Abstract
In the assembly of paramyxoviruses, interactions between viral proteins are presumed to be specific. The focus of this study is to elucidate the protein-protein interactions during the final stage of viral assembly that result in the incorporation of the viral envelope proteins into virions. To this end, we examined the specificity of HN incorporation into progeny virions by transiently transfecting HN cDNA genes into Sendai virus (SV)-infected cells. SV HN expressed from cDNA was efficiently incorporated into progeny Sendai virions, whereas Newcastle disease virus (NDV) HN was not. This observation supports the theory of a selective mechanism for HN incorporation. To identify the region on HN responsible for the selective incorporation, we constructed chimeric SV and NDV HN cDNAs and evaluated the incorporation of expressed proteins into progeny virions. Chimera HN that contained the SV cytoplasmic domain fused to the transmembrane and external domains of the NDV HN was incorporated to SV particles, indicating that amino acids in the cytoplasmic domain are responsible for the observed specificity. Additional experiments using the chimeric HNs showed that 14 N-terminal amino acids are sufficient for the specificity. Further analysis identified five consecutive amino acids (residues 10 to 14) that were required for the specific incorporation of HN into SV. These residues are conserved among all strains of SV as well as those of its counterpart, human parainfluenza virus type 1. These results suggest that this region near the N terminus of HN interacts with another viral protein(s) to lead to the specific incorporation of HN into progeny virions.
Envelope viruses are released from infected cells by budding at the cellular membrane. During the budding process, viral proteins are preferentially incorporated into progeny virions whereas cellular proteins are largely excluded. The protein-protein interaction during the stage of viral particle formation is not well understood. In particular, the molecular mechanisms that drive budding and guide the inclusion of spike proteins into the mature virus particle largely remain unresolved. The specificities of incorporation of envelope glycoproteins into virions appear to differ among RNA viruses. For example, foreign glycoproteins (e.g., the hemagglutinin [HA] and neuraminidase [NA] of influenza virus, the measles virus H protein, and the cellular glycoprotein CD4) are efficiently incorporated into virions of vesicular stomatitis virus, showing that incorporation of glycoprotein into progeny virion is not highly selective (20, 39). With influenza virus, HA molecules containing a foreign cytoplasmic tail and transmembrane domain failed to be incorporated into progeny virions (28). In contrast, a tailless HA molecule was efficiently incorporated into influenza virions, showing that the HA cytoplasmic tail is not required for HA incorporation into virions, although the possession of a cytoplasmic tail confers a growth advantage (17). Mitnaul et al. (26) reported that deleting the cytoplasmic tail of the NA molecule of influenza virus severely reduced NA incorporation into virions, although the cytoplasmic tail was not absolutely essential for virus replication. Regarding alphavirus, extensive molecular genetic and structural studies supported the theory of a direct, sequence-specific interaction between the cytoplasmic tail of envelope glycoprotein E2 and the nucleocapsid and the theory that this interaction directs virus budding (7, 41).
Sendai virus (SV), a prototype paramyxovirus, encodes at least six structural proteins: the two viral membrane glycoproteins HN and F, which are responsible for virus attachment, penetration, and release; three nucleocapsid proteins, NP, P, and L, which cover the genomic RNA and together are responsible for RNA transcription and replication; and one nonglycosylated internal membrane protein, M, which likely mediates packaging of the nucleocapsid into the viral envelope during virion assembly. The membrane glycoproteins are anchored in the viral envelope or the plasma membrane of an infected cell by a short hydrophobic transmembrane domain, with most of the protein (the external domain) extending extracellularly and a small tail region (the cytoplasmic domain) protruding from the inner surface of the membrane. The predicted SV HN protein includes a 35-residue cytoplasmic tail, a 25-residue transmembrane domain, and a large, 515-residue external domain, which contains five potential sites for addition of N-linked carbohydrate side chains (13). SV HN is highly homologous to other paramyxoviruses (e.g., 72% to human parainfluenza virus type 1 [hPIV1] and 62% to hPIV3) and moderately homologous to rubulaviruses (e.g., 35% to Newcastle disease virus [NDV] and 30% to simian virus 5) (13, 27). Synthesis of the viral glycoproteins (HN and F) occurs at the endoplasmic reticulum, where both proteins are cotranslationally inserted into membranes either as type I (F) or type II (HN) transmembrane proteins. After synthesis, each protein is transported via the exocytic pathway to the plasma membrane, where virus assembly occurs.
Paramyxovirus assembly can be viewed as a three-part process. First the NP subunits associate with the genomic RNA to form the helical nucleocapsid structure, and then the P and L proteins are added (34). Next the membrane proteins (HN, F, and M) accumulate at the plasma membrane. In the final assembly step, the nucleocapsid is enveloped during the budding process and progeny virus is produced. During budding at the plasma membrane, viral structural proteins are selectively incorporated into the nascent virion. It is generally thought that viral M protein plays a major role in assembly by forming a bridge between the assembled nucleocapsid core and the cytoplasmic domains of either HN or F or both (31). The M protein of SV may be cross-linked to the NP protein in fresh virions, suggesting that the M protein is complexed with the major nucleocapsid protein, NP (23). SV M protein was also reported to bind independently to either HN or F protein in vivo (37), although the results have not been confirmed (43). At present, the protein-protein interaction that regulates the incorporation of paramyxovirus glycoprotein into virions and the detailed sequence specificity involved are not known.
To understand the protein interactions in viral assembly, we used SV, NDV, and hPIV1 HNs to examine the sequences of HNs that were required for specific incorporation into progeny virions. In this report, we provide evidence that HN incorporation into SV is sequence specific in 14 N-terminal amino acids of the cytoplasmic domain of the HN molecule and that 5 consecutive amino acids conserved among SV and hPIV1 are required for the specificity.
MATERIALS AND METHODS
Viruses and cells.
293T cells, which constitutively express simian virus 40 large-T antigen (10), were maintained in Dulbecco’s modified Eagle’s medium supplemented with 10% fetal calf serum (FCS). The SV strain Enders, SV monoclonal antibody (MAb) escape mutant S2 (SVescS2; 46), and the NDV strain Kansas were grown in 11-day embryonated chicken eggs.
Plasmid constructs and construction of chimeric genes.
SV and hPIV1 HN genes were subcloned from pTF1 (5) into pCAGGS (29) by cleavage and religation at EcoRI and XhoI sites. The NDV HN gene was cloned from RNA extracted from infected cells with avian myeloblastosis virus reverse transcriptase (Boehringer Mannheim) and by PCR with specific primers located at the noncoding regions of the genes. Chimeric HN genes were constructed by using PCR for gene splicing by overlap extension (15).
MAbs.
Hybridoma cultures secreting antibody to NDV HN or SV NP were made by immunizing BALB/c mice with purified NDV or SV disrupted with the nonionic detergent n-octyl-d-glucopyranoside as described previously (9a). Ascites fluids of BALB/c mice injected with hybridomas were used for the experiments.
Incorporation of HN expressed from cDNA into SV.
293T cells were infected with SVescS2 at a multiplicity of infection of 5. After 1 h of incubation with virus, cells were washed and then transfected with pCAGGS plasmids containing the HN gene (2 μg) by using SuperFect (Qiagen). After a 4-h transfection period, the medium was replaced with Dulbecco’s modified Eagle’s medium–10% FCS and the cells were incubated at 34°C. Twenty-four hours later, cells were labeled with 1 ml of labeling medium (Sigma) containing 100 μCi of [35S]Trans-Label (ICN) for 16 h. The culture medium was harvested and centrifuged briefly, and the supernatants were used in radioimmunoprecipitation assays. SV containing HN molecules that had been expressed from cDNA was immunoprecipitated as follows. Ascitic fluid containing MAbs (2 μl) was incubated with 10 μl of Dynabeads (Dynal) in RIPA buffer (5) at 4°C for 30 min. The MAb-immunobead complexes were washed with RIPA buffer and then with phosphate-buffered saline containing 0.2% bovine serum albumin (PBS-BSA). The immunocomplexes, suspended in PBS-BSA (200 μl), were mixed with 100 μl of the culture supernatant containing the 35S-labeled virus described above for 30 min at 4°C, washed with PBS-BSA, and analyzed by sodium dodecyl sulfate–9% polyacrylamide gel electrophoresis (SDS-PAGE).
Western blotting.
293T cells were infected with SVescS2, transfected with cDNAs as described above, and cultured in the medium for 48 h. The culture supernatants were harvested and clarified to remove cell debris. The viruses in the media were purified by ultracentrifugation through 50% glycerol in PBS. Purified viruses were suspended in Laemmli sample buffer and were subjected to SDS-PAGE. Virus proteins were analyzed by Western immunoblotting with anti-NDV HN (N7) or anti-SV NP (M52) MAbs and revealed by peroxidase activity detection with a light-based ECL system as described by the manufacturer (Amersham Life Science).
Cell surface expression of HN proteins by fluorescence-activated cell sorting (FACS) analysis.
293T cells that were infected with SVescS2 and transfected with the appropriate pCAGGS construct were trypsinized to detach them from the plate and washed with PBS. Cells were suspended in 1 ml of PBS containing 10% FCS (PBS-FCS) and 2 μl of MAb. After incubation at room temperature for 30 min, cells were washed with PBS and suspended in 1 ml of PBS-FCS containing 1 μl of fluorescein isothiocyanate-conjugated anti-mouse immunoglobulin G (IgG; Boehringer Mannheim). After a 30-min incubation, cells were washed and fixed with 4% formalin (in PBS). Cell sorting was performed with a FACScan (Becton Dickinson) instrument.
HN protein oligomer formation.
Oligomer formation of SV, NDV, and chimeric HNs was determined by sucrose density gradient centrifugation as described previously (14). 293T cells transfected with pCAGGS containing the HN gene were labeled with 100 μCi of [35S]Trans-Label for 30 min and chased for 2 h. Cells were then solubilized in lysis buffer (1% Triton X-100, 50 mM Tris [pH 7.4], 100 mM NaCl). Postnuclear supernatants were laid over 10 ml of 5 to 25% (wt/vol) sucrose in 0.1% Triton X-100–50 mM Tris (pH 7.4)–100 mM NaCl. Gradients were centrifuged at 37,500 rpm at 20°C for 16 h in a SW41 rotor (Beckman). Twenty-seven fractions were collected, from which HN was immunoprecipitated. The polypeptides were analyzed on nonreducing 7.5% polyacrylamide gels.
NA activity and cell surface ELISA.
The NA activity of the chimeric HNs expressed at the cell surface was measured as reported previously (5). In brief, 24 h after transfection with cDNA, cells were washed with PBS and incubated with 0.2 M phosphate buffer (pH 5.9) containing N-acetylneuramin lactose for 2 h at 37°C. The supernatant was harvested, and the sialic acid in the buffer was measured. The same cells were used for measuring the amount of HN at the cell surface by cell surface enzyme-linked immunosorbent assay (ELISA) (5). Washed cells were reacted with an appropriate MAb, followed by incubation with horseradish peroxidase-conjugated sheep anti-mouse IgG (Bio-Rad) in PBS-BSA. The cells were then reacted with substrate, and the optical density was measured with a spectrophotometer at a wavelength of 405 nm.
Phosphorylation of HN proteins.
293T cells were transfected with the HN-containing pCAGGS constructs as described above and incubated overnight at 34°C. Cellular proteins were labeled at 34°C with either 50 μCi of [35S]Trans-Label in 1 ml of labeling medium or 100 μCi of [32P]orthophosphate (ICN) in 1 ml of phosphate-deficient medium (ICN). After a 4-h labeling period, cells were washed and then lysed by using cell lysis buffer (5). Labeled HN proteins in the lysates were immunoprecipitated with the appropriate MAb and analyzed by SDS-PAGE.
RESULTS
Specificity of HN incorporation into SV.
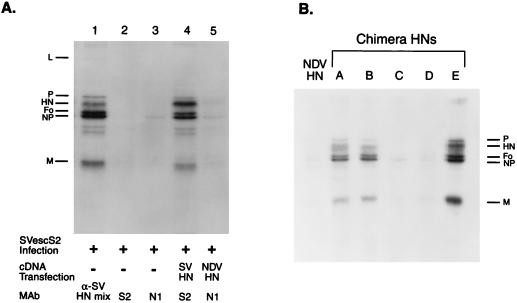
As a first step to characterize the specificity of HN incorporation into SV particles, we determined whether HN that was expressed from transfected cDNA could be incorporated into progeny virions. We infected 293T cells with the SV MAb escape mutant SVescS2 and transfected these cells with plasmids containing wild-type (wt) SV or NDV HN cDNA (pCAGGS-SVHN and pCAGGS-NDVHN, respectively). Labeled virions in the culture supernatant were immunoprecipitated by using MAb S2, which is specific for wt SV HN (32a), or N1, which is specific for NDV HN. Immunoprecipitations were done in the absence of detergent so that whole-virus particles containing the HN molecule expressed from cDNA could be precipitated. SVescS2 is the escape mutant of MAb S2; therefore, the HN of this virus does not react with either S2 or N1 (Fig. 1A). From the cell culture supernatant infected with SVescS2 and transfected with pCAGGS-SVHN, virus particles were immunoprecipitated by MAb S2, which showed that HN expressed from cDNA was incorporated into progeny virions. In contrast, N1 failed to immunoprecipitate any virus from cells infected with SVescS2 and transfected with pCAGGS-NDVHN, indicating that HN incorporation into SV is specific.
FIG. 1.
Incorporation of HN expressed from cDNA into progeny SV. 293T cells infected with an SV escape mutant (SVescS2) were transfected with a series of transient-expression vectors containing various HN genes. Cells were labeled with [35S]Trans-Label (100 μCi/ml) overnight, and the labeled progeny virions in the supernatants were immunoprecipitated with specific MAbs in the absence of detergent. (A) Cells were infected with SVescS2 and immunoprecipitated with an anti-SV HN MAb mixture (α-SV HN mix) (lane 1), anti-SV HN MAb S2 (lane 2), or anti-NDV HN MAb N1 (lane 3). Cells were infected with SVescS2, transfected with pCAGGS-SVHN (lane 4) or pCAGGS-NDV HN (lane 5), and immunoprecipitated with MAb S2 (lane 4) or N1 (lane 5). (B) Cells were infected with SVescS2 and transfected with pCAGGS containing the NDV HN or a chimeric HN gene (lanes A through E) and immunoprecipitated with MAb N1.
Because NDV HN was not incorporated into SV virions, we determined the level of NDV HN expression at the cell surface by FACS analysis. As shown in Fig. 2, NDV HN was expressed at the cell surface at a level sufficient to be detected. However, as shown in Fig. 1A, these NDV HN molecules were excluded from progeny SV virions, indicating the presence of a selective mechanism for incorporation of HN into SV.
FIG. 2.
Cell surface expression of HN expressed from cDNA. Cells were infected and transfected as described in the legend to Fig. 1. Live cells were immunostained by using a specific MAb against HN expressed from cDNA (MAb S2 for SV HN- or N1 for NDV and chimeric HN-expressing cells) followed by fluorescein isothiocyanate-conjugated anti-mouse IgG. Cells infected with SVescS2 but not transfected were reacted with a mixture of MAb S2 and N1 and used as a negative control. The solid portion of each histogram indicates results with positive cells.
Construction and characterization of chimeric HNs.
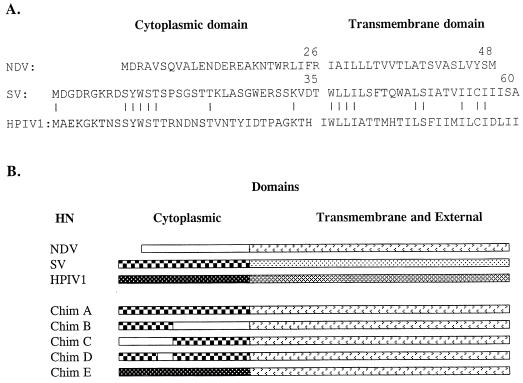
To identify which sequence on HN is required for specific incorporation of HN into SV virions, we used SV, NDV, and hPIV1 HNs (Fig. 3A) to construct chimeric HN molecules (Fig. 3B). SV or hPIV1 HN contains a cytoplasmic domain 9 or 8 amino acids longer (35 or 34 amino acids, respectively) than NDV HN (26 amino acids) (Fig. 3A). SV and hPIV1 HN are highly (72%) homologous (13). However, the sequences of their cytoplasmic domains are much less similar (23% identity), although there are five consecutive amino acids (SYWST) that are conserved between the two HNs. The cytoplasmic domain of NDV HN shows no homology with that of SV or hPIV1. Chimera A HN contains only the cytoplasmic domain from SV and the transmembrane and external domains from NDV. Chimera B HN contains the 14 N-terminal amino acids from SV, and the rest of its sequence is from NDV. Chimera C contains the 14 N-terminal amino acids from NDV and amino acids 15 to 35 from SV, and the rest of its sequence (transmembrane and external domains) is from NDV. Chimera D has the same sequence as chimera A, except the 5 amino acids (SYWST) at amino acids 10 to 14 were replaced with those of the corresponding region of the NDV HN (MDRAV). Chimera E contains the cytoplasmic domain from hPIV1 HN, and the rest of its sequence is from NDV.
FIG. 3.
Sequences and structures of chimeric HN proteins. (A) Comparison of the cytoplasmic and transmembrane domains of NDV, SV, and hPIV1 HN proteins. (B) Schematic diagram of chimera HN proteins. All chimera HNs contain transmembrane and external domains from NDV. Chimera A contains the whole cytoplasmic domain of SV. Chimera B has 14 N-terminal amino acids from SV, and the rest of its sequence is from NDV. Chimera C includes 14 N-terminal amino acids from NDV, followed by amino acids 15 to 35 of SV. Chimera D has the same structure as chimera A, but amino acids 10 to 14 were replaced with amino acids 1 to 5 of NDV HN. Chimera E contains the cytoplasmic domain of hPIV1.
These chimeric HNs were characterized for their expression at the cell surface, biological activities, and oligomer formation. Cell surface expression was determined by FACS analysis. All of the chimeras as well as NDV HN were expressed at the cell surface (Fig. 2). The levels of expression did not markedly differ among these proteins. To determine whether these chimeric HNs were biologically active, we measured and compared their NA activities expressed at the cell surface as described previously (5). All chimeric HNs had NA activities that were equivalent to that of the intact NDV HN (Table 1). This result was expected, because all of the chimeras contain the external domain of NDV HN, where NA activity is located. These results also show that the chimeric construction did not alter the antigenic or biologic activities of HN.
TABLE 1.
NA activities of chimera HNs
| Chimera HN | NA activity (%)a |
|---|---|
| NDV | 100.0 |
| A | 89.3 |
| B | 87.0 |
| C | 81.3 |
| D | 80.5 |
| E | 92.4 |
NA activities of the HNs expressed at cell surfaces were determined by the colorimetric assay. The values were standardized according to the amount of expressed HN as measured by cell surface ELISA (5). Values are averages of results from three experiments and are shown as percentages of NDV HN.
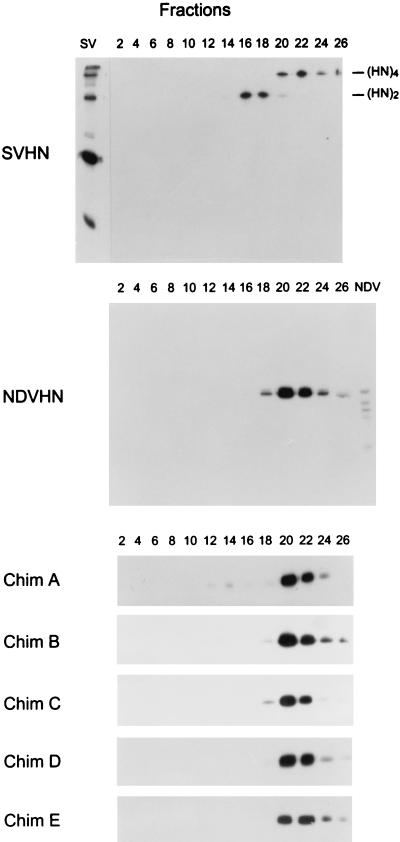
SV HN forms disulfide-linked dimers that noncovalently form homotetramers. Because the specific incorporation of HN into virions is presumed to be based on protein-protein interaction, changing the oligomeric form of HN may affect its specific incorporation into virions. Therefore, we next determined the oligomer formation of the wt and chimeric HNs by using sucrose gradient centrifugation. Consistent with previous findings (5), about 50% of SV HN sedimented at the positions of tetramers (Fig. 4) (fractions 20 to 26) and the remainder sedimented at the positions of dimers (fractions 16 to 18). The cysteine residue at position 123, located in the predicted stalk region in the external domain, is responsible for disulfide-linked homodimer formation of NDV HN (40). However, the HN protein of the NDV Kansas strain used in the present experiment has a tryptophan at position 123 and, therefore, does not form disulfide-linked dimers. However, as shown in Fig. 4, the majority of the HN molecules form homotetramers (fractions 20 to 26). Chimera A HN, in which the cytoplasmic tail sequence is that of SV HN, formed homotetramers, as was seen with the analysis of the NDV HN, suggesting that other regions (transmembrane and/or external domains) are responsible for formation of the HN homotetramers. All other chimera HNs were also detected on fractions 20 to 26, which correspond to tetramer forms of HNs, and no significant difference in homooligomer formation was observed between the HNs.
FIG. 4.
Sucrose gradient sedimentation analysis of oligomerization of chimera HN proteins. Cells transfected with HN-expressing vectors were labeled with [35S]Trans-Label (100 μCi/ml) for 30 min and chased for 2 h. Cell lysates were prepared and sedimented on 5 to 25% sucrose gradients. Twenty-seven fractions were harvested from the top of the gradients, and HN proteins in the fractions were immunoprecipitated with HN-specific MAbs and analyzed by using nonreducing polyacrylamide gels. The numeral above each lane corresponds to the fraction number. (HN)2, HN dimer; (HN)4, HN tetramer; Chim, chimera.
Incorporation of chimera HNs into SV.
The chimeric HNs that we constructed were biologically active (Table 1) tetramers (Fig. 4) and were expressed at the cell surface at levels similar to those of NDV and SV HNs (Fig. 2). We therefore determined whether the various HN chimeras were incorporated into progeny Sendai virions to identify the sequence responsible for the specificity of HN incorporation. Chimera A, in which only the cytoplasmic domain is from SV, was incorporated into Sendai virions (Fig. 1B), showing that the cytoplasmic domain is sufficient for the specific incorporation of HN into SV. Further, chimera B, which contains only the 14 N-terminal amino acids of SV, was incorporated into virions. In contrast, chimera C, which contains NDV sequences in its N-terminal region but SV sequences in the remainder of its cytoplasmic domain, was not incorporated into virions. These results indicate that the 14 N-terminal amino acids of the SV HN are sufficient for its specific incorporation into SV. Interestingly, chimera E, which contains the cytoplasmic domain of the hPIV1 HN, was also incorporated into SV (Fig. 1B). Despite the low (23%) homology between cytoplasmic domains of the SV and hPIV1 HNs, 5 consecutive amino acids (SYWST), located within the 14 N-terminal amino acids, are conserved (Fig. 3A). To determine whether these five amino acid residues were responsible for the incorporation of SV HN into virions, we examined the incorporation of chimeric HN D that replaces the five NDV amino acids in an otherwise SV cytoplasmic tail. Chimera D was not incorporated into virions (Fig. 1B), suggesting that these five consecutive amino acids are required for the specific incorporation of HN into SV. Quantitative analysis of HN incorporation into virions showed that the fraction of cDNA-derived HNs (wt SV HN and chimera A, B, and E HNs) incorporated into SV was about 1 to 3% of the HN expressed in cells (data not shown). In SV-infected cells, only 10 to 15% of the HN fraction was incorporated into virions. Therefore, in consideration of the transfection efficiency results shown in Fig. 2, the uptake of 1 to 3% of the cDNA-derived HN into virion is right in line with SV infection results.
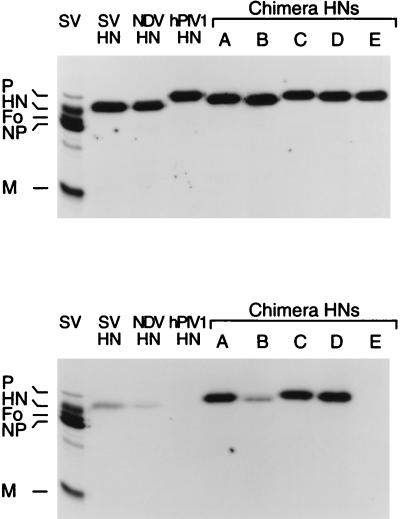
To determine whether HNs without the conserved sequences were completely excluded or were incorporated but to a limited extent, we performed Western blotting using a sensitive chemiluminescence method for protein detection. Culture supernatants of cells infected with SVescS2 and transfected with NDV or chimera HN cDNAs were collected, and the virions in the medium were purified by centrifugation through 50% glycerol. The HA titers in the culture supernatants were approximately the same among the cells infected with SV and transfected with or without various HN cDNAs (28 to 40 HAs). The amounts of cDNA-derived HN expressed at the cell surface were almost the same, except with chimera E, whose cDNA-derived HN was expressed at a level 1.5 to 2 times higher than those of the others (Fig. 5B). The amounts of cDNA-derived HNs in purified SV were determined by Western blotting with anti-NDV HN MAb N7, which reacts with denatured NDV HN. In a 5-s exposure, HN bands were clearly detected from cells transfected with chimera A, B, and E HNs but not NDV, chimera C, or chimera D HNs. However, a 10-min exposure revealed the presence of small amount of HN incorporated into SV from the samples transfected with NDV, chimera C, and chimera D HNs (Fig. 5). These results indicate that the HNs containing the five consecutive amino acids were incorporated into SV much more efficiently than those not containing the sequence.
FIG. 5.
Detection of cDNA-derived HNs in purified SV. Culture supernatants of cells infected with SV and transfected with various HN cDNAs were collected, and the SV virions in the medium were purified. The viruses were analyzed for amounts of cDNA-derived HNs by Western blotting. (A) NDV HN or chimera HNs were detected by MAb N7, which recognized denatured NDV HN molecules. Anti-SV NP MAb M52 was used to compare the amounts of virus in the samples. α, anti. (B) HA titers of culture supernatants of cells infected with SV and transfected with various HN cDNAs before purification. Amounts of cDNA-derived HNs at the cell surface were determined by cell surface ELISA and are shown in optical density units at a wavelength of 549 nm. −, no cDNA transfection.
Phosphorylation of the HN protein.
Of the five consecutive amino acids (SYWST) required for the specific incorporation of HN into progeny virions, four are potentially phosphorylatable. To establish whether phosphorylation of any of these amino acids might be related to the specific incorporation of HN into virions, we examined the in vivo phosphorylation of the chimeric HNs. To assess the level of HN expression and its phosphorylation, cells were transfected with the various HN expression vectors and labeled with [35S]Trans-Label (Fig. 6A) or [32P]orthophosphate (Fig. 6B) in parallel experiments and then the expressed HN was immunoprecipitated with a specific MAb and analyzed by SDS-PAGE. The wt SV and NDV HNs were phosphorylated, whereas hPIV1 HN was not. Among the chimeras, only chimera E (containing the cytoplasmic domain of hPIV1 and the transmembrane and external domains of NDV HN) was not phosphorylated. Together, these results show that residues in the NDV transmembrane and external domains were not phosphorylated. In addition, chimera A, which contains the cytoplasmic domain from SV but whose other regions are from NDV, was phosphorylated, indicating that the cytoplasmic domain of SV was phosphorylated. However, when considered in light of the nonphosphorylated status of chimera E, this finding suggests that none of the five consecutive amino acids (SYWST) was phosphorylated and that another residue(s) in the cytoplasmic domain led to the observed phosphorylation. The incorporation of chimera E into progeny virions indicates that phosphorylation of the cytoplasmic tail of HN is not required for its selective incorporation into SV.
FIG. 6.
Phosphorylation of the HN proteins expressed from cDNA. Cells transfected with the expression vector containing the HN gene were labeled with [35S]Trans-Label (A) or [32PO4]orthophosphate (B) in parallel, and labeled HN in the cell lysates was immunoprecipitated with specific MAbs. SV, purified 35S-labeled SV as a marker.
DISCUSSION
In this report, we showed that a five-residue sequence (SYWST) in the cytoplasmic domain of HN is required for this protein’s incorporation into SV virions. These amino acids are conserved between the cytoplasmic tails of HNs from SV and hPIV1, which otherwise are poorly homologous. Conserved consecutive amino acids (TYTLE) also occur in the cytoplasmic tails of the F glycoproteins of these two viruses (Fig. 7A), and other paramyxovirus glycoproteins display conserved amino acid motifs. For example, human and bovine PIV3 (hPIV3 and bPIV3, respectively) share a 5-residue sequence (PYVLT) in their F cytoplasmic tails and an 8-residue sequence (MEYWKHTN) in their HN cytoplasmic domains. Further comparison of these motifs revealed that 2 amino acids (YW) are conserved between the HNs of SV, hPIV1, hPIV3, and bPIV3, as are 2 amino acids (YXL) in the F protein (Fig. 7A). In addition, HN and F proteins derived from hPIV1 were incorporated into hPIV3 virions (45). We propose that the conserved sequences in the cytoplasmic tail regions of HN and F from these closely related viruses reveal a common strategy underlying glycoprotein incorporation and virion assembly.
FIG. 7.
Sequence comparison of the HN (H) and F cytoplasmic domains of paramyxoviruses (A) and morbilliviruses (B). The predicted cytoplasmic sequences of hPIV3 (12), bPIV3 (44), measles virus (MV) (1, 35), rinderpest virus (RP) (11, 47), dolphin morbillivirus (DM) (GenBank accession no. Z36978, 4), phocine distemper virus (PD) (19), and canine distemper virus (CD) (2, 8) are shown. Numbers above the sequence are those from the membrane. TM, transmembrane. Bold type indicates conserved amino acid residues.
The putative importance of the cytoplasmic domain of paramyxovirus glycoproteins is further supported. The distal half of the H protein cytoplasmic domain contains a 4-residue (AFYK) motif that is conserved among all morbilliviruses sequenced to date (Fig. 7B). Unlike the membrane-proximal portions, the C-terminal halves of the cytoplasmic domains of morbillivirus F proteins are almost completely homologous, suggesting the F protein’s involvement in virion formation. Further, extensive sequence analysis of viruses isolated from people with subacute sclerosing panencephalitis, a lethal disorder of the central nervous system that is causally related to persistent measles virus infection, revealed truncations and mutations in the C-terminal portion of the cytoplasmic domain of F. These changes may lead to the deficient virus formation observed in these cases (3, 6, 38).
These conserved motifs are distant from the membrane, perhaps suggesting their interaction with nucleocapsids during virus assembly. The cytoplasmic domain of the E2 glycoprotein of alphaviruses binds directly to the nucleocapsid (7, 22), and this interaction is required for virus budding (18, 30, 49). Further, Tyr400 and Leu402 (Sindbis virus numbering) bind to a hydrophobic pocket on the surface of the nucleocapsid protein. All of the motifs in paramyxovirus and morbillivirus glycoproteins that we have discussed contain a tyrosine residue. In particular, the paramyxovirus F glycoprotein contains the YXL motif identified in E2.
F may be essential for virus budding, but HN may enhance the efficiency of this process. At restrictive temperatures, a temperature-sensitive mutant of SV (SVts271) produces virions that lack HN (24, 32, 33, 42, 43, 48). Therefore, the HNs of paramyxoviruses seem to be dispensable in virus budding and likely play an auxiliary role. For example, virus particle production of G-deficient rabies virus mutants increased by about 30-fold in the presence of G (25). However, in paramyxovirus, levels of virion production by intact viruses and glycoprotein-deficient mutants have not been quantitatively compared, so the definitive role of glycoproteins in the budding process has not yet been established.
Most SV structural proteins (P, NP, L, M, and HN) are phosphorylated (21). Cellular protein kinase C ζ is responsible for the phosphorylation of SV and hPIV3 P proteins and is packaged into progeny virions (9, 16). In light of our results regarding the phosphorylation of the various wt and chimeric HNs, the HNs of NDV and SV are likely phosphorylated within the cytoplasmic domain. Chimera A, C, and D HNs were highly phosphorylated, while NDV and SV HNs were weakly phosphorylated. One possibility for the high phosphorylation of chimera A, C, and D HNs is that the combination of the SV cytoplasmic sequence adjacent to the NDV transmembrane region may enable additional amino acids to be phosphorylated. However, phosphorylation of HN cytoplasmic domain is apparently not required for the specific incorporation of HN into virions, since chimera E with its hPIV1 cytoplasmic domain was not phosphorylated and was incorporated into SV particles. The role of the phosphorylation of the HN cytoplasmic domain, therefore, remains unclear. It appears that the phosphorylation of SV or hPIV3 P proteins by protein kinase C ζ is required for virus replication (9, 16). However, phosphorylation of HN may not be required for virus replication; similarly, phosphorylation of the SV M protein also appears not to be essential for virus replication (36).
ACKNOWLEDGMENTS
This work was supported by grant AI-11949 from the National Institute of Allergy and Infectious Diseases, support grant CA-21765 from the Cancer Center (CORE), and the American Lebanese Syrian Associated Charities (ALSAC) of St. Jude Children’s Research Hospital.
We thank Amy L. Frazier for editorial assistance in preparing the manuscript.
REFERENCES
- 1.Alkhatib G, Briedis D J. The predicted primary structure of the measles virus hemagglutinin. Virology. 1986;150:479–490. doi: 10.1016/0042-6822(86)90312-0. [DOI] [PubMed] [Google Scholar]
- 2.Barrett T, Clarke D K, Evans S A, Rima B K. The nucleotide sequence of the gene encoding the F protein of canine distemper virus: a comparison of the deduced amino acid sequence with other paramyxoviruses. Virus Res. 1987;8:373–386. doi: 10.1016/0168-1702(87)90009-8. [DOI] [PubMed] [Google Scholar]
- 3.Billeter M A, Cattaneo R. Molecular biology of defective measles viruses persisting in the human central nervous system. In: Kingsbury D W, editor. The paramyxoviruses. New York, N.Y: Plenum; 1991. pp. 323–345. [Google Scholar]
- 4.Bolt G, Blixenkrone-Møller M, Gottschalck E, Wishaupt R G A, Welsh M J, Earle J A P, Rima B K. Nucleotide and deduced amino acid sequences of the matrix (M) and fusion (F) protein genes of cetacean morbilliviruses isolated from a porpoise and a dolphin. Virus Res. 1994;34:291–304. doi: 10.1016/0168-1702(94)90129-5. [DOI] [PubMed] [Google Scholar]
- 5.Bousse T, Takimoto T, Gorman W L, Takahashi T, Portner A. Region on the hemagglutinin-neuraminidase of human parainfluenza virus type-1 and Sendai virus important for membrane fusion. Virology. 1994;204:506–514. doi: 10.1006/viro.1994.1564. [DOI] [PubMed] [Google Scholar]
- 6.Cattaneo R, Schmid A, Spielhofer P, Kaelin K, Baczko K, ter Meulen V, Pardowitz J, Flanagan S, Rima B K, Udem S A, Billeter M A. Mutated and hypermutated genes of persistent measles viruses which caused lethal human brain diseases. Virology. 1989;173:415–425. doi: 10.1016/0042-6822(89)90554-0. [DOI] [PubMed] [Google Scholar]
- 7.Cheng R H, Kuhn R J, Olson N H, Rossmann M G, Choi H-K, Smith T J, Baker T S. Nucleocapsid and glycoprotein organization in an enveloped virus. Cell. 1995;80:621–630. doi: 10.1016/0092-8674(95)90516-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Curran M D, Clarke D K, Rima B K. The nucleotide sequence of the gene encoding the attachment protein H of canine distemper virus. J Gen Virol. 1991;72:443–447. doi: 10.1099/0022-1317-72-2-443. [DOI] [PubMed] [Google Scholar]
- 9.De B P, Gupta S, Gupta S, Banerjee A K. Cellular protein kinase C isoform ζ regulates human parainfluenza virus type 3 replication. Proc Natl Acad Sci USA. 1995;92:5204–5208. doi: 10.1073/pnas.92.11.5204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9a.Desphpande K L, Portner A. Structural and functional analysis of Sendai virus nucleocapsid protein NP with monoclonal antibodies. Virology. 1984;139:32–42. doi: 10.1016/0042-6822(84)90327-1. [DOI] [PubMed] [Google Scholar]
- 10.DuBridge R B, Tang P, Hsia H C, Leong P M, Miller J H, Calos M P. Analysis of mutation in human cells by using an Epstein-Barr virus shuttle system. Mol Cell Biol. 1987;7:379–387. doi: 10.1128/mcb.7.1.379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Evans S A, Baron M D, Chamberlain R W, Goatley L, Barrett T. Nucleotide sequence comparisons of the fusion protein gene from virulent and attenuated strains of rinderpest virus. J Gen Virol. 1994;75:3611–3617. doi: 10.1099/0022-1317-75-12-3611. [DOI] [PubMed] [Google Scholar]
- 12.Galinski M S, Mink M A, Pons M W. Molecular cloning and sequence analysis of the human parainfluenza 3 virus genes encoding the surface glycoproteins, F and HN. Virus Res. 1987;8:205–215. doi: 10.1016/0168-1702(87)90016-5. [DOI] [PubMed] [Google Scholar]
- 13.Gorman W L, Gill D, Scroggs R A, Portner A. The hemagglutinin-neuraminidase glycoproteins of human parainfluenza virus type 1 and Sendai virus have high structure-function similarity with limited antigenic cross-reactivity. Virology. 1990;175:211–221. doi: 10.1016/0042-6822(90)90201-2. [DOI] [PubMed] [Google Scholar]
- 14.Hogue B G, Nayak D P. Synthesis and processing of the influenza virus neuraminidase, a type II transmembrane glycoprotein. Virology. 1992;188:510–517. doi: 10.1016/0042-6822(92)90505-j. [DOI] [PubMed] [Google Scholar]
- 15.Horton R M, Hunt H D, Ho S N, Pullen J K, Pease L R. Engineering hybrid genes without the use of restriction enzymes: gene splicing by overlap extension. Gene. 1989;77:61–68. doi: 10.1016/0378-1119(89)90359-4. [DOI] [PubMed] [Google Scholar]
- 16.Huntley C C, De B P, Banerjee A K. Phosphorylation of Sendai virus phosphoprotein by cellular protein kinase C ζ. J Biol Chem. 1997;272:16578–16584. doi: 10.1074/jbc.272.26.16578. [DOI] [PubMed] [Google Scholar]
- 17.Jin H, Leser G P, Lamb R A. The influenza virus hemagglutinin cytoplasmic tail is not essential for virus assembly or infectivity. EMBO J. 1994;13:5504–5515. doi: 10.1002/j.1460-2075.1994.tb06885.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kail M, Hollinshead M, Ansorge W, Pepperkok R, Frank R, Griffiths G, Vaux D. The cytoplasmic domain of alphavirus E2 glycoprotein contains a short linear recognition signal required for viral budding. EMBO J. 1991;10:2343–2351. doi: 10.1002/j.1460-2075.1991.tb07773.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kövamees J, Blixenkrone-Möller M, Sharma B, Örvell C, Norrby E. The nucleotide sequence and deduced amino acid composition of the hemagglutinin and fusion proteins of the morbillivirus phocid distemper virus. J Gen Virol. 1991;72:2959–2966. doi: 10.1099/0022-1317-72-12-2959. [DOI] [PubMed] [Google Scholar]
- 20.Kretzschmar E, Buonocore L, Schnell M J, Rose J K. High-efficiency incorporation of functional influenza virus glycoproteins into recombinant vesicular stomatitis viruses. J Virol. 1997;71:5982–5989. doi: 10.1128/jvi.71.8.5982-5989.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lamb R A. The phosphorylation of Sendai virus proteins by a virus particle-associated protein kinase. J Gen Virol. 1975;26:249–263. doi: 10.1099/0022-1317-26-3-249. [DOI] [PubMed] [Google Scholar]
- 22.Lee S, Owen K E, Choi H-K, Lee H, Lu G, Wengler G, Brown D T, Rossmann M G, Kuhn R J. Identification of a protein binding site on the surface of the alphavirus nucleocapsid and its implication in virus assembly. Structure. 1996;4:531–541. doi: 10.1016/s0969-2126(96)00059-7. [DOI] [PubMed] [Google Scholar]
- 23.Markwell M A K, Fox C F. Protein-protein interactions within paramyxoviruses identified by native disulfide bonding or reversible chemical cross-linking. J Virol. 1980;33:152–166. doi: 10.1128/jvi.33.1.152-166.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Markwell M A K, Portner A, Schwartz A L. An alternative route of infection for viruses: entry by means of the asialoglycoprotein receptor of a Sendai virus mutant lacking its attachment protein. Proc Natl Acad Sci USA. 1985;82:978–982. doi: 10.1073/pnas.82.4.978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mebatsion T, König M, Conzelmann K-K. Budding of rabies virus particles in the absence of the spike glycoprotein. Cell. 1996;84:941–951. doi: 10.1016/s0092-8674(00)81072-7. [DOI] [PubMed] [Google Scholar]
- 26.Mitnaul L J, Castrucci M R, Murti K G, Kawaoka Y. The cytoplasmic tail of influenza A virus neuraminidase (NA) affects NA incorporation into virions, virion morphology, and virulence in mice but is not essential for virus replication. J Virol. 1996;70:873–879. doi: 10.1128/jvi.70.2.873-879.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Morrison T, Portner A. Structure, function, and intracellular processing of the glycoproteins of Paramyxoviridae. In: Kingsbury D W, editor. The paramyxoviruses. New York, N.Y: Plenum; 1991. pp. 347–382. [Google Scholar]
- 28.Naim H Y, Roth M G. Basis for selective incorporation of glycoproteins into the influenza virus envelope. J Virol. 1993;67:4831–4841. doi: 10.1128/jvi.67.8.4831-4841.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Niwa H, Yamamura K, Miyazaki J. Efficient selection for high-expression transfectants with a novel eukaryotic vector. Gene. 1991;108:193–200. doi: 10.1016/0378-1119(91)90434-d. [DOI] [PubMed] [Google Scholar]
- 30.Owen K E, Kuhn R J. Alphavirus budding is dependent on the interaction between the nucleocapsid and hydrophobic amino acids on the cytoplasmic domain of the E2 envelope glycoprotein. Virology. 1997;230:187–196. doi: 10.1006/viro.1997.8480. [DOI] [PubMed] [Google Scholar]
- 31.Peeples M E. Paramyxovirus M proteins: pulling it all together and taking it on the road. In: Kingsbury D W, editor. The paramyxoviruses. New York, N.Y: Plenum; 1991. pp. 427–456. [Google Scholar]
- 32.Portner A, Marx P A, Kingsbury D W. Isolation and characterization of Sendai virus temperature-sensitive mutants. J Virol. 1974;13:298–304. doi: 10.1128/jvi.13.2.298-304.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32a.Portner A, Scroggs R A, Kingsbury D W. Distinct functions of antigenic sites of the HN glycoprotein of Sendai virus. Virology. 1987;158:61–68. doi: 10.1016/0042-6822(87)90238-8. [DOI] [PubMed] [Google Scholar]
- 33.Portner A, Scroggs R A, Marx P A, Kingsbury D W. A temperature-sensitive mutant of Sendai virus with an altered hemagglutinin-neuraminidase polypeptide: consequences for virus assembly and cytopathology. Virology. 1975;67:179–187. doi: 10.1016/0042-6822(75)90415-8. [DOI] [PubMed] [Google Scholar]
- 34.Ray R, Roux L, Compans R W. Intracellular targeting and assembly of paramyxovirus proteins. In: Kingsbury D W, editor. The paramyxoviruses. New York, N.Y: Plenum; 1991. pp. 457–479. [Google Scholar]
- 35.Richardson C, Hull D, Greer P, Hasel K, Berkovich A, Englund G, Bellini W, Rima B, Lazzarini R. The nucleotide sequence of the mRNA encoding the fusion protein of measles virus (Edmonston strain): a comparison of fusion proteins from several different paramyxoviruses. Virology. 1986;155:508–523. doi: 10.1016/0042-6822(86)90212-6. [DOI] [PubMed] [Google Scholar]
- 36.Sakaguchi T, Kiyotani K, Kato A, Asakawa M, Fujii Y, Nagai Y, Yoshida T. Phosphorylation of the Sendai virus M protein is not essential for virus replication either in vitro or in vivo. Virology. 1997;235:360–366. doi: 10.1006/viro.1997.8701. [DOI] [PubMed] [Google Scholar]
- 37.Sanderson C M, Wu H-H, Nayak D P. Sendai virus M protein binds independently to either the F or the HN glycoprotein in vivo. J Virol. 1994;68:69–76. doi: 10.1128/jvi.68.1.69-76.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schmid A, Spielhofer P, Cattaneo R, Baczko K, ter Meulen V, Billeter M A. Subacute sclerosing panencephalitis is typically characterized by alterations in the fusion protein cytoplasmic domain of the persisting measles virus. Virology. 1992;188:910–915. doi: 10.1016/0042-6822(92)90552-z. [DOI] [PubMed] [Google Scholar]
- 39.Schnell M J, Buonocore L, Kretzschmar E, Johnson E, Rose J K. Foreign glycoproteins expressed from recombinant vesicular stomatitis viruses are incorporated efficiently into virus particles. Proc Natl Acad Sci USA. 1996;93:11359–11365. doi: 10.1073/pnas.93.21.11359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sheehan J P, Iorio R M, Syddall R J, Glickman R L, Bratt M A. Reducing agent-sensitive dimerization of the hemagglutinin-neuraminidase glycoprotein of Newcastle disease virus correlates with the presence of cysteine at residue 123. Virology. 1987;161:603–606. doi: 10.1016/0042-6822(87)90158-9. [DOI] [PubMed] [Google Scholar]
- 41.Strauss J H, Strauss E G, Kuhn R J. Budding of alphaviruses. Trends Microbiol. 1995;3:346–350. doi: 10.1016/s0966-842x(00)88973-8. [DOI] [PubMed] [Google Scholar]
- 42.Stricker R, Roux L. The major glycoprotein of Sendai virus is dispensable for efficient virus particle budding. J Gen Virol. 1991;72:1703–1707. doi: 10.1099/0022-1317-72-7-1703. [DOI] [PubMed] [Google Scholar]
- 43.Stricker R, Mottet G, Roux L. The Sendai virus matrix protein appears to be recruited in the cytoplasm by the viral nucleocapsid to function in viral assembly and budding. J Gen Virol. 1994;75:1031–1042. doi: 10.1099/0022-1317-75-5-1031. [DOI] [PubMed] [Google Scholar]
- 44.Suzu S, Sakai Y, Shioda T, Shibuta H. Nucleotide sequence of the bovine parainfluenza 3 virus genome: the genes of the F and HN glycoproteins. Nucleic Acids Res. 1987;15:2945–2958. doi: 10.1093/nar/15.7.2945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tao T, Durbin A P, Whitehead S S, Davoodi F, Collins P L, Murphy B R. Recovery of a fully viable chimeric human parainfluenza virus (PIV) type 3 in which the hemagglutinin-neuraminidase and fusion glycoproteins have been replaced by those of PIV type 1. J Virol. 1998;72:2955–2961. doi: 10.1128/jvi.72.4.2955-2961.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Thompson S D, Portner A. Localization of functional sites on the hemagglutinin-neuraminidase glycoprotein of Sendai virus by sequence analysis of antigenic and temperature-sensitive mutants. Virology. 1987;160:1–8. doi: 10.1016/0042-6822(87)90037-7. [DOI] [PubMed] [Google Scholar]
- 47.Tsukiyama K, Sugiyama M, Yoshikawa Y, Yamanouchi K. Molecular cloning and sequence analysis of the rinderpest virus mRNA encoding the hemagglutinin protein. Virology. 1987;160:48–54. doi: 10.1016/0042-6822(87)90042-0. [DOI] [PubMed] [Google Scholar]
- 48.Tuffereau C, Portner A, Roux L. The role of hemagglutinin-neuraminidase glycoprotein cell surface expression in the survival of Sendai virus-infected BHK-21 cells. J Gen Virol. 1985;66:2313–2318. doi: 10.1099/0022-1317-66-11-2313. [DOI] [PubMed] [Google Scholar]
- 49.Zhao H, Lindqvist B, Garoff H, von Bonsdorff C-H, Liljestorm P. A tyrosine-based motif in the cytoplasmic domain of the alphavirus envelope protein is essential for budding. EMBO J. 1994;13:4204–4211. doi: 10.1002/j.1460-2075.1994.tb06740.x. [DOI] [PMC free article] [PubMed] [Google Scholar]