Abstract
Genetically encoded circuits have been successfully utilized to assess and characterize target variants with desirable traits from large mutant libraries. Adenosylcobalamin is an essential coenzyme that is required in many intracellular physiological reactions and is widely used in the pharmaceutical and food industries. High-throughput screening techniques capable of detecting adenosylcobalamin productivity and selecting superior adenosylcobalamin biosynthesis strains are critical for the creation of an effective microbial cell factory for the production of adenosylcobalamin at an industrial level. In this study, we developed an RNA-protein hybrid biosensor whose input part was an endogenous RNA riboswitch to specifically respond to adenosylcobalamin, the inverter part was an orthogonal transcriptional repressor to obtain signal inversion, and the output part was a fluorescent protein to be easily detected. The hybrid biosensor could specifically and positively correlate adenosylcobalamin concentrations to green fluorescent protein expression levels in vivo. This study also improved the operating concentration and dynamic range of the hybrid biosensor by systematic optimization. An individual cell harboring the hybrid biosensor presented over 20-fold higher fluorescence intensity than the negative control. Then, using such a biosensor combined with fluorescence-activated cell sorting, we established a high-throughput screening platform for screening adenosylcobalamin overproducers. This study demonstrates that this platform has significant potential to quickly isolate high-productive strains to meet industrial demand and that the framework is acceptable for various metabolites.
Keywords: Adenosylcobalamin, Genetically encoded biosensor, Riboswitch, Transcriptional repressor, High-throughput screening
Graphical abstract
1. Introduction
Microbial metabolic engineering is a useful tool for designing industrial strains that relies on the rational redistribution of the metabolic flux to produce the maximum target products [1,2]. Rational approaches to improve target product yields require multiple cellular metabolic pathway modifications without disturbing cellular homeostasis and are often restricted by the limited knowledge of the highly complicated intracellular carbon and energy metabolism flux [3,4]. Target product yields are also impacted by unknown metabolic networks and regulation, which result in unwanted consequences of targeted strain modifications [5]. As a complement, non-rational high-throughput approaches based on efficient screening methods have been successful in achieving desired phenotypes in diverse random mutagenesis [6,7]. Nevertheless, this strategy is only available to objects with conspicuous phenotypes, such as growth rate, color and fluorescence. The majority of target chemicals, which lack a conspicuous phenotype, are difficult to screen for by these high-throughput approaches. To overcome the restricted conditions, genetically encoded biosensors provide a way to link the varied intracellular metabolite levels to fluorescent protein expression and enable fluorescence-based high-throughput screens such as flow cytometry [8,9], and many artificial biosensors have been utilized to screen inconspicuous phenotypes [10].
The development of a biosensor for high-throughput screening of inconspicuous products should satisfy the following conditions: one can specifically and sensitively respond to the target metabolites, and the other is to have a wide operating concentration range and a large dynamic range [11]. Such biosensors are powerful tools for the identification of target metabolites [[12], [13], [14], [15]]. Riboswitches are regulatory RNA elements that can respond to small ligands and have been identified in various microbes. They are most commonly located in the 5’ untranslated areas of mRNAs. The aptamer domains of the riboswitches enable selectivity and sensitivity to sense special intracellular metabolites and bind them particularly [16,17]. Most riboswitches regulate genetic expression by ligand-binding conformational alterations that translate target metabolite signals into the expression levels of corresponding downstream genes by affecting intrinsic transcription termination, translation initiation, or the rate of RNA degradation [18]. A riboswitch controlling target gene expression could be applied as a relatively noninvasive sensor to detect intracellular metabolites [[19], [20], [21], [22]].
Vitamin B12, also known as cobalamin, is a crucial cofactor for critical enzymes catalyzing numerous biochemical reactions and is an essential nutrient that has been broadly applied in food and pharmaceuticals [23,24]. Vitamin B12 has two active forms: adenosylcobalamin (AdoCbl) and methylcobalamin (MeCbl). The structure of cobalamin is complex, which makes it difficult to chemically synthesize [25]. Only a few microbes can de novo synthesize AdoCbl with more than 30 enzymes [26]. Currently, industrial AdoCbl production is primarily produced through microbial fermentation using Pseudomonas denitrificans, Propionibacterium freudenreichii, Ensifer adherents, and Sinorhizobium meliloti [[27], [28], [29], [30]]. Growing global demand for cobalamin has drawn much attention, but the lack of effective cobalamin synthesis methods has resulted in high market prices. Because of the limited understanding of cobalamin metabolic mechanisms and inefficient genetic manipulation tools, the rational modification of cobalamin biosynthesis is still difficult [31,32]. Recently, strategies to improve the industrial productivity of AdoCbl-producing strains have largely focused on traditional random mutagenesis [33,34]. However, the production of cobalamin is most commonly measured by liquid chromatography or mass spectrometry, which is restricted to approximately 102 tests per instrument per day, resulting in a heavy workload and severely restricting the efficiency of obtaining a superior overproducer from large genetic libraries. It is necessary to establish efficient screens to identify and select high producers from the vast library of natural or synthetic mutants. Many cobalamin-responsive riboswitches have been found in cobalamin-producing microbes that regulate cobalamin biosynthesis and metabolism [32,[35], [36], [37]]. All of the discovered cobalamin riboswitches act as negative regulators by inhibiting downstream gene expression via cobalamin feedback. This could be an off-type sensor. Some researchers found that the off-type riboswitch combined with an orthogonal transcriptional repressor could successfully result in signal inversion to become an on-type sensor [[38], [39], [40]].
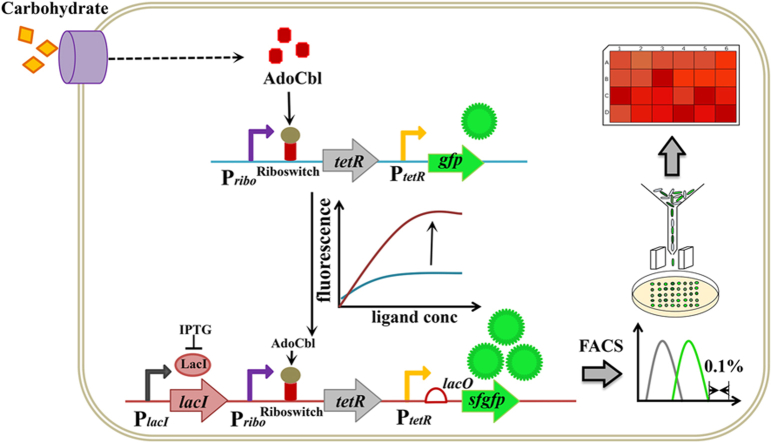
Sinorhizobium meliloti 320 (SM320) is a gram-negative alphaproteobacterium that contains all genes involved in AdoCbl de novo biosynthesis and has the capacity to be used for industrial production of AdoCbl. The lack of a comprehensive understanding of the AdoCbl metabolic biosynthesis mechanism and inefficient genetic manipulation tools make rational modifications difficult. In this study, we developed a genetically encoded RNA-protein hybrid device that was used to detect the different intracellular AdoCbl concentrations in SM320. The hybrid device comprises an endogenous riboswitch, which was found by transcriptome analysis and functions as an input part to specifically respond to AdoCbl, and an orthogonal transcriptional repressor, which functions as a signal inverter, converting the input signal from off to on [38,39]. Then, an RNA-protein hybrid biosensor was constructed to link the AdoCbl concentration to GFP expression levels. Then the dose-response parameters, including operating concentration and dynamic range, were optimized. We demonstrated that individual cells carrying this hybrid biosensor could present above 20-fold higher fluorescence intensity than the negative control. Then, we developed a generalizable in vivo HTS strategy based on the biosensor and fluorescence-activated cell sorting (FACS) to expedite the screening process depending on the amount of AdoCbl. The novel phenotypes of interest could be obtained from a large mutant library in a short period of time and might provide insight into the complex regulation of the AdoCbl biosynthesis pathway. Our work provides a strategy for developing biosensors to target metabolites that lack a conspicuous phenotype and confirms that the biosensor-based HTS platform is a potent tool to identify and isolate the high-performing strain.
2. Materials and methods
2.1. Bacterial strains and media
The strains and plasmids used in this study are listed in Table S1 and Table S2. Escherichia coli DH5α were utilized for plasmid construction. Sinorhizobium meliloti 320 (SM320) was used as the origin strain for AdoCbl production, which was stored in our laboratory [41]. DH5α was cultivated in Luria broth (LB) medium (10 g/L NaCl, 10 g/L tryptone and 5 g/L yeast extract) as well as 1.8 % agar for solid at 37 °C. SM320 was grown in TYC medium (5 g/L tryptone, 3 g/L yeast extract, 0.67 g/L CaCl2), as well as 1.8 % agar for solid at 32 °C. The minimal medium was TYC medium supplemented with 45 μg/L DMBI (5, 6-dimethylbenzimidazole) and 90 μg/L cobalt chloride. The fermentation culture and antibiotic utilization were the same as those in our previous study [42].
2.2. Plasmid construction
The plasmids used in this study are listed in Table S2. The high-copy number vector pKP was utilized as the backbone plasmid for AdoCbl biosensor construction [42]. All of the recombinant plasmids used in this study were constructed by Golden Gate Assembly. The hybrid biosensor pSRP was composed of a constitutive promoter P1888, and an AdoCbl riboswitch RS1888 was amplified from the SM320 genome. The sequences of promoter P1888 and AdoCbl riboswitch RS1888 are listed in Table S3. The transcriptional repressor TetR and its cognate promoter PtetR sequences from the literature [39] were synthesized by GENEWIZ Company (Suzhou, China). To construct the pSRPI1 plasmid, LacI repressor and lacO operate sequences were introduced into the pSRP plasmid. To construct the pSRPO1 plasmid, LacI repressor was deleted from the pSRPI1 plasmid. To construct plasmids pSRPI2 and pSRPI3, promoter P1888 was replaced by BBa_J23110 (P110) and BBa_J23100 (P100), respectively. To construct the pSRPI4 plasmid, the transcriptional repressor TetR and its cognate promoter PtetR were replaced by PhlF and its cognate promoter PPhlF [39], whose sequences were synthesized by GENEWIZ Company (Suzhou, China). To construct the pSRPI5 plasmid, the reporter gene gfp was replaced by sfgfp. Five genetically modified strains, SMWM1, SMWM2, SMPUM, SMBM, and SMPUBM, were constructed by the CRISPR/Cas12e genetic editing tool [43]. Successful knockouts or insertions were confirmed via colony PCR and Sanger sequencing with specific genomic primers. The primers used in this study are listed in Table S3.
2.3. Transcriptome sequencing of SM320
SM320 was grown in 5 mL TYC, and when the OD600 arrived about 0.6–0.8, vitamin B12 was added with the final concentration of 10 μM in the test groups. After 2 h, the cells were harvested, respectively. Then the cells were washed three times in phosphate buffered saline (PBS, pH 7.4) and the primary and whole transcriptomes of SM320 were characterized through sequencing of mRNA and ncRNA isolated from SM320 samples. Transcriptome sequencing performed on samples by GENEWIZ (Suzhou, China) using the Illumina platform, the fragments per kilo bases per million reads (FPKM) value of each gene was generated.
2.4. Characterization of the AdoCbl biosensor
SM320 strains harboring the AdoCbl biosensor were cultured overnight at 32 °C in TYC medium and then inoculated into 5 mL of fresh TYC medium containing SpeR (600 μg/mL) at an initial OD600 of 0.05. To identify the dose-response profile of the biosensor, Adenosylcobalamin (AdoCbl) (RiboBio, China) was added to the medium at different concentrations from 0 μM to 50 μM, and the strains were subjected to fluorescence detection after incubation at 32 °C for 20 h. In the induction situation, when the cell OD600 reached approximately 0.8, isopropyl-beta-d-thiogalactopyranoside (IPTG) was added at a final concentration of 1 mM and subsequently incubated at 32 °C for 20 h for fluorescence detection. To verify the specificity of the biosensor, 10 μM cyanocobalamin (CNCbl) (ruibio, China) or AdoCbl was added to the medium, and the strains were then incubated at 32 °C for 20 h for fluorescence detection. The fluorescence intensities of green fluorescent protein (GFP) and OD600 of the cells were measured by a Synergy Neo2 multimode microplate reader (BioTek, VT, USA). Fluorescence was detected using excitation and emission filters of 485 nm and 525 nm, respectively. The cellular OD600 was measured with a 600 nm filter. By OD600 normalizing the fluorescence intensity, the specific fluorescence was estimated. For biological investigations, each culture was carried out in triplicate. The fluorescence values of single cells were analyzed by FCM (flow cytometry, MoFlo XDP, Beckman, USA).
2.5. Validation of the AdoCbl biosensor response to endogenous AdoCbl
A single colony of the strain harboring the AdoCbl biosensor was inoculated into 5 mL of TYC containing SpeR (600 μg/mL) as seed cultures. After culturing at 32 °C for 18 h, the seed cultures were inoculated into minimal medium containing SpeR (600 μg/mL) with an initial OD600 of 0.1. A final dosage of 1 mM IPTG was applied when the cells’OD600 value reached approximately 0.8. OD600 and GFP fluorescence were measured after 18 h of incubation using a Synergy Neo2 multimode microplate reader (BioTek, VT, USA). All incubations were repeated biologically.
2.6. Fermentation and analytical methods
Colonies were cultured in 24-well deep microtiter plates (Corning Costar 3524, square U-bottom, 5 mL) to grow SM320 strains. Colonies were grown in 2 mL fermentation culture, and then 10 % (v/v) inoculum was transferred to 2 mL fresh fermentation culture using 24-well deep microtiter plates using a Microtron shaker at 32 °C, 800 rpm, and 60 % humidity for SM320 strains in 24-well deep microtiter plates (Corning Costar 3524, square U-bottom, 5 mL) (Infors, China). For shake flask fermentation, the seed was grown in fermentation culture at 32 °C for 48 h and was used to provide 10 % (v/v) inoculum for fermentation. Experiments were conducted in 250 mL flasks containing 25 mL of fermentation culture at 32 °C at an agitation rate of 200 rpm in rotary shakers for 7 days. The samples were used to determine the cell mass (OD600) and AdoCbl titer on the 7th day. The concentrations of the principal fermentation products were determined by high-performance liquid chromatography (HPLC) using the process described in our previous study [42].
2.7. Flow cytometry-based library screening
A single colony of the SM320 strain was inoculated into TYC medium to bring the cell OD600 to 0.8. The cells were then treated with ultraviolet radiation for 65 s. The cell mortality rate was approximately 80 %. Cells were inoculated in fresh TYC medium overnight. The sensors were introduced into the mutagenesis cells and cultivated in TYC medium until the OD600 of the cells reached 0.8. IPTG was added to a final concentration of 1 mM, and the strains were subsequently cultivated at 32 °C for 20 h. The collected strains were washed twice and resuspended in PBS buffer, and the cell suspension was diluted to an OD600 of 0.05 to avoid internal interference. Cells were sorted using a MoFlo XDP flow cytometer sorter (Beckman Coulter Inc., Fullerton, CA, USA). The GFP fluorescence signal was excited at 488 nm, and emission was detected using a 530/40 bandpass filter. The top 0.1 % of cells with the highest fluorescence intensity were isolated and cultured on TYC agar plates at 32 °C for 5 d for further analysis.
3. Results and discussion
3.1. Design of an in vivo active AdoCbl biosensor
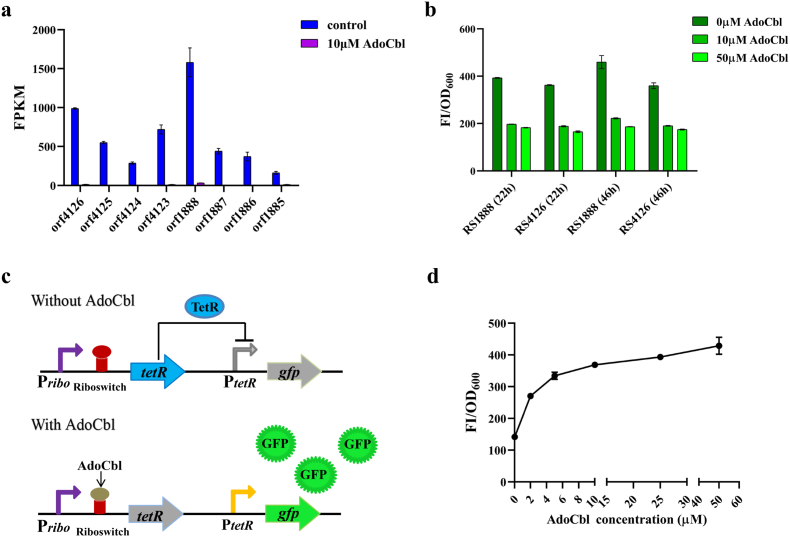
The AdoCbl-recognized riboswitches found in microbes are feedback-inhibited by AdoCbl. In this study, we did transcriptome analysis of the Sinorhizobium meliloti 320 (SM320) strain. Through transcriptome gene expression differentiation analysis between two groups, with exogenous AdoCbl added or not, we found that the expression of some genes underwent significant down-regulation with the addition of AdoCbl, and the expression of two operons, orf1888-1885 and orf4126-4123, was down-regulated the most (Fig. 1a). So we analyzed the 5′-untranslated region (5′-UTR) of these two operons, and using gfp as a reporter, we found that it could respond to AdoCbl and down-regulate the expression of gfp (Fig. 1b). The 5′-UTRs of the orf1888-1885 and orf4126-4123 operons from SM320 contained possible promoters and AdoCbl-responsive riboswitches (named RS1888 and RS4126) with sequences shown in Table S3. Sequence homology analysis showed that RS1888 and RS4126 had some sequence conservation compared with the AdoCbl riboswitch (PDB ID: 4GMA) of Thermoanaerobacter tengcongensis (Figure S1). The conserved sequences of RS1888 and RS4126 are shaded in yellow in Table S3. Transcriptome analysis is a convenient tool for obtaining target metabolite response riboswitches by gene expression differentiation analysis between two groups, with exogenous metabolites added or not. Furthermore, these riboswitches could be applied as a relatively noninvasive intracellular metabolite-recognizing component for biosensor development.
Fig. 1.
Design of a genetically encoded biosensor for measuring AdoCbl concentrations. (a) Transcriptome gene expression differentiation analysis showed that the expression of two operons, orf1888-1885 and orf4126-4123, underwent significant down-regulation with the addition of 10 μM AdoCbl. (b) RS1888 and RS4126 could respond to AdoCbl and down-regulate gfp expression. (c) The genetic circuit of the original RNA-protein hybrid AdoCbl biosensor pSRP. The hybrid biosensor contains a riboswitch RS1888 as a ligand-sense input part, which is expressed by its original promoter Pribo, the transcriptional repressor TetR and its cognate promoter PtetR as a signal inverter, and the fluorescence protein gfp as the output, which is expressed by PtetR. Lacking AdoCbl, TetR is fully expressed and binds to PtetR, suppressing the expression of the downstream gfp. In contrast, in the presence of AdoCbl, the expression of TetR will be inhibited, and reliving PtetR, the expression of gfp will be normal. (d) The dose-response profile of the strain containing the AdoCbl biosensor pSRP in different concentrations (0 μM–50 μM) of AdoCbl. The standard deviations are presented as error bars from three independent biological replicates.
To couple the in vivo AdoCbl level with a positive related and conspicuous readout, an RNA-protein hybrid biosensor that consists of three sections was designed: (1) the RNA riboswitch, which could respond to AdoCbl binding with specificity; (2) the transcriptional repressor and its cognate promoter to make input signal inversion; and (3) a select marker or the reporter gene for biosensor characterization. Its working principle is that the RNA riboswitch responds to elevated levels of AdoCbl and correspondingly downregulates transcriptional repressor transcription, relieving the expression of the gfp gene. Then, we chose the transcriptional repressor TetR and its cognate promoter PtetR, which have high orthogonality in cells [39,40]. In this constructed genetic circuit, tetR was expressed under the control of the AdoCbl riboswitch RS1888, and green fluorescence protein (GFP) was used as a reporter and expressed under the control of the TetR cognate promoter PtetR in the plsmid pKP. This initial sensor is named pSRP. A schematic representation of the hybrid biosensor is shown (Fig. 1c).
To characterize the designed AdoCbl sensor in vivo, exogenous AdoCbl ranging from 0 μM to 50 μM was added to SM320 strains harboring plasmid pSRP, and the resulting fluorescence intensity was measured after AdoCbl addition. An increase in specific GFP fluorescence (FI/OD600) was observed with increasing exogenous AdoCbl addition, but it was not applicable due to its narrow dose-response dynamic range and operating concentration (Fig. 1d).
3.2. Improving the dose-response parameters of the AdoCbl biosensor
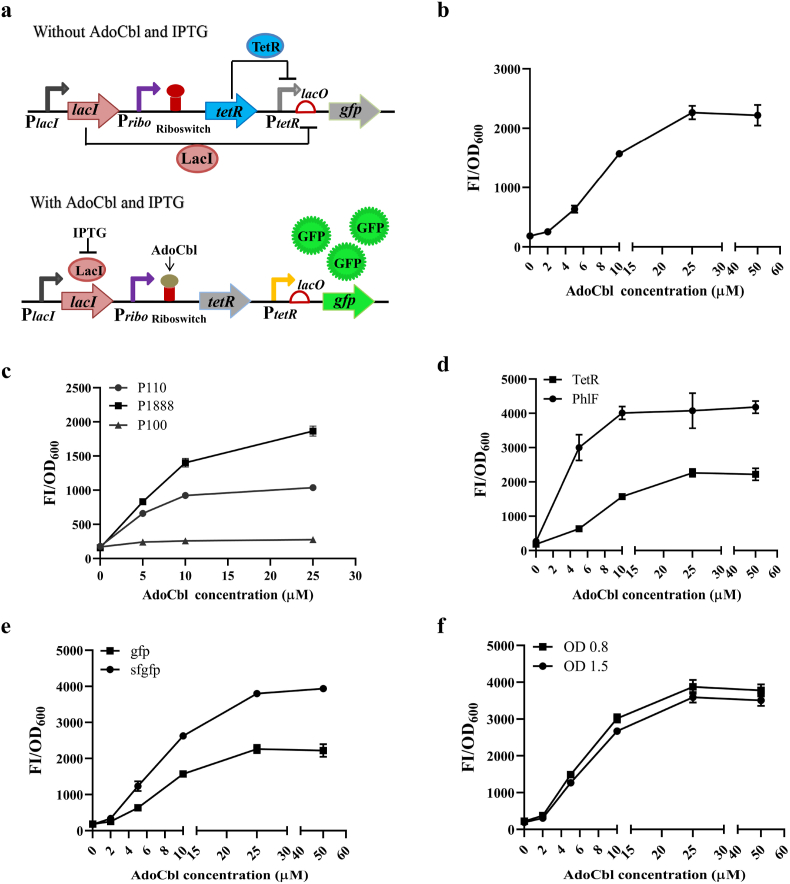
We have tried to increase the intensity of GFP expression in pSRP, but the results showed that it leads to severe leaky expression (Fig. S2). The wide operating concentrations and high dynamic range are also important principles for the development of a biosensor for high-throughput screening. The RNA-protein hybrid biosensor is generally faced with a similar problem where the low expression level of transcriptional repressor and high expression level of GFP cause serious GFP leaky expression, but the high expression level of transcriptional repressor and low expression level of GFP make the dynamic range narrow. Taking into consideration these situations, the transcriptional repressors lacI and lac operator (lacO) were introduced into pSRP. LacO was inserted downstream of PtetR to lower the leaky expression of GFP, which then produced the plasmid pSRPI1 (Fig. 2a). The dose-response dynamic range and operating concentration of pSRPI1 were controlled by both AdoCbl and IPTG (Fig. S3). To characterize pSRPI1, exogenous AdoCbl ranging from 0 μM to 50 μM was added to SM320 strains harboring plasmid pSRPI1. The optimal IPTG was added and the resulting fluorescence intensity was measured (Fig. S3). The results showed that our sensor exhibited increased GFP values in response to elevated concentrations of AdoCbl from 0 μM to 25 μM and appeared to plateau when the addition of AdoCbl was >25 μM (Fig. 2b). Additionally, the sensor's dynamic range and operating concentration were larger than those of pSRP. We further explored the reasons for the increase in gfp expression after insertion of LacI/lacO. After the insertion of lacO, the translation rate of the gfp gene changed from 7137.93 to 8211.21, as predicted by the De Novo DNA website [44]. Two plasmids pKP-PtetR-RBS-gfp (pRG) and pKP-PtetR-RBS/lacO-gfp (pROG) were constructed to determine the expression level of gfp. The results showed that there was no significant increase in gfp expression after the insertion of lacO (Fig. S4). We also identified LacI-free pSRPI1 (pSRPO1), and the dose-response parameters of pSRPO1 were similar to those of pSRP (Fig. S5). In summary, we hypothesize that the binding of LacI to lacO would prevent TetR from binding to its promoter PterR, leading to an increase in gfp fluorescence expression upon induction by the addition of IPTG. These results indicate that the high expression level of GFP could improve the dose-response dynamic range of the biosensor, with the premise of sufficient inhibition of its leaky expression.
Fig. 2.
Dose-response curves of AdoCbl biosensor tuning. (a) The genetic circuit of the IPTG-induced AdoCbl biosensor pSRPI1. In the absence of AdoCbl and IPTG, TetR and LacI are expressed and bind to PtetR and lacO, respectively, inhibiting the expression of gfp. In the presence of AdoCbl and IPTG, the expression of TetR will be inhibited, and reliving PtetR, LacI will bind to IPTG, and reliving lacO, the expression of gfp will be normal. (b) The dose-response profile of the strain containing the AdoCbl biosensor pSRPI1 in different concentrations (0 μM–50 μM) of AdoCbl. (c) Dose-response curves of pSRPI1 with promoters P110, P1888, and P100 in different concentrations (0 μM–25 μM) of AdoCbl. (d) Dose-response curves of pSRPI1 with TetR and PhlF in different concentrations (0 μM–25 μM) of AdoCbl. (e) Dose-response curves of pSRPI1 with gfp and sfgfp in different concentrations (0 μM–25 μM) of AdoCbl. (f) Dose-response curves of pSRPI5 with IPTG added at OD600 values of 0.8 and 1.5 in different concentrations (0 μM–25 μM) of AdoCbl. The standard deviations are presented as error bars from three independent biological replicates.
The riboswitch, transcriptional repressors, and output reporters are the main determinants for the dose-response dynamic range of the hybrid biosensor, and level matching between these elements is important. Then, three promoters with different strengths (P100>P1888>P110) were utilized to control transcriptional repressor expression for tuning the dose-response performance of the biosensor. The detail strengths of P100, P1888, and P110 were displayed in Fig. S6, and the sequences of P100, P1888, and P110 were shown in Table S3. As shown in Fig. 2c, the dynamic range of the AdoCbl biosensor was changed with different promoter strengths, and the moderate-strength promoter P1888 had the largest dynamic range and was used for further optimization. We thought the strong promoter might result in repressor overexpression that inhibited GFP expression irreversibly, and the weak promoter might result in an excessively low dynamic range of repressor expression. As the PhlF repressor has stronger inhibition than TetR [39], we replaced TetR and its cognate promoter with PhlF, as shown in Fig. 2d. The sensitivity of the sensor was improved, and the fold-activation was increased by approximately 2-fold, but the operating range was decreased a lot. Then, the output gfp gene was replaced by sfgfp, which has stronger fluorescence intensity than gfp. As shown in Fig. 2e, the fold change was increased by 2-fold, with the operational range remaining unchanged. In order to study the optimal timing of IPTG addition, two OD600s were chosen: OD600 at 0.8, when the strain was in the pre-logarithmic growth stage and AdoCbl was just beginning to be rapidly synthesized, and OD600 at 1.5, when the strain was in the late-logarithmic growth stage and AdoCbl was being rapidly synthesized. When the OD600 of the cells reached 0.8 and 1.5, IPTG was added to a final concentration of 1 mM. The experimental results showed that the fluorescence intensity was slight stronger when IPTG was added at an OD600 of approximately 0.8, as shown in Fig. 2f. To investigate whether the higher fluorescence intensity at OD600 0.8 than at OD600 1.5 was due to the low IPTG concentration, we examined the fluorescence intensity of the strains induced at 1, 2, 5, and 10 mM IPTG concentrations. The results showed that the fluorescence intensity at OD600 0.8 was always higher than that at OD600 1.5 at higher induction concentrations (Fig. S7). This may be due to the fact that at OD600 0.8 the strain is in a period of rapid growth with faster protein synthesis, which results in the expression of fluorescent proteins a bit more. Finally, we obtained an optimized biosensor, pSRPI5, whose concentration range covered by AdoCbl from 0 μM to 25 μM was converted into a dynamic range of more than 20-fold fluorescent signal output. These findings indicate that an in vivo active AdoCbl biosensor was successfully constructed.
3.3. Validation of the biosensor specificity in vivo
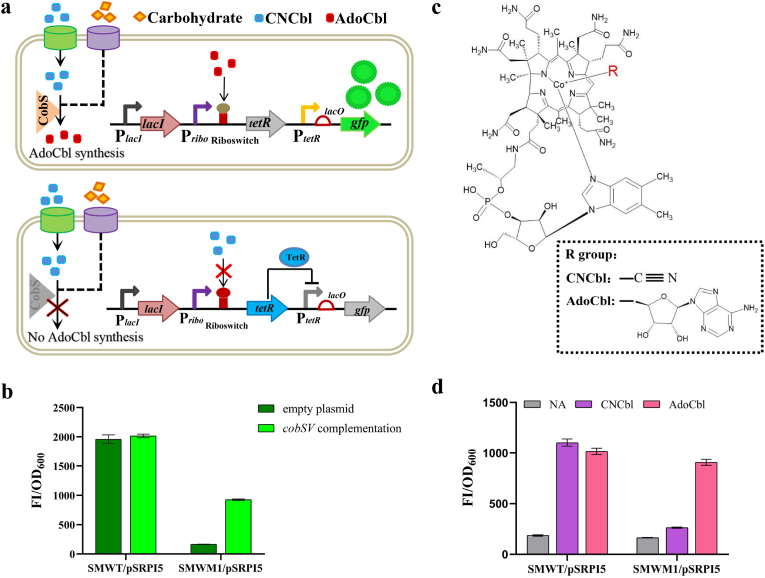
Specific responses to the target metabolites are one of the important principles for the development of a biosensor for high-throughput screening. To demonstrate the specificity of our sensor, we knocked out the cobSV gene and obtained the mutant strain SMWM1, which is unable to synthesize the end product AdoCbl (Fig. 3a and S8). The cobSV gene encodes adenosylcobinamide-GDP ribazoletransferase, which is the penultimate step enzyme of the AdoCbl biosynthesis pathway and is necessary for AdoCbl biosynthesis both in the de novo biosynthesis pathway and the cobalamin remodeling pathway [26,45]. Plasmid pSRPI5 was transformed into SMWM1 and the native strain SMWT. The SMWT/pSRPI5 strain had the ability to synthesize AdoCbl, resulting in a fluorescence response, but the SMWM1/pSRPI5 strain lost the ability to synthesize AdoCbl, which had no fluorescence response. The CobSV expression plasmid was constructed and complemented into the SMWM1/pSRPI5 strain, which made the SMWM1/pSRPI5 strain regain the AdoCbl synthesis capacity and have a fluorescence response (Fig. 3b). These results indicated with high probability that pSRPI5 could specifically respond to the end product AdoCbl. But not to intermediate metabolites produced during AdoCbl biosynthesis. To demonstrate the specificity of the biosensor, we exogenously added 10 μM CNCbl (cyanocobalamin) and AdoCbl to the medium. Because CNCbl and AdoCbl have the same structure except for the upper ligand, the upper ligand of CNCbl is cyan, while the upper ligand of AdoCbl is deoxyadenosine (Fig. 3c). CNCbl can be transformed into AdoCbl through the cobalamin remodeling pathway in SMWT, but it cannot in SMWM1 due to cobSV deletion. The result showed that the SMWT/pSRPI5 strain could respond to both AdoCbl and CNCbl. In contrast, the SMWM1/pSRPI5 strain was able to respond to AdoCbl, but very little to CNCbl. These results indicate that the biosensor is specific to AdoCbl (Fig. 3d). It was confirmed that an intracellularly active and specifically responsive AdoCbl biosensor was successfully constructed.
Fig. 3.
pSRPI5 specifically responds to AdoCbl. (a) Schematic of AdoCbl biosynthesis and biosensor workflow in SMWT (up) and SMWM1 (down). The cobSV gene is necessary for AdoCbl biosynthesis, both in de novo synthesis from glucose to AdoCbl and in the cobalamin remodeling pathway from CNCbl to AdoCbl. The mutant strain SMWM1 (cobSV deletion) leads to a lack of AdoCbl production. (b) pSRPI5 could specifically respond to the end product, AdoCbl. In the minimal medium, both SMWT complement empty plasmid and cobSV could produce AdoCbl, so pSRPI5 could respond to AdoCbl; SMWM1 complement empty plasmid could not produce AdoCbl, so pSRPI5 had no fluorescence response; but SMWM1 complement cobSV could produce AdoCbl, so pSRPI5 could respond to AdoCbl. (c) The molecular structure of cobalamin. The R residue is cyan to represent cyanocobalamin, and the R residue is adenosine to represent adenosylcobalamin. (d) The ligand specificity test for pSRPI5 with both CNCbl and AdoCbl. In minimal medium, SMWT could produce AdoCbl from CNCbl, so pSRPI5 had a fluorescence response to AdoCbl. SMWM1 could not produce AdoCbl from CNCbl, so pSRPI5 had no fluorescence response. NA represents no CNCbl or AdoCbl addition. The standard deviations are presented as error bars from three independent biological replicates.
3.4. Validation of the biosensor-based selection system
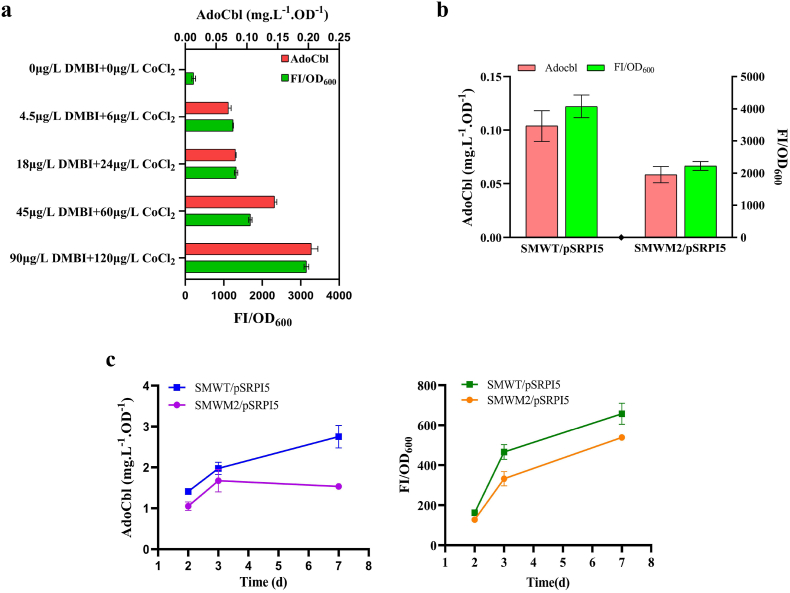
To verify that the biosensor can quantify the AdoCbl produced in SMWT strains, the relationship between GFP and intracellular AdoCbl concentrations was measured (Fig. 4a). The result showed that there is a correlation between GFP and AdoCbl produced intracellularly. To reveal the applicability of the AdoCbl biosensor for genetic screening, an enrichment experiment was carried out as a proof of concept. The RBS of the cobPWNO operon, which is necessary for AdoCbl synthesis, was replaced with the 5′-UTR of the orf6901-6904 operon (104 bases) from SM320 (Table S3), resulting in the SMWM2 strain, which has lower AdoCbl productivity than the SMWT strain in both minimal medium and fermentation culture (Fig. 4b and c). In SMWT and SMWM2 cells carrying pSRPI5, the specific GFP fluorescence increased with increasing AdoCbl production, and the specific GFP fluorescence of SMWT/pSRPI5 was higher than that of SMWM2/pSRPI5. However, during fermentation culture, AdoCbl production of the SMWM2/pSRPI5 strain reached a maximum on the third day, and the specific GFP fluorescence increased all the time. This is because the growth of the SMWM2/pSRPI5 strain reached a stable period on the third day, and the strain began to die in large numbers around the fifth day. By the end of the fermentation on the seventh day, the strain had died in large numbers, so the measured OD600 and fluorescence values may not be accurate.
Fig. 4.
Fermentation of SMWT/pSRPI5 and SMWM2/pSRPI5. (a) Relationship between GFP and intracellular AdoCbl concentration. As the intracellular production of AdoCbl increased, the fluorescence intensity was enhanced. (b) Fermentation and fluorescence responses of SMWT/pSRPI5 and SMWM2/pSRPI5 in minimal medium. (c) Fermentation and fluorescence responses of SMWT/pSRPI5 and SMWM2/pSRPI5 in fermentation medium. Error bars indicate standard errors from three independent biological replicates.
To separate single producers by flow cytometry, the cells must be isolated based on their fluorescence characteristics. To demonstrate our sensor in this way, SMWT/pSRPI5 and SMWM2/pSRPI5 cells were grown in minimal medium, and IPTG was added to a final concentration of 1 mM when the cell OD600 reached approximately 0.8. After 20 h, this cell population was analyzed via FACS. A clear distinction of groups was obtained according to the intensity of the GFP signal (Fig. S9a). The two species, SMWT/pSRPI5 and SMWM2/pSRPI5, were mixed at a ratio of 1:100 and co-cultured in minimal medium, and IPTG was added at a final concentration of 1 mM when the cell OD600 reached approximately 0.8. After 24 h, the mixture of SMWT/pSRPI5 and SMWM2/pSRPI5 was sorted by FACS based on GFP intensity at a rate of 2*104 per second to isolate the top 0.1 % GFP-intensity cell colonies. Colony PCR was adopted to assess the ratio of SMWT to SMWM2. In a total of 3 independent replicate experimental groups, the ratio of SMWT/pSRPI5 was 16/20, 14/20, and 14/20. In contrast, the ratios of SMWT/pSRPI5 were 0/24, 0/24, and 0/24 in the control group, which was randomly sorted by flow cytometry. This implies that the high AdoCbl producers SMWT/pSRPI5 were more than 70-fold enriched in the selection experiment (Fig. S9b). Here, a dosage of 1:100 was chosen to ensure an easily detectable enrichment rate within a single screening cycle. This result further supports the possibility of AdoCbl biosensor applications in the selection process of microbial evolution or pathway optimization.
3.5. Establishment of the high-throughput screening platform
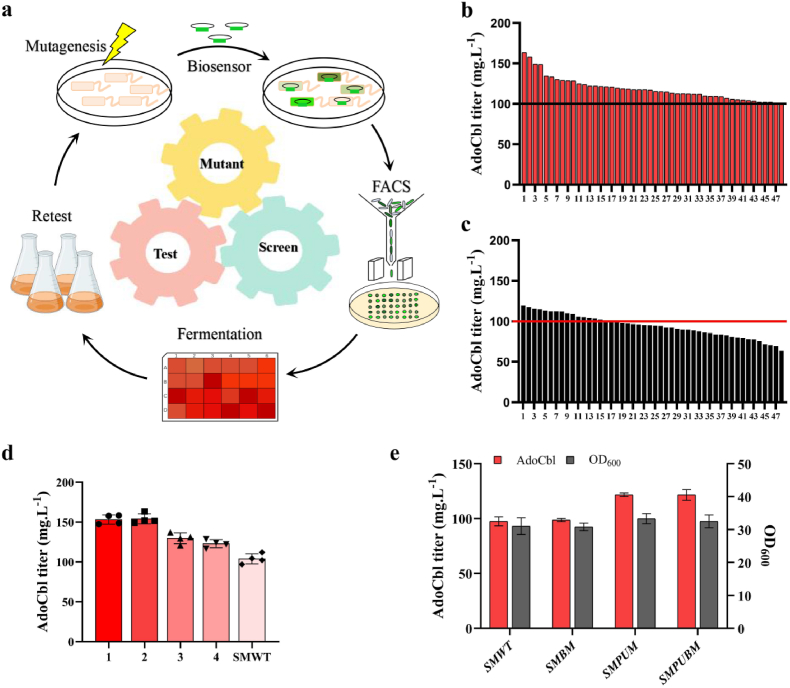
By combining the AdoCbl biosensor and FACS technology, a high-throughput screening (HTS) platform was created to screen high-performance production strains from a vast mutant library. The platform's workflow is illustrated in Fig. 5a. To generate random mutation libraries, the initial SM320 strain, SMWT, was exposed to UV radiation to obtain random mutagenesis. Based on a mortality rate of approximately 80 %, the optimal mutation time was 65 s (Fig. S10). Then, approximately 108 living cells were transformed with plasmid pSRPI5 to avoid unwanted mutations to the plasmid. After the mutation library was constructed, mutants with the top 0.1 % GFP signal intensity were sorted at a rate of 2*104 per second. The sorted single colonies were selected and incubated on agar plates. After incubation, 96 colonies were selected and cultured in 24-well microtiter plates. As a control, randomly selected 48 colonies from the mutagenesis library were incubated under the same conditions. The final AdoCbl titers of these candidate strains were analyzed by HPLC. The HPLC results showed that 45 % (43/96) of the strains had improved AdoCbl titers compared to the parental strains, and 10 % (10/96) of the strains obtained an access improvement of AdoCbl titers after screening on the HTS platform (Fig. 5b and c).
Fig. 5.
Construction of the HTS platform for AdoCbl high-producing strains. (a) Workflow of the biosensor-based HTS platform. (b) AdoCbl production of strains screened by FACS. Ninety-six colonies were randomly selected from the screened cells by FACS and incubated in 24-well microtiter plates, and the AdoCbl titers were analyzed by HPLC. The top 48/96 AdoCbl titers are shown. (c) AdoCbl production of strains randomly selected from the mutagenesis library. Forty-eight colonies were randomly selected from the screened cells by FACS and incubated in 24-well microtiter plates, and the AdoCbl titers were analyzed by HPLC. (d) AdoCbl production of the four highest strains from (b) and parent strain SMWT in 250 mL shake flask fermentation. AdoCbl titers were all determined by HPLC. (e) AdoCbl production of the SMPUM, SMBM, SMPUBM, and parent strain SMWT in 250 mL shake flask fermentation. AdoCbl titers were all determined by HPLC.
To further demonstrate the stability and reproducibility of the strains obtained by the HTS platform, the first four colonies with the highest AdoCbl titers and the initial strain SMWT were cultured and fermented in 250 mL shake flasks. The AdoCbl production capacity of these five strains in shake flasks was consistent with that in 24-well microplates (Fig. 5d). In order to identify gene mutations that contributed to the high production of AdoCbl, we sequenced the genomic DNA of the mutant strain 1 (Fig. 5d). We found that the genotype of the isolated mutant strain had lots of mutations that made it difficult to identify the most important mutagenesis. Among them, two genes of the AdoCbl synthesis pathway, cobPU and cobB, have mutations that result in cobPUmut (G59S, E100G, and Q153M) and cobBmut (A34V), which were not reported in S. meliloti. To investigate the roles of cobPUmut (G59S, E100G, and Q153M) and cobBmut (A34V), SMPUM, SMBM, and SMPUBM strains were constructed by replacing cobPU and cobB genes with cobPUmut and cobBmut on the genome alone or in combination. The results showed that, compared to the SMWT, there was a definite increase in AdoCbl yield in SMPUM, no significant increase in AdoCbl yield in SMBM, and no further increase in AdoCbl yield in SMPUBM (Fig. 5e).
These outcomes confirm that our high-throughput screening platform, a potent platform for obtaining desirable AdoCbl overproduction strains, has the advantages of a short cycle time, ease of operation and high efficiency. Although rational design is difficult due to the metabolic complexity of AdoCbl biosynthesis in SM320, genome-wide analysis of screened overproducers may shed new light on the mechanism of AdoCbl overproduction and the development of overproducing AdoCbl strains. With the replacement of target RNA elements, the biosensor could also be applicable to a broad range of organisms and metabolites. Therefore, the construction model of this biosensor is expected to be useful for efficient and high-throughput screening of other metabolite producers in diverse strains.
4. Conclusion
Biosensors play a crucial role in large-scale screening of mutant strains as a component linking genotype to phenotype. In this study, we developed an AdoCbl biosensor and, coupled with fluorescence-activated cell sorting (FACS) technology, established an HTS platform for screening of AdoCbl high-producing Sinorhizobium meliloti 320 strains, which could be a powerful platform for the application of monitoring and measuring AdoCbl. Almost all AdoCbl riboswitches have been found to be negatively regulated elements inhibited by AdoCbl feedback and cannot be utilized alone for high-throughput screening of industrial strains based on fluorescence signals. Because the selection of low fluorescence signals by FACS will result in a high sorting false rate in technology, we utilized RNA-protein hybridization elements for signal input and inversion to positively correlate intracellular AdoCbl levels with fluorescence readings. We also improved the operating concentration and dynamic range of the hybrid sensor by systematically optimizing its genetic components. We found that the hybrid biosensor could effectively and specifically respond to changes in the intracellular AdoCbl and transform them to an output signal corresponding to green fluorescent protein expression. Using the HTS platform could allow fast and efficient identification of improved mutants from a large library of mutant strains. Although rational design is difficult due to the metabolic complexity of AdoCbl biosynthesis in SM320, genome-wide analysis of screened overproducers may shed new light on the mechanism of AdoCbl overproduction and the development of overproducing AdoCbl strains. With the replacement of target RNA elements, the biosensor could also be applicable to a broad range of organisms and metabolites. Therefore, the construction model of this biosensor is expected to be useful for efficient and high-throughput screening of other metabolite producers in diverse strains.
Supplementary material
Strains used in this study (Table S1); Plasmids used in this study (Table S2). Oligonucleotides used in this study (Table S3). Sequence comparisons of RS1888 and RS4126 with the structurally resolved AdoCbl riboswitch (PDB ID: 4GMA) from Thermoanaerobacter tengcongensis (Fig. S1). The dose-response profile of the strain containing the AdoCbl biosensors pSRP and pSRP-RBSN in different concentrations (0, 10, and 25 μM) of AdoCbl (Fig. S2). Dynamic behavior of the AdoCbl biosensor pSRPI1 with 0, 0.05, 0.1, 0.5 or 1 mM IPTG concentrations under 0–25 μM AdoCbl (Fig. S3). Expression strength of the pRG (pKP-PtetR-RBS-gfp) and pROG (pKP-PtetR-RBS/lacO-gfp) (Fig. S4). The dose-response profile of the strain containing the AdoCbl biosensors pSRP and pSRO1 in different concentrations (0, 10, and 25 μM) of AdoCbl (Fig. S5). Expression strength of the three promoters (Fig. S6). Dynamic behavior of the AdoCbl biosensor pSRPI5 with 1, 2, 5, or 10 mM IPTG concentrations under 0–50 μM AdoCbl at OD600 0.8 and OD600 1.5 (Fig. S7). Identification of AdoCbl by HPLC (Fig. S8). Validation of the selection system using two different AdoCbl productivity strains, SMWT and SMWM2 (Fig. S9). Lethal rates of SM320 irradiated by UV mutagenesis with different mutation times (Fig. S10).
CRediT authorship contribution statement
Xia Yang: Conceptualization, Methodology, Investigation, Writing – review & editing. Huiying Wang: Conceptualization, Methodology, Investigation, Writing – review & editing, Writing – original draft. Dongqin Ding: Writing – review & editing. Huan Fang: Writing – review & editing, Funding acquisition. Huina Dong: Validation, Writing – review & editing. Dawei Zhang: Conceptualization, Resources, Writing – review & editing, Supervision, Funding acquisition.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
This work was supported by the National Key R&D Program of China [Grant number 2021YFC2100700]; the National Natural Science Foundation of China [Grant numbers 22178372, 32300069]; TIB-VIB Joint Center of Synthetic Biology [Grant number TSBICIP-IJCP-002]; the National Science Fund for Distinguished Young Scholars [Grant number 22325807]; the Tianjin Synthetic Biotechnology Innovation Capacity Improvement Project [Grant numbers TSBICIP-KJGG-011, TSBICIP-CXRC-055]; and the Yellow River Delta Industry Leading Talents [Grant number DYRC20190212].
Footnotes
Peer review under responsibility of KeAi Communications Co., Ltd.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.synbio.2024.04.008.
Contributor Information
Huina Dong, Email: dong_hn@tib.cas.cn.
Dawei Zhang, Email: zhang_dw@tib.cas.cn.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Keasling J.D. Synthetic biology and the development of tools for metabolic engineering. Metab Eng. 2012;14:189–195. doi: 10.1016/j.ymben.2012.01.004. [DOI] [PubMed] [Google Scholar]
- 2.Nielsen J., Keasling J.D. Engineering cellular metabolism. Cell. 2016;164:1185–1197. doi: 10.1016/j.cell.2016.02.004. [DOI] [PubMed] [Google Scholar]
- 3.Schellenberger J., Que R., Fleming R.M., Thiele I., Orth J.D., Feist A.M., Zielinski D.C., Bordbar A., Lewis N.E., Rahmanian S., Kang J., Hyduke D.R., Palsson B. Quantitative prediction of cellular metabolism with constraint-based models: the COBRA Toolbox v2.0. Nat Protoc. 2011;6:1290–1307. doi: 10.1038/nprot.2011.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen X., Gao C., Guo L., Hu G., Luo Q., Liu J., Nielsen J., Chen J., Liu L. DCEO Biotechnology: tools to design, construct, evaluate, and optimize the metabolic pathway for biosynthesis of chemicals. Chem Rev. 2018;118:4–72. doi: 10.1021/acs.chemrev.6b00804. [DOI] [PubMed] [Google Scholar]
- 5.Huang M., Bao J., Hallström B.M., Petranovic D., Nielsen J. Efficient protein production by yeast requires global tuning of metabolism. Nat Commun. 2017;8:1131. doi: 10.1038/s41467-017-00999-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rogers J.K., Taylor N.D., Church G.M. Biosensor-based engineering of biosynthetic pathways. Curr Opin Biotechnol. 2016;42:84–91. doi: 10.1016/j.copbio.2016.03.005. [DOI] [PubMed] [Google Scholar]
- 7.Zhang Y.X., Perry K., Vinci V.A., Powell K., Stemmer W.P., Del Cardayré S.B. Genome shuffling leads to rapid phenotypic improvement in bacteria. Nature. 2002;415:644–646. doi: 10.1038/415644a. [DOI] [PubMed] [Google Scholar]
- 8.Binder S., Schendzielorz G., Stäbler N., Krumbach K., Hoffmann K., Bott M., Eggeling L. A high-throughput approach to identify genomic variants of bacterial metabolite producers at the single-cell level. Genome Biol. 2012;13:R40. doi: 10.1186/gb-2012-13-5-r40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Becker S., Schmoldt H.U., Adams T.M., Wilhelm S., Kolmar H. Ultra-high-throughput screening based on cell-surface display and fluorescence-activated cell sorting for the identification of novel biocatalysts. Curr Opin Biotechnol. 2004;15:323–329. doi: 10.1016/j.copbio.2004.06.001. [DOI] [PubMed] [Google Scholar]
- 10.Dietrich J.A., Shis D.L., Alikhani A., Keasling J.D. Transcription factor-based screens and synthetic selections for microbial small-molecule biosynthesis. ACS Synth Biol. 2013;2:47–58. doi: 10.1021/sb300091d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Berens C., Suess B. Riboswitch engineering - making the all-important second and third steps. Curr Opin Biotechnol. 2015;31:10–15. doi: 10.1016/j.copbio.2014.07.014. [DOI] [PubMed] [Google Scholar]
- 12.Raman S., Rogers J.K., Taylor N.D., Church G.M. Evolution-guided optimization of biosynthetic pathways. Proc Natl Acad Sci U S A. 2014;111:17803–17808. doi: 10.1073/pnas.1409523111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tang S.Y., Qian S., Akinterinwa O., Frei C.S., Gredell J.A., Cirino P.C. Screening for enhanced triacetic acid lactone production by recombinant Escherichia coli expressing a designed triacetic acid lactone reporter. J Am Chem Soc. 2013;135:10099–10103. doi: 10.1021/ja402654z. [DOI] [PubMed] [Google Scholar]
- 14.Schendzielorz G., Dippong M., Grünberger A., Kohlheyer D., Yoshida A., Binder S., Nishiyama C., Nishiyama M., Bott M., Eggeling L. Taking control over control: use of product sensing in single cells to remove flux control at key enzymes in biosynthesis pathways. ACS Synth Biol. 2014;3:21–29. doi: 10.1021/sb400059y. [DOI] [PubMed] [Google Scholar]
- 15.Jha R.K., Kern T.L., Fox D.T., Ce M.S. Engineering an Acinetobacter regulon for biosensing and high-throughput enzyme screening in E. coli via flow cytometry. Nucleic Acids Res. 2014;42:8150–8160. doi: 10.1093/nar/gku444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nahvi A., Sudarsan N., Ebert M.S., Zou X., Brown K.L., Breaker R.R. Genetic control by a metabolite binding mRNA. Chem Biol. 2002;9:1043. doi: 10.1016/s1074-5521(02)00224-7. [DOI] [PubMed] [Google Scholar]
- 17.Mironov A.S., Gusarov I., Rafikov R., Lopez L.E., Shatalin K., Kreneva R.A., Perumov D.A., Nudler E. Sensing small molecules by nascent RNA: a mechanism to control transcription in bacteria. Cell. 2002;111:747–756. doi: 10.1016/s0092-8674(02)01134-0. [DOI] [PubMed] [Google Scholar]
- 18.Serganov A., Nudler E. A decade of riboswitches. Cell. 2013;152:17–24. doi: 10.1016/j.cell.2012.12.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Michener J.K., Smolke C.D. High-throughput enzyme evolution in Saccharomyces cerevisiae using a synthetic RNA switch. Metab Eng. 2012;14:306–316. doi: 10.1016/j.ymben.2012.04.004. [DOI] [PubMed] [Google Scholar]
- 20.Yang J., Seo S.W., Jang S., Shin S.I., Lim C.H., Roh T.Y., Jung G.Y. Synthetic RNA devices to expedite the evolution of metabolite-producing microbes. Nat Commun. 2013;4:1413. doi: 10.1038/ncomms2404. [DOI] [PubMed] [Google Scholar]
- 21.Lee S.W., Oh M.K. A synthetic suicide riboswitch for the high-throughput screening of metabolite production in Saccharomyces cerevisiae. Metab Eng. 2015;28:143–150. doi: 10.1016/j.ymben.2015.01.004. [DOI] [PubMed] [Google Scholar]
- 22.Marcano-Velazquez J.G., Lo J., Nag A., Maness P.C., Chou K.J. Developing riboswitch-mediated gene regulatory controls in thermophilic bacteria. ACS Synth Biol. 2019;8:633–640. doi: 10.1021/acssynbio.8b00487. [DOI] [PubMed] [Google Scholar]
- 23.Světnička M., Sigal A., Selinger E., Heniková M., El-Lababidi E., Gojda J. Cross-sectional study of the prevalence of cobalamin deficiency and vitamin B12 supplementation habits among vegetarian and vegan children in the Czech republic. Nutrients. 2022;14:535. doi: 10.3390/nu14030535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Green R., Miller J.W. Vitamin B12 deficiency. Vitam Horm. 2022;119:405–439. doi: 10.1016/bs.vh.2022.02.003. [DOI] [PubMed] [Google Scholar]
- 25.Kang Z., Zhang J., Zhou J., Qi Q., Du G., Chen J. Recent advances in microbial production of δ-aminolevulinic acid and vitamin B12. Biotechnol Adv. 2012;30:1533–1542. doi: 10.1016/j.biotechadv.2012.04.003. [DOI] [PubMed] [Google Scholar]
- 26.Martens H. Microbial production of vitamin B 12. Appl Microbiol Biotechnol. 2002;58:275–285. doi: 10.1007/s00253-001-0902-7. [DOI] [PubMed] [Google Scholar]
- 27.Cheng X., Chen W., Peng W.-F., Li K.-T. Improved vitamin B12 fermentation process by adding rotenone to regulate the metabolism of Pseudomonas denitrificans. Appl Biochem Biotechnol. 2014;173:673–681. doi: 10.1007/s12010-014-0878-2. [DOI] [PubMed] [Google Scholar]
- 28.Piwowarek K., Lipińska E., Hać-Szymańczuk E., Bzducha-Wróbel A., Synowiec A. Research on the ability of propionic acid and vitamin B12 biosynthesis by Propionibacterium freudenreichii strain T82. Antonie Leeuwenhoek. 2018;111:921–932. doi: 10.1007/s10482-017-0991-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhao T., Cheng K., Cao Y.-H., Ouwehand A.C., Jiao C.-F., Yao S. Identification and antibiotic resistance assessment of ensifer adhaerens YX1, a vitamin B12-producing strain used as a food and feed additive. J Food Sci. 2019;84:2925–2931. doi: 10.1111/1750-3841.14804. [DOI] [PubMed] [Google Scholar]
- 30.Dong H., Li S., Fang H., Xia M., Zheng P., Zhang D., Sun J. A newly isolated and identified vitamin B12 producing strain: Sinorhizobium meliloti 320. Bioproc Biosyst Eng. 2016;39:1527–1537. doi: 10.1007/s00449-016-1628-3. [DOI] [PubMed] [Google Scholar]
- 31.Zhou J., Yu X., Liu J., Qin W., He Z., Stahl D., Jiao N., Zhou J., Tu Q. VB(12)Path for accurate metagenomic profiling of microbially driven cobalamin synthesis pathways. mSystems. 2021;6 doi: 10.1128/mSystems.00497-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nguyen-Vo T.P., Ainala S.K., Kim J.R., Park S. Analysis and characterization of coenzyme B12 biosynthetic gene clusters and improvement of B12 biosynthesis in Pseudomonas denitrificans ATCC 13867. FEMS Microbiol Lett. 2018;365 doi: 10.1093/femsle/fny211. [DOI] [PubMed] [Google Scholar]
- 33.Xia W., Chen W., Peng W.-F., Li K.-T. Industrial vitamin B12 production by Pseudomonas denitrificans using maltose syrup and corn steep liquor as the cost-effective fermentation substrates. Bioproc Biosyst Eng. 2015;38:1065–1073. doi: 10.1007/s00449-014-1348-5. [DOI] [PubMed] [Google Scholar]
- 34.Milunovic B., Dicenzo G.C., Morton R.A., Finan T.M. Cell growth inhibition upon deletion of four toxin-antitoxin loci from the megaplasmids of Sinorhizobium meliloti. J Bacteriol. 2014;196:811–824. doi: 10.1128/jb.01104-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li J., Ge Y., Zadeh M., Curtiss R3rd, Mohamadzadeh M. Regulating vitamin B12 biosynthesis via the cbiMCbl riboswitch in Propionibacterium strain UF1. Proc Natl Acad Sci USA. 2020;117:602–609. doi: 10.1073/pnas.1916576116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Palou-Mir J., Musiari A., Sigel R.K., Barcelo-Oliver M. Characterization of the full-length btuB riboswitch from Klebsiella pneumoniae. J Inorg Biochem. 2016;160:106–113. doi: 10.1016/j.jinorgbio.2015.12.012. [DOI] [PubMed] [Google Scholar]
- 37.Vitreschak A.G., Rodionov D.A., Mironov A.A., Gelfand M.S. Regulation of the vitamin B12 metabolism and transport in bacteria by a conserved RNA structural element. Rna. 2003;9:1084–1097. doi: 10.1261/rna.5710303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jang S., Jang S., Noh M.H., Lim H.G., Jung G.Y. Novel hybrid input Part Using riboswitch and transcriptional repressor for signal inverting amplifier. ACS Synth Biol. 2018;7:2199–2204. doi: 10.1021/acssynbio.8b00213. [DOI] [PubMed] [Google Scholar]
- 39.Stanton B.C., Nielsen A.A., Tamsir A., Clancy K., Peterson T., Voigt C.A. Genomic mining of prokaryotic repressors for orthogonal logic gates. Nat Chem Biol. 2014;10:99–105. doi: 10.1038/nchembio.1411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fowler C.C., Brown E.D., Li Y. Using a riboswitch sensor to examine coenzyme B(12) metabolism and transport in E. coli. Chem Biol. 2010;17:756–765. doi: 10.1016/j.chembiol.2010.05.025. [DOI] [PubMed] [Google Scholar]
- 41.Dong H., Li S., Fang H., Xia M., Zheng P., Zhang D., Sun J. A newly isolated and identified vitamin B12 producing strain: Sinorhizobium meliloti 320. Bioproc Biosyst Eng. 2016;39:1527–1537. doi: 10.1007/s00449-016-1628-3. [DOI] [PubMed] [Google Scholar]
- 42.Cui Y., Dong H., Tong B., Wang H., Chen X., Liu G., Zhang D. A versatile Cas12k-based genetic engineering toolkit (C12KGET) for metabolic engineering in genetic manipulation-deprived strains. Nucleic Acids Res. 2022;50:8961–8973. doi: 10.1093/nar/gkac655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Liu G., Wang H., Tong B., Cui Y., Vonesch S.C., Dong H., Zhang D. An Efficient CRISPR/Cas12e System for Genome Editing in Sinorhizobium meliloti. ACS Synth Biol. 2023;12:898–903. doi: 10.1021/acssynbio.2c00629. [DOI] [PubMed] [Google Scholar]
- 44.Halper S.M., Hossain A., Salis H.M. Synthesis success calculator: predicting the rapid synthesis of DNA fragments with machine learning. ACS Synth Biol. 2020;9:1563–1571. doi: 10.1021/acssynbio.9b00460. [DOI] [PubMed] [Google Scholar]
- 45.Ma A.T., Kantner D.S., Beld J. Cobamide remodeling. Vitam Horm. 2022;119:43–63. doi: 10.1016/bs.vh.2022.01.004. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.