Abstract
The utility of recombinant herpes simplex virus type 1 (HSV-1) vectors may be expanded by manipulation of the virus envelope to achieve cell-specific gene delivery. To this end, an HSV-1 mutant virus deleted for glycoprotein C (gC) and the heparan sulfate binding domain of gB (KgBpK−gC−) was engineered to encode different chimeric proteins composed of N-terminally truncated forms of gC and the full-length erythropoietin hormone (EPO). Biochemical analyses demonstrated that one gC-EPO chimeric molecule (gCEPO2) was posttranslationally processed, incorporated into recombinant HSV-1 virus (KgBpK−gCEPO2), and neutralized with antibodies directed against gC or EPO in a complement-dependent manner. Moreover, KgBpK−gCEPO2 recombinant virus was specifically retained on a soluble EPO receptor column, was neutralized by soluble EPO receptor, and stimulated proliferation of FD-EPO cells, an EPO growth-dependent cell line. FD-EPO cells were nevertheless refractory to productive infection by both wild-type HSV-1 and recombinant KgBpK−gCEPO2 virus. Transmission electron microscopy of FD-EPO cells infected with KgBpK−gCEPO2 showed virus endocytosis leading to aborted infection. Despite the lack of productive infection, these data provide the first evidence of targeted HSV-1 binding to a non-HSV-1 cell surface receptor.
The development of vector systems suitable for the direct transfer of genes in vivo will be essential to the successful treatment of human disease by gene therapy. Highly attenuated or replication-defective recombinant viruses carrying novel transgenes represent potentially attractive vehicles for in vivo gene delivery because they efficiently utilize cell surface receptors to gain intracellular access where expression of the transgene can biochemically transduce the cell or function as a depot for local or systemic transgene product delivery to the host. Many viruses are also capable of persisting in a nonpathogenic, integrated, or episomal latent state requiring the expression of few, if any, viral gene products for maintenance of latency (25, 26, 77). Eliminating pathogenic virus properties while retaining the ability of the virus to establish latency or a latent-like state provides the core strategy for long-term gene therapy using viral vectors (42). However, to realize the full potential of viruses for in vivo gene transfer, tissue-specific and regulatable transgene expression from the latent viral genome may be required, a potentially achievable outcome particularly since many latent viruses possess latency-active promoters that function, often exclusively in specific cell types (11, 25, 77). Moreover, the application of viral vectors will, in some instances, exploit the natural virus host range, but in other cases, vectors must be designed to infect a novel tissue or cell type in order to achieve a second level of tissue specificity. Though difficult, this possibility may be realized through engineering the vector surface structures in a manner to control virus attachment and penetration. Finally, vectors must be designed to overcome vector-related immune responses that will impede either gene delivery or persistence. While considerable progress has been made in achieving many aspects of viral vector design, particularly the identification and removal of viral functions associated with pathogenesis (67, 80, 88), other impediments to viral vector-related gene delivery have proven more difficult to overcome.
Herpes simplex virus (HSV) has many features which make it a potentially attractive vehicle for gene transfer most particularly to neurons, where the virus naturally establishes a life-long latent state during which a neuronally active latency-specific promoter system can be used to express transgenes (8, 21). The cytotoxic features of this virus have been essentially eliminated by the systematic removal of immediate-early (IE) genes which prevent the expression of both early and late functions, and consequently virus replication or reactivation from latency can be achieved without virus replication (67). In addition, the virus genome structure, organization of essential and accessory functions, the near absence of spliced genes, and the development of techniques for creating recombinant viruses harboring multiple or large transgenes with considerable efficiency (38) have made feasible the rapid engineering of HSV vectors that can be propagated to relatively high titers in the appropriate complementing cell systems (46, 67). HSV vectors that express few or no lytic functions should prove useful in expanding the potential of HSV for gene transfer to cell types in addition to neurons since the highly defective viral DNA can persist in cells in a manner similar to latency (67). Despite these considerable advantages, the host range of HSV is quite broad, which can limit its use for direct targeting in vivo. Thus far, no attempts to modify the envelope components of HSV to achieve targeted infection have been reported.
The envelope glycoproteins mediate infection of the host cell through two identifiable stages: (i) attachment to the cell surface and (ii) fusion of the viral envelope with the cell surface membrane, resulting in virus entry. Virus attachment is initiated by binding of glycoproteins B and C (gB and gC) to cell surface glucosaminoglycans (GAGs) (30, 74), mainly heparan sulfate (HS) (26, 71, 89) but also dermatan sulfate (1). Together, this binding represents approximately 85% of the binding activity to Vero cells, the most commonly used cell type for HSV propagation, and the majority is associated with gC (29, 30). Deletion of gC and the HS binding domain of gB impairs binding to normal cells to an extent similar to the reduction in binding of wild-type virus to HS-deficient cells (26); however, these mutations do not prevent virus adsorption, indicating that other receptors must be involved (41). The initial binding of virus to cell surface HS is followed by gD-mediated tight binding to secondary receptors, one of which, the herpesvirus entry mediator (HVEM), was recently shown to be a member of the tumor necrosis factor/nerve growth factor receptor family (53). The sequential attachment steps in infection result in fusion of the viral envelope with the cell surface membrane and viral entry into the cell (50). Virus entry requires gB, gD, and gH, based on mutant virus studies (5, 14, 44), although how this is accomplished and what roles the different proteins play in this process are unclear.
Construction of targeted HSV type 1 (HSV-1) vectors is complicated by this highly evolved process of infection in which many of the molecular details remain unknown. In addition, most well-studied HSV gene products are multifunctional molecules including the glycoproteins (62, 74), and thus manipulation of the glycoproteins is complicated by the fact that some domains must be preserved while others must be replaced with a new functional element. The virus envelope is also complex, containing at least 10 glycoproteins (gB, gC, gD, gE, gG, gH, gI, gJ, gK, gL, and gM), of which gB, gD, gH, and gL are essential for viral infection (5, 14, 44, 63); although gI and gE are nonessential for cell culture infections, they nevertheless are required for cell-to-cell virus spread in vivo (16), and others (e.g., gC) are required for infection of particular cell surfaces (e.g., apical surface of MDCK cells [69]). Thus, depending on the need, the manipulation of both essential and accessory glycoproteins may prove useful for HSV vector targeting.
The alteration of the host-range of HSV-1 vectors will require at least two kinds of modifications of the virus envelope: (i) elimination of the natural tropism and (ii) expression of new viral ligands capable of binding to cell surface receptors present on the intended target cell in a manner to preserve the natural mechanism of virus entry through fusion of the virus envelope with the cell surface membrane. Our laboratory has accomplished, in part, the first step by deleting the HS-binding capacity of HSV-1 (41). A double-mutant virus (KgBpK−gC−) deleted for gC and the HS binding domain of gB (gBpK−) demonstrated an 80% reduction in binding to Vero cells compared to wild-type virus, although the virus remains infectious through recognition of a second receptor possibly mediated by gD. In an effort to alter the tropism of the mutant virus, the HS binding domain of gC (79) was genetically replaced by erythropoietin (EPO). Biochemical analysis demonstrated that two gC:EPO fusion proteins were incorporated into virions and that one recombinant virus was specifically retained on a glutathione S-transferase (GST)-soluble EPO receptor (EREx) column as well as neutralized by EREx. This gC:EPO recombinant virus stimulated the growth of an EPO-dependent cell line (FD-EPO) through binding to the EPO receptor encoded by these cells. FD-EPO cells are nonpermissive to HSV-1 infection, and gC:EPO recombinant virus was internalized following virus binding to the receptor. To our knowledge, these data provide the first evidence that HSV-1 tropism can be manipulated to recognize a nonherpesvirus receptor.
MATERIALS AND METHODS
Cells and viruses.
Vero cells were obtained from the American Type Culture Collection (Bethesda, Md.), and the mouse hematopoietic progenitor cell line FD-EPO (54) was obtained from Kazuo Todokono (University of Tsukuba, Tsukuba, Japan). Mouse L cells and Gag-negative derivatives (sog9 cells [1]) were kindly provided by Frank Tufaro, Vancouver, British Columbia, Canada. Vero, L, sog9, and VD60 (gD-complementing Vero cell line kindly provided by David C. Johnson, Portland, Oreg.) (44) cells were grown in Dulbecco’s modified essential medium, while FD-EPO cells were grown in RPMI 1640 medium (Gibco-BRL, Grand Island, N.Y.). All cell lines were maintained at 37°C, and the media were supplemented with 10% fetal bovine serum; EPO (1 U/ml; Amgen Inc., Thousand Oaks, Calif.) was added to FD-EPO medium. All mutant and recombinant virus strains used were derivatives of HSV-1 strain KOS. The gC−–human cytomegalovirus (HCMV)-lacZ KOS mutant (KCZ) and the HS-binding-defective virus double mutant expressing lacZ from the gC locus (KgBpK−gC−) were previously described (41).
Construction of plasmids.
Plasmid pgCEPO1 was created by inserting in frame the sequence encoding the polypeptide hormone EPO within a plasmid encoding the HSV-1 gC sequence (UL44) from KOS (pgC1 [32]). Plasmid pgC1 was partially digested with restriction endonucleases NaeI and NcoI in order to delete 244 bp of the gC coding sequence corresponding to residues 83 to 161. The human EPO cDNA sequence (kindly provided by Y. W. Kan, University of Southern California, Los Angeles), coding for the mature 166-amino-acid peptide hormone, without the 27-amino-acid signal peptide, was amplified by PCR with oligonucleotides encoding compatible ends (NaeI and NcoI at the 5′ and 3′ ends, respectively). The PCR product was digested with restriction endonucleases NaeI and NcoI and ligated with restriction enzyme-digested pgC1 while maintaining the proper reading frame of the fusion molecule. pgCEPO2 was constructed by digesting pgCEPO1 at unique sites with restriction endonucleases BglII and KpnI in order to delete the sequences encoding the signal peptide and the 82 N-terminal residues of gC, thereby deleting the coding sequences for 161 residues of the gC N terminus. The deleted sequence was replaced with the sequence encoding the signal peptide of EPO which was excised from the EPO plasmid with the endonucleases BstEII and KpnI. pgCEPO3 was constructed by removal of the DNA sequence between the two NcoI endonuclease sites of pgCEPO2. Removal of the 642 nucleotides maintained the reading frame of the fusion molecule and deleted gC sequence for nucleotides encoding amino acid residues 1 to 375. The wild-type gC promoter was excised from pgC1, pgCEPO1, pgCEPO2, and pgCEPO3 and replaced with the HCMV immediate-early promoter (IEp) (18) to create pHgC1, pHgCEPO1, pHgCEPO2, and pHgCEPO3, respectively.
A gD plasmid (pgD1) encoding the SacI sequence of HSV-1 KOS located between positions 137948 and 140791 was digested with NaeI in order to replace 480 bp of gD coding sequence with HCMV IEp driving lacZ (17). This plasmid (pgDLacZ) was used to create a gD-deleted virus as described below.
Immunofluorescence.
Thirty hours posttransfection or 24 h postinfection (p.i.), cell surface antigens were detected by indirect immunofluorescence of unfixed Vero cell monolayers, and cell-associated antigens were similarly detected in cells fixed with ice-cold methanol. The fixed or unfixed monolayers were incubated for 1 h at 4°C with a pool of monoclonal antibodies (MAbs) specific for gC (α-gC) (48) and an anti-EPO polyclonal antibody (α-EPO; Genzyme, Cambridge, Mass.), rinsed with Tris-buffered saline, pH 7.4 (TBS), and incubated for an additional hour with a Cy3-conjugated anti-rabbit antibody and fluorescein isothiocyanate (FITC)-conjugated anti-mouse antibody (Jackson Immunoresearch Laboratories, West Grove, Pa.). The monolayers were washed and treated with ice-cold methanol. Immunofluorescent cells were photographed with a Nikon model 211910 TMS microscope/camera.
Detection of gC:EPO fusion proteins from transfected cells.
Vero cells were transfected with 2 μg of pHgC1, pHgCEPO1, pHgCEPO2, and pHgCEPO3 in the presence of [35S]methionine-[35S]cysteine-supplemented medium. Thirty hours posttransfection, the monolayers were harvested and solubilized at 4°C with lysis buffer (50 mM Tris [pH 6.8], 150 mM NaCl, 1% [vol/vol] Nonidet P-40, 1 mM Nα-p-tosyl-l-lysine chloromethyl ketone [TLCK; Sigma, St. Louis, Mo.]) before clearance by centrifugation at 15,000 × g for 15 min. The supernatants were immunoprecipitated with α-gC or α-EPO (Genzyme) for a minimum of 6 h at 4°C. Protein A-Sepharose resuspended in lysis buffer was added to each sample for an additional hour, and the immune complexes were washed and separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). Polyacrylamide gels were dried and processed for autoradiography.
Far-Western blotting of gC:EPO fusion molecules with EREx.
Vero cells were transfected in absence of radiolabeled amino acid and processed as described above. The nonlabeled polyacrylamide gels containing the immunoprecipitated proteins were transferred to nitrocellulose membrane for EPO receptor binding analysis. The nitrocellulose membranes were blocked with BLOTTO (TBS supplemented with 3% [wt/vol] milk powder) and incubated for 1 h in the presence of 5 μg of EREx diluted in BLOTTO. EREx was purified from isopropyl-β-d-thiogalactopyranoside (IPTG)-induced bacteria as described by Harris et al. (28). Briefly, plasmid pGEX3X, encoding the EPO receptor extracellular domain fused to GST, was used to transform Escherichia coli JM109. A 100-ml overnight culture of transformed bacteria was used to inoculate a 1-liter culture for 1 h before addition of IPTG (1 mM). Four hours postinduction, the cell pellet was resuspended in 20 ml of phosphate buffer (50 mM phosphate buffer [pH 7.4], 10 mM EDTA, 10 mM 2-mercaptoethanol, 1 mM TLCK). The pellet was resuspended with 10 passages through an 18-gauge needle and incubated at 4°C for 30 min in presence of 30 mg of lysozyme (Sigma). The cells were lysed by three freeze-thaw cycles, and the lysate was centrifuged at 15,000 × g for 15 min at 4°C. The supernatant was purified on phosphate buffer-equilibrated glutathione-agarose beads (GAB; Sigma), and EREx was eluted with 10 mM reduced glutathione (Sigma) before being concentrated in a Centricon 30 concentrator (Amicon, Beverly, Mass.). Following incubation in the presence or absence of EREx, the membranes were incubated with a 1/100 dilution in BLOTTO of a mixture of polyclonal antibodies directed against the human EPO receptor and against the GST protein (Santa Cruz Biotechnology, Santa Cruz, Calif.). The membranes were then washed and incubated for 1 h at room temperature with an anti-rabbit horseradish peroxidase (HRP)-conjugated antibody (Sigma Immuno Chemical, St. Louis, Mo.) and revealed with ECL (enhanced chemiluminescence) substrates (Amersham).
Construction and isolation of recombinant viruses.
HSV-1 mutants were constructed by standard marker transfer procedures using LipofectAmine (Gibco-BRL) for cotransfection, and the resulting recombinant virus plaques were thrice plaque purified by limiting dilution prior to characterization. To construct the recombinant viruses KgBpK−gCEPO2 and KgBpK−gCEPO3, pgCEPO2 and pgCEPO3 plasmid DNAs were each cotransfected on Vero cells with the viral DNA from lacZ-expressing mutant recombinant KgBpK−gC− (41). The KgBpK−gCEPO2 and KgBpK−gCEPO3 recombinant viruses were selected by a clear-plaque phenotype following staining with β-galactosidase substract. The selected viruses were thrice plaque purified, and the viral DNAs were analyzed by Southern blotting for deletion of the wild-type gC sequence and insertion of the gCEPO2 and gCEPO3 coding sequences.
KgBpK−gCEPO2gD− recombinant virus was created by cotransfection of viral DNA from KgBpK−gCEPO2 and plasmid DNA from pgDLacZ on gD-complementing VD60 cells. Following 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal) staining, blue plaques were selected for plaque purification and viral DNA from recombinant virus was analyzed by Southern blotting for the deletion of gD and insertion of lacZ coding sequences.
Southern blot characterization of gC:EPO recombinant viruses.
Viral DNAs from plaque-purified virus were prepared, digested with BamHI, separated on agarose gels, and transferred to membranes for Southern blot analysis. A 496-bp NcoI fragment of a gB plasmid (pKBXX [6]) was 32P labeled and used as a gB probe to confirm the polylysine deletion within the gB sequence (41). A 61-bp PstI fragment from the EPO plasmid was 32P labeled and used as a probe for detection of the presence of the EPO coding sequence in recombinant viruses, while a 32P-labeled 642-bp NcoI fragment from pgC1 was used to confirm the deletion of this sequence in KgBpK−gC− and KgBpK−gCEPO3 viruses. Deletion of gD sequence from KgBpK−gCEPO2gD− recombinant virus was confirmed through absence of hybridization of a 32P-labeled 480-bp NaeI probe from pgD1 with BamHI-digested viral DNA from KgBpK−gCEPO2gD−, while hybridization with a 6,547-bp fragment from KgBpK−gCEPO2 was detected.
Purification of radiolabeled viruses.
Virions used for immunoprecipitation and binding assays were labeled and purified as follows. Confluent Vero cell monolayers in T150 flasks (Falcon; Becton Dickinson, Franklin Lakes, N.J.) were infected with viruses at a multiplicity of infection (MOI) of 10. Four hours p.i., 16 ml of minimal essential medium without methionine and cysteine (Gibco-BRL) supplemented with 1% dialyzed fetal calf serum was added to the infected cell monolayers. [35S]methionine-[35S]cysteine (ExpreSS; NEN-Dupont, Boston, Mass.) having a final specific activity of 50 μCi/ml was also added after 4 h. Twenty-four hours p.i., media containing radiolabeled virus was harvested and virions purified by centrifugation (SWTi-40 Beckman rotor) through sucrose gradients (30 to 65% sucrose in 0.5× TBS). The fractions containing the radiolabeled virus were pooled, diluted in sterile TBS, and centrifuged at 20,000 × g for 1 h at 4°C in an SWTi-40 rotor. The virus pellet was resuspended in TBS, and radioactivity was determined with a beta counter (Beckman, Fullerton, Calif.).
Immunoprecipitation analysis of surface glycoproteins.
Aliquots of radiolabeled virus were immunoprecipitated with a pool of gB-specific (5), gC-specific (48), and gD-specific (31) MAbs or α-EPO. Each virus aliquot was diluted in 200 μl of lysis buffer containing 2 μl of antibody and incubated at 4°C for a minimum of 4 h. The immune complexes were incubated with protein A-Sepharose (Sigma) for 1 h, centrifuged at 500 × g, and washed five times with 600 μl of lysis buffer. The protein A-Sepharose complexes were resuspended in Laemmli loading buffer (39), boiled for 2 min, and subjected to SDS-PAGE. After electrophoresis, the gels were fixed, treated with En3Hance solution (NEN-Dupont), vacuum dried, and exposed to X-Omat XAR film (Kodak, Rochester, N.Y.). Precipitated radiolabeled proteins were quantified with the 1-D Scan and Report program (Biomed Instruments, Fullerton, Calif.).
Titration of recombinant viruses on L and sog9 cells.
Different dilutions of wild-type KOS and recombinant viruses (KgBpK−, KCZ, KgBpK−gC−, KgBpK−gCEPO2, and KgBpK−gCEPO3) were used to infect mouse L cells and the Gag-deficient derivative sog9 cells. The cell monolayers were then overlaid with medium containing 0.5% methylcellulose and incubated for 48 h at 37°C to allow virus plaques to form. Cells were then fixed and stained with crystal violet to quantify plaque numbers. For the same dilution, the ratio of the number of plaques produced by each virus on sog9 cells was divided by that produced on parental L cells, expressed as a percentage and taken as the measure of HS-binding-independent activity of each virus.
Binding of purified virions to EREx.
EREx-transformed bacterial cell lysate was bound to GAB and washed with complete medium. As a control for EREx-specific binding, EREx-free GAB was produced by elution of EREx from GAB-bound EREx (EREx-GAB) with 10 mM reduced glutathione and washed with complete medium (GAB). Then 500 PFU of unlabeled purified virions was incubated in the presence of EREx-GAB or GAB for 1 h at room temperature, unbound virus was collected by using three washes with complete medium, and the pooled washes were titrated on Vero cell monolayers. The EREx-virus complexes were eluted with 10 mM reduced glutathione and titrated on Vero cells. Vero monolayers were then overlaid with medium containing 0.5% methylcellulose at 37°C to allow virus plaques to form. Cells were fixed and stained with crystal violet for plaque quantification; virus-specific binding to EREx was taken as the ratio of unabsorbed viruses from EREx-GAB to GAB expressed as a percentage.
Virus neutralization.
KOS, KgBpK−gC−, KgBpK−gCEPO2, and KgBpK−gCEPO3 (300 PFU of each) were incubated for 2 h at 37°C with EREx or with extract from bacteria transformed with the empty plasmid vector (input). For antibody neutralization, 500 PFU of each virus was incubated for 3 h at 37°C with 5 μl of rabbit complement (Gibco-BRL) and either 2 μl of different dilutions of a pool of mouse MAbs against gC site II (MAbs C9, C10, and C13 [87]) or site I (MAbs C2, C3, and C17 [87]) or a rabbit polyclonal antibody against EPO. The neutralized virus preparations were used to infect confluent monolayers of Vero cells for 2 h. The infected monolayers were washed twice with TBS and treated with low-pH glycine buffer to inactivate nonpenetrated virus. The cell monolayers were then overlaid with medium containing 0.5% methylcellulose, and virus plaques were allowed to form at 37°C. Cells were then fixed and stained with crystal violet for plaque number quantification. For EREx neutralization, the plaque number was expressed as the percentage of plaques formed following virus incubation in the absence (input) or presence of EREx divided by the average number of plaques formed in absence of EREx (input) and expressed as a percentage. For the antibody/complement neutralization analysis, the IC50 was determined as the dilution of antibody required to neutralize 50% of the input virus incubated with the rabbit complement only.
Binding of radiolabeled virus to cells.
Monolayers of confluent Vero cells and suspensions of FD-EPO cells intensively washed with medium without EPO were incubated at 4°C with radiolabeled purified virions. The viruses were allowed to bind to the cell surface for 10 to 320 min, after which the unbound viruses were removed by washing the cells three times with cold TBS. Cell-bound virions were quantified by liquid scintillation counting using a beta counter.
Western blot analysis of virus gene expression in infected FD-EPO cells.
Control Vero cells and test FD-EPO cells were infected at 37°C with wild-type KOS, KCZ (a gC-deleted virus [41]), and gC-EPO recombinant viruses at an MOI of 100; 2 and 12 h p.i., the cells were washed with TBS, resuspended in Laemmli buffer, sonicated, boiled for 2 min, cleared by centrifugation, and separated by SDS-PAGE. The gel proteins were transferred to nitrocellulose membranes and blotted with antibody specific for ICP0 (polyclonal antibody kindly provided by Richard J. Courtney, Pennsylvania State University), ICP4 (MAb kindly provided by David L. Thompson, Pennsylvania State College of Medicine), and ICP6 (MAb kindly provided by Bio-Méga Inc., Montréal, Québec, Canada). The membranes were then washed and incubated for 1 h at room temperature with an anti-rabbit or anti-mouse HRP-conjugated antibody (Sigma Immuno Chemical) and revealed with ECL substrates.
Virus growth curves.
FD-EPO cells were infected with wild-type KOS, KgBpK−gC−, KgBpK−gCEPO2, and KgBpK−gCEPO3 viruses at an MOI of 100. Every 4 h p.i. for a period of 36 h, viruses contained in infected cells and supernatants were combined and titrated on Vero cells. The quantity of produced infectious virus was plotted versus time p.i.
FD-EPO cell growth in the presence of purified virions.
FD-EPO cells were pelleted and rinsed three times with complete medium without EPO; 10,000 cells per well were incubated in the absence or presence of 1 U of EPO per ml or infected at MOIs of 100, 10, and 1.0 with KgBpK−gC−, KgBpK−gCEPO2, and KgBpK−gCEPO3 sucrose-purified virions or with equivalent preparations of sucrose-purified KgBpK−gCEPO2gD− grown on noncomplementing Vero cells. At different time points over a period of 4 days, the number of viable cells per well (determined by trypan blue exclusion) was counted in a hemacytometer (Fisher, Pittsburgh, Pa.) and plotted as the number of cells per well for each test condition.
Transmission electron microscopy.
A total of 10,000 FD-EPO cells were mock infected or infected with KgBpK−gC− or KgBpK−gCEPO2 virus at an MOI of 100; 90 min p.i., the cells were centrifuged, washed twice with TBS, and fixed overnight with 2.5% glutaraldehyde in phosphate-buffered saline (PBS). Samples were thrice washed with PBS and postfixed in 1% OsO4 in water for 1 h at room temperature. Samples were again thrice washed in PBS, dehydrated by using a graded series (30 to 95%) of ethanol (three changes of 100% ethanol and two changes of propylene oxide), and incubated in a 1:1 mixture of propylene oxide and Polybed 812 resin (Polysciences, Warrington, Pa.) for 1 h. The resin mixture was replaced with 100% resin and allowed to sit overnight at 4°C. The following day, the resin was changed twice, and samples were embedded in molds, cured overnight at 37°C, and hardened for an additional 2 days at 65°C. Thick (300-nm) sections, obtained by using a Reichert ultramicrotome fitted with a diamond knife, were heated onto glass slides, stained with 1% toluidine blue, and rinsed with water. Ultrathin (60-nm) sections were collected on Formvar-coated grids and stained with 2% uranyl acetate in 50% methanol for 10 min and then 1% lead citrate for 7 min. Sections were analyzed and photographed on a JEOL JEM 1210 transmission electron microscope at 80 kV.
RESULTS
Expression and processing of gC:EPO fusion molecules on transfected cells.
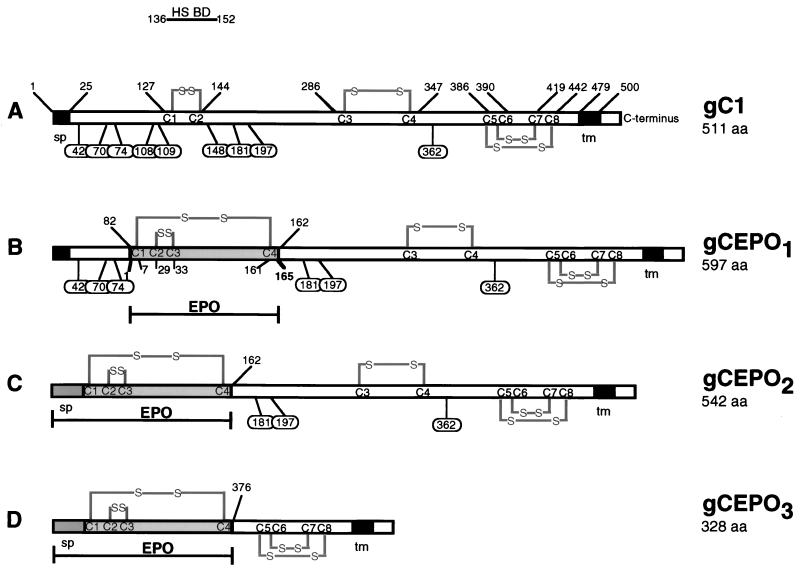
Infection of HSV-1 involves virus attachment through gB and gC to HS moieties present at the cell surface (26, 29, 30), followed by virus and cell membrane fusion and penetration of the nucleocapsid into the cytoplasm (62, 74). To redirect adsorption of HSV-1 to a receptor other than HS, we constructed three recombinant genes in which the HS binding domain of gC (79) was replaced with EPO (Fig. 1). In the first construct (gCEPO1), the EPO coding sequence (165 amino acids) was used to replace in frame the coding sequence for amino acids 83 to 161 of wild-type gC of HSV-1 (gC1). Removal of residues 83 to 161 deleted three predicted sites for N-linked oligosaccharides (N-CHO) (23) and most of antigenic site II of gC1 (site II is lies between gC1 residues 129 to 247 [87]) probably formed through a disulfide bridge between cysteines 1 and 2 (positions 127 and 144, respectively [66]). The more C-terminal antigenic site I, however, is left intact. In this construct, the coding sequence for the signal peptide (residues 1 to 25), the first 82 N-terminal residues, and the C-terminal residues 162 to 511 were left intact. We constructed a second fusion protein (gCEPO2) gene in which the sequence encoding the signal peptide and the first 161 N-terminal residues were replaced with the sequences encoding the signal peptide (27 amino acids) and the 165 residues from the EPO polypeptide. In this construct, the three predicted N-CHO sites of gC (positions 108, 109, and 148) were also deleted. In a third construct, the DNA coding for most of the external domain of gC1, residues 1 to 375, was deleted in order to create gCEPO3. In this construct, the sequence encoding the predicted remaining N-CHO sites of gC were deleted along with the DNA encoding the secondary loop structure produced by the disulfide bridge between cysteines 3 and 4 at positions 286 and 347, respectively. Wild-type gC1 and the three gC:EPO fusion molecules were cloned as expression cassettes in which either the HCMV IEp or the wild-type gC1 promoter was juxtaposed to the gC1 or gC:EPO recombinant gene.
FIG. 1.
Structures of gC:EPO fusion molecules. (A) Structure of wild-type HSV-1 gC strain KOS (gC1) depicting the HS binding domain (BD), transmembrane domain (tm), signal peptide (sp), cysteine bridges (S-S), predicted N-linked glycosylation sites (oval balloons), and the positions of these elements relative to the amino- and carboxy-terminal residues. The EPO protein (shaded box) was inserted in frame at three different positions within gC1, replacing amino acid residues (aa) 83 to 161 (B), 1 to 161 (C), and 1 to 375 (D) in order to create the recombinant fusion proteins gCEPO1, gCEPO2 and gCEPO3 respectively. gCEPO2 and gCEPO3 encode the EPO signal peptide (darkly shaded box).
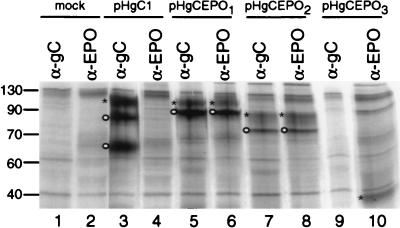
To examine the expression and processing of the recombinant gC:EPO glycoproteins, plasmids encoding wild-type gC1 and the gC:EPO fusion molecules were used to transfect Vero cells in presence of [35S]methionine-[35S]cysteine. Thirty hours posttransfection, the detergent-soluble proteins from transfected cells were immunoprecipitated with α-gC and α-EPO and visualized by autoradiography following SDS-PAGE. As demonstrated in Fig. 2, gC1 was immunoprecipitated with α-gC (lane 3) as precursors and the mature form of gC1 (24, 34); as expected, neither form was immunoprecipitated by α-EPO (lane 4). As predicted, the pre-gCEPO1 polypeptide (597 amino acids) was immunoprecipitated by α-gC or α-EPO (lane 5 or 6, respectively) and showed reduced mobility on SDS-PAGE compared to the immunoprecipitated pre-gC1 (511 amino acids) due to the additional 85 residues (addition of 166 residues from EPO and deletion of 80 residues from gC) encoded by gCEPO1. However, the mature gCEPO1 polypeptide showed a molecular ratio similar to that of mature gC1 due to the removal of three potential N-CHO sites at position 108, 109, and 148. Immunoprecipitation of gCEPO2 by α-gC or α-EPO (lane 7 or 8, respectively) showed a band representing pre-gCEPO2 migrating at a lower molecular ratio than pre-gCEPO1 due to the deletion of an additional 81 residues located at the N terminus of gCEPO1. The lower mobility of mature gCEPO2 than of pre-gCEPO2 is attributed to the three potential N-CHO sites remaining on the chimeric molecule at positions 181, 197, and 362. Taken together, these data demonstrate that gCEPO1 and gCEPO2 are recognized by anti-gC and anti-EPO antibodies and showed precursor-product relationships consistent with posttranslational modification in the endoplasmic reticulum (ER) and Golgi complex. As expected, α-gC1 complexed with protein A-Sepharose did not precipitate the gCEPO3 fusion molecule (lane 9) due to the absence of gC1 antigenic sites encoded by this fusion molecule and recognized by this pool of gC-specific MAbs. However, the fusion molecule was immunoprecipitated with α-EPO as a single band in the gel following SDS-PAGE (lane 10). The molecular size of this single band corresponded to the predicted molecular size of the fusion molecule in which the gC1-related sequence was not ER modified through N-CHO addition. Potential precursor forms resulting from EPO glycosylation were not detected. As expected, both α-gC and α-EPO failed to immunoprecipitate a specific protein from mock-transfected cells (lanes 1 and 2, respectively).
FIG. 2.
Immunoprecipitation of wild-type and gC:EPO fusion molecules from transfected cells. Vero cells were mock transfected (lanes 1 and 2) or transfected with the indicated plasmids in the presence of [35S]methionine-[35S]cysteine. The detergent-solubilized monolayers were immunoprecipitated with α-gC or α-EPO, as indicated. The protein A-bound immune complexes were separated by SDS-PAGE and visualized by autoradiography. Sizes are indicated in kilodaltons on the left. The mobility of each mature (∗) or premature (o) protein is indicated.
The gC:EPO recombinant molecules transported to the cell surface of transfected cells.
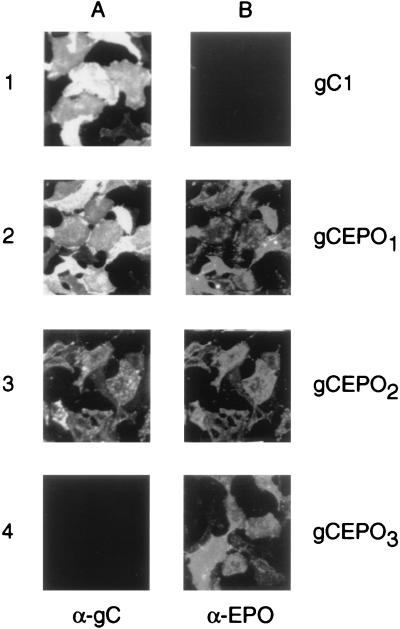
Modifications of HSV-1 glycoproteins can result in defective recombinant molecule processing and/or intracellular trafficking and consequently the failure to incorporate the modified protein into virus envelopes (14, 40). Data presented in Fig. 2 demonstrated that the chimeric gCEPO1 and gCEPO2 molecules were modified posttranscriptionally, as demonstrated by the presence of two distinct immunoprecipitated products from transfected cells. To examine the intracellular trafficking of the gC:EPO fusion molecules, we performed transient gene expression assays to determine whether the recombinant molecules were transported to the surface of infected cells (Fig. 3). Vero cells were transfected with pHgC1 (HCMV IEp driving gC1), and the plasmids encoding the three gC-EPO fusion molecules; 24 h posttransfection, we attempted to detect the presence of gC and EPO epitopes on the surfaces of unfixed transfected cells by indirect immunofluorescence using either α-gC or α-EPO. Bound MAbs were detected with an anti-mouse secondary antibody conjugated with FITC; bound polyclonal antibodies were detected with an anti-rabbit antibody conjugated with Cy3. The results shown in Fig. 3A demonstrate that wild-type gC1, gCEPO1, and gCEPO2 were each recognized by α-gC, demonstrating their presence on the cell surface, while this antibody did not detect gC epitopes from pHgCEPO3-transfected cells due to the absence of gC1-specific epitopes recognized by our pool of gC-specific MAbs on this chimeric molecule. Nevertheless, the presence of the gCEPO3 molecule at the surface of transfected Vero cells was demonstrated by the binding of α-EPO at the surface of the pHgCEPO3-transfected cells (Fig. 3B, panel 4). α-EPO recognized the EPO epitopes present at the surface of the pHgCEPO1- and pHgCEPO2-transfected cells but did not recognize these epitopes at the cell surface of wild-type gC1-transfected cells. These data demonstrate that the gC-EPO chimeric molecules were exposed on the plasma membrane of transfected cells, indicating that these recombinant proteins were processed and transported to the cell surface in a manner similar to wild-type gC.
FIG. 3.
Transfection of Vero cells with gC:EPO expression plasmids results in the appearance of these recombinant proteins at the cell surface. Vero cells were transfected with plasmids encoding gC1, gCEPO1, gCEPO2, and gCEPO3 fusion molecules, as indicated. Twenty-four hours posttransfection, the monolayers were incubated with α-gC MAbs (A) and α-EPO (B). An FITC-conjugated anti-mouse secondary antibody was used to detect α-gC, while α-EPO was detected with an anti-rabbit secondary antibody conjugated with Cy3. The monolayers were then fixed with ice-cold methanol, and the immunofluorescence signal was visualized and photographed with a Nikon model 211910 microscope.
Binding of gC:EPO fusion molecules to EREx.
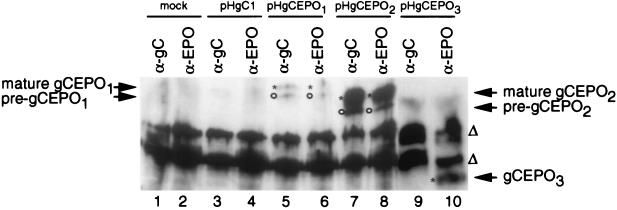
To determine if the gC:EPO chimeric molecules were capable of binding to the EPO receptor, we examined their ability to bind to EREx by far-Western blot analyses. In this assay, Vero cells were mock transfected (Fig. 4, lanes 1 and 2) or transfected with expression plasmids (lanes 3 to 10) as described for Fig. 2; 30 h posttransfection, the detergent-soluble proteins from transfected cells were immunoprecipitated with α-gC or α-EPO and far-Western blotted with EREx. The bound EREx proteins were detected with a rabbit anti-GST and anti-EPO receptor polyclonal antiserum in combination with an anti-rabbit secondary antibody conjugated with HRP. gCEPO2 (lanes 7 and 8) and gCEPO3 (lane 10) show greater binding to EREx than gCEPO1 (lanes 5 and 6), while gC1 (lanes 3 and 4) or immunoprecipitates from mock-transfected cells (lanes 1 and 2) failed to react with EREx. The absence of EREx binding in lane 9 is due to the failure of α-gC to immunoprecipitate gCEPO3 (Fig. 2) and not to the inability of gCEPO3 to bind EREx, as demonstrated by the presence of EREx binding in lane 10 when the fusion protein was immunoprecipitated by α-EPO. Nonspecific binding (probably due to the presence of antibody heavy chains present in the blotted samples) were detected in all samples to the same extent as in similar experiments performed in the absence of EREx (data not shown). These results demonstrated that gCEPO2 and gCEPO3 were each capable of binding to EREx.
FIG. 4.
Far-Western blot analysis using EREx distinguished the receptor-binding capabilities of the three gC:EPO recombinant molecules. Vero cells were mock transfected (lanes 1 and 2) or transfected with the indicated plasmids. The detergent-solubilized monolayers were immunoprecipitated with α-gC or α-EPO, as indicated. The protein A-bound immune complexes were separated by SDS-PAGE, transferred to a nitrocellulose membrane, and incubated with 5 μg of EREx, a mixture of polyclonal antibodies against the EPO receptor and GST, and a polyclonal antibody conjugated with HRP. Detection was performed using an ECL kit as described by the manufacturer. The mobility of each mature (∗) or premature (○) protein, as well as that of nonspecific protein binding (▵), is indicated.
Construction of gC:EPO recombinant viruses.
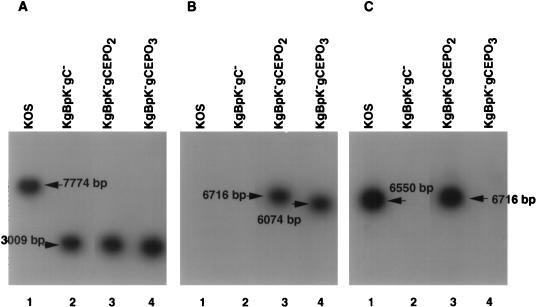
Since far-Western blot analyses demonstrated that gCEPO2 and gCEPO3 were capable of binding to EREx to a greater extent than gCEPO1, only the DNAs encoding gCEPO2 and gCEPO3 were recombined with viral DNA. To redirect virus infection to the EPO receptor, DNA from HSV-1 mutant KgBpK−gC− (41) was cotransfected with plasmids encoding gCEPO2 and gCEPO3 to produce KgBpK−gCEPO2 and KgBpK−gCEPO3 recombinant viruses, respectively. Recombinant viruses were selected by their clear-plaque phenotype following X-Gal staining. The recombinant viral plaques were analyzed by Southern blotting to confirm their genotypes. Viral DNAs were extracted from purified virions, digested with endonuclease BamHI, and Southern blotted with a 32P-labeled gB (Fig. 5A), EPO (Fig. 5B), or gC (Fig. 5C) probe to confirm the deletion of the HS binding domain within gB, the insertion of the EPO coding sequence, or the deletion of a coding sequence within the gC molecule, respectively. As shown in Fig. 5A, the 32P-labeled gB probe hybridized to the BamG fragment (7,774 bp) of HSV-1 DNA containing the wild-type gB coding sequence encoded by KOS (lane 1), while this probe hybridized to a 3,009-bp fragment in the KgBpK−gC− (lane 2), KgBpK−gCEPO2 (lane 3), and KgBpK−gCEPO3 (lane 4) recombinant viruses. These results confirmed the presence of the mutant gB gene since a BamHI recognition sequence was introduced at the site of the pK mutation (HS binding domain encoding a polylysine [pK] deletion) in the mutant viruses, resulting in the production of two subfragments (3,009 and 4,738 bp) after digestion (41). The insertion of the EPO coding sequence was confirmed by Southern blot hybridization of BamHI-digested viral DNA with a 32P-labeled EPO probe (125-bp PstI fragment of the EPO plasmid) that hybridized to an EPO sequence inserted within KgBpK−gCEPO2 and KgBpK−gCEPO3 recombinant viruses. As shown in Fig. 5B, the 32P-labeled EPO probe hybridized to 6,716- and 6,074-bp fragments in KgBpK−gCEPO2 and KgBpK−gCEPO3 digested viral DNAs (lanes 3 and 4, respectively) and failed to hybridize with KOS and KgBpK−gC− digested viral DNAs (lanes 1 and 2, respectively), demonstrating that the EPO coding sequence was present within both gC-EPO recombinant viruses. The difference in size in the hybridized viral DNA fragment from KgBpK−gCEPO2 and KgBpK−gCEPO3 is due to the additional 642-bp deletion of KgBpK−gCEPO3 compared to KgBpK−gCEPO2. The replacement of a gC coding sequence within the gC-deleted virus (KgBpK−gC−; lane 2) and KgBpK−gCEPO3 recombinant virus (lane 4) was confirmed by Southern blot hybridization of BamHI-digested viral DNA with a 32P-labeled gC probe (642-bp NcoI fragment of pgC1) that hybridized to a gC sequence deleted in both viruses. As shown in Fig. 5C, the 32P-labeled gC probe hybridized to a 6,650-bp fragment in KOS (lane 1) and a 6,717-bp fragment in KgBpK−gCEPO2 (lane 3), demonstrating the presence of this gC coding sequence in both viruses, while this same probe did not hybridize with digested viral DNAs from KgBpK−gC− (lane 2) and KgBpK−gCEPO3 (lane 4), demonstrating the deletion of this sequence from these two mutant viruses. Together, these data confirmed the isolation of KgBpK−gCEPO2 and KgBpK−gCEPO3 recombinant viruses deleted for the HS binding domain of gB as well as the predicted HS binding domain of gC and encoding the EPO coding sequence. The KgBpK−gCEPO2 virus recombinant was further modified by deletion of gD and replacement with a lacZ expression cassette to create KgBpK−gCEPO2gD− as described in Materials and Methods and confirmed by Southern blotting and gD-specific immunoprecipitation of purified radiolabeled virions grown on noncomplementing cells (data not shown).
FIG. 5.
Southern blot characterization of gC:EPO recombinant viruses. Viral DNAs from KOS (lanes 1), KgBpK−gC− (lanes 2), KgBpK−gCEPO2 (lanes 3), and KgBpK−gCEPO3 (lanes 4) were digested with endonuclease BamHI and subjected to Southern blot analysis using a gB (A), EPO (B), or gC (C) 32P-labeled probe. The molecular sizes of the bands are indicated relative to the molecular size standards.
Expression of gC:EPO fusion molecules from Vero-infected cells and their insertion into the envelope of recombinant viruses.
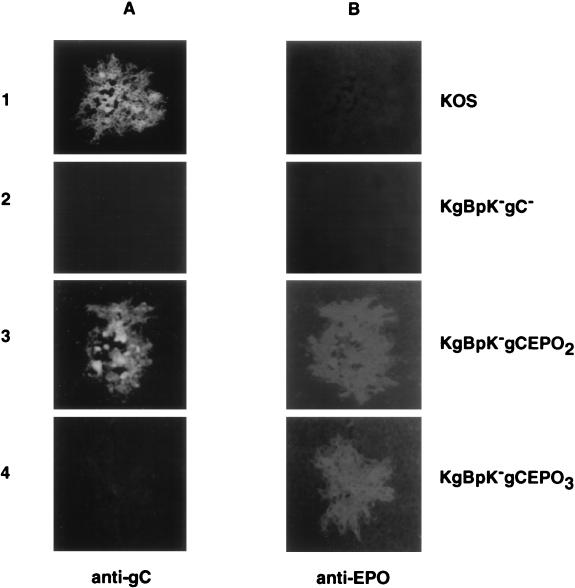
Recombination of the DNA encoding the gC:EPO fusion molecules with viral DNA may lead to redirected virus infection through recognition of the EPO receptor only if the recombinant glycoproteins are appropriately expressed and incorporated into the virus envelope. Accordingly, Vero cells infected with the recombinant viruses were analyzed for the presence of both cell- and virus-associated gC:EPO fusion proteins by immunofluorescence and radioimmunoprecipitation assays, respectively. For the immunofluorescence assays (Fig. 6), Vero cells were infected with wild-type KOS, KgBpK−gC−, KgBpK−gCEPO2, and KgBpK−gCEPO3 recombinant viruses, methanol fixed, and incubated with either α-gC or α-EPO. The bound antibodies were detected with an FITC-conjugated anti-mouse antibody (Fig. 3A) or a Cy3-conjugated anti-rabbit antibody (Fig. 3B). As shown in Fig. 6, wild-type KOS plaques were recognized by α-gC and, as expected, not by α-EPO; KgBpK−gC− plaques were negative for both antigens, since this mutant virus was deleted for the gC coding sequence and devoid of EPO-specific epitopes. Vero cells infected with KgBpK−gCEPO2 virus were detected with α-gC and α-EPO, demonstrating that the gCEPO2 fusion molecule was expressed from the recombinant KgBpK−gCEPO2 virus. As expected, only α-EPO reacted with the gCEPO3 fusion molecule from KgBpK−gCEPO3-infected Vero cells since the gCEPO3 fusion molecule was deleted of all antigenic sites recognized by α-gC. Taken together, these data demonstrate that gCEPO2 and gCEPO3 chimeric molecules were expressed from infected Vero cells with KgBpK−gCEPO2 and KgBpK−gCEPO3 viruses, respectively.
FIG. 6.
Expression of gC:EPO fusion molecules by recombinant viruses. Vero cell monolayers were infected with wild-type KOS, KgBpK−gC−, KgBpK−gCEPO2, and KgBpK−gCEPO3 viruses; 48 h p.i., the infected cell monolayers were fixed in ice-cold methanol and processed for immunofluorescence using α-gC (A) and α-EPO (B). An FITC-conjugated anti-mouse secondary antibody was used to detect α-gC, while α-EPO was detected with an anti-rabbit secondary antibody conjugated with Cy3. Fluorescent cells were visualized and photographed with a Nikon model 211910 microscope.
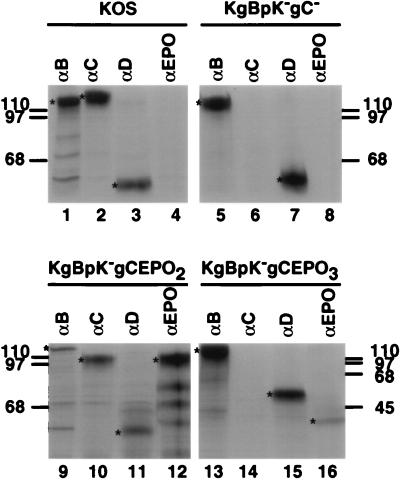
Incorporation of the recombinant molecules within the envelope of purified virions harvested from the supernatant of infected Vero cells in the presence of [35S]methionine-[35S]cysteine was detected by immunoprecipitation of detergent-soluble proteins with specific antibodies. The protein A-captured immune complexes were then analyzed by SDS-PAGE and autoradiography. As shown in Fig. 7, wild-type gB was immunoprecipitated from KOS (lane 1), KgBpK−gC− (lane 5), KgBpK−gCEPO2 (lane 9), and KgBpK−gCEPO3 (lane 13) viruses, demonstrating that the envelopes of wild-type and recombinant mutant viruses each contained gB. Similar analyses of immunoprecipitates derived from the use of α-gC demonstrated that gC was present in KOS (lane 2) and KgBpK−gCEPO2 (lane 10) viruses but absent in KgBpK−gC− (lane 6) and not precipitated from KgBpK−gCEPO3 solubilized virion proteins due to the lack of gC-specific epitopes contained within the gCEPO3 fusion molecule (lane 14). Immunoprecipitation performed with α-EPO demonstrated that the gCEPO2 and gCEPO3 fusion molecules were incorporated into KgBpK−gCEPO2 and KgBpK−gCEPO3 virus envelopes (lanes 12 and 16, respectively), while wild-type (lane 4) and KgBpK−gC− mutant (lane 8) viruses were not reactive with α-EPO. gD (lanes 3, 7, 11, and 15) was detected in all virion envelope preparations, and the ratio of the quantity (analyzed by densitometry) of immunoprecipitated gB, gBpK−, gC, or gC:EPO to gD demonstrated that (i) the pK mutation in gB did not affect the level of incorporation of the mutant gBpK− molecules into the virion envelopes, as demonstrated elsewhere (41), and the absence of gC did not increase the relative amount of gB incorporation into virus envelopes, and (ii) the gCEPO2 fusion molecules were incorporated into the envelopes of recombinant virus to a level similar to that of gC in wild-type virus; however, (iii) the gCEPO3 fusion molecule was an exception in that its level of incorporation relative to that demonstrated for wild-type gC into KOS virus was approximately 20%. Quantification of the level of recombinant proteins incorporated within mature virions was normalized for the number of methionine-cysteine-labeled residues per molecule.
FIG. 7.
Immunoprecipitation of gB, gC, gD, and gC:EPO fusion molecules from sucrose gradient-purified virions. [35S]methionine-[35S]cysteine-labeled sucrose-purified wild-type KOS, KgBpK−gC−, KgBpK−gCEPO2, and KgBpK−gCEPO3 viruses were detergent solubilized and immunoprecipitated with a pool of MAbs directed against gB (lanes 1, 5, 9, and 13), gC (lanes 2, 6, 10, and 14), and gD (lanes 3, 7, 11, and 15) or α-EPO (lanes 4, 8, 12, and 16). The protein A-bound immune complexes were separated by SDS-PAGE, and the dried gels were autoradiographed. Asterisks indicate the positions of proteins when present, and migration positions of molecular weight standards are indicated in kilodaltons at the left and right.
The incorporation of each gC:EPO fusion molecule into recombinant virus envelope was confirmed by complement-dependent neutralization assays using α-gC and α-EPO. As shown in Table 1, the 50% neutralization endpoint for wild-type virus in assays using MAb pools specific for gC antigenic sites I and II was 1/500, while the titer specific for site II only (MAbs C9, C10, and C13) was 1/200. α-EPO did not neutralize wild-type virus demonstrating that EPO and gC lacked epitopes cross-reactive with EPO. As expected, neither α-gC nor α-EPO neutralized KgBpK−gC− virus (Fig. 6 and 7). KgBpK−gCEPO2 retained the coding sequence for antigenic site I but not II and thus was resistant to neutralization by site II-specific antibodies. A 1/20 dilution of EPO-specific antibody was required to neutralize 50% of KgBpK−gCEPO2 recombinant virus particles, confirming the presence of EPO. KgBpK−gCEPO3 was also neutralized, albeit less efficiently, by this antibody, confirming that EPO was present but at a reduced level. These two recombinants were nevertheless equally sensitive to neutralization by gB-specific MAbs, indicating that virus preparations contained comparable amounts of infectious virus (data not shown).
TABLE 1.
Neutralization titer
| Virus | α-gC
|
α-EPO | |
|---|---|---|---|
| Sites I and II | Site II | ||
| KOS | 1/500 | 1/200 | —a |
| KgBpK−gC− | — | — | — |
| KgBpK−gCEPO2 | 1/250 | — | 1/20 |
| KgBpK−gCEPO3 | — | — | 1/2 |
—, no neutralization at highest concentration tested.
HS-independent binding of gC:EPO recombinant viruses.
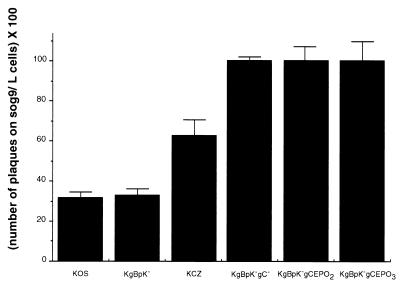
In a previous study, we reported that a KgBpK−gC− virus mutant was deleted for all detectable HS binding activity (41). Residues critical for the interaction of gC and HS were assigned to amino acids 143, 145, 147, and 150, although a distal residue at position 247 was found to be important for maintenance of this domain (79). Although the KgBpK−gCEPO2 and KgBpK−gCEPO3 recombinant viruses were deleted for the HS binding domain, we nevertheless carried out experiments to rule out any potential residual HS binding activity that might obscure the detection of EPO receptor binding. We compared the plaquing efficiencies of wild-type KOS virus, KOS derivative mutants KCZ, KgBpK, and KgBpK−gC (41), and the gC:EPO recombinant viruses on murine L and sog9 cells. As shown in Fig. 8, wild-type KOS and KOS mutants KgBpK− and KCZ possessed HS binding activity, since the ratios (expressed as percentages) of plaques produced on sog9 cells compared to L cells were 32, 33, and 63%, respectively. The gC:EPO recombinant viruses and HS-binding-defective mutant KgBpK− gC− formed comparable numbers of plaques on both cell lines, showing that the recombinants were devoid of HS binding function. These data were confirmed by assays of radiolabeled virus binding on HS+ Vero cells in which both the KgBpK−gC− and gC:EPO recombinant viruses bound similarly (data not shown).
FIG. 8.
HS binding activity of HSV-1 recombinant viruses. Wild-type KOS, KOS recombinant viruses (KgBpK−, KCZ, and KgBpK−gC−), and gC:EPO recombinants viruses (KgBpK−gCEPO2 and KgBpK−gCEPO3) were titrated on L and sog9 cells; 2 days p.i., plaques numbers were counted and expressed as a percentage of plaques produced on sog9 cells divided by that produced on L cells.
Binding of gC:EPO recombinant viruses to EREx.
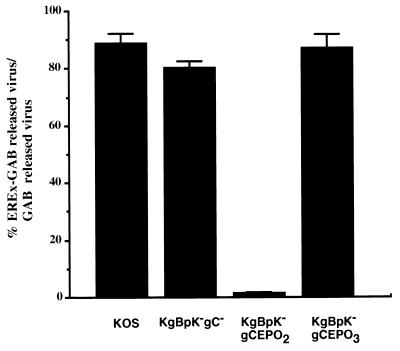
The experiments described above demonstrated that the recombinant mutants expressed the gC:EPO chimeric proteins and incorporated them into the envelopes of mature virus. It remained to be determined whether EPO was exposed on the virus envelopes and retained EPO receptor binding activity. Accordingly, we tested the abilities of recombinant gC:EPO viruses to bind a column containing EREx-GAB. KOS, KgBpK−gC−, KgBpK−gCEPO2, and KgBpK−gCEPO3 (200 PFU of unlabeled purified virions from each) were incubated with EREx-GAB or GAB for 1 h at room temperature. After the column was washed with complete medium, the quantity of virus collected in EREx-GAB flowthrough was expressed as a percentage of the virus collected in the flowthrough from GAB and taken as a measure of recombinant virus binding to EREx. As demonstrated in Fig. 9, wild-type KOS, KgBpK−gC−, and KgBpK−gCEPO3 mutant viruses were not specifically retained by EREx-GAB, compared with 98% retention of KgBpK−gCEPO2. These data demonstrated that KgBpK−gCEPO2 but not KgBpK−gCEPO3 recombinant virus was capable of binding to EREx. The inefficient binding of KgBpK−gCEPO3 to EREx-GAB was not the result of inefficient recognition of the EPO receptor by the gCEPO3 fusion molecule since it was readily recognized on EREx in far-Western blots (Fig. 4). The discrepancy between these binding activities might be explained by a substantially lower level of incorporation of gCEPO3 into mature virions (Fig. 7) or by the relative inaccessibility of the chimeric molecule in the context of the virus envelope. Elution of EREx-GAB-bound viruses with glutathione released 10 times more infectious KgBpK−gCEPO2 virus than elution of other viruses (data not shown); however, eluted virus represented only 10% of the calculated retained KgBpK−gCEPO2 virions. The low percentage of infectious EREx-KgBpK−gCEPO2 virions released from EREx-GAB by glutathione could be due either to the inactivation of virus infection by the reduced glutathione or to EREx neutralizing activity.
FIG. 9.
Binding of gC:EPO recombinant viruses to EREx-GAB. Wild-type KOS and recombinant KgBpK−gC−, KgBpK−gCEPO2, and KgBpK−gCEPO3 viruses (500 PFU of each) were incubated for 1 h at 37°C in the presence of EREx-GAB or to EREx-GAB washed with reduced glutathione in order to elute the bound EREx (GAB). Flowthrough and complete medium washes of the individual viruses were pooled and titrated on Vero cells; 2 days p.i., plaque numbers were counted and expressed as the percentage of unadsorbed viruses from EREX-GAB to GAB.
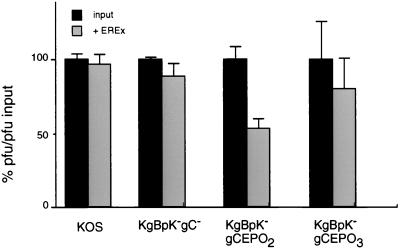
To distinguish between these two possibilities, a virus plaque reduction assay was carried out in the presence of reduced glutathione. The results demonstrated that the titers of each virus on Vero cells were similar in presence and absence of glutathione (data not shown), demonstrating that the reduced infectivity of released EREx-KgBpK−gCEPO2 was not due to free glutathione. To determine whether EREx had neutralizing activity, wild-type, KgBpK−gC−, and gC:EPO recombinant viruses were incubated with EREx or with control EREx-free eluent. The virus mixtures were used to infect Vero cells, and the number of plaques formed in the presence or absence of EREx was determined and expressed as a percentage of the control number (Fig. 10). The results demonstrated that wild-type KOS, KgBpK−gC−, and KgBpK−gCEPO3 viruses were not neutralized by EREx since the plaque numbers for EREx and control were not significantly different. However, KgBpK−gCEPO2 was neutralized with an IC50 of 0.1 mg of EREx. These data showed that KgBpK−gCEPO2 virus was capable of binding to EREx during infection and confirmed the column binding results shown in Fig. 9.
FIG. 10.
Neutralization of gC:EPO recombinant viruses by EREx. Wild-type KOS and recombinant KgBpK−gC−, KgBpK−gCEPO2, and KgBpK−gCEPO3 viruses (300 PFU of each) were incubated at 37°C for 2 h in the absence (input) or presence (+ EREx) of EREx and titrated on Vero cell monolayers; 2 h p.i., the monolayers were washed with low-pH glycine buffer, rinsed, and overlaid with methylcellulose; 2 days p.i., the monolayers were stained with crystal violet and the plaques were counted. Each count represents the average number of plaques formed following virus incubation in the absence or presence of EREx divided by the average number of plaques formed in absence of EREx and expressed as a percentage.
Binding of KgBpK−gCEPO2 recombinant virus to EPO receptor-bearing cells.
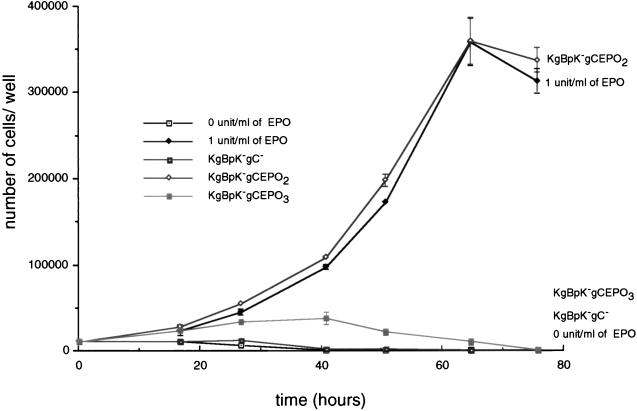
HSV-1 infection results in cell death which minimally requires IE gene expression (25, 37, 67). Incubation of FD-EPO cells with HSV-1 will not cause cell death since these cells are refractory to HSV-1 infection at high multiplicity (MOI of 100). Infections using a lacZ expression vector (41) failed to show β-galactosidase activity following X-Gal staining, and Western blot analysis of infected cells extracts failed to detect expression of viral IE gene products (data not shown). Moreover, attempts to grow wild-type, mutant, or recombinant virus on these cells were unsuccessful (data not shown). Since FD-EPO cells required stimulation by EPO for growth (54), we reasoned that if HSV-1 was not infectious and could not kill FD-EPO cells, then EPO-dependent FD-EPO cell proliferation might be stimulated by gC:EPO recombinant virus binding to the EPO receptor (Fig. 11). FD-EPO cells extensively washed with EPO-free medium were incubated with either KgBpK−gC−, KgBpK−gCEPO2, or KgBpK−gCEPO3 virus (MOI of 100) or soluble EPO (1 U/ml), and the change in number of FD-EPO cells was plotted as a function of time. KgBpK−gCEPO2 stimulated cell proliferation comparably to EPO at 1 U/ml, whereas both KgBpK−gC− and KgBpK−gCEPO3 were not stimulatory. The proliferative response to recombinant virus was dose dependent since incubation of FD-EPO cells with KgBpK−gCEPO2 virus at an MOI of 10 also stimulated cell growth at similar rate but for a shorter time period, whereas an MOI of 1.0 was insufficient to stimulate cell growth (data not shown).
FIG. 11.
Stimulation of FD-EPO cell growth upon binding of gC:EPO recombinant viruses to surface membrane EPO receptors. The number of FD-EPO cells was determined at different time points following incubation of 104 cells with 0 and 1 U of EPO per ml or with 106 PFU of KgBpK−gC− and gC:EPO recombinant viruses purified from the supernatant of infected Vero cells.
The possibility remained that virus-stimulated growth of FD-EPO cells was due to virus entry and intracellular stimulation of cell proliferation rather than virus binding to the receptor at the cell surface triggering a signaling pathway. To rule out intracellular signaling, similar experiments were performed with a KgBpK−gCEPO2 virus rendered defective for entry by removal of gD (KgBpK−gCEPO2gD−). Similar concentrations of defective particles stimulated FD-EPO cell proliferation, indicating that virus binding at the cell surface was responsible for cell growth (data not shown).
Virus internalization by FD-EPO cells following KgBpK−gCEPO2 recombinant virus binding to the EPO receptor.
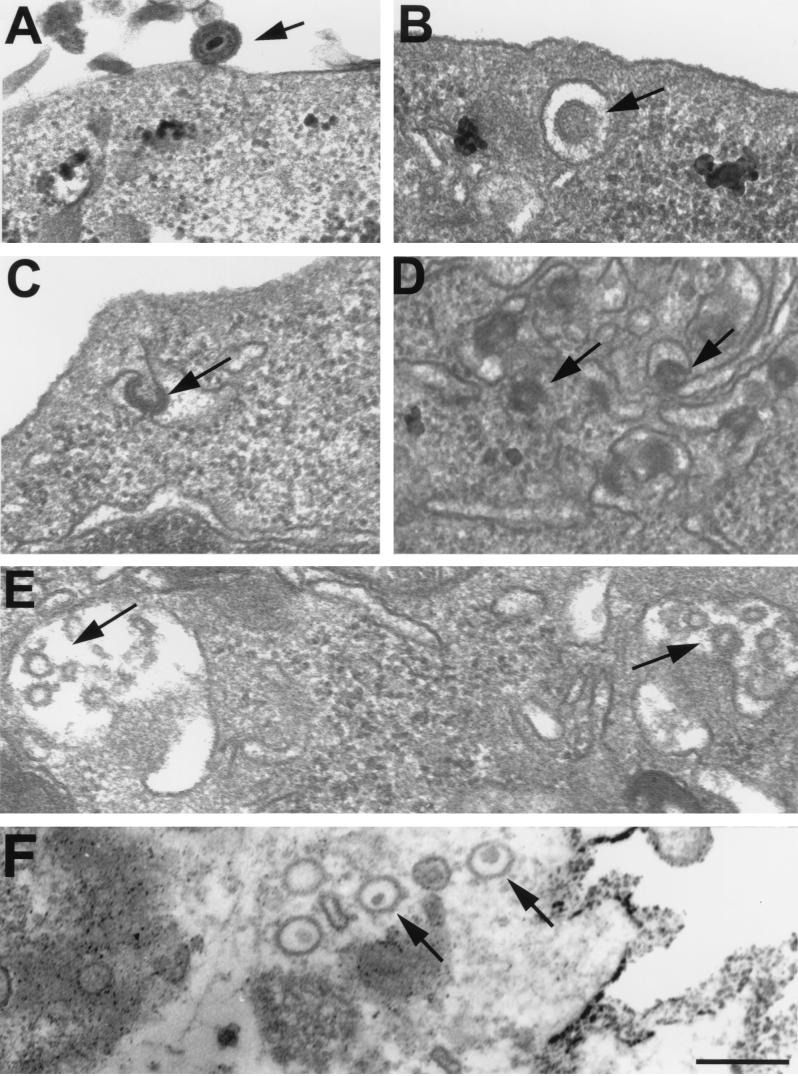
Binding of EPO to the EPO receptor activates a signaling event involving primarily phosphorylation of the receptor and other proteins involved in the JAK-STAT and ras pathways (22). The EPO receptor is then rapidly internalized by endocytosis and degraded (68). To determine if the KgBpK−gCEPO2 virus was similarly degraded following binding to the EPO receptor, transmission electron microscopy analysis of FD-EPO cells incubated with KgBpK−gC− and KgBpK−gCEPO2 viruses was performed to directly determine the fate of bound virus. Following binding of virus (Fig. 12A), internalized viruses were observed in FD-EPO cells incubated with KgBpK−gCEPO2 virus, suggesting that virus binding induced internalization of the EPO receptor (Fig. 12B to D). Moreover, the virus particles could be seen in prelysosomal vesicles, where particle degradation appeared to occur (Fig. 12E).
FIG. 12.
Transmission electron microscopy analysis of FD-EPO cells infected with KgBpK−gCEPO2 recombinant virus. FD-EPO cells were infected at an MOI of 100 with KgBpK−gCEPO2 (A to E) or wild-type HSV (F); 90 min p.i., cells were rinsed, fixed, and processed for transmission electron microscopy. (A) KgBpK−gCEPO2 bound to the surface of an FD-EPO cell; (B) KgBpK−gCEPO2 following endocytosis; (C and D) KgBpK−gCEPO2 displaying a condensing morphology within endocytotic vesicular structures; (E) KgBpK−gCEPO2 particles appearing as capsid-like structures undergoing degradation, as judged by the absence of a densely staining virus core typical of encapsulated virus DNA; (F) wild-type HSV capsid that has gained entry into Vero cells via the traditional infection route. Note that the capsid retains the densely staining DNA core. The bar equals 200 nm.
We observed within endosomal vesicles distinct changes in virus morphology that may be the result of acidification of the vesicles. Particles appeared to condense and become very dense within vesicles that were more distal to the surface (Fig. 12C and D). Internalization appeared to be specific for EPO receptor binding since internalization of KgBpK−gC− virus was rarely observed (data not shown). Wild-type virus entry of Vero cells followed the traditional infection route, releasing capsids into the cytoplasm (Fig. 12F).
DISCUSSION
Full implementation of in vivo gene therapy will require directed gene delivery using targetable vectors with the appropriate cell specificity. Viral vectors have considerable potential in this regard since the feasibility of targeted infection has already been demonstrated through evolutionary selection of particular cell types which best support individual virus life cycles. Progress in identifying receptors used by different virus groups and genetic and biochemical studies that define receptor-ligand interactions provide information essential to devising strategies for altering virus tropism (10, 27, 36, 72). In one approach, pseudotyped or recombinant hybrid vectors are constructed by substituting components from different viruses to achieve a new host cell specificity (20, 55). Although these vectors can be limited by the natural host range of the pseudotyping proteins, this method has improved vector utility in some instances by increasing the vector host range and enhancing vector stability during purification and storage (90, 91). For example, the vesicular stomatitis virus G glycoprotein has now been widely used to pseudotype Moloney murine leukemia virus (MoMLV)- and human immunodeficiency virus-based vectors with favorable results (2, 59). In a second approach, purified virions are modified by chemical cross-linking of a novel receptor binding ligand (e.g., MAb) (57, 64, 61). In a third approach, a targeting antibody is bound to virus by incorporating an engineered antibody binding ligand into the particle. For example, Sindbis virus can be modified to display a protein A-Env chimeric protein which has high affinity for the Fc region of various mammalian immunoglobulin G’s (58); thus, the virus could be armed with targeting antibodies. Although these procedures were highly efficient in vitro, they have not been tested in vivo. In a fourth approach, both enveloped and nonenveloped recombinant viruses are engineered to contain nonviral receptor ligands. A MoMLV variant expressing both wild-type Env and an Env-EPO chimeric protein displays infectivity for human cells bearing the EPO receptor (35). Likewise, it has been shown that a single-chain antibody fragment fused to the MoMLV env gene product recognizes its target cell epitope and that a corresponding viral vector can transduce cells resistant to infection by the parental virus (47, 65, 73). Recombinant adenoviruses (Ad) can also be engineered to encode modified hexon fiber (virus attachment) or penton base (virus entry) proteins (85). One vector was modified to recognize a novel αv-integrin (83) by substitution of a unique αv-integrin binding RGD peptide within the penton base protein, while another vector engineered to contain HS-binding polylysine sequences at the terminus of the Ad fiber was able to bind cell surface HS (84). Although binding to the natural coxsackievirus-Ad receptor (3) still occurred, these viruses demonstrated increased transduction of multiple cell types lacking high levels of the normal receptor.
Prior to the present research, reports of HSV recognition of a targeted receptor have been unavailable although various mutations or glycoprotein substitutions that alter the virus host range have been described. In one study, a novel cellular glycoprotein encoding the CD4 T-lymphocyte marker was introduced into the virus envelope (17); however, incorporation was inefficient and targeted virus infectivity was not tested. In other experiments, HSV gB was complemented by pseudorabies virus gB, but the reverse was not possible (52) and gC could not be substituted for gIII (82). While these experiments suggested the possibility of modifying the tropism of pseudorabies virus, this was not tested. Finally, HSV-1 and HSV-2 mixed infections have been used to pseudotype the two viruses, which acquire the ability to infect cells by using receptors used by the pseudotyping serotype. Although HSV-1–HSV-2 recombinants have been isolated in mixed infections, they were principally used for physical mapping of glycoprotein genes in combination with type-specific cytotoxic T-cell reactivities (9) or to induce cross-reactive immunity to the two virus serotypes (51) and were not studied for targeted infectivity or altered host range.
In this report, experiments were designed to meet three general objectives: (i) produce a virus particle carrying a novel envelope glycoprotein-ligand chimeric molecule, (ii) demonstrate that this chimeric molecule had a novel receptor binding function, and (iii) demonstrate that the chimeric molecule could selectively mediate virus attachment to cells expressing the targeted receptor. To achieve these objectives, we pursued a strategy in which the HS binding functions of the virus were removed and replaced by a novel chimeric molecule containing a new receptor binding function. The remaining glycoproteins were presumed to be retained in a functional state and capable of mediating virus infection. We selected for these studies the EPO molecule as the new binding ligand for several reasons. First, it has been sequenced for a number of species, and its receptor binding domains have been described (33, 45). Second, recent reports have also defined peptides that bind the EPO receptor with high affinity, providing the opportunity to introduce into the HSV glycoproteins smaller precise binding elements that may be less perturbing than the full-length EPO (34, 86). Third, the receptor is a single-chain cell surface molecule that has also been cloned (12) and thus is transferable to other cell types. Fourth, the distribution of the EPO receptor in animal tissues has been described and is primarily found on cells of erythroid origin (12, 68), although other tissues such as vascular endothelium (81) and brain neurons (15) are receptor positive. Fifth, the physiology and signaling pathway induced by EPO binding to its receptor are known (22). Binding of EPO to certain cell lines also delivers an essential signal for cell proliferation, affording a sensitive biological assay for EPO receptor binding (54), a feature that proved to be useful in this study, where EPO-mediated virus binding did not lead to productive infection. Finally, virus recombinants carrying EPO have been constructed for use in another virus system with some success (35), suggesting that it might be possible to make HSV glycoprotein-EPO recombinant molecules with EPO receptor binding activity. We selected gC as the EPO chimeric gene partner. Although gC has several accessory functions (e.g., binding to HS and the C3b component of complement [29, 70]), these functional domains are well characterized (23, 79), and we hypothesized that they might be deleted and replaced by EPO.
To satisfy our first objective, we engineered a series of gC-EPO chimeric molecules, one or more of which might prove stable, processed and incorporated into virus envelopes in amounts similar to the wild-type viral glycoprotein. In an attempt to conserve the secondary structure of gC, the sequences deleted included paired cysteine residues known to form disulfide bonds (66). As demonstrated by immunofluorescence (Fig. 3 and 6), immunoprecipitation (Fig. 2 and 7), and antibody neutralization (Fig. 8), both gC and EPO conformationally dependent antigenic structures were preserved. Immunoprecipitation analysis of cells transfected with pHgCEPO1 and pHgCEPO2 showed that the chimeric molecules were posttransductionally modified by ER and Golgi enzymes (Fig. 2), which resulted in transport of the recombinant glycoproteins to the cell surface (Fig. 3). The incorporation of mature gC:EPO chimeric molecules into recombinant virus envelopes was confirmed by glycoprotein immunoprecipitation from purified labeled viruses (Fig. 7).
The level of gCEPO2 incorporation was similar to that of wild-type gC, taking into account the number of radiolabeled residues per molecule. A comparative analysis of KgBpK−gCEPO2 and KgBpK−gCEPO3 demonstrated that incorporation of gCEPO3 was approximately fivefold less than that of gCEPO2, a finding supported by neutralization assay using α-EPO (Table 1). This difference might be attributed either to deletion of the external gC domain of gCEPO3, which may be required for efficient glycoprotein incorporation into virus, or to altered EPO antigenic determinants with consequent reduced antibody recognition. Nevertheless, the level of incorporation of at least two of the chimeric proteins was several orders of magnitude higher than for any previously reported non-HSV-1 protein incorporation into virus envelopes (17).
Our second objective was to demonstrate that a new binding specificity had indeed been introduced into the background of a virus mutant also deleted for the HS binding activity of gB (41). First, the absence of HS binding by KgBpK−gCEPO2 and KgBpK−gCEPO3 viruses was confirmed by similar recombinant virus titers obtained on L cells and Gag-deficient sog9 cells (Fig. 8), a conclusion also supported by binding assays using Vero cells, in which the binding capacity of the gC:EPO recombinant was similar to that of the KgBpK−gC− mutant virus. Second, the gC:EPO chimeric proteins had acquired binding activity for the EPO receptor, as shown by their recognition of EREx (Fig. 4). gCEPO1 proved to be a notable exception, probably because of its internally positioned EPO sequence. Four identifiable EPO domains have been reported to be important for EPO bioactivity (19), while two distinct receptor binding sites were identified (49). Binding of EPO to its receptor requires the collaboration of multiple EPO domains which could have been altered or masked by the gC N-terminal residues encoded by gCEPO1.
Despite binding of both gCEPO2 and gCEPO3 solubilized chimeric molecules to EREx (Fig. 4), only KgBpK−gCEPO2 recombinant virus bound immobilized EREx, a specific binding confirmed by EREx-dependent virus neutralization (48, 85) (Fig. 9 and 10). As anticipated by the results of studies showing that gCEPO1 did not bind EREx, a recombinant virus containing this recombinant gene product did not bind to immobilized EREx (unpublished finding). The neutralizing ability of EREx might have been facilitated by the dimerization of EREx known to occur following binding to EPO (56, 60), resulting in steric hindrance of virus infection, although we cannot rule out the possibility that receptor binding physically altered the envelope in a manner to prevent it from carrying out virus attachment or entry. The absence of gCEPO3 binding to EREx could be attributed to its reduced level of incorporation within the virus envelope or to a reduced accessibility to the EPO receptor. Indeed, immunoelectron microscopy studies of HSV-1 virions demonstrated that gC is present in extended spike structures (75). These 24-nm projections also contain the HS binding ligand (79), and thus replacement of 375 residues from gC with 165 EPO residues having a more compact conformation might have been sufficient to bury the molecule within the virus envelope and prevent its binding to soluble EPO receptor.
Our third objective was to demonstrate that HSV bearing a novel chimeric envelope protein could target binding to cells displaying the cognate receptor. Proliferation of FD-EPO cells is stimulated by receptor-mediated binding of EPO, providing a sensitive measure of EPO binding activity (54). Binding of the KgBpK−gCEPO2 recombinant virus to FD-EPO cells was demonstrated by virus particle stimulation of FD-EPO cell proliferation in a manner comparable to that for soluble EPO (Fig. 11). Erythroid cells infected with Friend spleen focus-forming virus are stimulated to proliferate through binding of the viral gp55 glycoprotein to cell surface EPO receptor (43), causing constitutive receptor signaling, a first stage event in the development of Friend erythroleukemia (13). A similar functional interaction between KgBpK−gCEPO2 with the EPO receptor was also observed at the cell surface since a derivative mutant of KgBpK−gCEPO2 virus deleted for the essential fusion glycoprotein, gD (KgBpK−gCEPO2gD−) was similarly mitogenic, indicating that virus attachment and not virus-mediated envelope fusion and penetration was responsible for this growth stimulation.
FD-EPO cells were not infectable by wild-type HSV-1, as demonstrated by the absence of IE gene expression following virus inoculation, which precluded an analysis of the impact of EPO receptor binding on the specificity of recombinant virus infection by the normal route. The refractory nature of these cells could be due to the lack of a suitable gD receptor or poor virus attachment. The latter possibility might have been overcome by recombinant virus binding to the EPO receptor; however, none of our recombinant viruses were able to productively infect these cells. Although this finding may favor the notion that FD-EPO cells lacked a functional gD receptor, the number of cell surface EPO receptor molecules was also low (ca. 500 to 1,000/cell [78]), and thus we were unable to demonstrate a difference in recombinant virus binding to FD-EPO cells compared with adsorption of our HS-binding-defective mutant virus. Although the recombinant virus did not productively infect FD-EPO cells, it might be expected that these particles would still enter cells by endocytosis of receptors occupied by virus-bound EPO, and indeed electron microscopy studies confirmed this prediction since EPO-deficient virus was rarely seen internalized in this manner (Fig. 12). Virus entry by inclusion into lysosomal vesicles has been reported to result in aborted virus infection (4, 7), an observation consistent with the failure of recombinant virus to initiate productive infection.
The results of our investigation clearly showed that one of our recombinant viruses, KgBpK−gCEPO2, achieved our objectives; however, we were unable to evaluate the potential utility of EPO receptor binding for targeted viral infection. Future studies are designed to engineer (i) additional EPO receptor-bearing cell lines which are permissive for HSV infection and (ii) additional recombinant viruses in which small EPO receptor-binding peptides are introduced into other viral glycoproteins to replace or compete for the normal receptor binding functions. These experiments should provide further information on methods to modify the host range of HSV-1 vectors.
ACKNOWLEDGMENTS
We thank J. C. Winkelman, K. Todokoro, and F. Tufaro for providing the EREx plasmid, FD-EPO cells, and murine L and derivative sog9 cells, respectively. We also thank S. Watkins for valuable assistance in carrying out the transmission electron microscopy experiments and N. Kasahara and Darren P. Wolfe for discussion of the data.
This work was supported by Public Health Service grant R01 CA66141-07 from the National Institutes of Health and by L’Association Française contre les Myopathies.
REFERENCES
- 1.Banfield B W, Leduc Y, Esford L, Schubert K, Tufaro F. Sequential isolation of proteoglycan synthesis mutants by using herpes simplex virus as a selective agent: evidence for a proteoglycan-independent virus entry pathway. J Virol. 1995;69:3290–3298. doi: 10.1128/jvi.69.6.3290-3298.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bartz S R, Vodicka M A. Production of high-titer human immunodeficiency virus type 1 pseudotyped with vesicular stomatitis glycoprotein. Methods. 1997;12:337–342. doi: 10.1006/meth.1997.0487. [DOI] [PubMed] [Google Scholar]
- 3.Bergelson J M, Cunningham J A, Droguett G, Kurt-Jones E A, Krithivas A, Hong J S, Horwitz M S, Crowell R L, Finberg R W. Isolation of a common receptor for coxsackie B viruses and adenoviruses 2 and 5. Science. 1997;275:1320–1323. doi: 10.1126/science.275.5304.1320. [DOI] [PubMed] [Google Scholar]
- 4.Butcher M, Raviprakash K, Ghosh H P. Acid pH-induced fusion of cells by herpes simplex virus glycoproteins gB and gD. J Biol Chem. 1990;265:5862–5868. [PubMed] [Google Scholar]
- 5.Cai W, Gu B, Person S. Role of glycoprotein B of herpes simplex virus type 1 in viral entry and cell fusion. J Virol. 1988;62:2596–2604. doi: 10.1128/jvi.62.8.2596-2604.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cai W, Person S, DebRoy C, Gu B. Functional regions and structural features of the gB glycoprotein of herpes simplex virus type 1: an analysis of linker insertion mutants. J Mol Biol. 1988;201:575–588. doi: 10.1016/0022-2836(88)90639-0. [DOI] [PubMed] [Google Scholar]
- 7.Campadelli-Fiume G, Avitabile E, Fini S, Stirpe D, Arsenakis M, Roizman R. Herpes simplex virus glycoprotein D is sufficient to induce spontaneous pH-independent fusion in a cell line that constitutively expresses the glycoprotein. Virology. 1988;166:598–602. doi: 10.1016/0042-6822(88)90533-8. [DOI] [PubMed] [Google Scholar]
- 8.Carpenter D E, Stevens J G. Long-term expression of a foreign gene from a unique position in the latent herpes simplex virus genome. Hum Gene Ther. 1996;7:1447–1454. doi: 10.1089/hum.1996.7.12-1447. [DOI] [PubMed] [Google Scholar]
- 9.Carter V C, Jennings S R, Rice P L, Tevethia S S. Mapping of a herpes simplex virus type 2-encoded function that affects the susceptibility of herpes simplex virus-infected target cells to lysis by herpes simplex virus-specific cytotoxic T lymphocytes. J Virol. 1984;49:766–771. doi: 10.1128/jvi.49.3.766-771.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cosset F L, Russell S J. Targeting retrovirus entry. Gene Ther. 1996;3:946–956. [PubMed] [Google Scholar]
- 11.Cruchley A T, Williams D M, Niedobitek G, Young L S. Epstein-Barr virus: biology and disease. Oral Dis. 1997;3:S156–S163. doi: 10.1111/j.1601-0825.1997.tb00351.x. [DOI] [PubMed] [Google Scholar]
- 12.D’Andrea A D, Lodish H F, Wong G G. Expression cloning of the murine erythropoietin receptor. Cell. 1989;57:277–285. doi: 10.1016/0092-8674(89)90965-3. [DOI] [PubMed] [Google Scholar]
- 13.D’Andrea A D, Moreau J F, Showers M O. Molecular mimicry of erythropoietin by the spleen focus-forming virus gp55 glycoprotein: the first stage of Friend virus-induced erythroleukemia. Biochim Biophys Acta. 1992;1114:31–41. doi: 10.1016/0304-419x(92)90004-i. [DOI] [PubMed] [Google Scholar]
- 14.Desai P, Homa F L, Person S, Glorioso J C. A genetic selection method for the transfer of HSV-1 glycoprotein B mutations from plasmid to the viral genome: preliminary characterization of transdominance and entry kinetics of mutant viruses. Virology. 1994;204:312–322. doi: 10.1006/viro.1994.1536. [DOI] [PubMed] [Google Scholar]
- 15.Digicaylioglu M, Bichet S, Marti H H, Wenger R H, Rovas L A, Bauer C, Gassmann M. Localization of specific erythropoietin binding site in defined area of the mouse brain. Proc Natl Acad Sci USA. 1995;92:3717–3720. doi: 10.1073/pnas.92.9.3717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dingwell K S, Brunetti C R, Hendricks R L, Tang Q, Tang M, Rainbow A J, Johnson D C. Herpes simplex virus glycoproteins E and I facilitate cell-to-cell spread in vivo and across junctions of cultured cells. J Virol. 1994;68:834–845. doi: 10.1128/jvi.68.2.834-845.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dolter K E, King S R, Holland T C. Incorporation of CD4 into virions by recombinant herpes simplex virus. J Virol. 1993;67:189–195. doi: 10.1128/jvi.67.1.189-195.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dolter K E, Goins W F, Levine M, Glorioso J C. Genetic analysis of type-specific antigenic determinants of herpes simplex virus glycoprotein C. J Virol. 1992;66:4864–4873. doi: 10.1128/jvi.66.8.4864-4873.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Elliot S, Lorenzini T, Chang D, Barzilay J, Delorme E. Mapping of the active site of recombinant human erythropoietin. Blood. 1997;89:493–502. [PubMed] [Google Scholar]
- 20.Emi N, Friedmann T, Yee J. Pseudotype formation of murine leukemia virus with the G protein of vesicular stomatitis virus. J Virol. 1991;65:1202–1207. doi: 10.1128/jvi.65.3.1202-1207.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fink D J, DeLuca N A, Goins W F, Glorioso J C. Gene transfer to neurons using herpes simplex virus-based vectors. Annu Rev Neurosci. 1996;19:265–287. doi: 10.1146/annurev.ne.19.030196.001405. [DOI] [PubMed] [Google Scholar]
- 22.Fisher J W. Erythropoietin: physiologic and pharmacologic aspects. Proc Soc Exp Biol Med. 1997;216:358–369. doi: 10.3181/00379727-216-44183. [DOI] [PubMed] [Google Scholar]
- 23.Frink R J, Eisenberg R J, Cohen G H, Wagner E K. Detailed analysis of the portion of the herpes simplex virus type 1 genome encoding glycoprotein C. J Virol. 1983;45:634–647. doi: 10.1128/jvi.45.2.634-647.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ghosh-Choudhury N, Butcher M, Reid E, Ghosh H P. Effect of tunicamycin and monensin on the transport to the cell surface and secretion of a viral membrane glycoprotein containing both N- and O-linked sugars. Biochem Cell Biol. 1994;72:20–25. doi: 10.1139/o94-004. [DOI] [PubMed] [Google Scholar]
- 25.Goins W F, Krisky D, Marconi P, Oligino T, Ramakrishnan R, Poliani P L, Fink D J, Glorioso J C. Herpes simplex virus vectors for gene transfer to the nervous system. J Neurovirol. 1997;3:S80–S88. [PubMed] [Google Scholar]
- 26.Gruenheid S, Gatzke L, Meadows H, Tufaro F. Herpes simplex virus infection and propagation in a mouse L-cell mutant lacking heparan sulfate proteoglycans. J Virol. 1993;67:93–100. doi: 10.1128/jvi.67.1.93-100.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gunzburg W H, Fleuchaus A, Saller R, Salmons B. Retroviral vector targeting for gene therapy. Cytokines Mol Ther. 1996;3:177–184. [PubMed] [Google Scholar]
- 28.Harris K W, Mitchell R A, Winkelmann J C. Ligand binding properties of the human erythropoietin receptor extracellular domain expressed in Escherichia coli. J Biol Chem. 1992;267:15205–15209. [PubMed] [Google Scholar]
- 29.Herold B C, WuDunn D, Soltys N, Spear P G. Glycoprotein C of herpes simplex virus type 1 plays a principal role in the adsorption of virus to cells and in infectivity. J Virol. 1991;65:1090–1098. doi: 10.1128/jvi.65.3.1090-1098.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Herold B C, Visalli R J, Sumarski N, Brandt C R, Spear P G. Glycoprotein C-independent binding of herpes simplex virus to cells requires cell surface heparan sulfate and glycoprotein B. J Gen Virol. 1994;75:1211–1222. doi: 10.1099/0022-1317-75-6-1211. [DOI] [PubMed] [Google Scholar]
- 31.Higlander S L, Sutherland S L, Gage P J, Johnson D C, Levine M, Glorioso J C. Neutralizing monoclonal antibodies specific for herpes simplex virus glycoprotein D inhibit virus penetration. J Virol. 1987;61:3356–3364. doi: 10.1128/jvi.61.11.3356-3364.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Holland T C, Homa F L, Marlin S D, Levine M, Glorioso J C. Herpes simplex virus type 1 glycoprotein C-negative mutants exhibit multiple phenotypes, including secretion of truncated glycoproteins. J Virol. 1984;52:566–574. doi: 10.1128/jvi.52.2.566-574.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Horton M A, Larson K A, Lee J J, Lee N A. Cloning of the murine eosinophil peroxidase gene (mEPO): characterization of a conserved subgroup of mammalian hematopoietic peroxidases. J Leukoc Biol. 1996;60:285–294. doi: 10.1002/jlb.60.2.285. [DOI] [PubMed] [Google Scholar]
- 34.Johnson D L, Farrell F X, Barbone F P, McMahon F J, Tullai J, Hoey K, Livnah O, Wrighton N C, Middleton S A, Loughney D A, Stura E A, Dower W J, Mulcahy L S, Wilson I A, Jolliffe L K. Identification of a 13 amino acid peptide mimetic of erythropoietin and description of amino acids critical for the mimetic activity of EMP1. Biochemistry. 1998;37:3699–3710. doi: 10.1021/bi971956y. [DOI] [PubMed] [Google Scholar]
- 35.Kasahara N, Dozy A M, Kan Y W. Tissue-specific targeting of retroviral vectors through ligand-receptor interactions. Science. 1994;266:1373–1376. doi: 10.1126/science.7973726. [DOI] [PubMed] [Google Scholar]
- 36.Kovesdi I, Brough D E, Bruder J T, Wickham T J. Adenovirus vector for gene transfer. Curr Opin Biotechnol. 1997;8:583–589. doi: 10.1016/s0958-1669(97)80033-x. [DOI] [PubMed] [Google Scholar]
- 37.Koyama A H, Adachi A. Induction of apoptosis by herpes simplex virus type 1. J Gen Virol. 1997;78:2909–2912. doi: 10.1099/0022-1317-78-11-2909. [DOI] [PubMed] [Google Scholar]
- 38.Krisky D M, Marconi P C, Oligino T, Rouse R J, Fink D J, Glorioso J C. Rapid method for construction of recombinant HSV gene transfer vectors. Gene Ther. 1997;4:1120–1125. doi: 10.1038/sj.gt.3300497. [DOI] [PubMed] [Google Scholar]
- 39.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature (London) 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 40.Laquerre S, Anderson D B, Argnani R, Glorioso J C. Herpes simplex virus type 1 glycoprotein B requires a cysteine residue at position 633 for folding, processing and incorporation into mature infectious virus particles. J Virol. 1998;72:4940–4949. doi: 10.1128/jvi.72.6.4940-4949.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Laquerre S, Argnani R, Anderson D B, Zucchini S, Manservigi R, Glorioso J C. Heparan sulfate proteoglycan binding by herpes simplex virus type 1 glycoproteins B and C differ in their contribution to virus attachment, penetration and cell-to-cell spread. J Virol. 1998;72:6119–6130. doi: 10.1128/jvi.72.7.6119-6130.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Laquerre, S., W. F. Goins, S. Moriuchi, T. J. Oligino, D. M. Krisky, P. Marconi, M. K. Soares, J. B. Cohen, D. J. Fink, and J. C. Glorioso. Gene transfer tool: herpes simplex virus vector. In The development of human gene therapy, in press. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 43.Li J P, Hu H O, Niu Q T, Fang C. Cell surface activation of the erythropoietin receptor by Friend spleen focus-forming virus gp55. J Virol. 1995;69:1714–1719. doi: 10.1128/jvi.69.3.1714-1719.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ligas M W, Johnson D C. A herpes simplex virus mutant in which glycoprotein D sequences are replaced by β-galactosidase sequences binds to but is unable to penetrate into cells. J Virol. 1988;62:1486–1494. doi: 10.1128/jvi.62.5.1486-1494.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lin F-K, Suggs S, Lin C-H, Browne J K, Smalling R, Egrie J C, Chen K K, Fox G M, Marlin F, Wasser Z. Cloning and expression of the human erythropoietin gene. Proc Natl Acad Sci USA. 1985;82:7850–7854. doi: 10.1073/pnas.82.22.7580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Marconi P, Krisky D, Oligino T, Poliani P L, Ramakrishnan R, Goins W F, Fink D J, Glorioso J C. Replication-defective herpes simplex virus vectors for gene transfer in vivo. Proc Natl Acad Sci USA. 1996;93:11319–11320. doi: 10.1073/pnas.93.21.11319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Marin M, Noel D, Valsesia-Wittman S, Brockly F, Etienne-Julan M, Russell S, Cosset F L, Piechaczyk J. Targeted infection of human cells via major histocompatibility complex class I molecules by Moloney murine leukemia virus-derived viruses displaying single-chain antibody fragment-envelope fusion proteins. Virology. 1996;70:2957–2962. doi: 10.1128/jvi.70.5.2957-2962.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Marlin S D, Holland T C, Levine M, Glorioso J C. Epitopes of herpes simplex virus type 1 glycoprotein gC are clustered in two distinct antigenic sites. J Virol. 1985;53:128–136. doi: 10.1128/jvi.53.1.128-136.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Matthews D J, Topping R S, Cass R T, Giebel L B. A sequential dimerization mechanism for erythropoietin receptor activation. Proc Natl Acad Sci USA. 1996;93:9471–9476. doi: 10.1073/pnas.93.18.9471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.McClain D S, Fuller A O. Cell-specific kinetics and efficiency of herpes simplex virus type 1 entry are determined by two distinct phases of attachment. Virology. 1994;198:690–702. doi: 10.1006/viro.1994.1081. [DOI] [PubMed] [Google Scholar]
- 51.Meignier B, Jourdier T M, Norrild B, Pereira L, Roizman B. Immunization of experimental animals with reconstituted glycoprotein mixtures of herpes simplex virus 1 and 2: protection against challenge with virulent virus. J Infect Dis. 1987;155:921–930. doi: 10.1093/infdis/155.5.921. [DOI] [PubMed] [Google Scholar]
- 52.Mettenleiter T C, Spear P G. Glycoprotein gB (gII) of pseudorabies virus can functionally substitute for glycoprotein gB in herpes simplex virus type 1. J Virol. 1994;68:500–504. doi: 10.1128/jvi.68.1.500-504.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Montgomery R I, Warner M S, Lum B J, Spear P G. Herpes simplex virus-1 entry into cells mediated by a novel member of the TNF/NGF receptor family. Cell. 1996;87:427–436. doi: 10.1016/s0092-8674(00)81363-x. [DOI] [PubMed] [Google Scholar]
- 54.Nagata Y, Nishida E, Todokoro K. Activation of JNK signaling pathway by erythropoietin, thrombopoietin and interleukin-3. Blood. 1997;89:2664–2669. [PubMed] [Google Scholar]
- 55.Naldini L, Blomer U, Gallay P, Mulligan R, Gage F H, Verma I M, Trono D. In vivo gene delivery and stable transduction of nondividing cells by a lentiviral vector. Science. 1996;272:263–267. doi: 10.1126/science.272.5259.263. [DOI] [PubMed] [Google Scholar]
- 56.Narhi L O, Aoki K H, Philo J S, Arakawa T. Changes in conformation and stability upon formation of complexes of erythropoietin (EPO) and soluble EPO receptor. J Protein Chem. 1997;16:213–225. doi: 10.1023/a:1026330909461. [DOI] [PubMed] [Google Scholar]
- 57.Neda H, Wu C H, Wu G Y. Chemical modification of an ecotropic murine leukemia virus results in redirection of its target cell specificity. J Biol Chem. 1991;266:14143–14146. [PubMed] [Google Scholar]
- 58.Ohno K, Sawai K, Iijima Y, Levin B, Meruelo D. Cell-specific targeting of Sindbis virus vectors displaying IgG-binding domains of protein A. Nat Biotechnol. 1997;15:763–767. doi: 10.1038/nbt0897-763. [DOI] [PubMed] [Google Scholar]
- 59.Ory D S, Neugeboren B A, Mulligan R C. A stable human-derived packaging cell line for production of high titer retrovirus/vesicular stomatitis virus G pseudotypes. Proc Natl Acad Sci USA. 1996;93:11400–11406. doi: 10.1073/pnas.93.21.11400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Philo J S, Aoki K H, Arakawa T, Narhi L O, Wen J. Dimerization of the extracellular domain of the erythropoietin (EPO) receptor by EPO: one high-affinity and one low-affinity interaction. Biochemistry. 1996;38:1681–1691. doi: 10.1021/bi9524272. [DOI] [PubMed] [Google Scholar]
- 61.Rogers B E, Douglas J T, Ahlem C, Buchsbaum D J, Frincke J, Curiel D T. Use of a novel cross-linking method to modify adenovirus tropism. Gene Ther. 1997;4:1387–1392. doi: 10.1038/sj.gt.3300541. [DOI] [PubMed] [Google Scholar]
- 62.Roizman B, Sears A E. Herpes simplex viruses and their replication. In: Fields B N, Knipe D M, Howley P M, et al., editors. Fields virology. 3rd ed. Philadelphia, Pa: Lippincott-Raven Publishers; 1996. pp. 2231–2295. [Google Scholar]
- 63.Roop C, Hutchinson L, Johnson D C. A mutant herpes simplex virus type 1 unable to express glycoprotein L cannot enter cells, and its particles lack glycoprotein H. J Virol. 1993;67:2285–2297. doi: 10.1128/jvi.67.4.2285-2297.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Roux P, Jeanteur P, Piechaczyk M. A versatile and potentially general approach to the targeting of specific cell types by retroviruses: application to the infection of human cells by means of major histocompatibility complex class I and class II antigens by mouse ecotropic murine leukemia virus-derived viruses. Proc Natl Acad Sci USA. 1989;86:9079–9083. doi: 10.1073/pnas.86.23.9079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Russell S J, Hawkins R E, Winter G. Retroviral vectors displaying functional antibody fragments. Nucleic Acids Res. 1993;21:1081–1085. doi: 10.1093/nar/21.5.1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Rux A H, More W T, Lambris J D, Abrams W R, Peng C, Friedman H M, Cohen G H, Eisenberg R J. Disulfide bond structure determination and biochemical analysis of glycoprotein C from herpes simplex virus. J Virol. 1996;70:5455–5465. doi: 10.1128/jvi.70.8.5455-5465.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Samaniego L A, Neiderhiser L, DeLuca N A. Persistence and expression of the herpes simplex virus genome in the absence of immediate-early proteins. J Virol. 1998;72:3307–3320. doi: 10.1128/jvi.72.4.3307-3320.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Sawyer S T. Receptors for erythropoietin. Distribution, structure and role in receptor-mediated endocytosis in erythroid cells. In: Harris M Jr, editor. Blood cell biochemistry. Vol. 1. New York, N.Y: Plenum Press; 1990. pp. 365–402. [Google Scholar]
- 69.Sears A E, McGwire B S, Roizman B. Infection of polarized MDCK cells with herpes simplex virus 1: two asymmetrically distributed cell receptors interact with different viral proteins. Proc Natl Acad Sci USA. 1991;88:5087–5091. doi: 10.1073/pnas.88.12.5087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Seidel-Dugan C, Ponce de Leon M, Friedman H M, Fries L F, Frank M M, Cohen G H, Eisenberg R J. C3b receptor activity on transfected cells expression glycoprotein C of herpes simplex virus types 1 and 2. J Virol. 1988;62:4027–4036. doi: 10.1128/jvi.62.11.4027-4036.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Shieh M-T, WuDunn D, Montgomery R I, Esko J D, Spear P G. Cell surface receptors for herpes simplex virus are heparan sulfate proteoglycans. J Cell Biol. 1992;116:1273–1281. doi: 10.1083/jcb.116.5.1273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Sochanik A, Szala S. On the strategy of using nonviral carriers in cancer gene therapy. Acta Biochim Pol. 1996;43:293–300. [PubMed] [Google Scholar]
- 73.Somia N V, Zoppe M, Verma I M. Generation of targeted retroviral vectors by using single-chain variable fragment: an approach to in vivo gene delivery. Proc Natl Acad Sci USA. 1995;92:7570–7574. doi: 10.1073/pnas.92.16.7570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Spear P G. Entry of alphaherpesviruses into cells. Semin Virol. 1993;4:167–180. [Google Scholar]
- 75.Stannard L M, Fuller A O, Spear P G. Herpes simplex virus glycoproteins associated with different morphological entities projecting from the virion envelope. J Gen Virol. 1987;68:715–725. doi: 10.1099/0022-1317-68-3-715. [DOI] [PubMed] [Google Scholar]
- 76.Steiner I, Kennedy P G J. Herpes simplex virus latent infection in the nervous system. Neurovirology. 1995;1:19–29. doi: 10.3109/13550289509111007. [DOI] [PubMed] [Google Scholar]
- 77.Stevenson M. Molecular mechanisms for the regulation of HIV replication, persistence and latency. AIDS. 1997;11:S25–S33. [PubMed] [Google Scholar]
- 78.Todokoro, K. Personal communication.
- 79.Trybala E, Berghtröm T, Svennerholm B, Jeansson S, Glorioso J C, Olofsson S. Localization of the functional site on herpes simplex virus type 1 glycoprotein C involved in binding to cell surface heparan sulfate. J Gen Virol. 1994;75:743–752. doi: 10.1099/0022-1317-75-4-743. [DOI] [PubMed] [Google Scholar]
- 80.Vile R G. Gene therapy. Curr Biol. 1998;8:R73–R75. doi: 10.1016/s0960-9822(98)70049-1. [DOI] [PubMed] [Google Scholar]
- 81.Vogel V, Kramer H J, Backer A, Meyer-Lehnert H, Jelkmann W, Fandrey J. Effects of erythropoietin on endothelin-1 synthesis and the cellular calcium messenger system in vascular endothelial cells. Am J Hypertens. 1997;10(3):289–296. doi: 10.1016/s0895-7061(96)00410-4. [DOI] [PubMed] [Google Scholar]
- 82.Whealy M E, Robbins A K, Enquist L W. Replacement of the pseudorabies virus glycoprotein gIII gene with its postulated homolog, the glycoprotein gC gene of herpes simplex virus type 1. J Virol. 1989;63:4055–4059. doi: 10.1128/jvi.63.9.4055-4059.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Wickham T J, Carrion M E, Kovesdi I. Targeting of adenovirus penton base to new receptors through replacement of its RGD motif with other receptor-specific peptide motifs. Gene Ther. 1995;2:750–756. [PubMed] [Google Scholar]
- 84.Wickham T J, Roelvink P W, Brough D E, Kovesdi I. Adenovirus targeted to heparan containing receptors increases its gene delivery efficiency to multiple cell types. Nat Biotechnol. 1996;14:1570–1573. doi: 10.1038/nbt1196-1570. [DOI] [PubMed] [Google Scholar]
- 85.Wickham T J, Tzeng E, Shears II L L, Roelvink P W, Li Y, Lee G M, Brough D E, Lizonova A, Kovesdi I. Increased in vivo gene transfer by adenovirus vector containing chimeric fiber proteins. J Virol. 1997;71:8221–8229. doi: 10.1128/jvi.71.11.8221-8229.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Wrighton N C, Farrell F X, Chang R, Kashyap A K, Barbone F P, Mulcahy L S, Johnson D L, Barrett R W, Jolliffe L K, Dower W J. Small peptides as potent mimetics of the protein hormone erythropoietin. Science. 1996;273:458–464. doi: 10.1126/science.273.5274.458. [DOI] [PubMed] [Google Scholar]
- 87.Wu C-T B, Levin M, Homa F, Highlander S L, Glorioso J C. Characterization of the antigenic structure of herpes simplex virus type 1 glycoprotein C through DNA sequence analysis of monoclonal antibody-resistant mutants. J Virol. 1990;64:856–863. doi: 10.1128/jvi.64.2.856-863.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Wu N, Watkins S C, Schaffer P A, DeLuca N A. Prolonged gene expression and cell survival after infection by a herpes simplex virus mutant defective in the immediate-early genes encoding ICP4, ICP27, and ICP22. J Virol. 1996;70:6358–6368. doi: 10.1128/jvi.70.9.6358-6369.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.WuDunn D, Spear P G. Initial interaction of herpes simplex virus with cells is binding to heparan sulfate. J Virol. 1989;63:52–58. doi: 10.1128/jvi.63.1.52-58.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Yang Y, Vanin E F, Whitt M A, Fornerod M, Zwart R, Schneiderman R D, Grosveld G, Nienhuis A W. Inducible, high-level production of infectious murine leukemia retroviral vector particles pseudotyped with vesicular stomatitis virus G envelope protein. Hum Gene Ther. 1995;6:1203–1213. doi: 10.1089/hum.1995.6.9-1203. [DOI] [PubMed] [Google Scholar]
- 91.Yee J K, Friedmann T, Burns J C. Generation of high-titer pseudotyped retroviral vectors with very broad host range. Methods Cell Biol. 1994;43:99–112. doi: 10.1016/s0091-679x(08)60600-7. [DOI] [PubMed] [Google Scholar]