Abstract
Despite technical and pharmacological advancements in recent years, including optimized therapies and personalized medicine, postoperative pain management remains challenging and sometimes undertreated. This review aims to summarize and update how genotype-guided therapeutics within personalized medicine can enhance postoperative pain management. Several studies in the area have demonstrated that genotype-guided therapy has the ability to lower opioid consumption and improve postoperative pain. Gene mutations, primarily OPRM1, CYP2D6, CYP2C9, COMT and ABCB1, have been shown to exert nuanced influences on analgesic response and related pharmacological outcomes. This review underscores the integration of pharmacogenetic-guided personalized medicine into perioperative care, particularly when there is uncertainty regarding opioid prescriptions. This approach leads to superior outcomes in terms of postoperative pain relief and reduced morbidity for numerous patients.
Keywords: pharmacogenetics, personalized medicine, postoperative pain, pain management, drug response, genetic markers, therapeutic efficacy
1. Introduction
Postoperative pain (POP) is a set of unpleasant sensory and emotional experiences that follows surgically induced tissue injury, being associated with physiological and behavioral responses [1,2]. Despite many technical and pharmacological advances over the last years, including pharmacogenetic within personalized medicine and optimized therapies, management of POP is still inadequate. High incidence of POP, around 65%, has been reported in the literature, with the median score for worst pain intensity varying from 5 to 9 on an 11-point numerical rating scale (0–10 NRS) [3,4,5,6,7,8,9,10]. Moreover, pain-induced complications are common and include pulmonary infections, increasing risks of cardiovascular, renal, and gastrointestinal dysfunction, immunodepression, postsurgical infection and poor wound healing [6,8,9,11]. Fear, anxiety, stress, and insomnia are among the psychological, physiological and behavioral responses that characterize the multidimensionality of the postoperative pain experience. It should also be stressed that POP, once established, is more resistant to analgesic treatment [6,8,12].
Despite the possibilities of POP management with different drugs and techniques, pain that persists after the surgical wound has healed is a major and largely unrecognized clinical problem. In fact, POP is followed by persistent pain in 10–50% of individuals in a wide variety of operations [12,13], and the intensity of the POP seems to be associated with the risk and severity of such pain chronification [12,14]. Therefore, ineffective POP management may have short- and long-term consequences, and lead to increased health care costs due to increases in length of stay, opioid abuse, higher morbidity, or residual disability [8,9,15].
Pharmacogenetics holds the promise of enhancing pain management by preemptively predicting an individual’s reaction to a particular analgesic before treatment beginning [16,17,18,19], achieved through analyzing polymorphisms in certain genes associated with altered drug metabolism. Moreover, it acknowledges that genes impacting drug receptors and other pain pathways can also play a significant role.
Over 90% of current medications, including analgesics, are metabolized by the cytochrome P450 (CYP450) enzymes. Thus far, the Human Genome Project has identified 57 CYP genes that influence drug metabolism [16,17]. Polymorphisms in CYP genes can alter enzyme function, leading to different phenotypes. Currently, we can categorize phenotypic changes in CYP enzymes in four groups: poor metabolizers, intermediate metabolizers, extensive metabolizers (normal) and ultra-rapid metabolizers [17,18]. The CYP1, CYP2, and CYP3 gene families are associated with drug metabolism, affecting the body’s response to pain. One of the most studied genes in this family is CYP2D6. This enzyme is responsible for the metabolism of commonly prescribed analgesics, such as codeine, tramadol and dihydrocodeine [17]. CYP3A4 has also been found to be involved in opioid metabolism [16]. Another important gene in analgesic metabolism is CYP2C9, as it is one of the metabolic enzymes for methadone and many NSAIDs [16,17].
Genes associated with specific drug receptors can also change drug efficacy. One example of this is gene OPRM1, which encodes μ-Opioid Receptor. This gene is associated with variable potency and effectiveness of morphine among patients [16].
In this context, pharmacogenetic advancements within personalized medicine offer promising strategies for optimizing therapies and improving outcomes in pain management [19,20]. Pharmacogenetics aims to refine and optimize the prescription process of medications tailored to each person by healthcare practitioners [16,17,19]. Consequently, this will lead to the development of best practice procedures in healthcare, resulting in better assistance and quality of life for the patients [4,6,8,21].
Therefore, this review summarizes and updates how the genotype-guided therapeutics within personalized medicine can enhance postoperative pain management in patients undergoing surgery, approaching important topics such as pain relief, morbidity, and quality of life.
2. Postoperative Pain Management and Pharmacogenetics
High percentages of moderate to severe POP can be found in the literature [3,4,5,6,7,8,9], leading to interferences in vital functions, causing adverse health effects, and affecting capacity of recovering [6,7,8,9,11,15]. The best POP management is ethical and imperative [22,23,24] not only for pain relief, but also to avoid the related morbidity with deleterious consequences in the short and long terms. Pre-emptive pharmacogenomic (PGx) testing emerges as an invaluable tool in guiding drug selection and determining the appropriate dosage, including in patients undergoing elective surgery [16,25,26].
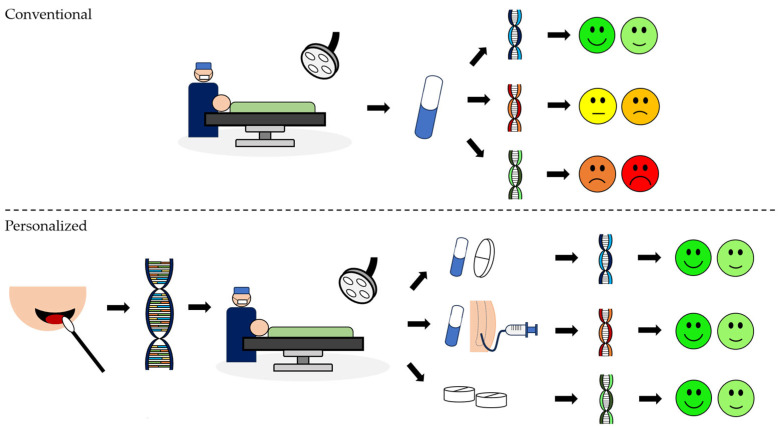
Genotype-guided therapeutics can lead to a more effective analgesic protocol, enhancing pain management and reducing related complications, particularly in high-risk patients. This procedure involves DNA extraction and evaluation to determine genetic factors relevant to pain response and medication metabolism. The next step is to tailor the analgesic protocol according to the results of the genetic tests, ensuring personalized and optimized pain relief for each patient (Figure 1).
Figure 1.
Differences between conventional and personalized analgesic approaches: Conventional approach: The same protocol in a conventional analgesic regimen can elicit different responses in different individuals. Personalized: In a programmed surgery, after an informed consent, buccal swabs provide a convenient means of obtaining a DNA sample. Following the collection, the DNA is extracted. Based on selected genes, the tailored analgesic protocol is developed to suit individual needs and optimize pain management. Legend: two green faces represent none to mild pain; two yellow faces represent mild to moderate pain, and two red faces represent moderate to severe pain.
Recent studies have indicated that implementing pharmacogenetic tests can reduce opioid consumption and side effects, together with lowering post-operative pain levels for most patients. Genetic variations, mainly in OPRM1, CYP2D6, CYP2C9, CYP3A4, COMT, ABCB1, and SLC22A1, were demonstrated to influence analgesic response, side effects and postoperative chronification, highlighting the importance of personalized medicine in POP management.
3. Comprehensive Analysis of Clinical Studies
A comprehensive literature review was conducted and divided into two analyses: 1. controlled clinical trials, and 2. prospective clinical trials. The inclusion criteria applied for the first analysis comprised adult human participants and randomized controlled clinical trials or controlled clinical trials published in English and with at least 10 patients. The inclusion criteria for the second analysis comprised adult human participants and only prospective studies published in English with at least 50 patients. The exclusion criteria for both analyses comprised studies in which participants did not undergo surgical procedures and those that investigated non-postoperative pain.
Three electronic bibliographic databases, Web of Science, PubMed, and Scopus, were used for the manuscript search, and this was carried out between December 2023 and January 2024. The search strategy was built up combining MeSH terms in PubMed and keywords in both Scopus and Web of Science.
The PubMed search yielded 49 articles using the following query: “Pharmacogenetics”[Mesh] AND “Pain, Postoperative”[Mesh]. A total of 47 articles were identified in Scopus using “Pharmacogenetics”, “Postoperative pain” and “Pain Management” as keywords. Also, 61 articles were identified in Web of Science using “Pharmacogenetics” and “Postoperative pain”. An additional manual search found 5 manuscripts.
One of the authors screened titles and abstracts to assess their relevance and alignment with the objective of this study. After this initial selection, a full-text review was conducted, and information from each selected study was extracted, including the characteristics of the participants and the conclusions drawn. Then, these were systematically compared and evaluated, ensuring that only studies with the appropriate methodology and outcomes were included in the systematic review and that the results were valid and reliable.
The data synthesis process was performed by the extraction of the relevant data from each study, including studied population, screened genes, pharmacogenetic approach, intervention, and main results. The other two authors critically reviewed the study selection process. Whenever discrepancies were present, the solution was found through consensus. Kappa test for agreement was 0.90.
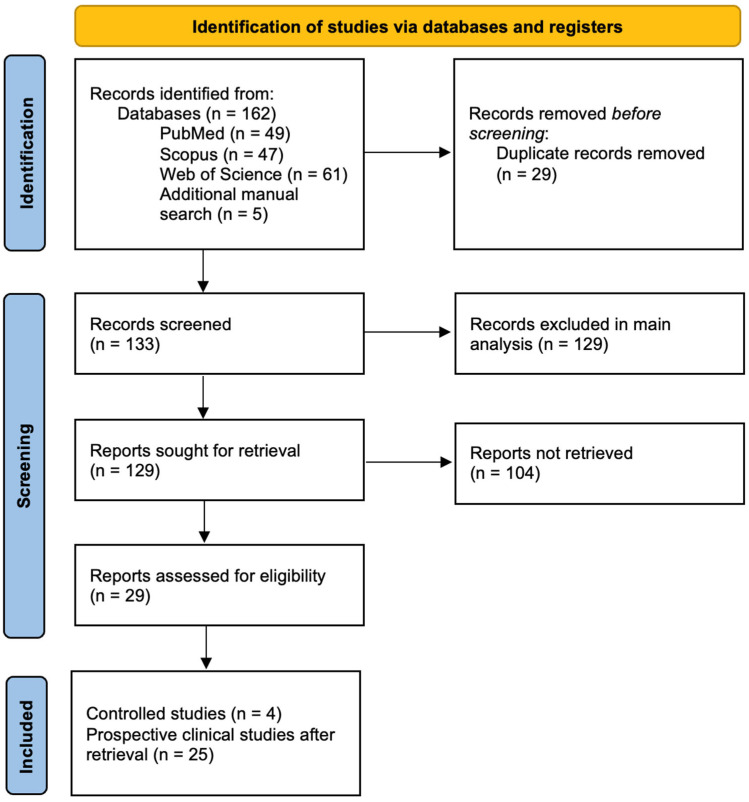
The initial search comprised 162 articles, of which 29 were removed since they were duplicated records, making a total of 133 articles. Records were first screened for inclusion in the main analysis, after which reports were retrieved for a secondary analysis of prospective clinical studies. Studies were excluded based on various criteria: study design (n = 108), population not meeting inclusion criteria (n = 13), assessment of non-post-operative pain (n = 4), full text not being available (n = 3), and publication language other than English (n = 1). For the secondary analysis, 25 non-controlled studies were retrieved, leaving 104 reports unretrieved. Referenced articles that were found pertinent through manual search, despite not being found with the set parameters of the initial search, were also included (Figure 2).
Figure 2.
PRISMA flowchart.
According to the established criteria, three randomized controlled clinical trials and one controlled clinical trial were included for the first analysis, and their descriptions are provided in Table 1.
Table 1.
Concise overview of relevant studies on pharmacogenetics for postoperative pain comparing genotype-guided versus standard care.
| Ref. | Participants/Country/Tools | Intervention/Treatment | Pharmacogenetic | Main Results |
|---|---|---|---|---|
| Hamilton 2022 [27] |
|
|
16 genes, including CYP2D6, CYP2C9, OPRM1, CYP3A4 and CYP1A2 |
|
| Thomas 2021 [28] |
|
|
CYP2D6 |
|
| Hamilton 2020 [29] |
|
|
7 genes, including CYP2D6, CYP2C9, OPRM1, CYP3A4 and CYP1A2 |
|
| Senagore 2017 [30] |
|
|
9 genes, including CYP2D6, CYP2C9, OPRM1, CYP3A4 and CYP1A2 |
|
Legend: Ref.—reference, %—percentage; USA—United States of America, PROMIS—Patient Reported Outcome Measurement Information System; OBAS—Overall benefit of analgesia score.
The main results in Table 1 demonstrated a significantly lower opioid consumption associated with employing genotype-guided therapeutics across all selected studies [27,28,29,30]. This intervention was also associated with lower pain levels in post-operative pain in two studies [27,29]. In the remaining two studies [28,30], no improvement in pain levels was reported. However, Senagore et al. [30] reported a lower incidence of analgesic-related side effects.
Given the limited quantity of controlled clinical studies, subsequent research was conducted, including exclusively prospective clinical trials. The goal was to compile clinical studies involving multiple pharmacogenetic tests for a more comprehensive analysis (Table 2).
Table 2.
Concise overview of findings from prospective cohort clinical trials on postoperative pain.
| Ref. | Participants/Country/Pain Assessm. | Intervention/Treatment | Pharmacogenetic | Main Results |
|---|---|---|---|---|
| Zhou 2023 [31] |
|
2 Groups based on OPRM1 A118G polymorphisms: wild-type (A/A) and heterozygous (G/A), and mutant homozygous (G/G) | OPRM1 A118G genotypes |
|
| Saiz-Rodríguez 2021 [32] |
|
3 Groups:
|
12 genes, including CYP2C9, CYP2C19, CYP3A4, CYP2D6, ABCB1 and OPRM1 |
|
| Matic 2020 [33] |
|
2 Groups based on intraoperative medication:
|
COMT and OPRM1 |
|
| Stamer 2016 [34] |
|
3 genotype groups: 0 active OCT1 allele (OCT1 poor transporter), 1 active OCT1 allele (heterozygous for 1 inactive allele), and 2 active OCT1 alleles (OCT1 extensive transporter). 4 CYP2D6 activity groups: CYP2D6 PM, CYP2D6 IM, CYP2D6 EM and CYP2D6 UM |
SLC22A1 (OCT1) and CYP2D6 |
|
| Tan 2016 [35] |
|
Groups were created based on COMT rs4633, rs4818, rs4680 polymorphisms and COMT haplotype | COMT |
|
| Dong 2015 [36] |
|
3 Groups according to CYP2D6*10 allele: wild-type (CYP2D6*1/*1), heterozygous (CYP2D6*1/*10) and mutant homozygous (CYP2D6*10/*10) | CYP2D6 |
|
| Seripa 2015 [37] |
|
4 Groups according to CYP2D6 activity: UM, EM, IM and PM POP treatment was multimodal, including opioids (tramadol, morphine), NSAIDs (ketoprofen) plus metoclopramide, and ranitidine |
CYP2D6 |
|
| Wu 2015 [38] |
|
3 Groups according to CYP2D6 polymorphisms: wild-type (W/W), heterozygous (M/W) and mutant homozygous (M/M) Cumulative Analgesic Consumption of Fentanyl during Postoperative Period |
CYP2D6 |
|
| Zhang 2015 [39] |
|
Groups were created based on polymorphisms in COMT SNPs (rs6269, rs4633, rs4818, rs4680) and their haplotypes | COMT |
|
| Boswell 2013 [40] |
|
2 Groups according to OPRM1 polymorphisms: wild-type (AA): both heterozygous and mutant homozygous (AG/GG) | OPRM1 |
|
| Candiotti 2013 [41] |
|
3 Groups according to ABCB1 C3435T genotype: wild-type (CC), heterozygous (CT) and mutant homozygous (TT) | ABCB1 |
|
| Henker 2013 [42] |
|
Groups were created based on polymorphisms in OPRM1 A118G rs1799971, rs1799972, COMT SNPs (rs6269, rs4633, rs4818, rs4680) and their haplotypes | COMT and OPRM1 |
|
| Slanar 2012 [43] |
|
3 groups according to ABCB1 genotype: wild-type (3435CC), heterozygous (3435CT) and mutant homozygous (3435TT) 4 CYP2D6 phenotype groups: CYP2D6 PM, homozygous CYP2D6 EM, heterozygous CYP2D6 EM and CYP2D6 UM |
CYP2D6 and ABCB1 (MDR1) |
|
| Tan 2012 [44] |
|
2 Groups according to CYP3A4 polymorphisms: wild-type (CYP3A4*1/*1) and mutant heterozygous (CYP3A4*1/*18) | CYP3A4 |
|
| De Gregori 2012 [45] |
|
Groups were created based on polymorphisms in OPRM1 A118G rs1799971, COMT SNPs (rs6269, rs4633, rs4818, rs4680) and their haplotypes, and 7 other UGT2B7 SNPs | COMT, OPRM1 and UGT2B7 |
|
| Zhang 2011 [46] |
|
3 groups according to CYP3A5 genotype: wild-type (CYP3A5*1/*1), heterozygous (CYP3A5*1/*3) and mutant homozygous (CYP3A5*3/*3) | CYP3A4 and CYP3A5 |
|
| Yuan 2011 [47] |
|
3 Groups according to CYP3A4 polymorphisms: wild-type (*1/*1), heterozygous (*1/*1G) and mutant homozygous (*1G/*1G) | CYP3A4 |
|
| Zwisler 2010 [48] |
|
2 Groups according to CYP2D6 activity: 8.9% Poor Metabolizers (PM), 91.1% Extensive Metabolizers (EM). | CYP2D6 genotypes |
|
| Fukuda 2009 [49] |
|
SNP genotypes: A118G and IVS3 + A8449G grouped into major allele homozygote (AA) and combined heterozygote/minor allele homozygote (AG + GG), resulting in 4 genotype groups. | OPRM1 |
|
| Tan 2009 [50] |
|
3 Groups according to OPRM1 polymorphisms: wild-type (AA) heterozygous (AG) and mutant homozygous (GG) | OPRM1 |
|
| Sia 2008 [51] |
|
3 Groups according to OPRM1 polymorphisms: wild-type A118 homozygous (AA), heterozygous (AG) and mutant G118 homozygous (GG) | OPRM1 |
|
| Chou 2006 [52] |
|
3 Groups according to OPRM1 polymorphisms: wild-type A118 homozygous (AA), heterozygous (AG) and mutant G118 homozygous (GG) | OPRM1 |
|
| Chou 2006 [53] |
|
3 Groups according to OPRM1 polymorphisms: wild-type A118 homozygous (AA), heterozygous (AG) and mutant G118 homozygous (GG) | OPRM1 |
|
| Wang 2006 [54] |
|
3 Groups were created according to CYP2D6*10 polymorphism: wild-type (Group I), heterozygous (Group II) and mutant homozygous (Group III) | CYP2D6 |
|
| Stamer 2003 [55] |
|
2 Groups according to CYP2D6 activity: CYP2D6 EM (mutant heterozygous) and CYP2D6 PM (mutant homozygous) | CYP2D6 |
|
Legend: Ref.—reference, PM—poor metabolizers, IM—intermediate metabolizers, UM—ultra-rapid metabolizers, SNP—single nucleotide polymorphism, NRS—Numeric Pain Rating Scale, VAS—Visual Analog Scale, RSS—Ramsay Sedation Scale, POP—postoperative pain.
Out of the 29 selected studies, buccal cheek swabs were utilized to collect DNA samples in five studies [27,28,29,30,42], while peripheral blood samples were employed in the remaining 24 studies. DNA extraction technique was not specified in four studies [27,28,29,42].
In the other studies, various DNA extraction methods were utilized: four studies used a conventional phenol–chloroform method [36,46,49,54], four studies employed a Gentra Puregene Blood Kit [34,35,50,51], three studies utilized a QIAamp DNA Blood Mini Kit [43,44,49], two studies used the salting-out method [37,48], two used E.Z.N.A SQ Blood DNA Kit [38,39], while one study each employed a Lab-Aid 820 Midi [31], a MagNA Pure LC DNA Isolation Kit [32], a Wizard Genomic-DNA Purification kit [45], a MagNA Pure LC 2.0 instrument [33], a QIAGEN EZ-1 BioRobot and Blood kit [40], a QIAamp DNA Blood Midi Kit [41], the Guanidinium isothiocyanate method [47], the Chelex method [52], and a PureGene DNA Purification Kit [53].
4. Main Genes Related to Postoperative Pain
4.1. OPRM1
OPRM1 encodes the mu opioid receptor, a pivotal drug target. Consequently, the significance of OPRM1 lies in its important involvement in opioid response and pharmacodynamics, as evidenced by several clinical studies [27,29,30,31,32,40,49,50,51,52,53], with significant improvement in POP outcomes, allowing for reduced opioid dosage and consequently fewer side effects [27,29,30].
It was demonstrated that OPRM1 mutant homozygote (A118G) patients required more morphine to reach analgesia [50,51,52,53] and reported higher pain scores [50,51]. On the other hand, these mutant homozygote patients were linked to a lower incidence of nausea [50,51] compared to OPRM1 wild-types (A118A). In one study, both mutant heterozygotes and homozygotes were found to have a better response to tramadol [32]. Mutation carriers were also associated with a higher incidence of opioid side effects [40]. Subjects carrying a IVS3 + A8449G SNP were also found to have a different analgesic response, requiring less fentanyl for post-operative pain control [49].
These findings underscore the potential of OPRM1 genetic testing to tailor opioid prescriptions for surgical patients, ensuring more effective pain management strategies while minimizing adverse effects based on individual genetic profiles.
4.2. CYP2D6
The CYP2D6 gene encodes an enzyme responsible for metabolizing a wide range of drugs, including antidepressants, antipsychotics, and opioids, affecting their efficacy and toxicity in individuals. Variations in the CYP2D6 gene can lead to differences in drug metabolism rates, influencing individual responses to medications.
When tailoring analgesic prescriptions based on genetic tests, it is also important to understand if people are poor or rapid metabolizers. Regarding codeine and tramadol, this enzyme has a crucial role, as it is responsible for the conversion to their active metabolites, morphine and O-desmethyltramadol, respectively. In these cases, poor metabolizers may be undermedicated under regular analgesic protocols and may be under increased risk for developing chronic pain due to the less effective POP management [17]. Furthermore, studies focusing on mutant homozygous CYP2D6 poor metabolizer phenotype patients revealed elevated pain levels [36,43], higher tramadol consumption [34,36,54,55], and an increased need for rescue medication [55]. Notably, mutant homozygous CYP2D6 patients required higher fentanyl dosages to achieve adequate analgesia [38]. Conversely, ultra-rapid metabolizers will quickly be under high systemic analgesic concentrations, a matter of concern when using opioids due to the high risk of side effects, including respiratory depression and potential for toxicity. It is also important to consider that patients will experience a very strong and short-lasting analgesic effect, and pain returns very fast [17,48]. Additionally, when a breastfeeding mother metabolizes codeine rapidly, it can significantly increase the risk of opioid overdose in infants who are nursing [56,57].
This emphasizes the role of CYP2D6 genetic testing when planning the use of opioids such as tramadol and fentanyl prescriptions for POP, allowing for more precise pain management strategies based on individual metabolic profiles.
4.3. CYP2C, CYP2C19, CYP2C9 and CYP2D6
Despite opioids being frequently measured in genetic tests, other analgesic drugs such as NSAIDs play a crucial role in managing postoperative pain. NSAIDs undergo primary metabolism associated with the CYP2C9 isoenzyme, with metabolizer categories similar to those of CYP2D6. This metabolism pattern can lead to increased plasma concentrations and extended half-life in poor or intermediate metabolizers, elevating the risk of side effects and toxicity.
The CYP2C gene locus also plays a complex role in predisposing individuals to peptic ulcer disease (PUD). This predisposition varies depending on the type and dosage of nonsteroidal anti-inflammatory drugs (NSAIDs) used and the patient’s complement of single nucleotide polymorphisms at the CYP2C gene locus, including those at CYP2C9 (where low-activity variants can increase exposure to NSAIDs) and the gain-of-function polymorphism at CYP2C19 (which enhances the metabolism of gastro-protective arachidonic acid). While CYP2C19 has a minor role in NSAID metabolism, it has been associated with peptic ulcer disease, particularly with the CYP2C19*17 variant, adding complexity and heightening the risk of gastrointestinal side effects, especially in PM patients exposed to NSAIDs for postoperative pain management [58,59,60,61,62,63].
Although not usually used in postoperative pain management, when employing multimodal analgesia, tricyclic antidepressants (TCAs) can be a valuable addition to the treatment arsenal. Amitriptyline, a TCA, undergoes primary metabolism through the CYP2C19 and CYP2D6 pathways. CYP2C19 produces active metabolites such as nortriptyline, while CYP2D6 forms a less active 10-hydroxy metabolite. Variations in these enzymes, such as “CYP2D6 ultrarapid metabolizers” or “CYP2C19 poor metabolizers,” significantly impact drug metabolism. Notably, the metabolism of TCAs, including amitriptyline, involves both CYP2D6 and CYP2C19, highlighting the crucial role of pharmacogenetics in tailoring antidepressant therapy to individual patients [64].
While genetic testing often prioritizes opioids in pain management, the inclusion of other analgesics is crucial for personalized pain management. Thus, genetic testing should encompass a broader spectrum of analgesics, recognizing the advantages of multimodal approaches over single-drug protocols.
4.4. ABCB1
The ABCB1 gene, also known as the multidrug resistance protein 1 (MDR1) gene, encodes a membrane transporter protein involved in the efflux of various drugs and toxins from cells, impacting their pharmacokinetics and efficacy. A significant association was demonstrated between the rs9282564 polymorphisms in the ABCB1 gene and opioid-induced respiratory depression in children. Additionally, ABCB1 SNP rs2229109 indicated a connection with postoperative morphine doses, suggesting that genetic influences play a role in individual responses to opioids. These associations emphasize the importance of considering multiple genetic factors in tailoring postoperative pain management strategies, especially in pediatric populations undergoing common surgical procedures like tonsillectomy [65].
Increasing evidence suggests that distinct genes and mechanisms play a role in each aspect of opioid response [66]; thus, analgesic effect is a result of the cumulative impact of multiple genes [18].
The impact of ABCB1 mutations on pain relief with tramadol presents conflicting outcomes. In one investigation [32], patients carrying ABCB1 mutated alleles demonstrated higher pain relief with tramadol, suggesting a potential correlation between specific genetic variations and enhanced analgesic response. However, in a different study [43], no discernible differences were observed in terms of drug consumption, adverse reactions, rescue analgesic usage, or pain levels among different ABCB1 genotype subgroups treated with tramadol. This discrepancy highlights the complexity of the genetic factors and others influencing the response to tramadol. It also underscores the need for further elucidation of the underlying mechanisms and variations in patient outcomes.
4.5. CYP3
Less directly associated genes may also contribute to enhancing POP management. It was demonstrated that CYP3A4*1G homozygote individuals exhibited a notable pattern, consuming less fentanyl, displaying higher plasma concentrations, and requiring less rescue medication compared to other groups, such as CYP3A4*18, where no significant differences were observed. Furthermore, the interaction between CYP3A53 and CYP3A4*1G demonstrated an additional layer of complexity, contributing to lower fentanyl consumption [44,46,47]. These insights underscore the potential of CYP3A4*1G and CYP3A4*18 in tailoring pain management strategies.
However, additional studies should explore the broader applicability of the cytochrome P450 gene variations in diverse populations and surgical settings, providing a comprehensive understanding of whether these genes should be included in the panel for effective and reliable personalized pain management strategies. Clarifying the role of these genes through expanded research will contribute crucial insights to guide the development of more targeted and efficient approaches in postoperative pain care.
4.6. COMT
The COMT gene is located at the gene map locus of 22q11.2 and encodes the enzyme Catechol-O-methyltransferase, responsible for the metabolism of catecholamines, such as adrenaline, noradrenaline and dopamine.
Evidence has demonstrated the significance of numerous COMT single nucleotide polymorphisms (SNPs) in enzymatic activity and in modulating an individual’s sensitivity to/perception of pain. Variations in COMT activity have been associated with pain sensitivity haplotypes. The most extensively studied SNP in the COMT gene is rs4680, identified as a functional polymorphism. This involves a guanosine (G) to adenosine (A) transition, resulting in a valine (Val) to methionine (Met) amino acid substitution. This substitution may lead to three possible SNP genotypes: GG (Val/Val) genotype, characterized by high enzymatic activity; AA (Met/Met) genotype, associated with defective enzymes; and GA (Val/Met) genotype, demonstrating moderate enzymatic activity [67,68]. In the existing literature, three primary haplotypes have shown a strong correlation with sensitivity to experimental pain [69]. These include low (LPS), average (APS), and high (HPS) pain sensitivity haplotypes.
The combination of COMT SNPs rs6269, rs4633, rs4818, and rs4680 determine LPS (GCGG), APS (ATCA) and HPS (ACCG) COMT haplotypes. These haplotypes contribute to the individual variation in postoperative opioid consumption: theoretically, under the same noxious stimulus, the greater the sensitivity to pain, the more opioid for analgesia [65].
Interestingly, there are contradictory findings for rs4680. The effects of the COMT gene haplotypes on the analgesic doses in pain are not so linear: it was demonstrated that a mutant homozygous rs4680 genotype was associated with higher pain scores [42] and had a lower opioid consumption [35,45]. The mutant G allele of rs4818 was also associated with higher pain levels [35], and mutant homozygous (G/G) were found to have a lower opioid consumption in the post-operative period [42].
Regarding COMT haplotypes, contradictory results were found: in one study, it was found that having at least one copy of the LPS haplotype resulted in higher pain scores and opioid consumption [42], whereas another study stated that LPS carriers had lower pain scores [35]. The APS/APS diplotype was also associated with a lower opioid consumption [45]. The HPS haplotype also had conflicting results, having been shown both to increase [39] and to lower opioid consumption [33].
Indeed, the reasons for the varied effects of COMT gene haplotypes on analgesic doses during acute and chronic pain remain unclear. It is plausible that conditions such as cancer and other chronic pain types entail prolonged or repetitive pain stimuli, potentially leading to increased neuronal enkephalin consumption and compensatory upregulation of mu opioid receptors. This process may result in the modulation of pain sensitivity, consequently reducing the requirement for opioids [33,39].
The impact of COMT haplotypes on pain also appears to vary with gender and ethnic background. Women carrying the COMT HPS haplotype (associated with low COMT activity) tend to experience heightened pain and exhibit decreased hepatic COMT levels [31,70], potentially linked to estrogenic levels.
The relationship between gene–gene networks, such as COMT and OPRM1, may also influence pain susceptibility and the efficacy of opioid analgesics. Patients having AA (Met/Met) of COMT rs4680 and AG of OPRM1 rs1799971 consumed the largest amount of opioid compared to those having other combinations [71]. Additionally, no association between different COMT haplotypes and symptoms such as nausea, vomiting, or dizziness was found [39].
Hence, this biomarker could prove invaluable in a multifactorial model integrating biological, physical, and social factors to predict both pain experience and opioid response, all while minimizing potential side effects. Its evaluation should be conducted in conjunction with other genetic and non-genetic factors.
5. Precision Personalized Medicine
To mitigate opioid misuse, alongside pharmacogenomic testing, precision personalized medicine for pain management should adopt an individualized approach, optimizing the control of postoperative pain while reducing associated side effects, especially with opioids. This rationale may justify initially targeting high-risk patients for poor pain control or chronification (e.g., major surgeries) for testing, with subsequent expansion to other patient groups [19,26].
Additionally, pain management should include a multimodal approach, since it is safer and better than single drugs in reducing POP, mainly in more invasive surgeries [72,73,74,75]. Evidence-driven multidisciplinary teams ensure optimal POP management with minimal morbidity, promoting enhanced analgesia. Tailored POP protocols aligned with the surgical procedure type effectively reduce pain and analgesic consumption, minimize side effects, discourage the use of non-recommended on-demand analgesics, and expedite recovery [27,28,29,30,72,73,74,76] (Table 2). To achieve this, it is recommended to utilize various analgesic agents, transition between opioids, adjust administration routes, implement patient-controlled analgesia, and administer local anesthetics whenever necessary. Frequent assessment and recording of pain should also be conducted [74,77,78,79,80,81,82].
6. Postoperative Pain and Pharmacogenetics Cost-Effectiveness
In addition to reducing POP, multimodal analgesia helps in decreasing the analgesic rate and severity of side effects. Moreover, pharmacy represents a very reduced percentage of the surgery costs, less than 5%. Much higher expenses are involved in the prolonged length of stay in the hospitals due to the undertreated POP that interferes with patients’ function and consequently recovery [9,15], or even worse, leads to chronic postoperative pain [1]. Since POP is more reported in young individuals [4,7,76,83], enhanced pain management can lead to indirect savings by reducing occurrences of work absenteeism. Furthermore, PGx testing will help clinicians choose treatments and doses objectively, rather than relying on only clinical judgement [19,84]. In this context, the expenses associated with genetic tests are relatively minimal when weighed against the substantial toll of adverse effects, hospitalizations, and enduring chronic pain. Moris et al. even suggested that the majority of PGx-guided treatment is cost-effective or cost-saving [85], although studies for analgesic PGx-guided treatment cost effectiveness are still lacking.
Therefore, it is expected that changes in the current model of analgesia, mainly by personalized medicine based on genetic tests, will contribute to better pain management and healthcare cost reductions [19,26]. Genetic material can be collected using saliva, reducing any unnecessary inconvenience for the patient while equipping the clinician with a potent resource to enhance the patient’s care by creating a pain-related gene panel [19]. Understanding these genetic differences becomes paramount, since small variations can represent a huge impact in several individuals’ responses to medications. Adverse drug reactions (ADRs) are noxious, unintended, and mostly due to genetic variations, leading to a huge burden on the healthcare system. ADRs are responsible for a high number of hospitalizations and deaths, as well as huge healthcare costs that cannot be precisely estimated due to poor reporting [78,80,81,86].
This is especially important when there is a family history of adverse reactions or when individuals do not respond favorably to certain medications. Specific populations may harbor higher occurrences of certain PGx variants, like the CYP2C19 loss-of-function variants that are more frequent in individuals with East Asian, South Asian, or Pacific Islander heritage [87,88]. Furthermore, African Americans report more postoperative pain than Caucasians, indicating widespread disparities. These stem from complex factors like communication, attitudes, and healthcare accessibility [89]. In this situation, PGx testing could benefit patients, allowing for more effective pain management.
Due to the significant impact of individual genetic data on both the individual and their family and descendants, ensuring the ethical use of genetic information becomes of great importance. Ethical concerns regarding PGx testing, such as privacy, confidentiality, discrimination, and incidental findings [90], should be included in PGx guidelines and regulations.
7. Platforms and Evidence-Based Protocols
One of the challenges in incorporating pharmacogenetic testing into clinical practice lies in the complexity of translating genetic laboratory test results into practical prescribing decisions for studied medications. To address this, there needs to be a standardized platform where clinicians can obtain evidence-based conclusions. The Clinical Pharmacogenetics Implementation Consortium (CPIC) is one of the currently available tools. CPIC focuses on creating, curating, and posting freely available, peer-reviewed, evidence-based, updated pharmacogenetic guidelines [91].
The CPIC guidelines assign levels to drugs/genes, and based on this score, advise whether genetic testing prior to prescription is recommended. Other resources available are the FDA-approved labels and PharmGKB Clinical Annotation Levels of Evidence, which also give information regarding drug pharmacogenomics. Some of the drug/gene pairs studied in this review, such as Tramadol and CYP2D6 [92], have been described in CPIC guidelines, which recommend that genetic data should inform drug prescriptions.
Pain management practices are increasingly relying on PGx testing to enhance their approach in determining the most effective relief strategy for patients while reducing risks and avoiding trial-and-error approaches [19]. This shift is especially evident in ongoing research initiatives such as the ImPreSS Trial [93]. Despite the limited evidence, the investigation on the feasibility and utility of pre-emptive PGx testing to guide medication decision-making in real time will contribute with valuable insights to the evolving landscape of personalized pain management strategies.
8. Future Directions
While pharmacogenomic tests are valuable, it is crucial not to overlook other significant factors that influence POP management. Patients should be treated comprehensively under personalized medicine to ensure a correct and appropriate analgesic approach. In this context, individuals differently respond to the same medication due to a multitude of factors, including weight and behavioral variations, sociocultural level, physiological function, and drug interaction including metabolism and elimination [17,19,76]. Age and genetic disparities are also important factors in pain management and pain perception [19,25]. POP, which is less frequently reported in the elderly, also correlates with the necessity of lower doses of analgesics such as morphine [4,7,83,94]. This underscores the importance of adjusting analgesic dosages, such as morphine, for elderly patients, acknowledging the impact of age-related factors on drug responses.
Future directions involving advancements in perioperative care through widespread adoption of better pain management strategies and the development of genetically tailored analgesic drugs could enhance healthcare practices. Integrating pharmacogenomics as an additional tool for postoperative pain management is expected to lead to improved outcomes and reduced risks, as demonstrated by several studies.
It is also important to underscore the imperative for conducting pharmacogenomic Genome-Wide Association Studies (GWAS) geared towards pinpointing novel genetic variants intricately associated with analgesic response in a comprehensive and unbiased manner. These initiatives hold significant promise in unlocking invaluable insights into personalized pain management strategies, thereby bolstering patient care and outcomes within the postoperative approach.
Additionally, it is essential to consider the heterogeneity of studies stemming from varying etiologies of postoperative pain, diverse analgesic protocols, and the genes under investigation. Conflicting results in prospective studies further underline the need for more extensive research in this area, emphasizing the importance of continued investigation and collaboration across multidisciplinary teams.
9. Conclusions
In summary, there is a need for better POP management. In this context, our study demonstrated the importance of pharmacogenetic-guided personalized medicine protocols for pain management in surgical patients. This approach has the capacity to yield superior outcomes in terms of enhanced pain relief, reduced morbidity, and overall improvement in patient-reported outcomes by improving standard pain management strategies.
Finally, it is expected that the results of this study will foster awareness for the review of educative programs based on the best practice in pain management. Thus, an exceptionally improved quality of life for surgical patients is anticipated, reducing healthcare and socio-economic costs in the near future.
Author Contributions
Conceptualization, M.L.F.d.C. and D.H.P.; methodology, M.L.F.d.C., S.F. and D.H.P.; writing—original draft preparation, M.L.F.d.C., S.F. and D.H.P.; writing—review and editing, M.L.F.d.C., S.F. and D.H.P.; supervision, S.F. and D.H.P.; project administration, D.H.P. All authors have read and agreed to the published version of the manuscript.
Conflicts of Interest
The authors declare no conflicts of interest.
Funding Statement
This research received no external funding.
Footnotes
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content.
References
- 1.Goldstein D.H., Ellis J., Brown R., Wilson R., Penning J., Chisom K., VanDenKerkhof E. Recommendations for improved acute pain services: Canadian collaborative acute pain initiative. Pain Res. Manag. 2004;9:123–130. doi: 10.1155/2004/397452. [DOI] [PubMed] [Google Scholar]
- 2.Hinrichs-Rocker A., Schulz K., Jarvinen I., Lefering R., Simanski C., Neugebauer E.A. Psychosocial predictors and correlates for chronic post-surgical pain (CPSP)—A systematic review. Eur. J. Pain. 2009;13:719–730. doi: 10.1016/j.ejpain.2008.07.015. [DOI] [PubMed] [Google Scholar]
- 3.Costantini M., Viterbori P., Flego G. Prevalence of pain in Italian hospitals: Results of a regional cross-sectional survey. J. Pain Symptom Manag. 2002;23:221–230. doi: 10.1016/S0885-3924(01)00405-5. [DOI] [PubMed] [Google Scholar]
- 4.Lin C.C. Applying the American Pain Society’s QA standards to evaluate the quality of pain management among surgical, oncology, and hospice inpatients in Taiwan. Pain. 2000;87:43–49. doi: 10.1016/S0304-3959(00)00267-0. [DOI] [PubMed] [Google Scholar]
- 5.Salomon L., Tcherny-Lessenot S., Collin E., Coutaux A., Levy-Soussan M., Legeron M.C., Bourgeois P., Cesselin F., Desfosses G., Rosenheim M. Pain prevalence in a French teaching hospital. J. Pain Symptom Manag. 2002;24:586–592. doi: 10.1016/S0885-3924(02)00528-6. [DOI] [PubMed] [Google Scholar]
- 6.Strohbuecker B., Mayer H., Evers G.C., Sabatowski R. Pain prevalence in hospitalized patients in a German university teaching hospital. J. Pain Symptom Manag. 2005;29:498–506. doi: 10.1016/j.jpainsymman.2004.08.012. [DOI] [PubMed] [Google Scholar]
- 7.Sommer M., de Rijke J.M., van Kleef M., Kessels A.G., Peters M.L., Geurts J.W., Gramke H.F., Marcus M.A. The prevalence of postoperative pain in a sample of 1490 surgical inpatients. Eur. J. Anaesthesiol. 2008;25:267–274. doi: 10.1017/S0265021507003031. [DOI] [PubMed] [Google Scholar]
- 8.Chung J.W., Lui J.C. Postoperative pain management: Study of patients’ level of pain and satisfaction with health care providers’ responsiveness to their reports of pain. Nurs. Health Sci. 2003;5:13–21. doi: 10.1046/j.1442-2018.2003.00130.x. [DOI] [PubMed] [Google Scholar]
- 9.Strassels S.A., Chen C., Carr D.B. Postoperative analgesia: Economics, resource use, and patient satisfaction in an urban teaching hospital. Anesth. Analg. 2002;94:130–137. doi: 10.1213/00000539-200201000-00025. [DOI] [PubMed] [Google Scholar]
- 10.Apfelbaum J.L., Chen C., Mehta S.S., Gan T.J. Postoperative pain experience: Results from a national survey suggest postoperative pain continues to be undermanaged. Anesth. Analg. 2003;97:534–540. doi: 10.1213/01.ANE.0000068822.10113.9E. [DOI] [PubMed] [Google Scholar]
- 11.Svensson I., Sjostrom B., Haljamae H. Influence of expectations and actual pain experiences on satisfaction with postoperative pain management. Eur. J. Pain. 2001;5:125–133. doi: 10.1053/eujp.2001.0227. [DOI] [PubMed] [Google Scholar]
- 12.Kehlet H., Jensen T.S., Woolf C.J. Persistent postsurgical pain: Risk factors and prevention. Lancet. 2006;367:1618–1625. doi: 10.1016/S0140-6736(06)68700-X. [DOI] [PubMed] [Google Scholar]
- 13.Macrae W.A. Chronic pain after surgery. Br. J. Anaesth. 2001;87:88–98. doi: 10.1093/bja/87.1.88. [DOI] [PubMed] [Google Scholar]
- 14.Wilder-Smith O.H., Tassonyi E., Arendt-Nielsen L. Preoperative back pain is associated with diverse manifestations of central neuroplasticity. Pain. 2002;97:189–194. doi: 10.1016/S0304-3959(01)00430-4. [DOI] [PubMed] [Google Scholar]
- 15.Gordon D.B., Pellino T.A., Miaskowski C., McNeill J.A., Paice J.A., Laferriere D., Bookbinder M. A 10-year review of quality improvement monitoring in pain management: Recommendations for standardized outcome measures. Pain Manag. Nurs. 2002;3:116–130. doi: 10.1053/jpmn.2002.127570. [DOI] [PubMed] [Google Scholar]
- 16.Awad M.E., Padela M.T., Sayeed Z., Abaab L., El-Othmani M.M., Saleh K.J. Pharmacogenomics Testing for Postoperative Pain Optimization Before Total Knee and Total Hip Arthroplasty. JBJS Rev. 2018;6:e3. doi: 10.2106/JBJS.RVW.17.00184. [DOI] [PubMed] [Google Scholar]
- 17.Schug S., Ting S. The pharmacogenomics of pain management: Prospects for personalized medicine. J. Pain Res. 2016;9:49. doi: 10.2147/JPR.S55595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhao M., Ma J., Li M., Zhang Y., Jiang B., Zhao X., Huai C., Shen L., Zhang N., He L., et al. Cytochrome P450 Enzymes and Drug Metabolism in Humans. Int. J. Mol. Sci. 2021;22:12808. doi: 10.3390/ijms222312808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kaye A.D., Garcia A.J., Hall O.M., Jeha G.M., Cramer K.D., Granier A.L., Kallurkar A., Cornett E.M., Urman R.D. Update on the pharmacogenomics of pain management. Pharmgenom. Pers. Med. 2019;12:125–143. doi: 10.2147/PGPM.S179152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Webster L.R., Belfer I. Pharmacogenetics and Personalized Medicine in Pain Management. Clin. Lab. Med. 2016;36:493–506. doi: 10.1016/j.cll.2016.05.007. [DOI] [PubMed] [Google Scholar]
- 21.Meredith P., Ownsworth T., Strong J. A review of the evidence linking adult attachment theory and chronic pain: Presenting a conceptual model. Clin. Psychol. Rev. 2008;28:407–429. doi: 10.1016/j.cpr.2007.07.009. [DOI] [PubMed] [Google Scholar]
- 22.Kennedy-Schwarz J. Pain management. A moral imperative. Am. J. Nurs. 2000;100:49–50. [PubMed] [Google Scholar]
- 23.Lome B. Acute pain and the critically ill trauma patient. Crit. Care Nurs. Q. 2005;28:200–207. doi: 10.1097/00002727-200504000-00012. [DOI] [PubMed] [Google Scholar]
- 24.Harmer M. Consent and ethics in postoperative pain management. Anaesthesia. 2002;57:1153–1154. doi: 10.1046/j.1365-2044.2002.02965.x. [DOI] [PubMed] [Google Scholar]
- 25.Nielsen L.M. Association Between Human Pain-Related Genotypes and Variability in Opioid Analgesia: An Updated Review. Pain Pract. 2015;15:580–594. doi: 10.1111/papr.12232. [DOI] [PubMed] [Google Scholar]
- 26.Yoshida K., Nishizawa D., Ide S., Ichinohe T., Fukuda K.I., Ikeda K. A pharmacogenetics approach to pain management. Neuropsychopharmacol. Rep. 2018;38:2–8. doi: 10.1002/npr2.12003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hamilton W.G. Prospective Randomized Study Using Pharmacogenetics to Customize Postoperative Pain Medication Following Hip and Knee Arthroplasty. J. Arthroplast. 2022;37:S76–S81. doi: 10.1016/j.arth.2022.02.037. [DOI] [PubMed] [Google Scholar]
- 28.Thomas C.D., Parvataneni H.K., Gray C.F., Deen J.T., Prieto H.A., Pulido L.F., Elsey A.R., Elwood E.N., Starostik P., Gong Y., et al. A hybrid implementation-effectiveness randomized trial of CYP2D6-guided postoperative pain management. Genet. Med. 2021;23:621–628. doi: 10.1038/s41436-020-01050-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hamilton W.G. Using pharmacogenetics to structure individual pain management protocols in total knee arthroplasty a randomized pilot study. Bone Jt. J. 2020;102:73–78. doi: 10.1302/0301-620X.102B6.BJJ-2019-1539.R1. [DOI] [PubMed] [Google Scholar]
- 30.Senagore A.J., Champagne B.J., Dosokey E., Brady J., Steele S.R., Reynolds H.L., Stein S.L., Delaney C.P. Pharmacogenetics-guided analgesics in major abdominal surgery: Further benefits within an enhanced recovery protocol. Am. J. Surg. 2017;213:467–472. doi: 10.1016/j.amjsurg.2016.11.008. [DOI] [PubMed] [Google Scholar]
- 31.Zhou Y., Cao L., Yang Y., Gao Y., Li Y., Wang B., Pan B., Huang J., Guo W. Is OPRM1 genotype a valuable predictor of VAS in patients undergoing laparoscopic radical resection of colorectal cancer with fentanyl? BMC Anesth. 2023;23:173. doi: 10.1186/s12871-023-02120-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Saiz-Rodríguez M., Valdez-Acosta S., Borobia A.M., Burgueño M., Gálvez-Múgica M., Acero J., Cabaleiro T., Muñoz-Guerra M.F., Puerro M., Llanos L., et al. Influence of Genetic Polymorphisms on the Response to Tramadol, Ibuprofen, and the Combination in Patients With Moderate to Severe Pain After Dental Surgery. Clin. Ther. 2021;43:e86–e102. doi: 10.1016/j.clinthera.2021.03.005. [DOI] [PubMed] [Google Scholar]
- 33.Matic M., de Hoogd S., de Wildt S.N., Tibboel D., Knibbe C.A., van Schaik R.H. OPRM1 and COMT polymorphisms: Implications on postoperative acute, chronic and experimental pain after cardiac surgery. Pharmacogenomics. 2020;21:181–193. doi: 10.2217/pgs-2019-0141. [DOI] [PubMed] [Google Scholar]
- 34.Stamer U.M., Musshoff F., Stüber F., Brockmöller J., Steffens M., Tzvetkov M.V. Loss-of-function polymorphisms in the organic cation transporter OCT1 are associated with reduced postoperative tramadol consumption. Pain. 2016;157:2467–2475. doi: 10.1097/j.pain.0000000000000662. [DOI] [PubMed] [Google Scholar]
- 35.Tan E.C., Lim E.C., Ocampo C.E., Allen J.C., Sng B.L., Sia A.T. Common variants of catechol-O-methyltransferase influence patient-controlled analgesia usage and postoperative pain in patients undergoing total hysterectomy. Pharmacogenomics J. 2016;16:186–192. doi: 10.1038/tpj.2015.33. [DOI] [PubMed] [Google Scholar]
- 36.Dong H., Lu S.-J., Zhang R., Liu D.-D., Zhang Y.-Z., Song C.-Y. Effect of the CYP2D6 gene polymorphism on postoperative analgesia of tramadol in Han nationality nephrectomy patients. Eur. J. Clin. Pharmacol. 2015;71:681–686. doi: 10.1007/s00228-015-1857-4. [DOI] [PubMed] [Google Scholar]
- 37.Seripa D., Latina P., Fontana A., Gravina C., Lattanzi M., Savino M., Gallo A.P., Melchionda G., Santini S.A., Margaglione M., et al. Role of CYP2D6 Polymorphisms in the Outcome of Postoperative Pain Treatment. Pain Med. 2015;16:2012–2023. doi: 10.1111/pme.12778. [DOI] [PubMed] [Google Scholar]
- 38.Wu S.B., Cai L.N., Yang X.H., Fu H.G., Sun K., Yuan F., Dong T.L. Impact of CYP2D6 Polymorphisms on Postoperative Fentanyl Analgesia in Gastric Cancer Patients. Genet. Test. Mol. Biomark. 2015;19:248–252. doi: 10.1089/gtmb.2014.0318. [DOI] [PubMed] [Google Scholar]
- 39.Zhang F., Tong J., Hu J., Zhang H., Ouyang W., Huang D., Tang Q., Liao Q. COMT gene haplotypes are closely associated with postoperative fentanyl dose in patients. Anesth. Analg. 2015;120:933–940. doi: 10.1213/ANE.0000000000000563. [DOI] [PubMed] [Google Scholar]
- 40.Boswell M.V., Stauble M.E., Loyd G.E., Langman L., Ramey-Hartung B., Baumgartner R.N., Tucker W.W., Jortani S.A. The role of hydromorphone and OPRM1 in postoperative pain relief with hydrocodone. Pain Physician. 2013;16:E227–E235. [PubMed] [Google Scholar]
- 41.Candiotti K., Yang Z., Xue L., Zhang Y., Rodriguez Y., Wang L., Hao S., Gitlin M. Single-nucleotide polymorphism C3435T in the ABCB1 gene is associated with opioid consumption in postoperative pain. Pain Med. 2013;14:1977–1984. doi: 10.1111/pme.12226. [DOI] [PubMed] [Google Scholar]
- 42.Henker R.A., Lewis A., Dai F., Lariviere W.R., Meng L., Gruen G.S., Sereika S.M., Pape H., Tarkin I.S., Gowda I., et al. The Associations between OPRM1 and COMT Genotypes and Postoperative Pain, Opioid Use, and Opioid-Induced Sedation. Biol. Res. Nurs. 2013;15:309–317. doi: 10.1177/1099800411436171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Slanar O., Dupal P., Matouskova O., Vondrackova H., Pafko P., Perlik F. Tramadol efficacy in patients with postoperative pain in relation to CYP2D6 and MDR1 polymorphisms. Bratisl. Med. J. 2012;113:152–155. doi: 10.4149/BLL_2012_036. [DOI] [PubMed] [Google Scholar]
- 44.Tan P.C., Hassan S.K., Mohamad N.A., Gan S.H. Cytochrome P450 3A4 genetic polymorphisms and post-operative fentanyl requirements. J. Clin. Pharm. Ther. 2012;37:100–104. doi: 10.1111/j.1365-2710.2010.01232.x. [DOI] [PubMed] [Google Scholar]
- 45.De Gregori M., Garbin G., De Gregori S., Minella C.E., Bugada D., Lisa A., Govoni S., Regazzi M., Allegri M., Ranzani G.N. Genetic variability at COMT but not at OPRM1 and UGT2B7 loci modulates morphine analgesic response in acute postoperative pain. Eur. J. Clin. Pharmacol. 2013;69:1651–1658. doi: 10.1007/s00228-013-1523-7. [DOI] [PubMed] [Google Scholar]
- 46.Zhang W., Yuan J.J., Kan Q.C., Zhang L.R., Chang Y.Z., Wang Z.Y., Li Z.S. Influence of CYP3A5*3 polymorphism and interaction between CYP3A5*3 and CYP3A4*1G polymorphisms on post-operative fentanyl analgesia in Chinese patients undergoing gynaecological surgery. Eur. J. Anaesthesiol. 2011;28:245–250. doi: 10.1097/EJA.0b013e3283438b39. [DOI] [PubMed] [Google Scholar]
- 47.Yuan R., Zhang X., Deng Q., Wu Y., Xiang G. Impact of CYP3A4*1G polymorphism on metabolism of fentanyl in Chinese patients undergoing lower abdominal surgery. Clin. Chim. Acta. 2011;412:755–760. doi: 10.1016/j.cca.2010.12.038. [DOI] [PubMed] [Google Scholar]
- 48.Zwisler S.T., Enggaard T.P., Mikkelsen S., Brosen K., Sindrup S.H. Impact of the CYP2D6 genotype on post-operative intravenous oxycodone analgesia. Acta Anaesthesiol. Scand. 2010;54:232–240. doi: 10.1111/j.1399-6576.2009.02104.x. [DOI] [PubMed] [Google Scholar]
- 49.Fukuda K., Hayashida M., Ide S., Saita N., Kokita Y., Kasai S., Nishizawa D., Ogai Y., Hasegawa J., Nagashima M., et al. Association between OPRM1 gene polymorphisms and fentanyl sensitivity in patients undergoing painful cosmetic surgery. Pain. 2009;147:194–201. doi: 10.1016/j.pain.2009.09.004. [DOI] [PubMed] [Google Scholar]
- 50.Tan E.C., Lim E.C., Teo Y.Y., Lim Y., Law H.Y., Sia A.T. Ethnicity and OPRM variant independently predict pain perception and patient-controlled analgesia usage for post-operative pain. Mol. Pain. 2009;5:32. doi: 10.1186/1744-8069-5-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sia A.T., Lim Y., Lim E.C., Goh R.W., Law H.Y., Landau R., Teo Y.Y., Tan E.C. A118G single nucleotide polymorphism of human mu-opioid receptor gene influences pain perception and patient-controlled intravenous morphine consumption after intrathecal morphine for postcesarean analgesia. Anesthesiology. 2008;109:520–526. doi: 10.1097/ALN.0b013e318182af21. [DOI] [PubMed] [Google Scholar]
- 52.Chou W.Y., Yang L.C., Lu H.F., Ko J.Y., Wang C.H., Lin S.H., Lee T.H., Concejero A., Hsu C.J. Association of μ-opioid receptor gene polymorphism (A118G) with variations in morphine consumption for analgesia after total knee arthroplasty. Acta Anaesthesiol. Scand. 2006;50:787–792. doi: 10.1111/j.1399-6576.2006.01058.x. [DOI] [PubMed] [Google Scholar]
- 53.Chou W.Y., Wang C.H., Liu P.H., Liu C.C., Tseng C.C., Jawan B. Human opioid receptor A118G polymorphism affects intravenous patient-controlled analgesia morphine consumption after total abdominal hysterectomy. Anesthesiology. 2006;105:334–337. doi: 10.1097/00000542-200608000-00016. [DOI] [PubMed] [Google Scholar]
- 54.Wang G., Zhang H., He F., Fang X. Effect of the CYP2D6*10 C188T polymorphism on postoperative tramadol analgesia in a Chinese population. Eur. J. Clin. Pharmacol. 2006;62:927–931. doi: 10.1007/s00228-006-0191-2. [DOI] [PubMed] [Google Scholar]
- 55.Stamer U.M., Lehnen K., Höthker F., Bayerer B., Wolf S., Hoeft A., Stuber F. Impact of CYP2D6 genotype on postoperative tramadol analgesia. Pain. 2003;105:231–238. doi: 10.1016/S0304-3959(03)00212-4. [DOI] [PubMed] [Google Scholar]
- 56.Koren G., Cairns J., Chitayat D., Gaedigk A., Leeder S.J. Pharmacogenetics of morphine poisoning in a breastfed neonate of a codeine-prescribed mother. Lancet. 2006;368:704. doi: 10.1016/S0140-6736(06)69255-6. [DOI] [PubMed] [Google Scholar]
- 57.Willmann S., Edginton A.N., Coboeken K., Ahr G., Lippert J. Risk to the breast-fed neonate from codeine treatment to the mother: A quantitative mechanistic modeling study. Clin. Pharmacol. Ther. 2009;86:634–643. doi: 10.1038/clpt.2009.151. [DOI] [PubMed] [Google Scholar]
- 58.Bagher A.M. Association of CYP2C9 *3 and CYP2C8 *3 Non-Functional Alleles with Ibuprofen-Induced Upper Gastrointestinal Toxicity in a Saudi Patient. Case Rep. Med. 2023;2023:6623269. doi: 10.1155/2023/6623269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zobdeh F., Eremenko I.I., Akan M.A., Tarasov V.V., Chubarev V.N., Schioth H.B., Mwinyi J. Pharmacogenetics and Pain Treatment with a Focus on Non-Steroidal Anti-Inflammatory Drugs (NSAIDs) and Antidepressants: A Systematic Review. Pharmaceutics. 2022;14:1190. doi: 10.3390/pharmaceutics14061190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Macias Y., Garcia-Menaya J.M., Marti M., Cordobes C., Jurado-Escobar R., Cornejo-Garcia J.A., Torres M.J., Blanca-Lopez N., Canto G., Blanca M., et al. Lack of Major Involvement of Common CYP2C Gene Polymorphisms in the Risk of Developing Cross-Hypersensitivity to NSAIDs. Front. Pharmacol. 2021;12:648262. doi: 10.3389/fphar.2021.648262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Frangakis S.G., MacEachern M., Akbar T.A., Bolton C., Lin V., Smith A.V., Brummett C.M., Bicket M.C. Association of Genetic Variants with Postsurgical Pain: A Systematic Review and Meta-analyses. Anesthesiology. 2023;139:827–839. doi: 10.1097/ALN.0000000000004677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.McEvoy L., Carr D.F., Pirmohamed M. Pharmacogenomics of NSAID-Induced Upper Gastrointestinal Toxicity. Front. Pharmacol. 2021;12:684162. doi: 10.3389/fphar.2021.684162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Musumba C.O., Jorgensen A., Sutton L., Van Eker D., Zhang E., O’Hara N., Carr D.F., Pritchard D.M., Pirmohamed M. CYP2C19*17 gain-of-function polymorphism is associated with peptic ulcer disease. Clin. Pharmacol. Ther. 2013;93:195–203. doi: 10.1038/clpt.2012.215. [DOI] [PubMed] [Google Scholar]
- 64.Dean L. Amitriptyline Therapy and CYP2D6 and CYP2C19 Genotype. In: Pratt V.M., Scott S.A., Pirmohamed M., Esquivel B., Kattman B.L., Malheiro A.J., editors. Medical Genetics Summarie. Medical Genetics Summaries; Bethesda, MD, USA: 2012. [Google Scholar]
- 65.Sadhasivam S., Chidambaran V., Zhang X., Meller J., Esslinger H., Zhang K., Martin L.J., McAuliffe J. Opioid-induced respiratory depression: ABCB1 transporter pharmacogenetics. Pharmacogenomics J. 2015;15:119–126. doi: 10.1038/tpj.2014.56. [DOI] [PubMed] [Google Scholar]
- 66.Branford R., Droney J., Ross J.R. Opioid genetics: The key to personalized pain control? Clin. Genet. 2012;82:301–310. doi: 10.1111/j.1399-0004.2012.01923.x. [DOI] [PubMed] [Google Scholar]
- 67.Korczeniewska O.A., Kuo F., Huang C.Y., Nasri-Heir C., Khan J., Benoliel R., Hirschberg C., Eliav E., Diehl S.R. Genetic variation in catechol-O-methyltransferase is associated with individual differences in conditioned pain modulation in healthy subjects. J. Gene Med. 2021;23:e3374. doi: 10.1002/jgm.3374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Vetterlein A., Monzel M., Reuter M. Are catechol-O-methyltransferase gene polymorphisms genetic markers for pain sensitivity after all? A review and meta-analysis. Neurosci. Biobehav. Rev. 2023;148:105112. doi: 10.1016/j.neubiorev.2023.105112. [DOI] [PubMed] [Google Scholar]
- 69.Diatchenko L., Slade G.D., Nackley A.G., Bhalang K., Sigurdsson A., Belfer I., Goldman D., Xu K., Shabalina S.A., Shagin D., et al. Genetic basis for individual variations in pain perception and the development of a chronic pain condition. Hum. Mol. Genet. 2005;14:135–143. doi: 10.1093/hmg/ddi013. [DOI] [PubMed] [Google Scholar]
- 70.Meloto C.B., Bortsov A.V., Bair E., Helgeson E., Ostrom C., Smith S.B., Dubner R., Slade G.D., Fillingim R.B., Greenspan J.D., et al. Modification of COMT-dependent pain sensitivity by psychological stress and sex. Pain. 2016;157:858–867. doi: 10.1097/j.pain.0000000000000449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Hu B., Zhang X., Xu G., Zhang Q., Qian P., Liu S., Zhu J., Shen R. Association between COMT Polymorphism Val158Met and Opioid Consumption in Patients with Postoperative Pain: A Meta-Analysis. Neurosignals. 2018;26:11–21. doi: 10.1159/000487038. [DOI] [PubMed] [Google Scholar]
- 72.Smietanska I., Adrian E., Smietanski M., Kitowski J. Does the Pain-free hospital certification improve the management of pain following hernioplasty? Anestezjol. Intens. Ter. 2010;42:190–193. [PubMed] [Google Scholar]
- 73.Skinner H.B., Shintani E.Y. Results of a multimodal analgesic trial involving patients with total hip or total knee arthroplasty. Am. J. Orthop. 2004;33:85–92; discussion 92. [PubMed] [Google Scholar]
- 74.Wells N., Pasero C., McCaffery M. Improving the Quality of Care through Pain Assessment and Management. Agency for Healthcare Research and Quality (US); Rockville, MD, USA: 2008. [PubMed] [Google Scholar]
- 75.American Society of Anesthesiologists Task Force on Acute Pain Management Practice guidelines for acute pain management in the perioperative setting: An updated report by the American Society of Anesthesiologists Task Force on Acute Pain Management. Anesthesiology. 2004;100:1573–1581. doi: 10.1097/00000542-200406000-00033. [DOI] [PubMed] [Google Scholar]
- 76.Gouveia B., Fonseca S., Pozza D.H., Xara D., Sa Rodrigues A. Relationship between Postoperative Pain and Sociocultural Level in Major Orthopedic Surgery. Adv. Orthop. 2022;2022:7867719. doi: 10.1155/2022/7867719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Bardiau F.M., Taviaux N.F., Albert A., Boogaerts J.G., Stadler M. An intervention study to enhance postoperative pain management. Anesth. Analg. 2003;96:179–185. doi: 10.1213/00000539-200301000-00038. [DOI] [PubMed] [Google Scholar]
- 78.Ernst F.R., Grizzle A.J. Drug-related morbidity and mortality: Updating the cost-of-illness model. J. Am. Pharm. Assoc. 2001;41:192–199. doi: 10.1016/S1086-5802(16)31229-3. [DOI] [PubMed] [Google Scholar]
- 79.Kalkman C.J., Visser K., Moen J., Bonsel G.J., Grobbee D.E., Moons K.G. Preoperative prediction of severe postoperative pain. Pain. 2003;105:415–423. doi: 10.1016/S0304-3959(03)00252-5. [DOI] [PubMed] [Google Scholar]
- 80.Severino G., Del Zompo M. Adverse drug reactions: Role of pharmacogenomics. Pharmacol. Res. 2004;49:363–373. doi: 10.1016/j.phrs.2003.05.003. [DOI] [PubMed] [Google Scholar]
- 81.Stephanie N., Schatz R.J.W. PSAP: CNS/Pharmacy Practice. American College of Clinical Pharmacy; Lenexa, KS, USA: 2015. [Google Scholar]
- 82.Pozza D.H., Azevedo L.F., Castro Lopes J.M. Pain as the fifth vital sign-A comparison between public and private healthcare systems. PLoS ONE. 2021;16:e0259535. doi: 10.1371/journal.pone.0259535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.McNeill J.A., Sherwood G.D., Starck P.L., Thompson C.J. Assessing clinical outcomes: Patient satisfaction with pain management. J. Pain Symptom Manag. 1998;16:29–40. doi: 10.1016/S0885-3924(98)00034-7. [DOI] [PubMed] [Google Scholar]
- 84.Bright D.R., Petry N., Roath E., Gibb T. Engaging pharmacogenomics in pain management and opioid selection. Pharmacogenomics. 2021;22:927–937. doi: 10.2217/pgs-2021-0044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Morris S.A., Alsaidi A.T., Verbyla A., Cruz A., Macfarlane C., Bauer J., Patel J.N. Cost Effectiveness of Pharmacogenetic Testing for Drugs with Clinical Pharmacogenetics Implementation Consortium (CPIC) Guidelines: A Systematic Review. Clin. Pharmacol. Ther. 2022;112:1318–1328. doi: 10.1002/cpt.2754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Lazarou J., Pomeranz B.H., Corey P.N. Incidence of adverse drug reactions in hospitalized patients: A meta-analysis of prospective studies. JAMA. 1998;279:1200–1205. doi: 10.1001/jama.279.15.1200. [DOI] [PubMed] [Google Scholar]
- 87.Ionova Y., Ashenhurst J., Zhan J., Nhan H., Kosinski C., Tamraz B., Chubb A. CYP2C19 Allele Frequencies in Over 2.2 Million Direct-to-Consumer Genetics Research Participants and the Potential Implication for Prescriptions in a Large Health System. Clin. Transl. Sci. 2020;13:1298–1306. doi: 10.1111/cts.12830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Alrajeh K.Y., Roman Y.M. The frequency of major CYP2C19 genetic polymorphisms in women of Asian, Native Hawaiian and Pacific Islander subgroups. Pers. Med. 2022;19:327–339. doi: 10.2217/pme-2021-0175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Green C.R., Anderson K.O., Baker T.A., Campbell L.C., Decker S., Fillingim R.B., Kalauokalani D.A., Lasch K.E., Myers C., Tait R.C., et al. The unequal burden of pain: Confronting racial and ethnic disparities in pain. Pain Med. 2003;4:277–294. doi: 10.1046/j.1526-4637.2003.03034.x. [DOI] [PubMed] [Google Scholar]
- 90.Salari P., Larijani B. Ethical Issues Surrounding Personalized Medicine: A Literature Review. Acta Med. Iran. 2017;55:209–217. [PubMed] [Google Scholar]
- 91.Relling M.V., Klein T.E. CPIC: Clinical Pharmacogenetics Implementation Consortium of the Pharmacogenomics Research Network. Clin. Pharmacol. Ther. 2011;89:464–467. doi: 10.1038/clpt.2010.279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Crews K.R., Monte A.A., Huddart R., Caudle K.E., Kharasch E.D., Gaedigk A., Dunnenberger H.M., Leeder J.S., Callaghan J.T., Samer C.F., et al. Clinical Pharmacogenetics Implementation Consortium Guideline for CYP2D6, OPRM1, and COMT Genotypes and Select Opioid Therapy. Clin. Pharmacol. Ther. 2021;110:888–896. doi: 10.1002/cpt.2149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Truong T.M., Apfelbaum J., Shahul S., Anitescu M., Danahey K., Knoebel R.W., Liebovitz D., Karrison T., Van Wijk X.M.R., Yeo K.T.J., et al. The ImPreSS Trial: Implementation of Point-of-Care Pharmacogenomic Decision Support in Perioperative Care. Clin. Pharmacol. Ther. 2019;106:1179–1183. doi: 10.1002/cpt.1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Coulbault L., Beaussier M., Verstuyft C., Weickmans H., Dubert L., Tregouet D., Descot C., Parc Y., Lienhart A., Jaillon P., et al. Environmental and genetic factors associated with morphine response in the postoperative period. Clin. Pharmacol. Ther. 2006;79:316–324. doi: 10.1016/j.clpt.2006.01.007. [DOI] [PubMed] [Google Scholar]