Abstract
Background & Aims
Liver sinusoidal endothelial cells (LSECs) are important in liver development, regeneration, and pathophysiology, but the differentiation process underlying their tissue-specific phenotype is poorly understood and difficult to study because primary human cells are scarce. The aim of this study was to use human induced pluripotent stem cell (hiPSC)-derived LSEC-like cells to investigate the differentiation process of LSECs.
Methods
hiPSC-derived endothelial cells (iECs) were transplanted into the livers of Fah−/−/Rag2−/−/Il2rg−/− mice and assessed over a 12-week period. Lineage tracing, immunofluorescence, flow cytometry, plasma human factor VIII measurement, and bulk and single cell transcriptomic analysis were used to assess the molecular and functional changes that occurred following transplantation.
Results
Progressive and long-term repopulation of the liver vasculature occurred as iECs expanded along the sinusoids between hepatocytes and increasingly produced human factor VIII, indicating differentiation into LSEC-like cells. To chart the developmental profile associated with LSEC specification, the bulk transcriptomes of transplanted cells between 1 and 12 weeks after transplantation were compared against primary human adult LSECs. This demonstrated a chronological increase in LSEC markers, LSEC differentiation pathways, and zonation. Bulk transcriptome analysis suggested that the transcription factors NOTCH1, GATA4, and FOS have a central role in LSEC specification, interacting with a network of 27 transcription factors. Novel markers associated with this process included EMCN and CLEC14A. Additionally, single cell transcriptomic analysis demonstrated that transplanted iECs at 4 weeks contained zonal subpopulations with a region-specific phenotype.
Conclusions
Collectively, this study confirms that hiPSCs can adopt LSEC-like features and provides insight into LSEC specification. This humanised xenograft system can be applied to further interrogate LSEC developmental biology and pathophysiology, bypassing current logistical obstacles associated with primary human LSECs.
Impact and implications
Liver sinusoidal endothelial cells (LSECs) are important cells for liver biology, but better model systems are required to study them. We present a pluripotent stem cell xenografting model that produces human LSEC-like cells. A detailed and longitudinal transcriptomic analysis of the development of LSEC-like cells is included, which will guide future studies to interrogate LSEC biology and produce LSEC-like cells that could be used for regenerative medicine.
Keywords: Liver, Liver sinusoidal endothelial cells, Tissue specification, Induced pluripotent stem cells, Cell transplantation
Graphical abstract
Highlights
-
•
iECs can engraft upon transplantation into the liver of mice to produce a humanised model of liver sinusoids.
-
•
iECs undergo tissue specification into LSEC-like cells over time in the in vivo liver microenvironment.
-
•
A novel network of 27 transcription factors is suggested to have a role in LSEC specification in this model.
Introduction
Liver sinusoidal endothelial cells (LSECs) line sinusoids in the liver and have a highly specialised tissue-specific phenotype. However, many aspects of LSEC biology remain unknown because of major obstacles to the study of LSECs, particularly those of human origin. These include the lack of LSEC-specific markers that can be used to identify and purify LSECs,1 controversy regarding the origin of LSECs,2,3 and the rapid dedifferentiation of LSECs in culture,4 which limits their use.
Human induced pluripotent stem cells (hiPSCs) provide a reliable source of personalised stem cells that could generate any cell type. They are similar to human embryonic stem cells (hESCs) but sidestep the ethical and logistical hurdles associated with these cells, implying greater potential for clinical translation and easier access. Many studies have generated endothelial cells (ECs) from hiPSCs, but the development of tissue-specific ECs is a new area of interest. LSEC-like cells have been derived from both hESCs and hiPSCs using in vitro protocols based on TGFβ inhibition, hypoxia, and adrenomedullin signalling,[5], [6], [7] and the overexpression of transcription factors (TFs), such as SPI1 and ETV2.8 However, LSEC-like cells generated in these studies exhibit an incomplete phenotype, suggesting that more complex cues are required. It is likely that the liver microenvironment has a key role in tissue specification, as shown when pluripotent stem cell (PSC)-derived venous angioblasts formed LSEC-like cells after transplantation into mouse livers.9,10 Liver-specific microenvironmental and spatiotemporal cues, such as extracellular matrix,11 paracrine signalling,12 and gut-derived cues,13 all contribute to the LSEC phenotype, but this complexity is difficult to recapitulate in vitro.
Although in vivo transplantation into the liver microenvironment appears to be the most effective strategy for producing LSEC-like cells, the temporal changes that underlie in vivo LSEC specification have not been investigated at the transcriptomic level. An in-depth analysis will guide future strategies to refine the production of LSEC-like cells for therapeutic purposes, such as in liver organoids or cell-based therapies for haemophilia A. It will also aid understanding of the ontology of LSECs.
In this study, hiPSC-derived ECs (iECs) were transplanted into the liver of Fah−/−/Rag2−/−/Il2rg−/− (FRG) mice14 to generate LSEC-like cells. iECs readily engrafted with long-term survival, and upregulated key gene signatures and pathways associated with LSECs. We provide insights into the developmental trajectory of transplanted iECs, and transcriptional regulators associated with LSEC specification, and compare these with primary human LSECs. Humanised mice repopulated with hiPSC-derived LSEC-like cells offer a unique PSC-based platform to study LSEC specification, with future applications in developmental biology, disease modelling, and drug testing.
Materials and methods
hLSEC isolation and culture
Human liver specimens were collected from patients who underwent liver resection at St Vincent’s Hospital Melbourne, with informed consent and approval by the St Vincent’s Hospital Human Research Ethics Committee (HREC protocol 52/03). Patients with a history of infection (such as HIV or HBV) were excluded. hLSECs were isolated as described previously for mouse LSECs,15,16 with a modification to the enzyme cocktail used [0.5% collagenase P (Roche, Basel, Switzerland), 0.125% hyaluronidase (Sigma-Aldrich. MO, USA), 0.05% DNAseI (Roche), and 1.25% Dispase II (Sigma-Aldrich)]. Isolated hLSECs were plated onto 8-well cell culture slides (Millicell® EZ Slides, Merck Millipore, Darmstadt, Germany) coated with human fibronectin (Sigma-Aldrich), in hLSEC medium [EGM-2-MV microvascular EC medium (Lonza, Basel, Switzerland) supplemented with 50 ng/ml recombinant human vascular endothelial growth factor-A (VEGF-A) (Peprotech), 50 ng/ml recombinant fibroblast growth factor 2 (Peprotech, NJ, USA), 10 mM HEPES, 10 μM Y-27832, 5 μM A83-01, and 1 μM SB-431542 (all Sigma-Aldrich)].
Immunocytochemistry of cultured hLSECs
Cells were fixed with 4% paraformaldehyde in PBS, and permeabilised with 0.3% Triton-X detergent in PHEM buffer (10 mM PIPES, 25 mM HEPES, 10 mM EGTA, 2 mM MgCl2, pH 6.9, all from Sigma-Aldrich). Primary antibodies against CD31 (1:50, JC70A, Dako, Glostrup, Denmark), LYVE-1 (1:100, AF2089, R&D Systems, MN, USA) CD32B (1:100, ab45143, Abcam, Cambridge, UK), Stabilin-2 (1:100, ab121893, Abcam), and Factor VIII (1:100, SAF8C-AP, Affinity Biologicals, ONT, Canada) were applied for 1 h, followed by secondary antibody for 30 min (either Alexa-Fluor 488 or 594 conjugated goat anti-mouse, goat anti-rabbit, or goat anti-sheep, all at 1:200, Thermo Fisher Scientific, MA, USA). After nuclear staining with DAPI, slides were mounted with fluorescence mounting medium (Dako) and a glass coverslip. Cells were imaged using a fluorescence microscope (Olympus BX61, Olympus, Tokyo, Japan).
hiPSC culture
hiPSCs expressing the fluorescent proteins tdTom or enhanced green fluorescent protein (eGFP) inserted into the GAPDH locus were established from RM3.5 hiPSCs derived from human foreskin fibroblasts.17 hiPSCs were maintained on Matrigel-coated tissue culture plates (non-growth factor reduced, hESC qualified, Corning, MA, USA) in TeSR-E8 medium (Stem Cell Technologies, Vancouver, BC, Canada) supplemented with 10% knockout serum replacement (Thermo Fisher Scientific).
hiPSC differentiation into iECs
tdTom and eGFP hiPSCs were differentiated into ECs using a published protocol,18 further optimised by our group.19 Dissociated hiPSCs were plated into hESC-qualified Matrigel-coated plates at a density of 1 × 105 cells/cm2 in hiPSC media with 10 μM Y-27632 (Sigma-Aldrich). After 24 h, medium was changed to DMEM/F-12 GlutaMAX medium with N2 and B27 supplements (Thermo Fisher Scientific), 8 μM CHIR99021, and 25 ng/ml BMP4 (Peprotech) for 3 days. From Day 4 to Day 6, medium was changed to StemPro-34 SFM medium (Thermo Fisher Scientific) with 200 ng/ml VEGF-A (Peprotech) and 2 μM forskolin (Sigma-Aldrich). At Day 6, cells were dissociated, labelled with either FITC or PE-conjugated mouse anti-human CD31 antibody (BD Bioscience, NJ, USA) and DAPI (to identify dead cells), and CD31+cells were purified using FACS (BD Influx cell sorter, BD Bioscience). Purified cells were plated onto human fibronectin (Sigma-Aldrich)-coated plates in iEC medium, which had the same composition as the hLSEC medium.
Endothelial tube formation assay with iECs
Tube formation assays were performed as described previously.15 Specimens were fixed in 4% paraformaldehyde, permeabilised, and immunolabelled for human CD31 (1:50, JC70A, Dako), VE-cadherin (1:100, clone 16B1, Thermo Fisher Scientific), von Willebrand factor (vWF) (1:100, A0082, Dako), and VEGFR2 (1:200, clone 55B11, Cell Signalling Technology, MA, USA), followed by Alexa-Fluor 488 conjugated anti-rabbit and Alexa-Fluor 647 conjugated anti-mouse secondary antibody (Thermo Fisher Scientific), and DAPI.
FRG mouse maintenance
All animal experiments conformed to the Australian National Health and Medical Research Council’s code for the care and use of animals for scientific purposes and were completed with prior approval from the St Vincent’s Hospital animal ethics committee. For routine breeding and maintenance, FRG mice14 were given drinking water comprising 3% dextrose water (Sigma-Aldrich) supplemented with 8 mg/L of NTBC (Yecuris).
iEC transplantation into FRG mice
Male mice 4–6-weeks old (∼15 g in weight) were used for all experiments. Before transplantation, FRG mice underwent a preconditioning regimen with weekly cyclical removal and administration of NTBC comprising 2 days with and 5–7 days without NTBC.16 After 3 weeks, the animals received a single intraperitoneal dose of monocrotaline (150 mg/kg, Sigma-Aldrich) the day before surgery to induce damage to the native liver microvasculature to facilitate engraftment of transplanted ECs. For surgery, after exposure of the spleen via open surgery to the left flank of each animal, cells or control media were injected into the distal pole of the spleen. The volume of injection was 50 μL, and contained 0.1% hyaluronic acid (Sigma-Aldrich) in hiPSC-EC media for control injections; for cell injections, the media/hyaluronic acid combination contained 1 × 106 cells. Animals were harvested at 1, 2, 4, and 12 weeks post transplantation.
Immunofluorescent analysis of mouse liver
Harvested mouse liver was cut into strips, fixed in 4% paraformaldehyde in PBS, snap frozen, and cryosectioned at –20 °C. Cryosections of 10-μm thickness were washed in PHEM buffer, permeabilised with 0.3% Triton-X in PHEM buffer, and incubated with primary antibodies against human CD31 (1:50, JC70A, Dako), PDGFRβ (1:100, AF385, R&D Systems), glutamine synthetase (1:200, ab73593, Abcam), CD32b (1:100, MA5-47232, Thermo Fisher Scientific), LYVE-1 (1:100, ab36993, Abcam), Stabilin-2 (1:100, ab121893, Abcam), CD36 (1:100, ab17044, Abcam), CLEC14A (1:50, PA5-58798, Thermo Fisher Scientific), CLEC4G (1:100, 18173-1-AP, Proteintech, IL, USA), AQP1 (1:100, AQP11-A, Thermo Fisher Scientific), EMCN (1:200, ab106100, Abcam), cytokeratin 19 [1:50, TROMA-III, Developmental Studies Hybridoma Bank (DSHB), Iowa, USA], A6 (1:50, DSHB), and RFP (labelling TdTomato, 1:200, 600-901-379, Rockland, PA, USA) for 1 h, followed by Alexa-fluor 488, 568 or 647 conjugated anti-mouse, anti-goat, anti-rabbit, anti-chicken or anti-rat secondary antibody (Thermo Fisher Scientific) for 30 min. Sections were incubated with DAPI, then mounted with fluorescence-mounting medium and glass coverslips. Immunofluorescence was imaged using a laser scanning confocal microscope (Nikon A1R confocal microscope, Nikon Corporation, Tokyo, Japan and Leica Stellaris 5, Leica, Wetzlar, Germany) and a Leica Thunder Imager (Leica).
Human coagulation factor VIII ELISA
During harvest, mouse blood was collected into 1-ml blood collection tubes containing 3.2% sodium citrate anti-coagulant (Sarstedt, Nümbrecht, Germany) and centrifuged to separate plasma. Control human blood was obtained from healthy patients undergoing elective surgical procedures (e.g. excision of skin lesions) and was processed in the same manner.
Human coagulation factor VIII levels were measured using a factor VIII ELISA kit (Affinity Biologicals).
Mouse liver function tests
Plasma from iEC-transplanted and sham control FRG mice was analysed by an accredited veterinary clinical pathology laboratory (ASAP Laboratories, VIC, Australia). ALT, AST, and ALP were measured with an AU680 Clinical Chemistry Analyser (Beckman Coulter, CA, USA).
Scanning electron microscopy of tdTom iECs isolated from FRG mouse liver
Livers were harvested from FRG mice transplanted with tdTom iECs at 4 weeks. After enzymatic digestion and initial centrifugation at 50 g, the cell pellet formed after centrifugation 300 g was resuspended in iEC medium and purified based on CD31 expression using mouse anti-human CD31 antibody-conjugated magnetic beads (MACS Miltenyi).
For scanning electron microscopy, purified cells were seeded onto human fibronectin-coated 10-mm glass coverslips (Proscitech) and fixed with 2.5% glutaraldehyde in 0.1 M sodium cacodylate buffer (Sigma-Aldrich). Cells were washed in 0.1 M sodium cacodylate in 2% sucrose buffer, osmicated, dehydrated, and treated with hexamethyldisilazane before being mounted and sputter coated with platinum as described previously.20 Specimens were imaged on a JEOL 6380 scanning electron microscope (JEOL Ltd, Tokyo, Japan), and n = 3 preparations were examined.
hiPSC-EC isolation after in vivo transplantation
hiPSC-ECs transplanted into mouse liver were isolated using FACS, at 1, 2, 4, and 12-weeks post transplantation. To obtain enough cells, n = 3 mouse livers were pooled for each preparation, with each preparation referred to as one biological replicate for subsequent analysis. Mouse livers were harvested, transported in Belzer UW® solution, minced, digested, and strained as previously described. The cell suspension was labelled with APC-conjugated anti-mouse CD31 antibody (BD Biosciences), FITC-conjugated anti-human CD31 antibody (BD Biosciences), and DAPI to exclude dead cells. Human umbilical vein ECs (HUVECs) were used as positive controls for human CD31 staining, and primary mouse LSECs from healthy FRG mice were used as positive controls for mouse CD31 staining. tdTom+ cells were purified from the cell isolation using FACS (BD Influx cell sorter), and the proportion of cells positive for human CD31 FITC and mouse CD31 APC was analysed. Cell debris was excluded based on scatter signals, and dead cells were excluded based on uptake of DAPI. Single antibody/DAPI-stained and unstained samples were used for fluorescent compensation. For positive controls, ECs differentiated from hiPSCs or human mammary epithelial cell (HMEC) primary cells were stained with FITC human CD31, and freshly isolated mouse LSECs from non-transplanted FRG mice stained with mouse CD31 APC were used.
Bulk RNA sequencing
iECs cultured in vitro and FACS purified cells (CD31+ cells from human liver and TdTom+ cells from iEC-transplanted livers) were lysed, genomic DNA was removed, and RNA was extracted using an RNeasy Plus Mini Kit (Qiagen, Hilden, Germany). cDNA libraries were prepared using a SMARTer® Stranded Total RNA-Seq Pico Input Mammalian kit v2 (Takara Bio, Shiga, Japan), and sequencing was completed using a HiSeq 2500 System (Illumina, CA, USA) to obtain 30 × 106 100-base pair (bp) paired-end reads per sample. Bulk RNAseq reads were aligned to the human reference genome (Homo sapiens.GRCh38.91) using the STAR aligner (v020201), and transcriptomic reads were counted using featureCounts (v1.5.2). All analyses were completed using R software (v3.6.3) with edgeR/limma (v3.28.1/3.42.2), Seurat (v3.1.5), and tidyverse packages. All plots were generated using the ggplot2 data visualisation package on R, unless specified otherwise. One publicly available bulk RNAseq data set (GSE43984)21 was integrated into the study using the same methodology, accessed via the Gene Expression Omnibus (GEO) database repository hosted by the National Center for Biotechnology Information (NCBI, MD, USA).
Novel markers of LSECs were investigated by comparing two separate DGE analyses: (i) between hLSEC FACS with iEC in vitro; and (ii) between iECs at 12 and 1 weeks. The intersection of upregulated genes from both comparisons were determined to be highly associated with LSEC specification. Comparison between iECs in vitro and iECs at 12 weeks was not used because this approach would select many genes associated with the change in environment (in vitro vs. ex vivo). The expression of these markers in human liver was assessed using the Human Protein Atlas, which shortlisted a subset of markers that were robustly and specifically expressed in LSECs.
Further analysis of markers and TFs involved in LSEC specification was completed using the proprietary Mogrify® platform (Mogrify Ltd, Cambridge, UK). Mogrify allows identification of key TFs to transition ECs to LSECs (expression data based on the FANTOM5 project). Mogrify selects TFs to obtain 95% gene network coverage with as few genes as possible. However, we selected all TFs considered by Mogrify as cell type defining, because we are interested in general differences between the cell types rather than an efficient trans-differentiation.
Single cell RNA sequencing
hLSECs were isolated from liver tissue obtained from a 55-year-old female undergoing hepatic resection for colorectal cancer liver metastasis. Tissue was obtained from the region furthest away from the tumour. No tumour deposits or pathology, such as hepatosteatosis, was present on gross and histopathological examination. The patient had no underlying liver disease and did not receive neoadjuvant chemotherapy. hLSECs were purified via FACS by labelling with mouse anti-human CD31-FITC conjugated antibody (clone WM-59, BD Biosciences). Two independent preparations of transplanted iECs were used for scRNAseq. Each preparation included the iECs isolated from the livers of three mice harvested at 4 weeks, and iECs were isolated based on their expression of tdTom fluorescence.
Isolated cells were processed using a Chromium™ Single Cell 3’ Library & Gel Bead Kit v2 and Chromium™ controller (10X Genomics, CA, USA), and sequencing was completed using a HiSeq 2500 System with 100-bp paired-end reads.
Single cell reads were aligned to the human reference genome (Homo sapiens.GRCh38.91), and transcriptomic reads counted using the 10X single cell software Cell Ranger (v3.1.0). Further analyses were completed using R software with associated packages. The curated gene list used for cell type annotations (Table S1) was sourced from recent single cell studies of the liver.[22], [23], [24], [25]
A publicly available data set was accessed through the NCBI GEO repository (GSE115469) for additional data integration. This data set is a human liver scRNAseq library constructed from the dissociation of five human livers.23
Statistical analysis
Apart from transcriptomic analysis, all data are expressed as mean ± SEM and were analysed using GraphPad Prism (Program version 8, GraphPad Software Inc, CA, USA) using one or two-way ANOVA with Bonferroni post hoc analysis, with p <0.05 considered statistically significant. Sample size was calculated to ensure adequate statistical power (0.8).
Results
iECs transplanted into regenerating mouse liver demonstrate long-term engraftment and function
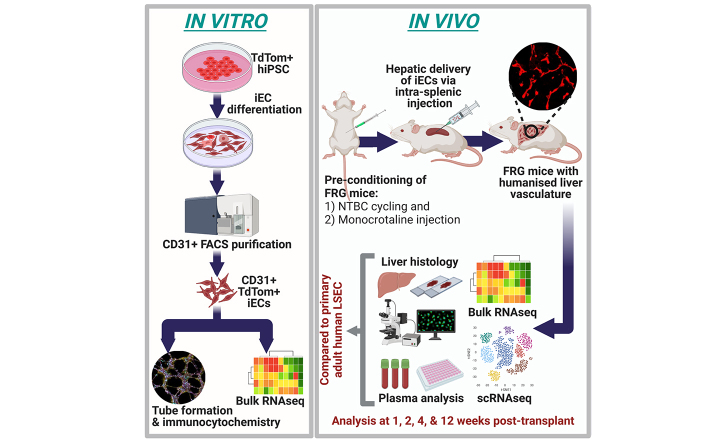
iECs used in this study were fluorescence activated cell sorting (FACS) purified based on human CD31 (hCD31) expression and confirmed to be endothelial in nature by Matrigel tube formation assays and the universal expression of the endothelial markers CD31, vWF, VEGFR, and VE-Cadherin (Fig. 1A and B).
Fig. 1.
Immunofluorescence characterisation of in vitro and transplanted iECs.
TdTom+ iECs formed an interconnected network of tubes in Matrigel, and expressed endothelial markers (A) vWF and CD31 and (B) VEGFR2 and VE-cadherin in vitro. (C) At 1 week post transplantation, a few sporadic CD31+ iECs were found around large blood vessels (yellow arrows). CD31+ iECs expanded along sinusoids over 2 weeks (D) and 4 weeks (E). (F) By 12 weeks, CD31+ iECs expanded extensively across the parenchyma, but there was also a subpopulation of spindle-shaped hCD31-/PDGFRβ+ mesenchymal cells located within the parenchyma adjacent to large vessels (yellow arrow). Using the bile duct marker CK19 to mark periportal Zone 1, this demonstrates that, at 1 week, iECs are mainly around the portal vein (G) and, by 4 weeks, they have expanded from the portal vein into the surrounding parenchyma along the sinusoids. (I,J) At 12 weeks, iECs have formed large tracts expanding from periportal Zone 1 (yellow arrows) near CK19+ bile ducts (green), streaming toward perivenous Zone 3, marked by GS+ hepatocytes (white). Scale bars: 50 μm (C–E,G,H), 100 μm (F), and 200 μm (A,B,I,J). CK19, cytokeratin 19; GS, glutamine synthetase; hCD31, human CD31; iEC, human induced pluripotent stem cell-derived endothelial cell; tdTom, tandem dimer Tomato; vWF, von Willebrand factor.
The FRG mouse model used in this study combines immunodeficiency and liver disease resulting from the lack of the liver enzyme fumarylacetoacetate hydrolase (FAH), which can be rescued by giving the protective drug 2-(2-nitro-4-trifluoromethylbenzoyl)-1,3cyclohexanedione (NTBC). Continuous administration of NTBC results in non-diseased liver, whereas periodical administration results in cycles of hepatotoxicity and liver regeneration. However, the withdrawal of NTBC every 5 days for a period of 2 or 3 days only results in very mild liver injury. Serological markers of liver injury, such as alanine transaminase (ALT), aspartate transaminase (AST), and alkaline phosphatase (ALP), were not elevated in animals on NTBC cycling at any time point and were at similar levels to animals on continuous NTBC (Fig. S1).
Initially, plated and expanded primary adult human LSECs (hLSECs) at Day 3 after isolation (Fig. S2A–F) were transplanted into both non-cycled and cycled FRG mice, but no survival was seen (n = 10). Similarly, there was very limited survival when iECs were transplanted into non-cycled FRG mice (i.e. no liver injury); however, when transplanted into NTBC-cycled FRG mice, iECs rapidly engrafted within the regenerating liver microenvironment (Fig. 1C). Cells injected into the spleen travelled into the liver via the portal vein and, at 1 week post transplantation, sporadic tandem dimer Tomato (tdTom)+ cells were seen exiting the portal vein (Fig. 1C and G). By 2 and 4 weeks after transplantation, flattened, elongated tdTom+ cells gradually infiltrated the surrounding parenchyma along sinusoids that traverse between hepatocytes (Fig. 1D and E). In some areas, iECs lined the entire lumen of portal veins (Fig. 1E and H). By 12 weeks, extensive areas of iECs were present (Fig. 1F and I, J). At all time points, most tdTom+ cells were hCD31+, and largely PDGFRβ negative. Occasionally, small clusters of tdTom+ cells were found in the vicinity of portal veins, with a more spindle-shaped morphology rather than the characteristic flattened endothelial morphology of tdTom+/hCD31+ cells in the sinusoids. These cells were only weakly hCD31+, but strongly PDGFRβ+ (Fig. 1F). At early time points, tdTom+ cells were usually associated with portal veins, which are major blood vessels directly adjacent to cytokeratin 19 (CK19)+ bile ducts (Fig. 1G and H). Starting at 4 weeks and increasingly so at 12 weeks, large tracts of tdTom+ cells extended from the portal region (containing portal veins and bile ducts) toward the centrilobular perivenous region, containing the central vein and glutamine synthetase (GS)+ hepatocytes (Fig. 1I and J). Similar engraftment and distribution were replicated using a second eGFP+ hiPSC line (Fig. S2G and H). These findings confirmed that iECs could repopulate the liver vasculature of FRG mice with increasing tissue distribution over time, and that a regenerating liver microenvironment was conducive to engraftment.
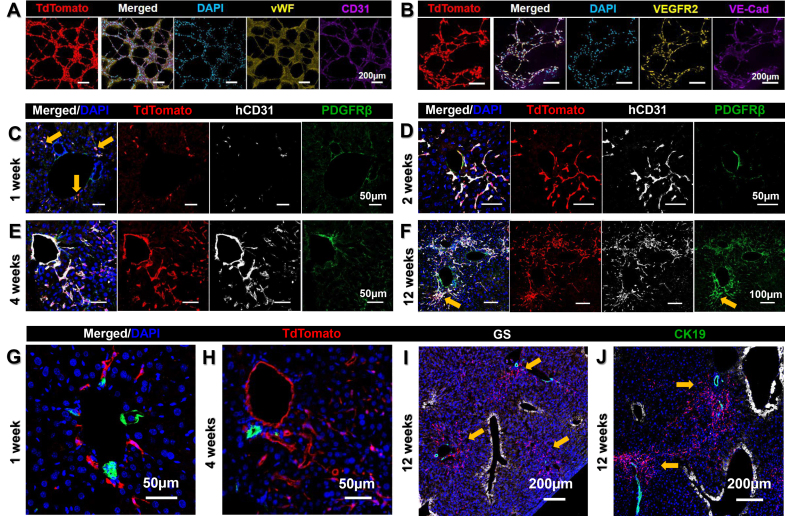
To quantify and characterise the temporal dynamics of iEC engraftment, tdTom+ cells were isolated at several time points after transplantation. Flow cytometric analysis of hCD31+ labelling within tdTom+ cells determined the proportion of ECs and the non-endothelial off-target population over time (Fig. 2A and B). This indicated that, at 1 week, 73.63 ± 23.38% of tdTom+ cells were hCD31+, compared with 96.12 ± 1.08% at 2 weeks, 74.23 ± 9.33 at 4 weeks %, and 61.77 ± 4.78% at 12 weeks (Fig. 2C). Concurrent analysis of mouse CD31+ (mCD31+) cells within the digested mouse liver tissue and calculation of the ratio of tdTom+ human cells to mCD31+ cells were used to quantify the repopulation of mouse liver vasculature by tdTom+ cells. Mouse liver vasculature repopulation increased almost fourfold from 1 to 4 weeks (2.74 ± 1.12% at 1 week, 4.86 ± 1.78% at 2 weeks, and 10.64 ± 3.66% at 4 weeks), but decreased by 12 weeks to 3.70 ± 0.41% (Fig. 2D). To assess functional specification, human factor VIII, a coagulation factor produced exclusively by LSECs, was measured in the mouse plasma. A significant 9.7-fold increase in human factor VIII was found between 1 and 12 weeks (p = 0.001). Plasma readings were compared between iEC-transplanted and sham surgery mice to obtain normalised readings of human-specific factor VIII, and demonstrated a steady increase over time from 0.018 ± 0.0009 IU/ml at 1 week to 0.066 ± 0.025 IU/ml at 2 weeks, 0.1 ± 0.025 IU/ml at 4 weeks, and 0.173±0.019 IU/ml at 12 weeks (Fig. 2E). At 12 weeks, human factor VIII concentration in mouse plasma was 11% of the concentration found in normal human plasma. Both iEC-transplanted and sham surgery (control) mice demonstrated a similar trend in weight gain over time, suggesting similar liver mass and validating the normalisation approach taken to measure human factor VIII and account for cross-reactivity with mouse factor VIII in the analysis. The weight of iEC-transplanted mice increased from 17.2 ± 0.37 g at 1 week to 29.8 ± 0.58 g at 12 weeks, and that of control mice increased from 17.8 ± 0.48 g at 1 week to 30.1 ± 0.52 g at 12 weeks (Fig. 2F). Collectively, these results indicate a spatiotemporal increase in iEC engraftment with a peak at 4 weeks, decreased but long-term engraftment up to 12 weeks, and increased functional specification over time into LSEC-like cells that secrete human factor VIII.
Fig. 2.
Analysis of iEC transplantations into FRG mice.
(A) Gating strategy for FACS purification of tdTom+ cells from FRG liver (red outline). Within TdTom+ cells, hCD31+ and hCD31– subsets were determined (divided by black line). (B) Flow cytometry of mCD31+ cells quantified mouse ECs within the digested liver sample (black outline). Representative plots for iEC transplantation at 4 weeks. (C) Percentage of hCD31+ ECs within the tdTom+ population isolated from FRG mouse livers at 1, 2, 4, and 12 weeks (n = 3–6 per group). One-way ANOVA showed significant overall difference between groups (p = 0.05), but not between individual groups using Bonferroni post hoc analysis. (D) Percentage repopulation of mouse vasculature by tdTom+ cells calculated from flow cytometry analysis of tdTom+ and mCD31+ cells in FRG mouse livers at 1, 2, 4, and 12, weeks (n = 3–6 per group). One-way ANOVA showed significant overall difference between groups (p = 0.001), but not between individual groups using Bonferroni post hoc analysis. (E) Levels of human coagulation factor VIII in FRG mice was calculated by subtracting readings for iEC transplanted mice at 1, 2, 4, and 12 weeks (iEC, dotted purple line) by readings for plasma from control mice treated with sham surgery for the same time points (Control, dotted lilac line, denoting ELISA species cross-reactivity), to produce normalised iEC readings (solid red line) (n = 4–8 per group). One-way ANOVA demonstrated a significant change over time (p = 0.0011), with significant difference between 1 week and 12 weeks (p = 0.001), where there was a 9.7-fold increase. (F) Similar trend in FRG mouse body weights between iEC-transplanted mice and sham surgery control mice. (G) Magnetic-bead isolation with hCD31-conjugated beads was used to purify human ECs from FRG mouse livers at 4 weeks. High purity of cells (∼98%) was achieved (tdTomato+/hCD31+). (H) A small proportion (∼5%) of TdTomato+ hCD31+ iECs purified from FRG mouse livers at 4 weeks demonstrated fenestrations, a morphological hallmark of LSECs. Scale bars: 1 μm (H), 50 μm (lower panel), and 100 μm (upper panel) (G). ECs, endothelial cells; FACS, fluorescence activated cell sorting; FRG, Fah−/−/Rag2−/−/Il2rg−/−; hCD31, human CD31; iEC, human induced pluripotent stem cell-derived endothelial cell; mCD31+, mouse CD31+; tdTom, tandem dimer Tomato.
To further evaluate the LSEC-like characteristics of transplanted iECs, scanning electron microscopy was completed on hCD31+ cells isolated from FRG mouse livers 4 weeks post iEC transplantation. Using magnetic bead separation, a highly purified population of tdTom+/hCD31+ cells was isolated (∼98%) (Fig. 2G), and a small population of these cells (∼5%) demonstrated the presence of membrane fenestrations, a morphological hallmark of LSECs (Fig. 2H). This indicates a small, but highly differentiated population of LSEC-like cells within the engrafted iECs.
Transplanted iECs progressively undergo tissue specification into LSEC-like cells
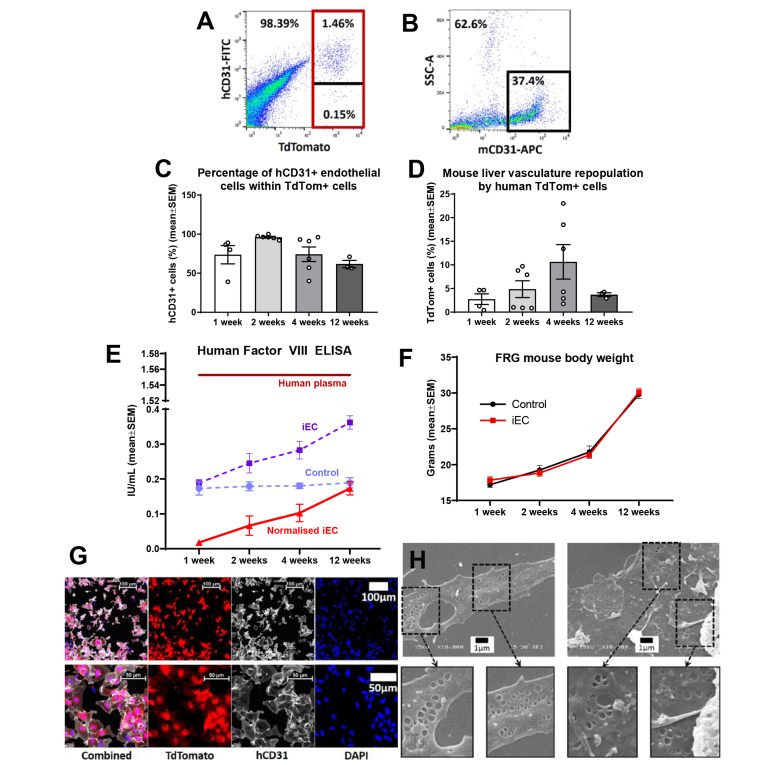
To characterise the temporal dynamics of iEC specification into LSECs at a molecular level, the bulk transcriptome of in vitro and ex vivo iECs was compared with that of primary human LSECs. Multidimensional scaling (MDS) plots of bulk RNA-sequencing (RNAseq) samples and hierarchical clustering demonstrated that all time points of transplanted iECs clustered closely together and, importantly, there was a strong distinguishing effect between in vitro samples (iECs in vitro and plated hLSECs) and all ex vivo samples (transplanted iECs and FACS-isolated LSECs) (Fig. 3A–C; Fig. S3C). MDS plots, particularly in dimensions 2 and 3, indicated the close relationship between transplanted iECs and FACS-isolated LSECs (Fig. 3B). Principal component analysis (PCA) yielded similar results to the MDS plots (Fig. S3A and B). This analysis indicated that the in vivo environment acts on iECs, inducing transcriptional changes that render them more similar to their in vivo counterparts. Samples within the same group clustered closely together, indicating biological reproducibility; the ex vivo hLSEC group was further validated by integrating a data set taken from a previous study on FACS-isolated hLSECs,23 which showed close clustering between hLSECs in the two studies (Fig. 3C,D; Fig. S3C and D).
Fig. 3.
Bulk RNAseq profiling of in vitro iEC and hLSECs (plated), ex vivo transplanted iECs, and FACS-purified hLSECs.
(A,B) Multidimensional scaling plots of (A) dimensions 1 and 2 and (B) dimensions 2 and 3. (C) Dendrogram of hierarchical clustering of bulk RNAseq samples. Note the clustering of a publicly available data set (hLSEC GSE43984) with samples in the hLSEC FACS group. (D) Heatmap and hierarchical clustering of samples based on their expression of the canonical LSEC genes CD14, CD36, F8, FCGR2B, LYVE1, MCAM, PECAM1, and STAB2. Compared with in vitro iECs, transplanted iECs at 12 weeks significantly upregulated the canonical LSEC genes CD14, CD36, F8, FCGR2B, LYVE1, PECAM1, and STAB2. (E) Quantitative expression of canonical LSEC genes derived from bulk RNAseq data. ∗p ≤0.05, ∗∗p ≤0.01, ∗∗∗∗p ≤0.0001 using one-way ANOVA; n = 3–4 per group. FACS, fluorescence activated cell sorting; iEC, human induced pluripotent stem cell-derived endothelial cell; (h)LSEC, (human) liver sinusoidal endothelial cell; RNAseq, RNA sequencing.
To further understand the transcriptional changes occurring in response to the in vivo environment, a canonical group of LSEC-associated genes was used to generate a heatmap (Fig. 3D), which demonstrated a stepwise acquisition of the LSEC signature. In vitro iECs minimally expressed canonical LSEC genes, whereas transplanted iECs at 12 weeks showed the strongest expression. The most robust expression of LSEC genes across all groups was found in FACS-isolated hLSECs, particularly F8, STAB2, and CD14 (although this group expressed lower levels of PECAM1 and MCAM). Quantitative analysis of the canonical LSEC genes demonstrated significant upregulation of seven out of eight genes (all p <0.0001, except MCAM) between in vitro iECs and transplanted iECs at 12 weeks. Comparison between iECs at 12 weeks and FACS-isolated hLSECs demonstrated higher expression of CD14, F8, and STAB2 in FACS-isolated hLSECs (relative to iECs at 12 weeks), higher expression of FCGR2B, MCAM, and PECAM1 in iECs at 12 weeks (relative to FACS-isolated hLSECs), and similar levels of expression of LYVE1 and CD36 between the two groups (Fig. 3E).
At the protein level, immunofluorescence staining of tissue sections was used to compare the expression of LSEC markers at early (2 week) and late (12 week) time points post transplantation. This demonstrated that LSEC markers, such as CD32b, LYVE-1, Stabilin-2, CD36, CLEC14A, and CLEC4G, were all absent at 2 weeks, but present in at least a subset of cells by 12 weeks (Fig. S4A–X). Overall, these results indicated that the in vivo liver microenvironment exerts a strong and time-dependent influence on LSEC specification, and further confirms that iECs can transition into LSEC-like cells.
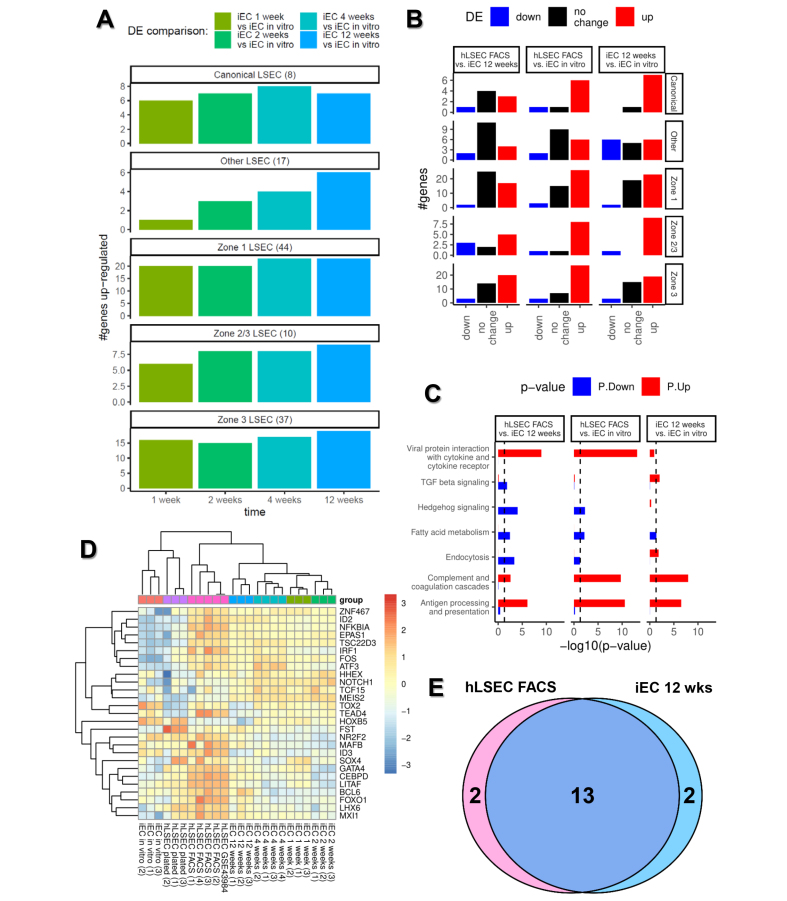
We next examined whether transplanted iECs acquire region-specific characteristics as they extend from their entry vessels into the surrounding tissue over time. The liver is anatomically organised in hexagonal-shaped tissue units called lobules, demarcated by the peripheral portal triads (hepatic artery, portal vein, and bile duct), where blood enters the lobule, and the centrilobular central vein, where blood exits. Conventionally, Zone 1 is defined as the region closest to the portal triad, Zone 3 is the region closest to the central vein, and Zone 2 lies in between. Hepatocytes exhibit phenotypic differences depending on their position within liver zones, with this patterning referred to as ‘metabolic zonation’. More recently, zonation has also been reported in LSECs and, using data from existing studies,[22], [23], [24], [25] we constructed categories of genes associated with Zone 1 LSECs, Zone 2/3 LSECs, and Zone 3 LSECs. Using these groupings together with the canonical LSEC genes described earlier, as well as a group of genes associated with LSECs but not known to be zonally expressed (‘other LSEC genes’) (Table S1), each iEC transplantation time point (1, 2, 4, and 12 weeks) was compared against in vitro iECs to examine the number of upregulated genes in each of the five gene groups. This analysis demonstrated that, at every time point, transplanted iECs acquired markers associated with all five gene categories, which furthermore demonstrated a progressive developmental profile over time, with upregulation of genes associated with Zone 1, 2/3, and 3 LSECs, and other non-zonal LSEC-associated genes (Fig. 4A). Differential gene analysis revealed that the highest number of differentially expressed genes occurred between in vitro iECs and FACS-isolated hLSECs (6,680 genes), and in vitro iECs and transplanted iECs at 12 weeks (6,723 genes) (Fig. S3E). Therefore, we compared the three sample groups across the five established LSEC gene categories, which reinforced the profile of developmental progression from in vitro iECs to transplanted iECs at 12 weeks then FACS-isolated hLSECs. Furthermore, FACS-isolated hLSECs and iECs at 12 weeks had many similarly expressed genes in the Canonical, Other, and Zone 1 LSEC categories (Fig. 4B). Overall, these data suggest that, as transplanted iECs increasingly extend from Zone 1 to Zone 3 over time, they also upregulate genes in a zonal pattern corresponding to their location and, hence, microenvironment. This is reflected by the upregulation of genes across all zones over time. Furthermore, at the protein level, expression of the Zone 1 LSEC marker aquaporin 1 (AQP1) (Fig. S5A and B) and Zone 3 LSEC marker endomucin (EMCN) (Fig. S5G and H) was compared between early (2 weeks) and late (12 weeks) time points post transplantation. AQP1 (Zone 1) was expressed at 12 weeks but absent at 2 weeks (Fig. S5C–F) whereas EMCN was expressed similarly at 2 and 12 weeks (Fig. S5I–L).
Fig. 4.
Characterising the developmental trajectory of transplanted iECs using bulk RNAseq data.
(A) Comparison of the transcriptome of transplanted iECs at 1, 2, 4, and 12 weeks with iECs in vitro to assess the number of upregulated genes in each of five curated groups of genes: (i) genes traditionally associated with LSECs (eight canonical LSEC genes); (ii) genes generally associated with LSECs regardless of their zonal location (other LSEC genes, 17 genes); (iii) genes associated with Zone 1 periportal LSECs (44 genes); (iv) Zone 2/3 mid to perivenous LSECs (10 genes); and (v) Zone 3 perivenous LSECs (37 genes). (B) Bar plot of the number of genes downregulated (blue), upregulated (red), and similarly expressed (black) within each of the five group of genes (Canonical, Other, Zone 1, Zone 2/3, and Zone 3), comparing hLSEC FACS with iECs at 12 weeks, hLSEC FACS with iECs in vitro, and iECs at 12 weeks with iECs in vitro. (C) Comparison of the upregulation (red) and downregulation (blue) of genes associated with key LSEC pathways in hLSEC FACS vs. iECs at 12 weeks, hLSEC FACS vs. iECs in vitro, and iECs at 12 weeks vs. iEC in vitro. (D) Heatmap and hierarchical clustering of samples based on their expression of 27 LSEC-specification transcription factors derived from the Mogrify webtool analysis. Note the clustering of a publicly available data set (hLSEC GSE43984) with samples in the hLSEC FACS group. (E) Venn diagram showing that, out of the 27 transcription factors predicted to be associated with LSEC specification, 13 were upregulated in both hLSEC FACS and iEC 12-week samples, two were upregulated only in hLSEC FACS samples, and two were upregulated only in iEC 12-week samples. FACS, fluorescence activated cell sorting; iEC, human induced pluripotent stem cell-derived endothelial cell; (h)LSEC, (human) liver sinusoidal endothelial cell; RNAseq, RNA sequencing.
Pathway analysis examined the top enriched pathways between FACS-sorted hLSECs, transplanted iECs at 12 weeks and in vitro iECs (Table S2), and pathways of relevance to LSEC function were selected (Fig. 4C). This demonstrated that FACS-sorted hLSECs upregulated the viral protein interaction, complement and coagulation cascade, and antigen processing and presentation pathways compared with both iEC groups, whereas transplanted iECs at 12 weeks had upregulated the viral protein, endocytosis, complement/coagulation, and antigen processing pathways compared with in vitro iECs. Pathways associated with LSEC injury and liver inflammation (TGFβ and Hedgehog signalling) were most highly enriched in iECs at 12 weeks compared with in vitro iECs or FACS hLSECs.
The same in vitro and ex vivo conditions were utilised with primary hLSECs as used for the iEC groups. Two types of hLSEC were used in the study: freshly FACS-isolated hLSECs, and hLSECs isolated through selective adhesion and culture in endothelial medium (plated hLSECs). While broad similarities in terms of the number of genes were observed in Canonical, Other, Zone 1, Zone 2/3, and Zone 3 categories of genes between the two sample types, freshly FACS-isolated hLSECs expressed many more genes compared with plated hLSECs across all five LSEC gene categories (Fig. S6A), and there were large differences in their transcriptome, with 3,696 differentially expressed genes (DEGs) (Fig. S3E). Similarly, fresh hLSECs were enriched in key functional pathways, including viral protein interaction with cytokine, complement and coagulation cascade, and antigen processing and presentation pathways (Fig. S6B). Again, similar to the in vitro and ex vivo comparisons with iEC groups, this demonstrated that the environment exerts a strong influence on the overall transcriptome.
scRNAseq defines subpopulations of isolated cell types and facilitates lineage tracing of transplanted iECs
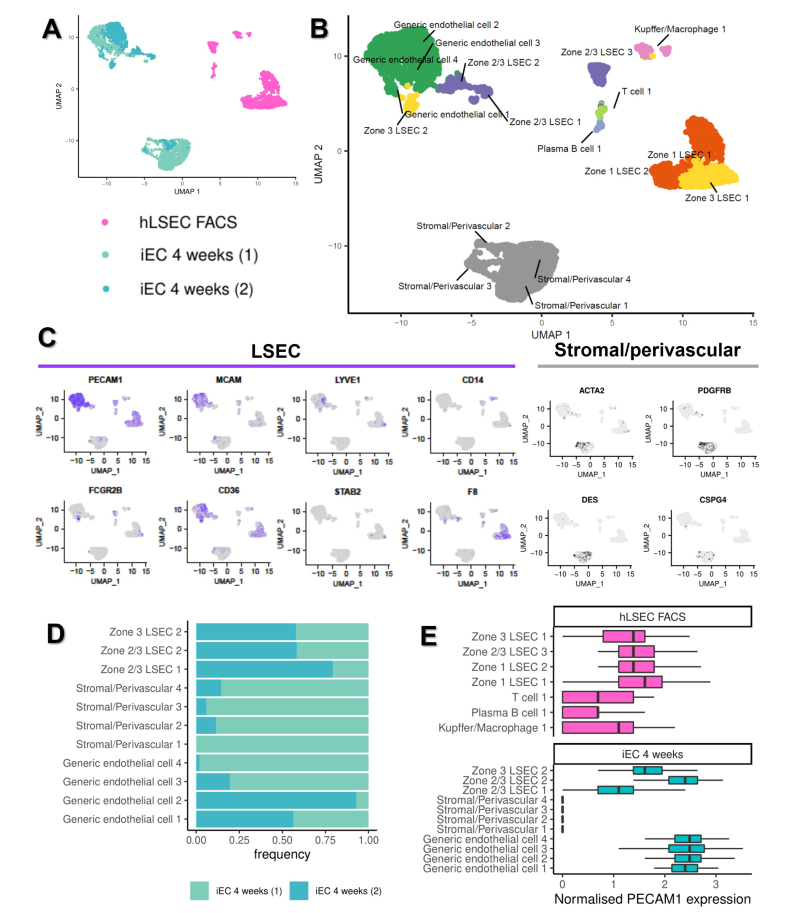
To assess the heterogeneity of the putative LSECs obtained upon iEC transplantation, single cell (sc)RNAseq was used to identify cell subpopulations within samples. The time point with peak engraftment (4 weeks) was used for ex vivo iEC samples. After quality control and filtering of cells (Fig. S7A), 1,643 FACS-isolated hLSECs and 2,809 iEC 4-week cells (1939 in Sample 1, and 870 in Sample 2) were analysed. UMAP plots of the samples demonstrated that the two iEC 4-week samples clustered together and were distinguishable from the FACS-isolated hLSEC sample (Fig. 5A). Using unsupervised clustering followed by cell type annotation using a list of genes associated with different cell types and LSEC zonation (Table S1), seven clusters were identified in the FACS-isolated hLSEC sample and 11 clusters in the iEC 4-week samples (Fig. 5B; Fig. S8). hCD31+ cells sorted from human liver tissue (presumed to be hLSEC) contained LSEC subpopulations from Zones 1, 2/3, and 3, as well as off-target populations, including Kupffer cells/macrophages, plasma B cells, and T cells.
Fig. 5.
ScRNAseq of ex vivo hLSEC FACS and iEC 4-week samples.
(A) UMAP plot of the relative positions of cells associated with three samples included in this analysis, one hLSEC FACS (1,643 cells), and two iEC 4-week samples (1,939 cells in sample 1, 870 cells in sample 2). (B) UMAP plot of clusters identified through automated clustering, with the identity of each cluster assigned based on the expression of key genes associated with different cell types found in the liver. (C) Expression plots of the relative expression of key LSEC (PECAM1, MCAM, LYVE1, CD14, FCGR2B, CD36, STAB2, and F8) and stromal/perivascular (ACTA2, PDGFRB, DES, and CSPG4) genes. Cells expressing the highest level of each gene are denoted by the darkest colour in each plot. (D) The composition of each of the iEC 4-week samples in terms of the relative size of each cluster present (based on number of cells, noted as frequencies) demonstrating biological variability. (E) Boxplots demonstrating the normalised expression of PECAM-1 in all clusters found in this analysis, validating the cell isolation strategy for scRNAseq. hLSEC FACS were purified from human liver based on PECAM-1 (CD31) expression, showing that all cells in this sample expressed PECAM-1, although T cell, plasma B cells, and Kupffer cells/macrophages had lower expression compared with LSECs. iEC 4-week samples (both samples pooled) were sorted based on their expression of TdTom, showing that stromal/perivascular cells did not express PECAM-1, whereas all LSECs and ECs did. EC, endothelial cell; FACS, fluorescence activated cell sorting; iEC, human induced pluripotent stem cell-derived endothelial cell; (h)LSEC, (human) liver sinusoidal endothelial cell; scRNAseq, single cell RNA sequencing; tdTom, tandem dimer Tomato.
Analysing all tdTom+ cells from mouse liver at 4 weeks facilitated lineage tracing of transplanted iECs within the in vivo liver microenvironment. In both iEC 4-week samples, two distinct cell types were observed, indicating that transplanted cells either maintained their EC phenotype, or formed mesenchymal cells with a stromal/perivascular signature (Fig. 5B). Endothelial subpopulations included four clusters that were largely generic ECs in nature, two clusters of Zone 2/3 LSEC-like cells, and one cluster of Zone 3 LSEC-like cells, which overall expressed the LSEC markers PECAM1, MCAM, LYVE1, FCGBR2B, C36, and F8 (Fig. 5C). The mesenchymal cells were categorised into four separate clusters of stromal/perivascular cells, and expressed genes including ACTA2, PDGFRB, DES, and CSPG4 (Fig. 5C), and upregulated genes associated with endothelial–mesenchymal transition, such as TGFB3, SNAI2, TWIST1, TWIST2, ZEB1, and FN1 (Fig. S9). Expression plots of the top-four DEGs associated with each cluster are outlined in Figs. S10 and S11, and Table S3.
Analysing the frequency of cell types associated with each cluster found in the iEC samples indicated that, although all clusters (except one stromal/perivascular cluster) were represented in both samples, there was some variation in frequency. iEC 4-week sample 1 had large clusters of stromal/perivascular cells and relatively fewer LSEC-like cells, whereas sample 2 had very fewer stromal/perivascular cells, and large subpopulations of LSEC-like cells (Fig. 5D). These findings were reflected in the overall differentiation status of the samples assessed using the CytoTRACE computational method, which indicated that iEC 4-week sample 1, with the large mesenchymal population, had the least differentiated state, compared with iEC 4-week sample 2, which had a larger population of LSEC-like cells. Primary hLSECs were much more differentiated compared with either iEC sample (Fig. S7B).
Comparison of normalised PECAM1 expression between all clusters indicated that primary hLSEC subpopulations and iEC-derived generic EC subpopulations highly expressed PECAM1, iEC-derived LSEC subpopulations expressed mid-range levels of PECAM1, T/B/macrophages expressed low levels of PECAM1 (but enough to be picked up through FACS), and iEC-derived stromal subpopulations did not express PECAM1 (Fig. 5E).
In summary, scRNAseq showed that transplanted iECs at 4 weeks contained multiple cellular subpopulations and confirmed the presence of LSEC-like cells and off-target stromal/perivascular cells. Additionally, there was biological variation in the frequency of cells in each of the subpopulations.
Transplanted iECs develop zonal subpopulations with regional differences in phenotype
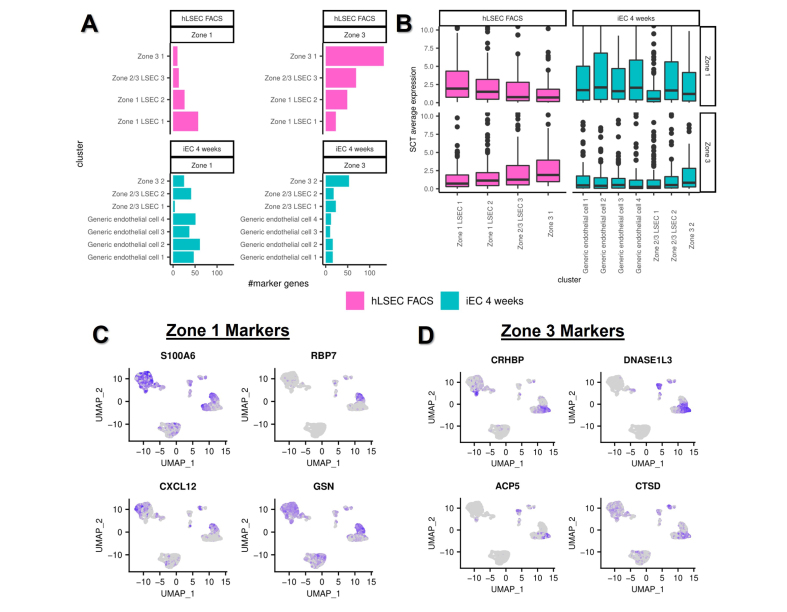
Having established Zone 1, 2/3, and 3 subpopulations in the FACS-isolated hLSEC sample in the scRNAseq data set, DEG analysis between Zone 1 and 3 was used to derive additional marker genes specific to these zones on opposite poles of the liver lobule (complementing the already-established zonation markers). This provided 118 genes specific to Zone 1 and 187 genes specific to Zone 3 (Table S4). This marker profile was then applied to all clusters in the scRNAseq data set, which confirmed that Zone 1 cells within the FACS-isolated hLSEC sample expressed the highest number and level of Zone 1 markers, and Zone 3 cells within the FACS-isolated hLSEC sample expressed the highest number and level of Zone 3 markers (Fig. 6A and B). Following this validation, the same analysis was applied to all endothelial subpopulations in the iEC 4-week samples to confirm that transplanted iECs also undergo zonation and to investigate the nature of generic EC subpopulations. This revealed that generic EC subpopulations expressed a high number and level of Zone 1 markers and a low number and level of Zone 3 markers, indicating they were located in the portal Zone 1 region (Fig. 6A and B). Similar to the FACS-isolated hLSEC sample, Zone 3 LSEC-like cells in the iEC 4-week samples expressed high numbers and levels of Zone 3 genes compared with other cell subpopulations, and relatively lower number and levels of Zone 1 genes (Fig. 6A and B). The top-four Zone 1 markers were then applied to a UMAP expression plot of the scRNAseq data set, which confirmed the high expression of S100A6, CXCL12, and GSN in the generic endothelial subpopulations of the iEC 4-week samples (Fig. 6C). Similarly, the top-four Zone 3 markers were applied to the UMAP expression plot, which demonstrated that CRHBP and ACP5 were expressed in the Zone 3 LSEC-like subpopulation of iEC 4-week samples (Fig. 6D).
Fig. 6.
Zonation of primary hLSECs and transplanted iECs at 4 weeks.
DEGs in Zone 1 and 3 clusters within the hLSEC sample were used to derive a Zone 1 hLSEC gene expression signature (118 genes) and Zone 3 signature (187 genes). (A) Number of genes of each signature (Zone 1 and 3) present in each cluster of the hLSEC and iEC 4-week samples. (B) Overall expression of Zone 1 and Zone 3 signatures in each cluster of the hLSEC and iEC samples. (C,D) UMAP expression plots of the top-four highly expressed markers (DEGs) in the (C) Zone 1 signature and (D) Zone 3 signature. DEG, differentially expressed gene; iEC, human induced pluripotent stem cell-derived endothelial cell; hLSEC, human liver sinusoidal endothelial cell.
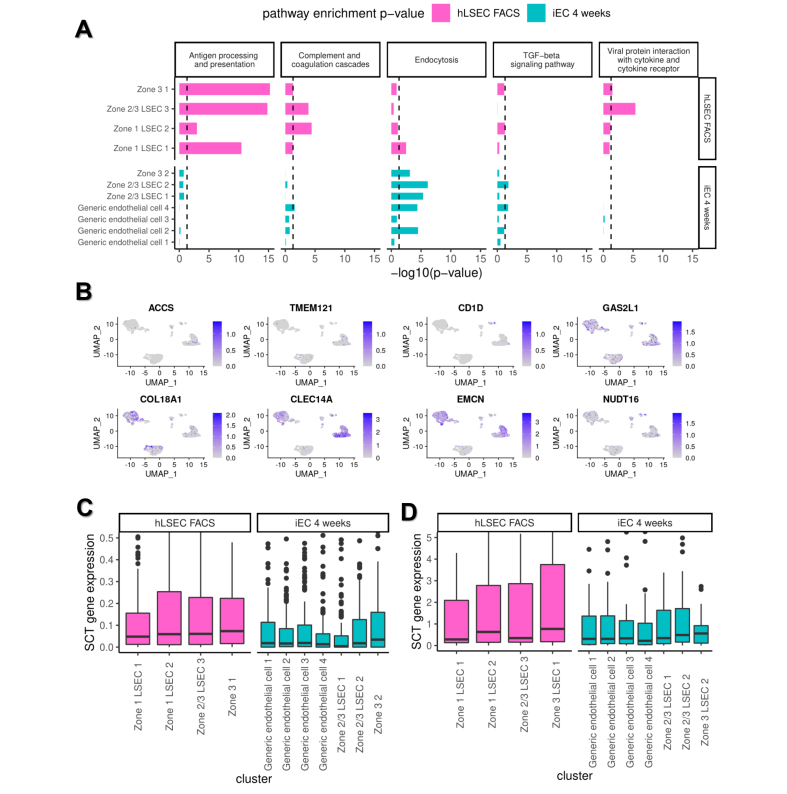
Pathway analysis of endothelial subpopulations in FACS-isolated hLSECs and iEC 4-week samples demonstrated high enrichment of antigen processing and presentation, complement and coagulation cascade, and viral protein interaction with cytokine and cytokine receptor pathways in FACS-isolated hLSEC subpopulations. LSEC-like subpopulations derived from iECs (Zone 2/3 and Zone 3) showed marked enrichment of the endocytosis pathway and, relative to the generic endothelial subpopulations, higher enrichment of the antigen processing and presentation pathway (Fig. 7A).
Fig. 7.
Pathway analysis of scRNAseq clusters and their expression of LSEC-specification markers.
(A) Enrichment of key LSEC pathways in each of the LSEC/endothelial clusters within the hLSEC FACS and iEC 4-weeks samples. Clusters within the hLSEC sample showed enrichment of the antigen processing and presentation, complement and coagulation cascade, and viral protein interaction with cytokine and cytokine receptor pathways. Clusters within the iEC 4-weeks samples showed enrichment in endocytosis and TGFβ signalling pathways. Within clusters in the iEC 4-week samples, LSEC clusters (Zones 3 and 2/3) showed enrichment of the antigen processing and presentation and endocytosis pathways compared with generic EC clusters. (B) Expression plots of eight LSEC marker genes derived from DEG analysis between the bulk transcriptome of hLSEC FACS and in vitro iECs and further short-listed (from an initial list of 213 genes) using the Human Protein Atlas. The relative expression of these markers within the scRNAseq data set is shown, with cells expressing the highest level of each gene being darkest in each plot. This demonstrates that CLEC14A and EMCN were the most robustly expressed in both hLSEC FACS and iEC 4-week samples. (C) Relative expression of the 213 genes associated with LSEC specification derived from DEG analysis within each of the clusters in hLSEC FACS and iEC 4-week samples. Zone 3 LSEC clusters in both hLSEC FACS and iEC 4-week samples expressed the highest level of LSEC specification genes. (D) Relative expression of the 27 transcription factors predicted to regulate LSEC specification (derived using the Mogrify webtool) within each of the clusters found in hLSEC FACS and iEC 4-week samples. Again, this shows that Zone 3 LSEC clusters in both hLSEC FACS and iEC 4 weeks expressed the highest level of these transcription factors. DEG, differentially expressed gene; EC, endothelial cell; FACS, fluorescence activated cell sorting; iEC, human induced pluripotent stem cell-derived endothelial cell; (h)LSEC, (human) liver sinusoidal endothelial cell; scRNAseq, single cell RNA sequencing.
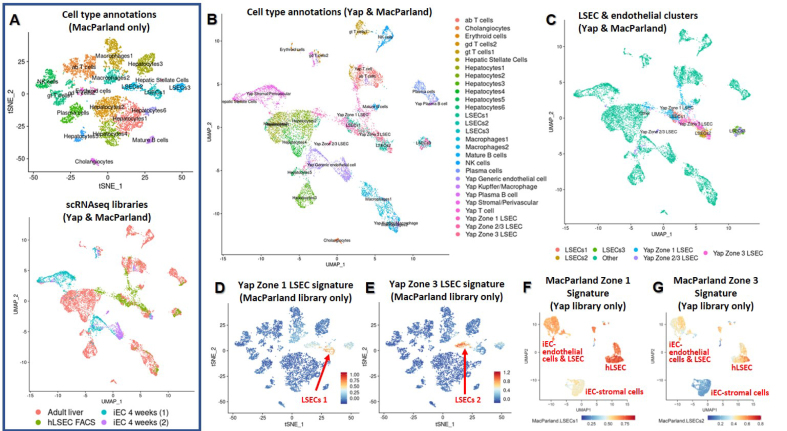
To further investigate zonation, the current scRNAseq library was integrated with a previously published data set containing all major cell types found in the human liver (MacParland et al.23) (Fig. 8A). This demonstrated that iEC-derived stromal/perivascular cells were similar to hepatic stellate cells, and iEC-derived LSECs (designated Zone 3 LSECs) were similar to Zone 3 LSECs in both data sets (i.e. ours and MacParland et al.’s) (Fig. 8B and C). When the Zone 1 and Zone 3 LSEC signatures derived in this study were applied to the MacParland library,23 they selectively identified Zone 1 and Zone 3 clusters, validating our zonation signatures (Fig. 8D and E). However, when the Zone 1 and Zone 3 signatures cited in the MacParland et al. study23 were applied to our library, they were not specific enough to identify specific zonated LSEC subpopulations (Fig. 8F and G).
Fig. 8.
Comparison of hLSECs and transplanted iECs with other cell types in the liver, and validation of LSEC zonation signatures.
(A) A previously published data set including all major cell types in the liver was used (MacParland et al.23) and is shown in the top plot. The bottom plot integrates the MacParland et al. scRNAseq library with the cells in the current study (referred to as the Yap scRNAseq library), and annotates the sample types. (B) Cell type annotations of all clusters within the integrated scRNAseq plot. iEC-derived stromal/perivascular cells cluster closely with hepatic stellate cells, and iEC-derived LSECs cluster closely with LSEC 2 (Zone 2/3 LSECs) from the MacParland et al. data set. Additionally, Zone 1 LSECs from both the Yap and MacParland et al. studies cluster together, as do Zone 3 LSECs from both studies. (C) Only the primary and iEC-derived LSECs are highlighted in the integrated plot to demonstrate the proximity of clusters. (D) Zone 1 LSEC signature identified in the current study was applied to the MacParland et al. library, confirming enrichment in Zone 1 LSECs. (E) Zone 3 LSEC signature identified in the current study was applied to the MacParland et al. library, which was enriched in Zone 3 LSECs. (F) Zone 1 signature identified in the MacParland et al. study was applied to our library and was expressed at higher levels in hLSECs than in iECs, but was not specific for any cluster, indicating a lack of zonal specificity. (G) Zone 3 signature identified in the MacParland et al. study was moderately expressed in iEC-derived ECs (both generic and LSECs) and in Zone 1 hLSECs, but was highest in Zone 3 hLSECs. iEC, human induced pluripotent stem cell-derived endothelial cell; (h)LSEC, (human) liver sinusoidal endothelial cell; scRNAseq, single cell RNA sequencing.
Overall, these data indicate that transplanted iECs at 4 weeks contain subpopulations reflecting their zonal location, and Zone 3 iEC-derived LSECs are similar to Zone 2/3 LSECs found in the human liver. This further confirms the effect of the regional microenvironment on transplanted cells.
Novel markers and transcriptional regulators of LSEC specification
Further analysis was performed to interrogate the process of LSEC specification and its associated markers, to address the current paucity of LSEC markers, and to also guide future strategies for differentiating and maintaining LSECs.
Initially, the expression of three TFs recently described to regulate LSEC specification were assessed: GATA4,26 MAF,27 and ZEB228 (Fig. S12A). Comparing the bulk transcriptome of in vitro iECs against transplanted iECs, GATA4 was most highly expressed at 1 week post transplantation, decreased at 2 weeks, then began increasing again over 4 and 12 weeks. MAF steadily increased over time from 1 to 12 weeks. Although ZEB2 was significantly higher in 1-week iECs compared with in vitro iECs, its expression remained relatively stable over time after an initial rise at 1 week. Additionally, scRNAseq expression plots of iECs at 4 weeks indicated that GATA4 was enriched in the iEC-derived Zone 3 LSEC cluster (Fig. S12B), MAF was enriched in a separate iEC-derived Zone 2/3 LSEC cluster (Fig. S12C), and ZEB2 was enriched in iEC-derived stromal cell subpopulations (Fig. S12D). Collectively, these data indicate that GATA4 and MAF are important in the tissue specification of iECs into LSEC-like cells. MAF steadily increased over time and was highly upregulated at long-term time points, whereas GATA4 was highly upregulated during the early phase post transplantation (1 week). Furthermore, MAF and GATA4 appeared to be upregulated in different subpopulations of iEC-derived LSEC-like cells, as shown via scRNAseq. ZEB2 did not appear to be strongly linked to the LSEC specification of iECs, but was associated with the endothelial–mesenchymal transition of iECs, similar to ZEB1 (Fig. S9H).
Subsequently, new markers associated with LSEC-specification were deduced using two strategies. In the first method, bulk RNAseq analysis of DEGs between transplanted iECs at 1 and 12 weeks and between hLSEC FACS and in vitro iECs were listed (top-100 DEGs are listed in Table S5). The intersection between these two DEG comparisons was taken to be genes associated with iEC specification into an LSEC-like phenotype, yielding 213 genes (Table S6). The expression of each of these markers in human liver was assessed using the Human Protein Atlas (www.proteinatlas.org), which showed only a few genes labelled as hLSEC-specific markers: ACCS, TMEM121, COL18A1, CLEC14A, CD1D, GAS2L1, EMCN, and NUDT16. Each of these genes was then used to construct UMAP expression plots of the scRNAseq data (Fig. 7B). This narrowed the list down to EMCN and CLEC14A being the most robustly expressed in endothelial/LSEC subpopulations in both primary hLSEC and iEC 4-week samples. Additionally, the expression of the 213 genes associated with LSEC specification was assessed within all endothelial subpopulations in the scRNAseq data set, which indicated higher expression in all LSEC (Zones 1, 2/3, and 3) subpopulations compared with all subpopulations in the iEC 4-week samples, and the highest expression in the Zone 3 subpopulation within hLSECs. Within the iEC 4-week samples (integrated together), Zone 3 LSEC-like cells also had the highest average expression of LSEC-specification genes (Fig. 7C).
A second strategy was adopted using the Mogrify predictive computational framework to compare the transcriptome of human LSECs and generic ECs (comprising macrovascular artery/vein and microvascular ECs) based on data from the FANTOM5 project and included in the webtool provided with the original publication.29 Using this webtool, 27 TFs were predicted to drive the tissue specification of ECs to LSECs. STRING analysis indicated that NOTCH1, GATA4, and FOS were the central factors in the network (Fig. S3F). Gene ontology (GO) terms denoting biological processes significantly enriched in this network included: ‘animal organ development’, ‘embryonic organ development’, ‘developmental process’, ‘cell differentiation’, ‘regulation of hemopoiesis’, ‘foregut morphogenesis’, ‘liver development’, and ‘vasculogenesis’. The expression of the 27 genes within the bulk RNAseq samples was assessed using a heatmap and hierarchical clustering, which confirmed that iECs at 12 weeks were closest to hLSEC FACS in terms of expression of these LSEC-specification factors (Fig. 4D). Both hLSEC FACS and iECs at 12 weeks shared 13 significantly upregulated genes compared with iEC in vitro samples, and two different upregulated genes in each group (Fig. 4E). This analysis was extended to the scRNAseq data set, which confirmed earlier findings that Zone 3 LSECs in both FACS-isolated hLSEC samples and iEC 4-week samples showed the highest expression of late LSEC-specification markers. Within endothelial subpopulations in the FACS-isolated hLSEC sample, Zone 1 LSECs had the lowest expression of these markers, whereas, within the iEC 4-week sample, generic ECs had the lowest expression (Fig. 7D).
This combined analysis not only yielded novel marker and transcriptional regulators, but also suggests that Zone 3 is the region associated with the highest upregulation of LSEC specification in both FACS-isolated hLSECs and transplanted iECs.
Discussion
By profiling the characteristics of iECs transplanted into the liver, this study confirmed that, given the appropriate microenvironment, ECs undergo spatiotemporal specification into LSEC-like cells. Transplanted iECs engrafted and expanded throughout the native sinusoids, and repopulation of the liver vasculature peaked at 4 weeks, where 11% of the mouse liver vasculature contained tdTom+ cells, of which 74% of these cells were CD31+ and endothelial in nature. Transplanted iECs demonstrated long-term survival of up to 12 weeks, with greatest maturation into LSEC-like cells at this time point. However, decreasing engraftment by 12 weeks suggests that iECs remodel with the native vasculature and become replaced over time.
iECs have been previously transplanted into mouse liver with long-term engraftment (12 weeks), but these hiPSCs were transduced to express factor VIII and the phenotype of cells post transplantation was not analysed.30 As a form of bioengineered liver constructs, iECs have also been transplanted into rodents,31,32 but whether iECs differentiate into LSEC-like cells is unknown. In 2020, Gage et al. transplanted hESC-derived venous angioblasts into the liver of neonatal and adult mice with some technical differences to this study, such as the different source of cells and double the cell dosage. Regardless, similar findings to our study included the widespread engraftment of LSEC-like cells, long-term but diminished survival at 12 weeks, and human factor VIII secretion at comparable levels.9 However, Gage et al. found a diverse off-target population comprising haemopoietic derivatives (macrophages and T cells), stellate/fibroblasts, and rare cholangiocytes, whereas we only found pericyte/mesenchymal stromal cells. This disparity is likely the result of the multipotent nature of immature angioblasts and also the rapidly transitioning neonatal liver microenvironment used by Gage et al.
Our study strengthens recent evidence that PSCs can generate LSEC-like cells and emphasises the importance of the liver microenvironment. Using our iEC xenograft system, we analysed the transcriptomic progression of iECs into LSEC-like cells upon transplantation to gather insights into the process of LSEC specification. This is of particular interest because the developmental origin and differentiation of LSECs remain unclear, and our current understanding stems largely from rodent studies. Several recent studies have highlighted important pathways and TFs involved in LSEC differentiation, such as the nitric oxide/cyclic guanosine-3′,5′-monophosphate (NO/cGMP) pathway,33 TGFβ1 and Rho/ROCK inhibition,34 Notch signalling,35 and key TFs, including GATA4, LMO3, TCFEC, MAF,26,36,37 ERG, SPI1, IRF1/2, PATZ1, KL4,6 and MEIS2.6,37 Much of this is reflected in our analysis, but we also identified new markers and TFs associated with LSEC differentiation, such as CLEC14A (associated with periportal LSECs in the regenerating human liver)38 and FOS. Furthermore, a portion of the genes we have listed correlates with recent scRNAseq data of several developmental stages of mouse and human embryonic/foetal liver.39 Out of the genes we identified, six are associated with primitive LSECs in the early embryonic liver (Week 5–7 in humans, embryonic day 11–13 in mice), namely GUK1, ID3, NR2F2, PDLIM1, SOX4, and HMCN1, and 11 are associated with LSECs in early–mid foetal liver (Week 7-19 in humans, embryonic day 13–17.5 in mice), namely CLEC14A, EPAS1, FOS, IL33, NFKBIA, ALDH2, BMP2, ID2, IL6ST, MEIS2, and RASGRP2. Collectively, this suggests that iECs undergo LSEC specification in a manner that reflects, at least partially, native LSEC differentiation and that this hiPSC-based platform presents a credible model to study LSEC biology.
Metabolic zonation in the liver is the hierarchical organisation and function of cells depending on anatomical location. While this has been well established in hepatocytes, LSEC zonation is a recently described phenomenon.[22], [23], [24] Intrahepatic transplantation of mouse LSECs via the portal vein of recipient mice has resulted in engrafted LSECs localising in the periportal region,40 and hESC-venous angioblast grafts in neonatal mouse liver demonstrated zonation 77 days after transplantation.9 In the current study, transplanted iECs upregulated zonated LSEC genes over time, suggesting increased zonation over time as transplanted iECs expand from the portal region (Zone 1) into the perivenous region (Zone 3). Single cell analysis was especially informative in confirming zonation, demonstrating that all three zones (1, 2/3, and 3) were represented in both primary hLSEC and transplanted iEC samples. Further analysis of individual clusters using DEGs between Zone 1 and Zone 3 hLSECs validated this pattern in iEC samples. Notably, we demonstrated that clusters labelled ‘generic endothelial cells’ expressed many genes associated with Zone 1 LSECs, and likely represent iECs located in the periportal region (Zone 1) and lining the portal veins, which could be described as ‘early transitional LSEC-like cells’. As iECs stream out from the periportal region, subpopulations of Zone 2/3 and Zone 3 cells are evident, corroborating our bulk RNAseq findings. The zonation of transplanted iECs provides further evidence that the interaction of transplanted cells with their local microenvironment has a crucial influence on their profile.
Several important findings indicate that iECs underwent tissue specification into LSEC-like cells. These include structural incorporation into the native liver vasculature, significantly increased secretion of human factor VIII over time, and the expression of canonical LSEC markers and TFs associated with LSEC specification. However, our unprecedented depth of transcriptomic analysis also revealed differences between transplanted iECs and primary hLSECs. Comparison of the bulk transcriptome between transplanted iECs and FACS-isolated hLSECs demonstrated a lack of overlap on the MDS plot, and a high number of DEGs, although this decreased over time. ScRNAseq showed that transplanted iECs at 4 weeks clustered separately from hLSECs on the UMAP plot and, although iECs contained zonated subpopulations, the number of zone-specific markers expressed in each subpopulation was less than the number expressed in hLSECs. However, when integrated with other cell types from the human liver, scRNAseq showed that a subpopulation of transplanted iECs, designated as Zone2/3 LSEC-like cells, overlapped with Zone 3 LSECs in the human liver. This small subpopulation likely represents the cells that were found to have membrane fenestrations (a morphological hallmark of LSECs), indicating that this subpopulation approximates LSECs. Taken together, our data indicate that iECs progressively transition into LSEC-like cells over time and transplanted cells in the perivenous region are most similar to hLSECs, particularly Zone 3 hLSECs.
There also are several limitations to note. Engraftment and LSEC specification of iECs was only seen when liver injury was present, and how the inflammatory and regenerating liver milieu specifically affects the LSEC phenotype post transplantation is unclear. One approach to explore this could be to compare transplanted iECs to primary human LSECs isolated from injured and regenerating human livers (such as during cirrhosis or after portal vein embolisation), rather than steady-state LSECs as in this study. Another approach would be to compare engrafted iECs in animals with varying degrees of liver injury, to understand injury-dependant changes in phenotype. Alternatively, to confirm whether the LSEC specification process would have still occurred had liver injury not been present, this could be explored in an in vitro model where iECs can be co-cultured within liver organoids to assess the exact influence of the liver microenvironment on iEC development. A second limitation is that, even though LSEC specification was observed, this resulted in a relatively small subpopulation of LSEC-like cells. Additional time points for scRNAseq will delineate whether the LSEC-like subpopulation increases over time, and help answer the question of whether longitudinal increases in LSEC marker expression and human factor VIII levels were a reflection of increased maturity, increased number of mature cells, or both. Lastly, the transcriptomic data need to be interpreted with caveats, which include that liver specification in this model was a result of a complex mixture of cues, including those associated with LSEC development and regeneration, endothelial–mesenchymal transition, and post-transplantation stressors, such as ischaemia. Xenotransplantation also occurs in a model with a compromised immune system and, therefore, the data do not account for the role of immune cells as mediators of the liver microenvironment, which could influence LSEC specification.
Future directions include the manipulation of signalling pathways highlighted in this study to further enhance the tissue specification of iECs into LSECs, to develop in vitro culture conditions that minimise capillarisation, further interrogation of the ontogeny of LSECs, and examining the effect of conditions, such as ageing, disease, and drug toxicity, on LSEC-like cells, using a humanised xenograft system.
Financial support
The authors received funding from the Australian National Health & Medical Research Council (GNT2011220 & GNT1125233), Stafford Fox Medical Research Foundation, O’Brien Foundation, St Vincent’s Institute Foundation, L.E.W. Carty Foundation, St Vincent’s Hospital Melbourne Research Endowment Fund, Australian Catholic University, University of Melbourne Centre for Stem Cell Systems, Australian and New Zealand Hepatic, Pancreatic and Biliary Association, and the Victorian State Government’s Department of Business Innovation Operational Infrastructure Support Program.
Authors’ contributions
KKY and GMM initially conceptualised the study, and KKY wrote the initial draft of this manuscript. Experiments and analyses were completed by KKY, JS, YG, AD, AK, GPL, VCC, JMP, and GMM. AMF, BK, SWB, AGE, EGS, and GCY were involved in the provision of study materials, including cell lines, animal models, and patient samples. KKY, GCY, WAM, and GMM were involved in the acquisition of funding related to this project. KKY, WAM, and GMM were responsible for project administration. All authors were involved in the review and editing of the manuscript before submission.
Data availability statement
Sequencing data have been deposited in the GEO repository under the accession numbers GSE157763 (bulk RNAseq) and GSE157767 (scRNAseq). All other data will be made available upon reasonable request to the corresponding author.
Conflicts of interest
None to declare.
Please refer to the accompanying ICMJE disclosure forms for further details.
Acknowledgements
The authors thank Liliana Pepe, Anna Deftereos, Amanda Rixon, and the late Sue McKay for assistance with animal experiments, and Chandana Herath (and Yecuris Corporation) for providing FRG mice. The authors also thank Dijana Miljkovic and Anthony Di Carluccio for assistance with flow cytometry and cell sorting. Access to a confocal microscopy was provided by the University of Melbourne Biological Optical Microscopy Platform. The authors are very grateful to Christine Wells for advice on sequencing experiments and bioinformatic analysis, and access to the Stem Cells Framework Data Initiative consortium, which assisted in the generation of data used in this publication. The Initiative is supported by funding from Bioplatforms Australia through the Australian Government National Collaborative Research Infrastructure Strategy, and we thank Mabel Lum for coordinating logistics. We also thank staff of the Australian Genome Research Facility for assistance in sample processing.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jhepr.2024.101023.
Supplementary data
The following are the supplementary data to this article:
References
- 1.Meyer J., Gonelle-Gispert C., Morel P., et al. Methods for isolation and purification of murine liver sinusoidal endothelial cells: a systematic review. PLoS One. 2016;11(3) doi: 10.1371/journal.pone.0151945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Goldman O., Han S., Hamou W., et al. Endoderm generates endothelial cells during liver development. Stem Cel Rep. 2014;3(4):556–565. doi: 10.1016/j.stemcr.2014.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhang H., Pu W., Tian X., et al. Genetic lineage tracing identifies endocardial origin of liver vasculature. Nat Genet. 2016;48(5):537–543. doi: 10.1038/ng.3536. [DOI] [PubMed] [Google Scholar]
- 4.Xie G., Choi S.S., Syn W.K., et al. Hedgehog signalling regulates liver sinusoidal endothelial cell capillarisation. Gut. 2013;62(2):299–309. doi: 10.1136/gutjnl-2011-301494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Koui Y., Kido T., Ito T., et al. An in vitro human liver model by iPSC-derived parenchymal and non-parenchymal cells. Stem Cel Rep. 2017;9(2):490–498. doi: 10.1016/j.stemcr.2017.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Danoy M., Poulain S., Koui Y., et al. Transcriptome profiling of hiPSC-derived LSECs with nanoCAGE. Mol Omics. 2020;16(2):138–146. doi: 10.1039/c9mo00135b. [DOI] [PubMed] [Google Scholar]
- 7.Arai T., Sakurai T., Kamiyoshi A., et al. Induction of LYVE-1/stabilin-2-positive liver sinusoidal endothelial-like cells from embryoid bodies by modulation of adrenomedullin-RAMP2 signaling. Peptides. 2011;32(9):1855–1865. doi: 10.1016/j.peptides.2011.07.005. [DOI] [PubMed] [Google Scholar]
- 8.De Smedt J., van Os E.A., Talon I., et al. PU.1 drives specification of pluripotent stem cell-derived endothelial cells to LSEC-like cells. Cell Death Dis. 2021;12(1):84. doi: 10.1038/s41419-020-03356-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gage B.K., Liu J.C., Innes B.T., et al. Generation of functional liver sinusoidal endothelial cells from human pluripotent stem-cell-derived venous angioblasts. Cell Stem Cell. 2020;27(2):254–269. doi: 10.1016/j.stem.2020.06.007. [DOI] [PubMed] [Google Scholar]
- 10.Gage B.K., Merlin S., Olgasi C., et al. Therapeutic correction of hemophilia A by transplantation of hPSC-derived liver sinusoidal endothelial cell progenitors. Cell Rep. 2022;39(1) doi: 10.1016/j.celrep.2022.110621. [DOI] [PubMed] [Google Scholar]
- 11.March S., Hui E.E., Underhill G.H., et al. Microenvironmental regulation of the sinusoidal endothelial cell phenotype in vitro. Hepatology. 2009;50(3):920–928. doi: 10.1002/hep.23085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.DeLeve L.D., Wang X., Hu L., et al. Rat liver sinusoidal endothelial cell phenotype is maintained by paracrine and autocrine regulation. Am J Physiol Gastrointest Liver Physiol. 2004;287(4):G757–G763. doi: 10.1152/ajpgi.00017.2004. [DOI] [PubMed] [Google Scholar]
- 13.Tripathi A., Debelius J., Brenner D.A., et al. The gut-liver axis and the intersection with the microbiome. Nat Rev Gastroenterol Hepatol. 2018;15(7):397–411. doi: 10.1038/s41575-018-0011-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Azuma H., Paulk N., Ranade A., et al. Robust expansion of human hepatocytes in Fah-/-/Rag2-/-/Il2rg-/- mice. Nat Biotechnol. 2007;25(8):903–910. doi: 10.1038/nbt1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dingle A.M., Yap K.K., Gerrand Y.W., et al. Characterization of isolated liver sinusoidal endothelial cells for liver bioengineering. Angiogenesis. 2018;21(3):581–597. doi: 10.1007/s10456-018-9610-0. [DOI] [PubMed] [Google Scholar]
- 16.Yap K.K., Gerrand Y.W., Dingle A.M., et al. Liver sinusoidal endothelial cells promote the differentiation and survival of mouse vascularised hepatobiliary organoids. Biomaterials. 2020;251 doi: 10.1016/j.biomaterials.2020.120091. [DOI] [PubMed] [Google Scholar]
- 17.Kao T., Labonne T., Niclis J.C., et al. GAPTrap: a simple expression system for pluripotent stem cells and their derivatives. Stem Cel Rep. 2016;7(3):518–526. doi: 10.1016/j.stemcr.2016.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Patsch C., Challet-Meylan L., Thoma E.C., et al. Generation of vascular endothelial and smooth muscle cells from human pluripotent stem cells. Nat Cel Biol. 2015;17(8):994–1003. doi: 10.1038/ncb3205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kong A.M., Yap K.K., Lim S.Y., et al. Bio-engineering a tissue flap utilizing a porous scaffold incorporating a human induced pluripotent stem cell derived endothelial cell capillary network connected to a vascular pedicle. Acta Biomater. 2019;94:281–294. doi: 10.1016/j.actbio.2019.05.067. [DOI] [PubMed] [Google Scholar]
- 20.Hunt N.J., Lockwood G.P., Warren A., et al. Manipulating fenestrations in young and old liver sinusoidal endothelial cells. Am J Physiol Gastrointest Liver Physiol. 2019;316(1):G144–G154. doi: 10.1152/ajpgi.00179.2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shahani T., Covens K., Lavend'homme R., et al. Human liver sinusoidal endothelial cells but not hepatocytes contain factor VIII. J Thromb Haemost. 2014;12(1):36–42. doi: 10.1111/jth.12412. [DOI] [PubMed] [Google Scholar]
- 22.Halpern K.B., Shenhav R., Massalha H., et al. Paired-cell sequencing enables spatial gene expression mapping of liver endothelial cells. Nat Biotechnol. 2018;36(10):962–970. doi: 10.1038/nbt.4231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.MacParland S.A., Liu J.C., Ma X.Z., et al. Single cell RNA sequencing of human liver reveals distinct intrahepatic macrophage populations. Nat Commun. 2018;9(1):4383. doi: 10.1038/s41467-018-06318-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Aizarani N., Saviano A., Sagar, et al. A human liver cell atlas reveals heterogeneity and epithelial progenitors. Nature. 2019;572(7768):199–204. doi: 10.1038/s41586-019-1373-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ben-Moshe S., Shapira Y., Moor A.E., et al. Spatial sorting enables comprehensive characterization of liver zonation. Nat Metab. 2019;1(9):899–911. doi: 10.1038/s42255-019-0109-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Geraud C., Koch P.S., Zierow J., et al. GATA4-dependent organ-specific endothelial differentiation controls liver development and embryonic hematopoiesis. J Clin Invest. 2017;127(3):1099–1114. doi: 10.1172/JCI90086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gómez-Salinero J.M., Izzo F., Lin Y., et al. Specification of fetal liver endothelial progenitors to functional zonated adult sinusoids requires c-Maf induction. Cell Stem Cell. 2022;29(4):593–609. doi: 10.1016/j.stem.2022.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.de Haan W., Dheedene W., Apelt K., et al. Endothelial Zeb2 preserves the hepatic angioarchitecture and protects against liver fibrosis. Cardiovasc Res. 2022;118(5):1262–1275. doi: 10.1093/cvr/cvab148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rackham O.J.L., Firas J., Fang H., et al. A predictive computational framework for direct reprogramming between human cell types. Nat Genet. 2016;48(3):331–335. doi: 10.1038/ng.3487. [DOI] [PubMed] [Google Scholar]
- 30.Olgasi C., Talmon M., Merlin S., et al. Patient-specific iPSC-derived endothelial cells provide long-term phenotypic correction of hemophilia A. Stem Cel Rep. 2018;11(6):1391–1406. doi: 10.1016/j.stemcr.2018.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Takeishi K., Collin de l'Hortet A., Wang Y., et al. Assembly and function of a bioengineered human liver for transplantation generated solely from induced pluripotent stem cells. Cel Rep. 2020;31(9) doi: 10.1016/j.celrep.2020.107711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Takebe T., Sekine K., Kimura M., et al. Massive and reproducible production of liver buds entirely from human pluripotent stem cells. Cel Rep. 2017;21(10):2661–2670. doi: 10.1016/j.celrep.2017.11.005. [DOI] [PubMed] [Google Scholar]
- 33.Xie G., Wang X., Wang L., et al. Role of differentiation of liver sinusoidal endothelial cells in progression and regression of hepatic fibrosis in rats. Gastroenterology. 2012;142(4):918–927. doi: 10.1053/j.gastro.2011.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Venkatraman L., Tucker-Kellogg L. The CD47-binding peptide of thrombospondin-1 induces defenestration of liver sinusoidal endothelial cells. Liver Int. 2013;33(9):1386–1397. doi: 10.1111/liv.12231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cuervo H., Nielsen C.M., Simonetto D.A., et al. Endothelial notch signaling is essential to prevent hepatic vascular malformations in mice. Hepatology. 2016;64(4):1302–1316. doi: 10.1002/hep.28713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Geraud C., Schledzewski K., Demory A., et al. Liver sinusoidal endothelium: a microenvironment-dependent differentiation program in rat including the novel junctional protein liver endothelial differentiation-associated protein-1. Hepatology. 2010;52(1):313–326. doi: 10.1002/hep.23618. [DOI] [PubMed] [Google Scholar]
- 37.de Haan W., Oie C., Benkheil M., et al. Unraveling the transcriptional determinants of liver sinusoidal endothelial cell specialization. Am J Physiol Gastrointest Liver Physiol. 2020;318(4):G803–G815. doi: 10.1152/ajpgi.00215.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Brazovskaja A., Gomes T., Körner C., et al. Cell atlas of the regenerating human liver after portal vein embolization. bioRxiv. 2021 doi: 10.1101/2021.06.03.444016. [DOI] [Google Scholar]
- 39.Wang X., Yang L., Wang Y.C., et al. Comparative analysis of cell lineage differentiation during hepatogenesis in humans and mice at the single-cell transcriptome level. Cell Res. 2020;30(12):1109–1126. doi: 10.1038/s41422-020-0378-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yadav N., Jaber F.L., Sharma Y., et al. Efficient reconstitution of hepatic microvasculature by endothelin receptor antagonism in liver sinusoidal endothelial cells. Hum Gene Ther. 2019;30(3):365–377. doi: 10.1089/hum.2018.166. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Sequencing data have been deposited in the GEO repository under the accession numbers GSE157763 (bulk RNAseq) and GSE157767 (scRNAseq). All other data will be made available upon reasonable request to the corresponding author.