Abstract
Human cytomegalovirus has two enhancer-containing immediate-early (IE) promoters with a cis repression sequence (CRS) positioned immediately upstream of the transcription start site, designated the major IE (MIE) promoter and the US3 promoter. The role of the CRS upstream of the US3 transcription start site in the context of the viral genome was determined by comparing the levels of transcription from these two enhancer-containing promoters in recombinant viruses with a wild-type or mutant CRS. Upstream of the CRS of the US3 promoter was either the endogenous enhancer (R2) or silencer (R1). The downstream US3 gene was replaced with the indicator gene chloramphenicol acetyltransferase (CAT). Infected permissive human fibroblast cells or nonpermissive, undifferentiated monocytic THP-1 cells were analyzed for expression from the US3 promoter containing either the wild-type or mutant CRS. With the wild-type CRS, the maximum level of transcription in permissive cells was detected within 4 to 6 h after infection and then declined. With the mutant CRS and the R2 enhancer upstream, expression from the US3 promoter continued to increase throughout the viral replication cycle to levels 20- to 40-fold higher than for the wild type. In nonpermissive or permissive monocytic THP-1 cells, expression from the US3 promoter was also significantly higher when the CRS was mutated. Less expression was obtained when only the R1 element was present, but expression was higher when the CRS was mutated. Thus, the CRS in the enhancer-containing US3 promoter appears to allow for a short burst of US3 gene expression followed by repression at early and late times after infection. Overexpression of US3 may be detrimental to viral replication, and its level of expression must be stringently controlled. The role of the CRS and the viral IE86 protein in controlling enhancer-containing promoters is discussed.
Human cytomegalovirus (HCMV) infections can result in congenital neurological complications in newborns and pneumonitis, retinitis, hepatitis, and gastroenteritis in immunosuppressed adults (1, 19). In healthy individuals, HCMV infections are normally asymptomatic, and the virus persists for the life-time of the host. Latent HCMV viral DNA can be detected in hematopoietic precursor cells in bone marrow and blood monocytes of infected individuals (24, 47). Uncharacterized stimulatory events in the host similar to allogenic T-cell stimulation and cytokines such as tumor necrosis factor alpha and gamma interferon can reactivate the latent viral genome. Productive viral replication occurs in monocyte-derived macrophages (39, 40).
During productive infection, the viral genes are temporally expressed in three broad categories designated immediate-early (IE; alpha), early (beta), and late (gamma). The transcription of the viral IE genes by RNA polymerase II does not require de novo viral protein synthesis. Viral receptor-mediated attachment to the host cell that involves viral glycoproteins gB and gH has been reported to activate viral transcription (56). Virion-associated tegument proteins that localize to the nucleus of the host cell, such as the viral proteins encoded by UL82 (pp71) and UL69, in combination with different host cell transcription factors enhance HCMV IE transcription (27, 53). Two classes of IE genes, designated major and ancillary IE genes, are expressed. The major IE genes, collectively referred to as UL123/122 (IE1/IE2), are transcribed and expressed at relatively high levels. This transcription unit is immediately downstream of a very strong enhancer-containing promoter referred to as the major IE (MIE) promoter. The ancillary IE genes (UL36 through UL38, UL115 through UL119, IRS1/TRS1, and US3) are expressed at lower relative levels (9). IE1/IE2, UL36 through UL38, and IRS1/TRS1 are required for HCMV ori-Lyt-mediated DNA replication. They are necessary for efficient activation of early viral promoters and subsequent expression of six early viral proteins required for viral DNA replication (2). Early viral genes are expressed prior to viral DNA replication, and late genes are expressed after viral DNA replication.
The IE1 and IE2 genes code for two regulatory proteins referred to as IE72 and IE86, respectively (42–44). The functions of the viral IE72 and IE86 proteins are not fully understood. IE72 has protein kinase activity, is independently a weak activator of some cellular and viral promoters, and has a significant synergistic effect on IE86-mediated activation of early viral promoters (8, 18, 20, 30, 34). IE86 is a multifunctional viral protein that interacts with the viral protein specified by the UL84 gene (11, 37, 41). While the IE1 gene is not essential for virus replication in tissue culture cells infected at a high multiplicity of infection (MOI) it is necessary for efficient replication at low MOIs (15, 33). IE86 may be essential for virus replication since attempts to delete this viral gene have been unsuccessful (50). The US3 gene is nonessential for replication in cell culture, but the viral gene product may be necessary for escape from immune surveillance in the host at the earliest stages of infection (21).
The characteristics of transcriptional regulation of the IE1/IE2 and US3 genes have the following similarities. Both transcription units have an upstream enhancer. Viral RNAs reach maximum steady-state levels at 4 to 6 h after infection and then decline (9, 42, 46). The MIE promoter has a very complicated upstream enhancer, while the US3 promoter has a simplified upstream enhancer with some similar cis-acting sites (32, 48, 52). The major cis-acting elements in the US3 enhancer, five NF-κB/Rel sites, are activated by the p65 subunit of NF-κB (48). The R1 element upstream of the US3 enhancer contains multiple pentanucleotide repeat sequences that bind cellular proteins (48). This element has been referred to as the R1 silencer because it represses downstream transcription from the US3 enhancer-containing promoter or the MIE promoter in transient transfection assays (6, 48). Finally, both the MIE and US3 promoters have a cis repression sequence (CRS) immediately upstream of the transcription start site and a consensus initiator-like (Inr) element immediately downstream (4, 5, 7, 26, 28, 29, 35). The CRS regulates transcription from the enhancer-containing promoter by presumably binding a repressor protein at early and late times after infection. The CRS, which is strategically positioned, does not function when placed upstream of the TATA box or 31 nucleotides downstream of the transcription start site (26). The viral IE86 protein binds to the CRS of the MIE promoter and significantly represses downstream in vitro transcription (29). The binding of the IE86 protein to the CRS may interfere with the binding of a 150-kDa cellular protein to the Inr sequence due to overlapping binding sites (28). This combination of events could prevent initiation of transcription by RNA polymerase II (29, 54). Therefore, the CRS may play an important role in regulating transcription from enhancer-containing promoters in the context of the viral genome during the productive replication cycle of HCMV.
Since the genes immediately downstream of the MIE promoter are essential for efficient replication in cell culture and the US3 gene is nonessential, we elected to test the role of the CRS in controlling expression from the US3 promoter. We tested recombinant viruses with either a wild-type or a mutant CRS upstream of the US3 transcription start site in which the gene downstream of the US3 promoter was replaced with the indicator gene chloramphenicol acetyltransferase (CAT). The data suggest that the transcription of the US3 gene, whose product is a type I glycoprotein believed to be involved in the early stages of escape from immune surveillance, is stringently controlled by the CRS at early and late times after productive virus infection.
MATERIALS AND METHODS
Cells and virus.
Primary human foreskin fibroblast (HFF) cells were maintained at 37°C in 5% CO2 and grown in Eagle’s minimal essential medium (Life Technologies, Gaithersburg, Md.) supplemented with 10% newborn bovine serum (Sigma, St. Louis, Mo.), penicillin (100 U/ml), and streptomycin (100 μg/ml). THP-1 cells were grown in RPMI 1640 medium (Life Technologies) containing 10% fetal bovine serum (HyClone, Logan, Utah) and 50 μg of gentamicin per ml. Stimulation and differentiation of THP-1 cells was with 100 ng of lipopolysaccharide (LPS; Sigma) per ml. HCMV Towne strain was propagated as described previously (45). Recombinant viruses isolated as described below were grown for at least one passage in the absence of mycophenolic acid and xanthine. Mutations in this region of the viral genome did not affect viral growth, as described previously (21). Titers of total infectious virus associated with the cells and extracellular fluid were determined on HFF cells by plaque assay as described previously (31).
Enzymes.
Restriction endonucleases were obtained from either Bethesda Research Laboratories Inc. (Gaithersburg, Md.) or New England Biolabs Inc. (Beverly, Mass.). T4 DNA ligase, the Klenow fragment of Escherichia coli DNA polymerase I, and calf intestinal phosphatase were acquired from Boehringer Mannheim Biochemicals (Indianapolis, Ind.). Taq DNA polymerase was obtained from Promega (Madison, Wis.). All enzymes were used according to the manufacturers’ specifications.
Plasmids.
The 5-kbp NcoI-to-XhoI DNA fragment (bp 193671 to 198622) of HCMV Towne containing the US3 and US9 genes was cloned into pBluescript II KS(+) (Stratagene, La Jolla, Calif.) to generate pKS+US3/9. The 700-bp SmaI-to-HindIII DNA fragment (bp 195838 to 195109) containing the US6 open reading frame (ORF) and R1 sequence was cloned into pBluescript II KS(+) to generate pKS+US6R1. The US3 enhancer-containing promoter and the CAT gene between the SmaI and BamHI site of p7R15R2CAT (48) were subcloned into the same sites of pKS+US6R1 to generate pUS6R1R2CAT.
The NdeI-to-MluI DNA (bp 194549 to 195544) in pKS+US3/9 was replaced with the 2,443-bp NdeI-to-MluI DNA fragment from pUS6R1R2CAT to generate pR1R2CAT. This plasmid contains the US3 promoter with the upstream R2 enhancer and the R1 silencer and the downstream CAT gene flanked by part of the HCMV US3 gene at the 3′ end and the US6 through US9 genes at the 5′ end.
The guanine phosphoribosyltransferase (gpt) gene under the control of a minimal simian virus 40 (SV40) promoter was isolated as a 2,121-bp MluI-to-BsrGI DNA fragment from pdlMSVgpt (31). This gpt-containing DNA fragment was cloned into the viral DNA contained in plasmid pR1R2CAT, replacing the US6 through US8 genes (bp 195544 to 197690) to generate plasmid pgptR1R2CAT.
SmaI and SnaBI restriction endonuclease digestion was used to delete the R2 enhancer from pgptR1R2CAT to generate pgptR1CAT. The R1 silencer was deleted from pgptR1R2CAT by digestion with restriction endonucleases SmaI and MluI. The MluI site was made blunt with Klenow polymerase, and the plasmid was religated to generate pgptR2CAT.
The CRS upstream of the US3 transcription start site was mutated by PCR using primer pair 5′-GCCAAGCTTGGGAGAAGTAGCtTGgccgggtaggctTGTTTTTG-3′ plus 5′-AGTCAGTGAGCGAGGAAGCG-3′. Nucleotide mutations are in lowercase, and the underline designates the location of the CRS. The 335-bp PCR-generated DNA fragment containing the mutant crs (CRS−) was inserted between the EcoRV and HindIII sites to generate pgptR1crs−CAT and pgptR2crs−CAT. All plasmid constructions and mutations used to generate recombinant viruses were analyzed by dideoxynucleotide sequencing prior to transfection of HFF cells as described below.
Recombinant viruses.
Recombinant viruses were isolated by the method of Greaves et al. (14) and as described previously (31). With plasmids pgptR1CAT, pgptR1crs−CAT, pgptR2CAT, and pgptR2crs−CAT as shuttle vectors, 10 μg of each plasmid was linearized by digestion with restriction endonuclease XhoI and used to transfect HFF cells by calcium phosphate precipitation (13). Twenty-four hours after transfection, the cells were infected with approximately 1 PFU of wild-type HCMV Towne strain per cell. At approximately 10 days after infection, the virus in the extracellular fluid was harvested, passed through a 0.45-μm-pore-size filter, and used, either undiluted or diluted 1:10, to infect HFF cells. Selection for recombinant virus was done with medium containing mycophenolic acid (40 μg/ml) and xanthine (200 μg/ml). Viruses were harvested as described above, and the enrichment cycle was repeated twice. Recombinant virus plaques were isolated on HFF monolayers grown under medium containing 0.8% agarose and mycophenolic acid and xanthine. Viral plaques were transferred to 48-well HFF culture units. Three to four days after the appearance of 100% cytopathic effect, cell-free virus from each well was mixed with an equal volume of 100% newborn calf serum and stored at −70°C. DNA was isolated from the infected cells in each well and analyzed by dot blot hybridization using a 32P-labeled CAT or 32P-labeled gpt DNA probe prepared as described previously (31). After identification of CAT and gpt-containing recombinant viruses by dot blot hybridization, the viruses were checked for CAT expression. Three different recombinant viruses from at least two different transfections were plaque purified three times.
Southern blots.
Culture medium containing cell-free virus was subjected to low-speed centrifugation to pellet particulate material and high-speed centrifugation to pellet virus as described previously (45). After solubilization of the viral envelope with 1% Sarkosyl and digestion of the viral proteins with 200 μg of proteinase K per ml in 0.1% sodium dodecyl sulfate, viral DNA was digested with restriction endonuclease BsrGI and subjected to agarose gel electrophoresis as described previously (31, 51). Southern blot analyses were done as described previously (31).
RNase protection assay.
Construction of the plasmid DNA templates and antisense CAT and IE1 riboprobe synthesis by the method of Krieg and Melton (25) have been described previously (17, 26, 31). Cytoplasmic RNA was harvested from two 100-mm-diameter plates of HFF cells either mock infected or infected with approximately 5 PFU of recombinant virus per cell as described previously (48). Twenty micrograms of RNA was hybridized with both 32P-antisense CAT and 32P-antisense IE1 riboprobes at room temperature overnight. The specific activities of the riboprobes were similar. Digestion with 150 U of RNase T1 (Boehringer Mannheim) was at 37°C for 1 h. RNAs protected from RNase digestion were subjected to electrophoresis in denaturing 6% polyacrylamide-urea gels. Signals were visualized by autoradiography on Hyperfilm MP (Amersham) and quantitated by image acquisition analysis (Packard Instant Imager, Meriden, Conn.).
CAT assay.
All infections with recombinant viruses on 100-mm-diameter plates of HFF cells were done in triplicate. After being infected with recombinant viruses and incubated for 1 h at 37°C, the THP-1 cells were washed, aliquoted, and then suspended in medium with or without LPS (100 ng/ml; Sigma). Transfections were done three times in duplicate on either 293-T or HFF cells by the calcium phosphate precipitation method of Graham and van der Eb (13). The CRS upstream of the transcription start site of the enhancer-containing MIE promoter is mutated in plasmids pSVIE2crs− and pSVIE2crs−HL. The mutation in pSVIE2crs−HL has been described previously (28, 55). CAT activities were determined in substrate excess as described by Gorman et al. (12). Acetylated derivatives were separated from nonacetylated 14C-chloramphenicol by thin-layer chromatography using a chloroform-methanol (95:5) solvent. The percentage of 14C-chloramphenicol acetylation was determined by image acquisition analysis. Protein concentration was determined by the Bradford method (Bio-Rad Laboratories, Richmond, Calif.).
Western blot analysis.
IE2 gene expression or mutant gene expression was detected with monoclonal antibody 810 (Chemicon, Temecula, Calif.) and the Pierce (Rockford, Ill.) chemiluminescence method.
EMSA.
Electrophoretic mobility shift assay (EMSA) was done with double-stranded wild-type (5′-TCAAAAACACCGTGCAGTCCACACGCTACTTCTCC-3′) and mutant (5′-TCAAAAACAagcctacccggcCAaGCTACTT-3′) CRS probes as described previously (29). The position of the CRS is underlined, and nucleotide mutations are in lowercase. Wild-type recombinant IE2 (rIE2) and mutant rIE2HL were purified as maltose fusion proteins as described previously (28, 29). Protein binding assay was done with 5.8 or 2.9 pmol of rIE2 and 5.8 pmol of rIE2HL.
RESULTS
Recombinant viruses with wild-type or mutant CRS.
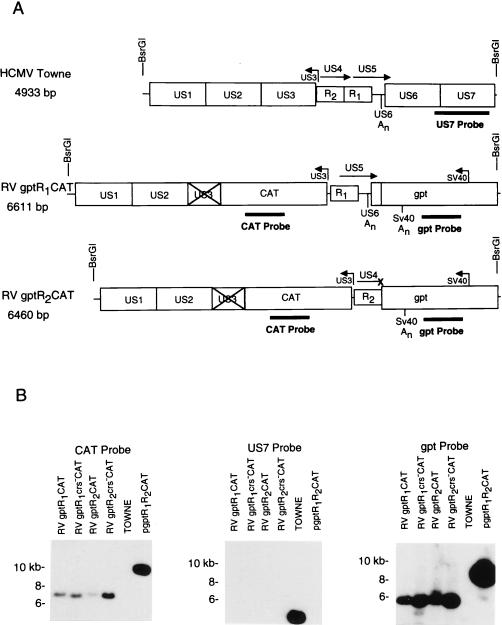
Transient transfection experiments have indicated that the CRS located immediately upstream of the transcription start site of the MIE and US3 promoters contribute to negative regulation of transcription (5, 26). The two CRS elements are similar in that the MIE promoter has a 5′-CGN10CG-3′ motif and the US3 CRS has a 5′-CGN11CG-3′ motif (Fig. 1). To test the role of the US3 CRS in the context of the viral genome, we mutated the CRS upstream of the transcription start site of the promoter as illustrated in Fig. 1. In addition, the viral US3 ORF was replaced with the indicator gene CAT for the following reasons: (i) overexpression of the US3 gene product was anticipated to be cytotoxic and to interfere with recombinant virus isolation, and (ii) antibodies to the US3 glycoprotein were considered unsatisfactory for quantitation of US3 gene expression. The gpt gene, driven by a minimal SV40 promoter, was used to facilitate selection of recombinant viruses as described by Greaves et al. (14).
FIG. 1.
DNA sequence of the CRS and Inr elements associated with the MIE and US3 promoters. The consensus sequences of the MIE CRS is CGN10CG; that of the US3 CRS is CGN11CG. Both IE promoters have an Inr element immediately downstream of the CRS and the transcription start site. The transcription start site is indicated by the arrow, and the base is in boldface. Mutation of the US3 CRS is indicated in lowercase.
Viral DNAs were digested with the restriction endonuclease BsrGI and analyzed by Southern blot hybridization using a 32P-labeled CAT, gpt, or Towne strain-specific DNA probe. The predicted sizes of the wild-type or recombinant viral DNA fragments after digestion with BsrGI are designated in Fig. 2A. The 32P-labeled CAT or gpt probe hybridized to DNA fragments of the appropriate size from the recombinant viruses RVgptR1CAT, RVgptR1crs−CAT, RVgptR2CAT, and RVgptR2crs−CAT (Fig. 2B). In contrast, 32P-labeled US7 DNA probe hybridized only to wild-type Towne strain DNA. The 9.3-kb linearized pgptR1R2CAT plasmid DNA, which contains the R1 and R2 elements as well as the CAT and gpt genes, was used as a positive control (Fig. 2B). These recombinant viruses with either the wild-type or mutant CRS upstream of the US3 transcription start site should have either the R1 silencer or the R2 enhancer upstream as illustrated in Fig. 2A. Recombinant viruses selected to have the gpt gene and both the R1 and R2 elements were unstable.
FIG. 2.
Autoradiogram of Southern blot hybridizations of wild-type and recombinant viral DNA fragments. Viral DNAs were isolated, digested with the restriction endonuclease BsrGI, and then subjected to electrophoresis followed by blotting and hybridization with 32P-labeled DNA probes as described in Materials and Methods. A shuttle DNA vector containing the gpt gene with the R1 and R2 elements upstream of the US3 promoter and the CAT gene was used as a hybridization control. (A) Maps of wild-type and recombinant viruses. Locations of the R2 and R1 elements and sizes of the DNA fragments after restriction endonuclease digestion with BsrGI are indicated. The × indicates an interruption of the US3 ORF. (B) Southern blot hybridization with the 32P-labeled CAT, US7, or gpt DNA probe.
Effect of a wild-type or mutant CRS with an upstream R2 enhancer.
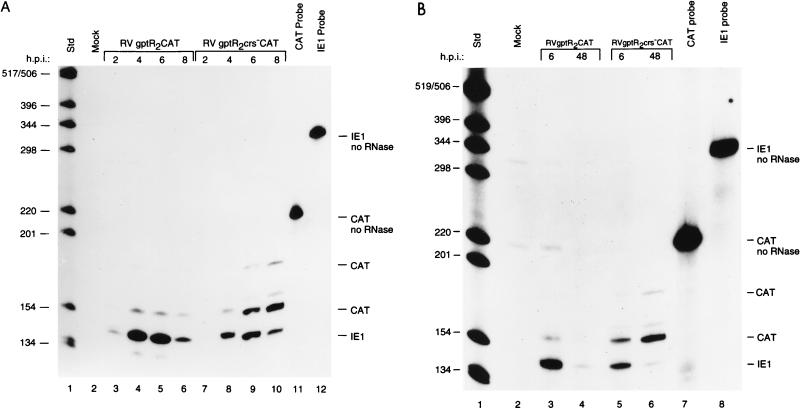
In the wild-type Towne and AD169 strains of HCMV, the MIE and US3 promoters follow similar patterns of transcription (9, 43, 46). Viral RNAs are detectable within 2 h after infection; peak levels are at 4 to 6 h, and then transcription is repressed. To compare the steady-state levels of viral RNA transcribed from the MIE and US3 promoters in the recombinant viruses, we prepared antisense RNA probes to the IE1 and CAT RNAs, respectively. RNase-protected CAT RNA levels of the expected size were then compared to IE1 RNA levels. After infection of permissive HFF cells at an MOI of approximately 5 PFU/cell, cytoplasmic RNA was analyzed at 2-h intervals after infection or at early (6 h) and late (48 h) times. In cells infected with RVgptR2CAT or RVgptR2crs−CAT, the viral RNA from the MIE promoter follows the typical pattern of peak levels at 4 to 6 h followed by a decline at 8 h (Fig. 3A; compare lanes 3 to 6 with lanes 7 to 10). Since transcription from the MIE promoter is influenced by the very strong upstream enhancer, the IE1 RNA levels, as expected, were approximately fivefold higher than the US3 RNA levels. When the wild-type CRS was present upstream of the US3 promoter transcription start site, the CAT RNA reached peak levels at 4 to 6 h and then declined (Fig. 3A, lanes 3 to 6). In contrast, when the mutant CRS was upstream of the US3 promoter transcription start site, CAT RNA levels increased at 6 h and continued to increase at 8 h after infection (Fig. 3A, lanes 9 and 10). Lower levels of RNA larger than the expected size were detected with both recombinant viruses and may represent incompletely RNase-digested RNA or an alternative transcription start site induced by the mutation. The majority of the RNA initiated at the US3 promoter start site.
FIG. 3.
Steady-state RNA levels transcribed either from the US3 promoter with a wild-type or mutant CRS downstream of the R2 enhancer element or from the enhancer-containing MIE promoter at early and late times after infection (h.p.i. [hours postinfection]). HFF cells were infected with approximately 5 PFU of RVgptR2CAT or RVgptR2crs−CAT per ml. Cytoplasmic RNA was harvested and analyzed by RNase protection assays as described in Materials and Methods. IE1-specific RNA from the MIE promoter and CAT-specific RNA from the US3 promoter are designated; IE1 and CAT probes not treated with RNase are also designated. Std, 32P-labeled DNA molecular weight markers, positions of which are indicated in nucleotides. (A) RNase protection assay at early times after infection; (B) RNase protection assay at early and late times after infection.
To determine the CAT RNA levels from the US3 promoter at early and late times after infection, cytoplasmic RNA was harvested at 6 and 48 h after infection. In cells infected with RVgptR2CAT or RVgptR2crs−CAT, the IE1 RNA steady-state level was high at 6 h and lower at 48 h (Fig. 3B). When the wild-type CRS was present, the CAT RNA was detected at 6 h but at relatively low levels at 48 h (Fig. 3B). In contrast, when the CRS was mutated, the CAT RNA level was relatively high at 48 h (Fig. 2B, lane 6). For the RNase-protected CAT RNA of the expected size, there was approximately a 20-fold-higher level of CAT RNA at 48 h when the CRS was mutated. We conclude that the CRS adjacent to the US3 promoter transcription start site has a critical role in controlling transcription of the US3 gene in the context of the viral genome at both early and late times during the viral replication cycle. When the wild-type CRS is present, viral RNA levels peak at 4 to 6 h and then decline. Repression of US3 gene transcription continued even after viral DNA replication.
Effect of wild-type and mutant CRS with an upstream R1 silencer.
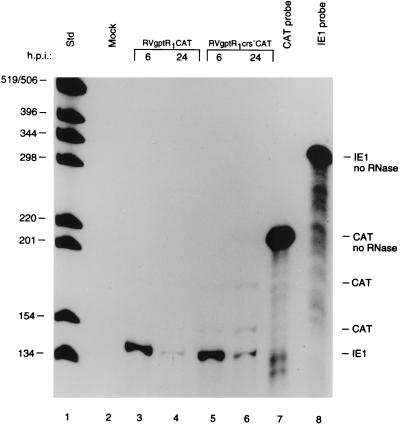
In transient transfection experiments, the R2 element is an enhancer and the R1 element is a silencer (6, 48). The effect of the R1 element alone on the US3 promoter in the context of the viral genome is not known. We isolated RVgptR1CAT and RVgptR1crs−CAT, which contain the wild-type and mutant CRS upstream of the US3 transcription start site, respectively. HFF cells were infected with approximately 5 PFU of the recombinant viruses per cell and analyzed for IE1 and CAT RNAs as described in Materials and Methods.
The steady-state level of IE1 RNA was high at 6 h and low at 24 h after infection, as expected (Fig. 4, lanes 3 and 4 and lanes 5 and 6, respectively). In the absence of the R2 element and in the presence of the R1 element, the level of CAT RNA from the US3 promoter containing the wild-type CRS was low. In contrast, the level of CAT RNA when the CRS was mutated was higher at 6 and 24 h after infection (Fig. 4; compare lanes 3 and 4 with lanes 5 and 6). In the absence of the R2 enhancer, the level of expression from the US3 promoter was significantly lower. However, the wild-type CRS still had a significant repressive effect on the US3 promoter.
FIG. 4.
Steady-state RNA levels transcribed from the US3 promoter with either the wild-type or mutant CRS downstream of the R1 silencer or the enhancer-containing MIE promoter at early and late times after infection (h.p.i. [hours postinfection]). HFF cells were infected with approximately 5 PFU of RVgptR1CAT or RVgptR1crs−CAT per cell. RNA samples were analyzed and designated as described in the legend to Fig. 3. Std, 32P-labeled DNA standard molecular weight markers, positions of which are indicated in nucleotides.
Cumulative effects on CAT gene expression downstream of the US3 promoter at various times after infection.
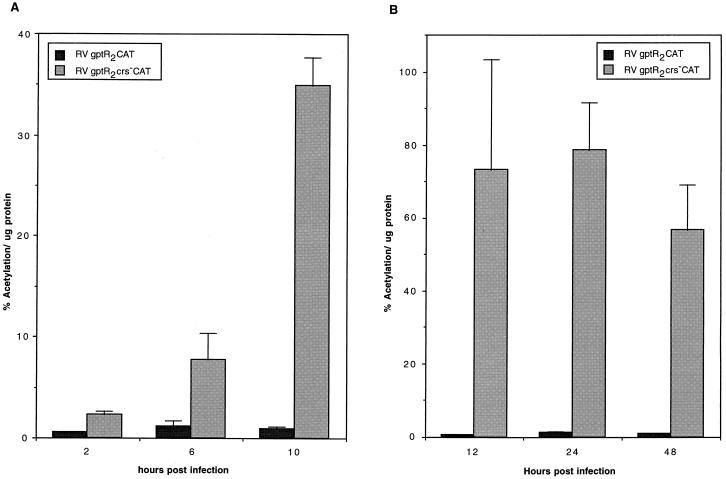
Since CAT RNA has a short half-life in the eucaryotic cell but the CAT enzyme is stable (12), we assayed CAT activity at various times after infection to determine the cumulative effect of a CRS mutation in the presence of the R2 enhancer or the R1 silencer. After infecting cells with approximately 5 PFU of RVgptR2CAT or RVgptR2crs−CAT per cell, we analyzed CAT activity at various times after infection as described in Materials and Methods.
When the R2 enhancer was present and the CRS was mutated, CAT gene product was detected within 2 h after infection and continued to increase at 6 and 10 h (Fig. 5A). When the wild-type CRS was present, CAT gene product was detectable but was maintained at a low level (Fig. 5A). Even at 12, 24, and 48 h after infection, the level of CAT activity was lower than in cells infected at the same MOI with a recombinant virus containing a mutated CRS (Fig. 5B). With recombinant viruses containing the mutated CRS and the upstream R2 enhancer, there was approximately a 40-fold increase in CAT between 2 and 24 h after infection. These data suggest that expression of the US3 gene product, which is involved in trapping major histocompatibility complex (MHC) class I molecules on the inner lumen of the endoplasmic reticulum, is rapidly induced after infection. The CRS immediately upstream of the US3 transcription start site functions to regulate the amount of US3 mRNA and type I glycoprotein gene product that is produced.
FIG. 5.
Effect of mutation of the CRS on cumulative CAT expression from the R2 enhancer-containing US3 promoter at various times after infection. HFF cells were infected with approximately 5 PFU/cell, and the amount of CAT activity per microgram of protein was determined at various times after infection as described in Materials and Methods. (A) CAT activity at early times after infection; (B) CAT activity at early and late times after infection.
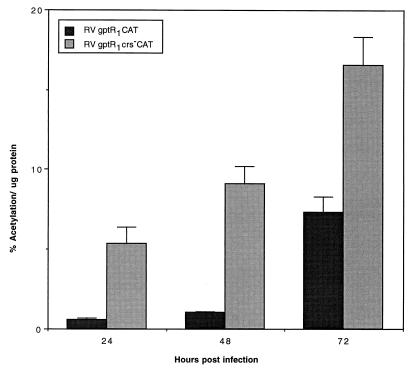
Since the cumulative effects of a CRS mutation resulted in a 40-fold difference in expression from the US3 promoter in the presence of the R2 enhancer, we tested the cumulative effect of a CRS mutation on the US3 promoter with only the R1 element upstream. After infection of HFF cells with approximately 5 PFU of RVgptR1CAT or RVgptR1crs−CAT per cell, the relative level of CAT gene product is low (Fig. 6). In the absence of the R2 enhancer and in the presence of the R1 silencer, CAT product is difficult to detect prior to 24 h. There is only a three- to sevenfold difference in the levels of CAT expression between 24 and 72 h after infection (Fig. 6). These data support the results of transient transfection experiments demonstrating that the R2 element functions as an enhancer (6, 48). Expression of CAT from the US3 promoter is higher when the CRS is mutated. Taken together, these data suggest that the R2 enhancer induces an early expression of the US3 type I glycoprotein and that the CRS, in turn, functions to regulate the level of type I glycoprotein expression.
FIG. 6.
Effect of mutation of the CRS on cumulative CAT expression from the R1 silencer-containing US3 promoter at various times after infection. HFF cells were infected with approximately 5 PFU of RVgptR1CAT or RVgptR1crs−CAT per ml, and the amount of CAT activity per microgram of protein was determined at various times after infection as described in Materials and Methods.
Effect of a CRS mutation in a nonpermissive cell type.
To test the role of the CRS in a nonpermissive cell type, we selected THP-1 cells. THP-1 is a monocytic cell line that is nonpermissive for HCMV replication unless stimulated and induced toward differentiation to macrophages. Since the R2 enhancer contains predominantly NF-κB/Rel cis-acting sites and is responsive to the p65 subunit of NF-κB (48), we tested the effect of THP-1 cell stimulation on US3 promoter activity with a wild-type or mutant CRS. Cell suspensions (6.8 × 106 cells/ml) were infected in parallel with approximately 3 PFU of either RVgptR2CAT or RVgptR2crs−CAT per cell for 1 h, washed, aliquoted, and then suspended in either medium or medium containing 100 ng of LPS per ml as described in Materials and Methods. In unstimulated cells infected with RVgptR2CAT, there was a very low level of CAT activity that did not increase significantly with time after infection (Fig. 7A). This activity is presumably due to the few cells in the culture that have differentiated spontaneously. With RVgptR2crs−CAT-infected THP-1 cells, CAT activity was detected at 12 h after infection and increased at 24 and 48 h (Fig. 7A). Without the wild-type CRS, there was less control over the viral US3 promoter. In contrast, in the cells infected with RVgptR2CAT or RVgptR2crs−CAT and then stimulated with LPS, higher levels of CAT activity were detected at 12 h after infection and increased with time after infection. The CAT activity with RVgptR2crs−CAT was approximately fivefold higher than that obtained with RVgptR2CAT at 24 and 48 h after infection (compare Fig. 7A and B). We conclude that the wild-type CRS element regulates the level of US3 promoter-directed gene expression in both the nonpermissive and the permissive and stimulated THP-1 cells.
FIG. 7.
Effect of a CRS mutation in a nonpermissive cell type. Undifferentiated THP-1 cells were infected with approximately 3 PFU of either RVgptR2CAT or RVgptR2crs−CAT per cell. After viral adsorption, the cells were washed in medium and suspended in medium with or without LPS. CAT activity per microgram of protein was determined at various times after infection as described in Materials and Methods. (A) THP-1 cells infected with RVgptR2CAT; (B) THP-1 cells infected with RVgptR2crs−CAT.
Effect of the viral IE86 protein.
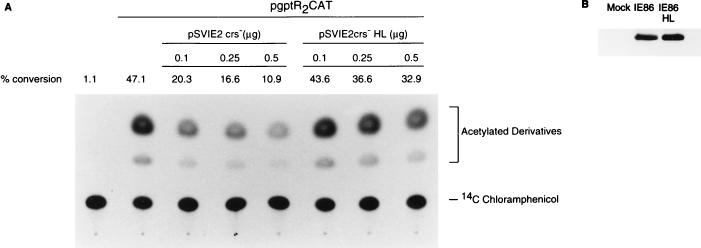
We and others have proposed that the IE2 gene product negatively autoregulates the MIE promoter by binding to the CRS and interfering with the transcription initiation complex (29, 54). Biegalke (4) proposed that the IE2 gene product was insufficient for repression of the US3 promoter. Using expression plasmids with the CRS mutated for high-level expression of the IE2 gene, we tested the effect of the IE86 protein or an IE86 protein mutated in the putative zinc finger motif (IE86HL) on CAT expression from plasmid pgptR2CAT in cotransfected 293-T cells as described in Materials and Methods. Under these conditions, there was a dose-dependent and repressive effect on the US3 promoter containing the wild-type CRS, while the mutant IE86HL protein had little effect. Western blot analysis determined that both the wild-type and mutant IE86 proteins were expressed efficiently in transfected 293-T cells (Fig. 8B).
FIG. 8.
CAT expression from the US3 promoter in 293-T cells cotransfected with a plasmid expressing either wild-type or mutant IE86. Cells were cotransfected by the calcium phosphate precipitation method and analyzed for CAT activity and for IE86 or IE86HL expression as described in Materials and Methods. (A) Autoradiogram of 14C-chloramphenicol and its acetylated derivatives; (B) Western blot of IE86 and IE86HL expression in 293-T cells.
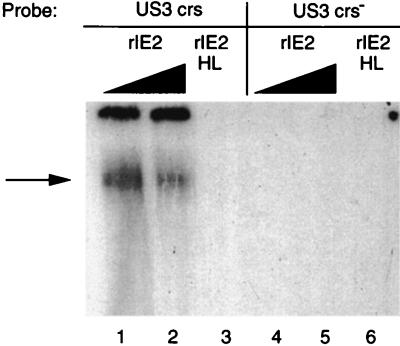
To determine whether the IE86 protein could act directly by binding to the CRS of the US3 promoter, like the MIE promoter, EMSAs were done with either wild-type or mutant IE2 gene product. One source of IE2 gene product was purified rIE2 protein where the maltose ORF was fused to the carboxyl half of the IE2 ORF at amino acid 290. A mutation in the putative zinc finger motif at histidine residues 446 and 452 of the IE2 ORF (rIE2HL) was also purified as a maltose fusion protein as described previously (28, 29). Figure 9 illustrates that rIE2 bound to the wild-type CRS of the US3 promoter, but rIE2HL failed to bind (Fig. 9; compare lanes 2 and 3 with lane 4). There was less binding of rIE2 to the wild-type US3 CRS with higher concentrations of protein due to aggregation of the rIE2. Both rIE2 and rIE2HL failed to bind to the mutant CRS (Fig. 9, lanes 5 to 7). For a positive control, rIE2 bound to the wild-type CRS of the MIE promoter as described previously (28, 29). We conclude that the viral IE86 protein can bind to the CRS of the US3 promoter and repress the US3 promoter. However, determination of whether this occurs in HCMV-infected cells will require a null mutation in the IE2 gene.
FIG. 9.
Binding of the IE2 gene product to the CRS of the US3 promoter. Wild-type (rIE2) and mutant (rIE2HL) maltose-IE2 fusion proteins were purified as described previously (28, 29) and used for EMSA as described in Materials and Methods. Lanes: 1, wild-type CRS plus 2.9 pmol of rIE2; 2, wild-type CRS plus 5.8 pmol of rIE2; 3, wild-type CRS plus 5.8 pmol of rIE2HL; 4, mutant CRS plus 2.9 pmol of rIE2; 5, mutant CRS plus 5.8 pmol of rIE2; 6, mutant CRS plus 5.8 pmol of rIE2HL.
DISCUSSION
After primary infection or reactivation from latency, the enhancer-containing US3 promoter is significantly activated. The US3 promoter, located in the small unique component of the viral genome, drives the expression of a type I glycoprotein thought to be involved in the early stages of escape from immune surveillance (21). The immediate and strong expression of the US3 viral glycoprotein is assumed to be important for the survival of the virus in the host. Strong enhancer-mediated activation of downstream IE gene expression followed by stringent repression of expression is a pattern that appears common to the MIE and US3 promoters. There are some sequence similarities between the CRSs of the two promoters, but whether they are functionally identical requires further investigation.
The US3 gene product prevents viral peptide presentation to the cell surface by class I MHC molecules (21). Overexpression of this viral gene product may be cytotoxic because it interacts with the MHC class I molecule and remains in the inner lumen of the endoplasmic reticulum. This may interfere with the normal functioning of the endoplasmic reticulum and the proper processing of early and late viral glycoproteins as well as cellular glycoproteins. Therefore, it is possible that overexpression of the US3 glycoprotein is cytotoxic.
The CRS associated with the MIE and US3 promoters has a critical role in the regulation of HCMV IE gene expression. The repression of these viral promoters at approximately 6 h after infection suggests that a protein is made de novo and accumulates in sufficient quantity to repress these strong enhancer-containing promoters. In the presence of an inhibitor of de novo protein synthesis, the viral promoters are not repressed (9, 44, 46, 51). In RNase protection assays, the level of RNA from the US3 promoter containing the wild-type CRS peaked at between 4 and 6 h after infection and then declined. In the context of the viral genome, the wild-type CRS had a very significant effect on controlling gene expression from the US3 promoter at early and late times after infection. There was a 20- to 40-fold difference in the levels of expression from the US3 promoter containing the wild-type CRS versus the mutant CRS.
Transient transfection experiments demonstrated that the CRS of the MIE promoter contributes to promoter repression and that the viral IE86 protein is involved in repression (7, 26, 35). The IE86 protein binds to the CRS on the MIE promoter that overlaps an adjacent binding site for a cellular protein of 150 kDa (28). This cellular protein may be similar to the human cellular initiation factor designated CIF150 and described by Kaufmann et al. (23). Binding of the IE86 protein to the CRS does not interfere with binding of the TATA-binding protein to the TATA box, but it does interfere with the initiation of transcription (22, 29, 54). The structural context of the DNA template around the CRS and the Inr plays a critical role in determining promoter activity. Therefore, the role of the CRS in either the MIE or the US3 promoter may be to block the assembly of the transcription initiation complex, which ultimately blocks the engagement of RNA polymerase II. Determination of whether the viral IE86 protein can independently regulate these enhancer-containing promoters during the viral replication cycle will require a recombinant HCMV with a IE2 gene deletion. The repressive effect of the wild-type IE86 protein on the CRS of the US3 promoter in transient transfection experiments and the binding of the wild-type rIE2 fusion protein to the wild-type CRS suggest that this viral promoter may also be negatively regulated by the IE86 protein. These data underscore what is functionally possible for the IE86 protein, but they do not necessarily reflect the function of the viral protein in the infected cell.
Control of transcription of the HCMV enhancer-containing MIE and US3 promoters at the transcription start site is different from that reported for papovaviruses. The SV40 large T antigen and the bovine papillomavirus E2 protein are thought to interfere with the formation of the preinitiation complex on DNA by preventing TFIID binding to the TATA box (10, 16, 49). Control of transcription from the HCMV enhancer-containing MIE promoter and possibly US3 promoter appears similar to that for the Drosophila alcohol dehydrogenase proximal promoter. In both systems, a zinc finger protein binds to the initiator region (29, 36). A CRS is critical for repressor binding, and when it is mutated, there is a high level of downstream expression. The AEF-1 repressor protein of Drosophila binds between positions −5 and +10 relative to the transcription start site, while the IE86 protein of HCMV binds between positions −15 and +2 of the MIE promoter. In vivo footprinting indicates that a protein(s) can bind to the CRS of the US3 promoter in HCMV-infected cells (3).
It was striking that so little CAT accumulated during the viral replication cycle when the US3 promoter contained the wild-type CRS. These results may explain, in part, why it is so difficult to detect the US3 type I glycoprotein in HCMV-infected cells (5a). The enhancer upstream of the US3 promoter has five consensus NF-κB/Rel binding sites and responds to the p65 subunit of NF-κB (48). When the R2 enhancer element is replaced by the R1 element, levels of both RNA and CAT expression from the US3 promoter are greatly reduced. The R1 element in the presence of the R2 element has a repressive effect on the US3 promoter in transient transfection experiments (6, 48). The role of the R1 element in the context of the viral genome is presently being investigated.
In the absence of stimulation and differentiation of monocytic THP-1 cells, there was little expression downstream of the US3 promoter containing the wild-type CRS. Expression downstream of the US3 promoter with the mutant CRS was approximately fivefold higher. As expected, expression from the US3 promoter containing the mutant CRS was higher when the THP-1 cells were stimulated with LPS. At the MOI used, there should be approximately 0.5 to 1.0 viral genome equivalent per nucleus (31). In similar experiments, the IE1 RNA from the MIE promoter was also not detectable in the undifferentiated THP-1 cells. Only low levels of the IE1 RNA were detected after treatment with cycloheximide. In contrast, significant levels of IE1 RNA were detected in the differentiated THP-1 cells (31). There was approximately a 12-fold difference in the levels of expression of CAT per microgram of protein in the HFF cells versus stimulated THP-1 cells. These results may reflect a slower entry and import of the viral chromosome to the nucleus of THP-1 cells, or the viral chromatin may affect transcription from the HCMV genome in THP-1 cells. There was a long delay between stimulation and expression from the US3 promoter. Since NF-κB is mobilized to the nucleus quickly after stimulation (38) and both the MIE and US3 promoters have NF-κB-responsive elements, these data suggest that other viral or cellular factors may suppress expression from the MIE or US3 promoter. Similar events may be associated with reactivation of HCMV from latency in blood monocytes. The appearance of infectious virus in monocyte-derived macrophages after allogeneic and cytokine stimulation of blood monocytes requires weeks (39, 40). An understanding of the factors that control transcription from the HCMV genome during latency and after reactivation from latency should contribute to our understanding of HCMV-induced pathogenesis in the host.
ACKNOWLEDGMENTS
We are grateful to Jeffrey Meier and Marty Stoltzfus for critical reading of the manuscript.
This work was supported by grant AI-13562 from the National Institutes of Health.
REFERENCES
- 1.Alford C A, Britt W J. Cytomegalovirus. In: Fields B N, Knipe D M, et al., editors. Virology. New York, N.Y: Raven Press, Ltd.; 1990. pp. 1981–2010. [Google Scholar]
- 2.Anders D G, McCue L A. The human cytomegalovirus genes and proteins required for DNA synthesis. Intervirology. 1996;39:378–388. doi: 10.1159/000150508. [DOI] [PubMed] [Google Scholar]
- 3.Biegalke B J. Characterization of the transcriptional repressive element of the human cytomegalovirus immediate-early US3 gene. J Virol. 1998;72:5457–5463. doi: 10.1128/jvi.72.7.5457-5463.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Biegalke B J. IE2 protein is insufficient for transcriptional repression of the human cytomegalovirus US3 promoter. J Virol. 1997;71:8056–8060. doi: 10.1128/jvi.71.10.8056-8060.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Biegalke B T. Regulation of human cytomegalovirus US3 gene transcription by a cis-repressive sequence. J Virol. 1995;69:5362–5367. doi: 10.1128/jvi.69.9.5362-5367.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5a.Bissell, J., and M. F. Stinski. Unpublished data.
- 6.Chan Y-J, Tseng W-P, Hayward G S. Two distinct upstream regulatory domains containing multicopy cellular transcription factor binding sites provide basal repression and inducible enhancer characteristics to the immediate-early IES (US3) promoter from human cytomegalovirus. J Virol. 1996;70:5312–5328. doi: 10.1128/jvi.70.8.5312-5328.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cherrington J M, Khoury E L, Mocarski E S. Human cytomegalovirus IE2 negatively regulates α gene expression via a short target sequence near the transcription start site. J Virol. 1991;65:887–896. doi: 10.1128/jvi.65.2.887-896.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cherrington J M, Mocarski E S. Human cytomegalovirus IE1 transactivates the α promoter-enhancer via an 18-base-pair repeat element. J Virol. 1989;63:1435–1440. doi: 10.1128/jvi.63.3.1435-1440.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Colberg-Poley A M. Functional roles of immediate early proteins encoded by the human cytomegalovirus UL36-38, UL115-119, TRS1/IRS1 and US3 loci. Intervirology. 1996;39:350–360. doi: 10.1159/000150506. [DOI] [PubMed] [Google Scholar]
- 10.Dostatni N, Lambert P F, Sousa R, Ham J, Howley P M, Yaniv M. The functional BPV-1 E2 trans-activating protein can act as a repressor by preventing formation of the initiation complex. Genes Dev. 1991;5:1651–1671. doi: 10.1101/gad.5.9.1657. [DOI] [PubMed] [Google Scholar]
- 11.Gebert S, Schmolke S, Sorg G, Floss S, Plachter B, Stamminger T. The UL84 protein of human cytomegalovirus acts as a transdominant inhibitor of immediate-early-mediated transactivation that is able to prevent viral replication. J Virol. 1997;71:7048–7060. doi: 10.1128/jvi.71.9.7048-7060.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gorman C M, Moffatt L F, Howard B H. Recombinant genomes which express chloramphenicol acetyltransferase in mammalian cells. Mol Cell Biol. 1982;2:1044–1051. doi: 10.1128/mcb.2.9.1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Graham F L, van der Eb A J. A new technique for the assay of infectivity of adenovirus 5 DNA. Virology. 1973;52:456–467. doi: 10.1016/0042-6822(73)90341-3. [DOI] [PubMed] [Google Scholar]
- 14.Greaves R F, Brown J M, Vieira J, Mocarski E S. Selectable insertion and deletion mutagenesis of the human cytomegalovirus genome using the E. coli guanosine phosphoribosyl transferase (gpt) gene. J Gen Virol. 1995;76:2151–2160. doi: 10.1099/0022-1317-76-9-2151. [DOI] [PubMed] [Google Scholar]
- 15.Greaves R F, Mocarski E S. Defective growth correlates with reduced accumulation of viral DNA replication protein after low-multiplicity infection by a human cytomegalovirus ie1 mutant. J Virol. 1998;72:366–379. doi: 10.1128/jvi.72.1.366-379.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hansen U, Tenen D G, Livingston D M, Sharp P A. T antigen repression of SV 40 early transcription form two promoters. Cell. 1981;27:603–613. doi: 10.1016/0092-8674(81)90402-5. [DOI] [PubMed] [Google Scholar]
- 17.Hermiston T W, Malone C L, Stinski M F. Human cytomegalovirus immediate-early two-protein region involved in negative regulation of the major immediate-early promoter. J Virol. 1990;64:3532–3536. doi: 10.1128/jvi.64.7.3532-3536.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hermiston T W, Malone C L, Witte P R, Stinski M F. Identification and characterization of the human cytomegalovirus immediate-early region 2 gene that stimulates gene expression from an inducible promoter. J Virol. 1987;61:3214–3221. doi: 10.1128/jvi.61.10.3214-3221.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ho M. Cytomegalovirus: biology and infection. New York, N.Y: Plenum Publishing Corp.; 1991. [Google Scholar]
- 20.Hunninghake G W, Monks B G, Geist L J, Monick M M, Monroy M A, Stinski M F, Webb A C, Dayer J M, Auron P E, Fenton M J. The functional importance of a cap site-proximal region of the human prointerleukin 1B gene is defined by viral protein transactivation. Mol Cell Biol. 1992;12:3439–3448. doi: 10.1128/mcb.12.8.3439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jones T R, Wiertz E J H J, Sun L, Fish K N, Nelson J A. Human cytomegalovirus US3 impairs transport and maturation of major histocompatibility complex class I heavy chains. Proc Natl Acad Sci USA. 1996;93:11327–11333. doi: 10.1073/pnas.93.21.11327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jupp R, Hoffmann S, Depto A, Stenberg R M, Ghazal P, Nelson J. Direct interaction of the human cytomegalovirus IE86 protein with the cis repression signal does not preclude TBP from binding to the TATA box. J Virol. 1993;67:5595–5604. doi: 10.1128/jvi.67.9.5595-5604.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kaufmann J, Ahrens K, Koop R, Smale S T, Muller R. CIF150, a human cofactor for transcription factor IID-dependent initiator function. Mol Cell Biol. 1998;18:233–239. doi: 10.1128/mcb.18.1.233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kondo K, Kaneshima H, Mocarski E S. Human cytomegalovirus latent infection of granulocyte-macrophage progenitors. Proc Natl Acad Sci USA. 1994;91:11879–11883. doi: 10.1073/pnas.91.25.11879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Krieg P A, Melton D A. In vitro RNA synthesis with SP6 RNA polymerase. Methods Enzymol. 1987;155:397–414. doi: 10.1016/0076-6879(87)55027-3. [DOI] [PubMed] [Google Scholar]
- 26.Liu B, Hermiston T W, Stinski M F. A cis-acting element in the major immediate-early (IE) promoter of human cytomegalovirus is required for negative regulation by IE2. J Virol. 1991;65:897–903. doi: 10.1128/jvi.65.2.897-903.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu B, Stinski M F. Human cytomegalovirus contains a tegument protein that enhances transcription from promoters with upstream ATF and AP-1 cis-acting elements. J Virol. 1992;66:4434–4444. doi: 10.1128/jvi.66.7.4434-4444.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Macias M P, Huang L, Lashmit P E, Stinski M F. Cellular and viral protein binding to a cytomegalovirus promoter transcription initiation site: effects on transcription. J Virol. 1996;70:3628–3635. doi: 10.1128/jvi.70.6.3628-3635.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Macias M P, Stinski M F. An in vitro system for human cytomegalovirus immediate early 2 protein (IE2)-mediated site-dependent repression of transcription and direct binding of IE2 to the major immediate early promoter. Proc Natl Acad Sci USA. 1993;90:707–711. doi: 10.1073/pnas.90.2.707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Malone C L, Vesole D H, Stinski M F. Transactivation of a human cytomegalovirus early promoter by gene products from the immediate-early gene IE2 and augmentation by IE1: mutational analysis of the viral proteins. J Virol. 1990;64:1498–1506. doi: 10.1128/jvi.64.4.1498-1506.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Meier J L, Stinski M F. Effect of a modulator deletion on transcription of the human cytomegalovirus major immediate-early genes in infected undifferentiated and differentiated cells. J Virol. 1997;71:1246–1255. doi: 10.1128/jvi.71.2.1246-1255.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Meier J L, Stinski M F. Regulation of human cytomegalovirus immediate-early gene expression. Intervirology. 1996;39:331–342. doi: 10.1159/000150504. [DOI] [PubMed] [Google Scholar]
- 33.Mocarski E S, Kemble G, Lyle J, Greaves R F. A deletion mutant in the human cytomegalovirus gene encoding IE1 491aa is replication defective due to a failure in autoregulation. Proc Natl Acad Sci USA. 1996;93:11321–11326. doi: 10.1073/pnas.93.21.11321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pajovic S, Wong E L, Black A R, Azizkhan J C. Identification of a viral kinase that phosphorylates specific E2Fs and pocket proteins. Mol Cell Biol. 1997;17:6459–6464. doi: 10.1128/mcb.17.11.6459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pizzorno M C, Hayward G S. The IE2 gene products of human cytomegalovirus specifically down-regulate expression from the major immediate-early promoter through a target located near the cap site. J Virol. 1990;64:6154–6165. doi: 10.1128/jvi.64.12.6154-6165.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ren B, Maniatis T. Regulation of Drosophila Adh promoter switching by an initiator-targeted repression mechanism. EMBO J. 1998;17:1076–1086. doi: 10.1093/emboj/17.4.1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Samaniego L A, Tevethia M J, Spector D J. The human cytomegalovirus 86-kilodalton immediate-early 2 protein: synthesis as a precursor polypeptide and interaction with a 75-kilodalton protein of probable viral origin. J Virol. 1994;68:720–729. doi: 10.1128/jvi.68.2.720-729.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sen R, Baltimore D. Inducibility of κ immunoglobulin enhancer-binding protein NF-κB by a posttranslational mechanism. Cell. 1986;47:921–928. doi: 10.1016/0092-8674(86)90807-x. [DOI] [PubMed] [Google Scholar]
- 39.Soderberg-Naucler C, Fish K N, Nelson J A. Interferon-gamma and tumor necrosis factor-alpha specifically induce formation of cytomegalovirus-permissive monocyte-derived macrophages that are refractory to the antiviral activity of these cytokines. J Clin Investig. 1997;100:3154–3163. doi: 10.1172/JCI119871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Soderberg-Naucler C, Fish K N, Nelson J A. Reactivation of latent human cytomegalovirus by allogeneic stimulation of blood cells from healthy donors. Cell. 1997;91:119–126. doi: 10.1016/s0092-8674(01)80014-3. [DOI] [PubMed] [Google Scholar]
- 41.Spector D J, Tevethia M J. Protein-protein interactions between human cytomegalovirus IE2-580aa and pUL84 in lytically infected cells. J Virol. 1994;68:7549–7553. doi: 10.1128/jvi.68.11.7549-7553.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Stenberg R M. The human cytomegalovirus major immediate-early gene. Intervirology. 1996;39:343–349. doi: 10.1159/000150505. [DOI] [PubMed] [Google Scholar]
- 43.Stenberg R M, Stinski M F. Autoregulation of the human cytomegalovirus major immediate-early gene. J Virol. 1985;56:676–682. doi: 10.1128/jvi.56.3.676-682.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Stenberg R M, Thomsen D R, Stinski M F. Structural analysis of the major immediate-early gene of human cytomegalovirus. J Virol. 1984;49:190–199. doi: 10.1128/jvi.49.1.190-199.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Stinski M F. Synthesis of proteins and glycoproteins in cells infected with human cytomegalovirus. J Virol. 1977;23:751–767. doi: 10.1128/jvi.23.3.751-767.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Stinski M F, Thomsen D R, Stenberg R M, Goldstein L M. Organization and expression of the immediate-early genes of human cytomegalovirus. J Virol. 1983;46:1–14. doi: 10.1128/jvi.46.1.1-14.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Taylor-Wiedeman J, Sissons J G, Borysiewicz L K, Sinclair J H. Monocytes are a major site of persistence of human cytomegalovirus in peripheral blood mononuclear cells. J Gen Virol. 1991;72:2059–2064. doi: 10.1099/0022-1317-72-9-2059. [DOI] [PubMed] [Google Scholar]
- 48.Thrower A R, Bullock G C, Bissell J E, Stinski M F. Regulation of a human cytomegalovirus immediate early gene (US3) by a silencer/enhancer combination. J Virol. 1996;70:91–100. doi: 10.1128/jvi.70.1.91-100.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tjian R. T antigen binding and the control of SV 40 gene expression. Cell. 1981;26:1–2. doi: 10.1016/0092-8674(81)90026-x. [DOI] [PubMed] [Google Scholar]
- 50.Wang, Y., and M. F. Stinski. Unpublished data.
- 51.Wathen M W, Stinski M F. Temporal patterns of human cytomegalovirus transcription: mapping the viral RNAs synthesized at immediate-early, early, and late times after infection. J Virol. 1982;41:462–477. doi: 10.1128/jvi.41.2.462-477.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Weston K. An enhancer element in the short unique region of human cytomegalovirus regulates the production of a group of abundant immediate early transcripts. Virology. 1988;162:406–416. doi: 10.1016/0042-6822(88)90481-3. [DOI] [PubMed] [Google Scholar]
- 53.Winkler M, Rice S A, Stamminger T. UL69 of human cytomegalovirus, an open reading frame with homology to ICP27 of herpes simplex virus, encodes a transactivator of gene expression. J Virol. 1994;68:3943–3954. doi: 10.1128/jvi.68.6.3943-3954.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wu J, Jupp R, Stenberg R M, Nelson J A, Ghazal P. Site-specific inhibition of RNA polymerase II preinitiation complex assembly by human cytomegalovirus IE86 protein. J Virol. 1993;67:7547–7555. doi: 10.1128/jvi.67.12.7547-7555.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yeung K C, Stoltzfus C M, Stinski M F. Mutations of the cytomegalovirus immediate-early 2 protein defines regions and amino acid motifs important in transactivation of transcription from the HIV-1 LTR promoter. Virology. 1993;195:786–792. doi: 10.1006/viro.1993.1431. [DOI] [PubMed] [Google Scholar]
- 56.Yurochko A D, Hwang E-S, Rasmussen L, Keay S, Pereira L, Huang E-S. The human cytomegalovirus UL55 (gB) and UL75 (gH) glycoprotein ligands initiate the rapid activation of Sp1 and NF-κB during infection. J Virol. 1997;71:5051–5059. doi: 10.1128/jvi.71.7.5051-5059.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]