Abstract
The thymidine kinases (TKs) of herpes simplex virus type 1 (HSV-1), HSV-2, and varicella-zoster virus (VZV) were expressed in human osteosarcoma cells as fusion proteins with the green fluorescent protein (GFP), and their intracellular localizations were determined. The three TK-GFP fusion products were localized in different subcellular compartments of the transfected tumor cells. HSV-1 TK-GFP was localized exclusively in the nucleus, HSV-2 TK-GFP was predominantly found in the cytosol, while VZV TK-GFP was localized in both the nucleus and the cytosol. In support of these findings, we identified a nuclear localization signal (NLS) in the N-terminal arginine-rich region of HSV-1 TK that was absent in HSV-2 and VZV TK. The first 34 amino acids proved necessary for the specific nuclear localization of HSV-1 TK and, when added to the VZV TK-GFP gene construct, also sufficed to specifically target VZV TK-GFP to the nucleus. Further analysis of this NLS through site-directed mutagenesis revealed that the basic amino acid-rich nonapeptide 25R-R-T-A-L-R-P-R-R33 is of crucial importance in the nuclear targeting of HSV-1 TK. In particular, we revealed that the presence of the arginine residues at positions 25, 26, 30, 32, and 33 is obligatory for efficient NLS functioning, whereas arginine and histidine residues outside of the nonapeptide (i.e., residues R18, R20, and H22) did not change the functional properties of the NLS.
The herpesviruses herpes simplex virus type 1 (HSV-1), HSV-2, and varicella-zoster virus (VZV) encode thymidine kinases (TKs), of which the broad substrate specificity is the basis for the treatment of herpesvirus infections. Indeed, the herpesvirus TKs phosphorylate several antiviral nucleoside analogues, including (E)-5-(2-bromovinyl)-2′-deoxyuridine (BVDU) and ganciclovir (17). The herpesvirus TKs also convert antiherpetic drugs to cytostatic agents in TK gene-transfected tumor cells (5–9, 18). The feasibility of a combined gene therapy-chemotherapy approach for the treatment of cancer with HSV-1 TK and ganciclovir is currently being investigated in clinical trials for the treatment of brain tumors (16, 33, 38–41) and ovarian cancer (22). In addition to their importance from a therapeutic viewpoint, herpesvirus thymidine kinases also appear to be necessary for reactivation from latency (15, 20).
The herpesvirus TKs and the mammalian 2′-deoxyguanosine kinase (dGK), 2′-deoxycytidine kinase (dCK), and TK2 are sequence related (4, 28–30). Johansson et al. (30) recently expressed the mammalian deoxyribonucleoside kinases as fusion proteins with the green fluorescent protein (GFP) (11, 48) to study protein localization in living cells (36, 43). Unexpectedly, they found dCK in the nuclear compartment of transfected cells (30), whereas it was formerly believed to localize in the cytosol (3). The literature on the intracellular distribution of the herpesvirus TKs is rather confusing. Cheng and Ostrander (14) found HSV-1 TK mainly in the cytosol of HSV-1 infected HeLa TK− cells. Others detected HSV-1 TK activity in nuclear extracts of HSV-1 TK gene-transfected cells but did not measure TK activity in cytosol extracts (35). Haarr and Flatmark (26), however, showed by an immunofluorescence technique that HSV-1 TK was localized in the cytosol of HSV-1 infected cells, and they found no TK activity in the nuclear extracts of these cells. HSV-2 TK was found in the cytosol of HeLa TK− cells infected with HSV-2 (14). Finally, fluorescent anti-VZV TK antibodies predominantly stained the nuclei of VZV-infected HEL cells (46).
We decided to study the intracellular localization of HSV-1, HSV-2, and VZV TK as fusion proteins with GFP. We found that the TKs of three herpesviruses, i.e., HSV-1, HSV-2, and VZV, were targeted to different cellular compartments after transfection of the cells with the corresponding TK-GFP fusion gene: HSV-1 TK was nuclear, HSV-2 TK was cytosolic, and VZV TK was spread over the nucleus and cytosol. We identified and further characterized a nuclear localization signal (NLS) in the N-terminal domain of HSV-1 TK (i.e., amino acids 1 to 34). We demonstrated that this newly identified NLS sequence was both necessary for specific nuclear localization of HSV-1 TK and sufficient to transport VZV TK (which we found localized in both the nucleus and the cytosol) to the nucleus of NLS-VZV TK-GFP fusion gene-transfected tumor cells. The HSV-1 TK NLS was further characterized in detail through site-directed mutagenesis experiments; the results of these experiments pointed to the crucial role of the individual arginine residues at positions 25, 26, 30, 32, and 33 in NLS functioning.
MATERIALS AND METHODS
Cell culture.
Adherent human osteosarcoma cells deficient in cytosol TK (designated OstTK−) were provided by M. Izquierdo (Universidad Autónoma de Madrid, Madrid, Spain). Cells were maintained at 37°C in a humidified CO2-controlled atmosphere in Eagle’s minimal essential medium (Gibco, Paisley, United Kingdom) supplemented with 10% heat-inactivated fetal calf serum (FCS) (Integro, Zaandam, The Netherlands), 2 mM l-glutamine (Gibco), 0.075% (wt/vol) NaHCO3 (Gibco), 0.5 μl of geomycine (Gentamycin; 40 mg/ml; Schering-Plough) per ml, and 0.5 μl of amphotericin B (Fungizone; 5 mg/ml; Bristol-Myers Squibb, Brussels, Belgium) per ml.
Plasmid construction.
The bacterial strain for plasmid constructions was Escherichia coli DH5α. The pRc/CMV/VZV TK plasmid containing the VZV TK gene coding sequence was kindly provided by J. Piette (University of Liège, Liège, Belgium). The pMCTK and pGR18 plasmids (42) harboring the HSV-1 TK and HSV-2 TK gene coding sequences, respectively, were kindly provided by D. Ayusawa (Yokohama City University, Yokohama, Japan). The following primers were obtained from Gibco or KEBO Lab (Stockholm, Sweden). Primers 1, 3, 5, and 7 introduce an EcoRI restriction site; primers 2, 4, and 6 introduce a SalI site; primers 8, 9, and 10 introduce a PstI site; and primer 11 introduces a NotI site in the amplified PCR product (primer 1, 5′-GAGGAATTCATGTCAACGGATAAAACCGATG; primer 2, 5′-CTCGTCGACAGGGAAGTGTTGTCCTGAACGGC; primer 3, 5′-GAGGAATTCATGGCTTCGTACCCCGGCCATC; primer 4, 5′-CTCGTCGACAGGTTAGCCTCCCCCATCTCCCG; primer 5, 5′-GAGGAATTCATGGCTTCTCACGCCGGCCAAC; primer 6, 5′-CTCGTCGACAGAACTCCCCCCACCTCGCGGGC; primer 7, 5′-GAGGAATTCATGCAAGAAGCCACGGAAGTCCG; primer 8, 5′-GAGCTGCAGCTGCCGGCGAGGGCGCAAC; primer 9, 5′-CTCCTGCAGTCAACGGATAAAACCGATGTAAA; primer 10, 5′-GAGCTGCAGGTGAGCAAGGGCGAGGAGCTG; and primer 11, 5′-GAGGCGGCCGCTTTACTTGTACAGC). The VZV TK (primers 1 and 2), HSV-1 TK (primers 3 and 4), and HSV-2 TK (primers 5 and 6) genes were amplified by PCR from the appropriate plasmids. Upon cloning of the PCR products in the pGEM-T vector (Promega Corp., Madison, Wis.), the EcoRI-SalI TK fragments were excised and cloned into the pEGFP-N1 N-Terminal Protein Fusion Vector (Clontech, Palo Alto, Calif.). To produce the Δ(AA1-34) HSV-1 TK-GFP construct, primers 7 and 4 were used to amplify the HSV-1 TK sequence lacking the coding sequence for amino acids 1 to 34, which was subsequently cloned into the pEGFP-N1 vector. The NLS-VZV TK-GFP vector was constructed by ligating an EcoRI-PstI fragment (derived from the HSV-1 TK gene by using primers 3 and 8) and a PstI-SalI fragment (derived from the VZV TK gene by using primers 9 and 2) together in an EcoRI-SalI-cut pEGFP-N1 vector. To construct the NLS-GFP vector, the aforementioned EcoRI-PstI fragment (derived from the HSV-1 TK gene by using primers 3 and 8) and a PstI-NotI fragment (derived from the GFP gene by using primers 10 and 11) were ligated together in the EcoRI-NotI-digested pEGFP-N1 vector.
Site-directed mutagenesis of putative NLS amino acids.
Eight mutant HSV-1 TK-GFP constructs were prepared according to the QuickChange site-directed mutagenesis kit (Stratagene, La Jolla, Calif.) protocol. The primers used were designed in such a way that the codons for the seven amino acid pairs were replaced by the BamHI restriction site 5′-GGATCC-3′ as follows: for the A17G-R18S mutations, sense primer 5′-CTGCGTTCGACCAGGCTGGATCCTCTCGCGGCCATAGCAAC and antisense primer 5′-GTTGCTATGGCCGCGAGAGGATCCAGCCTGGTCGAACGCAG; for the S19G-R20S mutations, sense primer 5′-CGACCAGGCTGCGCGTGGATCCGGCCATAGCAACCGAC and antisense primer 5′-GTCGGTTGCTATGGCCGGATCCACGCGCAGCCTGGTCG; for the H22S mutation, sense primer 5′-GCTGCGCGTTCTCGCGGATCCAGCAACCGACGTACG and antisense primer 5′-CGTACGTCGGTTGCTGGATCCGCGAGAACGCGCAGC; for the R25G-R26S mutations, sense primer 5′-CGGCCATAGCAACGGATCCACGGCGTTGCGCCC and antisense primer 5′-GGGCGCAACGCCGTGGATCCGTTGCTATGGCCG; for the L29G-R30S mutations, sense primer 5′-GCAACCGACGTACGGCGGGATCCCCTCGCCGGCAGC and antisense primer 5′-GCTGCCGGCGAGGGGATCCCGCCGTACGTCGGTTGC; for the R32G-R33S mutations, sense primer 5′-GGCGTTGCGCCCTGGATCCCAGCAAGAAGCCACG and antisense primer 5′-CGTGGCTTCTTGCTGGGATCCAGGGCGCAACGCC; and for the E36G-A37S mutations, sense primer 5′-CTCGCCGGCAGCAAGGATCCACGGAAGTCCGCC and antisense primer 5′-GGCGGACTTCCGTGGATCCTTGCTGCCGGCGAG. In addition, one insertion mutation was made, inserting 12 nucleotides between the L29 and R30 codons. The following two primers were used: sense primer 5′-GACGTACGGCGTTGGTGTCGACGGCTCGCCCTCGCCGGC and antisense primer 5′-GCCGGCGAGGGCGAGCCGTCGACACCAACGCCGTACGTC.
After linear amplification of the mutant primers with Pfu DNA polymerase (Stratagene) and wild-type HSV-1 TK-GFP vector as a template in a temperature cycler program (30 s at 95°C, followed by 20 cycles of 30 s at 95°C, 1 min at 55°C, and 12 min at 68°C), wild-type plasmid was digested with DpnI restriction enzyme (Stratagene), and the mutant DNA was transformed into competent E. coli DH5α. Kanamycin-resistant colonies were screened for mutant plasmids by SalI or BamHI restriction digestion of the plasmid preparations.
Stable and transient transfection of tumor cells.
The pEGFP-N1 vector and the herpesvirus TK-GFP fusion gene constructs were introduced into OstTK− cells via membrane fusion-mediated transfer by using plasmid-liposome complexes (LipofectAMINE reagent; Gibco) as described by the supplier. Briefly, 2 μg of plasmid DNA and 5 μl of LipofectAMINE reagent, diluted in Opti-MEM I reduced serum medium (Gibco) were used for each transfection of 500,000 cells in a 6-well plate (Nunc, Roskilde, Denmark). The stable TK-GFP fusion gene transfectants shown in Fig. 1 and 3 were isolated by maintaining the cell cultures in the presence of HAT medium (i.e., normal growth medium, supplemented with 100 μM hypoxanthine, 0.4 μM aminopterin, and 16 μM thymidine), while stably transfected GFP-expressing OstTK− cells were isolated after selection in the presence of 0.5 mg of Geneticin (Duchefa, Haarlem, The Netherlands) per ml. Monoclonal transfected OstTK− cell lines were obtained by plating the cells at clonal density in tissue culture plates (Corning, N.Y.), after which single colonies were isolated and expanded. The nuclear localization signal (NLS)-GFP fusion gene (Fig. 3) and the mutant HSV-1 TK-GFP fusion genes (Fig. 4) were only transiently transfected in OstTK−, as pictures were taken 24 h after the transfection procedure. A standard fluorescein isothiocyanate (FITC) filter-equipped fluorescence microscope was used.
FIG. 1.
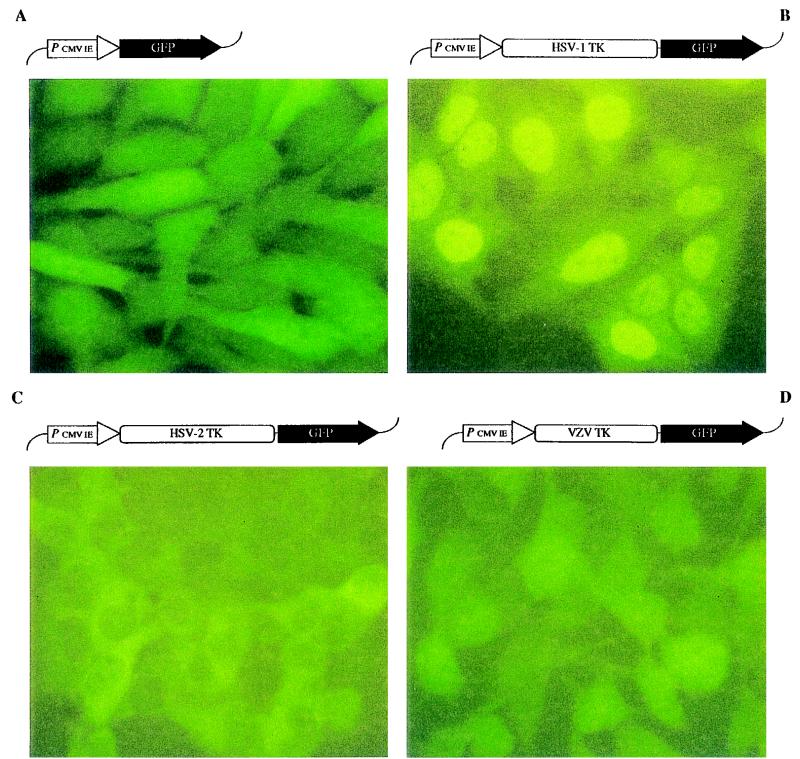
Herpesvirus TKs fused with GFP. The pEGFP-N1 vector, encoding for GFP, and the herpesvirus TK-GFP fusion constructs (shown at the top of each panel) were transfected into OstTK− cells. After selection of stable transfectants, the fluorescence pattern was evaluated by using an FITC filter-equipped fluorescence microscope. Panels: A, Nonfused GFP; B, HSV-1 TK-GFP; C, HSV-2 TK-GFP; D, VZV TK-GFP.
FIG. 3.
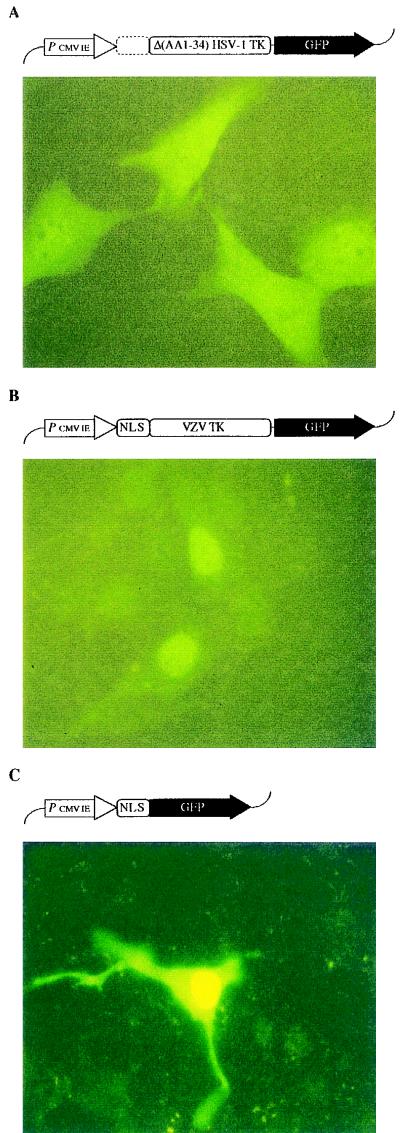
The N-terminal NLS of HSV-1 TK. The HSV-1 TK gene fragment encoding for the N-terminal 34 amino acids was deleted from the HSV-1 TK-GFP construct and transferred to both the VZV TK-GFP fusion gene and control GFP. The resulting GFP fusion constructs (shown on top of each picture) were transfected into OstTK− cells and evaluated by using an FITC filter-equipped fluorescence microscope. Panels: A, Δ(AA1-34) HSV-1 TK-GFP; B, NLS-VZV TK-GFP; C, NLS-GFP.
FIG. 4.
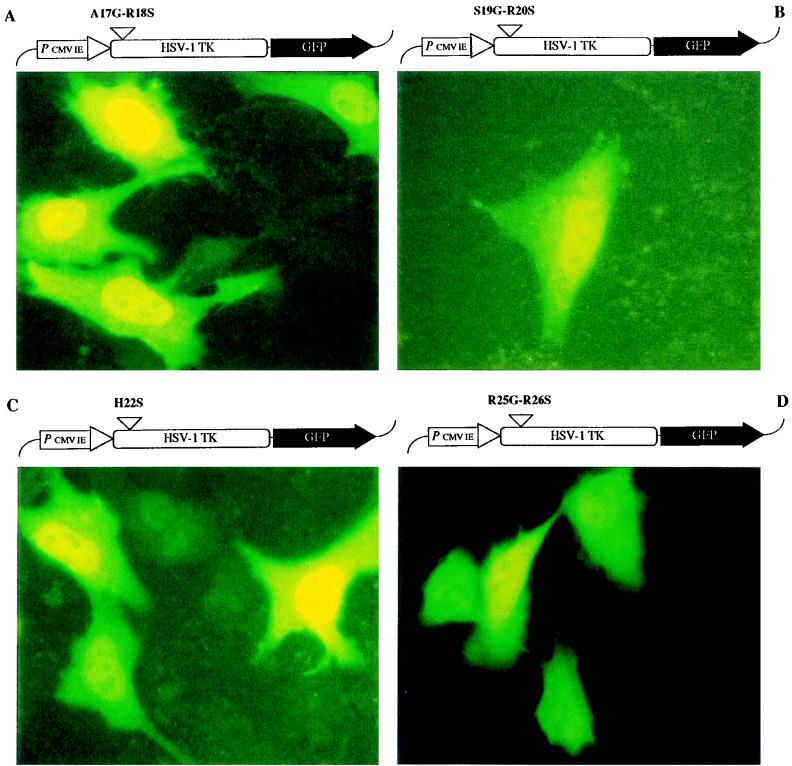
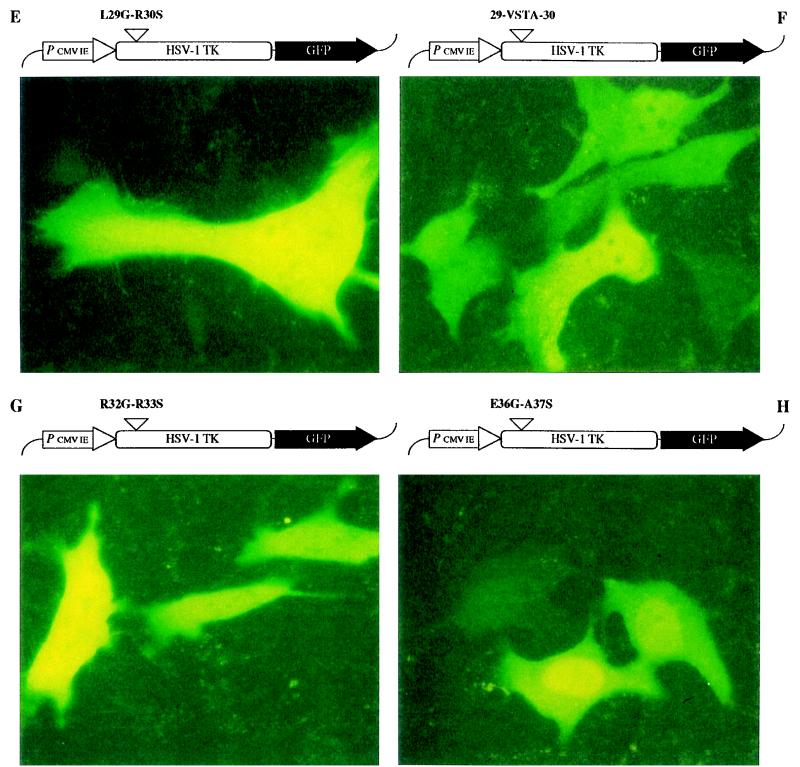
Site-directed mutagenesis in the NLS of HSV-1 TK. The mutant HSV-1 TK-GFP fusion constructs (shown at the top of each panel) were transfected into OstTK− cells and evaluated by using an FITC filter-equipped fluorescence microscope. Panels: A, A17G-R18S mutation; B, S19G-R20S mutation; C, H22S mutation; D, R25G-R26S mutation; E, L29G-R30S mutation; F, 29-VSTA-30 insertion; G, R32G-R33S mutation; H, E36G-A37S mutation.
HSV-1 and HSV-2 infection.
The procedure for the infection of Δ(AA1-34) HSV-1 TK-GFP and HSV-2 TK-GFP fusion gene-expressing OstTK− cells with HSV-1 (Lyons strain) and HSV-2 (G strain), respectively, was adapted from the method of Andrei et al. (2). Briefly, nearly confluent osteosarcoma cells, grown in 6-well plates (Nunc), were inoculated with various dilutions of virus stock (prepared on HEL cells) in 2% FCS-containing medium. After a 2-h adsorption period at 37°C, the medium was replaced by fresh 2% FCS-containing medium, and the cells were further incubated at 37°C in a humidified CO2-controlled atmosphere. Two days later, viral plaques were evaluated under an FITC filter-equipped microscope.
RESULTS
Intracellular localization of herpesvirus TKs.
To study the intracellular localization of HSV-1 TK, HSV-2 TK, and VZV TK, the corresponding genes, fused to the GFP coding sequence in the pEGFP-N1 vector (Clontech), were transfected in human osteosarcoma cells deficient in cytosolic TK (OstTK−). The fluorescence pattern was subsequently evaluated with a FITC filter-equipped fluorescence microscope. The wild-type pEGFP-N1 vector, encoding for GFP, was included as a control. Expression of GFP in the OstTK− cells showed, as expected, a strong fluorescence signal in both the nucleus and the cytosol (Fig. 1A). When the HSV-1 TK-GFP fusion gene was introduced into the OstTK− cells, fluorescence was mainly observed in the nucleus (panel B). The visualization of the nucleoli localize the presence of the HSV-1 TK-GFP fusion protein inside the nucleus and not in association with the nuclear envelope. The HSV-2 TK-GFP fusion product was localized in the cytosol, with no fluorescence in the nucleus (panel C). However, VZV TK fused to GFP showed essentially the same distribution pattern as that seen with control GFP (panel D), i.e., fluorescence was detected both in the nucleus and the cytosol to an equal degree.
HSV-1 TK contains an N-terminal NLS necessary for transport to the nucleus.
The observation that HSV-1 TK was localized in the nucleus, HSV-2 TK was localized in the cytosol, and VZV TK was spread over both the nucleus and cytosol suggested that an NLS was present in the HSV-1 TK protein. In our search of an NLS in the HSV-1 TK (GenBank database), we found that the N-terminal end (amino acids 1 to 34) of the HSV-1 TK consists of a basic amino acid cluster containing seven arginine residues (Fig. 2). Five of these arginine residues, which were localized within two clusters at positions 25 to 26 and at positions 30, 32, and 33 (with a proline residue present at position 31) closely resemble the bipartite consensus motif identified by Dingwall and Laskey (19) in several nuclear proteins, although with a 3-amino-acid spacer instead of a 10-amino-acid spacer. In the homologous HSV-2 TK sequence, only four arginine residues are present, while the VZV TK lacks the complete N-terminal region homologous to the HSV-1 and HSV-2 TK sequence (Fig. 2). No crystallographic data are available on the N-terminal domain (amino acids 1 to 34) of HSV-1 TK (10), and therefore it is unknown how the seven arginine residues are spatially organized in this part of the protein. However, due to the high content of charged amino acids, it is most likely that these residues are exposed on the outer surface of the enzyme, as is required for proper NLS functioning (45).
FIG. 2.
Alignment of N-terminal ends of herpesvirus TKs. The predicted amino acid sequences of HSV-1 TK, HSV-2 TK, and VZV TK were aligned and are shown together with their respective amino acid numbers. The amino acids are indicated by the one-letter code. Note that the region homologous to the N-terminal part of HSV-1 TK and HSV-2 TK is absent in the VZV TK sequence.
In order to delineate the role of the HSV-1 TK N-terminal basic amino acid cluster in the nuclear localization of the enzyme, an HSV-1 TK-GFP deletion mutant that lacks amino acids 1 to 34 [designated Δ(AA1-34) HSV-1 TK-GFP] was constructed. The mutant fusion protein was detected in both the nucleus and the cytosol, thereby resembling the control GFP and the VZV TK-GFP fluorescence pattern (Fig. 3A). This experiment provided evidence that the N-terminal 34 amino acids are indispensable for the specific nuclear localization of HSV-1 TK. To address the issue whether it could also target VZV TK to the nucleus, we linked the amino acid 1 to 34 coding sequence of HSV-1 TK to the 5′ end of the VZV TK-GFP fusion gene. The recombination mutant enzyme (designated NLS-VZV TK-GFP) was localized in the nucleus (Fig. 3B), whereas the wild-type VZV TK-GFP fusion protein was distributed in both the nucleus and the cytosol (compare with Fig. 1D). The recombinant NLS-GFP gene was constructed to confirm that the change to nucleoplasmic localization of the recombinant NLS-VZV TK-GFP depended solely on the HSV-1 TK NLS sequence and to ascertain that no specific sequences of VZV TK were involved. After transfection of the NLS-GFP fusion gene into OstTK− cells, fluorescence was predominantly observed in the nucleus (Fig. 3C).
Deletion of the nuclear localization fragment in HSV-1 TK-GFP is not complemented by viral infection.
The intracellular localization of HSV-1 TK, HSV-2 TK, and VZV TK raises the question as to the reason for this remarkable difference in TK localization. Expression of an individual viral protein in a transfected cell, however, greatly differs from the naturally occurring situation, i.e., viral infection, where a large amount of additional viral proteins is expressed. Morin et al. (37) showed complementation of a nuclear localization defect in a mutant adenovirus DNA binding protein by infection of cells with adenovirus encoding for the mutant protein. One could hypothesize that the three related herpes TKs are all transported to the nucleus by additional viral proteins during viral infection, utilizing an NLS-independent mechanism. Therefore, the Δ(AA1-34) HSV-1 TK-GFP and HSV-2 TK-GFP-expressing cell lines were infected with HSV-1 (Lyons) and HSV-2 (G), respectively, which gave rise to virus-induced plaques in the infected monolayers that were surrounded by rounded cells. However, virus infection did not result in the nuclear targeting of either Δ(AA1-34) HSV-1 TK-GFP or HSV-2 TK-GFP (data not shown).
Site-directed mutagenesis in the N-terminal NLS region.
In order to further characterize the NLS function of the N-terminal end of HSV-1 TK, a variety of mutations were introduced in this region, namely, one insertion between L29 and R30 (comprising 12 nucleotides), one single amino acid mutation at position 22, and six double amino acid mutations (five double amino acid mutations within the region of amino acids 1 to 34 and one double amino acid mutation just outside this region). Mutation of each of the five amino acid pairs in the identified NLS (i.e., A17G-R18S, S19G-R20S, R25G-R26S, L29G-R30S, and R32G-R33S) resulted in the loss of one or two arginine residues in the NLS. The double codons were designed in such a way that they were replaced by the BamHI restriction site, thus encoding for Gly-Ser and enabling rapid screening of mutants by restriction analysis. The single amino acid mutation affects the histidine residue at position 22, which is changed into a serine residue. The E36G-A37S mutation localized outside the NLS served as a control. The 12-nucleotide insertion mutation (designated 29-VSTA-30) was chosen to enlarge the distance between the R25-R26 and the R30-P31-R32-R33 basic amino acid clusters.
The intracellular distributions of the eight mutant HSV-1 TK-GFP constructs were determined after transfection into OstTK− cells (Fig. 4 and Table 1). The A17G-R18S, S19G-R20S, and H22S mutants (Fig. 4A, B, and C, respectively) were exclusively localized in the nuclei of transfected cells, in a way similar to the wild-type HSV-1 TK-GFP (Fig. 1B), indicating that the arginine residues at positions 18 and 20 and the histidine residue at position 22 are not involved in the nuclear signaling of HSV-1 TK. In contrast, the insertion mutation and all double mutations between positions 25 and 33 (i.e., R25G-R26S, panel D; L29G-R30S, panel E; 29-VSTA-30, panel F; and R32G-R33S, panel G) resulted in a diffuse distribution of both nuclear and cytoplasmic fluorescence, pointing to the importance of this whole amino acid segment for the specific nuclear targeting of HSV-1 TK. The control double mutant (E36G-A37S) outside the NLS domain gave fluorescence that was exclusively localized in the nucleus (Fig. 4H), as was the case for wild-type HSV-1 TK-GFP.
TABLE 1.
Overview of site-directed mutagenesis experimentsa
| HSV-1 TK-GFP type | Amino acid sequence (positions 15 to 40) | Locali-zation |
|---|---|---|
| Wild type | QAARSRGHSNRRTALRPRRQQEATEV | N |
| A17G-R18S mutation | QAGSSRGHSNRRTALRPRRQQEATEV | N |
| S19G-R20S mutation | QAARGSGHSNRRTALRPRRQQEATEV | N |
| H22S mutation | QAARSRGSSNRRTALRPRRQQEATEV | N |
| R25G-R26S mutation | QAARSRGHSNGSTALRPRRQQEATEV | C/N |
| L29G-R30S mutation | QAARSRGHSNRRTAGSPRRQQEATEV | C/N |
| 29-VSTA-30 insertion | QAARSRGHSNRRTALVSTARPRRQQEATEV | C/N |
| R32G-R33S mutation | QAARSRGHSNRRTALRPGSQQEATEV | C/N |
| E36G-A37S mutation | QAARSRGHSNRRTALRPRRQQGSTEV | N |
Six double amino acid mutations were carried out on the N-terminal end of HSV-1 TK, changing each targeted amino acid pair to Gly-Ser as indicated by the underscore. In addition, one single amino acid mutation and one insertion mutation of four amino acids were introduced. The intracellular localizations of the resulting HSV-1 TK-GFP fusion products are indicated (N, nuclear; C, cytosolic).
DISCUSSION
We found that the TKs of three evolutionarily related herpesviruses (i.e., the alpha-herpesviruses HSV-1, HSV-2, and VZV) fused with GFP were localized in different compartments of transfected OstTK− cells. Expression of HSV-1 TK-GFP in OstTK− cells gave rise to nuclear fluorescence. After transfection of the HSV-2 TK-GFP fusion gene in tumor cells, fluorescence was observed almost exclusively in the cytosol, whereas VZV TK fused to GFP was distributed throughout the cell (i.e., in both the nucleus and cytosol) and showed the same distribution pattern as the nonfused (control) GFP (Fig. 1).
The expression of the herpetic TKs in different cellular compartments was surprising given the fact that all herpesviruses replicate in the nuclear compartments of cells. Therefore, the localization of the TKs of the various herpesviruses was not expected to differ from one another. On the other hand, there is no evidence that a cytosolic 2′-deoxynucleotide (dNTP) pool physically separated from a nuclear dNTP pool may exist. Therefore, it may not be important whether the TK is expressed in the nucleus or in the cytosol. In fact, HSV TK-dependent nucleoside analogues, such as BVDU and GCV, were not found to differ in their cytostatic activity against HSV TK gene-transfected tumor cells, irrespective of the compartment in which the viral TK gene was expressed (unpublished observations). The same statement could hold for the sensitivity to antiviral nucleoside analogues of herpesviruses expressing either nuclear or cytosolic TK and will be the subject of further study. Johansson et al. (30) recently reached similar conclusions on dCK and the 2′-deoxycytidine (dCyd) nucleotide pools. They found that there was no difference in the cytostatic activity of dCK-dependent nucleoside analogues whether the dCK was expressed in the nuclear or in the cytosolic compartment. Thus, due to the fact that there is no evidence that the nucleus and the cytosol have separate dNTP pools, the biological meaning of the different localization of HSV-1, HSV-2, and VZV TKs in the cellular compartments is currently unclear. To obtain additional insights into the significance of compartmentation of the viral TKs, it would now also be of particular interest to reveal whether deletion of the HSV-2 TK N-terminal region has an effect on the intracellular location of the HSV-2 TK and whether an artificial NLS can be constructed at the N terminus by the substitution of nonbasic residues into arginine residues in a way that is analogous to that of the HSV-1 TK primary amino acid sequence.
HSV TK was demonstrated to play a pivotal role in latency. Indeed, TK-deficient HSV strains, although able to establish latent infections in mouse trigeminal ganglia, cannot reactivate from latency (15, 20). This dependence of reactivation on TK activity may be explained by the requirement for sufficiently high pools of pyrimidine deoxyribonucleotides to permit viral DNA replication in nonreplicating neurons. Recently, Chen and coworkers showed that human TK1 can functionally replace the HSV-1 TK for reactivation of latent virus (13). It is well known that human TK1 is located predominantly in the cytosol (3, 27), as was also shown recently by Johansson et al. with a TK1-GFP fusion construct (30). In addition, both HSV-1 and HSV-2 depend on TK activity for reactivation, yet HSV-1 TK and HSV-2 TK showed completely different intracellular targeting. Therefore, it seems unlikely that the nuclear localization of HSV-1 TK is crucial for reactivation and also that changing the intracellular localization of HSV-1 TK from the nucleus to the cytosol (by deleting the NLS) would affect the reactivation capacity of the virus.
Many nuclear targeting sequences have been identified. As for the targeting of proteins to cellular compartments other than the nucleus, there does not exist a strict NLS consensus motif. Some general characteristics have been deduced for nuclear localization signals (for an overview, see reference 23): (i) they usually consist of short amino acid sequences, containing a high proportion of positively charged amino acids (lysine and/or arginine), often flanked by a proline residue; (ii) unlike mitochondrial targeting signals, they are not localized at specific sites within the protein; (iii) they are not removed after nuclear entry; and (iv) they can coexist with other NLSs within the same protein. The seven-amino-acid stretch P-K-K-K-R-K-V of simian virus 40 large T antigen (SV40T-ag) has served as the prototype NLS (25, 31, 34), and many other nuclear import signals were identified on the basis of sequence homology with the SV40T-ag NLS. Chelsky et al. (12) proposed the four-residue consensus sequence K-R/K-X-R/K. Later, Robbins et al. (44) and Dingwall and Laskey (19) described a bipartite motif, defined as a pair of basic residues separated by a spacer region of any 10 amino acids from a second basic cluster downstream in which at least three of five amino acids are basic. Johansson and coworkers (30) identified a nuclear targeting signal of human dCK representing a “classical” bipartite consensus motif and showed that this signal was required for nuclear import of this protein. The nuclear localization of dCK is the first evidence of a mammalian deoxyribonucleoside kinase being localized in the nucleus.
We have now provided evidence that also HSV-1 TK is localized in the nucleus, and we have identified an amino acid stretch in HSV-1 TK that represents the nuclear targeting signal. The NLS that we identified in the N-terminal region of HSV-1 TK is different from the known “classical” NLS consensus motifs (Fig. 2). When the first 34 amino acids of the HSV-1 TK were deleted, the resulting mutant HSV-1 TK-GFP fusion protein [Δ(AA1-34) HSV-1 TK-GFP] became diffusely localized in both the nucleus and the cytosol, whereas the wild-type HSV-1 TK-GFP showed a specific nuclear localization. We also demonstrated that this NLS was sufficient to target VZV TK and GFP to the nucleus when linked to the 5′-terminal end of the VZV TK and GFP genes, respectively (Fig. 3). The site-directed mutagenesis studies that we have performed on the NLS revealed a nine-amino acid stretch within the NLS (25R-R-T-A-L-R-P-R-R33) that, following replacement of any of the arginine residues, lost its nuclear targeting function. In contrast, mutations of amino acids outside but near this nonapeptide sequence did not affect the nuclear signaling, pointing to the importance of the five-arginine cluster in the NLS identified above. In addition, we showed that the spatial configuration of these five arginines is important to the functioning of the NLS, since the insertion of four extra amino acids within the nonapeptide again destroyed the NLS-driven targeting of the TK to the nucleus.
It could be argued that the intracellular localization of the native herpesvirus TKs (not fused with GFP) should be visualized by using immunofluorescence techniques instead of visualizing the TK-GFP fusion proteins in transfected cells. However, since the introduction of GFP, numerous reports have been published showing the correct nuclear localization of proteins that are fused with GFP and known to reside in the nucleus (e.g., corticoid receptors [21, 24], histones [32, 47], topoisomerases [1], etc.). Moreover, it seems unlikely that the GFP moiety had a significant effect on overall protein folding. Indeed, GFP-linked HSV-1 TK proved to be still enzymatically active in the intact cells, since many antiherpes compounds were highly cytostatic in the HSV-1 TK-GFP gene-transfected tumor cells (unpublished results). Also, GFP-linked TK purified from TK-GFP overexpressing bacterial cell cultures did phosphorylate thymidine and BVDU to a marked extent (data not shown). Taken together with our conclusive data obtained on deletion, insertion, and mutagenesis of the HSV-1 TK NLS, we believe that the GFP fusion protein methodology for determining intracellular TK localization is consistent and reliable.
In conclusion, we have shown that, despite their close relationship, HSV-1 TK, HSV-2 TK, and VZV TK are localized in different intracellular compartments of TK-GFP fusion gene-transfected tumor cells. We have shown that the N-terminal 34 amino acids of HSV-1 TK are necessary and sufficient for its specific nuclear localization. This newly identified NLS does not represent a classical NLS motif but is nevertheless sufficient to specifically target the otherwise uniformly distributed VZV TK to the nucleus. Detailed analysis of the HSV-1 TK NLS by site-directed mutagenesis points to the nonapeptide 25R-R-T-A-L-R-P-R-R33 as being essential for the nuclear targeting of HSV-1 TK.
ACKNOWLEDGMENTS
We thank Graciela Andrei and Robert Snoeck for help with the herpesvirus infection experiments and Christiane Callebaut for dedicated editorial help.
This work was supported by Project 3.0180.95 from the Belgian Fonds Voor Geneeskundig Wetenschappelijk Onderzoek (B.D., E.D.C., and J.B.), Project 95/5 from the Belgian Geconcerteerde Onderzoeksacties (B.D., E.D.C., and J.B.), the Swedish Medical Research Council (A.K. and M.J.), the Medical Faculty of Karolinska Institute (A.K. and M.J.), and the Harald and Greta Jeansson Foundation (A.K. and M.J.). Bart Degrève is the recipient of an IWT fellowship from the Vlaams Instituut voor de bevordering van het Wetenschappelijk-Technologisch onderzoek in de Industrie.
REFERENCES
- 1.Adachi N, Miyaike M, Kato S, Kanamaru R, Koyama H, Kikuchi A. Cellular distribution of mammalian DNA topoisomerase II is determined by its catalytically dispensable C-terminal domain. Nucleic Acids Res. 1997;25:3135–3142. doi: 10.1093/nar/25.15.3135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Andrei G, Snoeck R, Goubau P, Desmyter J, De Clercq E. Comparative activity of various compounds against clinical strains of herpes simplex virus. Eur J Clin Microbiol Infect Dis. 1992;11:143–151. doi: 10.1007/BF01967066. [DOI] [PubMed] [Google Scholar]
- 3.Arnér E S J, Eriksson S. Mammalian deoxyribonucleoside kinases. Pharmacol Ther. 1995;67:155–186. doi: 10.1016/0163-7258(95)00015-9. [DOI] [PubMed] [Google Scholar]
- 4.Balasubramaniam N K, Veerisetty V, Gentry G A. Herpesviral deoxythymidine kinases contain a site analogous to the phosphoryl-binding arginine-rich region of porcine adenylate kinase: comparison of secondary structure predictions and conservation. J Gen Virol. 1990;71:2979–2987. doi: 10.1099/0022-1317-71-12-2979. [DOI] [PubMed] [Google Scholar]
- 5.Balzarini J, De Clercq E, Ayusawa D, Seno T. Murine mammary FM3A carcinoma cells transformed with the herpes simplex virus type 1 thymidine kinase gene are highly sensitive to the growth-inhibitory properties of (E)-5-(2-bromovinyl)-2′-deoxyuridine and related compounds. FEBS Lett. 1985;185:95–100. doi: 10.1016/0014-5793(85)80747-x. [DOI] [PubMed] [Google Scholar]
- 6.Balzarini J, De Clercq E, Verbruggen A, Ayusawa D, Shimizu K, Seno T. Thymidylate synthase Is the principal target enzyme for the cytostatic activity of (E)-5-(2-bromovinyl)-2′-deoxyuridine against murine mammary carcinoma (FM3A) cells transformed with the herpes simplex virus type 1 and type 2 thymidine kinase gene. Mol Pharmacol. 1987;32:410–416. [PubMed] [Google Scholar]
- 7.Balzarini J, Bohman C, De Clercq E. Differential mechanism of cytostatic effect of (E)-5-(2-bromovinyl)-2′-deoxyuridine, 9-(1,3-dihydroxy-2-propoxymethyl)guanine, and other antiherpetic drugs on tumor cells transfected by the thymidine kinase gene of herpes simplex virus type 1 or type 2. J Biol Chem. 1993;268:6332–6337. [PubMed] [Google Scholar]
- 8.Balzarini J, Bohman C, Walker R T, De Clercq E. Comparative cytostatic activity of different antiherpetic drugs against herpes simplex virus thymidine kinase gene-transfected tumor cells. Mol Pharmacol. 1994;45:1253–1258. [PubMed] [Google Scholar]
- 9.Balzarini J, Morin K W, Knaus E E, Wiebe L I, De Clercq E. Novel (E)-5-(2-iodovinyl)-2′-deoxyuridine derivatives as potential cytostatic agents against herpes simplex virus thymidine kinase gene transfected tumors. Gene Ther. 1995;2:317–322. [PubMed] [Google Scholar]
- 10.Brown D G, Visse R, Sandhu G, Davies A, Rizkallah P J, Melitz C, Summers W C, Sanderson M R. Crystal structures of the thymidine kinase from herpes simplex virus type-I in complex with deoxythymidine and ganciclovir. Nat Struct Biol. 1995;2:876–881. doi: 10.1038/nsb1095-876. [DOI] [PubMed] [Google Scholar]
- 11.Chalfie M, Tu Y, Euskirchen G, Ward W W, Prasher D C. Green fluorescent protein as a marker for gene expression. Science. 1994;263:802–805. doi: 10.1126/science.8303295. [DOI] [PubMed] [Google Scholar]
- 12.Chelsky D, Ralph R, Jonak G. Sequence requirements for synthetic peptide-mediated translocalization to the nucleus. Mol Cell Biol. 1989;9:2487–2492. doi: 10.1128/mcb.9.6.2487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen S-H, Cook W J, Grove K L, Coen D M. Human thymidine kinase can functionally replace herpes simplex virus type 1 thymidine kinase for viral replication in mouse sensory ganglia and reactivation from latency upon explant. J Virol. 1998;72:6710–6715. doi: 10.1128/jvi.72.8.6710-6715.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cheng Y-C, Ostrander M. Deoxythymidine kinase induced in HeLa TK− cells by herpes simplex virus type I and type II. II. Purification and characterization. J Biol Chem. 1976;251:2605–2610. [PubMed] [Google Scholar]
- 15.Coen D M, Kosz-Vnenchak M, Jacobson J G, Leib D A, Bogard C L, Schaffer P A, Tyler K L, Knipe D M. Thymidine kinase-negative herpes simplex virus mutants establish latency in mouse trigeminal ganglia but do not reactivate. Proc Natl Acad Sci USA. 1989;86:4736–4740. doi: 10.1073/pnas.86.12.4736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Culver K W, Van Gilder J, Link C J, Carlstrom T, Buroker T, Yuh W, Koch K, Schabold K, Doornbas S, Wetjen B. Gene therapy for the treatment of malignant brain tumors with in vivo tumor transduction with the herpes simplex thymidine kinase gene/ganciclovir system. Hum Gene Ther. 1994;5:343–379. doi: 10.1089/hum.1994.5.3-343. [DOI] [PubMed] [Google Scholar]
- 17.De Clercq E. Antivirals for the treatment of herpesvirus infections. J Antimicrob Chemother. 1993;32:121–132. doi: 10.1093/jac/32.suppl_a.121. [DOI] [PubMed] [Google Scholar]
- 18.Degrève B, Andrei G, Izquierdo M, Piette J, Morin K, Knaus E E, Wiebe L I, Basrah I, Walker R T, De Clercq E, Balzarini J. Varicella-zoster virus thymidine kinase gene and antiherpetic pyrimidine nucleoside analogues in a combined gene/chemotherapy treatment for cancer. Gene Ther. 1997;4:1107–1114. doi: 10.1038/sj.gt.3300502. [DOI] [PubMed] [Google Scholar]
- 19.Dingwall C, Laskey R A. Nuclear targeting sequences—a consensus? Trends Biochem Sci. 1991;16:478–481. doi: 10.1016/0968-0004(91)90184-w. [DOI] [PubMed] [Google Scholar]
- 20.Efstathiou S, Kemp S, Darby G, Minson A C. The role of herpes simplex virus type 1 thymidine kinase in pathogenesis. J Gen Virol. 1989;70:869–879. doi: 10.1099/0022-1317-70-4-869. [DOI] [PubMed] [Google Scholar]
- 21.Fejes-Toth G, Pearce D, Naray-Fejes-Toth A. Subcellular localization of mineralocorticoid receptors in living cells: effects of receptor agonists and antagonists. Proc Natl Acad Sci USA. 1998;95:2973–2978. doi: 10.1073/pnas.95.6.2973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Freeman S M, McCune C, Robinson W, Abboud C N, Abraham G N, Angel C, Marrogi A. The treatment of ovarian cancer with a gene modified cancer vaccine: a phase I study. Hum Gene Ther. 1995;6:927–939. doi: 10.1089/hum.1995.6.7-927. [DOI] [PubMed] [Google Scholar]
- 23.Garcia-Bustos J, Heitman J, Hall M N. Nuclear protein localization. Biochim Biophys Acta. 1991;1071:83–101. doi: 10.1016/0304-4157(91)90013-m. [DOI] [PubMed] [Google Scholar]
- 24.Georget V, Lobaccaro J M, Terouanne B, Mangeat P, Nicolas J C, Sultan C. Trafficking of the androgen receptor in living cells with fused green fluorescent protein-androgen receptor. Mol Cell Endocrinol. 1997;129:17–26. doi: 10.1016/s0303-7207(97)04034-3. [DOI] [PubMed] [Google Scholar]
- 25.Goldfarb D S, Gariepy J, Schoolnik G, Kornberg R D. Synthetic peptides as nuclear localization signals. Nature. 1986;332:641–644. doi: 10.1038/322641a0. [DOI] [PubMed] [Google Scholar]
- 26.Haarr L, Flatmark T. Evidence that deletion of coding sequences in the 5′ end of the thymidine kinase gene of herpes simplex virus type 1 affects the stability of the gene products. J Gen Virol. 1987;68:2817–2829. doi: 10.1099/0022-1317-68-11-2817. [DOI] [PubMed] [Google Scholar]
- 27.He Q, Skog S, Wang N, Eriksson S, Tribukait B. Characterization of a peptide antibody against a C-terminal part of human and mouse cytosolic thymidine kinase, which is a marker for cell proliferation. Eur J Cell Biol. 1996;70:117–124. [PubMed] [Google Scholar]
- 28.Johansson M, Karlsson A. Cloning and expression of human deoxyguanosine kinase cDNA. Proc Natl Acad Sci USA. 1996;93:7258–7262. doi: 10.1073/pnas.93.14.7258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Johansson M, Karlsson A. Cloning of the cDNA and chromosome localization of the gene for human thymidine kinase 2. J Biol Chem. 1997;272:8454–8458. doi: 10.1074/jbc.272.13.8454. [DOI] [PubMed] [Google Scholar]
- 30.Johansson M, Brismar S, Karlsson A. Human deoxycytidine kinase is localized in the cell nucleus. Proc Natl Acad Sci USA. 1997;94:11941–11945. doi: 10.1073/pnas.94.22.11941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kalderon D, Roberts B L, Richardson W D, Smith A E. A short amino acid sequence able to specify nuclear localization. Cell. 1984;39:499–509. doi: 10.1016/0092-8674(84)90457-4. [DOI] [PubMed] [Google Scholar]
- 32.Kanda T, Sullivan K F, Wahl G M. Histone-GFP fusion protein enables sensitive analysis of chromosome dynamics in living mammalian cells. Curr Biol. 1998;8:377–385. doi: 10.1016/s0960-9822(98)70156-3. [DOI] [PubMed] [Google Scholar]
- 33.Kun L E, Gajjar A, Muhlbauer M, Heideman R L, Sanford R, Brenner M, Walter A, Langston J, Jenkins J, Facchini S. Stereotactic injection of herpes simplex thymidine kinase vector producer cells (PA317-G1Tk1SvNa.7) and intravenous ganciclovir for the treatment of progressive or recurrent primary supratentorial pediatric malignant brain tumors. Hum Gene Ther. 1995;6:1231–1255. doi: 10.1089/hum.1995.6.9-1231. [DOI] [PubMed] [Google Scholar]
- 34.Lanford R E, Butel J S. Construction and characterization of an SV40 mutant defective in nuclear transport of T antigen. Cell. 1984;37:801–813. doi: 10.1016/0092-8674(84)90415-x. [DOI] [PubMed] [Google Scholar]
- 35.Lee C G, Kim C G, Namkung R, Lee S E, Park S D. Transfection of mouse cells with thymidine kinase gene of herpes simplex virus. Cytotechnology. 1990;3:141–147. doi: 10.1007/BF00143676. [DOI] [PubMed] [Google Scholar]
- 36.Levy J P, Muldoon R R, Zolotukhin S, Link C J. Retroviral transfer and expression of a humanized, red-shifted green fluorescent protein gene into human tumor cells. Nat Biotechnol. 1996;14:610–614. doi: 10.1038/nbt0596-610. [DOI] [PubMed] [Google Scholar]
- 37.Morin N, Delsert C, Klessig D F. Nuclear localization of the adenovirus DNA-binding protein: requirement for two signals and complementation during viral infection. Mol Cell Biol. 1989;9:4372–4380. doi: 10.1128/mcb.9.10.4372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Oldfield E H. Gene therapy for the treatment of brain tumors using intra-tumoral transduction with the thymidine kinase gene and intravenous ganciclovir. Clinical Protocols Hum Gene Ther. 1993;4:36–69. doi: 10.1089/hum.1993.4.1-39. [DOI] [PubMed] [Google Scholar]
- 39.Raffel C, Culver K, Kohn D, Nelson M, Siegel S, Gillis F, Link C J, Villablanca J G. Gene therapy for the treatment of recurrent pediatric malignant astrocytomas with in vivo tumor transduction with the herpes simplex thymidine kinase gene/ganciclovir system. Hum Gene Ther. 1994;5:863–890. doi: 10.1089/hum.1994.5.7-863. [DOI] [PubMed] [Google Scholar]
- 40.Ram Z, Culver K W, Walbridge S, Frank J A, Blaese R M, Oldfield E H. Toxicity studies of retroviral-mediated gene transfer for the treatment of brain tumors. J Neurosurg. 1993;79:400–407. doi: 10.3171/jns.1993.79.3.0400. [DOI] [PubMed] [Google Scholar]
- 41.Ram Z, Culver K W, Oshiro E, Viola J J, DeVroom H L, Otto E, Long Z, Chiang Y, McGarrity G J, Muul L M, Katz D, Blaese R M, Oldfield E H. Therapy of malignant brain tumors by intratumoral implantation of retroviral vector-producing cells. Nat Med. 1997;12:1354–1361. doi: 10.1038/nm1297-1354. [DOI] [PubMed] [Google Scholar]
- 42.Reyes G R, Jeang K-T, Hayward G S. Transfection with the isolated herpes simplex virus thymidine kinase gene. I. Minimal size of the active fragments from HSV-1 and HSV-2. J Gen Virol. 1982;62:191–206. doi: 10.1099/0022-1317-62-2-191. [DOI] [PubMed] [Google Scholar]
- 43.Rizzuto R, Brini M, Pizzo P, Murgia M, Pozzan T. Chimeric green fluorescent protein as a tool for visualizing subcellular organelles in living cells. Curr Biol. 1995;5:635–642. doi: 10.1016/s0960-9822(95)00128-x. [DOI] [PubMed] [Google Scholar]
- 44.Robbins J, Dilworth S, Laskey R A, Dingwall C. Two interdependent basic domains in nucleoplasmin nuclear targeting sequence: identification of a class of bipartite nuclear targeting sequence. Cell. 1991;64:615–623. doi: 10.1016/0092-8674(91)90245-t. [DOI] [PubMed] [Google Scholar]
- 45.Roberts B L, Richardson W D, Smith A E. The effect of protein context on nuclear localization signal function. Cell. 1987;50:465–475. doi: 10.1016/0092-8674(87)90500-9. [DOI] [PubMed] [Google Scholar]
- 46.Shiraki K, Ogino T, Yamanishi K, Takahashi M. Immunochemical characterization of pyrimidine kinase induced by varicella-zoster virus. J Gen Virol. 1985;66:221–229. doi: 10.1099/0022-1317-66-2-221. [DOI] [PubMed] [Google Scholar]
- 47.Ushinsky S C, Bussey H, Ahmed A A, Wang Y, Friesen J, Williams B A, Storms R K. Histone H1 in Saccharomyces cerevisiae. Yeast. 1997;13:151–61. doi: 10.1002/(SICI)1097-0061(199702)13:2<151::AID-YEA94>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- 48.Youvan D C, Michel-Beyerle M E. Structure and fluorescence mechanism of GFP. Nat Biotechnol. 1996;14:1219–1220. doi: 10.1038/nbt1096-1219. [DOI] [PubMed] [Google Scholar]