Abstract
Micro RNAs (miRs) have been implicated in various tumorigenic processes. Osteosarcoma (OS) is a primary bone malignancy seen in adolescents. However, the mechanism of miRs in OS has not been fully demonstrated yet. Here, miR-134-5p was found to inhibit OS progression and was also expressed at significantly lower levels in OS tissues and cells relative to normal controls. miR-134-5p was found to reduce vasculogenic mimicry, proliferation, invasion, and migration of OS cells, with miR-134-5p knockdown having the opposite effects. Mechanistically, miR-134-5p inhibited expression of the ITGB1/MMP2/PI3K/Akt axis, thus reducing the malignant features of OS cells. In summary, miR-134-5p reduced OS tumorigenesis by modulation of the ITGB1/MMP2/PI3K/Akt axis, suggesting the potential for using miR-134-5p as a target for treating OS.
Subject terms: Bone cancer, Non-coding RNAs
Introduction
Osteosarcoma (OS) is a primary tumor of bone, most frequent in young patients between the ages of 15 and 19 years, and originating from mesenchymal tissue [1–3]. OS is highly heterogeneous and carries a high risk of metastasis and recurrence [4]. The survival rates of patients with OS have not improved significantly over the past three decades [5]. This is due to difficulties in treatment, resulting from the complexity of the OS karyotype and a high degree of chromosomal and genomic instability [6]. Even with advances in the treatment of surgical resection under general anesthesia combined with chemotherapy and radiotherapy, only 65–70% of patients with OS achieve a curative outcome [7, 8]. Besides, little progress has been made in improving survival in OS patients over the past 40 years [9]. Thus, new developments are urgently required for OS treatment.
MicroRNAs (miRs) have been implicated in numerous physiological activities [10]. MiRNAs interact with the 3′ UTRs of mRNAs, inducing cleavage of the target mRNA or suppression of its translation [11, 12]. Abnormal expression of even one miRNA may influence tumor development [13]. Investigation of the specific roles of miRNAs in tumors is thus important [13]. Specific miRNAs have also been found to be useful biomarkers for different cancers [14, 15]. Vasculogenic mimicry (VM) is an alternative form of angiogenesis derived from tumor cells rather than vascular endothelial cells. VM positivity has been shown to be an independent prognostic factor of OS [16]. To date, the knockdown of lncRNA n340532 [17], MIG-7 [18], and VEGF [19] has been shown to inhibit VM in OS. Besides, Paris polyphylla extracts [20] and zoledronic acid [21] have been shown to inhibit VM in OS. However, the mechanism underlying OS VM requires further investigation.
A previous study reported the results of a global microarray analysis of miRNA levels in 19 human OS cell lines and four human bone tissue samples, finding that miR-134-5p was one of 154 miRNAs that were differentially expressed in OS [22]. Several studies have investigated the functions of miR-134-5p. In lung adenocarcinoma, miR-134-5p was shown to promote both chemoresistance and metastasis through the targeting of DAB2 [23], while in gastric cancer, miR-134-5p inhibited tumor progression and invasion by targeting YWHAZ [24]. Our team’s previous studies have demonstrated that miR-134-5p can inhibit the proliferation and metastasis of osteosarcoma [25, 26]. VM is an important malignant phenotype in tumor development. However, the relationship between miR-134-5p and VM has not yet been fully demonstrated. Here, it was demonstrated for the first time that miR-134-5p promoted VM, proliferation, invasion, and migration of OS via the ITGB1/MMP2/PI3K/Akt pathway.
Results
miR-134-5p inhibits VM, proliferation, migration, and invasion of OS cells
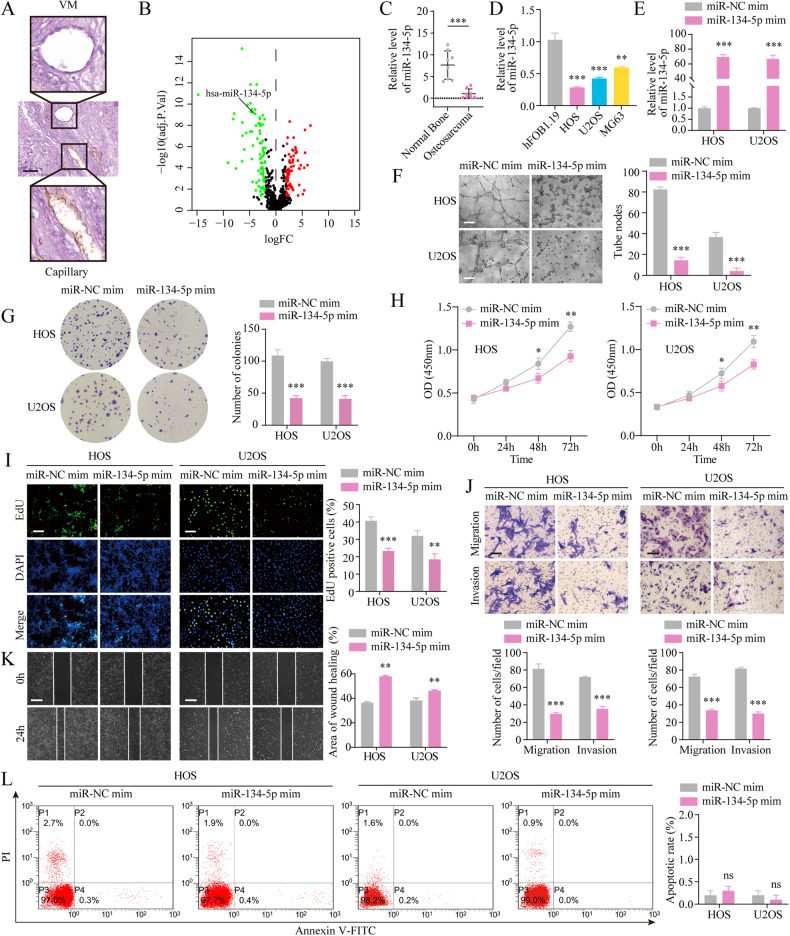
CD31/PAS staining showed the presence of characteristic VM structures in OS tissues, with the establishment of connections between cells forming a vascular lumen containing occasional erythrocytes, and the presence of a purple-colored net-like formation (Fig. 1A). To identify the critical miRNAs involved in OS progression, we examined the miRNA expression profiles in the GSE28423 dataset, which included information from 19 human OS cell lines and four human bone tissues [22]. Using the criteria of P value < 0.05 and log2|FC | ≥ 2, miR-134-5p was found to be one of 154 miRNAs identified in the database as differentially expressed (Fig. 1B). Furthermore, lower levels of miR-134-5p were observed in OS compared with normal tissue (Fig. 1C), as well as in OS cells relative to osteoblasts (Fig. 1D). Thus, miR-134-5p was selected for further analyses.
Fig. 1. miR-134-5p mimic blocks VM, proliferation, migration, and invasion in OS.
A CD31/PAS double-staining showing immunohistochemical staining for VM in OS. (Scale bar, 50 μm). B Gene expression profiles of GSE28423 are shown as volcano plots. Upregulated DEGs are represented in red and downregulated DEGs are represented in green. C Reduced miR-134-5p levels in OS tissue (n = 8) vs normal bone (n = 8), shown by qRT-PCR. Statistics using independent Student’s t-test. D Decreased miR-134-5p levels in OS cells versus osteoblasts. E Expression of miR-134-5p in transfected cells, shown by qRT-qPCR. F Tube formation assays to assess effects of the miR-134-5p mimic on VM. Scale bar, 200 μm. G–I Colony formation, CCK-8, and EdU assays (scale bar, 400 μm) to evaluate the effects of the miR-134-5p mimic on cell proliferation. J Transwell assays to evaluate the effects of the miR-134-5p mimic on cell migration and invasion. Scale bar, 100 μm. K Wound-healing assays to evaluate the effects of the miR-134-5p mimic on migration. Scale bar, 100 μm. L Flow cytometry to evaluate the effects of the miR-134-5p mimic on apoptosis. The data represent means ± SD. Statistics using one-way ANOVA with Tukey’s test. ns not significant; *p < 0.05; **p < 0.01; ***p < 0.001.
After transfection of cells with lentivirus (Fig. 1E), there were significantly fewer VM structures visible in cells overexpressing miR-134-5p (Fig. 1F), accompanied by a marked reduction in the proliferation of HOS and U2OS cells (Fig. 1G–I). In addition, both invasion and migration were dramatically reduced in OS cells overexpressing miR-134-5p (Fig. 1J, K) However, no significant change in apoptosis levels was observed after overexpression of miR-134-5p (Fig. 1L).
The miR-134-5p knockdown efficiency after transduction of OS cell lines with the miR-134-5p inhibitor or control lentivirus was evaluated using qRT-PCR (Fig. S1A). This showed that miR-134-5p knockdown resulted in markedly increased VM, proliferation, invasion, and migration in the cells, thus supporting (Fig. S1B–G) the anti-cancer function of miR-134-5p in OS.
Identification of miR-134-5p targets
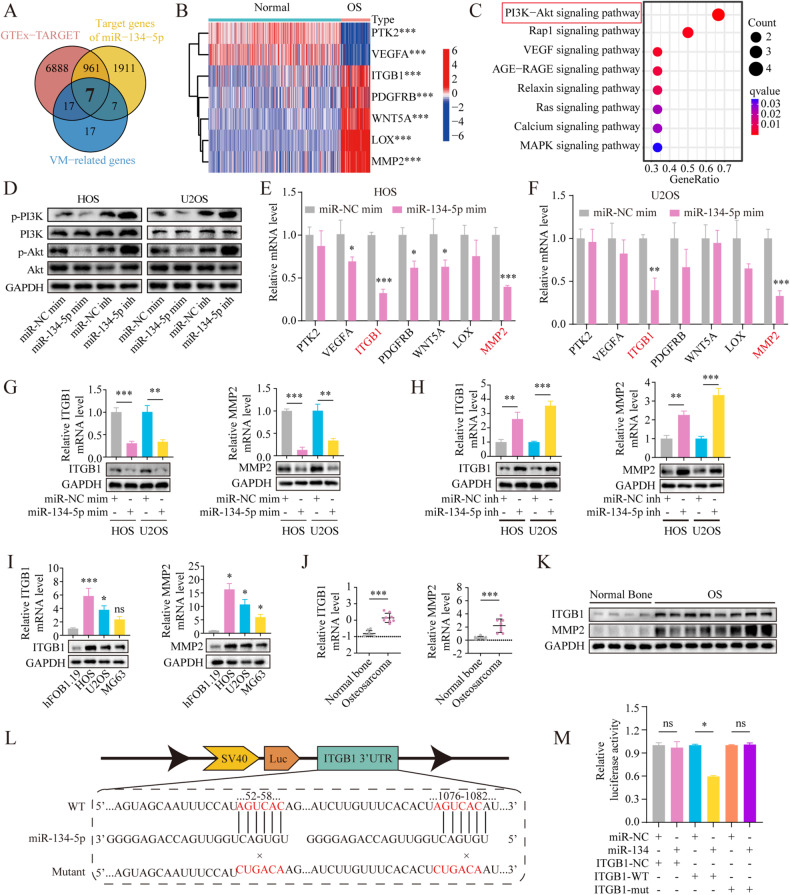
After merging three gene sets, including the 7873 DEGs, 48 VM-associated genes [27], and 2886 prediction target genes for miR-134-5p, seven candidate target genes were finally identified (Fig. 2A). The chromosomal localization of the seven genes is shown in Fig. S2A. PPI networks of the seven candidate target genes were constructed (Fig. S2B), and the diagnostic abilities of the genes were examined with receiver operating characteristic (ROC) curves. All seven genes were found to be potential markers for OS diagnosis (Fig. S2C). The comparative expression of these genes between normal and OS tissue in the GTEx-TARGET dataset is shown in the heatmap (Fig. 2B). KEGG analysis (Fig. 2C and Table S6) of the candidate genes showed significant enrichment in the PI3K-Akt pathway while GO analysis (Fig. S2D and Table S7) demonstrated significant enrichment in processes associated with cell chemotaxis and wound healing. Furthermore, MMP2 and ITGB1 were found to be strongly correlated (Fig. S2E). As shown by western blotting, miR-134-5p effectively blocked the phosphorylation of PI3K and Akt (Fig. 2D). All full and uncropped western blots were displayed in a file named “original western blots”. Evaluation of mRNA levels after miR-134-5p mimic transduction (Fig. 2E, F) indicated that MMP2 and ITGB1 were the most significantly downregulated; these two genes were thus used for further analyses. The results of GSEA pathway analysis show that high MMP2 levels were correlated with focal adhesion and oxidative phosphorylation, while high ITGB1 levels were enriched in pathways involved in focal adhesion and cytoskeletal regulation (Fig. S2F, G).
Fig. 2. Identification of miR-134-5p target genes in OS.
A The Venn diagram shows the intersection of three sets: 48 VM-associated genes, 2886 predicted miR-134-5p target genes, and 7873 DEGs. B Heatmap showing expression of seven genes between OS and control tissue in the GTEx-TARGET cohort. C KEGG enrichment analysis of the seven candidate miR-134-5p target genes. D Protein levels of p-PI3K, PI3K, p-Akt, and Akt in OS cells following transfection with the miR-134-5p mimic or miR-134-5p inhibitor or the corresponding negative control. E, F mRNA expression of seven candidate genes in cells transfected with the miR-134-5p mimic. G Protein and mRNA levels of MMP2 and ITGB1 in miR-134-5p mimic-transfected OS cells versus control. H Protein and mRNA levels of MMP2 and ITGB1 in miR-134-5p-knockdown OS cells versus control. I Protein and mRNA levels of MMP2 and ITGB1 in OS cells versus osteoblasts. Statistics using one-way ANOVA with Tukey’s test. J, K Protein and mRNA levels of MMP2 and ITGB1 in OS tissues versus normal bone. Statistics using independent Student’s t-test. L Wild-type and mutant binding sites for ITGB1 in miR-134-5p. M Luciferase assay of HOS cells transfected with miR-134-5p mimic (10 nM) or control miR-NC and plasmids containing 3′ UTRs of ITGB1. Statistics using one-way ANOVA with Tukey’s test. The data represent means ± SD. ns not significant; *p < 0.05; **p < 0.01; ***p < 0.001.
The influence of miR-134-5p on MMP2 and ITGB1 levels was then examined by cell transduction with an miR-134-5p mimic and subsequent measurement of MMP2 or ITGB1 levels. The qRT-PCR and western blotting analyses indicated that miR-134-5p decreased the levels of both MMP2 and ITGB1 (Fig. 2G, H). Besides, both MMP2 and ITGB1 levels are found to be significantly upregulated in human OS cells (Fig. 2I) and tissues (Fig. 2J, K). Potential binding between miR-134-5p, ITGB1, and MMP2 was then examined. By prediction, MMP2 was found to have no binding target for miR-134-5p, while ITGB1 had a binding target for miR-134-5 (Fig. 2L). Luciferase assays showed a decrease in luciferase activity after cell transfection of ITGB1-WT reporter vectors with the miR-134-5p mimic, while no changes were seen after transfection with the scrambled sequences. However, no luciferase activity was observed with mutated binding sequences (Fig. 2M).
miR-134-5p was found to regulate MMP2 expression by regulating the expression of ITGB1
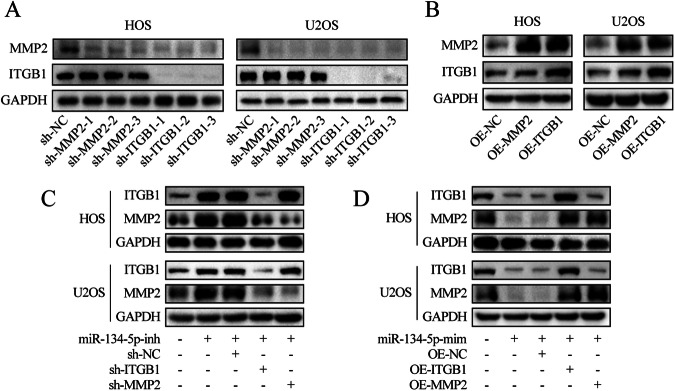
miR-134-5p cannot directly target MMP2 mRNA, but the addition of miR-134-5p mimics results in a decrease in MMP2 expression. We also found a significant positive correlation between MMP2 and ITGB1 by analyzing the correlation between the seven candidate target genes (Fig. S2E). So, we guessed that miR-134-5p may regulate MMP2 expression by regulating the expression of ITGB1. First, three short hairpin RNAs (shRNAs) for MMP2 and ITGB1 were designed. Of these, sh-ITGB1-1 and sh-MMP2-1 were found to be the most efficient and were thus used subsequently for knockdown experiments (Fig. 3A). Surprisingly, decreased MMP2 protein levels were observed in ITGB1 knockdown cells. Furthermore, western blotting showed that the knockdown of ITGB1 rescued the reduced MMP2 protein expression caused by miR-134-5p, although the knockdown of MMP2 was unable to rescue the decrease in ITGB1 caused by miR-134-5p (Fig. 3C). Conversely, overexpression of MMP2 and ITGB1 showed a similar trend (Fig. 3B, D). Together, these findings demonstrate that miR-134-5p reduced MMP2 expression by decreasing the expression of ITGB1.
Fig. 3. MMP2 changes with ITGB1 in OS cells.
A Protein levels of MMP2 and ITGB1 in OS cells transfected with knockdown lentivirus versus the corresponding negative control. B Protein levels of MMP2 and ITGB1 in OS cells transfected with overexpression lentivirus versus the corresponding negative control. C, D Protein levels of MMP2 and ITGB1 in different groups of OS cells.
miR-134-5p acts through the ITGB1/MMP2/PI3K/Akt axis
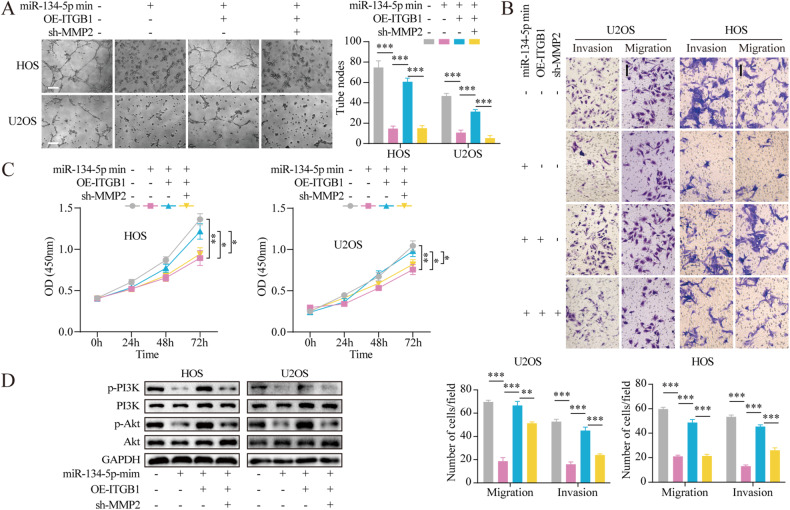
First, cells stably expressing the miR-134-5p mimic were transfected with the overexpression ITGB1 (OE-ITGB1) alone or together with sh-MMP2. The reductions in VM seen in the miR-134-5p mimic-expressing cells were significantly mitigated by OE-ITGB1. However, this effect was counteracted by co-transfection with sh-MMP2 (Fig. 4A). Transwell assay showed that the inhibition of cell invasion and migration induced by miR-134-5p was significantly attenuated by OE-ITGB1. In addition, in cells stably expressing the miR-134-5p mimic, the prevention of cell mobility induced by miR-134-5p recovered after co-transfection with OE-ITGB1 and sh-MMP2 (Fig. 4B). Furthermore, OE-ITGB1 abolished miR-134-5p mimic-induced inhibition of proliferation, while re-transfection with sh-MMP2 reduced these actions (Fig. 4C). Similar effects were seen in the regulatory actions of the miR-134-5p/ITGB1/MMP2 axis on components of the PI3K/Akt pathway (Fig. 4D). The results of functional rescue experiments in miR-134-5p-knockdown cells (Fig. S3) demonstrated that inhibition of miR-134-5p stimulated the PI3K/Akt pathway through activation of the ITGB1/MMP2 axis, thus facilitating the VM, proliferation, migration, and invasion of OS cells.
Fig. 4. Loss-of-function analyses verifying the involvement of the miR-134-5p/ITGB1/MMP2 axis in OS VM, proliferation, migration, and invasion.
A Tube formation assays to evaluate the effects of the miR-134-5p/ITGB1/MMP2 axis on VM. Scale bar, 200 μm. B Transwell assays to evaluate the effects of the miR-134-5p/ITGB1/MMP2 axis on migration and invasion. Scale bar, 100 μm. C CCK-8 assay were to evaluate the effect of miR-134-5p/ITGB1/MMP2 axis on cell proliferation. D Protein levels of p-PI3K, PI3K, p-Akt, and Akt, shown by western blotting. Statistics using one-way ANOVA with Tukey’s test. The data represent means ± SD. *p < 0.05; **p < 0.01; ***p < 0.001.
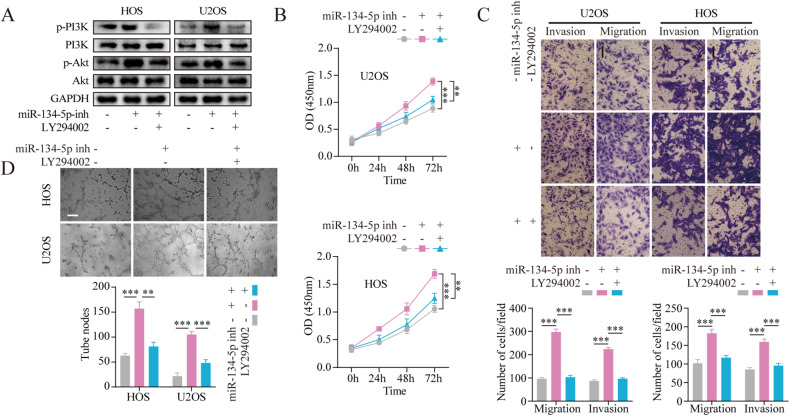
To confirm the observed molecular processes, the effects of miR-134-5p knockdown together with inhibition of PI3K (using LY294002 at a concentration of 20 μM) on the malignant phenotype of OS cells were evaluated. Treatment with the miR-134-5p inhibitor was found to raise phosphorylated PI3K and Akt levels in OS cells, while these effects were counteracted by exposure to LY294002 (Fig. 5A). It was also verified that the miR-134-5p inhibitor markedly enhanced the malignant phenotype, which was reversed by treatment with LY294002 (Fig. 5B–D).
Fig. 5. LY294002 rescues miR-134-5p-mediated promotion of OS cell VM, proliferation, migration, and invasion.
A Protein levels of p-PI3K, PI3K, p-Akt, and Akt in different groups. B CCK-8 assay to evaluate the effects of the miR-134-5p/PI3K/Akt axis on cell proliferation. C Transwell assays to evaluate the effects of the miR-134-5p/PI3K/Akt axis on migration and invasion. Scale bar, 100 μm. D Tube formation assays to evaluate the effects of the miR-134-5p/PI3K/Akt axis on VM. Statistics using one-way ANOVA with Tukey’s test. Scale bar, 200 μm. The data represent means ± SD. **p < 0.01; ***p < 0.001.
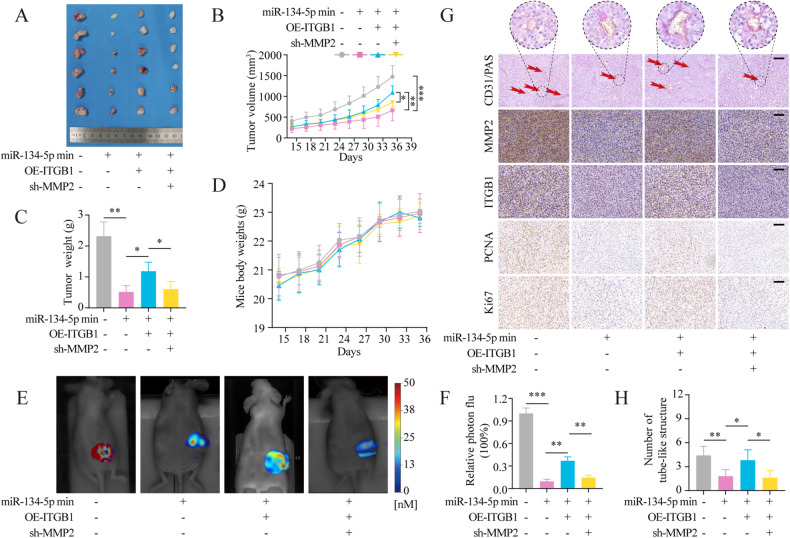
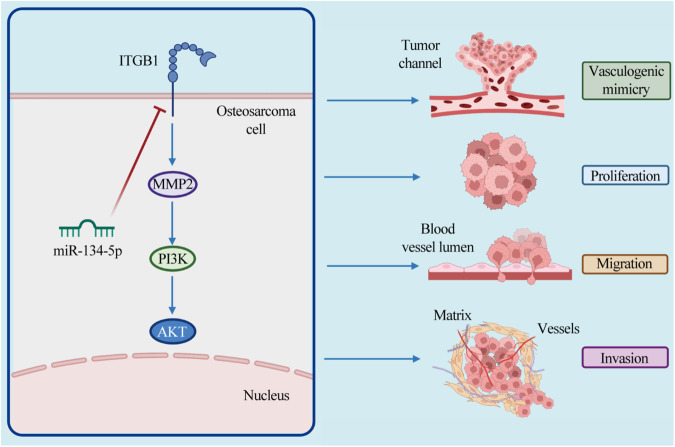
In the in vivo experiments, the miR-134-5p mimic was found to reduce the formation of tumors in mice receiving control cells. Furthermore, ITGB1 overexpression abolished these effects and promoted both growth in OS cells, while this was reversed by MMP2 knockdown (Fig. 6A-C). Mouse body weights were not significantly different between the three groups (Fig. 6D), and no significant toxicity was observed (Fig. S4). The results of in vivo fluorescence in animals followed the same trend as above (Fig. 6E, F). Besides, the results of IHC also showed the same trend in VM, proliferation, ITGB1, and MMP2 (Fig. 6G, H). The schematic shows the role of miR-134-5p in OS by affecting the ITGB1/MMP2/PI3K/Akt axis (Fig. 7).
Fig. 6. Loss-of-function analyses verifying the involvement of the miR-134-5p/ITGB1/MMP2 axis in mouse xenograft tumor proliferation and VM.
A Tumor images. B Tumor volumes. C Tumor weights. D Mice body weights. E, F Bioimaging of tumors using fluorescence molecular tomography and MMPSense 680 to quantify MMP activity in tumors. G, H Immunohistochemical staining of tumors; the red arrow indicates the VM. Scale bar, 100 μm. Statistics using one-way ANOVA with Tukey’s test. The data represent means ± SD. *p < 0.05; **p < 0.01; ***p < 0.001.
Fig. 7.
Schematic diagram of the proposed biological function of the miR-134-5p/ITGB1/MMP2/PI3K/Akt axis in OS VM, proliferation, migration, and invasion.
Discussion
OS is the most common bone malignancy and is linked to high levels of metastasis and mortality [28]. It has been estimated that about 80% of OS cases have subclinical metastases in the lungs at diagnosis [29]. Management of OS is impeded by an absence of accurate tumor markers. Recent demonstrations of dysregulated noncoding RNAs in OS have attracted much interest. miRNAs have been linked with a variety of functions [30–32] and their abnormal expression is linked to cancer-associated behaviors [33–44]. This highlights the importance of understanding the functions of miRs associated with tumors.
As an important epigenetic factor, miR-134-5p has been shown to inhibit the progression of gastric cancer [24], breast cancer [45], thyroid-like cancer [46], melanoma [47], and esophageal squamous cell carcinoma [48]. However, Zhang et al. reported that miR-134-5p, by targeting DAB2, can promote metastasis and chemoresistance in lung cancer [23]. The different roles played by miR-134-5p in different cancers may be due to the different tumor microenvironments in different cancers. Here, miR-134-5p was shown to contribute to OS tumorigenesis. This was seen in the downregulation of miR-134-5p in OS cells and tissues. Investigations into the function of miR-134-5p showed that it promoted VM, proliferation, migration, and invasion in OS cells. However, interactions between tumor-malignant phenotypes are possible. For example, miR-134-5p can directly inhibit VM by inhibiting ITGB1 and MMP2. Meanwhile, miR-134-5p’s role in indirectly inhibiting VM by inhibiting proliferation may also exist at the same time. Wang et al.‘s study indicates that proliferation can be inhibited by inhibiting tumor VM. So, it is also possible that miR-134-5p indirectly inhibits proliferation by inhibiting VM [49]. At present, we cannot distinguish whether there is a causal relationship between different malignant phenotypes, but our research concludes that miR-134-5p can simultaneously inhibit the proliferation of OS and VM.
MiR acts through its specific targets and target-mediated downstream pathways. Target genes regulated by the same miRNAs may also differ depending on cell and tissue type and different stress or disease environments, possibly due to different gene expression and regulatory profiles [10, 50, 51]. Integrins are cell membrane receptors that mediate mutual recognition and adhesion between cells and cells and between cells and the extracellular matrix, and have a role in linking the external role of cells to their internal structure [52]. ITGB1, as an important member of the integrin family, has previously been linked to VM [53–55], another mode of blood supply to tumors. Besides, miR-134-5p was shown to inhibit osteoclastogenesis by targeting the ITGB1/MAPK pathway [56]. In this study, binding between miR-134-5p and ITGB1 was demonstrated in OS. Overexpression of the ITGB1 was able to counteract the suppression of tumorigenic behaviors by miR-134-5p, whereas silencing of ITGB1 counteracted the effects of the miR-134-5p inhibitor.
Metalloproteinases have been shown to play a crucial role in the formation of VM networks. As a member of the matrix metalloproteinase gene family, MMP2 is a zinc-dependent enzyme and is the most widely recognized regulatory molecule involved in VM formation [57–60]. Microarray gene microarray analyses have shown a significant increase in MMP2 expression in aggressive melanoma cells compared to poorly invasive melanoma cells, which are required for increased MMP2 expression for VM [61, 62]. Our study found that ITGB1 regulates MMP2 expression, which modulates the malignant phenotype of osteosarcoma. Lei Zhang et al. also found that inhibition of ITGB1 suppressed the expression of MMP9 and MMP2 [63]. However, the exact mechanism through which ITGB1 regulates MMP2 expression has not yet been investigated. Zhu et al. reported that ITGB1 can regulate MMP9 expression by regulating the FAK/Src/β-Catenin axis [64]. Thus, the specific mechanism by which ITGB1 regulates MMP2 may have many intermediate variables that need to be explored in future studies.
In addition, we found that the tumorigenic effects mediated by the miR-134-5p/ITGB1/MMP2 axis in OS involved PI3K/Akt signaling. PI3K/Akt signaling contributes to many cellular processes and is often seen to be abnormally activated in cancers [65, 66]. Zhu et al. labeled that MMP2 promotes laryngeal squamous cell carcinoma in a PI3K/Akt/NFκB-dependent manner [67]. Furthermore, activation of the PI3K/ AKT pathway in the tumor microenvironment promotes the formation of VM in melanoma [68] and colorectal cancer [69].
In summary, the above findings together demonstrate that miR-134-5p can inhibit VM and other malignant phenotypes of osteosarcoma by inhibiting the ITGB1/MMP2/PI3K/AKT pathway. This novel axis can aid in appealing treatment modality concerning angiogenesis in osteosarcoma patients.
Materials and methods
Clinical tissue specimens
This study was approved by the local ethics committee of the Second Hospital of Shanxi Medical University and was carried out in accordance with the standards set by the Declaration of Helsinki. Eight osteosarcoma samples and eight normal bone tissue samples were obtained from patients. The study protocol was approved by the Ethics Review Committee of the Second Hospital of Shanxi Medical University (approval number DW2022054) in Taiyuan, Shanxi, China. The clinicopathological characteristics are shown in Table S1.
Cell lines and culture
Human OS cell lines (HOS, U2OS, and MG63) were purchased from the ATCC. Cells were maintained in DMEM containing 10% fetal bovine serum (FBS) and 1% penicillin/ streptomycin (Invitrogen, USA) in a humidified incubator at 37 °C with 5% CO2. The osteoblast cell line hFOB1.19 was grown in DMEM/F-12 (Gibco, USA) containing 10% FBS, 2.5 mM l-glutamine (Invitrogen), 0.3 mg/ml geneticin (Gibco), and antibiotics as above.
Cell transfection
Cells were grown in six-well plates until 40% confluent, after which they were transduced with lentiviral (Genechem) for 24 h and selected with puromycin (3 mg/mL) for 24 h. Transfections with lentivirus were performed with a MOI = 20 in the HOS cell line and a MOI = 10 in the U2OS cell line. The stability of expression was determined.
RNA isolation and qRT-PCR
RNA (both total and miRNA) was extracted from OS samples, cells, and transfected cells with TRIzol (Thermo Fisher Scientific) and the miRNeasy Mini Kit (Qiagen, MD, USA) according to the provided directions. Gene expression relative to 18S rRNA or U6 was measured by qRT-PCR using an IQ5 Multicolor Real-Time PCR Detection system (Bio-Rad Laboratories, CA, USA). PCR detection of miR-134-5p included reaction at 95 °C (3 min), 95 °C (15 s) for 40 cycles, 56.5 °C (15 s), and 72 °C (30 s). Primer sequences are provided in Table S2. Relative expression was determined by the 2−ΔΔCT method [70].
Immunohistochemistry
Tissue samples from xenograft tumors were fixed with 4% paraformaldehyde for 14 h, paraffin-embedded, and sectioned. After dewaxing, rehydration, and blocking (5% BSA), the sections were treated with antibodies against MMP2 (1:100) (ab92536, Abcam, UK), ITGB1 (1:50) (ab30394, Abcam), CD31 (1:100) (3528, Cell Signaling Technology, USA), Ki67 (1:1000) (ab15580, Abcam), and PCNA (1:10000) (2586, Cell Signaling Technology) overnight at 4 °C.
Periodic Acid-Schiff (PAS)/CD31 double-staining
Staining with CD31 was conducted first following the procedure described above. The reactions were visualized with DAB. PAS staining was subsequently performed, in accordance with the provided protocols in a PAS staining kit (G1281, Solarbio, China).
Measurement of tube formation
Plates (24-well) were coated with Matrigel (354234; Corning). Cells (105/well in medium) were inoculated and grown for 4 h at 37 °C. The numbers of tube nodes were determined using the Angiogenesis Analyzer in ImageJ.
Cell proliferation assays
CCK-8 assays were performed by seeding and maintaining 6000 cells per well in 96-well plates for 1, 2, or 3 days when the medium was replaced with 100 µL of fresh medium with 10 µL of the CCK-8 reagent (Sigma). After 2 h, absorbances at 450 nm were measured. Cells were fixed and permeabilized after the addition of 10 μM of EdU and incubated for 2 h. The Click-iT reaction was performed following the manufacturer’s manual (Thermo Fisher Scientific). Lastly, nuclei were counterstained with DAPI, and the cells were imaged under a fluorescence microscope (Thermo Fisher Scientific). In colony formation assays, 600 cells/well were inoculated and cultured for 10 days in 6-well plates. After washing with PBS, cells were fixed with 4% paraformaldehyde for 30 min and stained with 0.5% crystal violet for 1 h at room temperature. Colony numbers were assessed using a clone-counter program.
Wound-healing assay
Cells (106 per well) were inoculated in six-well plates after trypsinization and grown overnight. The cells were scraped with a sterile micropipette tip. At the indicated time point (0, 24 h), three different fields of each wound were randomly photographed using a light microscope. The wound area was measured by ImageJ software, set at 100% for 0 h, and the mean percentage of the total area was calculated.
Cell migration and invasion assays
Assays were conducted using 8-mm Transwell chamber (Corning). The chambers were precoated with growth factor-reduced Matrigel (354234; Corning) and 200 µL of cell suspension in DMEM without FBS was placed in the upper chamber, with 10% FBS placed in the lower chamber. After incubation for 24 h, the cells were fixed and stained as colony formation assay and examined under an inverted light microscope, evaluating the cells in three random fields.
Flow cytometry
A tube of digested, EDTA-free trypsin-treated OS cells was collected 48 h after transfection. Cells were treated with Annexin V/PI (Vazyme Biotech Co., Ltd, Nanjing, China) and analyzed on a flow cytometer (Beckman, USA).
Western blotting
Proteins were extracted from cells with RIPA buffer with a protease and phosphatase inhibitor cocktail (Keygen, Nanjing, China). After the determination of concentrations using BCA, the proteins were separated on SDS-PAGE and transferred to nitrocellulose membranes. The blots were treated with anti-ITGB1 (1:1000) (ab30394, Abcam), anti-MMP2 (1:2000) (ab92536, Abcam), anti-AKT (1:1000) (4685, CST), anti-P-AKT (1:1000) (4228, CST), anti-PI3K (1:1000) (4249, CST), anti-P-PI3K (1:1000) (4228, CST), and anti-GAPDH (1:5000) (ab181602, Abcam). The bands were visualized using enhanced chemiluminescence. The loading control was GAPDH.
Dual-luciferase reporter assay
HOS cells (2 × 104/well) were seeded in 24-well plates and cultured until 80% confluent after which they were transiently transfected with 0.4 µg of an miR-134-5p mimic or miR-control (NC) and co-transfected with 0.1 µg of plasmid containing the 3′-UTRs of ITGB1 or their mutant (Genechem, Shanghai, China) using Lipofectamine 3000 (Invitrogen) for 48 h. Proteins were extracted and measured as above. Luciferase activity was assessed with a Dual-Luciferase Reporter Assay System (Promega Corp, Madison, USA) and a GLOMAX 20/20 luminometer (Promega). Transfection efficiency was determined by firefly/Renilla luciferase ratios.
In vivo growth and bioimaging
Animal experiments were carried out in accordance with the Guide for the Care and Use of Laboratory Animals (Ministry of Science and Technology of China, 2006), and were approved by the animal ethics committee of Shanxi Medical University (approval number DW2022054). Five-week-old BALB/c nude mice were purchased from Charles River (China). Randomly selected mice received subcutaneous injections of 106 lentivirus-transfected HOS cells (n = 6 per group). Researchers were not blinded to the group of the animals. Tumor growth was assessed each day. After 3 weeks, MMP activity was evaluated by MMPSense 680 (PerkinElmer) and fluorescence molecular tomography (FMT) (PerkinElmer). The mice were euthanized after 5 weeks, and the tumors were removed and measured [71]. Samples were retained in 10% formalin, RNAlater solution, or lysis buffer and stored at − 80 °C.
Bioinformatics methods
Data collection
The functions of miR-134-5p in OS were examined using bioinformatics. Information on gene expression and patient characteristics was acquired from the TARGET database. Information on gene expression in 396 normal tissues was obtained from the Genotype-Tissue Expression (GTEx) database. The TARGET and GTEx datasets were combined and batch-to-batch variations were eliminated using the “sva” package in R. The RNA-seq data was converted from fragment per kilobase million to transcripts per million before analysis, and log2(x + 1) conversions were performed. The detailed information on 88 patients with OS is presented in Table S3. As described in our previous study [27], 48 VM-related genes were found to be expressed in the GTEx-TARGET dataset, and they are listed in Table S4. Potential miR-134-5p targets were predicted by miRWalk 3.0 using the criteria of scores >0.95 or experimental validation. Finally, 2886 candidate target genes were selected (Table S5).
Differential expression determination
The “limma” package was used to detect differentially expressed genes (DEGs) with the criteria of log2 |fold change (FC)| ≥1 and p value <0.05 in the GTEx-TARGET cohort. Furthermore, Kaplan–Meier curves were compiled using the “survival” and “survminer” packages.
Protein–protein interaction (PPI) networks
PPIs were created by STRING version 11.5 [72] using combined scores of 0.4 (medium confidence).
Functional enrichment analysis
DEG-enriched pathways were investigated using GO [73] and KEGG [74] enrichment analyses in the clusterProfiler package [75] using a false-discovery threshold of <0.05.
Gene set enrichment analysis (GSEA)
Differences in pathways and functions were assessed by GSEA [76] in the gene set “c2.cp.kegg.v7.4.symbols.gmt” from The Molecular Signatures Database (MSigDB), using P < 0.05 as the significance threshold.
Statistical analysis
All data were analyzed using the GraphPad Prism v7.0 software. Data were presented as mean ± standard deviation (SD). All in vitro experiments were repeated three times. The comparisons between experimental and control groups were made by unpaired Student’s t-test or one-way ANOVA with Tukey’s test. Pearson correlation analyses were used for assessing relationships between genes. All P values were two-sided, and p < 0.05 was the significance threshold.
Supplementary information
Fig. S1 Knockdown of miR-134-5p promotes the VM, proliferation, migration and invasion of OS.
Fig. S2 Bioinformatics analysis of the characteristics of seven candidate target genes of miR-134-5p.
Fig. S3 Gain-of-function analyses verifying the involvement of the miR-134-5p/ITGB1/MMP2 axis in OS VM, proliferation, migration, and invasion.
Acknowledgements
The authors would like to thank MJEditor (www.mjeditor.com) for its linguistic assistance during the preparation of this manuscript.
Author contributions
LY, ZL, ZT, and XS contributed to the conception and design of the study. LY, RZ, XQ, YP, CX, XN, and YL conducted experiments and maintained animals. LY, RL, YF, RZ, PL, XW, LZ, XJS, and RL participated in samples collection and data analysis. LY drafted the initial manuscript. XS, ZT, YF, and ZL revised the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the National Natural Science Foundation of China (No. U21A20353), the National Natural Science Foundation of China (No. 82172503 and No. 82302769), the Natural Science Foundation of Shanxi Province (Nos. 20210302123263, 20210302124410, and 202204041101023), Technology Innovation Teams of Shanxi Province (202304051001048), the Second Hospital of Shanxi Medical University, and four in-house research funding projects (Nos. 2020001-10 and SXEYRC202001).
Data availability
The data that support the findings of this study are available from the corresponding author upon reasonable request.
Competing interests
The authors declare no competing interests.
Ethical approval
This research was approved by the Ethics Review Committee of the Second Hospital of Shanxi Medical University (approval number DW2022054) in Taiyuan, Shanxi, China.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Lei Yan, Ruhao Zhou, Yi Feng.
Contributor Information
Xiaojuan Sun, Email: guzhongliusyz@163.com.
Zhi Lv, Email: zhilv2023@sxmu.edu.cn.
Zhi Tian, Email: drtianzh@sxmu.edu.cn.
Supplementary information
The online version contains supplementary material available at 10.1038/s41420-024-01946-z.
References
- 1.Kansara M, Teng MW, Smyth MJ, Thomas DM. Translational biology of osteosarcoma. Nat Rev Cancer. 2014;14:722–35. doi: 10.1038/nrc3838. [DOI] [PubMed] [Google Scholar]
- 2.Isakoff MS, Bielack SS, Meltzer P, Gorlick R. Osteosarcoma: current treatment and a collaborative pathway to success. J Clin Oncol. 2015;33:3029–35. doi: 10.1200/JCO.2014.59.4895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Roessner A, Lohmann C, Jechorek D. Translational cell biology of highly malignant osteosarcoma. Pathol Int. 2021;71:291–303. doi: 10.1111/pin.13080. [DOI] [PubMed] [Google Scholar]
- 4.Ferguson JL, Turner SP. Bone cancer: diagnosis and treatment principles. Am Fam Physician. 2018;98:205–13. [PubMed] [Google Scholar]
- 5.Corre I, Verrecchia F, Crenn V, Redini F, Trichet V. The osteosarcoma microenvironment: a complex but targetable ecosystem. Cells. 2020;9:976. doi: 10.3390/cells9040976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Martin JW, Squire JA, Zielenska M. The genetics of osteosarcoma. Sarcoma. 2012;2012:627254. doi: 10.1155/2012/627254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.PosthumaDeBoer J, Witlox MA, Kaspers GJ, van Royen BJ. Molecular alterations as target for therapy in metastatic osteosarcoma: a review of literature. Clin Exp Metastasis. 2011;28:493–503. doi: 10.1007/s10585-011-9384-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Whelan JS, Davis LE. Osteosarcoma, chondrosarcoma, and chordoma. J Clin Oncol. 2018;36:188–93. doi: 10.1200/JCO.2017.75.1743. [DOI] [PubMed] [Google Scholar]
- 9.Gill J, Gorlick R. Advancing therapy for osteosarcoma. Nat Rev Clin Oncol. 2021;18:609–24. doi: 10.1038/s41571-021-00519-8. [DOI] [PubMed] [Google Scholar]
- 10.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–97. doi: 10.1016/S0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 11.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–33. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shukla GC, Singh J, Barik S. MicroRNAs: processing, maturation, target recognition and regulatory functions. Mol Cell Pharm. 2011;3:83–92. [PMC free article] [PubMed] [Google Scholar]
- 13.Xu M, Jin H, Xu CX, Sun B, Song ZG, Bi WZ, et al. miR-382 inhibits osteosarcoma metastasis and relapse by targeting Y box-binding protein 1. Mol Ther. 2015;23:89–98. doi: 10.1038/mt.2014.197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li X, Shi Y, Yin Z, Xue X, Zhou B. An eight-miRNA signature as a potential biomarker for predicting survival in lung adenocarcinoma. J Transl Med. 2014;12:159. doi: 10.1186/1479-5876-12-159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zheng W, Zhao J, Tao Y, Guo M, Ya Z, Chen C, et al. MicroRNA-21: a promising biomarker for the prognosis and diagnosis of non-small cell lung cancer. Oncol Lett. 2018;16:2777–82. doi: 10.3892/ol.2018.8972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bao Z, Cheng Z, Chai D. The expressions of CD133, ALDH1, and vasculogenic mimicry in osteosarcoma and their clinical significance. Int J Clin Exp Pathol. 2018;11:3656–63. [PMC free article] [PubMed] [Google Scholar]
- 17.Ren K, Ni Y, Li X, Wang C, Chang Q, Li Y, et al. Expression profiling of long noncoding RNAs associated with vasculogenic mimicry in osteosarcoma. J Cell Biochem. 2019;120:12473–88. doi: 10.1002/jcb.28514. [DOI] [PubMed] [Google Scholar]
- 18.Ren K, Zhang J, Gu X, Wu S, Shi X, Ni Y, et al. Migration-inducing gene-7 independently predicts poor prognosis of human osteosarcoma and is associated with vasculogenic mimicry. Exp Cell Res. 2018;369:80–89. doi: 10.1016/j.yexcr.2018.05.008. [DOI] [PubMed] [Google Scholar]
- 19.Mei J, Gao Y, Zhang L, Cai X, Qian Z, Huang H, et al. VEGF-siRNA silencing induces apoptosis, inhibits proliferation and suppresses vasculogenic mimicry in osteosarcoma in vitro. Exp Oncol. 2008;30:29–34. [PubMed] [Google Scholar]
- 20.Yao N, Ren K, Wang Y, Jin Q, Lu X, Lu Y, et al. Paris polyphylla suppresses proliferation and vasculogenic mimicry of human osteosarcoma cells and inhibits tumor growth in vivo. Am J Chin Med. 2017;45:575–98. doi: 10.1142/S0192415X17500343. [DOI] [PubMed] [Google Scholar]
- 21.Fu D, He X, Yang S, Xu W, Lin T, Feng X. Zoledronic acid inhibits vasculogenic mimicry in murine osteosarcoma cell line in vitro. BMC Musculoskelet Disord. 2011;12:146. doi: 10.1186/1471-2474-12-146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Namløs HM, Meza-Zepeda LA, Barøy T, Østensen IH, Kresse SH, Kuijjer ML, et al. Modulation of the osteosarcoma expression phenotype by microRNAs. PLoS ONE. 2012;7:e48086. doi: 10.1371/journal.pone.0048086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang L, Huang P, Li Q, Wang D, Xu CX. miR-134-5p promotes stage I lung adenocarcinoma metastasis and chemoresistance by targeting DAB2. Mol Ther Nucleic Acids. 2019;18:627–37. doi: 10.1016/j.omtn.2019.09.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chi J, Liu T, Shi C, Luo H, Wu Z, Xiong B, et al. Long non-coding RNA LUCAT1 promotes proliferation and invasion in gastric cancer by regulating miR-134-5p/YWHAZ axis. Biomed Pharmacother. 2019;118:109201. doi: 10.1016/j.biopha.2019.109201. [DOI] [PubMed] [Google Scholar]
- 25.Zhang L, Lv Z, Xu J, Chen C, Ge Q, Li P, et al. MicroRNA-134 inhibits osteosarcoma angiogenesis and proliferation by targeting the VEGFA/VEGFR1 pathway. FEBS J. 2018;285:1359–71. doi: 10.1111/febs.14416. [DOI] [PubMed] [Google Scholar]
- 26.Chen CL, Zhang L, Jiao YR, Zhou Y, Ge QF, Li PC, et al. miR-134 inhibits osteosarcoma cell invasion and metastasis through targeting MMP1 and MMP3 in vitro and in vivo. FEBS Lett. 2019;593:1089–101. doi: 10.1002/1873-3468.13387. [DOI] [PubMed] [Google Scholar]
- 27.Yan L, Li R, Li D, Zhu Y, Lv Z, Wang B. Development of a novel vasculogenic mimicry-associated gene signature for the prognostic assessment of osteosarcoma patients. Clin Transl Oncol. 2023;25:3501–3518. [DOI] [PubMed]
- 28.Ottaviani G, Jaffe N. The epidemiology of osteosarcoma. Cancer Treat Res. 2009;152:3–13. doi: 10.1007/978-1-4419-0284-9_1. [DOI] [PubMed] [Google Scholar]
- 29.Jaffe N. Osteosarcoma: review of the past, impact on the future. The American experience. Cancer Treat Res. 2009;152:239–62. doi: 10.1007/978-1-4419-0284-9_12. [DOI] [PubMed] [Google Scholar]
- 30.Kelly EJ, Russell SJ. MicroRNAs and the regulation of vector tropism. Mol Ther. 2009;17:409–16. doi: 10.1038/mt.2008.288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Murphy MS, Tayade C, Smith GN. Maternal circulating microRNAs and pre-eclampsia: challenges for diagnostic potential. Mol Diagn Ther. 2017;21:23–30. doi: 10.1007/s40291-016-0233-0. [DOI] [PubMed] [Google Scholar]
- 32.Pourshafie N, Lee PR, Chen KL, Harmison GG, Bott LC, Katsuno M, et al. MiR-298 counteracts mutant androgen receptor toxicity in spinal and bulbar muscular atrophy. Mol Ther. 2016;24:937–45. doi: 10.1038/mt.2016.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wong HA, Fatimy RE, Onodera C, Wei Z, Yi M, Mohan A, et al. The cancer genome atlas analysis predicts microRNA for targeting cancer growth and vascularization in glioblastoma. Mol Ther. 2015;23:1234–47. doi: 10.1038/mt.2015.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sun CC, Li SJ, Zhang F, Zhang YD, Zuo ZY, Xi YY, et al. The novel miR-9600 suppresses tumor progression and promotes paclitaxel sensitivity in non-small-cell lung cancer through altering STAT3 expression. Mol Ther Nucleic Acids. 2016;5:e387. doi: 10.1038/mtna.2016.96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.D’Angelo B, Benedetti E, Cimini A, Giordano A. MicroRNAs: a puzzling tool in cancer diagnostics and therapy. Anticancer Res. 2016;36:5571–5. doi: 10.21873/anticanres.11142. [DOI] [PubMed] [Google Scholar]
- 36.Takahashi RU, Prieto-Vila M, Hironaka A, Ochiya T. The role of extracellular vesicle microRNAs in cancer biology. Clin Chem Lab Med. 2017;55:648–56. doi: 10.1515/cclm-2016-0708. [DOI] [PubMed] [Google Scholar]
- 37.Bertoli G, Cava C, Castiglioni I. MicroRNAs as biomarkers for diagnosis, prognosis and theranostics in prostate cancer. Int J Mol Sci. 2016;17:421. doi: 10.3390/ijms17030421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mamoori A, Gopalan V, Smith RA, Lam AK. Modulatory roles of microRNAs in the regulation of different signalling pathways in large bowel cancer stem cells. Biol Cell. 2016;108:51–64. doi: 10.1111/boc.201500062. [DOI] [PubMed] [Google Scholar]
- 39.Talekar M, Trivedi M, Shah P, Ouyang Q, Oka A, Gandham S, et al. Combination wt-p53 and MicroRNA-125b transfection in a genetically engineered lung cancer model using dual CD44/EGFR-targeting nanoparticles. Mol Ther. 2016;24:759–69. doi: 10.1038/mt.2015.225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hsieh TH, Hsu CY, Tsai CF, Long CY, Wu CH, Wu DC, et al. HDAC inhibitors target HDAC5, upregulate microRNA-125a-5p, and induce apoptosis in breast cancer cells. Mol Ther. 2015;23:656–66. doi: 10.1038/mt.2014.247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fujita Y, Yagishita S, Hagiwara K, Yoshioka Y, Kosaka N, Takeshita F, et al. The clinical relevance of the miR-197/CKS1B/STAT3-mediated PD-L1 network in chemoresistant non-small-cell lung cancer. Mol Ther. 2015;23:717–27. doi: 10.1038/mt.2015.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hiraki M, Nishimura J, Takahashi H, Wu X, Takahashi Y, Miyo M, et al. Concurrent targeting of KRAS and AKT by MiR-4689 is a novel treatment against mutant KRAS colorectal cancer. Mol Ther Nucleic Acids. 2015;4:e231. doi: 10.1038/mtna.2015.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Stiuso P, Potenza N, Lombardi A, Ferrandino I, Monaco A, Zappavigna S, et al. MicroRNA-423-5p promotes autophagy in cancer cells and is increased in serum from hepatocarcinoma patients treated with sorafenib. Mol Ther Nucleic Acids. 2015;4:e233. doi: 10.1038/mtna.2015.8. [DOI] [PubMed] [Google Scholar]
- 44.Gallo Cantafio ME, Nielsen BS, Mignogna C, Arbitrio M, Botta C, Frandsen NM, et al. Pharmacokinetics and pharmacodynamics of a 13-mer LNA-inhibitor-miR-221 in mice and non-human primates. Mol Ther Nucleic Acids. 2016;5:E326. [DOI] [PMC free article] [PubMed]
- 45.Ding Y, Huang Y, Zhang F, Gong L, Liang C, Ding K, et al. LncRNA TDRKH-AS1 promotes breast cancer progression via the miR-134-5p/CREB1 axis. J Transl Med. 2023;21:854. doi: 10.1186/s12967-023-04640-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhang H, Jiang J, He X, Zhou Q. Circ_0002111/miR-134-5p/FSTL1 signal axis regulates tumor progression and glycolytic metabolism in papillary thyroid carcinoma cells. J Endocrinol Invest. 2023;46:713–25. doi: 10.1007/s40618-022-01921-4. [DOI] [PubMed] [Google Scholar]
- 47.Cai G, Zou R, Yang H, Xie J, Chen X, Zheng C, et al. Circ_0084043-miR-134-5p axis regulates PCDH9 to suppress melanoma. Front Oncol. 2022;12:891476. doi: 10.3389/fonc.2022.891476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yang G, Zhang Y, Lin H, Liu J, Huang S, Zhong W, et al. CircRNA circ_0023984 promotes the progression of esophageal squamous cell carcinoma via regulating miR-134-5p/cystatin-s axis. Bioengineered. 2022;13:10578–93. doi: 10.1080/21655979.2022.2063562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang Y, Tong L, Wang J, Luo J, Tang J, Zhong L, et al. cRGD-functionalized nanoparticles for combination therapy of anti-endothelium dependent vessels and anti-vasculogenic mimicry to inhibit the proliferation of ovarian cancer. Acta Biomater. 2019;94:495–504. doi: 10.1016/j.actbio.2019.06.039. [DOI] [PubMed] [Google Scholar]
- 50.Singh RP, Massachi I, Manickavel S, Singh S, Rao NP, Hasan S, et al. The role of miRNA in inflammation and autoimmunity. Autoimmun Rev. 2013;12:1160–5. doi: 10.1016/j.autrev.2013.07.003. [DOI] [PubMed] [Google Scholar]
- 51.Jonas S, Izaurralde E. Towards a molecular understanding of microRNA-mediated gene silencing. Nat Rev Genet. 2015;16:421–33. doi: 10.1038/nrg3965. [DOI] [PubMed] [Google Scholar]
- 52.Bianconi D, Unseld M, Prager GW. Integrins in the spotlight of cancer. Int J Mol Sci. 2016;17:2037. [DOI] [PMC free article] [PubMed]
- 53.Kawahara R, Niwa Y, Simizu S. Integrin β1 is an essential factor in vasculogenic mimicry of human cancer cells. Cancer Sci. 2018;109:2490–6. doi: 10.1111/cas.13693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhang J, Liu K, Peng P, Li S, Ye Z, Su Y, et al. Upregulation of nectin-4 is associated with ITGB1 and vasculogenic mimicry and may serve as a predictor of poor prognosis in colorectal cancer. Oncol Lett. 2019;18:1163–70. doi: 10.3892/ol.2019.10417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Langer EM, Kendsersky ND, Daniel CJ, Kuziel GM, Pelz C, Murphy KM, et al. ZEB1-repressed microRNAs inhibit autocrine signaling that promotes vascular mimicry of breast cancer cells. Oncogene. 2018;37:1005–19. doi: 10.1038/onc.2017.356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Huang M, Wang Y, Wang Z, Qin Q, Zhang H, Liu S, et al. miR-134-5p inhibits osteoclastogenesis through a novel miR-134-5p/Itgb1/MAPK pathway. J Biol Chem. 2022;298:102116. doi: 10.1016/j.jbc.2022.102116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhuo M, Yuan C, Han T, Hu H, Cui J, Jiao F, et al. JQ1 effectively inhibits vasculogenic mimicry of pancreatic ductal adenocarcinoma cells via the ERK1/2-MMP-2/9 signaling pathway both in vitro and in vivo. Am J Transl Res. 2019;11:1030–9. [PMC free article] [PubMed] [Google Scholar]
- 58.Liu Y, Li F, Yang YT, Xu XD, Chen JS, Chen TL, et al. IGFBP2 promotes vasculogenic mimicry formation via regulating CD144 and MMP2 expression in glioma. Oncogene. 2019;38:1815–31. doi: 10.1038/s41388-018-0525-4. [DOI] [PubMed] [Google Scholar]
- 59.Cai HP, Wang J, Xi SY, Ni XR, Chen YS, Yu YJ, et al. Tenascin-cmediated vasculogenic mimicry formation via regulation of MMP2/MMP9 in glioma. Cell Death Dis. 2019;10:879. doi: 10.1038/s41419-019-2102-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wang Z, You D, Lu M, He Y, Yan S. Inhibitory effect of norcantharidin on melanoma tumor growth and vasculogenic mimicry by suppressing MMP-2 expression. Oncol Lett. 2017;13:1660–4. doi: 10.3892/ol.2017.5622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sood AK, Fletcher MS, Coffin JE, Yang M, Seftor EA, Gruman LM, et al. Functional role of matrix metalloproteinases in ovarian tumor cell plasticity. Am J Obstet Gynecol. 2004;190:899–909. doi: 10.1016/j.ajog.2004.02.011. [DOI] [PubMed] [Google Scholar]
- 62.Seftor RE, Seftor EA, Koshikawa N, Meltzer PS, Gardner LM, Bilban M, et al. Cooperative interactions of laminin 5 gamma2 chain, matrix metalloproteinase-2, and membrane type-1-matrix/metalloproteinase are required for mimicry of embryonic vasculogenesis by aggressive melanoma. Cancer Res. 2001;61:6322–7. [PubMed] [Google Scholar]
- 63.Zhang L, Zou W. Inhibition of integrin β1 decreases the malignancy of ovarian cancer cells and potentiates anticancer therapy via the FAK/STAT1 signaling pathway. Mol Med Rep. 2015;12:7869–76. doi: 10.3892/mmr.2015.4443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zhu X, Wang K, Chen Y. Ophiopogonin D suppresses TGF-β1-mediated metastatic behavior of MDA-MB-231 breast carcinoma cells via regulating ITGB1/FAK/Src/AKT/β-catenin/MMP-9 signaling axis. Toxicol Vitr. 2020;69:104973. doi: 10.1016/j.tiv.2020.104973. [DOI] [PubMed] [Google Scholar]
- 65.Ediriweera MK, Tennekoon KH, Samarakoon SR. Role of the PI3K/AKT/mTOR signaling pathway in ovarian cancer: biological and therapeutic significance. Semin Cancer Biol. 2019;59:147–60. doi: 10.1016/j.semcancer.2019.05.012. [DOI] [PubMed] [Google Scholar]
- 66.Alzahrani AS. PI3K/Akt/mTOR inhibitors in cancer: at the bench and bedside. Semin Cancer Biol. 2019;59:125–32. doi: 10.1016/j.semcancer.2019.07.009. [DOI] [PubMed] [Google Scholar]
- 67.Zhu Y, Yan L, Zhu W, Song X, Yang G, Wang S. MMP2/3 promote the growth and migration of laryngeal squamous cell carcinoma via PI3K/Akt-NF-κB-mediated epithelial-mesenchymal transformation. J Cell Physiol. 2019;234:15847–55. doi: 10.1002/jcp.28242. [DOI] [PubMed] [Google Scholar]
- 68.Xu X, Zong Y, Gao Y, Sun X, Zhao H, Luo W, et al. VEGF induce vasculogenic mimicry of choroidal melanoma through the PI3k signal pathway. Biomed Res Int. 2019;2019:3909102. doi: 10.1155/2019/3909102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Liu X, He H, Zhang F, Hu X, Bi F, Li K, et al. m6A methylated EphA2 and VEGFA through IGF2BP2/3 regulation promotes vasculogenic mimicry in colorectal cancer via PI3K/AKT and ERK1/2 signaling. Cell Death Dis. 2022;13:483. doi: 10.1038/s41419-022-04950-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-delta delta C(T)) method. Methods. 2001;25:402–8. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 71.Naito S, von Eschenbach AC, Giavazzi R, Fidler IJ. Growth and metastasis of tumor cells isolated from a human renal cell carcinoma implanted into different organs of nude mice. Cancer Res. 1986;46:4109–15. [PubMed] [Google Scholar]
- 72.Szklarczyk D, Gable AL, Lyon D, Junge A, Wyder S, Huerta-Cepas J, et al. STRING v11: protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019;47:D607–d613. doi: 10.1093/nar/gky1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wilkerson MD, Hayes DN. ConsensusClusterPlus: a class discovery tool with confidence assessments and item tracking. Bioinformatics. 2010;26:1572–3. doi: 10.1093/bioinformatics/btq170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kanehisa M, Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Yu G, Wang LG, Han Y, He QY. clusterProfiler: an R package for comparing biological themes among gene clusters. Omics. 2012;16:284–7. doi: 10.1089/omi.2011.0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102:15545–50. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1 Knockdown of miR-134-5p promotes the VM, proliferation, migration and invasion of OS.
Fig. S2 Bioinformatics analysis of the characteristics of seven candidate target genes of miR-134-5p.
Fig. S3 Gain-of-function analyses verifying the involvement of the miR-134-5p/ITGB1/MMP2 axis in OS VM, proliferation, migration, and invasion.
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.