Abstract
Prior to the recent discovery of the swine hepatitis E virus (swine HEV) in pigs from the midwestern United States, HEV was not considered endemic to this country. Since swine HEV is antigenically and genetically related to human strains of HEV, it was important to characterize this new virus further. The infectivity titer of a pool of swine HEV in pigs was determined in order to prepare a standardized reagent and to evaluate the dose response in pigs. Although the sequence of swine HEV varied extensively from those of most human strains of HEV, it was very closely related to the two strains of human HEV (US-1 and US-2) isolated in the United States. The U.S. strains which were recently recovered from two patients with clinical hepatitis E in the United States shared ≥97% amino acid identity with swine HEV in open reading frames 1 and 2. Phylogenetic analyses of different regions of the genome revealed that swine HEV and the U.S. strains grouped together and formed a distinct branch. These results suggested that swine HEV may infect humans. When we inoculated rhesus monkeys and a chimpanzee, experimental surrogates of humans, with swine HEV, the primates became infected. Furthermore, in a reciprocal experiment, specific-pathogen-free pigs were experimentally infected with the US-2 strain of human HEV that is genetically similar to swine HEV. These results provided experimental evidence for cross-species infection by the swine virus. Thus, humans appear to be at risk of infection with swine HEV or closely related viruses.
Hepatitis E virus (HEV), the causative agent of hepatitis E, is currently unclassified (29), although it was once considered to be a member of the family Caliciviridae (19, 30). In most subtropical and tropical developing countries of Asia and Africa, HEV is the primary cause of enterically transmitted non-A, non-B hepatitis, and, hence, hepatitis E is considered an important public health problem (1, 30, 33). The disease generally affects young adults and reportedly has a mortality rate of up to 20% in infected pregnant women (1, 2, 15, 30). Hepatitis E is rarely diagnosed in industrialized countries, although HEV antibodies (anti-HEV) have been found in a significant proportion of healthy individuals (17, 22, 27, 32, 37, 46).
HEV is transmitted primarily by the fecal-oral route, and water-borne epidemics are characteristic of hepatitis E (30, 33). Strains within large geographical regions tend to be closely related to each other (6, 30, 43) and distinct from those in distant geographical areas. Recently, we reported the discovery of HEV in pigs in the United States (23). Swine HEV is antigenically and genetically related to, but distinct from, most human strains of HEV (23). However, two cases of acute clinical hepatitis E recently reported in the United States (34) were caused by virus strains very closely related to the swine HEV. One of the cases (US-1) involved a patient who had no epidemiological evidence of exposure to HEV strains from endemic countries (20, 34). The patient in the second case (US-2) had traveled to Mexico prior to the diagnosis of the disease (10, 34).
In the present study, we extended genetic comparisons to confirm that swine HEV recovered from pigs in the midwestern United States and the U.S. strains of human HEV which caused hepatitis E were closely related. More importantly, we showed, under experimental conditions, that swine HEV can cross species barriers and infect nonhuman primates and that the US-2 strain of human HEV can infect specific-pathogen-free (SPF) pigs.
MATERIALS AND METHODS
Sources of HEV.
Crossbred SPF pigs (Sus scrofa domesticus) were used throughout this study (24). SPF pigs were experimentally infected with acute-phase serum from pigs naturally infected with swine HEV (23, 24). Fecal materials collected from one of the experimentally infected SPF pigs were used as the source of swine HEV for most of the subsequent experiments.
The source of the US-2 strain of human HEV was fecal materials from an experimentally infected macaque (10). The US-2 strain was originally recovered from the serum of a U.S. resident diagnosed with acute hepatitis in Memphis, Tenn. (34).
Generation of a standard pool of swine HEV.
Two SPF pigs, 4 weeks of age, were housed in the same room in a BL-2 facility. Blood and fecal samples were collected once prior to inoculation. The two pigs were each inoculated intravenously (i.v.) with 1 ml of eluate from a rectal swab collected from pig A1 in a previous study (24). Blood samples and fecal samples were collected weekly and daily, respectively. Bile was collected on days 13, 14, 17, and 19 postinoculation (p.i.) by an ultrasound-guided technique. Liver biopsies were obtained on days 13, 17, and 19 p.i. Serum, bile, and fecal samples were tested for HEV RNA by reverse transcription-polymerase chain reaction (RT-PCR) (see below). The titer of swine HEV genomes in fecal samples was determined by a sensitive RT-PCR as described previously (24). One genome equivalent (GE) is defined as the number of viral genomes present in the highest 10-fold dilution positive by RT-PCR. A standard infectious pool of swine HEV was prepared in phosphate-buffered saline (PBS) as a 10% suspension from feces of a pig collected at 14 days p.i.
Infectivity titration of swine HEV in SPF pigs.
Ten-fold dilutions of the standard pool of swine HEV were prepared in PBS buffer. Ten SPF pigs, 4 weeks of age, were housed in separate rooms in a BL-2 facility. Groups of two pigs each were inoculated i.v. with 1 ml of a 10−1, 10−4, 10−5, 10−6, or 10−7 dilution of the standard pool. Blood and fecal samples were collected twice before and weekly after inoculation. Serum and fecal samples were tested for viral RNA by a nested RT-PCR (24). The PCR titer of viral genomes in positive fecal samples was determined. Anti-HEV (immunoglobulin G [IgG] and IgM) was assayed by an enzyme-linked immunosorbent assay (ELISA) standardized for anti-HEV in swine (23, 24). For the two pigs inoculated with a 10−1 dilution, levels of liver enzymes in serum were tested weekly by standard methods, and liver biopsies were processed for routine histological examination. The liver biopsy slides were examined under code by a veterinary pathologist with experience in evaluating swine tissues (P.G.H.). The animals were monitored for up to 16 weeks p.i.
Experimental inoculation of nonhuman primates with swine HEV.
Two rhesus monkeys (Macaca mulatta), housed separately, were each inoculated by the i.v. route with 1 ml of the standard pool of swine HEV containing 104.5 50% pig infectious doses (PID50). Weekly serum samples were tested for viral RNA by nested RT-PCR, for anti-HEV by an ELISA, and for levels of liver enzymes in serum by standard methods. Weekly fecal samples were tested for viral RNA by a nested RT-PCR, and the titer of viral genomes in selected fecal samples was determined by a nested RT-PCR (reference 24; see below). Weekly liver biopsies were processed for routine histological examination. The liver biopsy slides were examined under code by a pathologist (S.G.) with experience in evaluating primate tissues. The animals were monitored for up to 16 weeks.
The standard pool of swine HEV prepared from swine feces appeared to be toxic to nonhuman primates (data not shown). Therefore, a first primate passage 10% fecal suspension containing about 106 GEs/ml of swine HEV was prepared from one of the infected rhesus monkeys (H400) in order to evaluate the susceptibility of chimpanzees (Pan troglodytes) to swine HEV infection. A chimpanzee (chimp 5835) was inoculated by the i.v. route with 1 ml of this suspension. Weekly fecal and serum samples were tested for viral RNA by a nested RT-PCR. Weekly serum samples were tested by an ELISA for anti-HEV and by standard methods for levels of liver enzymes in serum. The baseline liver enzymes of chimp 5835 were 30 U of alanine aminotransferase (ALT) and 248 U of isocitrate dehydrogenase (ICD) (an average of four preinoculation samples).
Experimental inoculation of pigs with the US-2 strain of human HEV.
Four SPF pigs, ≥4 weeks old, were housed as pairs in two rooms in a BL-3 facility. One pig in each pair was inoculated i.v. with 0.2 ml of the 10% simian stool suspension containing HEV strain US-2. The uninoculated pig in each pair served as a contact control. Blood samples from all four pigs were taken prior to inoculation and weekly after inoculation. Fecal samples were collected directly from the rectum twice prior to inoculation and weekly after inoculation. Samples were tested for anti-HEV, viral RNA, and levels of liver enzymes in serum as described above.
Serological analyses.
ELISAs for anti-HEV in swine and in nonhuman primates were standardized as previously described (23, 39). A high-performance liquid chromatography-purified, 55-kDa truncated form of the putative capsid protein, expressed from a recombinant baculovirus containing open reading frame 2 (ORF2) of a human strain of HEV (Sar-55), was used as the antigen for the ELISAs (38, 39). Sera taken from experimentally inoculated pigs or from primates were tested in duplicate for anti-HEV IgG and IgM. Levels of the liver enzymes alkaline phosphatase, sorbitol dehydrogenase, ALT, and gamma glutamyltransferase and bilirubin in serum were determined weekly for swine sera, and ICD, ALT, and gamma glutamyltransferase for primate sera were determined weekly by standard methods.
RNA extraction and RT-PCR.
Total RNA was extracted with TriZol reagent (GIBCO-BRL, Gaithersburg, Md.) from 100 μl of fecal suspension or serum. Total RNA was reverse transcribed with swine HEV-specific primers or HEV degenerate primers and SuperScript II reverse transcriptase (GIBCO-BRL) at 42°C for 1 h; cDNA was amplified by PCR with AmpliTaq Gold polymerase (Perkin-Elmer, Norwalk, Conn.). For titration of swine HEV viral genomes in serum, bile, and fecal samples, the PCRs consisted of 39 cycles of denaturation at 94°C for 1 min, annealing at 52°C for 1 min, and extension at 72°C for 1.5 min, followed by a nested PCR of 39 cycles with 10 μl of the first-round PCR product. The primers used for PCR titration of swine HEV were swine HEV specific (Table 1).
TABLE 1.
Degenerate or consensus primers used for genomic extension of swine HEV and for the detection and titration of the US-2 and swine HEV strains
| PCR fragment | External-set primersa | Nested-set primersa |
|---|---|---|
| A32-33 | F12-33 (5′-atatgtggtcgatgccatggag-3′) | F20-41 (5′-tcgatgccatggaggcccatca-3′) |
| R247-268 (5′-ctcggcagtactgttccagttc-3′) | R220-241 (5′-gtattgcccgctggataggatg-3′) | |
| 3487-88 | F49-70 (5′-aaggctcc[a]tggcatcactactg-3′) | F57-78 (5′-tggcatcactactgc[t]tattgag-3′) |
| R1102-1123 (5′-cagaggca[g]ttccagccttcatt-3′) | R1021-1042 (5′-gggagcagcaaaaggct[c]tggtc-3′) | |
| F6-F5 | F955-982 (5′-tctacatttcatgctgtcccggttcata-3′) | F1018-1037 (5′-gatgaccaagccttttgctg-3′) |
| R1447-1466 (5′-tcctgaccaagccacttcat-3′) | R1273-1292 (5′-taatcacggccggacttctc-3′) | |
| BRL3-4 | F1168-1193 (5′-tgccatcagcgttatcttcgcaccca-3′) | F1189-1214 (5′-acccaggcgatatccaagggcatgcg-3′) |
| R1189-1214 (5′-gcacggccaacatctgtggcagcatc-3′) | R2851-2876 (5′-gcatcgatctctaacgctaggttggc-3′) | |
| G4G8 | F2599-2618 (5′-tata[c]gg[a]ttggaacataaccc-3′) | F2722-2741 (5′-ttt[c]gac[t]gcctgggagcggaa-3′) |
| R3268-3287 (5′-cggtgtgtaacgtgccacca-3′) | R3196-3215 (5′-aaatcaatggcagggatctg-3′) | |
| 3506-7 | F3095-3116 (5′-ggcgc[t]c[a]gggttgtcattgatga-3′) | F3182-3203 (5′-ttggc[t]gacccgaat[c]cagatccc-3′) |
| R4295-4316 (5′-gggagtagggccagtatttctt-3′) | R4276-4297 (5′-ctttttcaatggcacggaacca-3′) | |
| 3154-77 | F4084-4106 (5′-gaggcc[g]atggtc(g)gagaagggcca-3′) | F4162-4184 (5′-accttc[t]ttccagaaa[g]gattgtaa-3′) |
| R5157-5178 (5′-aagagcaacaacagaacagccc-3′) | R5133-5154 (5′-ctagggcgcatggtgatcccat-3′) | |
| 3479-77 | F4563-4584 (5′-ctggaagaaa[g]cac[t]tct[c]ggtgag-3′) | F4600-4621 (5′-tggaatact[g]gtc[g]tggaac[t]atgg-3′) |
| R5157-5178 (5′-aagagcaacaacagaacagccc-3′) | R5133-5154 (5′-ctagggcgcatggtgatcccat-3′) | |
| 3301-2 | F6532-6553 (5′-ctcagttcttcgcgccaatgat-3′) | |
| R7180-7201 (5′-tttttttcagggagcgcggg[a]ac-3′) | ||
| 3331-2b | F5503-5526 (5′-agctcctgtacctgatgttgactc-3′) | F5572-5595 (5′-gctcacgtcatctgtcgctgctgg-3′) |
| R5907-5930 (5′-ctacagagcgccagccttgattgc-3′) | R5837-5860 (5′-gggctgaaccaaaatcctgacatc-3′) | |
| US1c-dc | F5503-5526 (5′-agctcctgtacctgatgttgactc-3′) | F5572-5595 (5′-gcttacatcatctgttgcttctgg-3′) |
| R5907-5930 (5′-caacagagcgccagccttggttgc-3′) | R5837-5860 (5′-gggctggaccaagatcctgacatc-3′) |
Degenerate bases are shown in brackets. Primer positions, relative to the Burmese strain (36), are indicated (F, forward primer; R, reverse primer).
PCR detection and titration of swine HEV; primers are based on the ORF2 sequence of swine HEV (23).
PCR detection and titration of the US-2 strain of HEV; primers are based on the published ORF2 sequence of the US-1 strain (34).
For detection of viral RNA in pigs infected with the US-2 strain of human HEV, a nested set of primers (Table 1) based on the published ORF2 sequence of the US-1 strain (34) was used. Reagent preparation, RNA extraction, cDNA synthesis, and PCR assembly were all performed in a laboratory separate from the one in which the PCR products were analyzed in order to avoid cross-contamination.
Amplification of nearly the full-length genome of swine HEV.
The sequence of ORF2 and ORF3 of swine HEV was previously determined (23). In order to extend this sequence to ORF1 and the terminal noncoding regions (NCRs), we utilized the genome-walking strategy. The complete ORF1 and the 5′ and 3′ NCRs of swine HEV were amplified by RT-PCR by using one swine HEV-specific primer and one HEV degenerate or consensus primer (Table 1). The PCR conditions used to amplify different regions of the genome varied.
Sequence and phylogenetic analysis.
The PCR products were purified by electrophoresis on a 1% agarose gel, followed by extraction with a Geneclean kit (Bio101, La Jolla, Calif.). Both strands were sequenced with an automated DNA Sequencer. The sequences were compiled and analyzed by the GeneWorks program (IntelliGenetics, Inc., Mountain View, Calif.). Sequence alignments were performed with the Clustal W program in the Genetics Computer Group (GCG) package. Phylogenetic analyses were conducted with the aid of the PAUP program in the same package (GCG version 9.1; David L. Swofford, Smithsonian Institute, Washington, D.C.). A bootstrap analysis with 1,000 replications was performed by branch-and-bound searching and midpoint rooting options to produce a 75% majority-rule consensus tree. The resulting phylograms were viewed with TREEVIEW program (26) to create the graphical outputs.
Nucleotide sequence accession number.
The genomic sequence of swine HEV reported in this paper has been deposited with the GenBank database under accession no. AF082843.
RESULTS
Infectivity titration of swine HEV in SPF pigs.
Fecal materials collected from an experimentally infected pig (24) were inoculated i.v. into two additional SPF pigs for amplification. The two inoculated pigs seroconverted to anti-HEV at 2 weeks p.i. Swine HEV RNA was first detected in feces at 3 or 4 days p.i. The standard infectious pool of swine HEV prepared from feces of one pig at 14 days p.i. had a PCR titer of 106 GEs/ml. Swine HEV RNA was also detected in bile in three of the four samples collected (days 13, 14, and 17, but not day 19 p.i.). Since the bile is produced by liver cells, the virus had to have replicated in the liver prior to accumulating in the bile of the gallbladder. One of the pigs had microscopic changes consistent with hepatitis on day 13 p.i., but liver lesions were not observed in the other two biopsy samples (days 17 and 19 p.i.).
The pool of swine HEV was characterized by titration in SPF pigs. Both pigs inoculated with dilutions of 10−1 and 10−4, respectively, of the virus pool seroconverted to anti-HEV and shed virus in feces (Table 2). None of the animals inoculated with dilutions of 10−5, 10−6, or 10−7 were infected. Therefore, the infectivity titer of the swine HEV standard pool was 104.5 PID50 per ml, which was approximately 30-fold lower than the GE titer measured by RT-PCR. In the two pigs each inoculated with the highest dose (104.5 PID50), seroconversion to anti-HEV occurred 3 weeks p.i., and fecal excretion of virus was first detected 1 week p.i. and lasted for 2 to 3 weeks. Mild-to-moderate diffuse hepatocellular swelling and vacuolation and mild increased numbers of mixed leukocytes within hepatic sinusoids were observed from 1 to 4 weeks p.i., but hepatic necrosis was not detected. Levels of liver enzymes in serum were not significantly elevated in either pig, and clinical signs of hepatitis were not observed. The pigs inoculated with the lowest infectious dose of swine HEV (100.5 PID50) took longer to seroconvert and to excrete virus in feces. In addition, the titer of viral genomes in the feces of pigs inoculated with 100.5 PID50 was lower than that of pigs inoculated with 104.5 PID50.
TABLE 2.
Infectivity titration of the swine HEV in pigs
| Pig | Dilution of virus stock | Wk p.i. at which fecal viral RNA was detected | Log10 peak genome titera | Liver enzyme level | Wk p.i. at which liver lesions detectedc | Wk p.i. at which anti-HEV IgG was first detected |
|---|---|---|---|---|---|---|
| 1 | 10−1 | 1–3 | 7 (2) | 0b | 1–4 (±)c | 3 |
| 2 | 10−1 | 1–2 | 6 (1) | 0 | 1–4 (±) | 3 |
| 3 | 10−4 | 2 | 3 (2) | NDd | ND | >8e |
| 4 | 10−4 | 2–4 | 5 (3) | ND | ND | 5 |
| 5–10 | 10−5 to 10−7 | —f | NAg | ND | ND | — |
GE/gram of feces (week at which peak viral genome titer was detected).
0, normal values throughout the study.
±, mild to moderate diffuse hepatocellular swelling and vacuolation; mild increased numbers of mixed leukocytes within hepatic sinusoids; no hepatic necrosis.
ND, not done.
This pig was seronegative at 8 weeks postinoculation but was seropositive when next tested at the end of the 16-week experiment.
—, RNA not detected.
NA, not applicable.
Sequence analysis of ORF1 and terminal NCRs of swine HEV.
The ORF1 of swine HEV has been very difficult to amplify, and its sequence has not been previously reported. The ORF1 of swine HEV was sequenced, and the putative functional domains and the hypervariable region (HVR) in the ORF1 were compared with the corresponding regions of other HEV strains (Table 3). The ORF1 of swine HEV and of the US-2 strain contains 5,127 nucleotides (nt), which is 3 nt less than those in the US-1 strain, but 45 and 51 nt more than those in the Asian and the Mexican strains, respectively (data not shown). Swine HEV varied extensively, both at the nucleotide and amino acid levels, from non-U.S. strains of HEV, although it was very similar to the two U.S. strains (see below). The sequence identity in the putative methyltransferase and RNA-dependent RNA polymerase (RDRP) regions between swine HEV and non-U.S. HEV strains varied from 74 to 76% at the nucleotide level and from 84 to 89% at the amino acid level. The GDD tripeptide motif found in all viral RDRP regions is conserved among different strains (data not shown). In the putative helicase region, a slightly higher sequence identity between swine HEV and non-U.S. HEV strains was observed, i.e., 74 to 77% at the nucleotide level and 91 to 92% at the amino acid level. Asian strains of HEV, including Burma (36), Myanmar (3), Pakistan (38), China (45), and Madras (GenBank accession no. X99441) are closely related to each other. The ORF1 of the Mexican strain of HEV (13), like ORF2 and ORF3 (23), also showed much greater sequence divergence from other HEV strains, ranging from 73 to 80% sequence identity at the nucleotide level and from 85 to 94% at the amino acid level. However, the sequence identity between swine HEV and the Mexican strain was as divergent as those between swine HEV or the Mexican strain and non-U.S. strains of HEV.
TABLE 3.
Pairwise comparisons of putative functional domains and the HVR in the ORF1 of HEV
| Virus strain | % Identity (nucleotide/amino acid)
|
||||||||
|---|---|---|---|---|---|---|---|---|---|
| Swine | US-1 | US-2 | Pakistan | Burma | Madras | China | Myanmar | Mexico | |
| Methyltransferase | |||||||||
| Swine | 91/97 | 90/97 | 74/86 | 75/86 | 74/86 | 74/86 | 74/86 | 76/89 | |
| US-1 | 92/100 | 93/98 | 74/88 | 73/88 | 74/88 | 73/88 | 73/87 | 77/91 | |
| US-2 | 93/99 | 93/99 | 74/88 | 73/88 | 74/88 | 74/88 | 73/88 | 77/91 | |
| Pakistan | 75/88 | 75/88 | 76/88 | 93/99 | 92/98 | 99/100 | 92/98 | 76/90 | |
| Burma | 75/88 | 75/88 | 75/88 | 94/99 | 95/98 | 93/99 | 98/99 | 75/90 | |
| Madras | 74/84 | 74/84 | 74/84 | 91/95 | 95/95 | 92/98 | 94/97 | 76/90 | |
| China | 76/88 | 75/88 | 76/88 | 99/100 | 94/99 | 91/95 | 92/98 | 77/90 | |
| Myanmar | 75/88 | 75/87 | 76/87 | 93/99 | 99/99 | 95/95 | 93/99 | 75/89 | |
| Mexico | 74/86 | 73/86 | 74/86 | 77/90 | 77/89 | 75/85 | 77/89 | 77/89 | |
| RNA-dependent RNA polymerase | |||||||||
| Helicase | |||||||||
| Swine | 93/99 | 92/100 | 75/92 | 75/91 | 74/91 | 75/92 | 75/91 | 77/91 | |
| US-1 | 87/83 | 93/99 | 75/91 | 74/90 | 74/90 | 75/91 | 74/90 | 75/90 | |
| US-2 | 87/88 | 86/83 | 74/92 | 73/91 | 74/91 | 75/92 | 74/91 | 75/91 | |
| Pakistan | 43/41 | 38/44 | 47/38 | 93/99 | 92/99 | 99/100 | 92/99 | 77/91 | |
| Burma | 38/40 | 31/43 | 33/39 | 92/89 | 97/98 | 93/99 | 99/98 | 75/91 | |
| Madras | 24/39 | 31/41 | 34/38 | 88/85 | 93/93 | 92/99 | 96/98 | 75/91 | |
| China | 44/40 | 49/43 | 39/40 | 97/96 | 92/90 | 89/87 | 92/99 | 76/91 | |
| Myanmar | 33/40 | 32/40 | 33/38 | 91/85 | 98/95 | 93/90 | 91/87 | 75/91 | |
| Mexico | 47/38 | 51/38 | 44/38 | 39/39 | 31/38 | 39/36 | 27/38 | 43/40 | |
| Hypervariable regiona | |||||||||
Because of the size differences in the HVRs and the limited homology among genotypes, extensive gaps were introduced to force an alignment. Therefore, the percentages of sequence homology listed between major genotypes are approximate.
The 3′ NCR of swine HEV was also amplified and sequenced. The primer sequence in the extreme 3′ end was excluded, and the 3′ NCR containing the remaining 54 bp of swine HEV was compared with the corresponding regions of other HEV strains (Fig. 1). The 3′ NCR of swine HEV appeared to be very divergent; it shared about 87% sequence identity with that of the US-1 strain but only about 58 to 70% sequence identity with the corresponding regions of the Asian strains. The 3′ NCR of the Mexican strain was the longest, and varied extensively from all other HEV strains, including US-1 and swine HEV. In contrast, the 3′ NCRs among the Asian strains were very conserved, ranging from 96 to 98% nucleotide sequence identity. The 5′ NCR of swine HEV was also amplified. However, only 9 nt were left in the 5′ NCR of swine HEV after excluding the primer sequence used for the amplification. Therefore, further analysis of this region was not performed.
FIG. 1.
Alignments of the 3′ NCR of swine HEV with the corresponding regions of other HEV strains. The consensus sequence is shown at the top, and only differences are indicated. −, deletions.
Additional evidence for a new and divergent genotype of HEV.
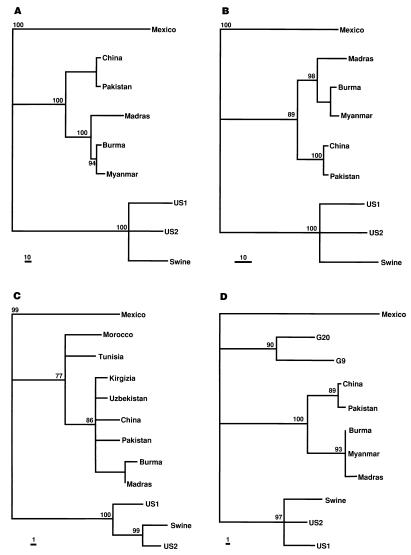
At least three genotypes of HEV have been previously identified, including the first genotype, represented by the Mexican strain; the second genotype, represented by Asian and African HEV strains; and the third genotype, represented by the swine HEV (23). A phylogenetic analysis was performed to determine the relationship of the U.S. and swine HEV strains to other strains. Since complete genomic sequences were not available for all HEV strains, phylogenetic analyses were based on partial regions of the genome (Fig. 2). All phylogenetic trees constructed from different regions of the genome were very similar. The swine HEV and the U.S. strains grouped very closely together and comprised a distinct genotype.
FIG. 2.
Phylogenetic trees based on the nucleotide sequences of different regions of the HEV genome. (A) RNA-directed RNA polymerase (1,462 bp); (B) methyltransferase (546 bp); (C) a stretch of 232 bp sequence in the overlap regions of ORF1, -2, and -3; (D) a 210-bp sequence in the putative RDRP region of ORF1 that corresponds to the only sequence available for strains G-20 and G-9. The trees were constructed by maximum parsimony methods with the aid of the PAUP program (GCG version 9.1; David L. Swofford, Smithsonian Institute, Washington, D.C.). Bootstrapping (1,000 replications) with branch-and-bound searching and midpoint rooting options was used to construct the trees. Bootstrap values of less than 75% are not shown. The scale bars representing the numbers of character state changes are shown. Branch lengths are proportional to the numbers of character state changes.
Also included in the analysis was a 210-bp sequence from the ORF1 region of two novel strains of HEV from patients in Guangzhou, China (13) (Fig. 2D). These two strains of HEV, G-20 and G-9, are unique in that they had only 11% sequence divergence from each other but varied extensively (22 to 24%) from other strains of HEV (14). Phylogenetic analysis of the 210-bp region suggested that the G-20 and G-9 strains do not group with the U.S. and swine HEV strains, the Mexican strain or Asian strains, including the other Chinese strains. The G-20 and G-9 strains may represent a fourth genotype. However, because of the limited sequence information available, a definitive phylogenetic relationship of G-20 and G-9 strains cannot be determined. Very recently, a novel strain of HEV in Taiwan from patients with sporadic acute hepatitis E was identified (12). This strain of HEV had only about 72 to 79% nucleotide sequence homology compared to other strains. However, the sequences available for this novel Taiwanese strain and the G-20 and G-9 strains were in different regions. Thus, it is not known if the novel strain from Taiwan is similar to the G-20 and G-9 strains from Southern China. However, the Taiwanese strain was different from the U.S. (human and swine) strains, and this finding provides further evidence for a fourth genotype of HEV.
Taxonomic evidence that the U.S. HEV strains infecting humans are very similar to swine HEV.
Further sequence analysis encompassing all three ORFs confirmed that the swine HEV and two U.S. strains were very closely related to each other and differed extensively from other strains of HEV (Table 3). In ORF1, the putative RDRP and the helicase of swine HEV and of the two U.S. strains had 92 to 93% sequence identity at the nucleotide level and 99 to 100% sequence identity at the amino acid level. Similarly, in the putative methyltransferase region, they shared 90 to 91% nucleotide sequence identity and 97% amino acid sequence identity. In the putative capsid gene (ORF2), swine HEV and the two U.S. strains shared 92% sequence identity at the nucleotide level and 98 to 99% sequence identity at the amino acid level. The small ORF3 of swine HEV had 95 to 98% nucleotide sequence and 93 to 97% amino acid sequence identity with the U.S. strains (data not shown). Interestingly, both swine HEV and the two U.S. strains had an in-frame deletion of 3 nt in the ORF3 region compared to other strains (data not shown).
In the putative HVR (37), extensive sequence variations were observed among the three major genotypes of HEV (U.S. strains, Mexican strain, and Asian strains), although the HVR was relatively conserved within each major genotype (Table 3). The lengths of the HVR varied among different HEV strains as follows: 105 amino acid residues for all Asian strains, 119 to 120 amino acids for the U.S. strains and swine HEV, and 103 amino acids for the Mexican strain (data not shown). The amino acid identities within a genotype were 85 to 96% among the Asian strains and 83 to 88% among swine HEV and the two U.S. strains. However, among the three major genotypes, the amino acid sequence identity was less than 50%. The significantly larger size of the HVRs of the swine and U.S. strains compared with those of all other strains, coupled with the high sequence identity among the swine and two U.S. strains, provides compelling evidence that the swine HEV and the U.S. strains are closely related. The very similar swine HEV and US-1 strains were isolated from the same small geographical area (swine HEV from pigs in Illinois [23] and the US-1 strain from a human in the neighboring state of Minnesota [20, 34]), suggesting that they may be variants of the same virus.
Infection of nonhuman primates with swine HEV.
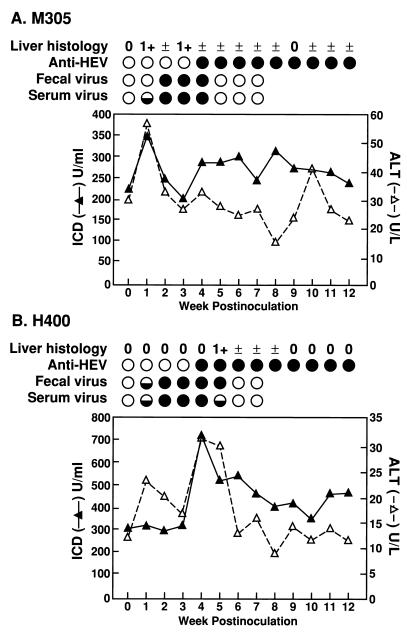
Because the swine HEV and U.S. strains of human HEV were so similar, it was of interest to determine if swine HEV could infect primates. Two rhesus monkeys were each inoculated i.v. with 104.5 PID50 of swine HEV (Fig. 3). Both primates seroconverted to anti-HEV 4 weeks p.i. and were still seropositive at the end of the 16-week experiment. Fecal excretion of swine HEV, indicative of replication, was detected 1 to 2 weeks p.i. and lasted for 3 to 5 weeks. The peak PCR titer of swine HEV in the feces was 106 GEs/g of feces (data not shown). Viremia lasting for 4 to 5 weeks was detected as early as 1 week p.i. There was a slight elevation of ALT and ICD in serum at 4 weeks p.i. in monkey H400 and at 1 week p.i. in monkey M305. However, the level of liver enzyme elevations in serum was low compared with that in primates experimentally infected with human strains of HEV (38–41). In both primates, focal necroinflammatory changes (1+), consistent with mild acute viral hepatitis were observed in liver biopsy specimens near the time of liver enzyme elevations in serum.
FIG. 3.
Experimental infection of rhesus monkeys (M305 and H400) with swine HEV. Levels of liver enzymes (ALT and ICD) in serum are plotted. The baseline liver enzymes were 15 U of ALT per liter and 472 U of ICD per ml for rhesus monkey H400 (an average of four preinoculation samples) and 28 U of ALT per liter and 323 U of ICD per ml for rhesus monkey M305 (an average of two preinoculation samples). Presence (•) or absence (○) of anti-HEV IgG in serum and swine HEV RNA in serum and feces is indicated. ◒, detected only by nested PCR. The degree of necroinflammatory changes (scale of 0 to 4+) observed in liver biopsy specimens is indicated.
The inoculated chimpanzee also became infected by swine HEV. Swine HEV RNA was detected in feces as early as 1 week p.i. and lasted for 3 weeks. Seroconversion to anti-HEV IgG occurred at 6 weeks p.i., although viremia was not detected by RT-PCR. The inoculated animal remained clinically normal, and levels of liver enzymes in serum were not significantly elevated. The PCR products amplified from feces of the infected rhesus monkeys and chimpanzee were sequenced and confirmed to be specific for swine HEV (data not shown).
Infection of pigs with the US-2 strain of human HEV.
Both pigs inoculated with strain US-2 seroconverted to anti-HEV at 2 weeks p.i. (Table 4). Fecal excretion of the virus was detected by RT-PCR in both inoculated pigs 1 week p.i. and lasted for about 2 weeks. One uninoculated contact control pig also became infected. This uninoculated pig seroconverted to anti-HEV about 2 weeks after the inoculated pig had seroconverted, and HEV RNA was detected in its feces about 2 weeks after the virus first appeared in the feces of the inoculated pig. The PCR product amplified from the feces of an infected pig was sequenced and confirmed to be specific for the US-2 isolate and distinct from swine HEV (data not shown). Clinically, both inoculated and contact control pigs appeared normal, and we did not observe significant elevation of any of the liver enzymes tested.
TABLE 4.
Experimental infection of SPF pigs with the US-2 strain of human HEV
| Room | Pig identificationa | Wks p.i. at which the following were found:
|
||
|---|---|---|---|---|
| Viral RNA in feces | Abnormal liver enzymes | Anti-HEV IgGb | ||
| A | I-1 | 1–2 | 0c | 2 |
| C-1 | 3–4 | 0 | 4 | |
| B | I-2 | 1–2 | 0 | 2 |
| C-2 | —d | 0 | — | |
I, inoculated; C, control.
Week first detected.
0, normal values throughout the study.
—, negative.
DISCUSSION
Symptomatic human infection with HEV in the United States and other developed countries is rare. Most of the few cases reported in these countries were in travellers returning from endemic regions (8, 11, 35), although in some cases in Western European countries this risk factor was absent (16, 17, 46). The recent identification of a case of acute hepatitis E in the United States in a patient with no history of travel to endemic regions (20, 34) reiterates that absence of travel to an endemic region does not necessarily exclude the diagnosis of acute hepatitis E infection. The need to explain the source of infection acquired in industrialized countries and to understand the ecology and epidemiology of HEV has led to a hypothesis that an animal reservoir may exist (33). In fact, antibodies reactive with HEV antigens have been detected in a number of animal species, including pigs, sheep, and rats (5, 7, 18, 21, 42). Unfortunately, either virus was not recovered from these species or virus recovered was not sequenced. Our discovery of the swine HEV as a ubiquitous agent in the swine population of the midwestern United States lends credence to the possibility of an animal reservoir.
In the present study, a standardized infectious pool of swine HEV was prepared and characterized. In our previous studies, we found only subclinical infections in naturally infected young pigs (23) or young pigs experimentally infected with an unquantified but relatively low dose of virus (24). In the present study, pigs infected with 104.5 PID50 also did not have clinical or biochemical evidence of hepatitis, although HEV was detected in bile, indicating that swine HEV replicated in the liver. In previous studies, cynomolgus macaques infected with a human strain of HEV developed biochemical evidence of significant hepatitis (ALT ≥100 U/liter) when the infecting dose was around 103.5 50% monkey infectious doses or higher but had little or no biochemical evidence of hepatitis when inoculated with lower doses (40). The rhesus macaques infected with 104.5 PID50 of swine HEV had mild focal necroinflammatory changes in liver biopsy specimens and a slight elevation of ALT and ICD levels in serum, which could indicate mild hepatitis and virus replication in the liver. The chimpanzee infected with 106 GEs of swine HEV was clinically normal, although fecal excretion of swine HEV and seroconversion to anti-HEV IgG were detected. This inoculum has not yet been titrated for infectivity; however, based on the ratio of GE to infectivity of the original pool of swine HEV in swine feces (106 GE/104.5 PID50), the infectious dose may have been the same as that given to the rhesus monkeys. Additional studies must be performed to determine if a larger dose will induce clinical disease. The patterns of appearance of viremia, anti-HEV, and fecal excretion of virus in primates infected with swine HEV were similar to those observed in infected pigs. Infection of primates with swine HEV demonstrated that swine HEV can cross species barriers, at least under experimental conditions, suggesting the possibility of human infection with swine HEV. The extremely high prevalence of swine HEV in pigs (23) and its ability to cross species barriers may put swine practitioners, swine producers, and other pig handlers at possible risk of zoonotic infection by the virus.
Balayan et al. (5) previously reported that Russian domestic swine were experimentally infected with a Central Asian strain of HEV isolated from a naturally infected patient. However, the virus infecting the pigs in this experiment was not sequenced to confirm its identity; thus, it is not clear whether the pigs were infected with the virus in the inoculum or were fortuitously infected by a swine virus circulating among the pigs at the time. We were unable to infect crossbred SPF pigs with strains of human HEV representing two major genotypes (24), Mex-14 (Mexican) and Sar-55 (Pakistan), even though the Sar-55 strain is from the same geographic region as the strain used by Balayan et al. and even though high doses of infectious virus (105 monkey infectious doses) were administered (24). Similarly, Platt et al. (28) failed to infect SPF pigs experimentally with the Mexican strain (Mex-14) of human HEV. Since the swine HEV shares only about 75% nucleotide sequence homology with either of the two human strains of HEV tested, our failure to infect swine with these strains (24) was likely due to a difference in susceptibility of pigs to different HEV strains. In contrast, when we inoculated SPF pigs with the US-2 strain of human HEV, the inoculated pigs became infected and, in one case, virus even spread to the uninoculated pig housed in the same room. Since the virus recovered from the infected pig had a unique sequence identical to that of the virus in the inoculum, these data provided conclusive experimental evidence for cross-species infection by HEV. Like pigs infected with swine HEV, swine infected with the US-2 strain of human HEV (either by inoculation or by contact) remained clinically normal, and levels of liver enzymes in serum were not significantly elevated. The rapid seroconversion of pigs infected with the US-2 strain of human HEV further suggested that the US-2 strain is already competent to replicate in swine and may be of swine origin. These data strongly suggest that a swine virus strain or one very similar to swine HEV infects and causes hepatitis in rare cases in humans in the United States and perhaps in other countries. The results suggested that pigs could well be an animal reservoir for HEV in the United States.
We have amplified and sequenced nearly the full-length genome of the swine HEV. Sequence analyses revealed that, in all three ORFs and the 3′ NCR, swine HEV and the two U.S. strains of HEV are very closely related. However, both swine HEV and the U.S. strains diverged extensively from other strains of HEV, especially in the HVR. Phylogenetic analyses have shown that genotypes of HEV generally have unique geographic distributions. Swine HEV and the closely related U.S. strains were all isolated in the United States. In general, the many Asian strains isolated are related both genetically and geographically. The African strains appear to be similar to but distinguishable from Asian strains. The Mexican strain is the only strain that forms a single branch. However, exceptions to this geographic distribution of HEV strains are the two unique strains of HEV, G-20 and G-9, recovered from patients in China (14). These two Chinese strains do not appear to group with the other Asian strains. The observed genetic diversities among different strains of HEV may affect the development of HEV vaccines. Although the experimental HEV vaccines are very promising (31, 41), their efficacy must be evaluated for protection against these novel and divergent strains.
The data presented in this study suggested that swine may serve as an animal reservoir for HEV. Young pigs infected naturally (23) or experimentally (24) with swine HEV appear not to have clinical symptoms, and primates showed negligible or minimal evidence of hepatitis. Therefore, subclinical infection of humans with swine HEV could explain the relatively high prevalence of anti-HEV in apparently healthy individuals in the United States and other industrialized countries (16, 17, 22, 37, 46). However, anti-HEV was also found in healthy individuals in the United States who seem to have had no contact with swine or other farm animals (22, 37). Thus, it is likely that other animal species may also serve as reservoirs for HEV. These putative animal reservoirs of HEV could be a source of contamination in regions where HEV is endemic and may also be responsible for the anti-HEV detected in healthy individuals in nonendemic regions.
Recently, xenotransplantation has become the focus of intensive research for a solution to the shortage of organ donors for transplantations (4). Swine are relatively easy to breed and maintain; therefore, xenotransplantation with pig organs has received considerable attention (4, 44). However, xenozoonoses, the transmission of pathogens from pigs to human recipients, is of major concern in xenotransplantation (25, 44). Many swine viruses are known to infect humans (9). Our results proved that swine HEV also has the ability to cross species barriers and therefore might infect humans. Although swine HEV appeared to be nonpathogenic for pigs and primates in experimental infections, it might become pathogenic in immunosuppressed xenotransplantation recipients. Therefore, it is important to develop sensitive and easy-to-perform assays to screen for swine HEV in donor pigs used for xenotransplantation. In addition, adequate diagnostic reagents are needed not only for screening of xenotransplantation donors but for epidemiologic studies as well. With the recent diagnosis in the United States of acute hepatitis E that was not associated with travel to endemic regions and the recovery from such a case of HEV that is closely related to swine HEV, HEV should now be considered as a possible etiologic agent in persons with acute hepatitis in the United States.
ACKNOWLEDGMENTS
We thank R. Royer for assistance in the swine transmission studies; E. Riedeseo for expert help in ultrasound-guided bile collection; and D. Wong, L. Rasmussen, Y. Huang, R. Engle, and H. Nguyen for assistance in the laboratory studies.
REFERENCES
- 1.Arankalle V A, Chadha M S, Tsarev S A, Emerson S U, Risbud A R, Banerjee K, Purcell R H. Seroepidemiology of water-born hepatitis in India and evidence for a third enterically-transmitted hepatitis agent. Proc Natl Acad Sci USA. 1994;91:3428–3432. doi: 10.1073/pnas.91.8.3428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arankalle V A, Tsarev S A, Chadha M S, Alling D W, Emerson S U, Banerjee K, Purcell R H. Age-specific prevalence of antibodies to hepatitis A and E viruses in Pune, India, 1982 and 1992. J Infect Dis. 1995;171:447–450. doi: 10.1093/infdis/171.2.447. [DOI] [PubMed] [Google Scholar]
- 3.Aye T T, Uchida T, Ma X, Iida F, Shikata T, Ichikawa M, Rikihisa T, Win K M. Sequence and gene structure of the hepatitis E virus isolated from Myanmar. Virus Genes. 1993;7:95–109. doi: 10.1007/BF01702352. [DOI] [PubMed] [Google Scholar]
- 4.Bach F H. Xenotransplantation: problems and prospects. Annu Rev Med. 1998;49:301–310. doi: 10.1146/annurev.med.49.1.301. [DOI] [PubMed] [Google Scholar]
- 5.Balayan M S, Usmanov R K, Zamyatina D I, Karas F R. Brief report: experimental hepatitis E infection in domestic pigs. J Med Virol. 1990;32:58–59. doi: 10.1002/jmv.1890320110. [DOI] [PubMed] [Google Scholar]
- 6.Chatterjee R, Tsarev S A, Pillot J, Coursaget P, Emerson S U, Purcell R H. African strains of hepatitis E virus that are distinct from Asian strains. J Med Virol. 1997;53:139–144. [PubMed] [Google Scholar]
- 7.Clayson E T, Innis B L, Myint K S A, Narupiti S, Vaughn D W, Giri S, Ranabhat P, Shrestha M P. Detection of hepatitis E virus infections among domestic swine in the Kathmandu Valley of Nepal. Am J Trop Med Hyg. 1995;53:228–232. doi: 10.4269/ajtmh.1995.53.228. [DOI] [PubMed] [Google Scholar]
- 8.Dawson G J, Mushahwar I K, Chau K H, Gitnick G L. Detection of long-lasting antibody to hepatitis E in a US traveller to Pakistan. Lancet. 1992;340:426–427. doi: 10.1016/0140-6736(92)91507-5. [DOI] [PubMed] [Google Scholar]
- 9.de Groen P C. Hepatitis E in the United States: a case of “hog fever”? Mayo Clin Proc. 1997;72:1197–1198. doi: 10.1016/S0025-6196(11)63687-2. [DOI] [PubMed] [Google Scholar]
- 10.Erker, J. C., S. M. Desai, G. G. Schlauder, G. J. Dawson, and I. K. Mushahwar. Hepatitis E variant in the United States: molecular characterization and transmission in cynomologus macaques. Submitted for publication. [DOI] [PubMed]
- 11.Herrera J L, Hill S, Shaw J, Fleenor M, Bader T, Wolfe M S. Hepatitis E among US travelers, 1989–1992. Mortal Morbid Weekly Rep. 1993;42:1–4. [PubMed] [Google Scholar]
- 12.Hsieh S Y, Yang P Y, Ho Y P, Chu C M, Liaw Y F. Identification of a novel strain of hepatitis E virus responsible for sporadic acute hepatitis in Taiwan. J Med Virol. 1998;55:300–304. doi: 10.1002/(sici)1096-9071(199808)55:4<300::aid-jmv8>3.0.co;2-4. [DOI] [PubMed] [Google Scholar]
- 13.Huang C-C, Nguyen D, Fernandez J, Yun K Y, Fry K E, Bradley D W, Tam A W, Reyes G R. Molecular cloning and sequencing of the Mexico isolate of hepatitis E virus (HEV) Virology. 1992;191:550–558. doi: 10.1016/0042-6822(92)90230-m. [DOI] [PubMed] [Google Scholar]
- 14.Huang R, Nakazono N, Ishii K, Kawamata O, Kawaguchi R, Tsukada Y. Existing variations on the gene structure of hepatitis E virus strains from some regions of China. J Med Virol. 1995;47:303–308. doi: 10.1002/jmv.1890470403. [DOI] [PubMed] [Google Scholar]
- 15.Hussaini S H, Skidmore S J, Richardson P, Sherratt L M, Cooper B T, O’Grady J G. Severe hepatitis E infection during pregnancy. J Viral Hepat. 1997;4:51–54. doi: 10.1046/j.1365-2893.1997.00123.x. [DOI] [PubMed] [Google Scholar]
- 16.Jardi R, Buti M, Rodriguez-Frias F, Esteban R. Hepatitis E infection in acute sporadic hepatitis in Spain. Lancet. 1993;341:1355–1356. doi: 10.1016/0140-6736(93)90874-g. [DOI] [PubMed] [Google Scholar]
- 17.Johansson P J, Mushahwar I K, Norkrans G, Weiland O, Nordenfelt E. Hepatitis E virus infections in patients with acute hepatitis non-A-D in Sweden. Scand J Infect Dis. 1995;27:543–546. doi: 10.3109/00365549509047064. [DOI] [PubMed] [Google Scholar]
- 18.Karetnyi Y V, Dzhumalieva D I, Usmanov R K, Titova I P, Litvak Y I, Balayan M S. Possible involvement of rodents in the spread of hepatitis E. Zh Mikrobiol Epidemiol Immunobiol. 1993;4:52–56. [PubMed] [Google Scholar]
- 19.Koonin E V, Gorbalenya A E, Purdy M A, Rozanov M N, Reyes G R, Bradley D W. Computer-assisted assignment of functional domains in the nonstructural polyprotein of hepatitis E virus: delineation of an additional group of positive-strand RNA plant and animal viruses. Proc Natl Acad Sci USA. 1992;89:8259–8263. doi: 10.1073/pnas.89.17.8259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kwo P Y, Schlauder G G, Carpenter H A, Murphy P J, Rosenblatt J E, Dawson G J, Mast E E, Krawczynski K, Balan V. Acute hepatitis E by a new isolate acquired in the United States. Mayo Clin Proc. 1997;72:1133–1136. doi: 10.4065/72.12.1133. [DOI] [PubMed] [Google Scholar]
- 21.Maneerat Y, Clayson E T, Myint K S A, Young G D, Innis B L. Experimental infection of the laboratory rat with the hepatitis E virus. J Med Virol. 1996;48:121–128. doi: 10.1002/(SICI)1096-9071(199602)48:2<121::AID-JMV1>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- 22.Mast E E, Kuramoto I K, Favorov M O, Schoening V R, Burkholder B T, Shapiro C N, Holland P V. Prevalence of and risk factors for antibody to hepatitis E virus seroreactivity among blood donors in Northern California. J Infect Dis. 1997;176:34–40. doi: 10.1086/514037. [DOI] [PubMed] [Google Scholar]
- 23.Meng X-J, Purcell R H, Halbur P G, Lehman J R, Webb D M, Tsareva T S, Haynes J S, Thacker B J, Emerson S U. A novel virus in swine is closely related to the human hepatitis E virus. Proc Natl Acad Sci USA. 1997;94:9860–9865. doi: 10.1073/pnas.94.18.9860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Meng X-J, Halbur P G, Haynes J S, Tsareva T S, Bruna J D, Royer R L, Purcell R H, Emerson S U. Experimental infection of pigs with the newly identified swine hepatitis E virus (swine HEV), but not with human strains of HEV. Arch Virol. 1998;143:1405–1415. doi: 10.1007/s007050050384. [DOI] [PubMed] [Google Scholar]
- 25.Murphy F A. The public health risk of animal organ and tissue transplantation into humans. Science. 1996;273:746–747. doi: 10.1126/science.273.5276.746. [DOI] [PubMed] [Google Scholar]
- 26.Page R D M. TREEVIEW: an application to display phylogenetic trees on personal computers. Comput Appl Biosci. 1996;12:357–358. doi: 10.1093/bioinformatics/12.4.357. [DOI] [PubMed] [Google Scholar]
- 27.Paul D A, Knigge M F, Ritter A, Gutierrez R, Pilot-Matias T, Chau K H, Dawson G J. Determination of hepatitis E virus seroprevalence by using recombinant fusion protein and synthetic peptides. J Infect Dis. 1994;169:801–806. doi: 10.1093/infdis/169.4.801. [DOI] [PubMed] [Google Scholar]
- 28.Platt K B, Yoon K-J, Zimmerman J J. 1998 Swine Research Report. Ames, IA: Iowa State University; 1998. Susceptibility of swine to hepatitis E virus and its significance to human health; pp. 125–126. [Google Scholar]
- 29.Pringle C. Minutes of the 27th International Committee on Taxonomy of Viruses Meeting. Arch Virol. 1998;143:1449–1459. [Google Scholar]
- 30.Purcell R H. Hepatitis E virus. In: Fields B N, Knipe D M, Howley P M, et al., editors. Fields virology. 3rd ed. Vol. 2. Philadelphia, Pa: Lippincott-Raven Publishers; 1996. pp. 2831–2843. [Google Scholar]
- 31.Purdy M A, McCaustland K A, Krawczynski K, Spelbring J, Reyes G R, Bradley D W. Preliminary evidence that a trpE-HEV fusion protein protects cynomolgus macaques against challenge with wild-type hepatitis E virus (HEV) J Med Virol. 1993;41:90–94. doi: 10.1002/jmv.1890410118. [DOI] [PubMed] [Google Scholar]
- 32.Quiroga J A, Cotonat T, Castillo I, Carreno V. Hepatitis E virus seroprevalence in acute viral hepatitis in a developed country confirmed by a supplemental assay. J Med Virol. 1996;50:16–19. doi: 10.1002/(SICI)1096-9071(199609)50:1<16::AID-JMV4>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- 33.Reyes G R. Overview of the epidemiology and biology of the hepatitis E virus. In: Willsen R A, editor. Viral hepatitis. New York, N.Y: Marcel Dekker, Inc.; 1997. pp. 239–258. [Google Scholar]
- 34.Schlauder G G, Dawson G J, Erker J C, Kwo P Y, Knigge M F, Smalley D L, Rosenblatt J E, Desai S M, Mushahwar I K. The sequence and phylogenetic analysis of a novel hepatitis E virus isolated from a patient with acute hepatitis reported in the United States. J Gen Virol. 1998;79:447–456. doi: 10.1099/0022-1317-79-3-447. [DOI] [PubMed] [Google Scholar]
- 35.Skidmore S J, Yarbough P O, Gabor K A, Tam A W, Reyes G R. Imported hepatitis E in UK. Lancet. 1991;337:1541. doi: 10.1016/0140-6736(91)93227-z. [DOI] [PubMed] [Google Scholar]
- 36.Tam A W, Smith M M, Guerra M E, Huang C C, Bradley D W, Fry K E, Reyes G R. Hepatitis E virus (HEV): molecular cloning and sequencing of the full-length viral genome. Virology. 1991;185:120–131. doi: 10.1016/0042-6822(91)90760-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Thomas D L, Yarbough P O, Vlahov D, Tsarev S A, Nelson K E, Saah A J, Purcell R H. Seroreactivity to hepatitis E virus in areas where the disease is not endemic. J Clin Microbiol. 1997;35:1244–1247. doi: 10.1128/jcm.35.5.1244-1247.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tsarev S A, Emerson S U, Reyes G R, Tsareva T S, Legters L J, Malik I A, Iqbal M, Purcell R H. Characterization of a prototype strain of hepatitis E virus. Proc Natl Acad Sci USA. 1992;89:559–563. doi: 10.1073/pnas.89.2.559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tsarev S A, Tsareva T S, Emerson S U, Kapikian A Z, Ticehurst J, London W, Purcell R H. ELISA for antibody to hepatitis E virus (HEV) based on complete open-reading frame-2 protein expressed in insect cells: identification of HEV infection in primates. J Infect Dis. 1993;168:369–378. doi: 10.1093/infdis/168.2.369. [DOI] [PubMed] [Google Scholar]
- 40.Tsarev S A, Tsareva T S, Emerson S U, Yarbough P O, Legters L J, Moskal T, Purcell R H. Infectivity titration of a prototype strain of hepatitis E virus in cynomolgus monkeys. J Med Virol. 1994;43:135–142. doi: 10.1002/jmv.1890430207. [DOI] [PubMed] [Google Scholar]
- 41.Tsarev S A, Tsareva T S, Emerson S U, Govindarajan S, Shapiro M, Gerin J L, Purcell R H. Recombinant vaccine against hepatitis E: dose response and protection against heterologous challenge. Vaccine. 1997;15:1834–1838. doi: 10.1016/s0264-410x(97)00145-x. [DOI] [PubMed] [Google Scholar]
- 42.Usmanov R K, Balayan M S, Dvoinikova O V, Alymbaeva D B, Zamiatina N A, Kazachkov LuA, Belov V I. An experimental infection in lambs by the hepatitis E virus. Vopr Virusol. 1994;39:165–168. [PubMed] [Google Scholar]
- 43.van Cuyck-Gandre H, Zhang H Y, Tsarev S A, Clements N J, Cohen S J, Caudill J D, Buisson Y, Coursaget P, Warren R L, Longer C F. Characterization of hepatitis E virus (HEV) from Algeria and Chad by partial genome sequence. J Med Virol. 1997;53:340–347. doi: 10.1002/(sici)1096-9071(199712)53:4<340::aid-jmv5>3.0.co;2-7. [DOI] [PubMed] [Google Scholar]
- 44.Weiss R A. Transgenic pigs and virus adaptation. Nature. 1998;391:327–328. doi: 10.1038/34772. [DOI] [PubMed] [Google Scholar]
- 45.Yin S, Purcell R H, Emerson S U. A new Chinese isolate of hepatitis E virus: comparison with strains recovered from different geographical regions. Virus Genes. 1994;9:23–32. doi: 10.1007/BF01703432. [DOI] [PubMed] [Google Scholar]
- 46.Zaaijer H L, Mauser-Bunschoten E P, ten Veen J H, Kapprell H P, Kok M, van den Berg H M, Lelie P N. Hepatitis E virus antibodies among patients with hemophilia, blood donors, and hepatitis patients. J Med Virol. 1995;46:244–246. doi: 10.1002/jmv.1890460313. [DOI] [PubMed] [Google Scholar]