Abstract
As an output effector of the Hippo signaling pathway, the TEAD transcription factor and co-activator YAP play crucial functions in promoting cell proliferation and organ size. The tumor suppressor NF2 has been shown to activate LATS1/2 kinases and interplay with the Hippo pathway to suppress the YAP-TEAD complex. However, whether and how NF2 could directly regulate TEAD remains unknown. We identified a direct link and physical interaction between NF2 and TEAD4. NF2 interacted with TEAD4 through its FERM domain and C-terminal tail and decreased the protein stability of TEAD4 independently of LATS1/2 and YAP. Furthermore, NF2 inhibited TEAD4 palmitoylation and induced the cytoplasmic translocation of TEAD4, resulting in ubiquitination and dysfunction of TEAD4. Moreover, the interaction with TEAD4 is required for NF2 function to suppress cell proliferation. These findings reveal an unanticipated role of NF2 as a binding partner and inhibitor of the transcription factor TEAD, shedding light on an alternative mechanism of how NF2 functions as a tumor suppressor through the Hippo signaling cascade.
Keywords: Hippo pathway, NF2, TEAD4, protein‒protein interaction, palmitoylation, tumor suppressor
In multicellular animals, cell proliferation and death must be precisely coordinated to ensure proper organ size and tissue homeostasis. The Hippo signaling pathway was initially identified as a key determinant of organ size (1, 2, 3, 4). This pathway is highly conserved from Drosophila to mammals (5, 6). The Hippo pathway constitutes a major kinase cascade, including the mammalian STE20-like protein kinase 1/2 (MST1/2) and large tumor suppressor kinase 1/2 (LATS1/2), which inhibit two transcriptional co-activators, Yes-associated protein (YAP) and transcriptional co-activator with PDZ-binding motif (TAZ), via phosphorylation (7). Dephosphorylated and activated YAP/TAZ translocate into the nucleus, where they interact with TEA domain transcription factors (TEADs) and induce the expression of target genes, such as CTGF and CYR61, to modulate cell proliferation, differentiation, and tumorigenesis (8, 9, 10). Unlike Drosophila, which expresses only one TEAD homolog, Scalloped (Sd), there are four TEAD homologs in mammals (TEAD1, TEAD2, TEAD3, and TEAD4). TEADs share a similar domain structure: a DNA-binding domain (DBD) at the N-terminus and a YAP-binding domain (YBD) at the C-terminus (9, 11). Dysregulation of the Hippo pathway has been linked to many human diseases, and targeted inhibition of the YAP-TEAD transcriptional complex for cancer therapy is being actively explored (5, 12, 13).
In addition to YAP/TAZ, the transcriptional activity of TEADs is regulated by different binding factors, including VGLL4, the glucocorticoid receptor (GR), TCF4, and AP-1 (14, 15, 16, 17). Specifically, VGLL4 directly competes with YAP/TAZ for binding to TEADs, thereby suppressing their transcriptional activity (18). p38 binding-dependent cytoplasmic translocation of TEADs provides spatial modulation of transcriptional activity (19). Post-translational modifications of TEADs, such as phosphorylation and palmitoylation, govern their protein stability and activity (20, 21, 22, 23). Four TEAD homologs are palmitoylated in mammalian cells (24, 25), and palmitoylation of TEAD is critical for protein stability and the YAP-TEAD interaction (Noland et al. 2016; Chan et al. 2016). Although targeting TEAD palmitoylation is considered a potential strategy for Hippo pathway molecular therapy (26, 27), the mechanisms regulating TEAD palmitoylation and depalmitoylation remain unclear.
Neurofibromin 2 (NF2), also called Merlin, is an Ezrin, Radixin, and Moesin (ERM) family protein that acts as a tumor suppressor, and the development of human cancer, schwannoma, meningioma, ependymoma, and malignant mesothelioma is strongly associated with the loss-of-function mutation of NF2 (28, 29, 30). NF2 functions in the Hippo pathway by responding to extracellular stimuli, such as cell density and osmotic stress (31, 32). NF2 associates with LATS1/2, thus activating the major kinase cascade of the Hippo pathway to inhibit YAP/TAZ and suppress cell proliferation and tumorigenesis (33). Several binding partners of NF2, including angiomotin (AMOT) and the E3 ubiquitin ligase CRL4DCAF1, are also involved in modulating the Hippo pathway (34, 35). However, whether NF2 directly regulates TEADs remains unclear and how NF2 modulates the Hippo pathway is not yet fully understood.
In this study, we identified the physical interaction between the tumor suppressor NF2 and transcription factor TEAD4. We found that NF2 directly interacted with TEAD4 through its FERM domain and the C-terminal tail, and weakened the protein stability of TEAD4 independently of LATS1/2 and YAP. We further revealed a molecular mechanism that NF2 inhibited TEAD4 palmitoylation and retained its cytoplasmic translocation via direct interaction, resulting in ubiquitination and dysfunction of TEAD4. Moreover, the TEAD4 interaction was required for NF2-mediated suppression of tumor cell proliferation. These findings suggest a new role for NF2 as a binding partner and inhibitor of TEADs and expand the molecular mechanism of how NF2 functions as a tumor suppressor.
Results
NF2 decreases the protein level of TEAD4 independently of LATS1/2 and YAP
As an upstream activator of the Hippo signaling pathway, the tumor suppressor NF2 has been shown to activate LATS1/2 kinases and suppress YAP function (33). NF2 also interacts with other regulators, AMOT, DDB1, and CUL4-associated factor homolog 1 (DCAF1), to modulate Hippo signaling (34, 35). We were curious whether other effectors might be directly involved in NF2 function. We first examined the protein levels of major Hippo pathway components in HEK293T cells overexpressing NF2. Consistent with a previous report (33), NF2 promoted YAP phosphorylation (Fig. 1A). Unexpectedly, the protein levels of TEAD4 were markedly reduced along with NF2 overexpression (Fig. 1A). We then confirmed this result by RNAi silencing and rescue assays. Depletion of NF2 by small-interfering RNA (siRNA) induced an increase in both TEAD2 and TEAD4 protein levels, and re-expression of NF2 decreased their protein levels again (Fig. S1A), indicating that TEAD2/4 protein levels are sensitive to the presence of NF2. Moreover, the mRNA levels of TEADs were unaffected by NF2 overexpression (Fig. 1B). These results suggest that NF2 may regulate the protein levels of TEADs without changes at their transcriptional levels.
Figure 1.
NF2 decreases the protein levels of TEADs independently of LATS1/2 and YAP.A, the protein levels of HA-NF2 (IB: HA), YAP, TAZ, p-YAP, TEAD4, and GAPDH were determined via western blotting in HEK293T cells overexpressing HA-NF2. B, RT‒PCR analysis of TEAD1/2/3/4, NF2, and YAP mRNA levels was performed in HeLa cells overexpressing HA-NF2. ns: not significant (one-way ANOVA). C, MCF-10A cells were transfected with control siRNA or NF2 siRNA-1. Then, 100 μg/ml cycloheximide (CHX) was added, and the cells were harvested at the indicated time points. The protein levels of endogenous TEAD4, YAP, p-YAP, LATS-1/2 and tubulin were determined via western blotting. D, Wild-type (WT) and YAP knockout (KO) HeLa cells were transfected with or without NF2 siRNA-1. The protein levels of NF2, TEAD4, YAP, Cyr61, and GAPDH were determined via western blotting. E, WT and LATS1/2 KO HeLa cells were transfected with the control siRNA or NF2 siRNA-1/2. The protein levels of NF2, TEAD4, LATS1, YAP, p-YAP, Cyr61 and GAPDH were determined via western blotting.
We then examined the protein stability of TEAD4 in cells treated with cycloheximide (CHX) to inhibit de novo protein synthesis. Compared with that in control cells, the half-life of the TEAD4 protein was significantly extended in cells with NF2 knockdown (Figs. 1C and S1B). However, the half-lives of other components, LATS1 and LATS2, were unaffected by NF2 expression or knockdown. Thus, NF2 decreases TEAD4 protein level by altering the posttranslational stability.
Since YAP is a well-known binding factor to TEAD4, we then asked whether YAP is involved in the regulation of TEAD4 protein stability by NF2. We generated YAP knockout (KO) HeLa cells by CRISPR/Cas9, in which successful YAP KO was verified by western blotting (Fig. 1D). The knockdown of NF2 still increased TEAD4 protein levels in both WT and YAP KO cells (Fig. 1D), and NF2 overexpression decreased TEAD4 protein levels in YAP KO cells (Fig. S1C), indicating that NF2 decreased the protein levels of TEAD4 independently of YAP. We also wondered whether LATS1/2 might be involved in the regulation of TEAD4 protein level by NF2. Again, in both WT and LATS1/2 KO cells, knockdown of NF2 increased TEAD4 protein expression levels (Fig. 1E). Additionally, the expression of Cyr61, a classic transcription target of TEADs, increased with NF2 knockdown (Fig. 1E). Therefore, our results suggest a direct regulation that NF2 directly regulates TEAD4 protein level and stability, irrespective of the presence or absence of LATS1/2 and YAP.
NF2 physically interacts with TEAD4 through its FERM domain and C-terminal tail
To further explore the direct connection between NF2 and TEAD4, we purified the YBD domain of TEAD4 (TEAD4-YBD) with a SUMO-His tag and full-length NF2 with a GST tag from Escherichia coli and examined their interaction. We found that TEAD4-YBD bound to GST-NF2 in vitro using a GST pull-down assay (Fig. 2A), suggesting a direct interaction and physical interaction between NF2 and TEAD4.
Figure 2.
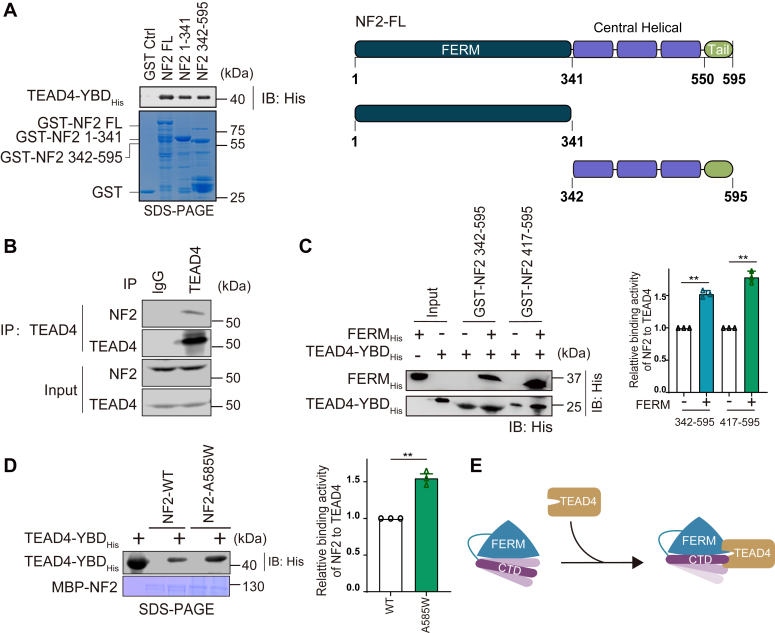
NF2 interacts with TEAD4 through the FERM domain and C-terminal tail.A, the GST pull-down assay was performed to assess the interaction between SUMO-tagged TEAD4-YBD and GST-tagged NF2 truncations. TEAD4 was indicated by western blotting, and schematic views of NF2 organization are shown in the right panel. B, coimmunoprecipitation of endogenous NF2 with TEAD4 was performed in HEK293T cells. The cell lysates were incubated with Protein-A Magnetic beads and TEAD4 antibodies or IgG. Immunoprecipitated samples were determined by western blotting. C, in vitro binding assay of GST-tagged NF2 C-terminal fragments with His-TEAD4-YBD. The GST-NF2 C-terminal fragments were pre-bound to GST beads, and beads were further incubated with TEAD4-YBD alone or the mixture of TEAD-YBD (His-tag) and FERM domain (His-tag). The FERM and TEAD4 on the beads were analyzed by western blotting. Quantitative analysis of the relative protein binding activity of NF2 to TEAD4 is shown in the right panel. Mean ± sd. N = 3, ∗∗p < 0.01 (unpaired two-tailed t test with Welch’s correction). D, in vitro pull-down assay of MBP-NF2 WT and the A585W mutant with His-TEAD4-YBD to assess the interaction between TEAD4 and NF2. The level of TEAD4 was determined by western blotting. Quantitative analysis of the relative protein binding activity of NF2 to TEAD4 is shown in the right panel. Mean ± sd. N = 3, ∗∗p < 0.01 (unpaired two-tailed t test with Welch’s correction). E, a cartoon model of NF2 binding to TEAD4 through the FERM domain and C-terminal tail.
According to the domain organization of the NF2 protein, which contains the N-terminal FERM domain, central helical domain and C-terminal tail (35), seven truncations of NF2 were constructed to determine minimal region involved in the TEAD4-YBD interaction (Figs. 2A and S2A). As shown by the results of the pull-down assay, both the N-terminal FERM domain (1–341 aa) and the C-terminal half (342–595 aa) remained to interact with TEAD4-YBD at a comparable level (Fig. 2A). Further, C-terminal tail (550–595 aa) was essential for the interaction instead of helical region (342–550 aa) of the C-terminal half (Fig. S2A). Additionally, the interaction between full-length NF2 and TEAD4 was verified by endogenous coimmunoprecipitation in HEK293T cells. Moreover, the interaction between endogenous NF2 and TEAD4 in vivo was also observed (Fig. 2B).
The intramolecular interaction of NF2 has been suggested to be maintained by the FERM domain and C-terminal tail (35, 36, 37). We then wondered whether the intramolecular interaction of NF2 affects its interaction with TEAD4. Compared to C-terminal fragments alone, co-incubation with the FERM domain markedly enhanced their binding to TEAD4 (Fig. 2C), indicating that the interaction between FERM and the C-terminal tail of NF2 promotes the interaction with TEAD4. Moreover, the A585W mutation of NF2, which can stabilize the intramolecular interaction and inactivate the LATS1/2 interaction (35), exhibited stronger binding activity to TEAD4 than NF2-WT (Fig. 2D). Taken together, these binding data demonstrate that NF2 directly interacts with TEAD4 through both the FERM domain and the C-terminal tail (Fig. 2E).
Characterization of the interaction between NF2 and TEAD4
We next investigated the interaction details between NF2 and TEAD4. Based on the crystal structure of human NF2 protein (35), single or combined point mutations on the structural surface of NF2 were designed for the binding screen (Fig. S2B). Overall, the binding results revealed that the L297, I301, and H304 residues in the FERM domain F3 lobe and the L582 and F591 residues in the C-terminal tail of NF2 were required for the interaction with TEAD4 (Figs. 3, A and B and S2C). We then generated two grouped mutants, NF2-5A (L297A/I301A/H304A/L582A/F591A) and NF2-4A-del (L297A/I301A/H304A/L582A and deletion of 590–595 aa) (Fig. S2, C and D), and found that both mutants strongly diminished their ability to interact with TEAD4 in a coimmunoprecipitation assay (Fig. 3C).
Figure 3.
NF2 decreases TEAD4 protein levels via direct interaction.A, GST pull-down assay of GST-NF2-FERM WT and mutants with His-TEAD4-YBD was performed to assess the interaction between TEAD4 and NF2-FERM. The level of TEAD4 was determined by western blotting. B, GST pull-down assay of GST-NF2-507-595 WT and mutants with His-TEAD4-YBD was performed to assess the interaction between TEAD4 and NF2 C-terminal tail. The level of TEAD4 was determined by western blotting. C, HEK293T cells were transfected with Myc-tagged NF2 WT/mutants and Flag-tagged TEAD4. Cell lysates were incubated with anti-Flag affinity beads, Protein-A magnetic beads and Myc antibodies. Myc-NF2 (IB: Myc) and Flag-TEAD4 (IB: Flag) on the beads was determined by western blotting. D, TEAD4 protein levels in NF2 KO HEK293A cells stably expressing GFP-NF2 WT or mutants. Protein expressions were determined via western blotting. Quantitative analysis of the relative protein levels of TEAD4 are shown in the right panel. Mean ± sd. N = 3, ∗∗∗∗p < 0.0001 (one-way ANOVA). E, competitive binding assay was performed to evaluate the binding effect of the YAP-TEAD4 complex with increasing doses of NF2. A schematic diagram of the competition assay is shown in the left panel. TEAD4 were pre-incubated with GST-YAP, after which NF2 was added as the competitor. The protein concentration on the beads was determined via SDS‒PAGE. ∗ Degradation.
To explore whether the binding-deficient mutant NF2-5A affects the role of NF2 in decreasing TEAD4 protein level, we subsequently generated NF2 KO stably expressing NF2 WT/mutants HEK293A cells and examined TEAD4 protein levels (Fig. S3A). As the results showed, both NF2-WT and NF2-A585W decreased TEAD4 protein levels compared to control cells, but NF2-5A did not (Fig. 3D). Further, the mRNA levels of TEADs were not significantly altered with NF2 depletion (Fig. S3B). Thus, NF2 decreased TEAD4 protein level via direct interaction.
Since both YAP and NF2 bind to the YBD domain of TEAD4, we next investigated whether NF2 and YAP bind to a similar surface on TEAD4. An in vitro competitive assay was performed and showed that NF2 gradually competed off TEAD4 from GST-YAP in a dose-dependent manner (Fig. 3E). Additionally, we screened the surface sites on TEAD4-YBD based on in vitro binding assay, and the results showed that the mutation of Y429 to histidine (Y429H), which lost the ability for YAP binding, impeded the interaction with NF2 (Fig. S2E). These results indicate that NF2 and YAP may occupy the overlapping interface on TEAD4-YBD and that NF2 inhibits the formation of the YAP-TEAD complex.
NF2 induces the cytoplasmic retention of TEAD4 via interaction
TEAD4 functions as a transcription factor in the nucleus, while NF2 is a plasma membrane-associated protein (33), raising the interesting points of how and where these two proteins interact in cellular environments. To determine the spatial localization of the NF2-TEAD4 complex within cells, we applied the bimolecular fluorescence complementation (BiFC) assay, in which two non-fluorescent half fragments of yellow fluorescent protein (n/cYFP) were fused with their respective binding partners. We observed no fluorescence upon co-expressing NF2-nYFP and NF2-cYFP in HEK293T cells, similar to control cells expressing NF2-cYFP or TEAD4-cYAP alone (Fig. 4A). Co-expression of well-known binding partners, NF2-cYFP and LATS2-nYFP, resulted in fluorescence at the plasma membrane (Fig. 4A). Co-expression of NF2-cYFP and TEAD4-nYFP allowed the observation of YFP signals at the plasma membrane, but more fluorescence signals were observed in the cytoplasm (Fig. 4A), suggesting that they form complexes in the cytoplasm rather than in the nucleus. Fluorescence signals were sequentially quantified by flow cytometry (Figs. 4B and S3C), confirming the strong interaction between NF2 and TEAD4 in cells, similar to the interaction between NF2 and LATS2. These findings indicate that the spatial organization of the NF2-TEAD4 complex is dominated by the extranuclear localization of NF2.
Figure 4.
NF2 induces the cytoplasmic retention of TEAD4 via interaction.A, BiFC assays were performed to determine the location of the NF2-TEAD4 complex. HEK293T cells were transfected with TEAD4-nYFP, NF2-cYFP, NF2-cYFP and TEAD4-nYFP, NF2-cYFP and LATS1 -nYFP, and NF2-cYFP and NF2-nYFP. The fluorescence signals of intact YFP were captured. Representative images are shown here. Scale bar = 10 μm. B, quantification of flow cytometry data from the BiFC assay (A). Mean ± sd. ∗∗∗∗p < 0.0001 (one-way ANOVA). C, immunofluorescence staining for endogenous TEAD4 was performed in NF2 KO HEK293A cells stably expressing WT or mutants GFP-NF2 WT and mutants at a high cell density. Scale bar = 10 μm. The co-localization analysis of NF2 and TEAD4 were shown in the right panel, and the fluorescence intensity and distance were measured using ImageJ.
A high cell density contributes to the cytoplasmic translocation of TEADs. To explore whether NF2 mediates the cytoplasmic retention of TEAD at high cell density, the subcellular localization of TEAD4 was examined. A high cell density indeed induced cytoplasmic localization in WT HEK293A cells but did not induce cytoplasmic localization in NF2 KO HEK293A cells (Fig. S3D), suggesting that NF2 is critical for TEAD4 subcellular localization. Further, we examined TEAD4 subcellular localization in NF2 KO stably expressing NF2-WT or mutants HEK293A cells. In contrast to the nuclear localization of TEAD4 in NF2 KO cells, a clear fluorescence signal of TEAD4 was observed in the cytoplasm and at the cell surface in NF2-WT and A585W expressing cells (Fig. 4C). Moreover, consistent with the above immunoblotting results, the overall expression level of TEAD4 generally decreased upon NF2-WT and A585W expression (Fig. 3D). Conversely, the fluorescence signal of TEAD4 was not detected in the cytoplasm of NF2-5A expressing cells (Fig. 4C), suggesting that NF2 induces the cytoplasmic retention of TEAD4 through direct interaction.
NF2 inhibits TEAD4 palmitoylation and presumably causes sequential ubiquitination
Palmitoylation of TEAD4 is required for its protein stability (20, 23, 24), prompting us to investigate whether NF2 decreases the protein stability of TEAD4 through palmitoylation. We performed in vitro auto-palmitoylation assays using click chemistry-based methods (38) (Fig. 5A). We observed that TEAD4 auto-palmitoylation significantly decreased during NF2 incubation but not during YAP incubation (Fig. 5, B and C), indicating that NF2 directly inhibited the auto-palmitoylation of TEAD4 in vitro.
Figure 5.
NF2 inhibits TEAD4 palmitoylation via direct interaction.A, schematic model of the in vitro auto-palmitoylation assay of recombinant TEAD4. B, TEAD4 was pre-incubated with NF2 or YAP. Then, in vitro auto-palmitoylation assay was performed to evaluate the palmitoylation of TEAD4 via streptavidin-mediated immunoblotting. C, quantitative analysis of the palmitoylated TEAD4 (Pal-TEAD4) panel in (B). D, schematic model of the in vivo palmitoylation assay with acyl resin-assisted capture methods in cells. E, in vivo palmitoylation assay to evaluate the palmitoylation levels of endogenous TEAD4 in NF2 KO HEK293A cells stably expressing the NF2 WT and mutants. The concentration of input TEAD4 was adjusted to a constant level for the palmitoylation capture assay, and the level of TEAD4 was determined via western blotting. The quantitative analysis of palmitoylated TEAD4 (Pal-TEAD4) is shown in the lower panel. Mean ± sd. N = 3, ∗∗p < 0.01, ∗∗∗∗p < 0.0001 (one-way ANOVA). F, in vitro ubiquitination assay was performed with purified recombinant E1, UbcH5b (the E2 ubiquitin-conjugating enzyme), and the E3 ligase CHIP to detect the ubiquitination levels of TEAD4 WT and 2CS. G, in vitro pull-down assay of MBP-NF2 with TEAD4 WT and 2CS. The two lanes were spliced from the same blot/gel, and the border was labeled with black dashed line.
Next, we examined the palmitoylation of TEAD4 in NF2 KO 293A cells using an acyl resin-assisted capture strategy (39) (Fig. 5D). Depletion of NF2 promoted the palmitoylation of TEAD4 compared to that in control cells (Fig. S4A), which is consistent with the findings of previous report (24). As expected, the stable expression of NF2-WT dramatically reduced TEAD4 palmitoylation, while the NF2-5A mutant had slight effects on TEAD4 palmitoylation in cells (Fig. 5E). In particular, NF2-A585W inhibited the palmitoylation of TEAD4 at a comparable level to NF2-WT, implying that the inhibitory effect of NF2 on TEAD4 palmitoylation was protein interaction dependent. Acyl-protein thioesterase 2 (APT2) and Alpha/beta hydrolase domain-containing protein 17A (ABHD17A) are proposed to be major depalmitoylases of TEAD family proteins (24). However, we found that the protein levels of APT2 and ABHD17A were not affected by the expression of NF2 (Fig. S4B), which excluded the possibility that NF2 reduced TEAD4 palmitoylation through APT2 and ABHD17A.
Previous studies have shown that depalmitoylation triggers the degradation of TEAD protein mediated by E3 ligase CHIP (24). In vitro ubiquitination assays confirmed that non-palmitoylated mutant TEAD4-2CS (C335S/C367S) exhibited markedly higher ubiquitination than TEAD4-WT (Fig. 5F). As positively relevant, TEAD4-2CS also exhibited stronger binding to NF2 than TEAD4-WT (Fig. 5G), indicating that NF2 preferentially binds to non-palmitoylated form of TEAD4 and triggers its ubiquitination. Thus, NF2 inhibits TEAD4 palmitoylation and presumably causes the sequential ubiquitination of TEAD4.
TEAD4 interaction is required for NF2-mediated suppression of cell proliferation
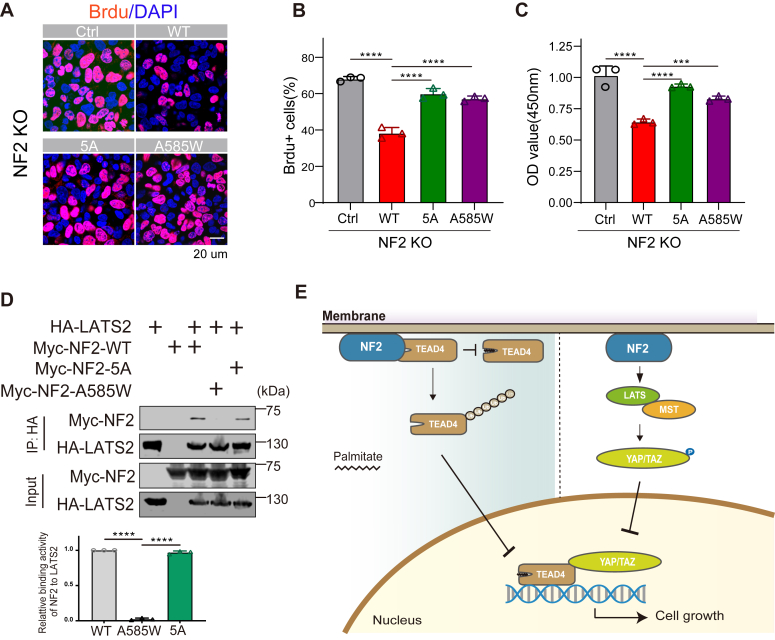
We then explored whether the direct interaction between NF2 and TEAD4 contributes to the tumor suppressor function of NF2. The BrdU incorporation assay was performed in NF2 KO HeLa cells to examine cell proliferation rates. In comparison with WT cells, BrdU incorporation efficiency was dramatically increased in NF2 KO cells, and the CCK-8 assay also showed increased cell viability after NF2 depletion (Fig. S5, A and B), confirming the inhibitory effect of NF2 on cell growth. As expected, the expression of NF2-WT significantly inhibited the incorporation of BrdU, while the expression of NF2-5A did not (Fig. 6, A and B). In addition, cells expressing NF2-5A exhibited higher cell viability than cells expressing NF2-WT (Fig. 6C). Further, the transcription of Cyr61 and CTGF, the downstream targets of TEAD4, was not effectively inhibited in cells expressing NF2-5A compared to those expressing NF2-WT, which was consistent with the observation in cell proliferation (Fig. S5, C and D). Therefore, the interaction with TEAD4 is required for NF2 to inhibit cell proliferation and plays a role in Hippo signaling.
Figure 6.
TEAD4 interaction is required for NF2 function to suppress tumor cell proliferation.A, NF2 KO HeLa or stably expressing NF2-WT/5A/A585W cells were subjected to BrdU incorporation assay. The fluorescence signals of BrdU and DAPI were detected. Representative images are shown here. Scale bar = 20 μm. B, the percentage of BrdU-positive cells in (A) was quantified. Mean ± sd. N = 3, ∗∗∗∗p < 0.0001 (one-way ANOVA). C, NF2 KO HeLa or stably expressing NF2-WT/5A/A585W cells were subjected to cell counting kit-8 (CCK-8), and the optical density at 450 nm was quantified. Mean ± sd. N = 3, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001 (one-way ANOVA). D, coimmunoprecipitation of Myc-tagged NF2 WT and mutants with HA-tagged LATS2 was performed in HEK293T cells. Cell lysates were incubated with Protein-A Magnetic beads and HA antibodies. Myc-NF2 (IB: Myc) and HA-LATS2 (IB: HA) expression on the beads was determined via western blotting. The quantitative analysis of the binding of NF2 to LATS2 is shown. Mean ± sd. N = 3, ∗∗∗∗p < 0.0001 (one-way ANOVA). E, working model for the molecular mechanism of NF2 function to suppress tumor cell proliferation through LATS1/2 (right) and directly through TEAD4 (left). NF2 directly binds to and downregulates TEAD4 activity could complement classical regulation through the Hippo pathway.
Notably, coimmunoprecipitation showed that, compared with NF2-WT, NF2-5A maintained a similar binding capacity to LATS2 (Fig. 6D). In addition, NF2-5A did not alter the LATS activity (Fig. S5E), indicating that the suppression defect of cell proliferation resulted by NF2-5A is independent of the conventional Hippo pathway in which NF2 binds and activates LATS kinase. NF2-A585W was introduced due to its inability to interact with LATS but binding to TEAD4, and the results showed that NF2-A585W expression also failed to effectively suppress cell growth (Fig. 6, B and C). These results highlight the fact that the ability of NF2 to suppress cell proliferation and function in Hippo signaling hinges on its binding activities to both TEAD and LATS proteins, and neither can be compromised.
Discussion
As a tumor suppressor, NF2 senses cell‒cell contact and regulates the Hippo pathway by activating the LATS1/2 kinases, resulting in phosphorylation and cytoplasmic retention of YAP. Phosphorylated YAP cannot form the complex with transcription factor TEADs in the nucleus, thereby inhibiting cell proliferation and suppressing tumor growth (33, 40, 41, 42). In contrast to the classic model of NF2, our findings suggest a straightforward regulatory mechanism in which NF2 directly associates with TEAD4 to promote cytoplasmic retention and inhibit palmitoylation of TEAD4, resulting in dysfunction of TEAD4 and suppression of cell proliferation (Fig. 6E). We further determined 5 key residues of NF2 that required for TEAD4 interaction and cell proliferation suppression. Missense mutations at these sites, such as L297V, H304Y, and F591L, are also found in various cancers (43, 44). The NF-5A mutant lost the binding ability to TEAD4 but still bound to LATS2, which indicates that the functional deficiency of NF2-5A is due to a defect in the binding of TEAD instead of the classical Hippo pathway. This straightforward regulation would shed light on an additional mechanism of how tumor suppressor NF2 functions and complement the regulation from LATS1/2 of Hippo pathway.
Palmitoylation is essential for protein stability and transcriptional activity of TEADs (20, 23), and NF2 has been shown to decrease the mRNA levels of fatty acid synthase (FASN) and induce depalmitoylation of TEADs (24). Alternatively, we found that palmitoylation of TEAD4 could also be inhibited by NF2 via direct interaction rather than through alteration of potential Acyl-protein thioesterases (APT2 and ABHD17A) activity in the downstream of FASN, which reflects a novel role of NF2 in regulating the palmitoylation and homeostasis of TEAD protein in cells.
As transcription factors, TEAD family proteins form transcriptional complexes with the major co-activators YAP/TAZ to activate the transcription of important target genes and promote cell proliferation and organ growth (5). Besides that, the inhibitory binding partners of TEADs, such as VGLL4, compete with YAP/TAZ and inhibit the transcriptional activity of TEAD4 to suppress cell proliferation and tumor growth in multiple cancers (15, 45). Our finding that the tumor suppressor NF2 inhibits TEAD4 palmitoylation via direct interaction suggested a new role for the classical protein NF2 as the inhibitory factor of TEAD4, which might update the functional understanding of NF2 in the Hippo pathway.
Nuclear localization is also required for TEAD transcription activity. Early studies have reported that the cytoplasmic translocation of TEADs can be induced by cell density and p38 (19). As a membrane-associated protein, NF2 has been shown to recruit and activate LATS1/2 kinases at the plasma membrane (33). Here, we show that NF2 induces the cytoplasmic translocation of TEAD4 via direct protein‒protein interactions. It is reminded that NF2 also triggers the LATS nucleus translocation to activate the Hippo pathway (46). The NF2-induced translocations of both LATS1/2 and TEADs reach the same goal of preventing TEAD transcription activity, suggesting that this straightforward regulation could complement the function of the Hippo pathway.
In summary, we identified the direct link and physical interaction between NF2 and TEAD4, which are important for NF2 function as tumor suppressor and TEADs protein homeostasis.
Experimental procedures
Protein purification
The human TEAD4 YBD domain (217–434) was cloned into a pET-28a vector with an N-terminal SUMO and a 6 × His tag, a pETa-29a vector with a C-terminal 6 × His tag, and a pGEX-6P-1 vector with an N-terminal GST tag. His-tagged and GST-tagged TEAD4-YBD were expressed in E. coli BL21 (DE3) cells, purified using Ni2+-NTA agarose resin (GE Healthcare) or GST agarose resin (GE Healthcare), and further purified via size-exclusion chromatography using a Superdex 200 column (GE Healthcare). Purified SUMO-TEAD4/TEAD4-6 × His and GST-TEAD4 were concentrated to 2 mg/ml in buffer containing 20 mM Tris (pH 8.0), 150 mM NaCl, and 1 mM DTT.
For TEAD4-YBD alone, the GST tag was removed with 3C protease overnight at 4 °C. The eluted TEAD4-YBD protein was purified via size-exclusion chromatography using a Superdex 200 column (GE Healthcare). Purified TEAD4-YBD was concentrated to 2 mg/ml in buffer containing 20 mM Tris (pH 8.0), 150 mM NaCl, and 1 mM DTT.
NF2 (18–595) was cloned into a pMAL-3C vector with an N-terminal MBP tag. MBP-NF2 was purified using amylose resin (New England Biolabs) and eluted using 10 mM maltose (BioFroxx). The elution was further purified via size exclusion chromatography using a Superdex 200 column (GE Healthcare) in buffer containing 20 mM Tris (pH 8.0), 150 mM NaCl, and 1 mM DTT. NF2-FERM was cloned into pET-29a with C-terminal 6 × His tag. FERM-6 × His was purified using Ni2+-NTA agarose resin (GE Healthcare) and a Superdex 200 column (GE Healthcare) in buffer containing 20 mM Tris (pH 8.0), 150 mM NaCl, and 1 mM DTT.
Human YAP (FL) and YAP (47–131) were cloned into a PGEX-6P-1 vector with an N-terminal GST tag. GST-YAP (FL) and GST-YAP (47–131) were purified using GST agarose resin (GE Healthcare). 3C protease was used to remove the GST tag overnight at 4 °C, GST-YAP (47–131) was eluted by 40 mM GSH (Biosharp). Then, YAP (FL) and GST-YAP (47–131) were further purified via size exclusion chromatography using a Superdex 200 column (GE Healthcare) in buffer containing 20 mM Tris (pH 8.0), 150 mM NaCl, and 1 mM DTT.
Human CHIP, Hsp70 and UbcH5b were cloned into PET-28a vector with an N-terminal SUMO tag. Ubiquitin and E1 were cloned into PET-28a vector with an N-terminal 6 × His tag. The proteins were purified using Ni2+-NTA agarose resin (GE Healthcare) and eluted with 200 mM imidazole (Sigma‒Aldrich). The proteins were then purified with a Superdex 200 column (GE Healthcare) in a buffer containing 20 mM Tris (pH 8.0), 150 mM NaCl, and 1 mM DTT.
Cell culture and transfection
HEK293T, HEK293A, HeLa, and MCF-10A cells were cultured in DMEM (Gibco) supplemented with 10% fetal bovine serum (FBS, Gibco) and 1% penicillin/streptomycin. The plasmids were transfected using HighGene Transfection Reagent (ABclonal). The siRNAs were transfected using Lipofectamine 2000 (Invitrogen). The sequences of the siRNAs used in this study:
| siCtrl: | TTCTCCGAACGTGTCACGT |
| siNF2-1: | CCGUGAGGAUCGUCACCAUTT |
| siNF2-2: | GGUACUGGAUCAUGAUGUUTT |
Western blotting and antibodies
The proteins in the samples were separated by 10% SDS‒PAGE and transferred to PVDF membranes (Bio-Rad) at 200 mA for 2 h. The membranes were washed with PBST (PBS and 0.05% Tween 20) and blocked by 5% skim milk in PBST at room temperature for 1 h. Then, the membranes were incubated with the primary antibodies at 4 °C overnight and washed 3 times by PBST. The membranes were further incubated with the secondary antibodies at room temperature in the dark for 1 h and washed 3 times by PBST. Finally, the membranes were developed with ChemiDoc XRS+ (Bio-Rad) or Odysse CLx (LI-COR).
Antibodies used: TEAD4 (abcam, ab58310, 1:1000), TEAD2 (abcam, ab273017, 1:1000), LATS1 (abcam, ab70561, 1:1000), LATS2 (abcam, ab243657, 1:1000), YAP (abcam, ab52771, 1:1000), TAZ (abcam, ab110239, 1:1000), p-YAP (abcam, ab76252, 1:500), Cyr61 (abcam, ab228592, 1:1000), GAPDH (Proteintech, 60004-1-Ig, 1:2000), beta-Actin (abcam, ab8227, 1:3000), NF2 (ABclonal, A2456, 1:1000), ATP2 (ABclonal, A15792, 1:000), ABHD17A (Proteintech, 15854-1-AP, 1:500), His×6 (Proteintech, 66005-I-Ig, 1:1000), GST (ABclonal, AE001, 1:1000), Flag (Proteintech, 20543-I-AP, 1:1000), Myc (Cell Signaling Technology, 71D10, 1:1000), HRP-Goat Anti-Mouse Recombinant Secondary Antibody (Proteintech, RGAM001, 1:3000), HRP-Goat Anti-Rabbit Recombinant Secondary Antibody (RGAR001, 1:3000), IRDye 800CW, Steptavidin (LI-COR, D10114–10, 1:2000), IRDye 800CW Goat anti-Mouse IgG Secondary Antibody (LI-COR, 926–32210, 1:5000), IRDye 680RD Goat anti-Rabbit IgG Secondary Antibody (LI-COR, 926–68071, 1:3000).
Protein immunoprecipitation
The cells were lysed in lysis buffer (20 mM Tris pH 7.5, 150 mM NaCl, 1 mM EDTA, 1% Triton, and phosphatase inhibitor cocktail) for 30 min at 0 °C. The supernatant was incubated with red anti-FLAG beads (Millipore) or Protein-A Magnetic beads (Bio-Rad) and 2 μg of Myc (Cell Signaling Technology, 71D10)/TEAD4 (abcam, ab58310) antibody overnight at 4 °C. The proteins on the beads were resolved by SDS‒PAGE and analyzed via western blotting.
In vitro protein-binding assay
5 μM (200 μl) recombinant GST-NF2 was bound to 20 μl GST resin (GE Healthcare), and 5 μM (200 μl) MBP-NF2 was bound to 20 μl MBP resin (New England Biolabs) in PBS for 1 h at 4 °C. After washing, the resin was incubated with 40 μM (200 μl) SUMO-TEAD4 in PBS for 1 h at 4 °C and washed four times. Proteins retained on the beads were analyzed using SDS‒PAGE and western blotting. SUMO-TEAD4 was detected using an antibody against 6 × His.
In vitro palmitoylation assay
Recombinant TEAD4 protein (500 ng) was incubated with 1 mM alkyne palmitoyl-CoA (Cayman Chemical) for 0.5 h in 20 mM Tris (pH 8.0) and 100 mM NaCl. A click reaction with biotin-azide (Sigma‒Aldrich) was performed for 1 h at 25 °C. The reactions were stopped using 2 × SDS sample buffer, followed by SDS‒PAGE analysis. Biotinylated TEAD4 was detected using streptavidin-IRDye (LI-COR, D10114–10, 1:2000).
In vivo palmitoylation assay
The cells were collected and subjected to the CAPTUREome S-Palmitoylated Protein Kit (Badrilla). Briefly, the cells were lysed and blocked with a blocking buffer at 40 °C for 4 h. The mixture was then subjected to ice-cold acetone precipitation. The precipitate was re-dissolved in the binding buffer and incubated with the thioester cleavage reagent and capture resin for 2 h. After washing, the capture resin was subjected to SDS-PAGE and analyzed via western blotting.
In vitro ubiquitination assay
In vitro ubiquitination reaction was performed using 0.5 μM E1, 4 μM UbcH5b, 2 μM CHIP, 1 μM Hsp70, 10 μM ubiquitin, and 1 μM recombinant GST-WT/2CS TEAD4 for 60 min at 37 °C in 20 mM Tris8.0, 100 mM NaCl, 5 mM ATP, 2.5 mM MgCl2, and 1 mM DTT. Ubiquitination reactions were stopped using 2 × SDS sample buffer, followed by detection via western blotting with the GST antibody (ABclonal, AE001, 1:1000).
Real-time PCR
Total RNA was extracted using TRIzol reagent (Invitrogen), and reverse transcription (RT) was performed using the iScript Reverse Transcription Supermix (Bio-Rad). RT‒PCR analysis was performed using SYBR Green Realtime PCR Master Mix (Toyobo) with the Applied Biosystems Step Two Real-Time PCR System (Applied Biosystems). GAPDH was used as a control. The standard comparative CT quantification method was used to analyze the RT‒PCR results.
The primers used:
| TEAD1 | F: ATGGAAAGGATGAGTGACTCTGC |
| TEAD1 | R: TCCCACATGGTGGATAGATAGC |
| TEAD2 | F: CTTCGTGGAACCGCCAGAT |
| TEAD2 | R: GGAGGCCACCCTTTTTCTCA |
| TEAD3 | F: TCATCCTGTCAGZCGAGGG |
| TEAD3 | R: TCTTCCGAGCTAGAACCTGTATG |
| TEAD4 | F: GAACGGGGACCCTCCAATG |
| TEAD4 | R: GCGAGCATACTCTGTCTCAAC |
| YAP | F: CACAGCATGTTCGAGCTCAT |
| YAP | R: GATGCTGAGCTGTGGGTGTA |
| NF2 | F: TGCGAGATGAAGTGGAAAGG |
| NF2 | R: GCCAAGAAGTGAAAGGTGAC |
| CTGF | F: AAAAGTGCATCCGTACTCCCA |
| CTGF | R: CCGTCGGTACATACTCCACAG |
| Cyr61 | F: GGTCAAAGTTACCGGGCAGT |
| Cyr61 | R: GGAGGCATCGAATCCCAGC |
| GADPH | F: GGCATCCTGGGCTACACTGA |
| GADPH | R: GAGTGGGTGTCGCTGTTGAA |
BiFC assay
N-terminal YFP (1–238) was divided into two insertions: nYFP (1–154) and cYFP (155–238). The pcDNA3.1-NF2/TEAD4/LATS vectors were receptions for C-terminal n/c-YFP fragments with the HindIII and BamHI sites. HEK-293T cells were plated in a 6-well plate for 24 h and transfected with 800 ng nYFP- and 800 ng cYFP-tagged constructs. The cells were treated at low temperature (30 °C) for 6 h for fluorophore maturation, and after 48 h, fluorescence was determined via flow cytometry using a BD FACS Calibur (BD Biosciences) or observed under a confocal laser scanning microscope (Olympus FV1200).
Stable cell line generation
Lentiviral infection was used to generate cells stably expressing NF2. NF2 WT/mutants were cloned into the pLVX-puro-GFP vector. Then, the viral vectors and packing plasmids were co-transfected into HEK293T cells. 72 h after transfection, the viral supernatants were collected by a 0.45 μM filter, and the NF2 KO 293A and HeLa cells were infected with polybrene. 48 h after infection, cells were selected by 2 μg/ml puromycin in a culture medium. Cells stably expressing NF2 were validated by western blotting and immunofluorescence.
Immunostaining
Cells at high density on coverslips were fixed in 4% paraformaldehyde (Aladin) for 30 min and then permeabilized with 0.1% Triton X-100 (Aladin) for 30 min. The cells were blocked in 3% BSA for 1 h and incubated overnight at 4 °C with TEAD4 antibody (Abcam, ab58310, 1:500) diluted in 3% BSA. The secondary antibody Alexa Fluor 647 (Jackson ImmunoResearch, 141562, 1:1000) was diluted in 3% BSA and incubated for 1 h. Then, the cells were stained with DAPI (Beyotime).
BrdU incorporation assay
Cells on coverslips were incubated with 10 μM BrdU (Beyotime) for 8 h at 37 °C, fixed with 4% paraformaldehyde for 30 min, and washed with PBS containing 1% Triton X-100 for 30 min. Then, the cells were incubated with 2 N HCl for 30 min at room temperature. After washing with PBS, the cells were blocked with PBS containing 1% Triton X-100 and 3% BSA. The cells were incubated with primary antibodies against BrdU (Abcam, ab6326, 1:50) overnight at 4 °C. Cells were further incubated with Alexa Fluor 647 (Jackson ImmunoResearch, 141562, 1:1000) for 1 h in the dark and then stained with DAPI (Beyotime). Each experiment was performed at least three times independently.
Cell counting Kit-8 (CCK-8) assay
Cells were seeded into 96-well plates. After 48 h, the cell counting kit-8 (Biosharp) was used to evaluate cell viability. The cells in each well were incubated with 10 μl CCK-8 reagent and 90 μl DMEM solution at 37 °C for 1 h. The absorbance at 450 nm was detected using a plate reader. Each experiment was performed at least three times independently.
Statistical analysis
All the data were analyzed with an unpaired two-tailed t test or one-way ANOVA with multiple comparisons in GraphPad Prism 6.0. ∗p <0.05 indicates a significant difference. ∗p <0.05, ∗∗p <0.01, ∗∗∗p <0.001, ∗∗∗∗p <0.0001. Quantitative data are presented as the mean ± sd. For quantification, fluorescence and WB intensities were measured using ImageJ (NIH).
Data availability
The original data presented in the study may be found in the article/Supporting information section. Further inquiries can be directed to the corresponding author.
Supporting information
This article contains supporting information.
Conflict of interest
The authors declare that they have no conflicts of interest with the contents of this article.
Acknowledgments
We thank Dr Faxing Yu (Children's Hospital of Fudan University) for providing the NF2 KO HEK293A cell line.
Author contributions
Liqiao Hu, L. Z., and X. H. conceptualization; Liqiao Hu, M. W., Lingli He, L. Y., B. Z., L. Z., and X. H. methodology; Liqiao Hu, M. W., Lingli He, and L. Y. investigation; Liqiao Hu and M. W. writing–original draft; Lingli He, B. Z., L. Z., and X. H. writing–review & editing; Liqiao Hu, L. Z., and X. H. funding acquisition; L. Z. and X. H. resources; X. H. supervision.
Funding and additional information
This work was supported by National Key R&D Program of China [2020YFA0803201 to X. H. and 2019YFA0802000 to L. Z.], the National Natural Science Foundation of China [31970672 and 32270877 to X. H., 32000496 to Liqiao Hu and 32030025 and 31625017 to L. Z.], the Shanghai Leading Talents Program to L. Z. and the China Postdoctoral Science Foundation [2019M662588 to Lingli He].
Reviewed by members of the JBC Editorial Board. Edited by Donita C. Brady
Contributor Information
Liqiao Hu, Email: liq@hust.edu.cn.
Xiaojing He, Email: hexj@hust.edu.cn.
Supporting information
References
- 1.Harvey K.F., Pfleger C.M., Hariharan I.K. The Drosophila Mst ortholog, hippo, restricts growth and cell proliferation and promotes apoptosis. Cell. 2003;114:457–467. doi: 10.1016/s0092-8674(03)00557-9. [DOI] [PubMed] [Google Scholar]
- 2.Huang J., Wu S., Barrera J., Matthews K., Pan D. The hippo signaling pathway coordinately regulates cell proliferation and apoptosis by inactivating Yorkie, the Drosophila homolog of YAP. Cell. 2005;122:421–434. doi: 10.1016/j.cell.2005.06.007. [DOI] [PubMed] [Google Scholar]
- 3.Pan D. The hippo signaling pathway in development and cancer. Dev. Cell. 2010;19:491–505. doi: 10.1016/j.devcel.2010.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wu S., Huang J., Dong J., Pan D. Hippo encodes a ste-20 family protein kinase that restricts cell proliferation and promotes apoptosis in conjunction with salvador and warts. Cell. 2003;114:445–456. doi: 10.1016/s0092-8674(03)00549-x. [DOI] [PubMed] [Google Scholar]
- 5.Yu F.-X., Zhao B., Guan K.-L. Hippo pathway in organ size control, tissue homeostasis, and cancer. Cell. 2015;163:811–828. doi: 10.1016/j.cell.2015.10.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhao B., Li L., Lei Q., Guan K.-L. The Hippo-YAP pathway in organ size control and tumorigenesis: an updated version. Genes Dev. 2010;24:862–874. doi: 10.1101/gad.1909210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhao B., Li L., Tumaneng K., Wang C.-Y., Guan K.-L. A coordinated phosphorylation by Lats and CK1 regulates YAP stability through SCF(beta-TRCP) Genes Dev. 2010;24:72–85. doi: 10.1101/gad.1843810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ota M., Sasaki H. Mammalian Tead proteins regulate cell proliferation and contact inhibition as transcriptional mediators of Hippo signaling. Development. 2008;135:4059–4069. doi: 10.1242/dev.027151. [DOI] [PubMed] [Google Scholar]
- 9.Zhang L., Ren F., Zhang Q., Chen Y., Wang B., Jiang J. The TEAD/TEF family of transcription factor Scalloped mediates Hippo signaling in organ size control. Dev. Cell. 2008;14:377–387. doi: 10.1016/j.devcel.2008.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhao B., Ye X., Yu J., Li L., Li W., Li S., et al. TEAD mediates YAP-dependent gene induction and growth control. Genes Dev. 2008;22:1962–1971. doi: 10.1101/gad.1664408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huh H., Kim D., Jeong H.-S., Park H. Regulation of TEAD transcription factors in cancer biology. Cells. 2019;8:600. doi: 10.3390/cells8060600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dey A., Varelas X., Guan K.-L. Targeting the Hippo pathway in cancer, fibrosis, wound healing and regenerative medicine. Nat. Rev. Drug Discov. 2020;19:480–494. doi: 10.1038/s41573-020-0070-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zheng Y., Pan D. The hippo signaling pathway in development and disease. Dev. Cell. 2019;50:264–282. doi: 10.1016/j.devcel.2019.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.He L., Yuan L., Sun Y., Wang P., Zhang H., Feng X., et al. Glucocorticoid receptor signaling activates TEAD4 to promote breast cancer progression. Cancer Res. 2019;79:4399–4411. doi: 10.1158/0008-5472.CAN-19-0012. [DOI] [PubMed] [Google Scholar]
- 15.Jiao S., Wang H., Shi Z., Dong A., Zhang W., Song X., et al. A peptide mimicking VGLL4 function acts as a YAP antagonist therapy against gastric cancer. Cancer Cell. 2014;25:166–180. doi: 10.1016/j.ccr.2014.01.010. [DOI] [PubMed] [Google Scholar]
- 16.Jiao S., Li C., Hao Q., Miao H., Zhang L., Li L., et al. VGLL4 targets a TCF4–TEAD4 complex to coregulate Wnt and Hippo signalling in colorectal cancer. Nat. Commun. 2017;8 doi: 10.1038/ncomms14058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu X., Li H., Rajurkar M., Li Q., Cotton J.L., Ou J., et al. Tead and AP1 coordinate transcription and motility. Cell Rep. 2016;14:1169–1180. doi: 10.1016/j.celrep.2015.12.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Deng X., Fang L. VGLL4 is a transcriptional cofactor acting as a novel tumor suppressor via interacting with TEADs. Am. J. Cancer Res. 2018;8:932–943. [PMC free article] [PubMed] [Google Scholar]
- 19.Lin K.C., Moroishi T., Meng Z., Jeong H.-S., Plouffe S.W., Sekido Y., et al. Regulation of Hippo pathway transcription factor TEAD by p38 MAPK-induced cytoplasmic translocation. Nat. Cell Biol. 2017;19:996–1002. doi: 10.1038/ncb3581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chan P., Han X., Zheng B., DeRan M., Yu J., Jarugumilli G.K., et al. Autopalmitoylation of TEAD proteins regulates transcriptional output of the Hippo pathway. Nat. Chem. Biol. 2016;12:282–289. doi: 10.1038/nchembio.2036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gupta M.P., Kogut P., Gupta M. Protein kinase-A dependent phosphorylation of transcription enhancer factor-1 represses its DNA-binding activity but enhances its gene activation ability. Nucleic Acids Res. 2000;28:3168–3177. doi: 10.1093/nar/28.16.3168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jiang S.W., Dong M., Trujillo M.A., Miller L.J., Eberhardt N.L. DNA binding of TEA/ATTS domain factors is regulated by protein kinase C phosphorylation in human choriocarcinoma cells. J. Biol. Chem. 2001;276:23464–23470. doi: 10.1074/jbc.M010934200. [DOI] [PubMed] [Google Scholar]
- 23.Noland C.L., Gierke S., Schnier P.D., Murray J., Sandoval W.N., Sagolla M., et al. Palmitoylation of TEAD transcription factors is required for their stability and function in hippo pathway signaling. Structure. 2016;24:179–186. doi: 10.1016/j.str.2015.11.005. [DOI] [PubMed] [Google Scholar]
- 24.Kim N.-G., Gumbiner B.M. Cell contact and Nf2/Merlin-dependent regulation of TEAD palmitoylation and activity. Proc. Natl. Acad. Sci. U. S. A. 2019;116:9877–9882. doi: 10.1073/pnas.1819400116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mesrouze Y., Meyerhofer M., Bokhovchuk F., Fontana P., Zimmermann C., Martin T., et al. Effect of the acylation of TEAD4 on its interaction with co-activators YAP and TAZ: TEAD acylation. Protein Sci. 2017;26:2399–2409. doi: 10.1002/pro.3312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bum-Erdene K., Zhou D., Gonzalez-Gutierrez G., Ghozayel M.K., Si Y., Xu D., et al. Small-molecule covalent modification of conserved cysteine leads to allosteric inhibition of the TEAD⋅Yap protein-protein interaction. Cell Chem. Biol. 2019;26:378–389.e13. doi: 10.1016/j.chembiol.2018.11.010. [DOI] [PubMed] [Google Scholar]
- 27.Pobbati A.V., Han X., Hung A.W., Weiguang S., Huda N., Chen G.-Y., et al. Targeting the central pocket in human transcription factor TEAD as a potential cancer therapeutic strategy. Structure. 2015;23:2076–2086. doi: 10.1016/j.str.2015.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen H., Xue L., Wang H., Wang Z., Wu H. Differential NF2 gene status in sporadic vestibular schwannomas and its prognostic impact on tumour growth patterns. Sci. Rep. 2017;7:5470. doi: 10.1038/s41598-017-05769-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cheng J.Q., Lee W.-C., Klein M.A., Cheng G.Z., Jhanwar S.C., Testa J.R. Frequent mutations of NF2 and allelic loss from chromosome band 22q12 in malignant mesothelioma: evidence for a two-hit mechanism ofNF2 inactivation. Genes Chromosomes Cancer. 1999;24:238–242. [PubMed] [Google Scholar]
- 30.Kalamarides M. Nf2 gene inactivation in arachnoidal cells is rate-limiting for meningioma development in the mouse. Genes Dev. 2002;16:1060–1065. doi: 10.1101/gad.226302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cooper J., Giancotti F.G. Molecular insights into NF2/Merlin tumor suppressor function. FEBS Lett. 2014;588:2743–2752. doi: 10.1016/j.febslet.2014.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hong A.W., Meng Z., Plouffe S.W., Lin Z., Zhang M., Guan K.-L. Critical roles of phosphoinositides and NF2 in Hippo pathway regulation. Genes Dev. 2020;34:511–525. doi: 10.1101/gad.333435.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yin F., Yu J., Zheng Y., Chen Q., Zhang N., Pan D. Spatial organization of hippo signaling at the plasma membrane mediated by the tumor suppressor merlin/NF2. Cell. 2013;154:1342–1355. doi: 10.1016/j.cell.2013.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li W., You L., Cooper J., Schiavon G., Pepe-Caprio A., Zhou L., et al. Merlin/NF2 suppresses tumorigenesis by inhibiting the E3 ubiquitin ligase CRL4DCAF1 in the nucleus. Cell. 2010;140:477–490. doi: 10.1016/j.cell.2010.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li Y., Zhou H., Li F., Chan S.W., Lin Z., Wei Z., et al. Angiomotin binding-induced activation of Merlin/NF2 in the Hippo pathway. Cell Res. 2015;25:801–817. doi: 10.1038/cr.2015.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chinthalapudi K., Mandati V., Zheng J., Sharff A.J., Bricogne G., Griffin P.R., et al. Lipid binding promotes the open conformation and tumor-suppressive activity of neurofibromin 2. Nat. Commun. 2018;9:1338. doi: 10.1038/s41467-018-03648-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sher I., Hanemann C.O., Karplus P.A., Bretscher A. The tumor suppressor merlin controls growth in its open state and is converted by phosphorylation to a less-active more-closed state. Dev. Cell. 2012;22:703. doi: 10.1016/j.devcel.2012.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zheng B., Zhu S., Wu X. Clickable analogue of cerulenin as chemical probe to explore protein palmitoylation. ACS Chem. Biol. 2015;10:115–121. doi: 10.1021/cb500758s. [DOI] [PubMed] [Google Scholar]
- 39.Forrester M.T., Hess D.T., Thompson J.W., Hultman R., Moseley M.A., Stamler J.S., et al. Site-specific analysis of protein S-acylation by resin-assisted capture. J. Lipid Res. 2011;52:393–398. doi: 10.1194/jlr.D011106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Morrison H., Sherman L.S., Legg J., Banine F., Isacke C., Haipek C.A., et al. The NF2 tumor suppressor gene product, merlin, mediates contact inhibition of growth through interactions with CD44. Genes Dev. 2001;15:968–980. doi: 10.1101/gad.189601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Okada T., Lopez-Lago M., Giancotti F.G. Merlin/NF-2 mediates contact inhibition of growth by suppressing recruitment of Rac to the plasma membrane. J. Cell Biol. 2005;171:361–371. doi: 10.1083/jcb.200503165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Meng Z., Moroishi T., Guan K.-L. Mechanisms of Hippo pathway regulation. Genes Dev. 2016;30:1–17. doi: 10.1101/gad.274027.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zehir A., Benayed R., Shah R.H., Syed A., Middha S., Kim H.R., et al. Mutational landscape of metastatic cancer revealed from prospective clinical sequencing of 10,000 patients. Nat. Med. 2017;23:703–713. doi: 10.1038/nm.4333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bonilla X., Parmentier L., King B., Bezrukov F., Kaya G., Zoete V., et al. Genomic analysis identifies new drivers and progression pathways in skin basal cell carcinoma. Nat. Genet. 2016;48:398–406. doi: 10.1038/ng.3525. [DOI] [PubMed] [Google Scholar]
- 45.Zhang W., Gao Y., Li P., Shi Z., Guo T., Li F., et al. VGLL4 functions as a new tumor suppressor in lung cancer by negatively regulating the YAP-TEAD transcriptional complex. Cell Res. 2014;24:331–343. doi: 10.1038/cr.2014.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li W., Cooper J., Zhou L., Yang C., Erdjument-Bromage H., Zagzag D., et al. Merlin/NF2 loss-driven tumorigenesis linked to CRL4DCAF1-mediated inhibition of the hippo pathway kinases Lats1 and 2 in the nucleus. Cancer Cell. 2014;26:48–60. doi: 10.1016/j.ccr.2014.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The original data presented in the study may be found in the article/Supporting information section. Further inquiries can be directed to the corresponding author.