Summary
Deep proteomic profiling of rare cell populations has been constrained by sample input requirements. Here, we present DROPPS (droplet-based one-pot preparation for proteomic samples), an accessible low-input platform that generates high-fidelity proteomic profiles of 100–2,500 cells. By applying DROPPS within the mammary epithelium, we elucidated the connection between mitochondrial activity and clonogenicity, identifying CD36 as a marker of progenitor capacity in the basal cell compartment. We anticipate that DROPPS will accelerate biology-driven proteomic research for a multitude of rare cell populations.
Keywords: low-input, proteomics, progenitor, mammary epithelium, CD36, metabolism
Graphical abstract
Highlights
-
•
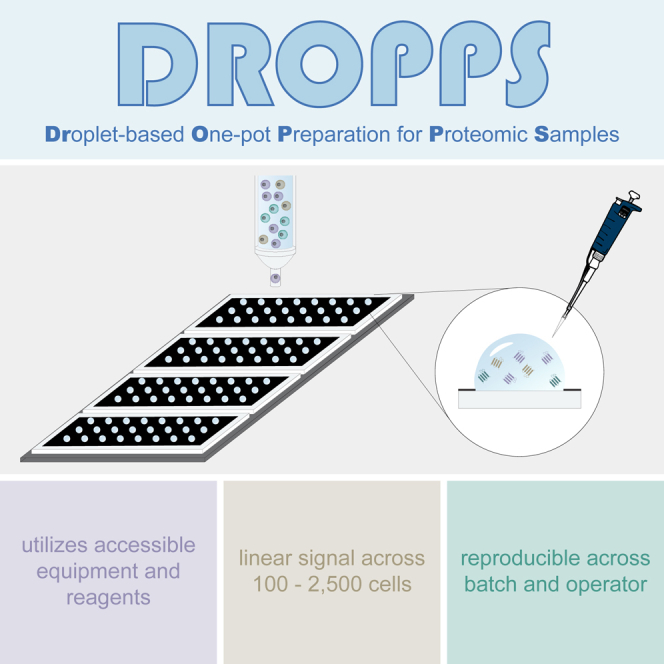
An accessible low-input proteomic method for rare populations of cells
-
•
Easy to implement, relying exclusively on common laboratory equipment
-
•
Performance is robust across operators and sample processing batches
-
•
Identifies CD36 as marker for mammary basal epithelial cells with progenitor capacity
Motivation
Proteomics allows for direct measurement of cellular machinery, allowing for interrogation of a variety of biological systems. Many low-input proteomic methods only support specific study designs or require specialized equipment. To address this limitation, we have developed an accessible low-input proteomic method for studying rare populations of cells: droplet-based one-pot preparation for proteomic samples (DROPPS).
Waas et al. develop and validate DROPPS, a proteomic workflow designed to democratize the capacity to profile rare cell populations without compromising data quality. DROPPS generates high-fidelity proteomic profiles from samples comprising as low as 100 cells and is amenable to facile integration with cell sorting.
Introduction
The mammary gland is a dynamic organ that substantially transforms during pubertal development, sex-hormone-triggered reproductive cycling, and pregnancy.1,2,3 The mammary epithelium that forms the branching system of ducts and alveolae constitutes 10%–15% of the overall volume of the mammary gland and contains cells of heterogeneous function including hormone-sensing, secretory, stem, and progenitor activity.1,2,3 Stem and progenitor cells essential for mammary ductal expansion constitute <5% of the epithelium.4 These rare cells are the cells of origin for the most aggressive, deadly subtypes of breast cancers (i.e., triple-negative, basal-like, and claudin-low subtypes).5,6,7 An attractive prevention strategy for breast cancer is to develop therapeutic modalities to monitor and target the oncogenic precursors as they arise.8 Though various markers have been reported to enrich stem/progenitor capacity,9,10,11,12 there remains no consensus regarding the exact trajectory of mammary epithelial cells and how the cells enriched by distinct markers relate to each other. Improving our knowledge of the features and regulation of stem/progenitor cells has the potential to improve clinical outcomes for patients with breast cancer.
In-depth proteomic interrogation of cells, tissues, and fluids is often performed with bottom-up mass spectrometry (MS) workflows where proteins are extracted and enzymatically digested to obtain peptides.13,14,15 Despite advances in instrument sensitivity—which enable deep proteomic profiling from <1 μg—the analysis of rare cell populations or other sample-limited systems is impeded by the loss of material incurred by sample preparation. Accordingly, proteomic methods often call for 10,000s–1,000,000s of cells (roughly 10s–100s μg) as starting material.16 Recent single-cell or low-input proteomic methods typically leverage low-volume and minimal manual manipulation to reduce losses during sample preparation.17,18,19 Despite these technical advances, many low-input methods only support specific study designs (e.g., isobaric labeling with booster channel) or require specialized equipment (e.g., microfabrication capabilities, microfluidic cell isolation systems). To address this limitation, we have developed an accessible low-input proteomic method for studying rare populations of cells, droplet-based one-pot preparation for proteomic samples (DROPPS), that is simple to implement and easily integrates with fluorescence-activated cell sorting (FACS). DROPPS utilizes commercially available and economical materials to enable reproducible and biologically informative proteomic analysis of small numbers of cells (100s–1,000s of cells, or 20 ng–1 μg) from in vitro and in vivo systems.
We previously generated proteomic profiles of FACS-purified luminal and basal compartments of the mouse mammary epithelium and discovered differences in preferred metabolic pathways.20 Investigating the functional consequences of these differences revealed a positive relationship between mitochondrial membrane potential (ΔΨM) and clonogenicity, particularly in the basal compartment.20 Here, we expand upon those findings by applying DROPPS to subpopulations of mammary epithelial cells FACS purified according to ΔΨM to investigate the proteomic features of enhanced clonogenic activity. Strikingly, in basal cells, high mitochondrial activity is associated with proliferation pathways and adhesion-dependent signaling. We identify CD36 as a marker for basal cells with high mitochondrial potential, increased fatty acid abundance, and enhanced clonogenicity. These findings further define the relationship between energy metabolism and progenitor cell capacity, providing insights into mammary basal cell compartment functional heterogeneity. Overall, we demonstrate that DROPPS is an accessible proteomics workflow to enhance and accelerate biological discovery for rare cell populations.
Results
Droplet digestion for proteomic profiling of low-input samples
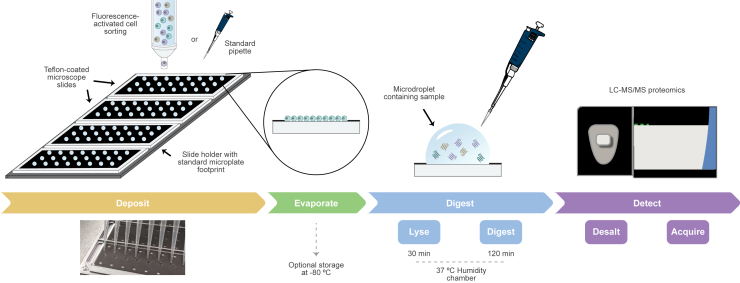
Recognizing the benefits that could be attained from digestion in a droplet (e.g., reduction in surface area contact and enhanced enzyme kinetics), we sought to develop an accessible platform for low-input proteomic sample preparation (Figure 1). We designed a microscope slide with a ∼20 μm coating of polytetrafluoroethylene (i.e., Teflon)—a hydrophobic, chemically inert polymer—to contain aqueous droplets of ≤10 μL. The slides were procured from a commercial vendor with well spacing to match 96-well plates, enabling the use of standard multichannel pipettes. We designed and 3D printed an adapter composed of polylactic acid that has the same footprint as a standard 96-well plate (available at NIH 3D: 3DPX-020483; https://3d.nih.gov/entries/3DPX-020483). This allowed us to easily transfer and position four DROPPS slides within plate-compatible flow cytometers, allowing parallel processing of 96 samples.
Figure 1.
DROPPS workflow for low-input protein digestion
Cells are deposited onto Teflon-coated slides by fluorescence-activated cell sorting or standard pipette. Cell transfer buffer is evaporated, and slides can be optionally stored at −80°C. Cell lysis and protein digestion are performed with MS-compatible reagents in a simple humidity chamber. Samples are transferred to standard sample vials or plates and analyzed by LC-MS for quantitative proteomic analysis.
To determine the range of sample input that can be processed by DROPPS with high reproducibility, we evaluated the performance of DROPPS over a range of starting materials (10–2,500 FAC-sorted cells, n = 3) (Figure S1A) from MCF10A, a human mammary epithelial cell line. As expected, the number of input cells drastically affected the depth of proteomic coverage and variability in measurements (Figures S1B–S1E). Protein intensity was linear across the 100–2,500 cell range, supporting the utility of DROPPS for quantitatively profiling rare populations of cells (plot shown for the luminal epithelial marker KRT18, median R2 among all proteins was 0.97; Figures S1F–S1G). We evaluated DROPPS reproducibility by having three operators perform independent tryptic digestion of 500 MCF10A cells (n = 8) (Figure S1H). To emulate typical workflows used for cellular biology, cells were counted by hemocytometer and deposited on the glass slide by pipette. The method was highly reproducible, with similar numbers of proteins and peptides detected in samples prepared by each operator (Figures S1I and S1J). Technical variability from sample handling was low (median peptide coefficients of variation [CVs] ranging from 12.4% to 14.5% and protein CVs ranging from 8.3% to 9.9% per operator) (Figures S1K and S1L). The proteomic depth and variance obtained by DROPPS are significant improvements over surfactant-assisted one-pot sample processing at the standard volume coupled with MS (SOPs-MS)19—a comparable one-pot low input proteomic method performed in a standard 96-well plate—emphasizing the advantage of the droplet-based digestion (Figure S1M–S1P). Notably, DROPPS does not need pre-incubation of sample plates and requires far less processing time (2.5 h incubation compared to two overnight incubations for SOP). As for accessibility, SOP and DROPPS both utilize commercially available consumables—the estimated additional cost per sample for DROPPS is <$0.03. Overall, these results demonstrate that DROPPS can be used to reproducibly generate quantitative proteomic profiles of 100–2,500 cells using common methods of sample handling.
Biological differences of breast cancer cell lines are recapitulated by DROPPS profiles
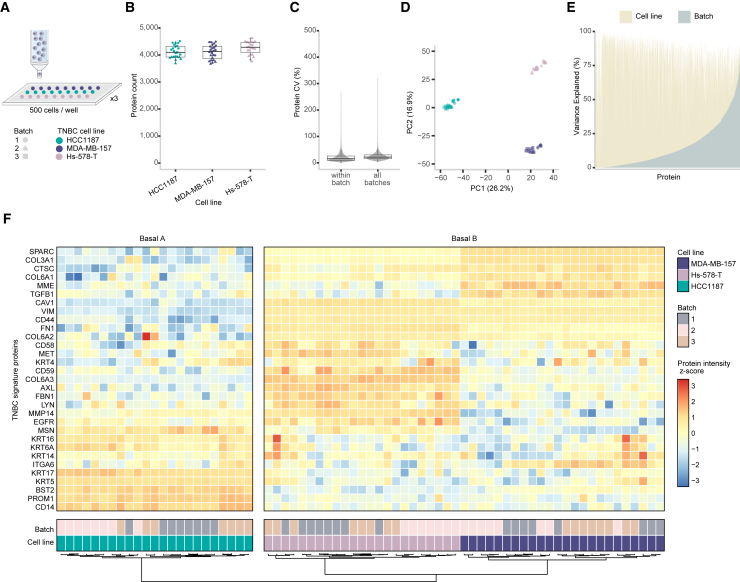
We next evaluated the ability of DROPPS to repeatably detect biological differences among three human triple-negative breast cancer (TNBC) cell lines (HCC1187, MDA-MB-157, and Hs-578-T) across three batches of 500 cell replicates (n = 7–8 per batch) (Figure 2A). To rigorously assess DROPPS performance across batches, every step of the workflow (i.e., cell culture, cell sorting, protein digestion, MS) was completed on different days. Variance in quantitation was low across batches despite differences in the depth of proteomic coverage (Figures 2B, 2C, S2A, and S2B). TNBC cell line protein profiles were highly similar within and between batches with median Spearman’s ρ values of 0.984 and 0.970, respectively (Figure S2C). Principal-component analysis (PCA) distinctly separates the cell lines (Figure 2D). Differences between cell lines were larger than differences between batches for the majority (80%) of proteins (Figure 2E). These results show that most of the observed protein variance is due to biological differences between cell lines rather than experimental variance. Importantly, proteomic profile differences acquired by DROPPS were highly similar to proteomes acquired in two independent bulk proteomic datasets13,14 (Spearman’s ρ values of 0.62 and 0.64, both p < 2.2 × 10−16) (Figures S2D and S2E). Furthermore, our data also successfully clustered breast cancer cell lines into molecular subtypes of TNBC,21 indicating that the profiles generated by DROPPS accurately reflect previously described phenotypic differences (Figures 2F and S2F; Table S1). These data indicate that our platform consistently and accurately recapitulates biological features of TNBC from 500 cells.
Figure 2.
Evaluation of DROPPS for profiling TNBC cell line differences across batches
(A) Workflow for testing inter-batch variation by DROPPS. Sorted cells (500 cells) from three TNBC cell lines were processed separately on different days. Each cell line had seven or eight sample replicates processed each day.
(B) The protein counts of the individual runs.
(C) Protein CVs for runs within a batch or for combining batches.
(D) First two principal components of PCA.
(E) The percentage of variance explained by cell line and batch from two-way ANOVA (i.e., omega-squared).
(F) Z scored log2(intensity) of TNBC signature proteins21 using data acquired by DROPPS.
Boxplots show the median, interquartile ranges, and 95% confidence interval estimate. TNBC, triple negative breast cancer.
Profiles of mammary epithelial compartments can be obtained from individual mice
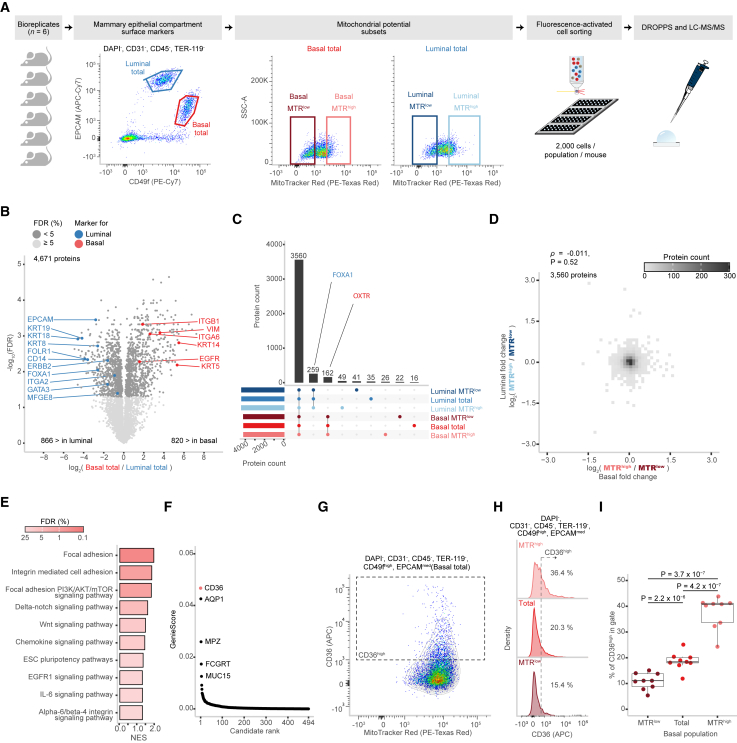
The mammary epithelium is a ductal structure composed of an inner layer of luminal cells encompassed by basal cells—the cells from luminal and basal compartments have distinct molecular phenotypes, which reflect their different functions.20,22 Here, by applying DROPPS, we obtained improved depth of proteomic profiles for FACS-purified luminal and basal compartments from individual mice than had previous been achieved22 by pooled samples (Figures 3A and S3A–S3D). Proteins uniquely detected in DROPPS were less abundant and detected in fewer mice, highlighting heterogeneity among mice uncovered by the capacity to analyze ∼60-fold fewer cells than our previous study22 (Figures S3E and S3F). As expected, differential expression analysis revealed pronounced differences between luminal and basal cells that were highly correlated to our previous study22 (Figures 3B and S3G). The cell surface proteins used to distinguish luminal from basal cells,23,24,25 ITGA6 (CD49f) and EPCAM, were among the most significantly different, along with other described lineage markers22 (e.g., KRT5/14 and KRT8/18/19) (Figure 3B). These results show that DROPPS enables the investigation of mammary epithelial subpopulations (of at least 2,000 cells) from individual mice—a significant advance enabling the investigation of proteomic heterogeneity within and between samples.
Figure 3.
Proteomic profiling of primary mammary epithelial cells sorted by mitochondrial potential
(A) Schematic of experimental design and FACS gating scheme used to sort subsets of mammary epithelial cells of basal and luminal compartments based on their mitochondrial potential as marked by MTR dye (n = 6 each) that were subsequently processed using DROPPS.
(B) Differences in log2(intensities) of proteins from the total basal and total luminal populations. Points are colored by significance, and known compartment markers are labeled.
(C) Overlap in proteins detected between select subsets of epithelial cells.
(D) 2D bin plot depicting the distribution of log2 fold changes between MTRhigh and MTRlow for the basal and luminal compartments. Spearman correlation and associated p value are shown.
(E) Select gene set enrichment analysis (GSEA)-enriched pathways in the MTRhigh population compared to the total population for the basal and luminal compartments.
(F) Rank plot depicting the distribution of GenieScores for proteins with surface prediction consensus >0 calculated using SurfaceGenie26 on mean log2(intensities).
(G) Representative scatterplot depicting the relationship between CD36 and MTR fluorescent intensity.
(H) Representative histograms depicting the distribution of CD36 flow cytometry signal in MTRlow, total, and MTRhigh basal populations.
(I) The percentage of CD36high cells in different MTR fractions of basal cells shown with paired t test p values (n = 8).
Boxplots show the median, interquartile ranges, and 95% confidence interval estimate. MTR, MitoTracker Red.
CD36 identified as a marker of basal mitochondrial potential
The ability to monitor and target oncogenic precursors could improve clinical outcomes for patients with breast cancer.8 As stem/progenitor cells give rise to the deadliest breast cancer subtypes,5,6,7 it is of particular interest to identify biomarkers and molecular vulnerabilities of these cell types. We previously identified a positive correlation between ΔΨM—measured by MitoTracker Red (MTR)—and clonogenicity within mammary epithelial compartments.20 Luminal and basal compartments both had significantly diminished clonogenicity within MTRlow fractions. MTRhigh fractions demonstrated a modest increase of clonogenicity in the luminal compartment and a significant increase in the basal compartment. As clonogenicity is an established trait of stem/progenitor cells, we applied DROPPS to FACS-purified MTRhigh, MTRlow, and total cell fractions from basal and luminal compartments to interrogate how ΔΨM heterogeneity relates to clonogenicity within the mouse mammary epithelium (2,000 cells per fraction, n = 6) (Figures 3A and S3A). Overall, we detected a total of 5,576 proteins (a mean of 4,423 proteins per sample) (Figures S3B and S3D). As expected, we revealed a substantial overlap in the proteomes with some known compartment markers demonstrating compartment-specific detection (e.g., FOXA1,23,27 OXTR28,29) (Figures 3C; Table S1). Unsurprisingly, the epithelial compartment was the largest source of sample variance (Figure S3H).
The proteins correlated with ΔΨM were completely independent between the luminal and basal cells (Spearman’s ρ of −0.011, p = 0.52), suggesting compartment-specific pathways associated with MTRhigh and MTRlow populations (Figure 3D). We performed differential expression analysis to interrogate which pathways are associated with MTRhigh in each compartment. Luminal MTRhigh cells are enriched in oxidative phosphorylation—the cellular process that generates the proton gradient component of ΔΨM (Table S2). In contrast, basal MTRhigh cells are enriched in various proliferation, survival, adhesion-sensitive growth, and pluripotency pathways (Table S2; select terms in Figure 3E). These results align with the previously described20 differences, namely a larger effect on clonogenicity in the basal compartment for MTRhigh populations. The accessibility of surface proteins makes them attractive markers and drug targets. As basal MTRhigh cells are highly clonogenic and have progenitor associated pathways, we used SurfaceGenie26 to rank potential cell surface markers, revealing CD36 as a high-priority, differentially expressed candidate (Figures 3F and S3I). We validated that CD36 expression was positively correlated with ΔΨM by live-cell flow cytometry (Figure 3G) and observed significantly different frequencies of CD36high cells between MTRlow, total, and MTRhigh subsets (Figures 3H and 3I). These results clearly depict a relationship between ΔΨM and CD36 in basal epithelial cells. Overall, these results demonstrate how DROPPS can be applied to generate insights into mammary biology and to identify cell surface markers for desired epithelial subpopulations.
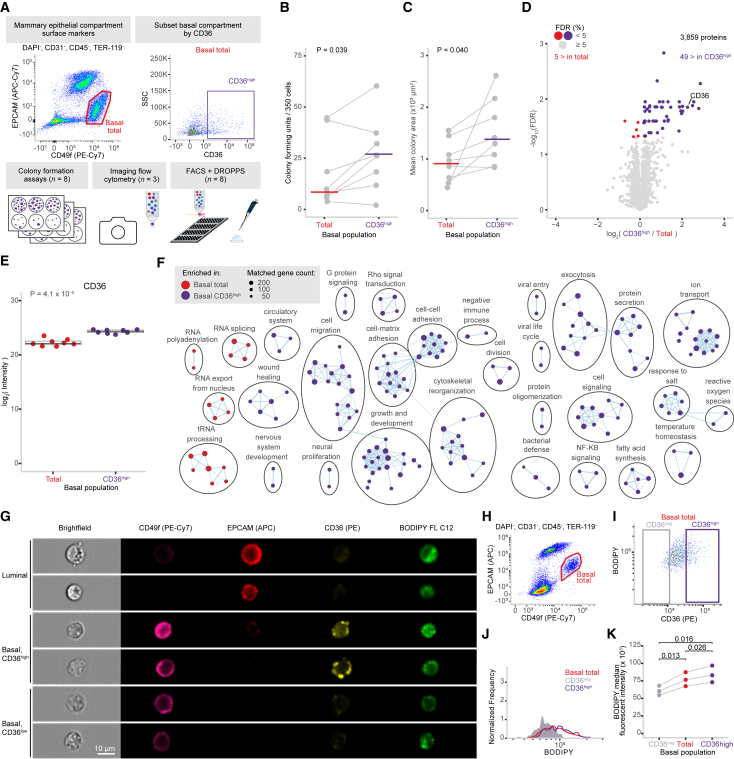
Basal progenitors are enriched by CD36
We next investigated whether CD36 was associated with enhanced clonogenicity, or solely ΔΨM, by comparing CD36high cells to the total basal population using a colony-forming assay (Figure 4A). CD36high cells had enhanced clonogenicity shown by significantly more colonies (paired t test, p = 0.039) and larger colonies (paired t test, p = 0.048) (Figures 4A–4C and S4A). We then analyzed CD36high basal cells by DROPPS to determine which pathways were associated with the enhanced clonogenicity detecting a mean of 4,187 proteins per sample (2,000 cells per mouse, n = 8) (Figure S4B). We detected 49 proteins significantly enriched in CD36high compared to the total basal population—including CD36 (Figure 4D). CD36 expression was associated with enriched proliferation (e.g., growth and development, cell division, stem cell proliferation), various signaling pathways (e.g., Notch, MAPK, Rho, nuclear factor (NF) κB), and metabolism (e.g., organic hydroxy compound transport, monocarboxylic acid transport, and lipid homeostasis)—all potentially contributing to the enhanced clonogenicity of the CD36high population (Table S2; clustered terms in Figure 4E). A major function of CD36 is the uptake of long-chain fatty acid, which has been tied to survival, metabolic rewiring, and epithelial-to-mesenchymal transition in breast cancer.30,31,32 We investigate whether CD36 was associated with increased fatty acid accumulation in basal epithelial cells using imaging flow cytometry with a fluorescent long-chain fatty acid analog, BODIPY-dodecanoic acid (Figures 4G–4I and S4D). Our analyses revealed that CD36 expression in basal cells was associated with significantly increased BODIPY signals, indicating a potential mechanism by which CD36 contributes to increased ΔΨM and enhanced progenitor capacity (Figures 4J and 4K). Taken together, these discoveries exhibit how DROPPS can provide insights into the underlying biology of rare cell populations.
Figure 4.
Investigating the phenotype of CD36high basal mammary epithelial cells
(A) Schematic of experimental design and FACS gating scheme used to sort subsets of mammary epithelial cells of basal and luminal compartments based on CD36 expression that were subsequently used for CFC, processed using DROPPS, or analyzed by imaging flow cytometry.
(B and C) Line plots depicting the (B) number and (C) size of the colonies formed by basal cells sorted by CD36 expression shown with median value and paired t test p values.
(D) Differential protein expression results from total basal and CD36high basal populations. Points are colored by significance and directionality.
(E) The log2(intensity) of CD36 from individual runs shown with the paired t test p value (n = 8).
(F) Cytoscape enrichment map for significantly enriched GO Biological Processes between total basal and CD36high basal populations.
(G) Representative images from imaging cytometry highlighting the surface markers used to distinguish epithelial compartments along with CD36 and the fluorescent fatty acid analog BODIPY.
(H) Scatterplot depicting the gating scheme used to identify basal epithelial cells.
(I) Scatterplot depicting the relationship between BODIPY and CD36 intensities along with CD36neg and CD36high gates.
(J) Representative histogram depicting the BODIPY intensity distributions among CD36neg, total, and CD36high basal populations.
(K) Dot plot depicting the median BODIPY fluorescent intensity in CD36neg, total, and CD36high basal populations along with paired t test p values.
CFC, colony-forming capacity.
Discussion
Unraveling the mechanisms underlying development, proper function, and disease will require understanding the dynamic changes and interactions of myriad cell types including those that are rare. Here, we developed and validated DROPPS, a proteomic workflow designed to democratize the capacity to profile rare cell populations without compromising data quality. DROPPS generates high-fidelity proteomic profiles from samples comprising as low as 100 cells and is amenable to facile integration with FACS. DROPPS performance is robust across operators and sample processing batches, and it is easy to implement—relying exclusively on common or affordable laboratory equipment. Notably, DROPPS outperforms other recently described accessible low-input proteomic methods19,33,34,35,36,37,38 (Table S3). Provided access to liquid handling systems, automation of DROPPS should be practical, as it involves very limited sample handling and straightforward transfer steps. The capacity to rapidly generate protein profiles of rare cell populations enables experimental designs that were previously inaccessible or impractical.
There remains a discrepancy regarding the exact differentiation hierarchy of mammary epithelial cells and how to identify and enrich for populations of different functions along the lineage trajectory. Here, we applied DROPPS to examine the proteomic correlates of a previously described20 relationship between ΔΨM and clonogenic potential in lineage-defined mouse mammary epithelial cells. Specifically, basal MTRhigh cells demonstrated a significant increase in clonogenicity compared to the total basal population. MTRlow cells in both basal and luminal compartments had significantly diminished clonogenicity. DROPPS revealed that differences in ΔΨM were reflected in the proteome in a compartment-specific manner. Basal MTRhigh cells were enriched in pathways associated with proliferation and adhesion-dependent signaling. We uncovered CD36 as a candidate marker for basal cells with elevated ΔΨM and enhanced clonogenicity. This work contributes to our understanding of the relationship between molecular phenotype and clonogenicity in the mammary basal epithelium and provides a marker, CD36, by which primary cells with progenitor capacity can be enriched and studied.
CD36 is a multifunction cell surface glycoprotein that acts as a receptor for a broad range of ligands including proteins and lipids. In this study, we found that CD36 was associated with clonogenicity, increased fatty acid abundance, and enrichment of myriad growth, signaling, and metabolic pathways in primary basal epithelial cells. The putative association between increased fatty acid uptake and progenitor capacity aligns with our single-cell RNA sequencing analysis of human mammary basal epithelial cells in which higher oxidative phosphorylation was associated with a less differentiated cell state.20 In another study that characterized features of mouse mammary stem cells, CD36 is among the proteins associated with the mammary stem cell signature.39 The expression of CD36 is associated with increased free fatty acid uptake and metabolic rewiring in a variety of biological systems including hepatocellular carcinoma, hematopoietic stem cells, and breast cancer.30,40,41 CD36 has also been implicated in enhancing proliferation, migration, and epithelial-to-mesenchymal transition in multiple types of cancer.32,42,43 In an MMTV-PyMT mouse breast cancer model, deletion of CD36 diminished high-fat-diet-induced metastasis.44 CD36 positivity in circulating epithelial tumor cells is highly correlated with disease progression in patients with breast cancer.45 Our study identifies CD36 as a marker for progenitor capacity in the basal mammary epithelium that may function to metabolically reprogram cells toward a less differentiated and proliferative state. Our findings contribute to the growing body of evidence that CD36 represents a target of interest for therapeutic intervention in breast cancer.
Limitations of the study
For biological questions that require experiments of <100 cells, further experimentation would be required to quantify whether the increased variance in measurement beneath 100 cells is due to technical limitations of the materials/methods or random sampling of biological variability. Though a benefit of DROPPS is its accessibility, more sophisticated equipment could potentially improve performance. For example, access to an improved humidity chamber could extend digestion beyond the 2 h currently used to minimize evaporation. Future efforts will aim to increase the specificity and selectivity for desired cell populations (e.g., multipotent mammary stem cells) by identification of additional candidate markers and will continue to characterize the relationship between CD36 and the functional potential of mammary epithelial cells. Improving our knowledge of the proteomic features of mammary epithelial progenitor and stem cells may enable us to understand the relationship between the various populations of cells enriched by previously reported markers (e.g., TSPAN8 and PROCR).
Conclusion
In summary, we present an accessible proteomic platform, DROPPS, and demonstrate its utility to uncover biological insights of rare epithelial cell populations. DROPPS affords the capacity to assess mouse-to-mouse variability, differences between subsets of cells within each mouse, and rare populations of cells, thus enabling proteomic insights into the functional heterogeneity that were precluded by previous sample requirements. DROPPS has widespread applicability to various sample-limited systems and is well positioned to transform our ability to map the proteomic landscapes of rare cell populations.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| APC-Cy7 rat anti-mouse CD326 clone G8.8 (EpCAM) | BioLegend | Cat# 118217; RRID: AB_1501158 |
| PE-Cy7 rat anti-human CD49f clone GoH3 | BioLegend | Cat# 313622; RRID: AB_2561705 |
| eFluor™ 450 rat anti-mouse CD31 clone 390 | ThermoFisher | Cat# 48-0311-82; RRID: AB_10598807 |
| eFluor™ 450 rat anti-mouse CD45 clone 30-F11 | ThermoFisher | Cat# 48-0451-82; RRID: AB_1518806 |
| eFluor™ 450 rat anti-mouse TER-119 clone TER-119 | ThermoFisher | Cat# 48-5921-82; RRID: AB_1518808 |
| APC Mouse Anti-Mouse CD36 clone CRF D-2712 | BD Biosciences | Cat# 562744; RRID: AB_2737763 |
| APC rat anti-mouse CD326 clone G8.8 (EpCAM) | BioLegend | Cat# 118214; RRID: AB_1134102 |
| PE Mouse Anti-Mouse CD36 clone CRF D-2712 | BD Biosciences | Cat# 562702; RRID: AB_2737732 |
| Critical commercial assays | ||
| MTR CM-H2Xros | ThermoFisher | M7513 |
| BODIPY™ FL C12 | ThermoFisher | D3822 |
| Deposited data | ||
| Raw mass spectrometry data | This study | MassIVE: MSV000092829 |
| Processed mass spectrometry data | This study | See Table S4 |
| Experimental models: Cell lines | ||
| Human: Hs 578T | ATCC | HTB-126 |
| Human: HCC1187 | ATCC | CRL-2322 |
| Human: MCF10A | ATCC | CRL-10317 |
| Human: MDA-MB-157 | ATCC | HTB-24 |
| Experimental models: Organisms/strains | ||
| Mouse: 8- to 12-week-old virgin female FVB wild-type mice | The Jackson Laboratory or Charles River | 001800 or 207 |
| Software and algorithms | ||
| MSFragger (v.3.8) run using FragPipe (v.20.0) | Kong et al.46 and Teo et al.47 | N/A |
| ProteinProphet | Nesvizhskii et al.48 | N/A |
| Percolator | Käll et al.49 | N/A |
| IonQuant v.1.9.8: | Yu et al.50 | N/A |
| Philosopher v.5.0.0 | da Veiga Leprevost et al.51 | N/A |
| Other | ||
| Orbitrap Fusion Tribrid mass spectrometer | Thermo Scientific | N/A |
| EASY-nLC 1000 HPLC system | Thermo Scientific | N/A |
| Teflon-coated microscope slides | Tekdon | N/A |
| Microscrope slide holder | This study | N/A |
| ImageStream AMNIS MKII imaging flow cytometer | Cytek Biosciences | N/A |
| FACS ARIA Fusion Flow Cytometer | BD Biosciences | N/A |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Thomas Kislinger (thomas.kislinger@utoronto.ca).
Materials availability
The template used to order the DROPPS slides is available upon request. Slide holder template available at https://3d.nih.gov/entries/3DPX-020483.
Data and code availability
-
•
All mass spectrometry raw files acquired for this study are publicly available from UCSD’s MassIVE database (MassIVE: MSV000092829) under dataset identifier MSV000092829. Processed proteomics data are available in this paper’s Table S4.
-
•
This paper does not report original code.
-
•
Any additional information required to reanalyze the data reported in this work paper is available from the lead contact upon request.
Experimental model and study participant details
Cell lines
All cell lines used in this study were purchased from ATCC, were authenticated using short tandem repeat (STR) DNA profiling identity at TCAG Facilities (Sick Kids Hospital, Toronto), and were tested for mycoplasma contamination using the ATCC universal mycoplasma detection kit according to the instructions of the manufacturer. Growth media and supplements were purchased from Gibco unless otherwise specified. Hs 578T (ATCC HTB-126) and MDA-MB-157 (ATCC HTB-24) cells were cultured in DMEM media supplemented with 10% fetal bovine serum (FBS) and penicillin-streptomycin (100 U/mL penicillin, 100 μg/mL streptomycin, Gibco). HCC1187 (ATCC CRL-2322) cells were cultured in RPMI media supplemented with 10% FBS and penicillin-streptomycin (100 U/mL penicillin, 100 μg/mL streptomycin). MCF10A (ATCC CRL-10317) cells were grown in DMEM:F12 media supplemented with 5% horse serum, 20 ng/mL epidermal growth factor (EGF), 10 μg/mL insulin, 500 ng/mL hydrocortisone, 100 ng/mL cholera toxin, and penicillin-streptomycin (100 U/mL penicillin, 100 μg/mL streptomycin). All cell lines were incubated at 37°C, 5% CO2.
Mouse experiments
All experiments were performed using 8- to 12-week-old virgin female FVB wild-type mice (The Jackson Laboratory or Charles River). Mice were ovariectomized bilaterally, then allowed 10-14 days to recover. A slow-release 0.14 mg 17-β oestradiol plus 14 mg progesterone pellet (Innovative Research of America) was then placed subcutaneously near the thoracic mammary gland for 12–14 days. This was done to obtain large quantities of viable mammary stem/progenitor cells for subsequent analysis, as previously reported.22,52 The light cycle in the rooms was set to 12 h on and 12 h off with a 30-min transition. Temperature was set to 21–23°C and humidity at 30–60%. All animal procedures were in compliance with ethical guidelines established by the Canadian Council for Animal Care under protocols approved by the Animal Care Committee of the Ontario Cancer Institute.
Method details
Mouse mammary single-cell suspensions
Mammary glands were collected and manually minced with scissors for 2 min, and then enzymatically dissociated using 750 U/mL collagenase and 250 U/mL hyaluronidase (STEMCELL Technologies, 07912) and diluted in DMEM:F12 for 1.5 h. Samples were vortexed after 1 h and 1.5 h. Red blood cells were lysed using ammonium chloride (STEMCELL Technologies, 07850). Cells were then triturated in prewarmed (37°C) trypsin-EDTA (0.25%; STEMCELL Technologies, 07901) with a 1 mL pipette for 2 min. Next, they were washed in HBSS without calcium or magnesium plus 2% FBS and centrifuged at 350 x g. Finally, cells were further dissociated in Dispase 5 U/mL (STEMCELL Technologies, 07913) plus 50 μg/mL DNase I (Sigma, D4513) for 2 min, washed in HBBS +2% FBS and filtered using a 40 μm cell strainer to obtain single cells.
Cell line single-cell suspensions
Single cell suspensions were generated by washing cells with PBS followed by 3–5 min incubation in TryplE. Digestion was quenched with equal volume of growth media and collected by centrifugation at 300 x g. Cells were washed twice with PBS before incubation with DAPI. After 10 min incubation, cells were collected by centrifugation at 300 x g and filtered through a 35 μm cell strainer.
Fluorescent activated cell sorting
Cell lines
Dead cells were excluded using DAPI following doublet exclusion.
Mouse experiments
Staining for mitochondrial activity (250 nM MTR CM-H2Xros; ThermoFisher, M7513) was performed before cell-surface-marker staining protocol by incubating cells at 37 °C for 30 min following the manufacturer’s protocols. For FACS staining, APC-Cy7 rat anti-mouse CD326 clone G8.8 (EpCAM; BioLegend, 118217; 1:200 dilution), PE-Cy7 rat anti-human CD49f clone GoH3 (BioLegend, 313622; 1:100 dilution), eFluor 450 rat anti-mouse CD31 clone 390 (ThermoFisher, 48-0311-82; 1:200 dilution), eFluor 450 rat anti-mouse CD45 clone 30-F11 (ThermoFisher, 48-0451-82; 1:800 dilution), eFluor 450 rat anti-mouse TER-119 clone TER-119 (ThermoFisher, 48-5921-82; 1:100 dilution), APC Mouse Anti-Mouse CD36 clone CRF D-2712 (BD Biosciences, 562744; 1:20 dilution) were used. Dead cells were excluded following doublet exclusion using DAPI. Lineage-positive cells were defined as Ter119+CD31+CD45+. Mouse mammary cell subpopulations were defined as: total basal (Lin−EpCAMlo-medCD49fhi), total luminal (Lin−EpCAMhiCD49flo) as previously described.23,24,25 MTRhigh and MTRlow mitochondrial activity populations were defined as top 10% and bottom 30% of MTR signal, respectively, and applied after gating for total luminal and basal populations. Cell sorting was performed on a BD FACS ARIA Fusion with FACSDiva (v.8.0.1 and v.6.1.3). Flow data analysis was performed using FlowJo (v.10.8.1).
Imaging flow cytometry
Incubation of samples in 1 μM BODIPY FL C12 (D3822, ThermoFisher) was performed for 30 min at 37°C for quantification of fatty acid prior to incubation with cell surface antibodies. Cell surface marker staining was performed using the same antibodies against CD49f, CD31, CD45, and TER-119 as in fluorescent activated cell sorting experiments. In addition, APC rat anti-mouse CD326 clone G8.8 (EpCAM; BioLegend, 118214; 1:200 dilution) and PE Mouse Anti-Mouse CD36 clone CRF D-2712 (BD Biosciences, 562702; 1:20 dilution) were used. CD36high was annotated as the top ∼20% CD36 expressing cells and CD36neg was set using fluorescent-minus-one control. Raw imaging flow cytometry files were captured on a Cytek ImageStream AMNIS MKII using INSPIRE software and data analysis performed using IDEAS 6.3 software.
Mouse colony-forming cell assay
In total, 350 cells of the specified FACS-purified population were seeded together with 20,000 irradiated NIH 3T3 mouse fibroblasts per well in six-well plate format. Cells were cultured for 7 days at 5% O2 in EpiCult-B mouse medium (STEMCELL Technologies, 05610) supplemented with 5% FBS, 10 ng/mL human epidermal growth factor (STEMCELL Technologies, 78006), 20 ng/mL basic fibroblast growth factor (STEMCELL Technologies, 78003), 4 μg/mL heparin (STEMCELL Technologies, 07980) and 5 μM ROCK inhibitor (Millipore). Colonies were stained with Wright-Giesma and plates were scanned using the Biotek Cytation 5 Imaging Multimodal Reader (Agilent). From scanned images, colonies were counted and sized using Biotek Gen5 software (v3.11).
Proteomic sample preparation
DROPPS
Cells were deposited into wells of Teflon-coated slides (Tekdon, template available upon request) by pipette or directly by FACS. The solution accompanying the cells was allowed to evaporate at room temperature in a biosafety cabinet and then slides were stored at −80°C until further processing. For lysis and protein reduction, 2.5 μL of buffer consisting of 30% (v/v) Invitrosol (ThermoFisher, MS10007), 15% (v/v) acetonitrile, 0.06% (w/v) n-dodecyl-β-D-maltoside (Sigma, 850520P), 5 mM Tris(2-carboxyethyl)phosphine, and 100 mM ammonium bicarbonate in HPLC grade water. Slides were placed in a 37°C humidity chamber and allowed to incubate for 30 min. For alkylation and protein digestion, 1 μL of buffer containing 35 mM iodoacetamide and Trypsin/Lys-C (Promega, V5072) was added to each sample. Samples were allowed to digest for 2 h in a 37°C humidity chamber. Digestion was quenched by bringing samples to 0.1% (v/v) formic acid with 8 μL of 0.14% (v/v) formic acid in 37°C HPLC grade water and then samples were transferred to 96-well plate. The wells were washed with an additional 8 μL of 0.1% (v/v) formic acid and then the plate was transferred to EASY-nLC 1000 System chilled to 7°C.
SOP
SOP-MS protocol was adapted from Martin et al.19 A 96 well PCR plate was pretreated by addition of 200uL 0.1% DDM for 5 min and then allowed to dry in chemical fume hood after removal of the 0.1% DDM. Cells were deposited into wells by FACS and then collected by centrifugation for 3 min at 3000 x g at 4°C. Volumes were brought to 50 μL of 25 mM ammonium bicarbonate in HPLC grade water. 2 μL of 0.2% DDM in 25 mM ABC was added to each well and then plates were incubated overnight at 4°C. After DDM incubation, cells were sonicated five times at 1 min intervals in water batch placing plate on ice for 1 min in between cycles. The plate was then centrifuged for 3 min at 3000 x g at 4°C and then placed in Eppendorf Thermomixer at 75°C for 1 h. After heating, the plate was centrifuged for 3000 x g at 4°C. 10 ng and 20 ng of Trypsin/Lys-C were added to 100 and 1000 cell replicates, respectively, and then digested for 16 h at 37°C with mixing at 500 rpm. To quench digestion, 2 μL of 5% formic acid was added to each well. The sample volume was reduced to ∼20 μL by SpeedVac. Samples were then centrifuged for 1 h at 3000 x g at 4°C. The 96-well plate was sealed with PCR sealing tape and stored at −20°C before analysis.
Mass spectrometry data acquisition
LC-MS/MS analysis was performed on an Orbitrap Fusion MS (ThermoFisher) coupled to EASY-nLC 1000 System (ThermoFisher). Peptides were washed on pre-column (Acclaim PepMap 100 C18, ThermoFisher) with 60 μL of mobile phase A (0.1% FA in HPLC grade water) at 3 μL/min separated using a 50 cm EASY-Spray column (ES803, ThermoFisher) ramping mobile phase B (0.1% FA in HPLC grade acetonitrile) from 0% to 5% in 2 min, 5%–27% in 160 min, 27%–60% in 40 min interfaced online using an EASY-Spray source (ThermoFisher). The Orbitrap Fusion MS was operated in data dependent acquisition mode using a 2.5 s cycle at a full MS resolution of 240,000 with a full scan range of 350–1550 m/z with RF Lens at 60%, full MS AGC at 200%, and maximum inject time at 40 ms. MS/MS scans were recorded in the ion trap with 1.2 Th isolation window, 100 ms maximum injection time, MS/MS AGC at 200%, with a scan range of 200–1400 m/z using Normal scan rate. Ions for MS/MS were selected using monoisotopic peak detection, intensity threshold of 1,000, positive charge states of 2–5, 40 s dynamic exclusion, and then fragmented using HCD with 31% NCE.
Mass spectrometry raw data analysis
Raw files were analyzed using FragPipe (v.20.0) using MSFragger46,47 (v.3.8) to search against a human (Uniprot, 43,392 sequences, accessed 2023-02-08) or mouse (Uniprot, 25,474 sequences, accessed 2021-06-03) proteomes – canonical plus isoforms. Default settings for LFQ workflow48,49 were used using IonQuant50 v.1.9.8 and Philosopher51 v.5.0.0 with the following modifications: Precursor and fragment mass tolerance were specified at −50 to 50 ppm and 0.15 Da, respectively; parameter optimization was disabled; Pyro-Glu or loss of ammonia at peptide N-terminal was included as a variable modification; MaxLFQ min ions was set to 1; MBR RT tolerance was set to 2 min, and MBR top runs was set to 10. The cell titration and SOP-MS comparison experiment was searched without MaxLFQ and without normalization. No carbamidomethylation on cysteine was used for SOP-MS runs.
Quantification and statistical analysis
Statistical details of experiments (e.g., statistical tests used, exact value of n) can be found in the text and figure legends. Definition of center, and dispersion and precision measures can be found in figure legends. Significance levels are stated in the main text where they apply.
Mass spectrometry statistical analysis
All analysis was performed using R programming language (v.4.2.2) with Tidyverse pacakge (tidyverse_1.3.2) unless otherwise specified. All correlation estimates and p values were calculated using the “cor.test” function. For all experiments except the titration and SOP-MS comparison experiment, the “MaxLFQ Intensity” columns were extracted from the “combined_protein.tsv” and “combined_peptide.tsv” output files from FragPipe (Table S4). For the titration and SOP-MS comparison experiment the “Intensity” columns were used. Proteins were filtered out if the Uniprot Accession ID matched one from the MaxQuant contaminants list unless they belonged to a set of IDs being used for comparison (e.g., TNBC Basal subtype markers or epithelial compartment markers, Table S1). Coefficients of variation were calculated on intensity measurements with three or more replicates without imputation. Run-to-run correlations were calculated using Spearman’s correlation on log2-transformed intensities without imputation. Principal component analysis was performed on log2-transformed intensities imputing missing values to 0. Results from statistical tests are reported in Table S5.
TNBC cell line analysis
First proteins were filtered for presence in >50% of replicates for at least one cell line and subsequently imputed with lower tail imputation (downshift of 1.8 s.d. and width of 0.5 s.d.).53 TNBC basal markers21 heatmap generation was clustered using Pretty Heatmap package (pheatmap_1.0.12) using correlation as a distance metric for proteins, Euclidean distance for samples, and Ward D2 linkage The GSVA54 package (GSVA_1.46.0) was used to calculated similarity between cell lines and TNBC Basal subtypes. Omega-squared values were calculated fitting log2-transformed intensities to a two-way ANOVA with batch and cell line as independent factors. Differential expression was calculated using the Estimated Marginal Means package (emmeans_1.8.4–1) to calculate group means from a linear model fit with batch as cell line as factors with an interaction term. Mean fold-changes were calculated from supplementary files from previous bulk proteomic studies13,14 and plotted against mean fold changes from this study.
Primary total epithelial compartments
First proteins were filtered for presence in >50% of replicates for at least one epithelial compartment and subsequently imputed with lower tail imputation.53 Differential expression was calculated using Estimated Marginal Means package using a linear mixed model with compartment as a fixed effect and allowing for a different intercept per mouse. Genes in volcano plot were annotated with canonical compartment markers as previously described.22 Data from Casey et al. were accessed from Table S5 and fold changes were calculated using the “LFQ or adj iBAQ” values.
Primary epithelial MTR subsets
Data for basal and luminal compartments were analyzed independently. First proteins were filtered for presence in >50% of replicates for total, MTRhigh and MTRlow subsets and subsequently imputed with random forest algorithm using the MissForest package (missForest_1.5). Significant differences were detected using repeated measures ANOVA with mitochondrial subset as variable within mouse as a grouping variable. Fold changes were estimated using Estimated Marginal Means package using a linear mixed model (lme4_1.1–31) with mitochondrial subset as a fixed effect and allowing for a different intercept per mouse. GSEA was applied using a preranked list of Gene Symbols sorted based on estimated fold changes against the Mouse Wikipathways gene sets with minimum and maximum sizes of 25 and 200, respectively. GenieScore was calculated as described26 using mitochondrial subset means as input values.
Basal CD36 population
First proteins were filtered for presence in >50% of replicates for total and CD36high subsequently imputed using the MissForest package. Fold changes and significance were estimated using Estimated Marginal Means package using a linear mixed model with CD36 subset as a fixed effect and allowing for a different intercept per mouse. GSEA was applied using a preranked list of Gene Symbols sorted based on estimated fold changes against the Mouse Gene Ontology Biological Processes gene sets with minimum and maximum sizes of 25 and 200, respectively. Outputs from GSEA were used as inputs to Cytoscape (v.3.9.1). GSEA gene sets were visualized in EnrichmentMap (v.3.3.4) and AutoAnnotate (v.1.3.5).
Data visualization
Unless otherwise specified, plots were generated using R programming language (v.4.2.2) with Tidyverse pacakge (tidyverse_1.3.2) with ggthemes_4.2.4, ggpubr_0.5.0, ggplot2_3.4.0, and ggbeeswarm_0.6.0 packages.
Acknowledgments
Flow cytometry was performed using instrumentation in the Princess Margaret Flow facility. We are especially thankful to Frances Tong, Aleks Spurmanis, and Arian Khandani for feedback on slide and plate design and for assistance with sample collection. Special thanks to Patrik Lanzhotsky for assistance with 3D printing. Work in the Kislinger and Khokha labs was supported by operating grants from the Canadian Institutes of Health Research (CIHR), the Terry Fox Research Institute PPG, the Canadian Cancer Society, and the Canadian Research Chair program. A.K. was supported by the Ontario Graduate Scholarship and Ontario Student Opportunity Trust Fund Awards. C.W.M. was supported by a Banting Postdoctoral Fellowship. M.W. was supported by a CIHR Postdoctoral Fellowship.
Author contributions
M.W., A.K., P.T., C.W.M., M.G., B.Z., S.K., and P.D.W. designed experiments, performed experiments, and contributed to data acquisition. M.W. and S.K. acquired the MS data. M.W., A.K., P.T., C.W.M., and B.Z. analyzed the data. M.W. and A.K. generated figures. R.K. and T.K. supervised the study. M.W., A.K., and M.G. wrote the manuscript, which all other authors edited and approved.
Declaration of interests
The authors declare no competing interests.
Published: April 2, 2024
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.crmeth.2024.100741.
Contributor Information
Rama Khokha, Email: rama.khokha@uhnresearch.ca.
Thomas Kislinger, Email: thomas.kislinger@utoronto.ca.
Supplemental information
References
- 1.Pandya S., Moore R.G. Breast development and anatomy. Clin. Obstet. Gynecol. 2011;54:91–95. doi: 10.1097/GRF.0b013e318207ffe9. [DOI] [PubMed] [Google Scholar]
- 2.Macias H., Hinck L. Mammary Gland Development. Wiley Interdiscip. Rev. Dev. Biol. 2012;1:533–557. doi: 10.1002/wdev.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hassiotou F., Geddes D. Anatomy of the human mammary gland: Current status of knowledge. Clin. Anat. 2013;26:29–48. doi: 10.1002/ca.22165. [DOI] [PubMed] [Google Scholar]
- 4.Visvader J.E. Keeping abreast of the mammary epithelial hierarchy and breast tumorigenesis. Genes Dev. 2009;23:2563–2577. doi: 10.1101/gad.1849509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fu N.Y., Nolan E., Lindeman G.J., Visvader J.E. Stem Cells and the Differentiation Hierarchy in Mammary Gland Development. Physiol. Rev. 2020;100:489–523. doi: 10.1152/physrev.00040.2018. [DOI] [PubMed] [Google Scholar]
- 6.Prat A., Parker J.S., Karginova O., Fan C., Livasy C., Herschkowitz J.I., He X., Perou C.M. Phenotypic and molecular characterization of the claudin-low intrinsic subtype of breast cancer. Breast Cancer Res. 2010;12:R68. doi: 10.1186/bcr2635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lim E., Vaillant F., Wu D., Forrest N.C., Pal B., Hart A.H., Asselin-Labat M.L., Gyorki D.E., Ward T., Partanen A., et al. Aberrant luminal progenitors as the candidate target population for basal tumor development in BRCA1 mutation carriers. Nat. Med. 2009;15:907–913. doi: 10.1038/nm.2000. [DOI] [PubMed] [Google Scholar]
- 8.Tharmapalan P., Mahendralingam M., Berman H.K., Khokha R. Mammary stem cells and progenitors: targeting the roots of breast cancer for prevention. EMBO J. 2019;38 doi: 10.15252/embj.2018100852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cai S., Kalisky T., Sahoo D., Dalerba P., Feng W., Lin Y., Qian D., Kong A., Yu J., Wang F., et al. A Quiescent Bcl11b High Stem Cell Population Is Required for Maintenance of the Mammary Gland. Cell Stem Cell. 2017;20:247–260.e5. doi: 10.1016/j.stem.2016.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.dos Santos C.O., Rebbeck C., Rozhkova E., Valentine A., Samuels A., Kadiri L.R., Osten P., Harris E.Y., Uren P.J., Smith A.D., Hannon G.J. Molecular hierarchy of mammary differentiation yields refined markers of mammary stem cells. Proc. Natl. Acad. Sci. USA. 2013;110:7123–7130. doi: 10.1073/pnas.1303919110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fu N.Y., Rios A.C., Pal B., Law C.W., Jamieson P., Liu R., Vaillant F., Jackling F., Liu K.H., Smyth G.K., et al. Identification of quiescent and spatially restricted mammary stem cells that are hormone responsive. Nat. Cell Biol. 2017;19:164–176. doi: 10.1038/ncb3471. [DOI] [PubMed] [Google Scholar]
- 12.Wang D., Cai C., Dong X., Yu Q.C., Zhang X.O., Yang L., Zeng Y.A. Identification of multipotent mammary stem cells by protein C receptor expression. Nature. 2015;517:81–84. doi: 10.1038/nature13851. [DOI] [PubMed] [Google Scholar]
- 13.Nusinow D.P., Szpyt J., Ghandi M., Rose C.M., McDonald E.R., 3rd, Kalocsay M., Jané-Valbuena J., Gelfand E., Schweppe D.K., Jedrychowski M., et al. Quantitative Proteomics of the Cancer Cell Line Encyclopedia. Cell. 2020;180:387–402.e16. doi: 10.1016/j.cell.2019.12.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lawrence R.T., Perez E.M., Hernández D., Miller C.P., Haas K.M., Irie H.Y., Lee S.I., Blau C.A., Villén J. The Proteomic Landscape of Triple-Negative Breast Cancer. Cell Rep. 2015;11:630–644. doi: 10.1016/j.celrep.2015.03.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang D., Eraslan B., Wieland T., Hallström B., Hopf T., Zolg D.P., Zecha J., Asplund A., Li L.H., Meng C., et al. A deep proteome and transcriptome abundance atlas of 29 healthy human tissues. Mol. Syst. Biol. 2019;15 doi: 10.15252/msb.20188503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Waas M., Kislinger T. Addressing Cellular Heterogeneity in Cancer through Precision Proteomics. J. Proteome Res. 2020 doi: 10.1021/acs.jproteome.0c00338. [DOI] [PubMed] [Google Scholar]
- 17.Leduc A., Huffman R.G., Cantlon J., Khan S., Slavov N. Exploring functional protein covariation across single cells using nPOP. Genome Biol. 2022;23:261. doi: 10.1186/s13059-022-02817-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhu Y., Piehowski P.D., Zhao R., Chen J., Shen Y., Moore R.J., Shukla A.K., Petyuk V.A., Campbell-Thompson M., Mathews C.E., et al. Nanodroplet processing platform for deep and quantitative proteome profiling of 10–100 mammalian cells. Nat. Commun. 2018;9:882. doi: 10.1038/s41467-018-03367-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Martin K., Zhang T., Lin T.T., Habowski A.N., Zhao R., Tsai C.F., Chrisler W.B., Sontag R.L., Orton D.J., Lu Y.J., et al. Facile One-Pot Nanoproteomics for Label-Free Proteome Profiling of 50–1000 Mammalian Cells. J. Proteome Res. 2021;20:4452–4461. doi: 10.1021/acs.jproteome.1c00403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mahendralingam M., Kim H., McCloskey C.W., Aliar K., Casey A.E., Tharmapalan P., Pellacani D., Ignatchenko V., Garcia-Valero M., Palomero L., et al. Mammary epithelial cells have lineage-rooted metabolic identities. Nat. Met. 2021 doi: 10.1038/s42255-021-00388-6. [DOI] [PubMed] [Google Scholar]
- 21.Dai X., Cheng H., Bai Z., Li J. Breast Cancer Cell Line Classification and Its Relevance with Breast Tumor Subtyping. J. Cancer. 2017;8:3131–3141. doi: 10.7150/jca.18457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Casey A.E., Sinha A., Singhania R., Livingstone J., Waterhouse P., Tharmapalan P., Cruickshank J., Shehata M., Drysdale E., Fang H., et al. Mammary molecular portraits reveal lineage-specific features and progenitor cell vulnerabilities. J. Cell Biol. 2018;217:2951–2974. doi: 10.1083/jcb.201804042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shehata M., Teschendorff A., Sharp G., Novcic N., Russell I.A., Avril S., Prater M., Eirew P., Caldas C., Watson C.J., Stingl J. Phenotypic and functional characterisation of the luminal cell hierarchy of the mammary gland. Breast Cancer Res. 2012;14:R134. doi: 10.1186/bcr3334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Joshi P.A., Jackson H.W., Beristain A.G., Di Grappa M.A., Mote P.A., Clarke C.L., Stingl J., Waterhouse P.D., Khokha R. Progesterone induces adult mammary stem cell expansion. Nature. 2010;465:803–807. doi: 10.1038/nature09091. [DOI] [PubMed] [Google Scholar]
- 25.Stingl J., Eirew P., Ricketson I., Shackleton M., Vaillant F., Choi D., Li H.I., Eaves C.J. Purification and unique properties of mammary epithelial stem cells. Nature. 2006;439:993–997. doi: 10.1038/nature04496. [DOI] [PubMed] [Google Scholar]
- 26.Waas M., Snarrenberg S.T., Littrell J., Jones Lipinski R.A., Hansen P.A., Corbett J.A., Gundry R.L. SurfaceGenie: a web-based application for prioritizing cell-type-specific marker candidates. Bioinformatics. 2020;36:3447–3456. doi: 10.1093/bioinformatics/btaa092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bernardo G.M., Lozada K.L., Miedler J.D., Harburg G., Hewitt S.C., Mosley J.D., Godwin A.K., Korach K.S., Visvader J.E., Kaestner K.H., et al. FOXA1 is an essential determinant of ERα expression and mammary ductal morphogenesis. Development. 2010;137:2045–2054. doi: 10.1242/dev.043299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sanborn B.M., Dodge K., Monga M., Qian A., Wang W., Yue C. In: Zingg H.H., Bourque C.W., Bichet D.G., editors. Vol. 449. Springer US; 1998. Molecular Mechanisms Regulating the Effects of Oxytocin on Myometrial Intracellular Calcium; pp. 277–286. (Vasopressin and Oxytocin). [DOI] [PubMed] [Google Scholar]
- 29.Stevenson A.J., Vanwalleghem G., Stewart T.A., Condon N.D., Lloyd-Lewis B., Marino N., Putney J.W., Scott E.K., Ewing A.D., Davis F.M. Multiscale imaging of basal cell dynamics in the functionally mature mammary gland. Proc. Natl. Acad. Sci. USA. 2020;117:26822–26832. doi: 10.1073/pnas.2016905117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Feng W.W., Wilkins O., Bang S., Ung M., Li J., An J., Del Genio C., Canfield K., DiRenzo J., Wells W., et al. CD36-Mediated Metabolic Rewiring of Breast Cancer Cells Promotes Resistance to HER2-Targeted Therapies. Cell Rep. 2019;29:3405–3420.e5. doi: 10.1016/j.celrep.2019.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gyamfi J., Yeo J.H., Kwon D., Min B.S., Cha Y.J., Koo J.S., Jeong J., Lee J., Choi J. Interaction between CD36 and FABP4 modulates adipocyte-induced fatty acid import and metabolism in breast cancer. npj Breast Cancer. 2021;7:129. doi: 10.1038/s41523-021-00324-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liang Y., Han H., Liu L., Duan Y., Yang X., Ma C., Zhu Y., Han J., Li X., Chen Y. CD36 plays a critical role in proliferation, migration and tamoxifen-inhibited growth of ER-positive breast cancer cells. Oncogenesis. 2018;7:98. doi: 10.1038/s41389-018-0107-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sielaff M., Kuharev J., Bohn T., Hahlbrock J., Bopp T., Tenzer S., Distler U. Evaluation of FASP, SP3, and iST Protocols for Proteomic Sample Preparation in the Low Microgram Range. J. Proteome Res. 2017;16:4060–4072. doi: 10.1021/acs.jproteome.7b00433. [DOI] [PubMed] [Google Scholar]
- 34.Vitrinel B., Iannitelli D.E., Mazzoni E.O., Christiaen L., Vogel C. Simple Method to Quantify Protein Abundances from 1000 Cells. ACS Omega. 2020;5:15537–15546. doi: 10.1021/acsomega.0c01191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang S., Ghalandari B., Wang A., Li S., Chen Y., Wang Q., Jiang L., Ding X. Superparamagnetic Composite Nanobeads Anchored with Molecular Glues for Ultrasensitive Label-free Proteomics. Angew. Chem. Int. Ed. Engl. 2023;62:e202309806. doi: 10.1002/anie.202309806. [DOI] [PubMed] [Google Scholar]
- 36.Koschade S.E., Tascher G., Parmar B.S., Brandts C.H., Münch C. SpinTip: A Simple, Robust, and Versatile Preanalytical Method for Microscale Suspension Cell Proteomics. J. Proteome Res. 2022;21:2827–2835. doi: 10.1021/acs.jproteome.2c00478. [DOI] [PubMed] [Google Scholar]
- 37.Amon S., Meier-Abt F., Gillet L.C., Dimitrieva S., Theocharides A.P.A., Manz M.G., Aebersold R. Sensitive Quantitative Proteomics of Human Hematopoietic Stem and Progenitor Cells by Data-independent Acquisition Mass Spectrometry. Mol. Cell. Proteomics. 2019;18:1454–1467. doi: 10.1074/mcp.TIR119.001431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Myers S.A., Rhoads A., Cocco A.R., Peckner R., Haber A.L., Schweitzer L.D., Krug K., Mani D.R., Clauser K.R., Rozenblatt-Rosen O., et al. Streamlined Protocol for Deep Proteomic Profiling of FAC-sorted Cells and Its Application to Freshly Isolated Murine Immune Cells. Mol. Cell. Proteomics. 2019;18:995–1009. doi: 10.1074/mcp.RA118.001259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Soady K.J., Kendrick H., Gao Q., Tutt A., Zvelebil M., Ordonez L.D., Quist J., Tan D.W.M., Isacke C.M., Grigoriadis A., Smalley M.J. Mouse mammary stem cells express prognostic markers for triple-negative breast cancer. Breast Cancer Res. 2015;17:31. doi: 10.1186/s13058-015-0539-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nath A., Li I., Roberts L.R., Chan C. Elevated free fatty acid uptake via CD36 promotes epithelial-mesenchymal transition in hepatocellular carcinoma. Sci. Rep. 2015;5 doi: 10.1038/srep14752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mistry J.J., Hellmich C., Moore J.A., Jibril A., Macaulay I., Moreno-Gonzalez M., Di Palma F., Beraza N., Bowles K.M., Rushworth S.A. Free fatty-acid transport via CD36 drives β-oxidation-mediated hematopoietic stem cell response to infection. Nat. Commun. 2021;12:7130. doi: 10.1038/s41467-021-27460-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang J., Wen T., Li Z., Che X., Gong L., Jiao Z., Qu X., Liu Y. CD36 upregulates DEK transcription and promotes cell migration and invasion via GSK-3β/β-catenin-mediated epithelial-to-mesenchymal transition in gastric cancer. Aging (Albany NY) 2020;13:1883–1897. doi: 10.18632/aging.103985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Deng M., Cai X., Long L., Xie L., Ma H., Zhou Y., Liu S., Zeng C. CD36 promotes the epithelial–mesenchymal transition and metastasis in cervical cancer by interacting with TGF-β. J. Transl. Med. 2019;17:352. doi: 10.1186/s12967-019-2098-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Terry A.R., Nogueira V., Rho H., Ramakrishnan G., Li J., Kang S., Pathmasiri K.C., Bhat S.A., Jiang L., Kuchay S., et al. CD36 maintains lipid homeostasis via selective uptake of monounsaturated fatty acids during matrix detachment and tumor progression. Cell Metabol. 2023;0 doi: 10.1016/j.cmet.2023.09.012. [DOI] [PubMed] [Google Scholar]
- 45.Schott D., Pizon M., Pachamnn U., Pachmann K. Correlation of fatty acid receptor (CD36) positive circulating epithelial tumor cells (CETCs) with aggressiveness of breast cancer disease. J. Clin. Orthod. 2018;36 [Google Scholar]
- 46.Kong A.T., Leprevost F.V., Avtonomov D.M., Mellacheruvu D., Nesvizhskii A.I. MSFragger: ultrafast and comprehensive peptide identification in mass spectrometry–based proteomics. Nat. Methods. 2017;14:513–520. doi: 10.1038/nmeth.4256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Teo G.C., Polasky D.A., Yu F., Nesvizhskii A.I. Fast Deisotoping Algorithm and Its Implementation in the MSFragger Search Engine. J. Proteome Res. 2021;20:498–505. doi: 10.1021/acs.jproteome.0c00544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nesvizhskii A.I., Keller A., Kolker E., Aebersold R. A Statistical Model for Identifying Proteins by Tandem Mass Spectrometry. Anal. Chem. 2003;75:4646–4658. doi: 10.1021/ac0341261. [DOI] [PubMed] [Google Scholar]
- 49.Käll L., Canterbury J.D., Weston J., Noble W.S., MacCoss M.J. Semi-supervised learning for peptide identification from shotgun proteomics datasets. Nat. Methods. 2007;4:923–925. doi: 10.1038/nmeth1113. [DOI] [PubMed] [Google Scholar]
- 50.Yu F., Haynes S.E., Nesvizhskii A.I. IonQuant Enables Accurate and Sensitive Label-Free Quantification With FDR-Controlled Match-Between-Runs. Mol. Cell. Proteomics. 2021;20 doi: 10.1016/j.mcpro.2021.100077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.da Veiga Leprevost F., Haynes S.E., Avtonomov D.M., Chang H.Y., Shanmugam A.K., Mellacheruvu D., Kong A.T., Nesvizhskii A.I. Philosopher: a versatile toolkit for shotgun proteomics data analysis. Nat. Methods. 2020;17:869–870. doi: 10.1038/s41592-020-0912-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Shiah Y.-J., Tharmapalan P., Casey A.E., Joshi P.A., McKee T.D., Jackson H.W., Beristain A.G., Chan-Seng-Yue M.A., Bader G.D., Lydon J.P., et al. A Progesterone-CXCR4 Axis Controls Mammary Progenitor Cell Fate in the Adult Gland. Stem Cell Rep. 2015;4:313–322. doi: 10.1016/j.stemcr.2015.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tyanova S., Temu T., Sinitcyn P., Carlson A., Hein M.Y., Geiger T., Mann M., Cox J. The Perseus computational platform for comprehensive analysis of (prote)omics data. Nat. Methods. 2016;13:731–740. doi: 10.1038/nmeth.3901. [DOI] [PubMed] [Google Scholar]
- 54.Hänzelmann S., Castelo R., Guinney J. gene set variation analysis for microarray and RNA-Seq data. BMC Bioinf. 2013;14:7. doi: 10.1186/1471-2105-14-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
All mass spectrometry raw files acquired for this study are publicly available from UCSD’s MassIVE database (MassIVE: MSV000092829) under dataset identifier MSV000092829. Processed proteomics data are available in this paper’s Table S4.
-
•
This paper does not report original code.
-
•
Any additional information required to reanalyze the data reported in this work paper is available from the lead contact upon request.