Abstract
Most equine infectious anemia virus (EIAV)-infected horses have acute clinical disease, but they eventually control the disease and become lifelong carriers. Cytotoxic T lymphocytes (CTL) are considered an important immune component in the control of infections with lentiviruses including EIAV, but definitive evidence for CTL in the control of disease in carrier horses is lacking. By using retroviral vector-transduced target cells expressing different Gag proteins and overlapping synthetic peptides of 16 to 25 amino acids, peptides containing at least 12 Gag CTL epitopes recognized by virus-stimulated PBMC from six long-term EIAV-infected horses were identified. All identified peptides were located within Gag matrix (p15) and capsid (p26) proteins, as no killing of target cells expressing p11 and p9 occurred. Each of the six horses had CTL recognizing at least one Gag epitope, while CTL from one horse recognized at least eight different Gag epitopes. None of the identified peptides were recognized by CTL from all six horses. Two nonamer peptide epitopes were defined from Gag p26; one (18a) was likely restricted by class I equine leukocyte alloantigen A5.1 (ELA-A5.1) molecules, and the other (28b-1) was likely restricted by ELA-A9 molecules. Sensitization of equine kidney target cells for CTLm killing required 10 nM peptide 18a and 1 nM 28b-1. The results demonstrated that diverse CTL responses against Gag epitopes were generated in long-term EIAV-infected horses and indicated that ELA-A class I molecules were responsible for the diversity of CTL epitopes recognized. This information indicates that multiple epitopes or whole proteins will be needed to induce CTL in horses with different ELA-A alleles in order to evaluate their role in controlling EIAV.
Equine infectious anemia virus (EIAV) belongs to the Lentivirus genus, which includes human immunodeficiency virus type 1 (HIV-1), simian immunodeficiency virus (SIV), and several other animal viruses. EIAV causes disease in horses which is characterized by recurrent febrile episodes associated with viremia, anemia, and thrombocytopenia (10). Most infected horses are able to eventually control the disease and become lifelong EIAV carriers (9). The ability of horses to restrict EIAV replication to very low levels and to remain free of clinical disease provides an opportunity to determine the immunologic mechanisms involved in this lentivirus control.
Immune responses are required for the termination of the acute viremia during EIAV infection since foals with severe combined immunodeficiency cannot control the initial viremia following EIAV infection, in contrast to normal foals (41). Results suggesting that immune responses are involved in the control of EIAV in carrier horses include the observation that corticosteroid- and cyclophosphamide-treated carrier horses have recurrent viremia and disease (24). Neutralizing antibody can be an important component of the protective immune response against lentiviral infections (12). Type-specific neutralizing antibody appears following the episodes of plasma viremia in EIAV-infected horses (25); however, there is evidence suggesting that the presence of the neutralizing antibody does not necessarily relate to the occurrence and control of viremic episodes (8, 25). Detectable neutralizing antibodies to the variant isolated during a disease episode can appear after the episode is controlled (8). Neutralizing antibody-escape variants are isolated from EIAV carrier horses as early as 5 days after corticosteroid treatment, when the antibody levels have not significantly changed (24). Further, the viremic episode induced by corticosteroid treatment can be terminated before the appearance of neutralizing antibody to the variant causing viremia (24). Other evidence implicating immune responses other than neutralizing antibody in EIAV control includes the following: (i) EIAV carrier horses can resist challenge with a heterologous strain in the absence of detectable neutralizing antibody to the challenge virus (23), and (ii) some horses immunized with an inactivated virus vaccine resist homologous strain challenge without detectable levels of neutralizing antibody but with virus-specific cell-mediated immune responses (17).
Accumulating evidence suggests that major histocompatibility complex (MHC) class I-restricted virus-specific cytotoxic T lymphocytes (CTL) may play an important role in the immune control of diseases caused by HIV-1 and SIV infection (5, 26, 51). CTL appear to be involved in both the clearance of the primary viremia in HIV-1 infection (26) and the prevention of disease progression to AIDS (42). In EIAV infection, the appearance of activated CD8+ CTL (effectors) correlated with the control of the initial viremic episodes (33). Although the CTL effectors decline to low levels when plasma viremias become undetectable, a high frequency of memory CTL (CTLm) has been detected in some carrier horses (34), and these CTLm recognize either EIAV Env or Gag/Pr proteins or both (15, 34). Both CD8+ and CD4+ CTL activities have been detected in some EIAV-infected horses (15), but their roles in disease control are not known.
The epitopes recognized by CD8+ CTL are usually peptides of 8 to 11 amino acids (aa) presented by MHC class I molecules on the target cell surface. Identifying the CTL epitopes and the MHC class I molecules that restrict responses is necessary in order to determine how CTL are involved in the control of disease and to stimulate CTL by vaccination. However, the occurrence of escape mutants which are no longer recognized by CTL is one of the major difficulties for inducing effective CTL responses against different variants (6). Gag protein epitopes recognized by CTL may be of importance because Gag proteins are relatively conserved among EIAV strains (21, 32, 40, 48). In this study, at least 12 peptides with CTL epitopes were recognized by stimulated peripheral blood mononuclear cells (PBMC) from six long-term EIAV-infected horses with different ELA-A alleles. These peptides were identified by using retroviral vectors expressing individual Gag proteins and synthetic overlapping peptides from recognized proteins. We identified two nonamer peptides, one apparently restricted by ELA-A5.1, and another by ELA-A9, molecules.
MATERIALS AND METHODS
Animals.
Six mixed-breed ponies (horses) experimentally infected with EIAVWSU5 were used as sources of PBMC. The derivation of EIAVWSU5 and the infection of five of these horses were previously described (33, 34). A sixth horse, H513, was similarly infected with 108 50% tissue culture infective doses (TCID50) of EIAVWSU5. At the time of this study, H507 had been infected for 5 years, H521 had been infected for 4 years, H529, H532, and H540 had been infected for 3 years, and H513 had been infected for 1.5 years.
Cell lines.
Equine kidney (EK) cells for CTL targets were expanded from kidney biopsies of the six horses obtained before EIAV infection (33). Other cell lines used in this study included PA317 amphotropic retroviral packaging cells (ATCC CRL 9078) and PG13 packaging cells (ATCC CRL 10686). All cell lines were maintained in Dulbecco modified Eagle medium (DMEM; Gibco BRL, Grand Island, N.Y.) containing 5% fetal bovine serum (FBS).
ELA-A typing.
PBMC were typed for ELA-A molecules by microcytotoxicity assays using described antibody reagents (2, 27). The ELA-A locus, the only well-defined MHC class I locus in the horse, contains 17 internationally accepted alleles (A1 to A10, A14, A15, A19, W16, W17, W18, and W20) (31).
Construction of retroviral vectors.
The EIAVWSU5 gag gene was amplified from pEIA5G (32) into five DNA segments by PCR using specific primers (Table 1). These segments encoded p15, the N-terminal half of p26 (p26a), the C-terminal half of p26 (p26b), p11, and p9. The optimal primers for cloning resulted in overlaps of 12 bp of the p26a with p26b segments and overlaps of 6 bp on both ends of the p11 segment with the p26b and p9 gene segments. EcoRI and XhoI restriction enzyme sites designed in the forward and backward primers, respectively, were used for inserting the segments into retroviral vector plasmid pLXSN (provided by A. Dusty Miller, Fred Hutchinson Cancer Center, Seattle, Wash.), using previously described procedures (30). pLXSN contains the neomycin phosphoribosyltransferase gene under control of the simian virus 40 early gene promoter. Expression of the gag gene segments was driven by the Moloney murine sarcoma virus long terminal repeat promoter (35). The orientation of the inserts was determined by restriction enzyme analysis, and the nucleotide sequences were determined. Transfection of the retroviral packaging PA317 cells with the vector plasmids with or without the gag gene segments was accomplished by using LipofectAMINE (Gibco BRL). Forty-eight hours after transfection, the supernatants were collected, filtered through a 0.45-μm-pore-size syringe filter, and used to transduce the PG13 packaging cell line, which incorporates gibbon ape leukemia virus Env into vector virion membranes for cell entry with a wide host range (35). The transduced PG13 cells were selected with medium containing 700 μg of G418 sulfate (Calbiochem, La Jolla, Calif.) per ml. Regular medium (DMEM with 10% FBS) was used on the resulting G418-resistant PG13 cell line for collecting supernatant containing vector virus. The supernatant was titrated on EK cells, and the titer ranged from 1 × to 6 × 105 transducing particles/ml.
TABLE 1.
Primers used for PCR, RT-PCR, and sequencing
| Primera | Sequence |
|---|---|
| p15 | |
| F | GCGCGAATTCAAGATGGGAGACCCTTTGACA |
| B | CGCGCTCGAGTTAATATTCTTCAGAGGGCTCAGA |
| p26a | |
| F | GCGCGAATTCAAGATGGCACCAATCATGATAGATGGG |
| B | CGCGCTCGAGTTAAGCAGGCTCCATCTGTCTTTC |
| p26b | |
| F | GCGCGAATTCAAGATGGAGCCTGCTTTTGATCAG |
| B | CGCGCTCGAGTTAAAGTGCTTTTGCCAATAACAT |
| p11 | |
| F | GCGCGAATTCAACATGGCACTTCAGACTGGTCTTGC |
| B | GCGCCTCGAGTTATATCGGGAAAGTTTGTTTCTG |
| p9 | |
| F | GCGCGAATTCAAGATGGGACCGATACAACAGAAGAGTCAG |
| B | GCGCCTCGAGCATTACTCCCACAAACTGTCCAG |
F, forward; B, backward.
Synthetic peptides.
Peptides 16 to 25 aa in length and overlapping by 8 aa corresponding to p15 and p26 (Table 2) were synthesized by the Laboratory for Biotechnology and Bioanalysis I, Washington State University, Pullman. Additional peptides (9 to 12 aa) were synthesized for fine mapping of two CTL epitopes. High-pressure liquid chromatography analysis indicated that the 9- to 12-aa peptides were >95% pure. The peptides were dissolved at 2 mg/ml in sterile phosphate-buffered saline with 10% dimethyl sulfoxide before dilution with medium and pulsing of EK target cells.
TABLE 2.
Gag p15 and p26 overlapping synthetic peptidesa
| Gag protein | Peptide no. | Position in Gag | Sequenceb |
|---|---|---|---|
| p15 | 1 | 1–20 | MGDPLTWSKALKKLEKVTVQ |
| 2 | 13–32 | KLEKVTVQGSQKLTTGNCNW | |
| 3 | 25–44 | LTTGNCNWALSLVDLFHDTN | |
| 4 | 37–56 | VDLFHDTNFVKEKDWQLRDV | |
| 5 | 49–68 | KDWQLRDVIPLLEDVTQTLS | |
| 6 | 61–80 | EDVTQTLSGQEREAFERTWW | |
| 7 | 73–92 | EAFERTWWAISAVKMGLQIN | |
| 8 | 85–104 | VKMGLQINNVVDGKASFQLL | |
| 9 | 97–116 | GKASFQLLRAKYEKKTANKK | |
| 10 | 109–124 | EKKTANKKQSEPSEEY | |
| p26a | 11 | 125–144 | PIMIDGAGNRNFRPLTPRGY |
| 12 | 137–156 | RPLTPRGYTTWVNTIQTNGL | |
| 13 | 149–168 | NTIQTNGLLNEASQNLFGIL | |
| 14 | 161–180 | SQNLFGILSVDCTSEEMNAF | |
| 15 | 173–192 | TSEEMNAFLDVVPGQAGQKQ | |
| 16 | 185–204 | PGQAGQKQILLDAIDKIADD | |
| 17 | 197–216 | AIDKIADDWDNRHPLPNAPL | |
| 18 | 209–228 | HPLPNAPLVAPPQGPIPMTA | |
| 19 | 221–245 | QGPIPMTARFIRGLGVPRERQMEPA | |
| p26b | 20 | 242–261 | MEPAFDQFRQTYRQWIIEAM |
| 21 | 254–273 | RQWIIEAMSEGIKVMIGKPK | |
| 22 | 266–285 | KVMIGKPKAQNIRQGAKEPY | |
| 23 | 278–297 | RQGAKEPYPEFVDRLLSQIK | |
| 24 | 290–309 | DRLLSQIFSEGHPQEISKFL | |
| 25 | 302–321 | PQEISKFLTDTLTIQNANEE | |
| 26 | 314–333 | TIQNANEECRNAMRHLRPED | |
| 27 | 326–345 | MRHLRPEDTLEEKMYACRDI | |
| 28 | 338–359 | KMYACRDIGTTKQKMMLLAKAL |
Peptides within p15, p26a, and p26b were overlapped by 8 aa.
Derived from EIAVWSU5. Sequences unique to the various peptides are underlined.
Generation of CTLm.
PBMC were separated from the blood of EIAV-infected horses by layering leukocyte-rich plasma over Histopaque and centrifuging. To generate CTLm, 106 PBMC were cultured with 106 irradiated, EIAV-infected or peptide-pulsed autologous PBMC in each well of a 24-well plate with 1 ml of DMEM containing 10% FBS and 20 mM HEPES. The stimulator PBMC were prepared by irradiating 108 autologous PBMC with 3 krad of gamma irradiation, mixing with 5 ml of 106 TCID50 of EIAVWSU5 per ml or various peptides at 20 μg/ml and incubated at 37°C for 1 h. After 7 days, 5 × 105 viable cells were restimulated with 106 stimulator cells in 1 ml of medium/well containing 10 U of recombinant human interleukin-2 (Gibco BRL) per ml. After 4 days, the restimulated PBMC were split from one well to two wells with fresh medium and recombinant human interleukin-2. The cells were further expanded for another 3 to 4 days before use as effector cells in CTL assays (34).
Cytotoxicity assay.
EK cells were transduced with retroviral vectors with or without the gag gene segments in DMEM containing 10% FBS for 24 h at a multiplicity of infection of 2 to 5. The transduced EK cells were then placed under selection in DMEM containing 700 μg of G418 per ml for at least 1 week before the CTL assay. EK target cells (3 × 104/well) were incubated in collagen-coated wells of 96-well plates at 37°C with 5% CO2 for 24 h before 51Cr labeling. Peptide-sensitized EK target cells were prepared by pulsing the normal EK cells with synthetic peptides at concentrations indicated in the figure legends for 2 to 16 h in 96-well plates at 37°C in a 5% CO2 and humidified atmosphere before 51Cr labeling. Target cells were labeled for 2 h with 2.5 μCi of 51Cr per well in 50 μl of DMEM with 5% FBS. Different effector-to-target cell ratios were incubated at 37°C with 5% CO2 for 17 h (33), and then 100 μl of supernatant was removed from each well to determine 51Cr release. Percent specific lysis was calculated as [(E − S)/(M − S]) × 100, where E is the mean of six test wells, S is the mean spontaneous release from six wells without effector cells, and M is the mean maximal release from six wells with 3% Triton X-100. The standard error (SE) of percent specific lysis was calculated as described previously (33). Only assays with a spontaneous lysis of <30% were used.
RT-PCR amplification of RNA from transduced EK cells.
Total RNA was isolated from retroviral vector-transduced EK cells by using a commercial kit (Qiagen Inc., Hilden, Germany). One microgram of the RNA sample was treated with DNase I for 15 min at room temperature, and the reverse transcription (RT) reactions were performed with 300 ng of DNase-treated total RNA by using an RNA-PCR kit (Perkin-Elmer Cetus, Norwalk, Conn.). Briefly, the cDNA was synthesized in a total volume of 20 μl, using backward primers specific to the sequences of the gag segments (Table 1) and reaction conditions of 42°C for 15 min, 95°C for 10 min, and then 5°C for 5 min. Reactions without RT were used as negative controls. Then PCR was performed by adding the forward primers and DNA Taq polymerase to the cDNA followed by 35 cycles of 95°C for 1 min, 60°C for 1 min, and 72°C for 7 min. Ten-microliter aliquots of the PCR products were electrophoresed on 1.5% agarose gels stained with ethidium bromide to visualize the reaction products.
RESULTS
Retroviral vector transcription of EIAV gag genes in transduced EK cells.
As an initial step to map CTL epitopes from Gag proteins, retroviral vectors were constructed to express segments of the gag gene encoding p15, the N-terminal half of p26 (p26a), the C-terminal half of p26 (p26b), p11, and p9. To verify transcription of the gene segments, total RNA was extracted from each of the retroviral vector-transduced EK cells and used for RT-PCR. The predicted size of cDNA was amplified from each RNA sample (data not shown). No cDNA amplification was detected without RT or when RNA was extracted from EK cells transduced with the control retroviral vector, vLXSN (data not shown). The DNA sequences of the RT-PCR products were identical to the EIAVWSU5 gag gene (32). These results indicated that the mRNAs for the five gag gene segments were transcribed and had the correct RNA sequence. The transduced EK cells were then used as targets in CTL assays.
Recognition of Gag proteins by CTLm from EIAV-infected horses.
To determine which Gag proteins were recognized by CTLm, EK cells transduced with the five retroviral vectors were used as CTL targets. PBMC from six long-term EIAVWSU5-infected horses were used as sources of CTL for mapping. These six horses were monitored by taking daily body temperature and determining platelet counts, packed cell volumes, and plasma viremia twice per week. Four horses (H507, H521, H529, and H540) were considered inapparent carriers because they had no fever, anemia, or thrombocytopenia and no detectable viremia during the 6-month period when these experiments were done. H532 occasionally had thrombocytopenia and low-level viremia detected by assay for virus on EK cell cultures (38) but lacked fever. H513 had two febrile episodes with associated viremia during the 6-month period. EIAVWSU5-stimulated PBMC from all the horses except H513 were previously reported to have CTLm activity against target cells either infected with a recombinant vaccinia virus expressing EIAV Gag/Pr (34) or transduced with a retroviral vector expressing EIAV Gag/Pr (30). CTLm from four horses (H507, H513, H521, and H529) recognized p15, CTLm from three horses (H513, H529, and H540) recognized p26a, and CTLm from four horses (H507, H521, H529, and H532) recognized p26b (Fig. 1). CTLm from H-513 were assayed with the transduced target cells at an effector-to-target cell ratio of 20:1 (data not shown). None had CTLm recognizing p11 and p9, and none killed transduced EK cells which were mismatched at the ELA-A locus and expressing Gag protein segments. The results demonstrated that ELA-A-restricted CTLm recognized epitopes in p15, p26a, and p26b but not in p11 and p9. These CTL epitopes were then further mapped by using overlapping peptides.
FIG. 1.
Recognition of EIAV Gag proteins by CTLm from EIAV-infected horses. Autologous and ELA-A-mismatched EK cells were transduced with retroviral vectors expressing p15, p26a, p26b, p11, p9, and the vector (vLXSN) expressing no EIAV proteins as a negative control (NC). Effector-to-target cell ratios of 20:1, 10:1, and 5:1 were used in these assays. Vertical lines on columns are SEs, and asterisks represent significant specific lysis ≥3 SE above the negative control value.
CTLm killing of EK target cells pulsed with synthetic peptides from p15.
Ten peptides, 16 to 20 aa in length and overlapping by 8 aa, were made from p15 and numbered 1 to 10 from N to C terminus (Table 2). PBMC from the four horses with CTLm recognizing p15 (H513, H507, H521, and H529) were stimulated with EIAVWSU5 and used in CTL assays against EK targets pulsed with the p15 peptides (Fig. 2). The results indicated that at least one p15 epitope was recognized by CTLm from H513 and H507. Because the possibility of a single epitope in the overlapping region of two positive adjacent peptides existed, at least two p15 epitopes were recognized by CTLm from H521, and at least five epitopes were recognized by CTLm from H529. Further, CTLm from H513, H507, and H529 recognized peptide 1, while CTLm from both H521 and H529 recognized peptide 8.
FIG. 2.
Recognition of overlapping p15 peptides by CTLm from H507, H513, H521, and H529. Autologous EK target cells were pulsed with p15 peptides 1 to 10 at a final concentration of 200 μg/ml, pulsed with no peptide as a negative control (NC), or transduced with a retroviral vector expressing p15. The effector-to-target cell ratio was 20:1. Vertical lines on columns are SEs, and asterisks represent significant specific lysis ≥3 SE above the negative control value.
CTLm killing of EK target cells pulsed with synthetic peptides from p26a.
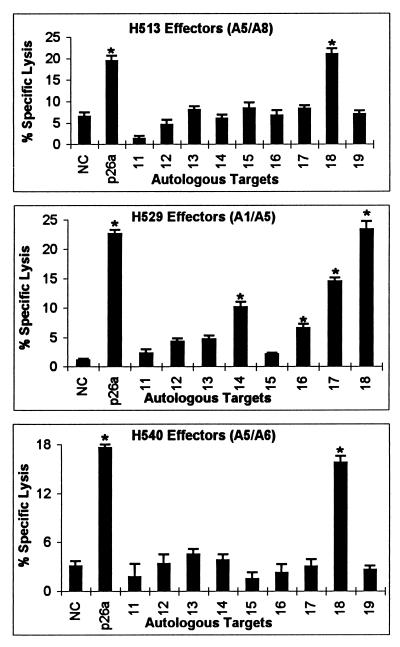
Nine peptides, 20 to 25 aa in length and overlapping by 8 aa, were made from p26a and numbered 11 to 19 from N to C terminus (Table 2). CTLm from H513, H529, and H540, which recognized retroviral vector-transduced targets expressing p26a, were used to identify p26a peptides. CTLm from all three horses recognized targets pulsed with peptide 18, while CTLm from H529 also recognized peptides 14, 16, and 17 (Fig. 3). Even though the killing of target cells pulsed with peptides 14 and 16 was relatively low in the assay presented in Fig. 3, in another assay, target cells pulsed with peptides 14, peptide 16, and no peptide had 29, 25, and 3% specific lysis, respectively. Differences in killing between assays may relate to the effectiveness of the in vitro stimulation of the CTLm. Therefore, at least one epitope in peptide 18 was recognized by CTLm from H513, H529, and H540, and CTLm from H529 recognized at least two additional epitopes.
FIG. 3.
Recognition of overlapping p26a peptides by CTLm from H513, H529, and H540. Autologous EK target cells were pulsed with p26a peptides 11 to 19 at a final concentration of 200 μg/ml, pulsed with no peptide as a negative control (NC), or transduced with retroviral vector expressing p26a. The effector-to-target cell ratio was 20:1. Vertical lines on columns are SEs, and asterisks represent significant specific lysis ≥3 SE above the negative control value.
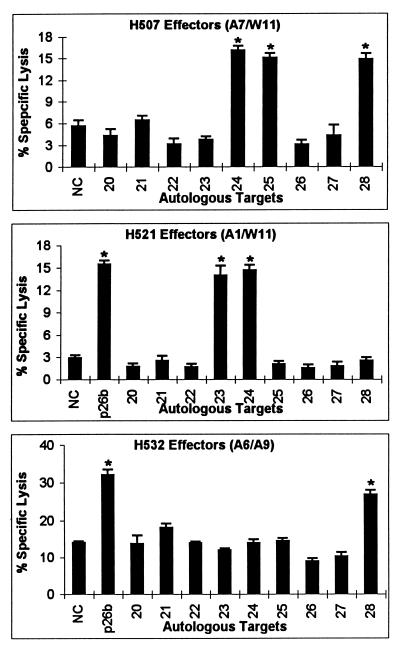
CTLm killing of EK target cells pulsed with synthetic p26b peptides.
Nine peptides, 20 to 22 aa in length and overlapping by 8 aa, were made from p26b and numbered 20 to 28 from N to C terminus (Table 2). CTLm from H507, H521, and H532, which recognized p26b, were used against EK target cells pulsed with these peptides. CTLm from H532 recognized peptide 28, CTLm from H507 recognized peptides 24, 25, and 28, and CTLm from H521 recognized peptides 23 and 24 (Fig. 4). Because peptides 23 and 24 overlapped by 8 aa, it was possible that CTLm from H521 recognized an epitope located in the overlapping region of peptides 23 and 24. To test this possibility, a nonamer peptide (sequence VDRLLSOIK) that covered the overlapping region was made and used in CTLm assays. However, CTLm from H521 did not recognize this nonamer peptide (data not shown). These results indicated that at least two different epitopes were recognized by CTLm from H507 and H521. Finally, CTLm from both H507 and H532 recognized peptide 28, and CTLm from both H507 and H521 recognized peptide 24.
FIG. 4.
Recognition of overlapping p26b peptides by CTLm from H507, H521, and H532. Autologous EK cells were pulsed with p26b peptides 20 to 28 at a final concentration of 200 μg/ml, pulsed with no peptide as a negative control (NC), or transduced with a retroviral vector expressing p26b. The effector-to-target cell ratio was 20:1. Vertical lines on columns are SEs, and asterisks represent significant specific lysis ≥3 SE above the negative control value.
Fine mapping of two CTLm epitopes located within p26.
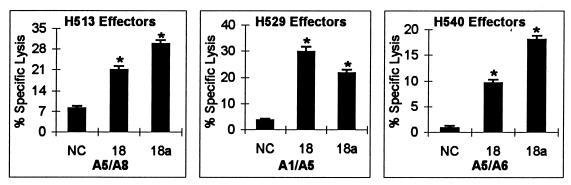
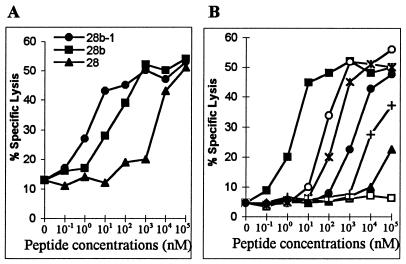
Because CTLm from H529 recognized both p26 peptides 17 and 18, a nonamer peptide (18a; sequence HPLPNAPLV) covering the overlapping region was made. This nonamer peptide was recognized by CTLm from H529 and by CTLm from H513 and H540, which also recognized peptide 18 (Fig. 5). CTL assays using PBMC from H513 stimulated with peptide 18a demonstrated that about 10 nM peptide 18a was required to sensitize EK cell targets for lysis. Peptide 18a was recognized at 103- to 104-fold-lower concentrations than the 20-aa peptide 18 (Fig. 6A). To further determine if peptide 18a was the optimal epitope, additional 8- to 10-aa peptides with deletion or addition of residues to peptide 18a were made (Fig. 6B) and compared in CTL assays with CTLm from H513. Deletion of the first-position amino acid H from 8-, 9-, and 10-aa peptides resulted in no killing of pulsed target cells (Fig. 6B). This observation and the finding that there was no killing with CTLm from H513 when the C-terminal V was missing from peptide 17 (Fig. 3) demonstrated that the first-position H and the ninth-position V were critical residues for this CTL epitope. Further, a 10-aa peptide (HPLPNAPLVA) required a 100-fold increase in concentration to be recognized by CTLm, further indicating that peptide 18a was the optimal epitope.
FIG. 5.
Recognition of p26a nonamer peptide 18a by CTLm from H513, H529, and H540. Autologous EK cells were pulsed with p26a peptides 18 and 18a at a final concentration of 200 μg/ml or with no peptide as a negative control (NC). The effector-to-target cell ratio was 20:1. Vertical lines on columns are SEs, and asterisks represent significant specific lysis ≥3 SE above the negative control value.
FIG. 6.
Optimal peptide epitope recognized by CTLm from H513. Effectors were peptide 18a-stimulated CTLm from H513, and the effector-to-target cell ratio was 20:1. (A) Efficiencies of recognition of peptide 18a (HPLPNAPLV) and peptide 18 (HPLPNAPLVAPPQGPIPMTA); (B) comparison of peptide 18a (▪) with 8-aa, 9-aa, and 10-aa versions of the peptide. (○, HPLPNAPLVA; ∗, PLPNAPLVAP; □, PLPNAPLVA; ▴, LPNAPLVAP; •, PLPNAPLV; +, LPNAPLVA).
Fine mapping of the p26b peptide 28 epitope recognized by CTLm from H532 was also performed. Peptides 28a and 28b but not 28c were recognized (Table 3, experiment 1). Peptides 28a and 28b overlapped by 8 aa, and the lysis of target cells pulsed with peptide 28b was significantly higher than the lysis of cells pulsed with peptide 28a (Table 3, experiment 1). Therefore, a nonamer peptide epitope located in the N-terminal end of peptide 28b was made and designated 28b-1 (sequence GTTKQKMML). Peptides 28b and 28b-1 were equally killed by CTLm from H532 (Table 3, experiment 2). In dose-response experiments, 1 nM peptide 28b-1 was required to be recognized by CTLm from H532, and peptide 28b-1 was recognized at 103- to 104-fold-lower concentrations than the 20-aa peptide 28 (Fig. 7A). Additional 8- to 10-aa peptides with deletion and addition of peptide 28b-1 residues were compared (Fig. 7B). Peptides with a deletion of the first-position G or ninth-position L were less efficiently recognized by CTLm from H532, indicating that these two residues were needed in the epitope (Fig. 7B). Moreover, adding an I to the N terminus or an L to the C terminus of peptide 28b-1 did not increase the efficiency of target cell lysis by CTLm (Fig. 7B). These results further confirmed that the nonamer peptide (GTTKQKMML) was the optimal epitope recognized by CTLm from H532.
TABLE 3.
Fine mapping of the p26 epitope recognized by CTLm from H532a
| Expt | Peptide
|
% Specific lysis | |
|---|---|---|---|
| Name | Sequence | ||
| 1 | None | 6.0 | |
| 27 | MRHLRPEDTLEEKMYACRDI | 9.8 | |
| 28 | KMYACRDIGTTKQKMMLLAKAL | 41.1 | |
| 28a | CRDIGTTKQKMM | 23.6 | |
| 28b | GTTKQKMMLLA | 39.8 | |
| 28c | KQKMMLLAKAL | 11.8 | |
| 2 | None | 5.2 | |
| 28b | GTTKQKMMLLA | 25.8 | |
| 28b-1 | GTTKQKMML | 26.0 | |
Target cells were autologous EK cells pulsed with peptides or with no peptide as a negative control. The effector-to-target cell ratio was 20:1.
FIG. 7.
Optimal peptide epitope recognized by CTLm from H532. Effectors were peptide 28b-1-stimulated CTLm from H532, and the effector-to-target cell ratio was 20:1. (A) Efficiencies of recognition of peptides 28b-1 (GTTKQKMML), 28 (KMYACRDIGTTKQKMMLLAKAL), and 28b (GTTKQKMMLLA); (B) comparison of peptide 28b-1 (■) with 8-, 9-, and 10-aa versions of the peptide (○, IGTTKQKMML; ∗, GTTKQKMMLL; □, TTKQKMMLL; ▴, IGTTKQKMM; •, GTTKQKMM; +, TTKQKMML).
ELA-A restriction of the CTL responses to EIAV Gag epitopes.
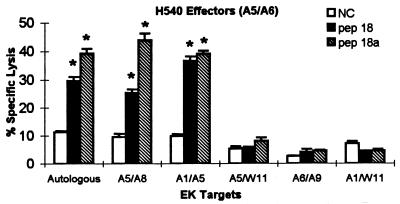
The ELA-A type restricting H540 CTLm killing of target cells pulsed with p26a peptide 18a was evaluated in assays using EK target cells that were half-matched or mismatched at the ELA-A locus and autologous EK target cells (Fig. 8). Two EK target cells that shared (half-matched) ELA-A5 were lysed by CTLm from H540, while one EK target cell that shared ELA-A5 was not lysed. This unexpected observation suggested that while the ELA-A5 molecules from these four horses were recognized by the same typing serum (44), one was not recognized by the T-cell receptor (TCR) on H540 CTLm. Similar subtypes of human HLA-A2 and Cw3 alleles have been reported for CTL from HIV-1- and influenza A virus-infected patients (4, 29). Because CTLm from H513 and H529 also recognized peptide 18a, ELA-A restriction of epitope 18a from H513 and H529 was also evaluated, and the results were identical to those for H540 (data not shown). Therefore, it is likely that epitope HPLPNAPLV was restricted by a subtype of ELA-A5 molecules designated ELA-A5.1 (CTL1). CTLm from H532 were tested against different EK target cells pulsed with p26b peptide 28 (Fig. 9). The results provided preliminary data that the p26b peptide 28 epitope recognized by CTL from H532 was restricted by ELA-A9 molecules.
FIG. 8.
ELA-A5 restriction of CTLm recognizing p26a nonamer peptide 18a. Autologous or heterologous EK target cells with half-matched or mismatched ELA-A alleles were pulsed with p26a peptides 18 (pep 18) and 18a (pep 18a) or with no peptide as a negative control (NC). The effector-to-target cell ratio was 20:1. Vertical lines on columns are SEs, and asterisks represent significant specific lysis ≥3 SE above the negative control value.
FIG. 9.
ELA-A9 restriction of CTLm recognizing p26b peptide 28. Autologous EK target cells or heterologous EK target cells with half-matched or mismatched ELA-A alleles were pulsed with p26b peptide 28 (pep 28) or with no peptide as a negative control (NC). The effector-to-target cell ratio was 20:1. Vertical lines on columns are SEs, and asterisks represent significant specific lysis ≥3 SE above the negative control value.
DISCUSSION
Identifying EIAV CTL epitopes provides the detailed information necessary to evaluate the role of CTL responses in controlling infection and perhaps to induce CTL in horses. In this study, several ELA-A class I-restricted EIAV Gag epitopes were identified with CTLm from six long-term EIAV-infected horses. The CTL epitope mapping was facilitated by using target cells transduced with retroviral vectors expressing gag gene segments. Retroviral vectors have several advantages for use in CTL studies. (i) Gene expression and thus epitope presentation are relatively stable in the transduced target cells (46). (ii) Unlike the case for other viral vectors, retroviral vector-transduced cells express only the target gene and a neomycin phosphoribosyltransferase gene (50). (iii) Retroviral vectors do not have a cytopathic effect on transduced cells (19). (iv) Proteins expressed by retroviral vectors undergo endogenous pathway processing and presentation, thereby facilitating recognition by CTL (20). Retroviral vectors have been used extensively for gene therapy (19) but have had limited use for mapping CTL epitopes and for induction of CTL responses in vitro (47) and in vivo (16). These reports are extended by the results presented in this paper which demonstrated the utility of transduced EK target cells expressing EIAVWSU5 gag segments for preliminary mapping of CTL epitopes.
The Gag protein epitopes recognized by CTL from four EIAV-infected carrier horses and two other long-term EIAV-infected horse were all located on the p15 and p26 proteins and included at least 12 different epitopes. These results are consistent with the location of the Gag epitopes identified by CTL from HIV-1-infected patients, which are almost exclusively located in HIV-1 p17 and p24 proteins (14, 18, 36, 37, 49). Moreover, the majority of HIV-1 patients have CTL recognizing Gag epitopes. For instance, the Gag epitopes are the protein epitopes most frequently recognized by CTL from HIV-1-infected nonprogressors, with CTL from 16 of 25 (64%) nonprogressors recognize HIV-1 Gag epitopes, while CTL from 11 of 25 (44%) recognize Env epitopes, CTL from 9 of 25 (36%) recognize RT, and CTL from 1 of 25 (4%) recognize Tat (28). Our results demonstrating at least one EIAV Gag epitope identified by CTL from each of the six horses indicate that Gag epitopes may be recognized by CTL from most EIAV-infected horses.
Stimulation of MHC class I-restricted CTL responses to viral epitopes is determined by several molecular interactions in addition to the requirement for recognition by TCR (11). These other interactions include cleavage of the viral proteins by proteases and transport of the viral peptides into the endoplasmic reticulum by transport-associated proteins (11). Among these interactions, presentation of epitopes by MHC class I molecules are critical (45) because the epitopes have to bind to MHC class I molecules to be recognized by TCR on CTL for both stimulation and killing (11). It has been shown that the number of viral CTL epitopes is restricted in virus-infected animals with limited MHC class I polymorphism (7, 13). In contrast, in most outbred populations, MHC class I genes are very polymorphic, leading to the diversity of CTL responses in such populations. For example, more than 266 different human MHC class I alleles have been identified by nucleotide sequence analysis (39), and diverse CTL responses have been found in HIV-1-infected patients (37). Our results agree with the findings for HIV-1-infected patients in that diverse CTL responses were generated from the six infected horses with different HLA-A types.
The results also indicated that the Gag protein nonamer epitopes were presented by products of ELA-A alleles. One nonamer epitope recognized by CTLm from three horses was probably presented by the product of the ELA-A5.1 allele. Another epitope recognized by CTLm from a different horse was likely ELA-A9 restricted. This is because the restriction of CTL responses detected in PBMC from EIAV-infected or equine herpesvirus-infected horses with defined ELA-A alleles has so far been associated with the ELA-A alleles (1, 15, 34). Moreover, in this study, no target cells mismatched at both ELA-A alleles were killed by CTL from the infected horses. Nevertheless, the possibility of restriction by a second ELA class I locus (B locus) (3) or another undefined locus exists.
In comparing the CTL responses in the slow and rapid progressors of the HIV-1-infected patients, the responses cannot be distinguished by the frequency of the CTL (22), and so it is assumed that the quality of the CTL responses may differ in these patients (14). If the majority of CTL recognize conserved epitopes, it may be harder for viruses to escape the CTL responses. The CTL epitopes on the HIV-1 Gag proteins are reported to be highly conserved among HIV-1 isolates (18, 49), and CTL responses to HIV-1 Gag epitopes are associated with decreased risk of progression to AIDS (43). In EIAV infection, most infected horses eventually control the disease and become lifelong carriers (9). This outcome may occur because the majority of carrier horses have CTL recognizing highly conserved epitopes. The predicted amino acid sequences of EIAV matrix protein (p15) and capsid protein (p26) are identical among the five EIAV strains examined (21, 32, 40, 48). However, these strains are all derived from the Wyoming wild-type strain and do not reflect the possible diversity of other horse-passaged strains. Data from five of the horses in this study suggested that despite possible diversity of Gag epitopes, CTL to Gag epitopes could still be an important immune response for maintaining the carrier state of long-term EIAV-infected horses.
In conclusion, the use of retroviral vectors expressing segments of the gag gene greatly facilitated the mapping of CTL epitopes by reducing the number of synthetic peptides needed and the number of peptides evaluated for each horse. Results from the six horses in this study indicated that Gag epitopes are consistently recognized by CTL from long-term EIAV-infected horses. Even though peptides with a minimum of 12 different epitopes were identified, none were recognized by CTLm from all horses. The CTLm were ELA-A restricted, and different peptides were identified by CTLm from horses expressing different ELA-A molecules. The nonamer peptides which were likely restricted by ELA-A5.1 or ELA-A9 molecules could be used to induce CTL in horses expressing these molecules. However, a successful EIAV vaccine for horse populations with polymorphic MHC class I molecules will need to include a large number of CTL epitopes in order to induce CTL in all individuals. The use of whole viral proteins expressed by retroviral or other vectors may present the diversity of epitopes needed to stimulate CTL in horse populations. Gag proteins should be included in the proteins expressed by such vectors.
ACKNOWLEDGMENTS
This work was supported in part by Public Health Service NIAID grant AI-24291 and U.S. Department of Agriculture NRICGP grant 96-35204-3427.
We thank Wendy C. Brown, William C. Davis, and William P. Cheevers for many valuable discussions, and we thank Steve Leib, Eldon Libstaff, and Emma Karel for technical assistance.
REFERENCES
- 1.Allen G, Yeargan M, Costa L R, Cross R. Major histocompatibility complex class I-restricted cytotoxic T-lymphocyte responses in horses infected with equine herpesvirus 1. J Virol. 1995;69:606–612. doi: 10.1128/jvi.69.1.606-612.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bailey E. Population studies on the ELA system in American standardbred and thoroughbred mares. Anim Blood Groups Biochem Genet. 1983;14:201–211. doi: 10.1111/j.1365-2052.1983.tb01073.x. [DOI] [PubMed] [Google Scholar]
- 3.Bernoco D, Byrns G, Bailey E, Lew A M. Evidence of a second polymorphic ELA class I (ELA-B) locus and gene order for three loci of the equine major histocompatibility complex. Anim Genet. 1987;18:103–118. doi: 10.1111/j.1365-2052.1987.tb00749.x. [DOI] [PubMed] [Google Scholar]
- 4.Biddison W E, Krangel M S, Strominger J L, Ward F E, Shearer G M, Shaw S. Virus-immune cytotoxic T cells recognize structural differences between serologically indistinguishable HLA-A2 molecules. Hum Immunol. 1980;1:225–232. doi: 10.1016/0198-8859(80)90017-8. [DOI] [PubMed] [Google Scholar]
- 5.Borrow P, Lewicki H, Hahn B H, Shaw G M, Oldstone M B. Virus-specific CD8+ cytotoxic T-lymphocyte activity associated with control of viremia in primary human immunodeficiency virus type 1 infection. J Virol. 1994;68:6103–6110. doi: 10.1128/jvi.68.9.6103-6110.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Borrow P, Lewicki H, Wei X P, Horwitz M S, Peffer N, Meyers H, Nelson J A, Gairin J E, Hahn B H, Oldstone M B A, Shaw G M. Antiviral pressure exerted by HIV-1-specific cytotoxic T lymphocytes (CTLs) during primary infection demonstrated by rapid selection of CTL escape virus. Nat Med. 1997;3:205–211. doi: 10.1038/nm0297-205. [DOI] [PubMed] [Google Scholar]
- 7.Braciale T J, Sweetser M T, Morrison L A, Kittlesen D J, Braciale V L. Class I major histocompatibility complex-restricted cytolytic T lymphocytes recognize a limited number of sites on the influenza hemagglutinin. Proc Natl Acad Sci USA. 1989;86:277–281. doi: 10.1073/pnas.86.1.277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Carpenter S, Evans L H, Sevoian M, Chesebro B. Role of the host immune response in selection of equine infectious anemia virus variants. J Virol. 1987;61:3783–3789. doi: 10.1128/jvi.61.12.3783-3789.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Coggins L. Carriers of equine infectious anemia virus. J Am Vet Med Assoc. 1984;184:279–281. [PubMed] [Google Scholar]
- 10.Crawford T B, Wardrop K J, Tornquist S J, Reilich E, Meyers K M, McGuire T C. A primary production deficit in the thrombocytopenia of equine infectious anemia. J Virol. 1996;70:7842–7850. doi: 10.1128/jvi.70.11.7842-7850.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Deng Y P, Yewdell J W, Eisenlohr L C, Bennink J R. MHC affinity, peptide liberation, T cell repertoire, and immunodominance all contribute to the paucity of MHC class I-restricted peptides recognized by antiviral CTL. J Immunol. 1997;158:1507–1515. [PubMed] [Google Scholar]
- 12.Emini E A, Schleif W A, Nunberg J H, Conley A J, Eda Y, Tokiyoshi S, Putney S D, Matsushita S, Cobb K E, Jett C M, et al. Prevention of HIV-1 infection in chimpanzees by gp120 V3 domain-specific monoclonal antibody. Nature. 1992;355:728–730. doi: 10.1038/355728a0. [DOI] [PubMed] [Google Scholar]
- 13.Evans D T, Piekarczyk M S, Allen T M, Boyson J E, Yeager M, Hughes A L, Gotch F M, Hinshaw V S, Watkins D I. Immunodominance of a single CTL epitope in a primate species with limited MHC class I polymorphism. J Immunol. 1997;159:1374–1382. [PubMed] [Google Scholar]
- 14.Goulder P J, Sewell A K, Lalloo D G, Price D A, Whelan J A, Evans J, Taylor G P, Luzzi G, Giangrande P, Phillips R E, McMichael A J. Patterns of immunodominance in HIV-1-specific cytotoxic T lymphocyte responses in two human histocompatibility leukocyte antigens (HLA)-identical siblings with HLA-A0201 are influenced by epitope mutation. J Exp Med. 1997;185:1423–1433. doi: 10.1084/jem.185.8.1423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hammond S A, Cook S J, Lichtenstein D L, Issel C J, Montelaro R C. Maturation of the cellular and humoral immune responses to persistent infection in horses by equine infectious anemia virus is a complex and lengthy process. J Virol. 1997;71:3840–3852. doi: 10.1128/jvi.71.5.3840-3852.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Irwin M J, Laube L S, Lee V, Austin M, Chada S, Anderson C G, Townsend K, Jolly D J, Warner J F. Direct injection of a recombinant retroviral vector induces human immunodeficiency virus-specific immune responses in mice and nonhuman primates. J Virol. 1994;68:5036–5044. doi: 10.1128/jvi.68.8.5036-5044.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Issel C J, Horohov D W, Lea D F, Adams W V, Jr, Hagius S D, McManus J M, Allison A C, Montelaro R C. Efficacy of inactivated whole-virus and subunit vaccines in preventing infection and disease caused by equine infectious anemia virus. J Virol. 1992;66:3398–3408. doi: 10.1128/jvi.66.6.3398-3408.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Johnson R P, Trocha A, Yang L, Mazzara G P, Panicali D L, Buchanan T M, Walker B D. HIV-1 gag-specific cytotoxic T lymphocytes recognize multiple highly conserved epitopes. Fine specificity of the gag-specific response defined by using unstimulated peripheral blood mononuclear cells and cloned effector cells. J Immunol. 1991;147:1512–1521. [PubMed] [Google Scholar]
- 19.Jolly D J, Warner J F. Retroviral vectors as vaccines and immunotherapeutics. Semin Immunol. 1990;2:329–339. [PubMed] [Google Scholar]
- 20.Jolly D J, Warner J F. Induction of anti-HIV-1 immune responses by retroviral vectors. Biotechnol Ther. 1991;2:179–193. [PubMed] [Google Scholar]
- 21.Kawakami T, Sherman L, Dahlberg J, Gazit A, Yaniv A, Tronick S R, Aaronson S A. Nucleotide sequence analysis of equine infectious anemia virus proviral DNA. Virology. 1987;158:300–312. doi: 10.1016/0042-6822(87)90202-9. [DOI] [PubMed] [Google Scholar]
- 22.Klein M R, van-Baalen C A, Holwerda A M, Kerkhof-Garde S R, Bende R J, Keet I P, Eeftinck-Schattenkerk J K, Osterhaus A D, Schuitemaker H, Miedema F. Kinetics of Gag-specific cytotoxic T lymphocyte responses during the clinical course of HIV-1 infection: a longitudinal analysis of rapid progressors and long-term asymptomatics. J Exp Med. 1995;181:1365–1372. doi: 10.1084/jem.181.4.1365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kono Y, Hirasawa K, Fukunaga Y, Taniguchi T. Resistance of horses infected chronically with equine infectious anemia virus against reinfection. Natl Inst Anim Health Q. 1973;13:173–181. [PubMed] [Google Scholar]
- 24.Kono Y, Hirasawa K, Fukunaga Y, Taniguchi T. Recrudescence of equine infectious anemia by treatment with immunosuppressive drugs. Natl Inst Anim Health Q. 1976;16:8–15. [PubMed] [Google Scholar]
- 25.Kono Y, Kobayashi K, Fukunaga Y. Antigenic drift of equine infectious anemia virus in chronically infected horses. Arch Gesamte Virusforsch. 1973;41:1–10. doi: 10.1007/BF01249923. [DOI] [PubMed] [Google Scholar]
- 26.Koup R A, Safrit J T, Cao Y, Andrews C A, McLeod G, Borkowsky W, Farthing C, Ho D D. Temporal association of cellular immune responses with the initial control of viremia in primary human immunodeficiency virus type 1 syndrome. J Virol. 1994;68:4650–4655. doi: 10.1128/jvi.68.7.4650-4655.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Langemeier J L, Bailey E, Henney P J. Linkage studies between the Tcp-1, Tcp-10, and Mhc-Eqca-A loci in the horse. Immunogenetics. 1993;38:359–362. doi: 10.1007/BF00210478. [DOI] [PubMed] [Google Scholar]
- 28.Lieberman J, Fabry J A, Fong D M, Parkerson G R. Recognition of a small number of diverse epitopes dominates the cytotoxic T lymphocyte response to HIV type 1 in an infected individual. AIDS Res Hum Retroviruses. 1997;13:383–392. doi: 10.1089/aid.1997.13.383. [DOI] [PubMed] [Google Scholar]
- 29.Littaua R A, Oldstone M B, Takeda A, Debouck C, Wong J T, Tuazon C U, Moss B, Kievits F, Ennis F A. An HLA-C-restricted CD8+ cytotoxic T-lymphocyte clone recognizes a highly conserved epitope on human immunodeficiency virus type 1 Gag. J Virol. 1991;65:4051–4056. doi: 10.1128/jvi.65.8.4051-4056.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lonning, S. M., W. Zhang, S. R. Leib, and T. C. McGuire. Equine infectious anemia virus-specific memory cytotoxic T lymphocytes efficiently lyse retroviral vector transduced target cells. Submitted for publication.
- 31.Marti E, Szalai G, Antczak D F, Bailey E, Gerber H, Lazary S. The equine major histocompatibility complex. In: Schook L B, Lamont S J, editors. The major histocompatibility complex region of domestic animal species. Boca Raton, Fla: CRC Press; 1996. pp. 245–268. [Google Scholar]
- 32.McGuire T C, O’Rourke K I, Baszler T V, Leib S R, Brassfield A L, Davis W C. Expression of functional protease and subviral particles by vaccinia virus containing equine infectious anaemia virus gag and 5′ pol genes. J Gen Virol. 1994;75:895–900. doi: 10.1099/0022-1317-75-4-895. [DOI] [PubMed] [Google Scholar]
- 33.McGuire T C, Tumas D B, Byrne K M, Hines M T, Leib S R, Brassfield A L, O’Rourke K I, Perryman L E. Major histocompatibility complex-restricted CD8+ cytotoxic T lymphocytes from horses with equine infectious anemia virus recognize Env and Gag/PR proteins. J Virol. 1994;68:1459–1467. doi: 10.1128/jvi.68.3.1459-1467.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McGuire T C, Zhang W, Hines M T, Henney P J, Byrne K M. Frequency of memory cytotoxic T lymphocytes to equine infectious anemia virus proteins in blood from carrier horses. Virology. 1997;238:85–93. doi: 10.1006/viro.1997.8795. [DOI] [PubMed] [Google Scholar]
- 35.Miller A D, Garcia J V, von Suhr N, Lynch C M, Wilson C, Eiden M V. Construction and properties of retrovirus packaging cells based on gibbon ape leukemia virus. J Virol. 1991;65:2220–2224. doi: 10.1128/jvi.65.5.2220-2224.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nakamura Y, Kameoka M, Tobiume M, Kaya M, Ohki K, Yamada T, Ikuta K. A chain section containing epitopes for cytotoxic T, B and helper T cells within a highly conserved region found in the human immunodeficiency virus type 1 Gag protein. Vaccine. 1997;15:489–496. doi: 10.1016/s0264-410x(96)00224-1. [DOI] [PubMed] [Google Scholar]
- 37.Nixon D F, McMichael A J. Cytotoxic T-cell recognition of HIV proteins and peptides. AIDS. 1991;5:1049–1059. [PubMed] [Google Scholar]
- 38.O’Rourke K, Perryman L E, McGuire T C. Antiviral, anti-glycoprotein and neutralizing antibodies in foals with equine infectious anemia virus. J Gen Virol. 1988;69:667–674. doi: 10.1099/0022-1317-69-3-667. [DOI] [PubMed] [Google Scholar]
- 39.Parham P, Ohta T. Population biology of antigen presentation by MHC class I molecules. Science. 1996;272:67–74. doi: 10.1126/science.272.5258.67. [DOI] [PubMed] [Google Scholar]
- 40.Perry S T, Flaherty M T, Kelley M J, Clabough D L, Tronick S R, Coggins L, Whetter L, Lengel C R, Fuller F. The surface envelope protein gene region of equine infectious anemia virus is not an important determinant of tropism in vitro. J Virol. 1992;66:4085–4097. doi: 10.1128/jvi.66.7.4085-4097.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Perryman L E, O’Rourke K I, McGuire T C. Immune responses are required to terminate viremia in equine infectious anemia lentivirus infection. J Virol. 1988;62:3073–3076. doi: 10.1128/jvi.62.8.3073-3076.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rinaldo C, Huang X L, Fan Z F, Ding M, Beltz L, Logar A, Panicali D, Mazzara G, Liebmann J, Cottrill M. High levels of anti-human immunodeficiency virus type 1 (HIV-1) memory cytotoxic T-lymphocyte activity and low viral load are associated with lack of disease in HIV-1-infected long-term nonprogressors. J Virol. 1995;69:5838–5842. doi: 10.1128/jvi.69.9.5838-5842.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Riviere Y, McChesney M B, Porrot F, Tanneau S F, Sansonetti P, Lopez O, Pialoux G, Feuillie V, Mollereau M, Chamaret S. Gag-specific cytotoxic responses to HIV type 1 are associated with a decreased risk of progression to AIDS-related complex or AIDS. AIDS Res Hum Retroviruses. 1995;11:903–907. doi: 10.1089/aid.1995.11.903. [DOI] [PubMed] [Google Scholar]
- 44.Schuberth H J, Anders I, Pape U, Leibold W. One-dimensional isoelectric focusing and immunoblotting of equine major histocompatibility complex class I antigens. Anim Genet. 1992;23:87–95. [PubMed] [Google Scholar]
- 45.Shirai M, Kozlowski S, Margulies D H, Berzofsky J A. Degenerate MHC restriction reveals the contribution of class I MHC molecules in determining the fine specificity of CTL recognition of an immunodominant determinant of HIV-1 gp160 V3 loop. J Immunol. 1997;158:3181–3188. [PubMed] [Google Scholar]
- 46.Song E S, Lee V, Surh C D, Lynn A, Brumm D, Jolly D J, Warner J F, Chada S. Antigen presentation in retroviral vector-mediated gene transfer in vivo. Proc Natl Acad Sci USA. 1997;94:1943–1948. doi: 10.1073/pnas.94.5.1943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Song W, Collisson E W, Li J, Wolf A M, Elder J H, Grant C K, Brown W C. Feline immunodeficiency virus (FIV)-specific cytotoxic T lymphocytes from chronically infected cats are induced in vitro by retroviral vector-transduced feline T cells expressing the FIV capsid protein. Virology. 1995;209:390–399. doi: 10.1006/viro.1995.1271. [DOI] [PubMed] [Google Scholar]
- 48.Stephens R M, Casey J W, Rice N R. Equine infectious anemia virus gag and pol genes: relatedness to visna and AIDS virus. Science. 1986;231:589–594. doi: 10.1126/science.3003905. [DOI] [PubMed] [Google Scholar]
- 49.van-Baalen C A, Klein M R, Huisman R C, Dings M E, Kerkhof-Garde S R, Geretti A M, Gruters R, van-Els C A, Miedema F, Osterhaus A D. Fine-specificity of cytotoxic T lymphocytes which recognize conserved epitopes of the Gag protein of human immunodeficiency virus type 1. J Gen Virol. 1996;77:1659–1665. doi: 10.1099/0022-1317-77-8-1659. [DOI] [PubMed] [Google Scholar]
- 50.Warner J F, Anderson C G, Laube L, Jolly D J, Townsend K, Chada S, St. Louis D. Induction of HIV-specific CTL and antibody responses in mice using retroviral vector-transduced cells. AIDS Res Hum Retroviruses. 1991;7:645–655. doi: 10.1089/aid.1991.7.645. [DOI] [PubMed] [Google Scholar]
- 51.Yasutomi Y, Reimann K A, Lord C I, Miller M D, Letvin N L. Simian immunodeficiency virus-specific CD8+ lymphocyte response in acutely infected rhesus monkeys. J Virol. 1993;67:1707–1711. doi: 10.1128/jvi.67.3.1707-1711.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]