Abstract
The putative core protein of hepatitis C virus (HCV) regulates cellular growth and a number of cellular promoters. To further understand its effect, we investigated the role of the core protein in the endogenous regulation of two distinct transcription factors, nuclear factor-κB (NF-κB) and activating protein-1 (AP-1), and the related mitogen-activated protein kinase kinase (MAPKK) and c-Jun N-terminal kinase (JNK). Stable cell transfectants expressing the HCV core protein suppressed tumor necrosis factor (TNF)-induced NF-κB activation. Supershift analysis revealed that NF-κB consists of p50 and p65 subunits. This correlated with inhibition of the degradation of IκBα, the inhibitory subunit of NF-κB. The effect was not specific to TNF, as suppression in core protein-expressing cells was also observed in response to a number of other inflammatory agents known to activate NF-κB. In contrast to the effect on NF-κB, the HCV core protein constitutively activated AP-1, which correlated with the activation of JNK and MAPKK, which are known to regulate AP-1. These observations indicated that the core protein targets transcription factors known to be involved in the regulation of inflammatory responses and the immune system.
Hepatitis C virus (HCV) is a major causative agent of the development of acute and chronic hepatitis, which often leads to liver cirrhosis and hepatocellular carcinoma (46). Although humoral and cellular immune responses to HCV have been detected, a proper understanding of viral persistence is lacking. Viral infection may often induce defense responses in host cells, and many viruses, in turn, encode proteins which inhibit this defense mechanism (53). These alterations in cell survival contribute to the pathogenesis of a number of human diseases, including viral oncogenesis (44).
HCV contains a single-stranded positive-sense RNA genome which encodes a precursor polypeptide of approximately 3,000 amino acids. This precursor polypeptide is cleaved by both viral and host proteases into its functional units, i.e. at least 10 individual proteins: core, E1, E2, p7, NS2, NS3, NS4A, NS4B, NS5A, and NS5B. The diverse functional activities of the HCV putative core protein have already been noted by a number of investigators. These include nucleocytoplasmic localization (reviewed in reference 55), the regulation of cellular and unrelated viral promoters in in vitro studies (36, 39, 41, 49), inhibition of apoptosis under certain conditions (38), a physical association with apolipoprotein II (3) and with the cytoplasmic tail of the lymphotoxin β-receptor (9, 32), and the promotion of normal cells to a transformed phenotype (7, 37). Furthermore, mutations and truncations in the genomic region encoding the HCV core protein have been observed and implicated in hepatocellular carcinogenesis in infected humans (45, 50, 56).
Tumor necrosis factor (TNF) is a major inflammatory cytokine secreted primarily by activated macrophages and T lymphocytes in response to viral infections, which may inhibit viral replication or induce apoptosis, and some viruses have evolved strategies to block the antiviral effect of TNF (4, 5, 12, 21). This proinflammatory cytokine binds to receptors to initiate intracellular signaling cascades leading to apoptosis. Cells respond to the cytokine by selectively expressing a wide range of genes. TNF is known to activate both the nuclear factor-κB (NF-κB)- and activating protein-1 (AP-1)-specific recognition sequences present in the promoters of a large number of genes known to be involved in inflammation control and immune regulation (2, 13, 22, 29, 35, 54). The activation of NF-κB requires phosphorylation, ubiquitination, and degradation of IκBα, which occurs through the activation of a family of kinases (1, 2). The activation of AP-1 occurs through the c-Jun N-terminal kinase (JNK) (35). A number of transcription factors are activated through phosphorylation by signal-responsive protein kinases (22). Phosphorylation can affect DNA binding activity, transcriptional activity, and subcellular localization of transcription factors. Several studies indicate that c-Jun is a component of dimeric-sequence-specific activator protein AP-1. JNK mediates phosphorylation of c-Jun at the N terminus for transcriptional activity (15), whereas another mitogen-activated protein kinase (MAPK), ERK41/42, leads to phosphorylation and activation of the Elk-1 transcription factor required for transcriptional activation of Fos (15, 31). The activity of MAPK, ERK1 and ERK2 is rapidly stimulated by activation of the dual-specificity protein kinase MAPKK (11), which phosphorylates ERK1 and ERK2.
We have recently reported that the HCV core protein inhibits TNF-mediated apoptosis by using sensitive human breast carcinoma MCF-7 cells as a model system (40). In the present study, we investigated the role of the HCV core protein in the regulation of cellular transcription factors. The results suggested that the HCV core protein differentially regulates NF-κB and AP-1, thereby affecting the signaling cascades involved in cell growth regulation.
MATERIALS AND METHODS
Cell lines.
A pool of MCF-7 cells transfected with the HCV core gene, three individual clones derived from the transfectants, and vector-transfected MCF-7 cells (control) were used. Characterization of the cells has been recently described (40). The cells were routinely grown in Dulbecco modified Eagle medium supplemented with 10% heat-inactivated fetal bovine serum and antibiotics.
EMSA.
The electrophoretic mobility shift assay (EMSA) was carried out by folloiwng the standard procedure (8, 48). Briefly, nuclear extracts were prepared from 2 × 106 cells following treatment with the indicated reagent(s) and used immediately or stored at −70°C. Reactions were performed by incubation of 4 μg of the nuclear extracts with 8 fmol of a 32P-end-labeled 45-mer double-stranded NF-κB oligonucleotide from the human immunodeficiency virus long terminal repeat (5′-TTGTTACAAGGGACTTTCCGCTGGGGACTTTCCAGGGAGGCGTGG-3′ [underlined sequences represent the NF-κB binding site]) for 15 min at 37°C. The incubation mixture contained 2 to 3 μg of poly(dI-dC) in a binding buffer (25 mM HEPES [pH 7.9], 0.5 mM EDTA, 0.5 mM dithiothreitol [DTT], 1% Nonidet P-40 [NP-40], 5% glycerol, 50 mM NaCl). The DNA-protein complex was separated from the free oligonucleotide by electrophoresis on a 5% native polyacrylamide gel using buffer containing 50 mM Tris, 200 mM glycine (pH 8.5), and 1 mM EDTA and detected by autoradiography.
The specificity of binding was analyzed by competition with the unlabeled oligonucleotide and an oligonucleotide with a mutated NF-κB binding site in a supershift assay. Nuclear extracts were incubated with an antibody to either the p50 or the p65 subunit of NF-κB for 30 min at 37°C and then subjected to an EMSA. An antibody to cyclin D1 or c-Rel (Santa Cruz Biotechnology, Inc.) was included as the negative control. The EMSA for AP-1 was performed as described for NF-κB, by using 32P-end-labeled double-stranded oligonucleotides. Specificity of binding was determined by using an excess of the unlabeled oligonucleotide for competition as described earlier (51). The radioactive intensity of bands was analyzed by a PhosphorImager (Molecular Dynamics, Sunnyvale, Calif.).
Western blotting.
IκBα was analyzed by Western blot analysis as described previously (42). Briefly, the cytoplasmic extracts from treated cells were resolved on a sodium dodecyl sulfate (SDS)–10% polyacrylamide gel, transferred to nitrocellulose filters, probed with a rabbit polyclonal antibody to IκBα (Santa Cruz Biotechnology), and detected by chemiluminescence (ECL; Amersham).
JNK assay.
The JNK assay was performed by using a method similar to one described earlier (27). Briefly, cells (3 × 106) were treated with TNF (Genentech, Inc.) for different times, lysed in a buffer containing 20 mM HEPES (pH 7.4), 2 mM EDTA, 250 mM NaCl, 1% NP-40, 2-μg/ml leupeptin, 2-μg/ml aprotinin, 1 mM phenylmethylsulfonyl fluoride, 0.5-μg/ml benzamidine, and 1 mM DTT. Cell extracts (150 to 250 μg/sample) were subjected to immunoprecipitation with 0.03 μg of anti-JNK antibody (Santa Cruz Biotechnology) for 60 min at 4°C. The immune complex was separated by incubation with protein A/G Sepharose beads for 45 min at 4°C. The beads were washed with lysis buffer (4 × 400 μl) and kinase buffer (2 × 400 μl; 20 mM HEPES [pH 7.4], 1 mM DTT, 25 mM NaCl). An in vitro kinase assay was performed for 15 min at 30°C with glutathione S-transferase (GST)–Jun(1-79) as the substrate in 20 mM HEPES (pH 7.4)–10 mM MgCl2–1 mM DTT–10 μCi of [γ-32P]ATP. The reaction was terminated by adding 15 μl of 2× SDS sample buffer. The sample was boiled for 5 min and subjected to SDS–9% polyacrylamide gel electrophoresis. The GST-Jun(1-79) band was visualized by staining with Coomassie blue. The gel was analyzed by densitometric scanning (Molecular Dynamics).
MAPK assay.
MCF-7 cells were stimulated with different concentrations of TNF. After incubation for 30 min at 37°C, cells were washed with Dulbecco’s phosphate-buffered saline and lysed with 20 mM HEPES (pH 7.4)–2 mM EDTA–250 mM NaCl–0.1% NP-40–2-μg/ml leupeptin–2-μg/ml aprotinin–1 mM phenylmethylsulfonyl fluoride–0.5%-μg/ml benzamidine–1 mM DTT–1 mM sodium orthovanadate. The protein concentration in the supernatant was determined, and the proteins were resolved by SDS–10% polyacrylamide gel electrophoresis. After electrophoresis, the proteins were transferred onto nitrocellulose and probed with the antibody to p44/42 MAPK (Thr 202/Tyr 204; New England Biolabs, Inc.). The membrane was incubated with peroxidase-conjugated anti-rabbit immunoglobulin G, and the bands were detected by chemiluminescence (ECL; Amersham).
In vitro assay for regulation of AP-1.
HepG2 cells were plated at a density of 5 × 105 in a 60-mm-diameter plate in minimal essential medium containing 10% fetal bovine serum and antibiotics. Cells were cotransfected with 0.5 μg of the reporter plasmid (phARE-CAT, having the AP-1 binding site, or phARE-Ap-1, with a mutant AP-1 binding site [25]) and the effector plasmid (pHCV-core) by using the standard Lipofectamine method (Life Technologies). The total amount of the expression vector was adjusted to 2.5 μg with an appropriate amount of empty expression vector. The cells were harvested after 48 h of transfection, and cytoplasmic extracts were prepared by three cycles of freezing and thawing. Cytoplasmic extracts were analyzed for chloramphenicol acetyltransferase (CAT) activity by thin-layer chromatography. The acetylated [14C]chloramphenicol was quantitated by measuring the radioactivity with a PhosphorImager (Molecular Dynamics). β-Galactosidase activity was measured as an internal control for transfection.
RESULTS
Suppression of TNF-mediated NF-κB activation in HCV core protein-expressing cells.
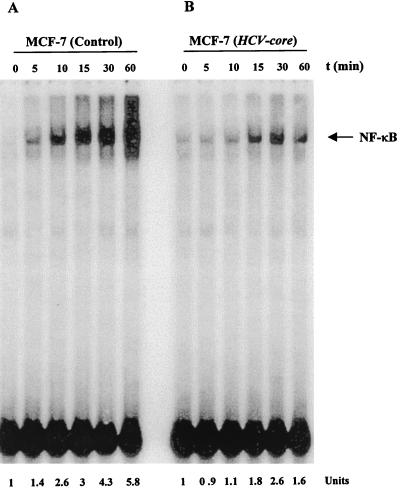
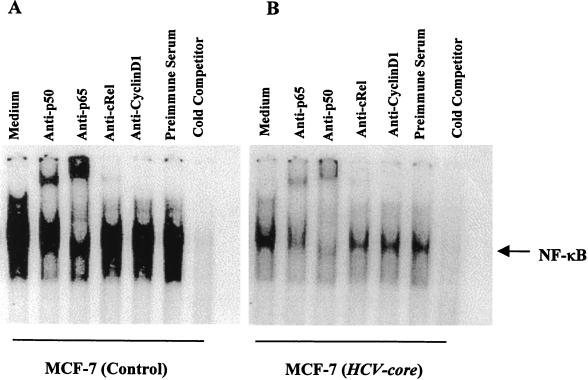
Empty-vector-transfected control and cloned HCV core plasmid DNA-transfected MCF-7 cells were stimulated with a predetermined optimum dose of TNF (100 pM) for different times at 37°C, and NF-κB activation was determined by EMSA. TNF activated NF-κB 5.8-fold within 60 min in control cells, compared to a 1.6-fold increase in core DNA-transfected cells (Fig. 1A and B). These results suggested that the HCV core protein represses TNF-mediated NF-κB activation. We also examined three different clones of HCV core DNA-transfected MCF-7 cells. All of these clones exhibited reduced NF-κB activation compared to the control cells under identical conditions (data not shown). NF-κB is a family of proteins, and various combinations of Rel/NF-κB protein constitute an active NF-κB heterodimer which binds to a specific sequence of DNA (3). We ascertained the authenticity of the NF-κB band following incubation of the nuclear extracts with antibodies to the p50 (NF-κB1) or p65 (RelA) subunit by EMSA. Antibodies to either of the subunits shifted the band to a higher molecular weight (Fig. 2). On the other hand, antibodies to c-Rel or unrelated antibodies did not shift the NF-κB band, suggesting that the TNF-activated complex consists of the p50 and p65 subunits. The disappearance of the NF-κB band with the unlabeled oligonucleotide further revealed the specificity of the NF-κB band. Mutation of the NF-κB binding site in the oligonucleotide failed to bind the target sequence and exhibited the specificity of the target protein (data not shown).
FIG. 1.
Effect of HCV core protein expression on TNF-induced NF-κB activation. Empty-vector-transfected (A) and HCV core DNA-transfected (B) MCF-7 cells were incubated at 37°C for 0, 5, 10, 15, 30, and 60 min with TNF (100 pM). Nuclear extracts were prepared and analyzed for NF-κB activation. The units at the bottom show fold increases in TNF-induced NF-κB activation with time compared to that in untreated control cells.
FIG. 2.
Supershift analysis and specificity of TNF-induced NF-κB activation. The nuclear extracts from TNF-treated (100 pM, 30 min) control (A) and HCV core DNA-transfected (B) cells were incubated with different antibodies and a 50-fold excess of an unlabeled competitor for 30 min at 37°C. The reaction was monitored by EMSA with a labeled NF-κB probe.
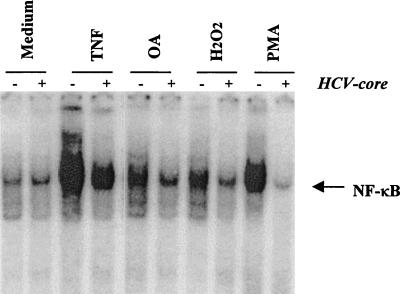
Suppression of NF-κB activation by OA, H2O2, and PMA in HCV core protein-expressing cells.
A wide variety of agents, including okadaic acid (OA), H2O2, and phorbol myristate acetate (PMA), are known to activate NF-κB by different mechanisms (34). To examine whether the HCV core protein affects the NF-κB activation induced by these agents, cells were treated with an optimal dose of the inducers and analyzed for NF-κB activation. Core protein-expressing cells significantly blocked the activation of NF-κB induced by these agents (Fig. 3). The results from this set of experiments suggested that the core protein has a suppressor effect on NF-κB activation and is likely to act at or following a step at which all of these agents converge in the signal transduction pathway leading to NF-κB activation.
FIG. 3.
Effect of HCV core protein on OA-H2O2-, and PMA-mediated activation of NF-κB. HCV core DNA-transfected and empty-vector-transfected MCF-7 cells were incubated with TNF (100 pM), PMA (100 ng/ml), H2O2 (250 μM), or OA (500 nM) for 30 min at 37°C and tested for NF-κB activation.
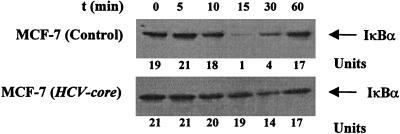
Inhibition of TNF-dependent degradation of IκBα in HCV core protein-expressing cells.
The translocation of NF-κB to the nucleus is preceded by the phosphorylation and proteolytic degradation of IκBα (52). To determine whether core protein expression affects IκBα degradation, the kinetics of TNF-induced IκB degradation in HCV core DNA-transfected cells was studied. TNF treatment of control cells produced disappearance of IκBα within 10 to 15 min, and IκBα reappeared at around 30 min (Fig. 4). A faster rate of IκBα degradation is expected in the absence of a protein synthesis inhibitor. Because cycloheximide is known to activate NF-κB by itself, we examined the rate of degradation of NF-κB in the absence of this agent (Fig. 1). However, the disappearance of the IκBα protein was not observed in HCV core DNA-transfected cells. Thus, our results clearly show that the HCV core decreases the TNF-induced IκBα degradation which may account for inhibition of NF-κB activation. As TNF treatment leads to serine phosphorylation of IκBα, which is necessary for its ubiquitination and degradation, it is possible that the HCV core alters some of the phosphorylation events involved in IκBα and NF-κB activation.
FIG. 4.
Time-dependent IκBα expression following TNF-α induction in control and HCV core-expressing cells. MCF-7 cells were incubated for different times with TNF (100 pM). Cytoplasmic extracts were prepared and analyzed for IκBα by Western blotting. The units at the bottom reflect densitometric scanning of the IκBα level. Similar results were obtained in three independent experiments.
Activation of AP-1 in HCV core protein-expressing cells.
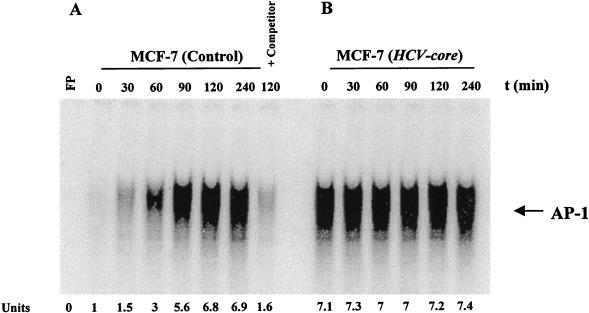
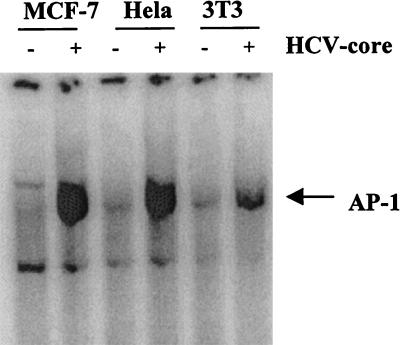
TNF is known to activate transcription factor AP-1. We therefore investigated the role of the HCV core protein in a AP-1 activation mediated by a predetermined optimum dose of TNF (100 pM) in MCF-7 cells. In a time course experiment, AP-1 activation in the control cells was noted within 30 min and reached a maximum by 2 h (Fig. 5A). The disappearance of the band in an EMSA using an unlabeled oligonucleotide further suggested the specificity of AP-1 activation. On the other hand, activation of AP-1 had achieved its optimum in the presence of the HCV core prior to exposure to TNF, and prolonged exposure to TNF was unable to enhance AP-1 activation in cells (Fig. 5B). To determine whether AP-1 activation is cell type specific, HeLa and NIH 3T3 cells expressing the HCV core protein were also analyzed similarly for AP-1 activation. Vector-transfected cells were included in the experiment as a control. Results from this study further suggested that core protein-expressing cells have increased AP-1 activity compared to control cells (Fig. 6).
FIG. 5.
TNF-induced AP-1 activation in control and HCV core-expressing cells. Empty-vector-transfected (A) and HCV core gene-transfected (B) MCF-7 cells were incubated at 37°C for 0, 30, 60, 90, 120, and 240 min with TNF (100 pM). Nuclear extracts were prepared and analyzed for AP-1 activation as described in the text. Results obtained with an unlabeled oligonucleotide as an unlabeled competitor are shown in panel A. The units at the bottom indicate fold increases in TNF-induced AP-1 activation compared to that in untreated control cells. FP, free probe.
FIG. 6.
Effect of HCV core protein expression on AP-1 activation in different cell lines. MCF-7, HeLa, and NIH 3T3 cells were stably transfected with the HCV core gene, and nuclear extracts from vector-transfected control and core DNA-transfected cells were analyzed for AP-1 activity by gel shift assay.
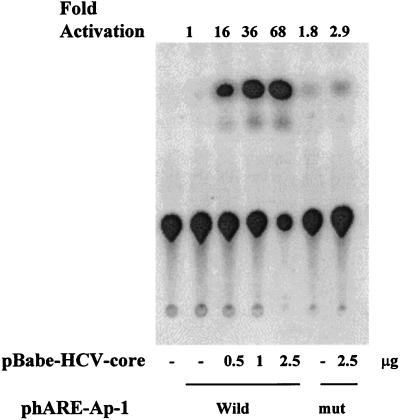
To determine whether the HCV core protein affects AP-1-dependent gene transcription, cells were transfected with a plasmid containing a human antioxidant response element (hARE) having an AP-1 binding site linked to a CAT gene. A dose-dependent increase in CAT activity was observed with HCV core expression (Fig. 7). On the other hand, the core protein did not induce CAT activity on pmut-phARE, having a mutant AP-1 binding site, in a similar assay. The results from this study suggested that the core protein has an augmenting effect on AP-1-dependent gene transcription, and this corroborated the DNA binding results.
FIG. 7.
Autoradiograph of a representative CAT assay in which HepG2 cells were cotransfected with 0.5 μg of a reporter gene construct (phARE or pmut-hARE) or 2.5 μg of an empty vector (pPAC) and different concentrations of pHCV-core. After 48 h of transfection, the cytoplasmic extracts were analyzed for CAT activity. Fold CAT activation is indicated at the top of each lane, and the plasmids used in the transfection are shown below. mut, mutant.
Activation of JNK in HCV core-expressing cells.
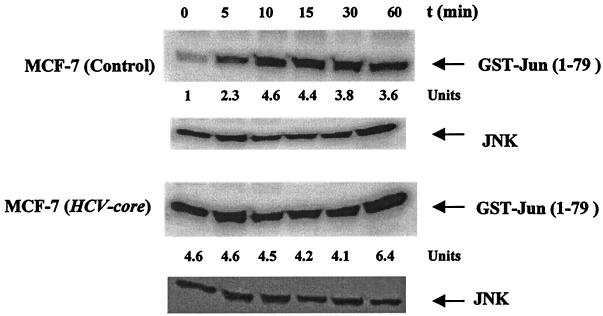
Since TNF activates JNK (17), we investigated the role of HCV core protein expression on TNF-mediated JNK activation. Empty-vector-transfected control cells and HCV core DNA-transfected cells were stimulated with TNF (100 pM) for different times, and activation of JNK was examined (Fig. 8). An increase in JNK activity was detected in control MCF-7 cells 10 to 15 min after the introduction of TNF. On the other hand, HCV core DNA-transfected cells had a high basal c-Jun kinase activity which remained unchanged upon TNF treatment. The results indicated that the HCV core protein induces the activation of JNK. The high basal JNK activity observed in core DNA-transfected cells may account for the similar AP-1 basal activity in these cells.
FIG. 8.
Effect of HCV core protein on TNF-induced JNK activation. Empty-vector-transfected and HCV core DNA-transfected MCF-7 cells were treated with 1 nM TNF at 37°C for the indicated times. Cells were washed and lysed, and the JNK was immunoprecipitated from extracts by a specific antibody. JNK activity was measured by using the immune complex in a kinase assay with GST–C-Jun(1-79) as the substrate. Total JNK protein expression was determined by Western blot analysis. The units at the bottom indicate fold increases in JNK activity induced by TNF compared to that in untreated control cells.
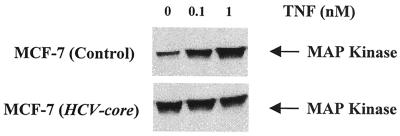
MAPKK activity in HCV core-expressing cells.
MAPKK is an upstream effector of JNK activation. Defective MAPKK activity may manifest itself downstream as a defect in JNK and AP-1 function. MAPKK activity, known to be induced by TNF, was investigated in HCV core protein-expressing cells. Control and experimental cells were stimulated with TNF (100 pM) for different times, and phosphorylated MAPK was examined by Western blot analysis. Enhanced phosphorylation was detected within 30 and 60 min of TNF treatment (data not shown). Subsequently, control and HCV core DNA-transfected cells were treated with 0.1 and 1 nM TNF for 30 min, and the phosphorylated MAPK was analyzed by Western blot analysis (Fig. 9). In control MCF-7 cells, activation of MAPKK occurred at 0.1 nM TNF, but cells transfected with HCV core DNA had a high basal activity and did not further activate MAPKK following treatment with TNF.
FIG. 9.
TNF-induced MAPKK activity in control and HCV core-expressing cells. Empty-vector- and HCV core gene-transfected MCF-7 cells were stimulated with different concentrations of TNF for 30 min. Cell lysates were analyzed for MAPKK activity by Western blot analysis with a specific antibody to phosphorylated MAPK (phosphospecific anti-p44/42 MAPK).
DISCUSSION
In this study, we investigated whether stable expression of the HCV core protein in mammalian cells has an effect on the regulation of transcription factors with or without the induction of TNF. The results indicated the status of NF-κB, AP-1, the protein kinase JNK, and MAPKK in relation to the HCV core protein. Suppression of NF-κB in HCV core-expressing cells was observed following treatment with TNF, PMA, OA, and H2O2. These agents activate NF-κB through steps which are overlapping and nonoverlapping. The exact pathway required to activate NF-κB is not fully understood. Our results, however, suggest that the HCV core must act at a step common to all of these inducers of NF-κB. Besides the NF-κB inducers tested in this study, there are several other agents known to activate NF-κB. Thus, whether the HCV core is a generalized inhibitor of NF-κB activation remains to be established. The results that we obtained by ectopic expression of the HCV core protein are the first to demonstrate endogenous regulation of cellular genes.
The mechanism of NF-κB suppression and AP-1 activation in HCV core-expressing cells is not known. Generation of superoxide radicals is common to most inducers of NF-κB. Overexpression of the antioxidative enzymes thioredoxin, manganese superoxide dismutase, and catalases impairs NF-κB activity (30, 47). High-level manganese superoxide dismutase expression in adenovirus E1B19K-expressing cells has been implicated in NF-κB-inhibiting activity (18, 26). Like the HCV core, the thiol antioxidants pyrrolidine dithiocarbamate and N-acetyl-l-cysteine activate AP-1 and suppress TNF-mediated induction of NF-κB (34). Wether activation of AP-1 and suppression of NF-κB by the HCV core are linked to activation of the MAPK pathway is not clear. Similar to the HCV core, however, antioxidants have also been shown to activate AP-1 and suppress NF-κB (34). This suggests that the HCV core produces its effects by acting as an antioxidant.
NF-κB is one of the major components induced by TNF-α, and its role in the signalling of TNF-induced cell death remains controversial (19). Although NF-κB appears to stimulate the expression of specific protective genes, neither the identities of these genes nor their precise roles as inhibitors of the apoptotic process are known (10). A number of recent studies have suggested that the inhibition of NF-κB does not alter TNF-induced apoptosis (6, 16, 18, 19, 43). In core DNA-transfected MCF-7 cells, NF-κB and AP-1 appear to have different requirements for activation and AP-1 activity is regulated positively by the core protein. This study extended our previous observations that the HCV core protein enhances or suppresses the transcription of various viral and cellular genes by using transfected reporter constructs (36, 39, 41). Taken together, these observations suggest an involvement of the HCV core protein and the common transcription factor(s) leading to diverse gene regulatory effects, which merits further investigations.
The activity of AP-1 is elevated in response to a large number of environmental stimuli. The increase in basal AP-1 activity suggests that the core protein promotes cellular proliferation for the maintenance of replication and survival (17, 24). This is further supported by the observed enhancement of the basal activity of JNK and MAPKK in core-expressing cells. The effect of the HCV core on AP-1 transcriptional activity may result, in part, from the enhanced phosphorylation of the c-Jun NH2-terminal activation domain, as well as from the induction of Fos and Jun gene transcription. Together with NF-κB, AP-1 is likely to mediate the induction of other cytokines and immunoregulatory molecules by TNF, leading to a variety of inflammatory responses (27). To determine whether results obtained with MCF-7 cells are applicable to cells of hepatic origin, some of the studies were also performed with HepG2 cells, and the results were comparable. Future studies with other mammalian cell lines, especially primary hepatocytes, should further clarify the importance of these results.
The HCV core protein does not appear to induce a strong cytotoxic T-lymphocyte response. Even after multiple immunizations, <10% (spontaneous) lysis of unprimed mouse splenocytes and <30% specific lysis of in vitro-stimulated mouse splenocytes occurred (14, 23, 28). HCV infection is likely to induce a selective defect in antigen-presenting cells, which may enhance the ability of HCV to establish a persistent infection. Indeed, expression of TNF and interleukin-1 requires NF-κB activation, and both of these cytokines are suppressed in cells of HCV-infected humans (20, 33). HCV often causes persistent infection and silent disease which may lead to hepatocellular carcinoma. A recent study demonstrated a defective HCV genome comprising the major virus population of the ascitic fluid from a patient with hepatocellular carcinoma (56). In the genome, deletions and frame shifts were suggested to terminate the open reading frame and produce a truncated viral core protein. In persistently infected cells, arising defective virus particles could express a subset of the viral gene, or the continued presence of the viral core protein would likely have a detrimental effect on cellular genes. It is appealing to speculate that suppression of NF-κB in TNF-induced cells and endogenous activation of AP-1 may have a profound negative effect on the appropriate immune regulation and the normal function of HCV core-expressing cells.
ACKNOWLEDGMENTS
We thank Robert B. Belshe for helpful discussion, Ratna B. Ray and Keith Meyer for critical review of the manuscript, Michael Houghton for providing HCV cDNA (Blue4/C5p-1), and Suz Ann Price for preparation of the manuscript.
This research was supported by The Clayton Foundation for Research, internal funding from Saint Louis University, and AI-45250 from the NIAID.
REFERENCES
- 1.Baeuerle P A, Baltimore D. NF-κB: ten years after. Cell. 1996;87:13–20. doi: 10.1016/s0092-8674(00)81318-5. [DOI] [PubMed] [Google Scholar]
- 2.Baldwin A S. The NF-κB and IκB proteins: new discoveries and insights. Annu Rev Immunol. 1996;14:649–681. doi: 10.1146/annurev.immunol.14.1.649. [DOI] [PubMed] [Google Scholar]
- 3.Barba G, Harper F, Harada T, Kohara M, Goulinet S, Matsuura Y, Eder G, Schaff Z, Chapman M J, Miyamura T, Brechot C. Hepatitis C virus core protein shows a cytoplasmic localization and associates to cellular lipid storage droplets. Proc Natl Acad Sci USA. 1997;94:1200–1205. doi: 10.1073/pnas.94.4.1200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Beidler D R, Tewari M, Friesen M, Poirier G, Dixit V M. The baculovirus p35 protein inhibits Fas- and tumor necrosis factor-induced apoptosis. J Biol Chem. 1995;270:16526–16528. doi: 10.1074/jbc.270.28.16526. [DOI] [PubMed] [Google Scholar]
- 5.Bertin J, Armstrong R C, Ottilie S, Martin D A, Wang Y, Banks S, Wang G H, Senkevich T G, Alnemri E S, Moss B, Lenardo M J, Tomaselli K J, Cohen J I. Death effector domain-containing herpesvirus and poxvirus proteins inhibit both Fas- and TNFR1-induced apoptosis. Proc Natl Acad Sci USA. 1997;94:1172–1176. doi: 10.1073/pnas.94.4.1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cai Z, Korner M, Tarantino N, Chouaib S. IkappaB alpha overexpression in human breast carcinoma MCF7 cells inhibits nuclear factor-kappaB activation but not tumor necrosis factor-alpha-induced apoptosis. J Biol Chem. 1997;272:96–101. doi: 10.1074/jbc.272.1.96. [DOI] [PubMed] [Google Scholar]
- 7.Chang J, Se-Hwan Y, Cho Y-G, Hwang S B, Hahn Y S, Sung Y C. Hepatitis C virus core from two different genotypes has an oncogenic potential but is not sufficient for transforming primary rat embryo fibroblasts in cooperation with the H-ras oncogene. J Virol. 1998;72:3060–3065. doi: 10.1128/jvi.72.4.3060-3065.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chaturvedi M M, LaPushin R, Aggarwal B B. Tumor necrosis factor and lymphotoxin. Qualitative and quantitative differences in the mediation of early and late cellular response. J Biol Chem. 1994;269:14575–14583. [PubMed] [Google Scholar]
- 9.Chen C, You L, Hwang L, Lee Y W. Direct interaction of hepatitis C virus core protein with the cellular lymphotoxin-β receptor modulates the signal pathway of the lymphotoxin-β receptor. J Virol. 1997;71:9417–9426. doi: 10.1128/jvi.71.12.9417-9426.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chu Z L, Mckinsey T A, Liu L, Gentry J J, Malim M H, Ballard D W. Suppression of tumor necrosis factor-induced cell death by inhibitor of apoptosis c-IAP2 is under NF-kappaB control. Proc Natl Acad Sci USA. 1997;94:10057–10062. doi: 10.1073/pnas.94.19.10057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Crews C M, Alessandrini A, Erikson R L. The primary structure of MEK, a protein kinase that phosphorylates the ERK gene product. Science. 1992;258:478–480. doi: 10.1126/science.1411546. [DOI] [PubMed] [Google Scholar]
- 12.Dimitrov T, Krajesi P, Hermiston T W, Tollefson A E, Hannink M, Wold W S. Adenovirus E3-10.4K/14.5K protein complex inhibits tumor necrosis factor-induced translocation of cytosolic phospholipase A2 to membranes. J Virol. 1997;71:2830–2837. doi: 10.1128/jvi.71.4.2830-2837.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Foletta V C, Segal D H, Cohen D R. Transcriptional regulation in the immune system: all roads lead to AP-1. J Leukoc Biol. 1998;63:139–152. doi: 10.1002/jlb.63.2.139. [DOI] [PubMed] [Google Scholar]
- 14.Geissler M, Gesien A, Tokushige K, Wands J. Enhancement of cellular and humoral immune responses to hepatitis C virus core protein using DNA-based vaccines augmented with cytokine-expressing plasmids. J Immunol. 1997;158:1231–1237. [PubMed] [Google Scholar]
- 15.Gille H, Sharrocks A D, Shaw P E. Phosphorylation of transcription factor p62TCF by MAP kinase stimulates ternary complex formation at c-fos promoter. Nature. 1992;358:414–417. doi: 10.1038/358414a0. [DOI] [PubMed] [Google Scholar]
- 16.Guo Y L, Kang B, Williamson J R. Inhibition of the expression of mitogen activated protein phosphatase-1 potentiates apoptosis induced by tumor necrosis factor-alpha in rat mesangial cells. J Biol Chem. 1998;273:10362–10366. doi: 10.1074/jbc.273.17.10362. [DOI] [PubMed] [Google Scholar]
- 17.Guo Y L, Baysal K, Kang B, Yang L J, Williamson J R. Correlation between sustained c-Jun N-terminal protein kinase activation and apoptosis induced by tumor necrosis factor-alpha in rat mesangial cells. J Biol Chem. 1998;273:4027–4034. doi: 10.1074/jbc.273.7.4027. [DOI] [PubMed] [Google Scholar]
- 18.Hashimoto S, Ishi A, Yonehara S. The E1B oncogene of adenovirus confers cellular resistance to cytotoxicity of tumor necrosis factor and monoclonal anti-Fas antibody. Int Immunol. 1991;3:343–351. doi: 10.1093/intimm/3.4.343. [DOI] [PubMed] [Google Scholar]
- 19.Hehner S P, Hofmann T G, Ratter F, Dumont A, Droge W, Schimitz M L. Tumor necrosis factor-α-induced cell killing and activation of transcription factor NF-κB are uncoupled in L929 cells. J Biol Chem. 1998;273:18117–18121. doi: 10.1074/jbc.273.29.18117. [DOI] [PubMed] [Google Scholar]
- 20.Holtmann H, Hahn T, Wallach D. Interrelated effects of tumor necrosis factor and interleukin 1 on cell viability. Immunobiology. 1988;177:7–22. doi: 10.1016/S0171-2985(88)80087-1. [DOI] [PubMed] [Google Scholar]
- 21.Hu S, Vincenz C, Buller M, Dixit V M. A novel family of viral death effector domain-containing molecules that inhibit both CD-95- and tumor necrosis factor receptor-1-induced apoptosis. J Biol Chem. 1997;272:9621–9624. doi: 10.1074/jbc.272.15.9621. [DOI] [PubMed] [Google Scholar]
- 22.Karin M, Liu Z, Zandi E. AP-1 function and regulation. Curr Opin Cell Biol. 1997;9:240–246. doi: 10.1016/s0955-0674(97)80068-3. [DOI] [PubMed] [Google Scholar]
- 23.Lagging L M, Meyer K, Hoft D, Houghton M, Belshe R B, Ray R. Immune responses to plasmid DNA encoding the hepatitis C virus core protein. J Virol. 1995;69:5859–5863. doi: 10.1128/jvi.69.9.5859-5863.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lenczowski J M, Dominguez L, Eder A M, King L B, Zacharchuk C M, Ashwell J D. Lack of a role for Jun kinase and AP-1 in Fas-induced apoptosis. Mol Cell Biol. 1997;17:170–181. doi: 10.1128/mcb.17.1.170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li Y, Jaiswal A K. Regulation of human NAD(P)H:quinone oxidoreductase gene. Role of AP1 binding site contained within human antioxidant response element. J Biol Chem. 1992;267:15097–15104. [PubMed] [Google Scholar]
- 26.Limbourg F P, Stadtler H, Chinnadurai G, Baeuerle P A, Schmitz M L. A hydrophobic region within the adenovirus E1B 19 kDa protein is necessary for the transient inhibition of NF-kappaB activated by different stimuli. J Biol Chem. 1996;271:20392–20398. doi: 10.1074/jbc.271.34.20392. [DOI] [PubMed] [Google Scholar]
- 27.Liu Z, Hsu H, Goeddel D V, Karin M. Dissection of TNF receptor 1 effector functions: JNK activation is not linked to apoptosis while NF-κB activation prevents cell death. Cell. 1996;87:565–576. doi: 10.1016/s0092-8674(00)81375-6. [DOI] [PubMed] [Google Scholar]
- 28.Major M E, Vitvitski L, Mink M A, Schleef M, Whalen R G, Trepo C, Inchauspe G. DNA-based immunization with chimeric vectors for the induction of immune responses against the hepatitis C virus nucleocapsid. J Virol. 1995;69:1798–5805. doi: 10.1128/jvi.69.9.5798-5805.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Maniatis T, Whittemore L A, Du W, Fan C M, Keller A, Palombella V, Thanos D. Positive and negative control of human interferon-β gene expression. In: Mcknight S L, Yamamoto K, editors. Transcriptional regulation, part 2. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1992. pp. 1193–1220. [Google Scholar]
- 30.Manna S K, Zhang H J, Yan T, Oberley L W, Aggarwal B B. Overexpression of manganese superoxide dismutase suppresses tumor necrosis factor-induced apoptosis and activation of nuclear transcription factor-κB and activated protein-1. J Biol Chem. 1998;273:13245–13254. doi: 10.1074/jbc.273.21.13245. [DOI] [PubMed] [Google Scholar]
- 31.Marais R, Wynne J, Treisman R. The SRF accessory protein Elk-1 contains a growth factor-regulated transcriptional activation domain. Cell. 1993;73:381–383. doi: 10.1016/0092-8674(93)90237-k. [DOI] [PubMed] [Google Scholar]
- 32.Matsumoto M, Hsieh T-Y, Zhu N, VanArsdale T, Hwang S B, Jeng K-S, Gorbalenya A E, Lo S-Y, Ou J-S, Ware C F, Lai M M C. Hepatitis C virus core protein interacts with the cytoplasmic tail of lymphotoxin-β receptor. J Virol. 1997;71:1301–1309. doi: 10.1128/jvi.71.2.1301-1309.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mendoza E C, Paglieroni T G, Zeldis J B. Decreased phorbol myristate acetate-induced release of tumor necrosis factor-alpha and interleukin-1 beta from peripheral blood monocytes of patients chronically infected with hepatitis C virus. J Infect Dis. 1996;174:842–844. doi: 10.1093/infdis/174.4.842. [DOI] [PubMed] [Google Scholar]
- 34.Meyer M, Schreck R, Baeuerle P A. H2O2 and antioxidants have opposite effects on activation of NF-kappaB and AP-1 in intact cells: AP-1 as secondary antioxidant-responsive factor. EMBO J. 1993;12:2005–2015. doi: 10.1002/j.1460-2075.1993.tb05850.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Minden A, Lin A, McMahon M, Lange-Carter C, Derijard B, Davis R J, Johnson G L, Karin M. Differential activation of ERK and JNK mitogen-activated protein kinases by Raf-1 and MEKK. Science. 1994;266:1719–1723. doi: 10.1126/science.7992057. [DOI] [PubMed] [Google Scholar]
- 36.Ray R B, Lagging L M, Meyer K, Steele R, Ray R. Transcriptional regulation of cellular and viral promoters by the hepatitis C virus core protein. Virus Res. 1995;37:209–220. doi: 10.1016/0168-1702(95)00034-n. [DOI] [PubMed] [Google Scholar]
- 37.Ray R B, Lagging L M, Meyer K, Ray R. Hepatitis C virus core protein cooperates with ras and transforms primary rat embryo fibroblasts to tumorigenic phenotype. J Virol. 1996;70:4438–4443. doi: 10.1128/jvi.70.7.4438-4443.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ray R B, Meyer K, Ray R. Suppression of apoptotic cell death by hepatitis C virus core protein. Virology. 1996;226:176–182. doi: 10.1006/viro.1996.0644. [DOI] [PubMed] [Google Scholar]
- 39.Ray R B, Steele R, Meyer K, Ray R. Transcriptional repression of p53 promoter by hepatitis C virus core protein. J Biol Chem. 1997;272:10983–10986. doi: 10.1074/jbc.272.17.10983. [DOI] [PubMed] [Google Scholar]
- 40.Ray R B, Meyer K, Steele R, Shrivastava A, Aggarwal B B, Ray R. Inhibition of tumor necrosis factor (TNF-α) mediated apoptosis by hepatitis C virus core protein. J Biol Chem. 1998;273:2256–2259. doi: 10.1074/jbc.273.4.2256. [DOI] [PubMed] [Google Scholar]
- 41.Ray R B, Steele R, Meyer K, Ray R. Hepatitis C virus core protein represses p21 WAF1/Cip1/Sid1 promoter activity. Gene. 1998;208:331–336. doi: 10.1016/s0378-1119(98)00030-4. [DOI] [PubMed] [Google Scholar]
- 42.Reddy S A, Chaturvedi M M, Darnay B G, Chan H, Higuchi M, Aggarwal B B. Reconstitution of nuclear factor kappa B activation induced by tumor necrosis factor requires membrane-associated components. Comparison with pathway activated by ceramide. J Biol Chem. 1994;269:25369–25372. [PubMed] [Google Scholar]
- 43.Roulston A, Reinhard C, Amiri P, Williams L T. Early activation of c-Jun N-terminal kinase and p38 kinase regulate cell survival in response to tumor necrosis factor-α. J Biol Chem. 1998;273:10232–10239. doi: 10.1074/jbc.273.17.10232. [DOI] [PubMed] [Google Scholar]
- 44.Rudin C M, Thompson C B. Apoptosis and disease: regulation and clinical relevance of programmed cell death. Annu Rev Med. 1997;48:267–281. doi: 10.1146/annurev.med.48.1.267. [DOI] [PubMed] [Google Scholar]
- 45.Ruster B, Zeuzem S, Roth W K. Hepatitis C virus sequences encoding truncated core proteins detected in a hepatocellular carcinoma. Biochem Biophys Res Commun. 1996;219:911–915. doi: 10.1006/bbrc.1996.0340. [DOI] [PubMed] [Google Scholar]
- 46.Saito I, Miyamura T, Ohbayashi A, Harada H, Katayama T, Kikuchi S, Watanabe Y, Koi S, Onji M, Choo Q-L, Houghton M, Kuo G. Hepatitis C virus infection is associated with the development of hepatocellular carcinoma. Proc Natl Acad Sci USA. 1990;87:6547–6549. doi: 10.1073/pnas.87.17.6547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schmidt K N, Traenckner E B, Beier B, Baeuerle P A. Induction of oxidative stress by okadaic acid is required for activation of transcription factor NF-κB. J Biol Chem. 1995;270:27136–27142. doi: 10.1074/jbc.270.45.27136. [DOI] [PubMed] [Google Scholar]
- 48.Schreiber E, Matthias P, Muller M M, Schaffner W W. Rapid detection of octamer binding proteins with ’mini-extracts’ prepared from a small number of cells. Nucleic Acids Res. 1989;17:6419. doi: 10.1093/nar/17.15.6419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shib C M, Lo S J, Miyamura T, Chen S Y, Lee Y W. Suppression of hepatitis B virus expression and replication by hepatitis C virus core protein in HuH-7 cells. J Viol. 1993;67:5823–5832. doi: 10.1128/jvi.67.10.5823-5832.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shimizu I, Yao D-F, Horie C, Yasuda M, Shiba M, Horie T, Nishikado T, Meng X-Y, Ito S. Mutations in a hydrophilic part of the core gene of hepatitis C virus in patients with hepatocellular carcinoma in China. J Gastroenterol. 1997;32:47–55. doi: 10.1007/BF01213296. [DOI] [PubMed] [Google Scholar]
- 51.Singh S, Aggarwal B B. Protein-tyrosine phosphatase inhibitors block tumor necrosis factor-dependent activation of the nuclear transcription factor NF-kappa B. J Biol Chem. 1995;270:10631–10639. doi: 10.1074/jbc.270.18.10631. [DOI] [PubMed] [Google Scholar]
- 52.Thonos D, Maniathis T. NF-κB: a lesson in family values. Cell. 1995;80:529–532. doi: 10.1016/0092-8674(95)90506-5. [DOI] [PubMed] [Google Scholar]
- 53.White E. Life, death, and the pursuit of apoptosis. Genes Dev. 1996;10:1–15. doi: 10.1101/gad.10.1.1. [DOI] [PubMed] [Google Scholar]
- 54.Wright K L, White L C, Kelly A, Beck S, Trowsdale J, Ting J P. Coordinate regulation of the human TAP1 and LMP2 genes from a shared bidirectional promoter. J Exp Med. 1995;181:1459–1471. doi: 10.1084/jem.181.4.1459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yasui K, Wakita T, Tsukiama-Kohara K, Funahashi S, Ichikawas M, Kajita T, Moradpour D, Wands J R, Kohara M. The native form and maturation process of hepatitis C virus core protein. J Virol. 1998;72:6048–6055. doi: 10.1128/jvi.72.7.6048-6055.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yeh C-T, Lu S-C, Chu C-M, Liaw Y-F. Molecular cloning of a defective hepatitis C virus genome for the ascitic fluid of a patient with hepatocellular carcinoma. J Gen Virol. 1997;78:2761–2770. doi: 10.1099/0022-1317-78-11-2761. [DOI] [PubMed] [Google Scholar]