: 10.1105/tpc.7.8.1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Scholthof H B, Scholthof K-B G, Kikkert M, Jackson A O. Tomato bushy stunt virus spread is regulated by two nested genes that function in cell-to-cell movement and host-dependent systemic invasion. Virology. 1995;213:425–438. doi: 10.1006/viro.1995.0015. [DOI] [PubMed] [Google Scholar]
- 23.Scholthof K-B G, Scholthof H B, Jackson A O. The tomato bushy stunt virus replicase proteins are coordinately expressed and membrane associated. Virology. 1995;208:365–369. doi: 10.1006/viro.1995.1162. [DOI] [PubMed] [Google Scholar]
- 24.Scholthof K-B G, Scholthof H B, Jackson A O. The effect of defective interfering RNAs on the accumulation of tomato bushy stunt virus proteins and implications for disease attenuation. Virology. 1995;211:324–328. doi: 10.1006/viro.1995.1410. [DOI] [PubMed] [Google Scholar]
- 25.Simon A E, Bujarski J J. RNA-RNA recombination and evolution in virus-infected plants. Annu Rev Phytopathol. 1994;32:337–362. [Google Scholar]
- 26.White K A. Formation and evolution of Tombusvirus defective interfering RNAs. Semin Virol. 1997;7:409–416. [Google Scholar]