Abstract
BACKGROUND
Acute pancreatitis (AP) encompasses a spectrum of pancreatic inflammatory conditions, ranging from mild inflammation to severe pancreatic necrosis and multisystem organ failure. Given the challenges associated with obtaining human pancreatic samples, research on AP predominantly relies on animal models. In this study, we aimed to elucidate the fundamental molecular mechanisms underlying AP using various AP models.
AIM
To investigate the shared molecular changes underlying the development of AP across varying severity levels.
METHODS
AP was induced in animal models through treatment with caerulein alone or in combination with lipopolysaccharide (LPS). Additionally, using Ptf1α to drive the specific expression of the hM3 promoter in pancreatic acinar cells transgenic C57BL/6J- hM3/Ptf1α(cre) mice were administered Clozapine N-oxide to induce AP. Subsequently, we conducted RNA sequencing of pancreatic tissues and validated the expression of significantly different genes using the Gene Expression Omnibus (GEO) database.
RESULTS
Caerulein-induced AP showed severe inflammation and edema, which were exacerbated when combined with LPS and accompanied by partial pancreatic tissue necrosis. Compared with the control group, RNA sequencing analysis revealed 880 significantly differentially expressed genes in the caerulein model and 885 in the caerulein combined with the LPS model. Kyoto Encyclopedia of Genes and Genomes enrichment analysis and Gene Set Enrichment Analysis indicated substantial enrichment of the TLR and NOD-like receptor signaling pathway, TLR signaling pathway, and NF-κB signaling pathway, alongside elevated levels of apoptosis-related pathways, such as apoptosis, P53 pathway, and phagosome pathway. The significantly elevated genes in the TLR and NOD-like receptor signaling pathways, as well as in the apoptosis pathway, were validated through quantitative real-time PCR experiments in animal models. Validation from the GEO database revealed that only MYD88 concurred in both mouse pancreatic tissue and human AP peripheral blood, while TLR1, TLR7, RIPK3, and OAS2 genes exhibited marked elevation in human AP. The genes TUBA1A and GADD45A played significant roles in apoptosis within human AP. The transgenic mouse model hM3/Ptf1α(cre) successfully validated significant differential genes in the TLR and NOD-like receptor signaling pathways as well as the apoptosis pathway, indicating that these pathways represent shared pathological processes in AP across different models.
CONCLUSION
The TLR and NOD receptor signaling pathways play crucial roles in the inflammatory progression of AP, notably the MYD88 gene. Apoptosis holds a central position in the necrotic processes of AP, with TUBA1A and GADD45A genes exhibiting prominence in human AP.
Keywords: Acute pancreatitis, RNA-sequencing, Experimental acute pancreatitis models, Inflammatory, Apoptosis, TLR and NOD-like signaling pathways
Core Tip: AP is a critical emergency condition with no effective targeted therapeutic interventions currently available. Therefore, RNA sequencing (RNA-seq) was employed to investigate the molecular alterations in acute pancreatitis (AP), aiming to identify novel therapeutic strategies. The RNA-seq analysis showed a significant upregulation of TLR and NOD-like signaling pathways in AP, with crucial involvement of genes such as TLR7, IRF7, and MYD88. Notably, the TUBA1A and GADD45A genes were identified as key players in the apoptosis signaling pathway. Substantial evidence was provided through comprehensive validation using Gene Expression Omnibus Series datasets from human peripheral blood and mouse pancreatic tissues, as well as transgenic mouse models to examine inflammation and apoptosis-related molecules. This study offers fresh insights for future therapeutic approaches in managing AP and establishes new directions for subsequent fundamental investigations.
INTRODUCTION
Acute pancreatitis (AP), defined as an inflammatory disorder of the pancreas, is characterized by early activation of digestive enzymes followed by self-digestion of the pancreas[1]. The disease ranges from a mild and self-limiting condition to severe AP (SAP), which is associated with high mortality due to pancreatic necrosis and persistent organ failure[2]. Although the etiology varies in different countries and regions, gallstone disease, alcohol intake, and hyperglyceridemia are common causes of AP. Generally, the initial management of AP is nutritional support, and effective treatment strategies are limited[3].
Currently, the exact pathogenic mechanism remains unclear. Due to the unpredictable progression of this disease and limited access to human pancreatic tissues, there are still great challenges in research on the molecular mechanism of AP. Therefore, experimental animal models have been extensively used to explore the pathogenic mechanism of AP, which could be helpful for the development of novel preventive and therapeutic strategies for AP.
In 1977, Lampel and Kern were the first to report that the intravenous administration of high concentrations of the gut hormone cholecystokinin homolog caerulein could induce a mild and reversible form of AP in rats[4]. Since then, the caerulein model has emerged as one of the most extensively employed experimental models for investigating the molecular mechanisms underpinning AP[5,6]. The premature activation of digestive enzymes, such as trypsinogen, and their intrapancreatic activation are distinctive traits of pancreatic hyperstimulation, which are also fundamental mechanisms underlying the caerulein model.
Furthermore, the caerulein model aids in unraveling processes related to autophagic dysfunction, aberrant calcium signaling, and endoplasmic reticulum stress, all of which constitute the central pathogenic mechanisms of AP[7]. Key characteristics of the caerulein-induced AP model include prominent pancreatic edema, extensive inflammation, and a degree of acinar cell necrosis. Notably, this model exhibits a pronounced self-limiting nature, with recovery typically occurring within approximately one week. However, it is firmly established clinically that the severity of AP is linked to significantly different clinical outcomes. While mild cases may spontaneously resolve, severe cases are associated with high mortality rates.
To investigate the shared molecular changes underlying the development of AP across varying severity levels, a combination of caerulein and lipopolysaccharide (LPS) has been employed to induce SAP[5]. The inclusion of LPS treatment was linked to substantial damage to the intestinal barrier, resembling peripheral organ injuries observed in patients. This exacerbated the pancreatic inflammatory responses and acinar cell injuries[2].
The TLR and NOD-like receptor signaling pathways are widely recognized as playing pivotal roles in the inflammatory immune response. Activation of these signaling pathways results in the release of various inflammatory mediators, fostering the advancement of tissue inflammation. Additionally, the release of chemokines facilitates the migration of immune cells from the bloodstream into tissues[8,9]. The discovery of crucial genes within TLR and NOD-like receptor signaling pathways offered fresh perspectives on mitigating the inflammatory response in AP and introduced novel therapeutic targets.
Apoptosis is a regulated cell death process critical for preserving tissue and organ stability and overall health[10]. In the progression of AP, apoptosis plays a role in the elimination of damaged pancreatic cells, thereby averting additional inflammation and tissue damage. This function can be perceived as a protective mechanism that curtails the disease's severity[11]. However, excessive apoptosis may lead to severe damage to pancreatic tissues[12]. Hence, comprehending the molecular mechanisms governing acinar cell apoptosis in AP is of paramount significance, as it has the potential to open up novel avenues and therapeutic strategies aimed at halting the progression of necrotic alterations in AP or obstructing its necrotic pathways.
To explore the molecular biological changes in pancreatic tissue as AP progresses, we conducted RNA sequencing (RNA-seq) on AP animal models exposed to both caerulein alone and in conjunction with LPS. This analysis unveiled the extensive activation of inflammatory and apoptotic signaling pathways, along with the genes implicated in the progression of AP. RNA-seq data from AP mouse pancreatic tissue and AP patient blood samples in the Gene Expression Omnibus (GEO) database corroborated the transcriptional changes in these key genes. Ptf1a serves as a pancreatic acinar-specific promoter[13], whereas hM3 is a subtype of human cloned muscarinic receptors[14]. Specific activation of hM3 within the pancreatic acini of mice triggers excessive secretion of pancreatic enzymes, leading to the onset of AP. The transgenic mouse hM3/Ptf1α(Cre) AP model further emphasized the key role of the apoptosis pathway in the development of AP. These molecular biological changes provide comprehensive insights and evidence, offering directions for future disease treatment and fundamental research.
MATERIALS AND METHODS
Experimental animals
Male wild-type mice at the age of 6-8 wk were obtained from Nanjing Gempharmatech Co. Ltd. Ptf1α(cre) mice and hM3-/- mice were both acquired from Shanghai Southern Model Biological Technology Co., Ltd. hM3/Ptf1α (cre) mice were generated by breeding hM3-/- mice and Ptf1α(cre) mice. All animal experiments were approved by the Institutional Animal Care and Use Committee of The First Affiliated Hospital of Nanchang University and complied with the national and international guidelines for the Care and Use of Laboratory Animals. In each group of every batch of animal models, there were at least 6 mice included.
Construction of AP experimental models
The induction of AP was achieved by administering ten consecutive intraperitoneal injections of caerulein (Sigma-Aldrich, St. Louis, Missouri, United States, at a dose of 50 μg/kg), with one-hour intervals between each injection, followed immediately by a single dose of LPS (Sigma-Aldrich, St. Louis, Missouri, United States, at a dose of 10 mg/kg). The control group received intraperitoneal injections of saline. hM3/Ptf1α(cre) mice were pre-treated one month in advance by daily intragastric administration of tamoxifen (Sigma-Aldrich, 10 mg/kg) for one week to activate Ptf1α(cre) expression. Subsequently, a single intraperitoneal injection of Clozapine N-oxide (Sigma-Aldrich, 1 mg/kg) was administered to induce spontaneous AP.
Histopathology
The mice were euthanized 24 h after AP modeling, and fresh pancreatic tissue was retrieved. The pancreatic tissues were fixed in 4% formalin for 24 h, followed by embedding in paraffin and sectioning into 4 μm slices for H&E staining. Pancreatic injury was blindly assessed by two pathologists according to previously described criteria. In brief, the pancreatic histopathological assessment included four categories: edema, inflammatory cell infiltration, necrosis, each scored from 0-3[15].
RNA isolation
The RNA was extracted using the MolPure® Cell Tissue Kit (Yeasen Biotechnology Shanghai Co., Ltd., 19221ES50). Add the processed homogenate into Mol Pure DNA Removal/RNA Binding Column A2 (column placed in a 2 mL Collection Tube), then centrifuge and collect the filtrate containing RNA. Proceed to protein removal, clear away proteins through protein removal solution, wash the column twice with wash solution, and elute the collected RNA using RNase-free H2O.
Real-time PCR analysis
The quantitative real-time PCR (qRT-PCR) assay was performed to measure the mRNA expression levels of the target genes using the Hifair® III 1st Strand cDNA Synthesis SuperMix (Yeasen Biotechnology Shanghai Co., Ltd., 11141ES10) for qPCR Kit and the Hieff UNICON® Universal Blue qPCR SYBR Green Master Mix Kit (Yeasen Biotechnology Shanghai Co., Ltd., 11211ES03). GAPDH was employed as the internal reference. The primer sequences used are listed in Supplementary Table 1.
Database
The validation data were sourced from the GEO database, with GSE109227 comprising RNA-seq data from pancreatic samples of normal mice and pancreatitis-induced mice[16], and GSE194331 containing RNA-seq data from peripheral blood of healthy individuals and AP patients[17].
RNA-seq
The libraries were constructed using VAHTS Universal V6 RNA-seq Library Prep Kit according to the manufacturer’s instructions. The transcriptome sequencing and analysis were conducted by OE Biotech Co., Ltd. (Shanghai, China). The libraries were sequenced on a llumina Novaseq 6000 platform and 150 bp paired-end reads were generated. About 55 raw reads for each sample were generated. Raw reads of fastq format were firstly processed using fastp and the low-quality reads were removed to obtain the clean reads. Then about 50 clean reads for each sample were retained for subsequent analyses. The clean reads were mapped to the reference genome using HISAT2 FPKM3 of each gene was calculated and the read counts of each gene were obtained by HT Seq-count principal component analysis (PCA) analysis were performed using R (v 3.2.0) to evaluate the biological duplication of samples.
Differential expression analysis was performed using the DESeq2 Q value < 0.05 and foldchange > 2 or foldchange < 0.5 was set as the threshold for significantly differential expression genes (DEGs). Hierarchical cluster analysis of DEGs was performed using R (v 3.2.0) to demonstrate the expression pattern of genes in different groups and samples. The radar map of top 30 genes was drawn to show the expression of up-regulated or down-regulated DEGs using R packet ggradar.
Based on the hypergeometric distribution, Gene Ontology (GO), Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway, Reactome and Wiki Pathways enrichment analysis of DEGs were performed to screen the significant enriched term using R (v 3.2.0), respectively. R (v 3.2.0) was used to draw the column diagram, the chord diagram and bubble diagram of the significant enrichment term.
Gene Set Enrichment Analysis analysis
Gene Set Enrichment Analysis (GSEA) was performed using GSEA software. The analysis was used a predefined gene set, and the genes were ranked according to the degree of differential expression in the two types of samples. Then it is tested whether the predefined gene set was enriched at the top or bottom of the ranking list.
Statistical analysis
The data were expressed as the mean ± SEM. Statistical analysis was performed using the SPSS 13.0 software. Differences between two groups with normal distributions were assessed by Student’s t test, and one-way analysis of variance was used to compare differences between more than two groups. The least significant difference post hoc test was performed when analysis of variance indicated significance. The value of P < 0.05 was regarded as the cutoff for statistical significance.
RESULTS
Treatment with caerulein alone or in combination with LPS induced varying degrees of experimental AP
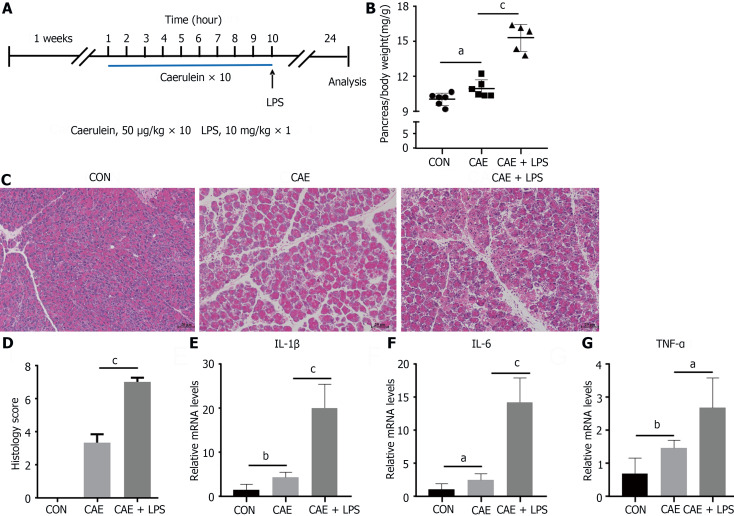
The drug AP models mainly include the caerulein model and the arginine model, among which the caerulein model has become the most widely used AP model due to its close similarity to human pancreatitis pathogenesis and its feasibility. To investigate the common characteristics of AP, treatment with caerulein alone or in combination with LPS was performed to induce the AP model (Figure 1A). After stimulation with caerulein, pancreatic acinar cell edema was markedly evident, manifesting as a significant increase in the pancreas-to-body weight ratio. This edema was even more pronounced when caerulein was combined with LPS (Figure 1B).
Figure 1.
Treatment with caerulein alone or in combination with lipopolysaccharide induced varying degrees of experimental acute pancreatitis. Modeling methods for the control group, caerulein (CAE) group, and CAE combined with lipopolysaccharide (LPS; CAE + LPS) group, evaluation of pancreatic tissue changes, and relevant parameters. A: Schematic representation of the modeling methods for the CAE group and CAE + LPS group; B: Pancreatic weight ratios in the three groups; C: H&E staining of pancreatic tissue; D: Scores for edema, inflammation, and necrosis in pancreatic pathology; E-G: Quantitative real-time PCR expression values of pancreatic inflammatory factors. n = 6 per group, aP < 0.05, bP < 0.01, cP < 0.001. CAE: Caerulein; CON: Control; LPS: Lipopolysaccharide.
H&E staining analysis showed an increased interstitial space between pancreatic acini in mice with AP, with infiltration of inflammatory cells and pronounced pancreatic necrosis following the coadministration of caerulein with LPS (Figure 1C). Edema and inflammation were notably enhanced when caerulein was combined with LPS, as reflected in the pathological scoring (Figure 1D).
qRT-PCR analysis was used to assess the expression of the proinflammatory cytokines TNF-α, Interleukin-6, and Interleukin-1beta (Figure 1E-G). These inflammatory factors were significantly elevated in AP, with a more pronounced increase observed in mice treated with LPS. This finding aligned well with the clinical features of AP, especially in patients with concurrent intestinal dysfunction, who had a higher risk of pancreatic necrosis[18,19]. Despite variations in the severity of the AP model, the observed pancreatic inflammatory characteristics remain consistent with the clinical manifestations, featuring prominent edema, inflammation, and necrosis.
RNA-seq identified the molecular changes during the development of AP
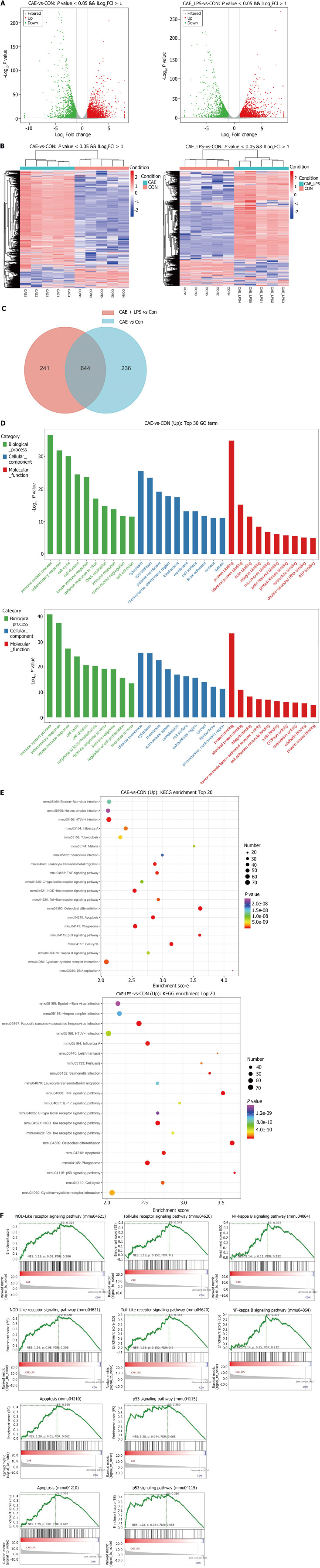
To investigate the molecular changes during the onset of AP, RNA-seq was performed on pancreatic tissues from normal pancreas and two AP models. PCA and bar charts revealed significant differentially expressed (SDE) gene expression at the transcriptional level among the three groups (Supplementary Figure 1). In comparison to normal pancreas, there were 2268 upregulated and 1787 downregulated genes in the AP experimental model induced by caerulein alone. There were 2400 upregulated and 1878 downregulated genes compared to normal pancreas in the AP model induced by caerulein in conjunction with LPS. The number of differentially expressed genes between the two AP models was relatively low. Using volcano plots and heatmaps, we displayed the SDE genes between the two AP models and the normal pancreas (Figure 2A and B).
Figure 2.
RNA-seq identified the molecular changes during the development of acute pancreatitis. Comparative analysis of the caerulein (CAE) group and CAE + lipopolysaccharide (LPS) group with the control (Con) group. A: Volcano plot of significant differentially expressed (SDE) genes (P value < 0.05 and |log2FC| > 1), with red indicating significant upregulation and green indicating significant downregulation; B: Venn diagram, where blue represents CAE group vs Con group, and red represents CAE + LPS group vs Con group, with the gray area representing commonly differentially expressed genes in both groups compared to the Con group; C: Heatmap displaying the expression of SDE genes in each pancreatic sample, with red indicating upregulated genes and blue indicating downregulated genes; D: Gene Ontology (GO) functional annotation of the top 30 significantly different genes, with the GO classification chart showing the distribution of entries related to biological processes, cellular components, and molecular functions; E: Enrichment analysis results of the top 20 significantly different Kyoto Encyclopedia of Genes and Genomes pathways; F: Gene Set Enrichment Analysis for the NOD-like receptor, TLR, NF-κB, and P53 signaling pathways, as well as apoptosis. CAE: Caerulein; CON: Control; LPS: Lipopolysaccharide.
In comparison to a normal pancreas, the AP induced solely by caerulein contained 880 SDE genes, while the AP model induced by caerulein combined with LPS had 885 SDE genes. It was noteworthy that there were 644 common SDE genes between the two AP models, indicating a high degree of overlap. This high overlap in SDE genes suggested that although these models differ in severity, their pathogenic mechanisms are remarkably similar (Figure 2C).
GO enrichment analysis revealed that AP mice were enriched in biological processes related to immune system pathways and inflammatory responses. The cytoplasm and cytoskeleton were the major cellular components affected. In terms of molecular function, there was a significant influence on protein binding (Figure 2D). KEGG analysis showed that pathways associated with apoptosis processes in AP showed significant enrichment. Consistent with previous studies, both groups were enriched in immune- and inflammation-related signaling pathways, such as the NOD-like receptor, TLR, and NF-κB signaling pathways (Figure 2E)[8,20]. GSEA also showed upregulation of the NOD-like receptor, TLR, NF-κB, apoptosis, and P53 signaling pathways in both AP models. Taken together, these data suggest that inflammation, immune response and cell apoptosis play crucial roles during the onset of AP.
NOD-like receptor and TLR signaling pathways play a crucial role in the progression of AP
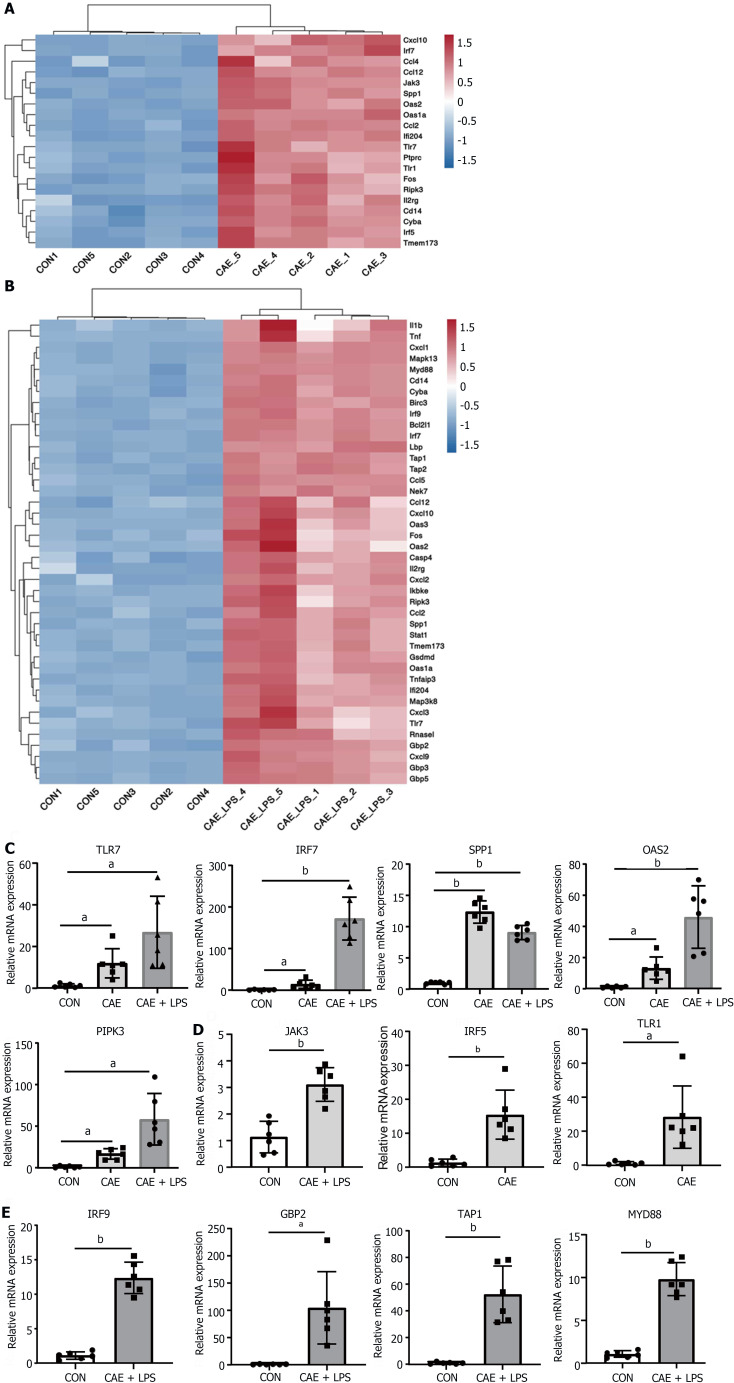
The TLR and NOD-like receptor signaling pathways, which are critical for the regulation of immune and inflammatory responses, have been reported to play important roles in AP pathogenesis[20-22]. As shown in Figure 3A and B, the SDE genes in these two pathways were identified. TLR7, IRF7, SPP1, OAS2, and RIPK3 were the common SDE genes observed when comparing the two models to normal pancreas. Among these genes, TLR7 is often reported in conjunction with IL22 to mitigate various diseases and is thought to have a similar role in AP[23]. Furthermore, interferon IRF7 may play a crucial role in anti-inflammation and immune activation[24], while SPP1 is often reported for its central role in modulating the immunological microenvironment of cancer[25].
Figure 3.
NOD-like receptor and TLR signaling pathway played a crucial role in the progression of acute pancreatitis. Significant differentially expressed genes in the NOD-like receptor and TLR signaling pathways after comparing the caerulein (CAE) group and CAE + lipopolysaccharide (LPS) group to the control (Con) group. A: Heatmap of significant differentially expressed (SDE) genes in the CAE Group compared to the Con group; B: Heatmap of SDE genes in the CAE + LPS group compared to the Con group; C: Quantitative real-time PCR (qRT-PCR) expression values of commonly significantly different genes in both groups; D: qRT-PCR expression values of specifically significantly different genes in the CAE Group compared to the Con group; E: qRT-PCR expression values of specifically significantly different genes in the CAE + LPS group compared to the Con group. n = 6 per group, aP < 0.01, bP < 0.001. CAE: Caerulein; CON: Control; LPS: Lipopolysaccharide.
To confirm that these genes are involved in AP pathogenesis, we generated two experimental AP animal models with caerulein or in combination with LPS treatment. qRT-PCR analysis showed that the mRNA levels of TLR7, IRF7, and SPP1 were significantly upregulated in both AP models compared with the control group (Figure 3C). Interestingly, not all genes showed an association with disease severity. In the model involving caerulein and LPS, TLR7, IRF7, OAS2, and RIPK3 exhibited more pronounced upregulation, possibly linked to the intestinal damage caused by LPS. SPP1 did not show a similar trend, possibly due to its close association with ductal growth and development in the pancreas[26]. Additionally, the unique SDE genes in each of the two AP models were validated. Some genes, including JAK3, IRF5, and TLR1, were uniquely upregulated in the experimental model treated with caerulein alone, while other genes, including IRF9, GBP2, TAP1 and MYD88, were upregulated in the model treated with caerulein combined with LPS (Figure 3D and E). In conclusion, these data suggested that genes in the TLR and NOD-like receptor signaling pathways, including TLR7, IRF7, OAS2, and RIPK3, may be involved in the progression of AP and are strongly associated with the severity of AP.
Public RNA-seq data reveal the significant role of TLR and NOD-like signaling pathways in AP pathogenesis
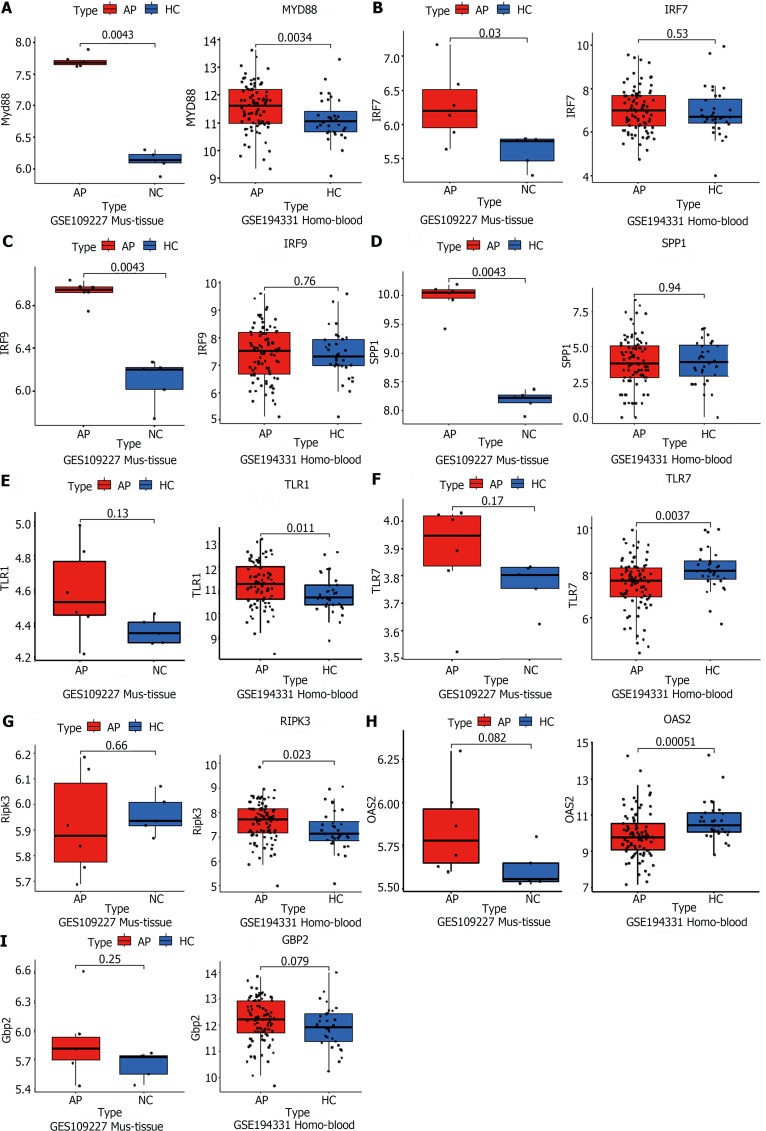
To validate the RNA-seq results, two GSE datasets (GSE109227, GSE194331) from the GEO database were downloaded. The GSE109277 dataset comprised RNA sequencing results of mouse pancreatic tissue. Wild-type C57BL6J mice were injected intraperitoneally with 9 hourly injections of 50 mg/kg caerulein (n = 6) to induce experimental AP, while sodium chloride was used as a control (n = 4). The data from GSE194331 consists of human peripheral blood RNA sequencing, encompassing 32 healthy volunteers and samples taken within 24 h of presentation from 87 patients admitted to the hospital with AP. These data indicated that the mRNA levels of MYD88 were upregulated, both in AP experimental models and in serum from patients with AP (Figure 4A). This observation suggested a significant degree of similarity in AP development between different species, with MYD88 genes playing key roles in the development of AP in animal models and in humans. However, the IRF7, IRF9, and SPP1 genes were only validated in the mouse pancreatic tissue sequencing (Figure 4B-D). Interestingly, despite not being successfully validated in mouse pancreatic tissue sequencing data, TLR1, TLR7, RIPK3 and OAS2 were validated in human peripheral blood sequencing data (Figure 4E-H). This may be attributed to the limited sample size of mouse pancreatic tissue and substantial individual variations. Unfortunately, GBP2 was slightly elevated in both humans and mice, but there was no statistical difference (Figure 4I). In summary, these data further indicated that the TLR and NOD-like receptor signaling pathways play an important role in the development of AP through the regulation of some key genes, such as MYD88.
Figure 4.
Public RNA-seq data reveals that the significant role of TLR and NOD-like signaling pathways in acute pancreatitis pathogenesis. A-I: The acute pancreatitis (AP) animal RNA-seq dataset (GSE109227) and human AP patient blood RNA-seq dataset (GSE194331) were downloaded from the Gene Expression Omnibus database for the external validation of significant differentially expressed genes in the successfully validated NOD-like receptor and TLR signaling pathways in the animal model, using quantitative real-time PCR. n = 6 per group.
Apoptosis serves as a crucial pathway mediating pancreatic necrosis in AP
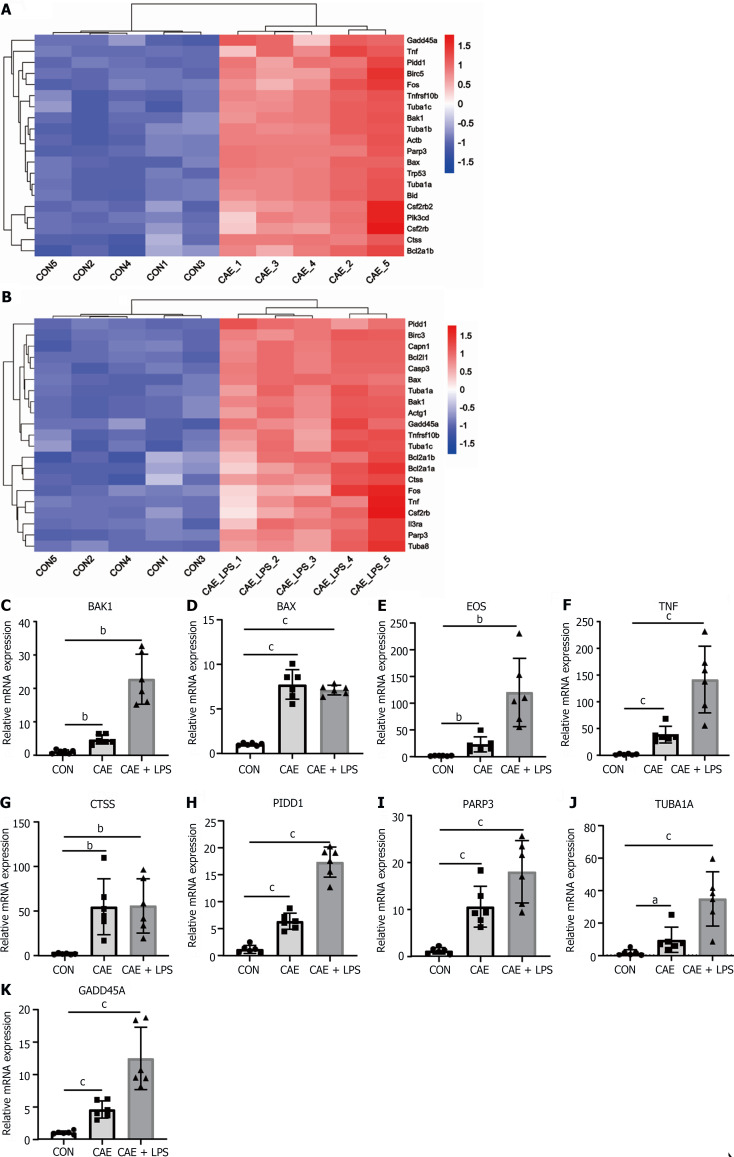
Apoptosis, a programmed cell death process, has been reported to play a key role in AP and can clear damaged cells in the early stages of inflammation. However, it can also become excessively activated in the later stages, leading to significant acinar cell death[7,11]. Given that the apoptosis pathway was significantly activated in the AP mouse models (Figure 2F), we further identified the SDE genes involved in this pathway. The heatmap plot showed SDE genes in the two AP models vs normal pancreatic tissue, including apoptosis-related genes such as BAK1, BAX, PIDD1, the FOS and the factor TNF (Figure 5A and B).
Figure 5.
Apoptosis served as a crucial pathway mediating pancreatic necrosis in acute pancreatitis. Significant differentially expressed (SDE) genes in the apoptotic signaling pathway after comparing the caerulein (CAE) group and CAE + lipopolysaccharide (LPS) group to the control (Con) group. A: Heatmap of SDE genes in the CAE group compared to the Con group; B: Heatmap of SDE genes in the CAE + LPS group compared to the Con group; C-K: Quantitative real-time PCR expression values of commonly significantly different genes in both groups, n = 6 per group, aP < 0.05, bP < 0.01, cP < 0.001. CAE: Caerulein; CON: Control; LPS: Lipopolysaccharide.
To validate the RNA-seq results, two additional batches of AP animal models were generated. Consistently, compared with the normal group, the mRNA levels of these genes were significantly upregulated in the experimental model of AP treated with caerulein or in combination with LPS treatment, as detected by qRT-PCR (Figure 5C-K). Notably, several apoptosis-related genes, such as BAK1, FOS, and TNF, showed a more pronounced increase in expression in the group with AP induced by the combination of caerulein and LPS, indicating a close correlation between apoptosis genes and the severity of AP. These data indicated that apoptosis is considered an important biological feature of AP.
TUBA1A and GADD45A are genes closely associated with apoptosis in AP
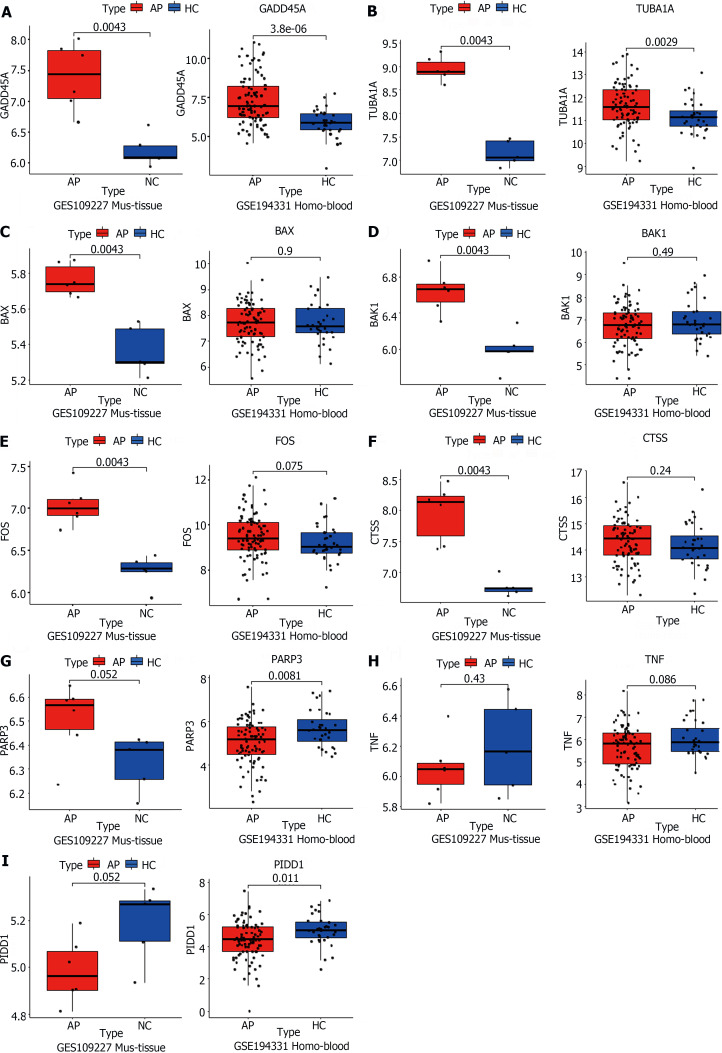
Similarly, the validation of apoptosis-related genes was conducted using RNA-seq data from both animal pancreatic tissue and human peripheral blood samples. In contrast to the confirmation of inflammation and immune-related genes mentioned earlier, most apoptosis-related genes were not confirmed in the human peripheral blood sequencing results. Only two genes, TUBA1A and GADD45A, were successfully validated (Figure 6A and B). This discrepancy may be attributed to species differences and variations in the sequenced samples. Apoptosis primarily acts on cells within tissues and has a smaller impact on the entire systemic circulation, making it challenging to detect in peripheral blood samples.
Figure 6.
TUBA1A and GADD45A were genes closely associated with apoptosis in acute pancreatitis. A-I: The acute pancreatitis (AP) animal RNA-seq dataset (GSE109227) and human AP patient blood RNA-seq dataset (GSE194331) were downloaded from the Gene Expression Omnibus database for the external validation of the genes successfully validated in the animal model and related to the apoptotic signaling pathway, using quantitative real-time PCR. AP: Acute pancreatitis; HC: Healthy control.
As expected, the mouse pancreatic sequencing results were highly consistent with our results, such as BAX, BAK1, FOS, CTSS, TUBA1A, and GADD45A (Figure 6A-E). Unfortunately, while the expression of the PARP3 gene aligned with our anticipated trend, it did not exhibit significant differences (Figure 6F). Further exploration revealed that the TNF and PIDD1 genes demonstrated a completely contrary expression pattern to our expectations (Figure 6G and H). Unfortunately, PIDD1 had the exact opposite trend, suggesting that it may not be representative of the variation in AP (Figure 6I). Therefore, we believe that further efforts are warranted in the exploration of apoptosis-related processes in AP to identify targets that also play roles in humans.
Inflammatory response in hM3/Ptf1α(cre) transgenic animal models consistent with the caerulein model
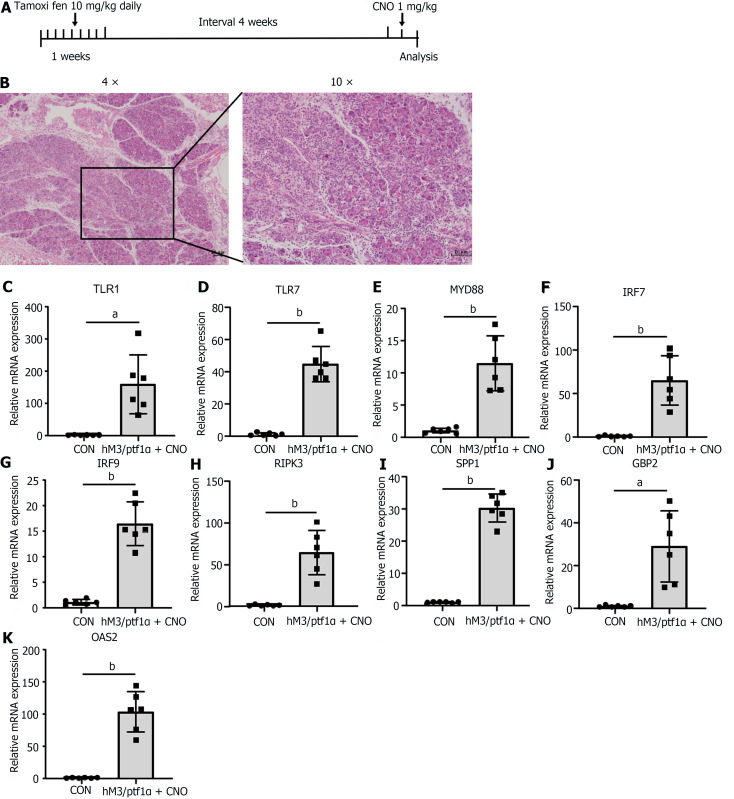
To gather further evidence to substantiate the significance and universality of apoptosis in AP, we decided to employ a transgenic animal model for validation. The hM3/Ptf1α(cre) model involves the activation of M3 receptors in acinar cells of the pancreas, induced by a pancreas-specific promoter, Ptf1α, leading to spontaneous pancreatitis. The hM3 cholinergic receptor is a component of the cholinergic nervous system and is widely distributed across various organs and tissues. Within the digestive system, the hM3 receptor plays a significant role in the contraction of gastrointestinal smooth muscles, secretion of digestive fluids, and gastrointestinal motility[27]. The Ptf1α gene functions as a transcription factor that is crucial in regulating the development of pancreatic acinar cells. The cre recombinase driven by the Ptf1α gene promoter marked the hM3 gene specifically expressed in acinar cells[28]. Transgenic mice were administered tamoxifen orally to activate Ptf1α(cre), followed by intraperitoneal injection of Clozapine N-oxide (CNO) after a one-month interval to activate the Ptf1α gene (Figure 7A).
Figure 7.
Inflammatory response in hM3/Ptf1α(cre) transgenic animal models consistent with the caerulein model. hM3/Ptf1α(cre) model and its pathological images with inflammatory gene validation. A: Schematic representation of the hM3/Ptf1α(cre) model; B: H&E staining of pancreatic tissue schematic representation of the modeling methods; C-K: Validation of the significant differentially expressed genes in the transgenic animal hM3/Ptf1α(cre) model after inducing acute pancreatitis, which were found in the NOD-like receptor and TLR signaling pathways. n = 6 per group, aP < 0.01, bP < 0.001. CON: Control; CNO: Clozapine N-oxide.
Activation of the hM3 gene led to excessive secretion of pancreatic enzymes, causing AP. More severe than the caerulein model, the mice exhibited extensive diffuse necrosis and abundant inflammatory infiltration in the pancreas (Figure 7B). After inducing pancreatitis, qRT-PCR was utilized to assess the inflammatory genes mentioned above. The results revealed that despite variations in modeling approaches, all genes exhibited a consistent trend with the caerulein model: notably elevated during AP (Figure 7C-K). This underscores once more the pivotal role of the genes we have identified as key players in AP inflammation.
The apoptosis in AP in the hM3/Ptf1α(cre) transgenic animal model was consistent with the caerulein models
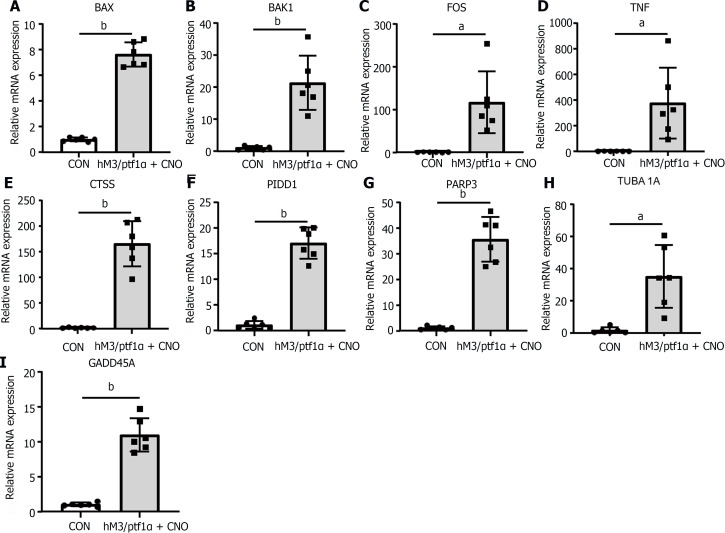
To further investigate the expression changes in apoptotic genes after AP induction in transgenic mice, qRT-PCR was employed to assess their expression. Surprisingly, apoptotic gene expression was confirmed in the hM3/Ptf1α(cre) mouse model and these findings confirmed the similarity in the regulatory mechanisms of the apoptotic pathway across different AP models (Figure 8). Identifying key targets that influence apoptosis and promote a positive role in AP could aid in reducing pancreatic necrosis.
Figure 8.
The apoptosis in acute pancreatitis in the hM3/Ptf1α(cre) transgenic animal model was consistent with the caerulein models. A-I: Validation of the significant differentially expressed genes in the NOD-like receptor and TLR signaling pathways in the transgenic animal hM3/Ptf1α(cre) model after inducing acute pancreatitis. n = 6 per group, aP < 0.01, bP < 0.001. CON: Control; CNO: Clozapine N-oxide.
DISCUSSION
In our effort to unravel the molecular changes underlying the development of AP, we adopted the widely recognized and physiologically relevant caerulein model. To closely mimic clinical scenarios and to avoid the limitations of a single severity model, we employed a combination of caerulein and LPS to induce SAP. Transcriptome analysis utilizing RNA-seq revealed dynamic transcriptional alterations in AP. Comparative analysis between the caerulein group and the caerulein combined with LPS group revealed a highly congruent set of SDE genes. GO and KEGG analyses highlighted the enrichment of pathways predominantly associated with inflammation, immunity, and apoptosis. Notably, the NOD-like receptor and TLR signaling pathways exhibited robust concordance of SDE genes in our induced animal samples, bolstering the internal validity of our experimental results. However, this congruence was not fully recapitulated in external datasets. Importantly, genes including TLR1, MYD88, RIPK3, and GBP2, which were differentially expressed in our model, were also validated in human blood samples, underscoring the reliability of our findings and offering novel therapeutic targets for future studies in the realms of inflammation and immunity. Within the apoptosis-related pathways, the atypical expression of the TUBA1A and GADD45A genes stood out remarkably. Their dysregulation not only held true in our animal model but also withstood scrutiny in publicly available GEO datasets and transgenic animal experiments. This presents a promising outlook for TUBA1A and GADD45A as novel intervention targets for the necrosis of AP in future clinical treatment.
In our quest to decipher the intricate mechanisms underlying AP, we judiciously opted not to employ all available modeling approaches. This decision was rooted in the realization that certain models fail to faithfully represent the pathophysiological mechanisms and exhibit unique, albeit nonrepresentative, conditions associated with AP. For instance, the AP model induced by intraperitoneal injection of L-arginine, while being informative in specific contexts, does not provide clear mechanistic insights into AP and may be related to amino acid imbalances and oxidative stress[29-31]. Importantly, this model does not closely mimic the etiology observed in clinical AP patients, thus limiting its translational relevance. Similarly, the retrograde ductal infusion-induced AP model primarily manifests lesions in the pancreatic head characterized by extensive necrosis, with the pancreatic tail displaying primarily edematous changes[32]. The non-uniform distribution of pathological alterations in this model, while recapitulating certain aspects of gallstone-induced pancreatitis, deviates from the clinical course of AP[33]. Nevertheless, this model bears semblance to the mechanistic underpinnings of biliary pancreatitis, offering a prospective avenue for investigating the molecular mechanisms underlying the development of gallstone-induced pancreatitis.
The early-phase inflammatory response in AP plays a pivotal role in exacerbating disease progression[34]. Consequently, our study placed particular emphasis on the major inflammatory signaling pathways: the TLR and NOD-like receptor signaling pathways. High-quality research has reported a close association between TLR-like receptors and inflammatory damage in AP[19]. Notably, the absence of the toll-like receptor 2 receptor effectively alleviates AP in animal models[17], while the TLR4 receptor is closely linked to lung injury in AP[18]. Evidently, TLR-like receptor signaling pathways are intricately linked to the progression of AP.
This study verified key genes, such as TLR1, MYD88, RIPK3, and GBP2, through multiple models and datasets, offering valuable insights and directions for future research endeavors. MYD88 functions as a downstream molecule of TLR-like receptors, participating jointly in the regulation of immunity and inflammatory responses while also having the capability to modulate RIPK3 involvement in cellular apoptosis pathways[35,36]. GBP2 is a highly regarded molecule within the field of oncology research, as it has the capacity to enhance tumor invasion and proliferation abilities[37]. NOD receptors often play a supplementary role alongside TLR-like receptors, and numerous reports have associated them with intestinal injury in AP. For instance, they can assist Paneth cells in mitigating intestinal damage in the absence of TLR4[38]. Therefore, these two crucial inflammatory signaling pathways warrant further exploration, as they may serve as critical points of initiation in the inflammatory cascade of AP.
Apoptosis, a form of programmed cell death, is critically important for the development and maintenance of the immune system, as well as its response to external and internal stimuli[11]. In the course of AP, apoptosis of acinar cells is often closely associated with oxidative stress and inflammation. Numerous reports have highlighted genes that can modulate and influence the progression of apoptosis, including recent research findings implicating genes such as those involved in the ATF6/P53/IFM2 and Sirt1/Nrf2/TNF pathways[39,40]. Many reviews have emphasized the role of key genes such as caspases; however, significant differential expression of caspases at the transcriptional level was not observed in our study, possibly due to minimal changes in their transcription and more pronounced alterations at the protein level[7,41]. TNF-α, an apoptosis-responsive protein present in pancreatic acinar cells, exhibited notable transcriptional changes[40]. Among the successfully validated genes, TUBA1A and its association with apoptosis have received limited attention[42], while GADD45A, as a critical apoptosis-related gene, plays a significant role in promoting apoptosis through interactions with various molecules[43,44]. Subsequent research efforts may shift the focus toward TUBA1A to explore novel avenues of investigation. This research underscores the intricate regulatory mechanisms of apoptosis in the context of AP, shedding light on key genes and their transcriptional dynamics, with potential implications for therapeutic intervention.
The etiology of AP was diverse, and we have yet to explore other causative factors leading to AP, such as hyperlipidemic pancreatitis and alcoholic pancreatitis. Furthermore, there was a wide range of animal models available for AP, including models induced by L-arginine, pancreatic duct ligation, and retrograde ductal infusion, but comparative experiments in this area were lacking. Despite the identification of numerous genes playing important roles in inflammation and apoptosis, further validation experiments in animal models and in-depth exploration of their mechanisms will be finished in future work. Transgenic mouse models of AP certainly exhibit unique molecular changes, but we have not extensively investigated the differences between these models and the commonly used caerulein model. The aforementioned unfinished aspects will be the focus of our subsequent work. Nonetheless, our study provides insights into the molecular alterations in AP and identifies genes that play crucial roles in inflammation and apoptosis processes, offering potential therapeutic targets.
CONCLUSION
This study investigated the molecular changes associated with the development of AP. RNA-seq analysis identified a significant overlap in the gene expression patterns between AP and normal pancreas, primarily involving inflammatory, immune, and apoptotic pathways. The validation of the TLR and NOD-like receptor signaling pathways using animal tissues and two GEO datasets highlighted several potential key genes, including TLR7, IRF7, SPP1, OAS2, and RIPK3. Moreover, we emphasized the importance of apoptotic pathways in AP. By incorporating transgenic animal models into the validation process, TUBA1A and GADD45A were identified as the most important expressed genes, suggesting their potential as critical targets for future interventions and therapies in AP. Both wild type and the hM3/Ptf1α(cre) mice shared the same pattern of inflammation. These discoveries provide new avenues for the treatment of necrosis in AP.
ARTICLE HIGHLIGHTS
Research background
Acute pancreatitis (AP) is a severe abdominal condition with an increasing incidence rate. Currently, there are no specific therapeutic approaches targeting the underlying causes of this disease. Research on AP is still in its early stages, and this study focuses on the molecular changes associated with inflammation and apoptosis, two major pathological events in AP. The aim is to identify new potential targets for treatment interventions.
Research motivation
This study primarily focused on the molecular changes in AP, indicating significant alterations in inflammation and apoptosis. The research also identified key genes involved in the TLR and NOD signaling pathways, as well as in the apoptotic signaling pathway, highlighting new research and intervention targets for future investigations in this field.
Research objectives
The purpose of this research was to investigate the parthenogenesis and molecular changes associated with AP. In fact, we have identified genes that play important roles in the inflammatory and apoptotic signaling pathways. These findings provide directions for future studies aimed at reducing inflammation and alleviating pancreatic necrosis in AP, as well as discovering new therapeutic approaches for AP.
Research methods
In this study, RNA sequencing analysis was employed to investigate the molecular changes associated with AP and identify key genes involved. Additionally, external GSE from human peripheral blood samples and mouse pancreatic tissues were downloaded and used for validation purposes. Transgenic mice models were also utilized to further validate the findings after induction of AP.
Research results
The molecular changes in inflammation and apoptosis are consistent between different animal models of AP and transgenic AP models. The TLR and NOD signaling pathways play important roles in the inflammatory response of AP, with key genes identified as TLR1, TLR7, RIPK3, and OAS2. TUBA1A and GADD45A have been identified as crucial molecules involved in regulating acinar cell apoptosis in AP. However, further analysis is still needed to investigate AP associated with various etiologies and different modeling method.
Research conclusions
New theories: (1) TUBA1A and GADD45A are key molecules involved in regulating apoptosis of vesicular cells in AP; and (2) Transgenic mice, hM3/Ptf1a(cre) with AP induced by caerulein, exhibit similar molecular changes. New method: Transgenic mice carrying the hM3/Ptf1α(cre) construct were generated and successfully developed AP.
Research perspectives
Using the latest single-cell sequencing technology to investigate the pathogenic mechanisms of AP in-depth.
Footnotes
Institutional animal care and use committee statement: The First Affiliated Hospital of Nanchang University approved experimental animal welfare ethics Approved by the Experimental Animal Welfare Ethics Committee of the First Affiliated Hospital of Nanchang University, Zhu Yin, Gastroenterology Department of our Hospital, "Research on the mechanism of Bifidobacterium pseudolongum Protecting intestinal mucosal Barrier in acute pancreatitis by activating NOD2-autophagy signal axis" (Approval number: CDYFYIACUC-202306QR022), does not violate the ethical principles of experimental animal welfare, and agrees to carry out the research under the approved protocol. Experimental animal Welfare Ethics Committee of the First Affiliated Hospital of Nanchang University
Conflict-of-interest statement: The authors declare that they have no competing interests.
ARRIVE guidelines statement: The authors have read the ARRIVE Guidelines, and the manuscript was prepared and revised according to the ARRIVE Guidelines.
Provenance and peer review: Unsolicited article; Externally peer reviewed.
Peer-review model: Single blind
Peer-review started: December 8, 2023
First decision: January 4, 2024
Article in press: March 1, 2024
Specialty type: Gastroenterology and hepatology
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): B, B
Grade C (Good): 0
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Hakim GD, Turkey; Rodrigo L, Spain S-Editor: Lin C L-Editor: A P-Editor: Cai YX
Contributor Information
Pan Zheng, Department of Gastroenterology, Digestive Disease Hospital, The First Affiliated Hospital, Jiangxi Medical College, Nanchang University, Nanchang 330006, Jiangxi Province, China.
Xue-Yang Li, Department of Gastroenterology, Digestive Disease Hospital, The First Affiliated Hospital, Jiangxi Medical College, Nanchang University, Nanchang 330006, Jiangxi Province, China.
Xiao-Yu Yang, Department of Gastroenterology, Digestive Disease Hospital, The First Affiliated Hospital, Jiangxi Medical College, Nanchang University, Nanchang 330006, Jiangxi Province, China.
Huan Wang, Department of Gastroenterology, Digestive Disease Hospital, The First Affiliated Hospital, Jiangxi Medical College, Nanchang University, Nanchang 330006, Jiangxi Province, China.
Ling Ding, Department of Gastroenterology, Digestive Disease Hospital, The First Affiliated Hospital, Jiangxi Medical College, Nanchang University, Nanchang 330006, Jiangxi Province, China.
Cong He, Department of Gastroenterology, Digestive Disease Hospital, The First Affiliated Hospital, Jiangxi Medical College, Nanchang University, Nanchang 330006, Jiangxi Province, China.
Jian-Hua Wan, Department of Gastroenterology, Digestive Disease Hospital, The First Affiliated Hospital, Jiangxi Medical College, Nanchang University, Nanchang 330006, Jiangxi Province, China.
Hua-Jing Ke, Department of Gastroenterology, Digestive Disease Hospital, The First Affiliated Hospital, Jiangxi Medical College, Nanchang University, Nanchang 330006, Jiangxi Province, China.
Nong-Hua Lu, Department of Gastroenterology, Digestive Disease Hospital, The First Affiliated Hospital, Jiangxi Medical College, Nanchang University, Nanchang 330006, Jiangxi Province, China.
Nian-Shuang Li, Department of Gastroenterology, Digestive Disease Hospital, The First Affiliated Hospital, Jiangxi Medical College, Nanchang University, Nanchang 330006, Jiangxi Province, China.
Yin Zhu, Department of Gastroenterology, Digestive Disease Hospital, The First Affiliated Hospital, Jiangxi Medical College, Nanchang University, Nanchang 330006, Jiangxi Province, China. ndyfy01977@ncu.edu.cn.
Data sharing statement
The first author obtained two files, GSE109227 and GSE194331, from the public database GEO for validation purposes.
References
- 1.Habtezion A, Gukovskaya AS, Pandol SJ. Acute Pancreatitis: A Multifaceted Set of Organelle and Cellular Interactions. Gastroenterology. 2019;156:1941–1950. doi: 10.1053/j.gastro.2018.11.082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Garg PK, Singh VP. Organ Failure Due to Systemic Injury in Acute Pancreatitis. Gastroenterology. 2019;156:2008–2023. doi: 10.1053/j.gastro.2018.12.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boxhoorn L, Voermans RP, Bouwense SA, Bruno MJ, Verdonk RC, Boermeester MA, van Santvoort HC, Besselink MG. Acute pancreatitis. Lancet. 2020;396:726–734. doi: 10.1016/S0140-6736(20)31310-6. [DOI] [PubMed] [Google Scholar]
- 4.Lampel M, Kern HF. Acute interstitial pancreatitis in the rat induced by excessive doses of a pancreatic secretagogue. Virchows Arch A Pathol Anat Histol. 1977;373:97–117. doi: 10.1007/BF00432156. [DOI] [PubMed] [Google Scholar]
- 5.Lerch MM, Gorelick FS. Models of acute and chronic pancreatitis. Gastroenterology. 2013;144:1180–1193. doi: 10.1053/j.gastro.2012.12.043. [DOI] [PubMed] [Google Scholar]
- 6.Zhan X, Wang F, Bi Y, Ji B. Animal models of gastrointestinal and liver diseases. Animal models of acute and chronic pancreatitis. Am J Physiol Gastrointest Liver Physiol. 2016;311:G343–G355. doi: 10.1152/ajpgi.00372.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lee PJ, Papachristou GI. New insights into acute pancreatitis. Nat Rev Gastroenterol Hepatol. 2019;16:479–496. doi: 10.1038/s41575-019-0158-2. [DOI] [PubMed] [Google Scholar]
- 8.Santoni M, Andrikou K, Sotte V, Bittoni A, Lanese A, Pellei C, Piva F, Conti A, Nabissi M, Santoni G, Cascinu S. Toll like receptors and pancreatic diseases: From a pathogenetic mechanism to a therapeutic target. Cancer Treat Rev. 2015;41:569–576. doi: 10.1016/j.ctrv.2015.04.004. [DOI] [PubMed] [Google Scholar]
- 9.Caruso R, Warner N, Inohara N, Núñez G. NOD1 and NOD2: signaling, host defense, and inflammatory disease. Immunity. 2014;41:898–908. doi: 10.1016/j.immuni.2014.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Han J, Zhong CQ, Zhang DW. Programmed necrosis: backup to and competitor with apoptosis in the immune system. Nat Immunol. 2011;12:1143–1149. doi: 10.1038/ni.2159. [DOI] [PubMed] [Google Scholar]
- 11.Bhatia M. Apoptosis versus necrosis in acute pancreatitis. Am J Physiol Gastrointest Liver Physiol. 2004;286:G189–G196. doi: 10.1152/ajpgi.00304.2003. [DOI] [PubMed] [Google Scholar]
- 12.Wang G, Qu FZ, Li L, Lv JC, Sun B. Necroptosis: a potential, promising target and switch in acute pancreatitis. Apoptosis. 2016;21:121–129. doi: 10.1007/s10495-015-1192-3. [DOI] [PubMed] [Google Scholar]
- 13.Ogawa S, Fukuda A, Matsumoto Y, Hanyu Y, Sono M, Fukunaga Y, Masuda T, Araki O, Nagao M, Yoshikawa T, Goto N, Hiramatsu Y, Tsuda M, Maruno T, Nakanishi Y, Hussein MS, Tsuruyama T, Takaori K, Uemoto S, Seno H. SETDB1 Inhibits p53-Mediated Apoptosis and Is Required for Formation of Pancreatic Ductal Adenocarcinomas in Mice. Gastroenterology. 2020;159:682–696.e13. doi: 10.1053/j.gastro.2020.04.047. [DOI] [PubMed] [Google Scholar]
- 14.Ruiz de Azua I, Gautam D, Guettier JM, Wess J. Novel insights into the function of β-cell M3 muscarinic acetylcholine receptors: therapeutic implications. Trends Endocrinol Metab. 2011;22:74–80. doi: 10.1016/j.tem.2010.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tian R, Tan JT, Wang RL, Xie H, Qian YB, Yu KL. The role of intestinal mucosa oxidative stress in gut barrier dysfunction of severe acute pancreatitis. Eur Rev Med Pharmacol Sci. 2013;17:349–355. [PubMed] [Google Scholar]
- 16.Norberg KJ, Nania S, Li X, Gao H, Szatmary P, Segersvärd R, Haas S, Wagman A, Arnelo U, Sutton R, Heuchel RL, Löhr JM. RCAN1 is a marker of oxidative stress, induced in acute pancreatitis. Pancreatology. 2018;18:734–741. doi: 10.1016/j.pan.2018.08.005. [DOI] [PubMed] [Google Scholar]
- 17.Nesvaderani M, Dhillon BK, Chew T, Tang B, Baghela A, Hancock RE, Eslick GD, Cox M. Gene Expression Profiling: Identification of Novel Pathways and Potential Biomarkers in Severe Acute Pancreatitis. J Am Coll Surg. 2022;234:803–815. doi: 10.1097/XCS.0000000000000115. [DOI] [PubMed] [Google Scholar]
- 18.Zhu Y, He C, Li X, Cai Y, Hu J, Liao Y, Zhao J, Xia L, He W, Liu L, Luo C, Shu X, Cai Q, Chen Y, Lu N. Gut microbiota dysbiosis worsens the severity of acute pancreatitis in patients and mice. J Gastroenterol. 2019;54:347–358. doi: 10.1007/s00535-018-1529-0. [DOI] [PubMed] [Google Scholar]
- 19.Li XY, He C, Zhu Y, Lu NH. Role of gut microbiota on intestinal barrier function in acute pancreatitis. World J Gastroenterol. 2020;26:2187–2193. doi: 10.3748/wjg.v26.i18.2187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li L, Liu Q, Le C, Zhang H, Liu W, Gu Y, Yang J, Zhang X. Toll-like receptor 2 deficiency alleviates acute pancreatitis by inactivating the NF-κB/NLRP3 pathway. Int Immunopharmacol. 2023;121:110547. doi: 10.1016/j.intimp.2023.110547. [DOI] [PubMed] [Google Scholar]
- 21.Sharif R, Dawra R, Wasiluk K, Phillips P, Dudeja V, Kurt-Jones E, Finberg R, Saluja A. Impact of toll-like receptor 4 on the severity of acute pancreatitis and pancreatitis-associated lung injury in mice. Gut. 2009;58:813–819. doi: 10.1136/gut.2008.170423. [DOI] [PubMed] [Google Scholar]
- 22.Hoque R, Farooq A, Ghani A, Gorelick F, Mehal WZ. Lactate reduces liver and pancreatic injury in Toll-like receptor- and inflammasome-mediated inflammation via GPR81-mediated suppression of innate immunity. Gastroenterology. 2014;146:1763–1774. doi: 10.1053/j.gastro.2014.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Abt MC, Buffie CG, Sušac B, Becattini S, Carter RA, Leiner I, Keith JW, Artis D, Osborne LC, Pamer EG. TLR-7 activation enhances IL-22-mediated colonization resistance against vancomycin-resistant enterococcus. Sci Transl Med. 2016;8:327ra325. doi: 10.1126/scitranslmed.aad6663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Minaga K, Watanabe T, Arai Y, Shiokawa M, Hara A, Yoshikawa T, Kamata K, Yamashita K, Kudo M. Activation of interferon regulatory factor 7 in plasmacytoid dendritic cells promotes experimental autoimmune pancreatitis. J Gastroenterol. 2020;55:565–576. doi: 10.1007/s00535-020-01662-2. [DOI] [PubMed] [Google Scholar]
- 25.Storrs EP, Chati P, Usmani A, Sloan I, Krasnick BA, Babbra R, Harris PK, Sachs CM, Qaium F, Chatterjee D, Wetzel C, Goedegebuure SP, Hollander T, Anthony H, Ponce J, Khaliq AM, Badiyan S, Kim H, Denardo DG, Lang GD, Cosgrove ND, Kushnir VM, Early DS, Masood A, Lim KH, Hawkins WG, Ding L, Fields RC, Das KK, Chaudhuri AA. High-dimensional deconstruction of pancreatic cancer identifies tumor microenvironmental and developmental stemness features that predict survival. NPJ Precis Oncol. 2023;7:105. doi: 10.1038/s41698-023-00455-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hendley AM, Rao AA, Leonhardt L, Ashe S, Smith JA, Giacometti S, Peng XL, Jiang H, Berrios DI, Pawlak M, Li LY, Lee J, Collisson EA, Anderson MS, Fragiadakis GK, Yeh JJ, Ye CJ, Kim GE, Weaver VM, Hebrok M. Single-cell transcriptome analysis defines heterogeneity of the murine pancreatic ductal tree. Elife. 2021;10 doi: 10.7554/eLife.67776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tolaymat M, Sundel MH, Alizadeh M, Xie G, Raufman JP. Potential Role for Combined Subtype-Selective Targeting of M(1) and M(3) Muscarinic Receptors in Gastrointestinal and Liver Diseases. Front Pharmacol. 2021;12:786105. doi: 10.3389/fphar.2021.786105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Datta J, Dai X, Bianchi A, De Castro Silva I, Mehra S, Garrido VT, Lamichhane P, Singh SP, Zhou Z, Dosch AR, Messaggio F, Ban Y, Umland O, Hosein PJ, Nagathihalli NS, Merchant NB. Combined MEK and STAT3 Inhibition Uncovers Stromal Plasticity by Enriching for Cancer-Associated Fibroblasts With Mesenchymal Stem Cell-Like Features to Overcome Immunotherapy Resistance in Pancreatic Cancer. Gastroenterology. 2022;163:1593–1612. doi: 10.1053/j.gastro.2022.07.076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Siriviriyakul P, Sriko J, Somanawat K, Chayanupatkul M, Klaikeaw N, Werawatganon D. Genistein attenuated oxidative stress, inflammation, and apoptosis in L-arginine induced acute pancreatitis in mice. BMC Complement Med Ther. 2022;22:208. doi: 10.1186/s12906-022-03689-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kaur J, Sidhu S, Chopra K, Khan MU. Calendula officinalis ameliorates l-arginine-induced acute necrotizing pancreatitis in rats. Pharm Biol. 2016;54:2951–2959. doi: 10.1080/13880209.2016.1195848. [DOI] [PubMed] [Google Scholar]
- 31.Perides G, Laukkarinen JM, Vassileva G, Steer ML. Biliary acute pancreatitis in mice is mediated by the G-protein-coupled cell surface bile acid receptor Gpbar1. Gastroenterology. 2010;138:715–725. doi: 10.1053/j.gastro.2009.10.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gardner TB. Acute Pancreatitis. Ann Intern Med. 2021;174:ITC17–ITC32. doi: 10.7326/AITC202102160. [DOI] [PubMed] [Google Scholar]
- 33.Jakkampudi A, Jangala R, Reddy R, Mitnala S, Rao GV, Pradeep R, Reddy DN, Talukdar R. Acinar injury and early cytokine response in human acute biliary pancreatitis. Sci Rep. 2017;7:15276. doi: 10.1038/s41598-017-15479-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen L, Zheng L, Chen P, Liang G. Myeloid Differentiation Primary Response Protein 88 (MyD88): The Central Hub of TLR/IL-1R Signaling. J Med Chem. 2020;63:13316–13329. doi: 10.1021/acs.jmedchem.0c00884. [DOI] [PubMed] [Google Scholar]
- 35.Lawlor KE, Feltham R, Yabal M, Conos SA, Chen KW, Ziehe S, Graß C, Zhan Y, Nguyen TA, Hall C, Vince AJ, Chatfield SM, D'Silva DB, Pang KC, Schroder K, Silke J, Vaux DL, Jost PJ, Vince JE. XIAP Loss Triggers RIPK3- and Caspase-8-Driven IL-1β Activation and Cell Death as a Consequence of TLR-MyD88-Induced cIAP1-TRAF2 Degradation. Cell Rep. 2017;20:668–682. doi: 10.1016/j.celrep.2017.06.073. [DOI] [PubMed] [Google Scholar]
- 36.Ren Y, Yang B, Guo G, Zhang J, Sun Y, Liu D, Guo S, Wu Y, Wang X, Wang S, Zhang W, Guo X, Li X, Li R, He J, Zhou Z. GBP2 facilitates the progression of glioma via regulation of KIF22/EGFR signaling. Cell Death Discov. 2022;8:208. doi: 10.1038/s41420-022-01018-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yu S, Yu X, Sun L, Zheng Y, Chen L, Xu H, Jin J, Lan Q, Chen CC, Li M. GBP2 enhances glioblastoma invasion through Stat3/fibronectin pathway. Oncogene. 2020;39:5042–5055. doi: 10.1038/s41388-020-1348-7. [DOI] [PubMed] [Google Scholar]
- 38.Qi-Xiang M, Yang F, Ze-Hua H, Nuo-Ming Y, Rui-Long W, Bin-Qiang X, Jun-Jie F, Chun-Lan H, Yue Z. Intestinal TLR4 deletion exacerbates acute pancreatitis through gut microbiota dysbiosis and Paneth cells deficiency. Gut Microbes. 2022;14:2112882. doi: 10.1080/19490976.2022.2112882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tan JH, Cao RC, Zhou L, Zhou ZT, Chen HJ, Xu J, Chen XM, Jin YC, Lin JY, Zeng JL, Li SJ, Luo M, Hu GD, Yang XB, Jin J, Zhang GW. ATF6 aggravates acinar cell apoptosis and injury by regulating p53/AIFM2 transcription in Severe Acute Pancreatitis. Theranostics. 2020;10:8298–8314. doi: 10.7150/thno.46934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Abdelzaher WY, Ahmed SM, Welson NN, Marraiki N, Batiha GE, Kamel MY. Vinpocetine ameliorates L-arginine induced acute pancreatitis via Sirt1/Nrf2/TNF pathway and inhibition of oxidative stress, inflammation, and apoptosis. Biomed Pharmacother. 2021;133:110976. doi: 10.1016/j.biopha.2020.110976. [DOI] [PubMed] [Google Scholar]
- 41.Wang X, Cai H, Chen Z, Zhang Y, Wu M, Xu X, Yang L. Baicalein alleviates pyroptosis and inflammation in hyperlipidemic pancreatitis by inhibiting NLRP3/Caspase-1 pathway through the miR-192-5p/TXNIP axis. Int Immunopharmacol. 2021;101:108315. doi: 10.1016/j.intimp.2021.108315. [DOI] [PubMed] [Google Scholar]
- 42.Zenón-Meléndez CN, Carrasquillo Carrión K, Cantres Rosario Y, Roche Lima A, Meléndez LM. Inhibition of Cathepsin B and SAPC Secreted by HIV-Infected Macrophages Reverses Common and Unique Apoptosis Pathways. J Proteome Res. 2022;21:301–312. doi: 10.1021/acs.jproteome.1c00187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang Y, Wu J, Fu Y, Yu R, Su H, Zheng Q, Wu H, Zhou S, Wang K, Zhao J, Shen S, Xu G, Wang L, Yan C, Zou X, Lv Y, Zhang S. Hesperadin suppresses pancreatic cancer through ATF4/GADD45A axis at nanomolar concentrations. Oncogene. 2022;41:3394–3408. doi: 10.1038/s41388-022-02328-4. [DOI] [PubMed] [Google Scholar]
- 44.Wang M, Tian B, Shen J, Xu S, Liu C, Guan L, Guo M, Dou J. Bavachin induces apoptosis in colorectal cancer cells through Gadd45a via the MAPK signaling pathway. Chin J Nat Med. 2023;21:36–46. doi: 10.1016/S1875-5364(23)60383-8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The first author obtained two files, GSE109227 and GSE194331, from the public database GEO for validation purposes.