Abstract
Previous studies of a group of mutants of the murine coronavirus mouse hepatitis virus (MHV)-A59, isolated from persistently infected glial cells, have shown a strong correlation between a Q159L amino acid substitution in the S1 subunit of the spike gene and a loss in the ability to induce hepatitis and demyelination. To determine if Q159L alone is sufficient to cause these altered pathogenic properties, targeted RNA recombination was used to introduce a Q159L amino acid substitution into the spike gene of MHV-A59. Recombination was carried out between the genome of a temperature-sensitive mutant of MHV-A59 (Alb4) and RNA transcribed from a plasmid (pFV1) containing the spike gene as well as downstream regions, through the 3′ end, of the MHV-A59 genome. We have selected and characterized two recombinant viruses containing Q159L. These recombinant viruses (159R36 and 159R40) replicate in the brains of C57BL/6 mice and induce encephalitis to a similar extent as wild-type MHV-A59. However, they exhibit a markedly reduced ability to replicate in the liver or produce hepatitis compared to wild-type MHV-A59. These viruses also exhibit reduced virulence and reduced demyelination. A recombinant virus containing the wild-type MHV-A59 spike gene, wtR10, behaved essentially like wild-type MHV-A59. This is the first report of the isolation of recombinant viruses containing a site-directed mutation, encoding an amino acid substitution, within the spike gene of any coronavirus. This technology will allow us to begin to map the molecular determinants of pathogenesis within the spike glycoprotein.
Various strains of mouse hepatitis virus (MHV) induce different patterns of pathogenesis, including enteritis, hepatitis, encephalitis, and demyelination in the mouse (12). The A59 strain of MHV (MHV-A59) produces severe hepatitis as well as mild acute meningoencephalitis and chronic demyelination in C57BL/6 weanling mice (16, 17). One of the major determinants of pathogenic properties of MHV is the spike glycoprotein, S (4, 6, 23). The S protein, found on the virion envelope and on the plasma membrane of infected cells, is responsible for attachment to viral receptor and virus-cell fusion during viral entry and for cell-to-cell fusion later during infection. S is an 180-kDa glycoprotein, which is cleaved into two noncovalently associated 90-kDa subunits, the amino-terminal S1 and carboxy-terminal S2 subunits (5, 20). It is believed that the S1 subunit forms the globular head of the spike and the S2 subunit forms the membrane-bound stalk portion (2). Recently, a receptor binding activity has been demonstrated in studies using a recombinant protein containing the amino-terminal 330 residues of the S1 subunit of MHV-JHM (15). S2 is believed to contain the domain that mediates fusion of viral and cell membranes (1, 2).
We have previously characterized a group of fusion-defective mutants of MHV-A59 derived from persistently infected glial cell cultures (8); these mutants are attenuated and display an altered pathogenesis including decreased hepatitis and demyelination (9, 11, 19). All mutants displaying these phenotypes have two, and only two, amino acid substitutions in the spike gene, H716D and Q159L. Analysis of fusion-competent revertant viruses demonstrated that the H716D amino acid substitution, located in the putative signal sequence for cleavage of S into S1 and S2, is responsible for the fusion phenotype but not for the alteration in pathogenesis (8, 11). Through the analysis of other mutant and revertant viruses, we have found a correlation between Q159L and a greatly reduced ability to induce hepatitis as well as demyelination (11, 19). We have further shown that the genome of one of these mutants, C12, has only three additional amino acid substitutions, all within the replicase gene (19).
Studies of the determinants of pathogenesis of coronaviruses in general have been limited by the lack of a technology with which to introduce mutations into the genome. To date there are no full-length cDNA clones of any coronavirus genome, probably due to the large (31 kb in the case of MHV) size of the genome. The best method currently available for the introduction of amino acid substitutions into the genome is targeted recombination. Targeted RNA recombination has been developed and used by Masters and colleagues to introduce mutations into the nucleocapsid (N) gene (14, 23a) and more recently into more upstream genes (3). We have extended this technology to the S gene. To determine whether Q159L alone is sufficient to cause the altered hepatotropism and demyelination phenotypes exhibited by the mutants such as C12, we have introduced this mutation into the wild-type MHV-A59 genome. Two recombinant viruses, containing the Q159L amino acid substitution, display a markedly reduced hepatotropism, while control recombinant viruses, containing the wild-type sequences of S, display pathogenesis similar to that of wild-type MHV-A59. The data suggest that Q159L also contributes to attenuation and loss of the ability to induce demyelination efficiently.
MATERIALS AND METHODS
Virus and cells.
MHV-A59 was obtained from Lawrence Sturman (Albany, N.Y.). C12 is a fusion-delayed, attenuated, weakly hepatotropic and weakly demyelinating mutant of MHV-A59 that was isolated from a persistently infected glial cell culture at 16 weeks after infection and characterized as previously described (8, 11, 19). Alb4, obtained from Paul Masters, contains an 87-nucleotide deletion (resulting in a 29-amino-acid in-frame deletion) in a nonessential spacer region in the N gene; it produces small plaques at the nonpermissive temperature (39°C) and is thermolabile (14). Murine L2 cells and 17Cl-1 cells were maintained on plastic tissue culture dishes in Dulbecco’s minimal essential medium (MEM) with 10% fetal bovine serum (FBS). Spinner cultures of L2 cells were maintained in Joklik’s MEM with 10% FBS at densities of between 2 × 105 and 2 × 106 cells per ml.
Plasmids and PCR mutagenesis.
The construction of pFV1 (obtained from Paul Masters) was described previously (3). Briefly, pFV1 was derived by addition of the wild-type MHV-A59 genes 3 to 6 to pB36 (22). PFV1 contains, downstream of a T7 RNA bacteriophage polymerase promoter, an approximately 500-nucleotide 5′ segment of open reading frame (ORF) 1a fused in frame through a 54-nucleotide linker to the 5′ end of the spike ORF. Downstream of the S gene is the rest of the MHV-A59 genome through the 3′ end, including a sequence encoding an approximately 115-residue poly(A) tail (Fig. 1). The sequence of the S gene in pFV1 is the same as that of our wild-type MHV-A59 strain (11) except for silent changes in codons 173 and 174 resulting in the loss of a HindIII site and the concomitant addition of an AseI site. The elimination of the HindIII site was necessary to allow the use of a HindIII site following the poly(A) tail for linearization of the plasmid for RNA transcription. This HindIII/AseI restriction site difference is additionally useful because it allows us to distinguish the S gene derived from pFV1 from the Alb4 S gene in the recombinant viruses (3).
FIG. 1.
Schematic diagram of targeted recombination. The synthetic RNA transcribed from the vector pFV1 and the corresponding 3′ end of the Alb4 genome are shown. Viral genes are indicated. There is a 54-nucleotide spacer sequence between the gene 1 fragment and S in pFV1. The sites of the Alb4 deletion, the mutation encoding Q159L, and the relevant restriction sites are shown. The curved line between the genome and the pFV-1 RNA indicates the region in which the crossover must have occurred.
Mutagenesis to introduce Q159L into the S gene of pFV1 was performed by using recombinant PCR (10) with Vent polymerase and the primers listed in Table 1. A restriction fragment containing the Q159L mutation was generated by amplification of two shorter fragments. A 5′ end fragment was generated with primers FS36 and WZL66, and a 3′ end was generated with primers FSQ159L and RS415. The 391- and 811-bp PCR fragments were gel purified and used as templates for a third PCR with primers FS36 and RS415. The 1,161-bp fragment was gel purified, digested with restriction enzymes SwaI and PstI, and then cloned into the corresponding sites in pFV1. This clone was designed pFV1-Q159L. The entire SwaI/PstI fragment of pFV1-Q159L was sequenced. This verified the presence, at codon 159, of CTG (encoding L) rather than the wild-type CAG (encoding Q) as well as the absence of other mutations.
TABLE 1.
Primers used for mutagenesis of pFV1 and for recombinant genome sequencing
| Primer | Sequence (5′→3′) | Genome location (nucleotide no.) |
|---|---|---|
| FIJ79 | GCGAATTATAGTGGTGGCACAC | 944–966 (hemagglutinin esterase ORF) |
| FS36 | GCATTAGCACTGAGACCGTTGAAG | 107–130 (S gene) |
| RIJ82 | CTCAGTTAGCTTGTCGTGG | 255–273 (S gene) |
| FSQ159L | TGCCAGTATACCATTTGTCTGTTACCTTACACTGATTGTAAG | 456–497 (S gene) |
| WZL66 | CTTACAATCAGTGTAAGGTAACAGACAAATGGTATACTGGCA | 456–497 (S gene) |
| RS415 | GTGGCAGCTGTATCAATCTTATAA | 1244–1267 (S gene) |
| RS221 | GTTTATCCGCATAGTACGCAT | 641–661 (S gene) |
| IZJ5 | GCTCCAACAGTTGGTGCC | 952–969 (N gene) |
| IZJ6 | ACGTAGGACCTTGCTAACTTC | 168–188 (3′ nontranslated region) |
Targeted RNA recombination.
Targeted RNA recombination was carried out between synthetic capped RNAs transcribed from wild-type pFV1 or pFV1-Q159L, using a T7 polymerase transcription kit (Ambion). Infection of L2 cells with Alb4 and transfection with the synthetic RNA were carried out as described previously (22). Briefly, L2 cells in spinner culture were harvested and concentrated to a density of between 4 × 106 and 107 cells per ml, washed in Joklik MEM, and infected at a multiplicity of infection (MOI) of 1 PFU of Alb4 per cell at 33°C. Two hours after infection, the cells were washed in phosphate-buffered saline (PBS) without calcium and magnesium and then transfected by electroporation with the synthetic RNA (10 to 30 μg), using two consecutive pulses from a Bio-Rad Gene Pulser apparatus set at 0.3 kV and 960 μF. Infected and transfected L2 cells were plated onto a monolayer of 17Cl-1 cells, and released virus was harvested when extensive cytopathic effect appeared (approximately 24 h after plating). Recombinant viruses were selected by heat treatment for 24 h at 40°C in 50 mM Tris-maleate (pH 6.5)–100 mM NaCl–1 mM EDTA–10% FBS and then identified by their large-plaque morphology (compared to the parental Alb4) after plaquing at 39°C on L2 cells. Candidate recombinants were plaque purified one more time, and viral stocks were grown on 17Cl-1 cells for further analysis (3).
Genome sequencing.
For sequencing of the viral genomes, reverse transcriptase-mediated PCR (RT-PCR) amplification was carried out, using as templates cytoplasmic RNA extracted from virus-infected L2 cell monolayers or RNA extracted from brain and liver lysates from infected animals. The oligonucleotides listed in Table 1 were used for amplification as designated for each experiment. Double-stranded PCR products were gel purified and analyzed by automated sequencing by the Taq dye terminator procedure according to the manufacturer’s protocol (Taq DyeDeoxy Terminator Cycle Sequencing kit; Applied Biosystems). The primers used for amplification were also used for sequencing, and each fragment was sequenced in both directions (19).
Viral growth curves.
L2 cell monolayers were prepared in 24-well plates in Dulbecco’s modified Eagle’s medium (DMEM) with 10% FBS. Confluent monolayers were infected with each virus (5 PFU/cell) in duplicate wells and incubated for 1 h at 37°C. Following adsorption, the cells were washed with Tris-buffered saline three times and then fed with 1 ml of DMEM–10% FBS. At the times indicated, the cells were lysed by three cycles of freeze-thawing, and the supernatants were removed and titered by plaque assay on L2 cells as previously described (8).
Inoculation of mice.
All animal experiments used 4-week-old MHV-free C57BL/6 mice (The Jackson Laboratory, Bar Harbor, Maine). Viruses were diluted into PBS containing 0.75% bovine serum albumin. Mice were anesthetized with methoxyflurane (Metofane; Pittman-Moore, Mundelein, Ill.). For intracerebral inoculations, 20 μl of diluted virus was injected into the left cerebral hemisphere. Mock-infected controls were inoculated similarly but with an uninfected cell lysate at a comparable dilution. For intrahepatic inoculations, mice were infected directly into the liver by injection below the sternum and below the right side of the inferior margin of the rib cage, using 5,000 PFU/0.05 ml of PBS-bovine serum albumin as described previously (11). In a previous study, we verified, using India ink in the inoculum, that this method of inoculation introduces virus directly into the liver (11).
Virulence assay.
Fifty percent lethal dose (LD)50 assays were carried out as described previously (11). Mice were inoculated intracerebrally with a 5- or 10-fold serial dilution of wild-type or mutant MHV-A59, five mice per dilution. Mice were examined for signs of disease or death on a daily basis up to 21 days postinfection. LD50 values were calculated by the Reed-Muench method (17, 25).
Virus replication in mice.
For measurement of virus replication in the liver and the brain, at selected times postinfection, mice were sacrificed and brains and livers were removed. Brains were placed directly into 4 ml of isotonic saline with 0.167% gelatin (gel saline), while livers were first rinsed with PBS and then placed in 10 ml of gel saline. All organs were weighed and stored frozen at −80°C until titered for virus (24). Organs were homogenized, and virus titers were determined by plaque assay on L2 cell monolayers all as previously described (11).
Histology.
For analysis of encephalitis and hepatitis, animals were infected intracerebrally and sacrificed at the indicated times, and the brains and livers were removed. One half of the brain and a small piece of the liver were fixed overnight in formalin. (The rest of the brain and liver were used for virus titration as described above.) Formalin-fixed tissue was embedded in paraffin, sectioned, and stained with hematoxylin and eosin (H&E). H&E-stained sections were used for pathologic evaluation. Slides were coded and read in a blinded fashion.
Demyelination.
Mice were infected intracerebrally with three doses of virus (50, 500, and 5,000 PFU per mouse), using five mice per dose. At 30 days postinfection, infected mice were perfused with PBS, followed by 10% buffered formalin. Brains and spinal cords were removed; tissue was embedded in paraffin and sectioned for staining with luxol fast blue to detect plaques indicative of demyelination. Demyelination was quantitated by examining one spinal cord section (four quadrants) from each of five levels of spinal cord for each mouse; thus, approximately 100 quadrants were examined for each dose of virus. Slides were coded and read in a blinded fashion. Statistical analyses of the demyelination values was performed, using SPSS version 7.5 for Windows (SPSS, Inc., Chicago, Ill.). The Mann-Whitney U test was used to compare the demyelination levels for each pair of viruses at each dose; data generating values of P < 0.05 were considered significantly different.
RESULTS
Selection of viruses with recombinant spike genes.
We used the scheme depicted in Fig. 1 to select recombinant viruses containing either the wild-type MHV-A59 S gene or an S gene encoding a Q159L amino acid substitution. Thus, we carried out recombination (as described in Materials and Methods) between synthetic RNAs transcribed from either pFV1 or pFV1-Q159L and the recipient virus Alb4, previously used in recombination experiments (3). Alb4 is thermolabile and displays a small-plaque morphology at 39°C; thus, putative recombinants were selected as large-plaque viruses after treatment for 24 h at 40°C and plating on L2 cell monolayers at 39°C. These viruses were further plaque purified as described in Materials and Methods. Because Alb4 is thermolabile (14) and does not replicate efficiently in animals (unpublished observations), it was important to select recombinant viruses with the wild-type S and N genes, in the Alb4 background, to be used as controls instead of Alb4.
To verify that large-plaque viruses were indeed recombinants, these viruses (15 each for both mutant and wild-type spike genes) were analyzed for repair of the Alb4 deletion. RT-PCR amplification was carried out, using as templates cytoplasmic RNA extracted from L2 cell monolayers infected with wild-type MHV-A59, Alb4, or candidate recombinants. A 602-nucleotide region surrounding the 87-nucleotide Alb4 deletion in the N gene was amplified with primers IZJ5 and IZJ6 (Table 1). The PCR product fragment derived from Alb4 is shorter than that from wild-type virus; thus, recombinants in which the deletion has been repaired are easily identified since the fragment is the same length as that amplified from wild-type virus. All large-plaque viruses generated the larger fragment (data not shown), demonstrating that plaque morphology is a reliable indicator of the presence of the recombinant N gene. These viruses were then screened for the presence of the 5′ portion of the recombinant S gene. This screen depends on the introduction of an AseI site and the simultaneous loss of the HindIII site, which was accomplished by silent mutations within codons 173 and 174 of the recombinant S gene (Materials and Methods). A 555-nucleotide fragment surrounding the AseI/HindIII site was amplified from the genomes of the recombinant viruses, as well as wild-type and Alb4, using primers FS36 and RS221. By digestion of this fragment separately with HindIII or AseI prior to electrophoresis, we were able to screen for the loss of the HindIII site and the simultaneous acquisition of an AseI site (the AseI+/HindIII− phenotype). The results are shown for five putative recombinants as well as wild-type MHV-A59 (Fig. 2); R1 and R10 are derived from wild-type pFV1, while R36, R40, and R45 are derived from pFV1-Q159L. R1, R10, R36, and R40 all generated AseI+/HindIII− fragments, indicating that recombination occurred to the 5′ side of codons 173 and 174 of the spike gene. R45, however, generated an AseI−/HindIII+ fragment (as did wild-type MHV-A59), indicating that recombination in this virus must have occurred between codons 173 and 174 of the spike gene and the Alb4 deletion site. For each of the Q159L and wild-type recombinants, 2 of 15 had the AseI+/HindIII− phenotype. Since the AseI site is only 14 codons away from the Q159L site, it was likely that all viruses with the AseI+/HindIII− phenotype would also contain the residue at position 159 from the recombinant gene.
FIG. 2.
RT-PCR analysis of putative recombinant viruses. A 555-nucleotide fragment of the S gene was amplified from cytoplasmic RNA extracted from L2 cell monolayers infected with recombinants and wild-type MHV-A59, using oligonucleotide primers FS36 and RS221 (Table 1). These DNAs were digested with either AseI or HindIII, as designated, prior to electrophoresis. M, size markers.
The S genes in the four recombinant virus genomes were sequenced from the 3′ portion of the hemagglutinin esterase ORF into the S gene and past the sequences encoding amino acid 159. This was accomplished by amplification and sequencing of two overlapping DNA fragments, one by using primers FIJ79 and RIJ82 and the other by using FS36 and RS221 (Table 1). The presence of the wild-type spike Q159 was verified in R1 and R10, which were designated wtR1 and wtR10; the presence of Q159L was verified in R36 and R40, which were designated 159R36 and 159R40. All other sequences were the same as in wild-type A59. Thus, there were no extraneous mutations introduced into the region in which the crossovers occurred in these or seven other recombinants examined (data not shown).
Replication of recombinant viruses in cultured cells in vitro.
Before infecting animals with the recombinant viruses, we wanted to determine whether they replicate efficiently in tissue culture. Thus, we compared the replication of both wild-type and Q159L-containing recombinant viruses to that of wild-type MHV-A59 in cultured L2 cells. Growth curves were performed, and the results shown in Fig. 3. Both wild-type (wtR10 and wtR1) and Q159L-containing (159R36 and 159R40) recombinant viruses replicated with similar kinetics and to a similar final extent as wild-type MHV-A59. All recombinant viruses displayed wild-type plaque morphology and induced cell-to-cell fusion with similar kinetics and to a similar extent as wild-type MHV-A59 (data not shown). Thus, genes 1 and 2, derived from Alb4, do not cause alterations in these phenotypes in vitro. Furthermore, Q159L also does not affect fusion phenotype or plaque morphology. This finding is consistent with our previous conclusions that the plaque morphology and fusion phenotypes of C12 and other glial cell mutants were determined not by Q159L but rather by the H716D amino acid substitution within the cleavage signal of the S protein (11, 19).
FIG. 3.
Replication of recombinant viruses in vitro in L2 cells. L2 cells were infected in duplicate with wild-type MHV-A59 (■), wtR10 (•), wtR1 (▴), 159R36 (□), and 159R40 (○) at an MOI of 5 PFU/cell. Cells and media above them were freeze-thawed at the indicated times, and virus titers in the lysates were determined. Each point represents the mean titer of the duplicate samples. Duplicate values were within ±25% of each other.
Recombinant virus replication and pathogenesis in brains and livers of infected mice.
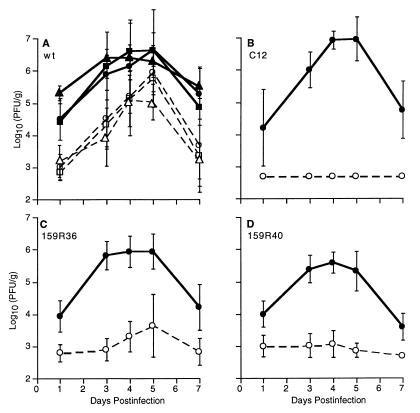
Previous studies demonstrated a strong correlation between the Q159L mutation and the weakly hepatotropic phenotype of mutants of MHV-A59 (11, 19). However, since the mutants contain amino acid substitutions other than Q159L, we needed to analyze the recombinant viruses to determine the effects on hepatotropism of the Q159L amino acid substitution alone. Thus, we inoculated mice intracerebrally with 159R36 and 159R40 as well as C12, wtR10, wtR1, and wild-type A59. The titers of virus in the brain and livers at various times postinfection were determined as described in Materials and Methods and shown in Fig. 4. Wild-type recombinant viruses wtR10 and wtR1 replicated with similar kinetics and to a similar final extent as parental wild-type MHV-A59 (Fig. 4A), with replication in the brain and liver peaking at 4 to 5 days postinfection as we have observed previously (11). Also, as reported previously (11) and as shown in Fig. 4B, the C12 mutant replicated with similar kinetics and to a similar titer as wild-type virus in the brain, while replication in the liver was at or below the level of detection (500 PFU/g in this experiment). The Q159L-containing recombinants 159R36 (Fig. 4C) and 159R40 (Fig. 4D) resembled C12 in their replication patterns. Virus replicated efficiently in the brain, while replication in the liver was near or at the level of detection in most animals.
FIG. 4.
Viral replication in the brains and livers of animals following intracerebral inoculation. C57BL/6 weanling mice were infected intracerebrally with 5,000 PFU of wild-type MHV-A59 (■, □), wtR10 (•, ○), or wtR1 (▴, ▵) (collectively designated wt) (A) C12 (B), 159R36 (C), or 159R40 (D). Animals were sacrificed at the indicated times, and virus titers in the brains (solid lines) or livers (dotted lines) were determined by plaque assay. The limit of detection was 500 PFU/g of tissue. The data shown represent the means (and standard deviations) of the titers from six (C12) or 8 to 12 (all other viruses) animals.
We previously noted that while most of the Q159L-containing mutants that we examined could not replicate in the liver following either intracerebral or intrahepatic inoculation, one mutant, B11, was able to replicate to a similar level as wild-type MHV-A59 after delivery directly into the liver (11). Thus, we wanted to determine whether Q159L-containing recombinant viruses could replicate in the liver following intrahepatic inoculation. As we have previously demonstrated and as illustrated in Fig. 5, wild-type virus replicates efficiently after inoculation by this route, and while C12 replicates to somewhat higher levels after this route of inoculation than when given intracerebrally (Fig. 4), it still replicates to levels of at least 2 log10 less than wild-type A59 (11). After inoculation directly into the liver, wtR10 and wtR1 (Fig. 5A) replicated to similar levels as wild-type MHV-A59; the recombinants 159R36 and 159R40 replicated to a similar extent as C12. Thus, these data demonstrate that the Q159L amino acid substitution is sufficient to confer on the recombinants a phenotype similar to that of C12, that is, the inhibition of replication in the liver following direct inoculation into the liver.
FIG. 5.
Viral replication in the livers of animals following intrahepatic inoculation. C57BL/6 weanling mice were infected intrahepatically with 5,000 PFU of wild-type MHV-A59 (■), wtR10 (•), or wtR1 (▴) (collectively designated wt) (A) C12 (B), 159R36 (C), or 159R40 (D). Animals were sacrificed at the indicated times, and virus titers in the livers were determined by plaque assay. The limit of detection was 500 PFU/g of tissue. The data shown represent the averages (and standard deviations) of the titers from four (wtR10 and wtR1) or six (A59, C12, R36, and R40) animals.
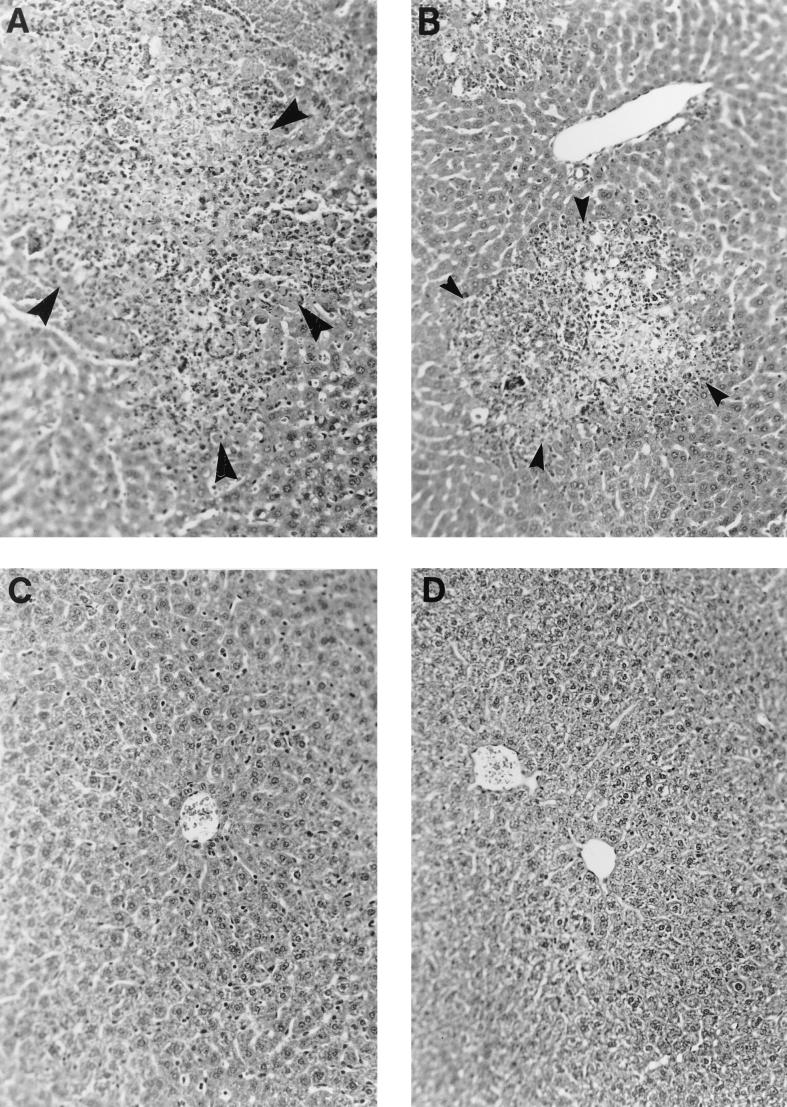
Liver sections from animals infected intracerebrally with wild-type, C12, and recombinant viruses were prepared and screened for hepatitis following staining with H&E. Figure 6 illustrates representative liver sections from animals infected with wild-type MHV-A59, C12, wtR10, and 159R36. The livers from mice infected with either wild-type MHV-A59 (Fig. 6A) or wtR10 (Fig. 6B) as well as wtR1 (data not shown) showed evidence of acute hepatitis, while the livers from mice infected with C12 (Fig. 6C), 159R36 (Fig. 6D) or 159R40 (data not shown) looked normal, as in uninfected mice. Each one of the images is representative of the entire section examined; there is very little variation in histopathologic changes among various parts of the same liver and among different mice infected with the same virus. As expected, mice infected with either wild-type or Q159L containing recombinant viruses exhibited mild to severe meningoencephalitis (data not shown).
FIG. 6.
Histopathology of the livers of C57BL/6 mice infected with recombinant viruses. Mice were infected intracerebrally as for Fig. 4 and sacrificed at 4 (or, in the case of the wtR10-infected mouse, 5) days postinfection. Livers were removed, fixed, sectioned, and stained with H&E as described in Materials and Methods. (A) Wild-type MHV-A59-infected liver shows acute hepatitis. The arrowheads shows a lesion exhibiting necrosis surrounded by inflammatory cells including lymphocytes, histiocytes, and polymorphonuclear granulocytes. (B) wtR10-infected liver shows similar pathological changes similar to those in panel A. (C) C12-infected liver shows normal liver histology without evidence of hepatitis. Normal liver parenchyma, including a central vein, is surrounded by hepatocytes and Kupffer cells arranged in a normal sinusoidal architecture. (D) 159R36-infected liver appears normal as in panel C. Magnification, ×200.
To verify that the replication of 159R36 or 159R40 in the brains and livers of infected mice does not select for revertant viruses (with the wild-type S gene) and that virus isolated from infected animals still encodes the Q159L amino acid substitution, RNA was isolated from the brains and livers of mice infected with wtR10 as well as 159R36 and 159R40. RT-PCR was used to amplify DNA fragments containing the region encoding Q159, using primers FS36 and RS221 (Table 1); these fragments were then sequenced by using the same primers. The Q159L substitution was maintained in virus recovered from the brains and livers of mice infected with 159R36 and 159R40, while virus recovered from the brains and livers of mice infected with wtR10 encoded Q159. Thus, we did not observe the emergence of wild-type revertants encoding Q159 in either the brains or livers of mice infected with Q159L-containing recombinants.
Virulence of recombinant viruses.
Most of the weakly hepatotropic, Q159L-containing mutants that we have analyzed (for example B10, B11, and C12) are also attenuated relative to wild-type MHV-A59 (11), suggesting that the loss of hepatotropism may be responsible for the attenuation. However, the observation that one mutant, B12, is as virulent as the wild type (11) suggests that loss of hepatotropism and the Q159L mutation do not necessarily result in loss of virulence. To determine the effect of Q159L on the virulence of recombinant viruses, we inoculated animals intracerebrally with serial dilutions of wild-type, C12, and recombinant viruses, observed them for lethality, and then calculated LD50s. The results are shown in Table 2. As expected, wtR10 displays the same LD50 as wild-type MHV-A59; 159R36 and 159R40, however, display a degree of attenuation similar to that found for C12. These data suggest that Q159L confers attenuation on the recombinant viruses. The second wild-type recombinant virus (wtR1) was intermediate in virulence between wild-type MHV-A59 and C12 (see Discussion).
TABLE 2.
Virulence of recombinant viruses
| Virus | Virulence, log10 (LD50)a |
|---|---|
| MHV-A59 | 3.87 |
| wtR10 | 3.74 |
| wtR1 | 4.64 |
| 159R36 | 5.15 |
| 159R40 | 5.26 |
| C12 | 5.14 |
Virulence assays were carried out as described in Materials and Methods. Values represent means obtained from four to five experiments with each virus.
Demyelination in recombinant infected mice.
Since we observed in a previous study that C12 and other mutants encoding the Q159L S protein amino acid substitution had lost the ability to efficiently induce demyelination (19), we wanted to test the effects of Q159L alone on demyelination. Thus, we measured the ability of recombinant viruses to induce demyelination as a function of dose inoculated (as described in Materials and Methods) compared to wild-type MHV-A59 and C12. Figure 7 shows data pooled from several experiments. Wild-type recombinant virus wtR10 demyelinated to a similar extent as wild-type parental virus; this level of demyelination is similar to that we have previously reported for wild-type virus (19). C12 displayed a level of demyelination that was low but, on average somewhat higher than previously observed (19). Statistical analysis (Materials and Methods) showed that the difference between these wild types and C12 was significant (P < 0.001 at a dose of 5,000 PFU). The demyelination levels observed for the Q159L-containing recombinant viruses were also variable from experiment to experiment but, on average, intermediate between levels for wild-type and C12. The values were not significantly different between the Q159L-containing recombinants and C12 (P > 0.113 at a dose of 5,000 PFU); they were, however, significantly different from the wild-type value (P < 0.003 at a dose of 5,000 PFU), suggesting that the Q159L amino acid substitution does result in lower levels of demyelination compared with the wild type. The second wild-type recombinant, wtR1, however, demyelinated more like Q159LR36 and R40 (see Discussion).
FIG. 7.
Demyelination induced by recombinant viruses. C57BL/6 mice were inoculated intracerebrally with various doses of wild-type (■), C12 (▵), and recombinant wtR10 (•), wtR1 (▴), 159R36 (□), and 159R40 (○) viruses. Animals surviving the acute infection were sacrificed at 30 days postinfection, and spinal cords were analyzed for demyelination by staining with luxol fast blue as described in the text. Data represent the mean values for 15 mice (5,000-PFU dose), 5 to 15 mice (500-PFU dose), and 5 mice (50-PFU dose).
DISCUSSION
Targeted recombination has previously been used to select murine hepatitis viruses with recombinant N genes (14) and more recently with recombination sites in more upstream genes (3). We have extended the technology to select viruses with recombinant S genes. Analysis of three sets of recombinants showed that 8 of 46 (17%) of the recombinants with the wild-type N gene also had the AseI+/HindIII− phenotype. This demonstrates that the Alb4 mutant, in which the temperature-sensitive lesion is approximately 6 kb from the AseI/HindIII site in the S gene, is a useful parent for the selection of S recombinants. Furthermore, these data suggest that Alb4 may also be useful for the selection of recombinants in more upstream portions of the genome, such as the hemagglutinin esterase gene. In a recent study using a similar recombination strategy, Zhang et al. (28) were able to detect recombination within the spike gene of MHV; however, they did not isolate viruses containing these recombinant S genes, probably due the requirement for double crossovers to occur as well as an inability to select against the parental wild-type virus.
We have previously reported the correlation between the Q159L amino acid substitution and altered hepatotropism and demyelination in a group of mutants isolated from persistently infected glial cells (11, 19). However, there were other mutations in the genomes of this group of mutants. B11, C12, and B12 all had the H716D mutation in the protease cleavage signal in the spike; the data suggested that this mutation, however, did not correlate with altered organ tropism but was responsible for a delayed-fusion phenotype (8, 11). The entire C12 genome was sequenced and found to encode three additional amino acid substitutions, outside the S gene. These substitutions were not present in the B11 or B12 genome and appeared also not to be necessary for altered hepatotropism and demyelination phenotypes (19); however, it is likely that these substitutions contribute to differences in the pathogenic phenotypes among these mutants. These conclusions were drawn from correlations of the presence of amino acid substitutions with phenotypes; targeted recombination has allowed us to take this a step further and definitively demonstrate the effects of the Q159L substitution alone in the wild-type background.
We have characterized a pair of recombinant viruses containing the Q159L amino acid substitution (159R36 and 159R40). These two recombinant viruses were recovered from separate transfection events and do indeed demonstrate the same phenotypic properties both in cell culture and in vivo in mice. This finding indicates that the phenotypes exhibited by these recombinants are a result of the Q159L amino acid substitution rather than to spurious mutations that may be present in the recombinant viruses. This influence is further supported by the observation that the wild-type recombinant virus wtR10 has in vitro and in vivo phenotypes indistinguishable from those of wild-type MHV-A59. Since the parental virus, Alb4, of recombinants generated in this study is a temperature-sensitive derivative of MHV-A59 (derived from the same wild-type isolate that we use), it does not replicate well in animals. Therefore recombinant virus in which we have reintroduced the wild-type S sequence into Alb4 is an important control for possible effects of Alb4-derived genes upstream of S; all genes downstream of S contain wild-type A59 sequences derived from pFV1. However, since the wild-type recombinant virus wtR10 is indistinguishable from wild-type A59 in its in vitro (Fig. 3) and in vivo properties (Figs. 4 to 7), we feel confident that the mutant phenotypes we observe do indeed reflect the Q159L substitution. The observation that wild-type recombinant virus was phenotypically similar to MHV-A59 supports an earlier observation that in vivo and in vitro phenotypes are largely determined by the structural genes at the 3′ end of the genome (18).
A second wild-type recombinant virus that we characterized, wtR1, also has the same in vitro and in vivo replication phenotypes as wild-type MHV-A59 and wtR10 (Figs. 3 to 5); however, wtR1 appears less virulent than wild-type MHV-A59 and wtR10 (Table 2); it also appears to induce demyelination to a lower extent than wild-type MHV-A59 and wtR10 (Fig. 7). It is possible that wtR1 has acquired mutation(s) which affects complex properties such as virulence and demyelination. Sequencing of crossover regions in the genomes of the recombinants described here as well as other recombinants (data not shown), in addition to the sequence analyses of recombinants by Peng et al. (23a), indicate that the recombination process does not result in spurious mutation in the region at or around the crossovers. We are examining additional wild-type recombinant viruses to determine if the difference between wtR1 and wild-type MHV-A59 is a common feature for wild-type recombinant viruses.
The pathogenesis of the acute disease induced by Q159L-containing recombinants is similar to that of the C12 mutant (11). While wild-type MHV-A59 and wtR10 replicate efficiently in the brains and the livers of animals, 159R36 and 159R40 are severely inhibited in the ability to replicate in the liver following either intracerebral (Fig. 4) or intrahepatic (Fig. 5) inoculation. Consistent with this finding, we observed little if any hepatitis in the livers of infected animals following intracerebral inoculation (Fig. 6). Thus, Q159L alone results in the loss of the ability of MHV-A59 to efficiently induce hepatitis. As we published previously, the level of replication of C12 in the liver following intracerebral inoculation is at or below the level of detection in these experiments. The amount of replication of the Q159L-containing recombinants in the liver following intracerebral inoculation was, in a few animals, above the level of detection (Fig. 4). A similar pattern was observed with the B11 mutant (11), suggesting that other amino acid substitution(s) in the C12 genome may further contribute to the loss of the ability to replicate in the liver. Replication patterns of the recombinant viruses following intrahepatic inoculation, however, indicate that 159R36 and 159R40 behave more like C12 than B11, which replicates to wild-type levels following intrahepatic inoculation. Thus, while Q159L clearly has a major effect on hepatotropism following either route of inoculation, there must be other mutations in the genomes of B11 and C12 that further influence this phenotype. This observation is an example of how the recombination technique will be useful in assigning phenotypes to individual amino acid substitutions.
The Q159L amino acid substitution is located within the amino-terminal portion of the S1 subunit of the spike, within the region of the protein believed to contain the receptor binding domain of the S protein (15, 26). The location of Q159L within this domain suggests the possibility that this amino acid substitution can lead to altered interaction with a viral receptor and thus change its tropism and result in altered pathogenic properties. The observation that Q159L alone can alter the ability of the virus to replicate efficiently in the liver adds support to this idea. We compared the replication of wild-type MHV-A59 and the C12 mutant in hepatocyte cultures in vitro and found that neither virus replicated efficiently (data not shown); thus, it is unlikely that the hepatocyte is the cell that differentiates between wild-type and Q159L-containing virus. This is consistent with studies which demonstrated that differences in the ability of various strains of MHV to replicate in the liver are due to differences in the ability to replicate either in endothelial cells or Kupffer cells, cells that are likely involved in the uptake of virus into the liver, rather than hepatocytes (13, 21, 27). Since viruses expressing a Q159L-containing S protein can replicate efficiently in cultured cell lines expressing the receptor MHV-R, we can conclude that Q159L does not significantly alter the interaction with this receptor. Thus, we speculate that perhaps entry into Kupffer cells, which do not express detectable levels of MHV-R (7), requires interaction with an alternate receptor which prefers amino acid residue 159 of the spike to be glutamine rather than leucine.
Since, we observed a correlation between the presence of Q159L and greatly reduced levels of demyelination in all of five glial cell mutants examined (19), we wanted to determine whether this could be due to the Q159L amino acid substitution alone. As shown in Fig. 7, which contains data averaged from several experiments, the recombinant viruses exhibited statistically significant reduced levels of demyelination relative to wild-type and wtR10, suggesting that Q159L does indeed influence the demyelination phenotype. This conclusion, however, must be considered tentative until we can further analyze more wild-type recombinant viruses or more completely understand the phenotype of wtR1. We are also investigating the replication and spread of C12 within the central nervous system in order to understand why mutants such as C12 do not demyelinate well.
We suggested in an earlier publication (17) that mortality of MHV-A59-infected mice is due to a combination of the effects of hepatitis and encephalitis. In the analysis of the glial cell mutants, we observed a general correlation between a loss of the ability to replicate in the liver and an attenuated phenotype (11). However, one nonhepatotropic mutant, B12, is as virulent as wild-type virus. We used the weakly hepatotropic recombinant viruses to investigate the determinants of virulence further and to examine whether the Q159L substitution alone is enough to cause an attenuated phenotype. The results, shown in Table 2, demonstrate that Q159L alone results in attenuation of recombinant viruses. This finding suggests that the B12 genome may have compensatory mutations outside the Spike gene that result in virulence. We plan to investigate whether B12 causes some other pathology not observed in C12 and B11. Virulence as measured by LD50 reflects the total pathogenic properties of the virus, and it is not surprising that many mutations may compromise virulence.
We have shown that targeted recombination can be used to introduce specific mutations into the S gene of MHV-A59. These studies will pave the way for further studies on the pathogenesis of MHV-A59 and the beginnings of understanding the structure and function of the S gene.
ACKNOWLEDGMENTS
This work was supported by Public Health Service grants NS-21954 and NS-30606 (S.R.W.) as well as grant RG-2615A1/2 (E.L.) from the National Multiple Sclerosis Society. J.P. was supported in part by training grant GM-07229.
We thank Paul Masters for pFV1, Alb4, and advice throughout the project. We acknowledge Françoise Fischer for help with the recombination technology and Kelly Rudnick for technical assistance.
REFERENCES
- 1.Chambers P, Pringle C R, Easton A J. Heptad repeat sequences are located adjacent to hydrophobic regions in several types of virus fusion glycoproteins. J Gen Virol. 1990;71:3075–3080. doi: 10.1099/0022-1317-71-12-3075. [DOI] [PubMed] [Google Scholar]
- 2.DeGroot R J, Luytjes W, Horzinek M C, van der Zeijst B A M, Spaan W J M, Lenstra J A. Evidence for a coiled-coil structure in the spike proteins of coronaviruses. J Mol Biol. 1987;196:963–966. doi: 10.1016/0022-2836(87)90422-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fischer F, Stegen C F, Koetzner C A, Masters P S. Analysis of a recombinant mouse hepatitis virus expressing a foreign gene reveals a novel aspect of coronavirus transcription. J Virol. 1997;71:5148–5160. doi: 10.1128/jvi.71.7.5148-5160.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fleming J O, Trousdale M D, El-Zaatari F A K, Stohlman S A, Weiner L P. Pathogenicity of antigenic variants of murine coronavirus JHM selected with monoclonal antibodies. J Virol. 1986;58:869–875. doi: 10.1128/jvi.58.3.869-875.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Frana M F, Behnke J N, Sturman S, Holmes K V. Proteolytic cleavage of the E2 glycoprotein of murine coronavirus: host-dependent differences in proteolytic cleavage and cell fusion. J Virol. 1985;56:912–920. doi: 10.1128/jvi.56.3.912-920.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gallagher T M, Parker S E, Buchmeier M J. Neutralization resistant variants of a neurotropic coronavirus are generated by deletions within the amino-terminal half of the spike glycoprotein. J Virol. 1990;64:731–741. doi: 10.1128/jvi.64.2.731-741.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Godfraind C, Langreth S, Cardellichio C, Knobler R, Coutelier J P, Dubois-Dalcq M, Holmes K V. Tissue and cellular distribution of an adhesion molecule in the carcinoembryonic antigen family that serves as a receptor for mouse hepatitis virus. Lab Investig. 1995;73:615–627. [PubMed] [Google Scholar]
- 8.Gombold J L, Hingely S T, Weiss S R. Fusion-defective mutants of mouse hepatitis virus A59 contain a mutation in the spike protein cleavage signal. J Virol. 1993;67:4504–4512. doi: 10.1128/jvi.67.8.4504-4512.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gombold J L, Sutherland R, Lavi E, Paterson Y, Weiss S R. Mouse hepatitis virus-induced demyelination can occur in the absence of CD8+ T cells. Microb Pathog. 1995;18:211–221. doi: 10.1016/S0882-4010(95)90058-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Higuchi R. PCR protocols: a guide to methods and applications. San Diego, Calif: Academic Press; 1990. pp. 177–183. [Google Scholar]
- 11.Hingley S T, Gombold J L, Lavi E, Weiss S R. MHV-A59 fusion mutants are attenuated and display altered hepatotropism. Virology. 1994;200:1–10. doi: 10.1006/viro.1994.1156. [DOI] [PubMed] [Google Scholar]
- 12.Houtman J J, Fleming J O. Pathogenesis of mouse hepatitis virus-induced demyelination. J Neurovirol. 1996;2:361–376. doi: 10.3109/13550289609146902. [DOI] [PubMed] [Google Scholar]
- 13.Joseph J, Kim R, Siebert K, Lublin F D, Offenbach C, Knobler R L. Organ specific endothelial cell heterogeneity influences differential replication and cytopathogenicity of MHV-3 and MHV-4. In: Talbot P J, Levy G A, editors. Corona- and related viruses. New York, N.Y: Plenum Press; 1995. pp. 43–50. [DOI] [PubMed] [Google Scholar]
- 14.Koetzner C A, Parker M M, Ricard C S, Sturman L S, Masters P S. Repair and mutagenesis of the genome of a deletion mutant of the murine coronavirus mouse hepatitis virus by targeted RNA recombination. J Virol. 1992;66:1841–1848. doi: 10.1128/jvi.66.4.1841-1848.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kubo H, Yamada Y K, Taguchi F. Localization of neutralizing epitopes and the receptor binding site within the amino-terminal 330 amino acids of the murine coronavirus spike protein. J Virol. 1994;68:5404–5410. doi: 10.1128/jvi.68.9.5403-5410.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lavi E, Fishman P S, Highkin M K, Weiss S R. Limbic encephalitis after inhalation of a murine coronavirus. Lab Investig. 1988;58:31–36. [PubMed] [Google Scholar]
- 17.Lavi E, Gilden D H, Wroblewska Z, Rorke L B, Weiss S R. Experimental demyelination produced by the A59 strain of mouse hepatitis virus. Neurology. 1984;34:597–603. doi: 10.1212/wnl.34.5.597. [DOI] [PubMed] [Google Scholar]
- 18.Lavi E, Murray E M, Makino S, Stohlman S A, Lai M M C, Weiss S R. Determinants of coronavirus MHV pathogenesis are localized to 3′ portions of the genome as determined by ribonucleic acid-ribonucleic acid recombination. Lab Investig. 1990;62:570–578. [PubMed] [Google Scholar]
- 19.Leparc-Goffart I, Hingley S T, Chua M M, Jiang X, Lavi E, Weiss S R. Altered pathogenesis of a mutant of the murine coronavirus MHV-A59 is associated with a Q159L amino acid substitution in the spike protein. Virology. 1997;239:1–10. doi: 10.1006/viro.1997.8877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Luytjes W, Sturman L, Bredenbeek P J, Charite J, van der Zeijst B A M, Horzinek M C, Spaan W J M. Primary structure of the glycoprotein E2 of coronavirus MHV-A59 and identification of the trypsin cleavage site. Virology. 1987;161:479–487. doi: 10.1016/0042-6822(87)90142-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Martin J P, Chen W, Koehren F, Pereira C A. The virulence of mouse hepatitis virus 3, as evidenced by permissivity of cultured hepatic cells toward escape mutants. Res Virol. 1994;145:297–302. doi: 10.1016/S0923-2516(07)80034-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Masters P S, Koetzner C A, Kerr C A, Heo Y. Optimization of targeted RNA recombination and mapping of a novel nucleocapsid gene mutation in the coronavirus mouse hepatitis virus. J Virol. 1994;68:328–337. doi: 10.1128/jvi.68.1.328-337.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Parker S E, Gallagher T M, Buchmeier M J. Sequence analysis reveals extensive polymorphism and evidence of deletions within the E2 glycoproteins of several strains of murine hepatitis virus. Virology. 1989;173:664–673. doi: 10.1016/0042-6822(89)90579-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23a.Peng D, Koetzner C A, McMahon T, Zhao Y, Masters P S. Construction of murine coronavirus mutants containing interspecies chimeric nucleocapsid proteins. J Virol. 1995;69:5475–5484. doi: 10.1128/jvi.69.9.5475-5484.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ramig R F. Isolation and genetic characterization of temperature sensitive mutants of simian rotavirus SA11. Virology. 1982;120:93–135. doi: 10.1016/0042-6822(82)90009-5. [DOI] [PubMed] [Google Scholar]
- 25.Reed L J, Muench H. A simple method of estimating fifty per cent points. Am J Hyg. 1938;27:493–497. [Google Scholar]
- 26.Suzuki H, Taguchi F. Analysis of the receptor-binding site of murine coronavirus spike protein. J Virol. 1996;70:2632–2635. doi: 10.1128/jvi.70.4.2632-2636.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Taguchi F, Kawamura S, Fujiwara K. Replication of mouse hepatitis viruses with high and low virulence in cultured hepatocytes. Infect Immun. 1983;39:955–959. doi: 10.1128/iai.39.2.955-959.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang L, Homberger F, Spaan W, Luytjes W. Recombinant genomic RNA of coronavirus MHV-A59 after co-replication with a DI RNA containing the MHV-RI spike gene. Virology. 1997;230:93–102. doi: 10.1006/viro.1997.8460. [DOI] [PMC free article] [PubMed] [Google Scholar]