Abstract
Physical interactions of simian virus 40 (SV40) large tumor (T) antigen with cellular DNA polymerase α-primase (Pol/Prim) and replication protein A (RPA) appear to be responsible for multiple functional interactions among these proteins that are required for initiation of viral DNA replication at the origin, as well as during lagging-strand synthesis. In this study, we mapped an RPA binding site in T antigen (residues 164 to 249) that is embedded within the DNA binding domain of T antigen. Two monoclonal antibodies whose epitopes map within this region specifically interfered with RPA binding to T antigen but did not affect T-antigen binding to origin DNA or Pol/Prim, ATPase, or DNA helicase activity and had only a modest effect on origin DNA unwinding, suggesting that they could be used to test the functional importance of this RPA binding site in the initiation of viral DNA replication. To rule out a possible effect of these antibodies on origin DNA unwinding, we used a two-step initiation reaction in which an underwound template was first generated in the absence of primer synthesis. In the second step, primer synthesis was monitored with or without the antibodies. Alternatively, an underwound primed template was formed in the first step, and primer elongation was tested with or without antibodies in the second step. The results show that the antibodies specifically inhibited both primer synthesis and primer elongation, demonstrating that this RPA binding site in T antigen plays an essential role in both events.
Simian virus 40 (SV40) DNA replication is carried out entirely by host cell replication proteins, with the exception of one essential viral protein, large tumor (T) antigen (4, 6, 29, 40). The use of a cell-free SV40 DNA replication system and fractionated cell extracts has led to the identification and characterization of 10 cellular factors necessary and sufficient to reconstitute the process (5, 6, 79, 87, 88). Two of these essential cellular proteins, replication protein A (RPA) (27, 95, 97) and DNA polymerase α-primase complex (Pol/Prim) (42, 50, 63, 95), act together with T antigen and topoisomerase I or II (100) during the initiation step (56, 84, 89). RPA and Pol/Prim, probably guided by physical protein-protein interactions with T antigen (2, 15, 23–25, 30, 31, 60, 67, 69, 72), are thought to form a preinitiation complex (66, 69, 71) after or perhaps concomitantly with assembly of T antigen as a double hexamer on its recognition site (18, 22, 55, 93). T antigen distorts the origin region locally and catalyzes bidirectional unwinding of the template DNA, forming an underwound intermediate that represents the template for the first primer synthesis (4, 6–8, 40). In the absence of other replication proteins, RPA can be replaced in the unwinding reaction by Escherichia coli single-stranded DNA (ssDNA) binding protein (SSB) or other ssDNA binding proteins that do not support SV40 DNA replication, except for T4 gene 32 protein, implying that its DNA binding activity is probably required simply to stabilize the single-stranded regions (3, 27, 95–97). However, in the presence of crude cellular protein extracts, unwinding is limited to the origin-proximal region, and subsequent primer synthesis initiates on the lagging-strand template in sequences outside but very close to the core origin (7–10, 20). These studies and others (65, 83) suggested that unwinding and initiation of DNA synthesis are coupled, but the mechanisms and factors that limit the extent of unwinding in crude extracts have not been determined. As unwinding becomes more extensive, primer synthesis on the lagging-strand template occurs at sites progressively farther away from the core origin (20).
The fact that RPA of metazoan origin is required to support SV40 DNA replication (96) suggests that specific protein-protein interactions between RPA and other replication proteins are responsible for functional interactions among these proteins during replication. RPA specifically stimulates Pol/Prim during elongation (26, 45, 46, 96). RPA inhibits primer synthesis by Pol/Prim on M13 template, and T antigen partially relieves the inhibition (14, 60, 64). The presence of Pol/Prim was also reported to stimulate assembly of T antigen on the origin, and together, Pol/Prim and RPA slowed T-antigen translocation during unwinding, an interaction that is likely to play a role in coupling unwinding with primer synthesis (65, 66).
The sites of interaction of T antigen with Pol/Prim have been localized to two regions in T antigen, a weak site at the amino terminus that is not essential for viral DNA replication and a strong site in the carboxy-terminal region (4, 6, 25, 29, 30, 72, 90). SV40 T antigen was shown to bind directly to specific sequences in both the p180 and p68 subunits of Pol/Prim (15, 23, 25). Monoclonal antibodies against T antigen (Pab414 and Pab204) abrogate its physical interaction with Pol/Prim, its ability to stimulate primer synthesis and elongation by purified Pol/Prim, and also SV40 DNA replication in crude extracts (14, 25, 31, 72). Hence, T-antigen association with Pol/Prim was concluded to be essential for viral replication. Human RPA subunits p70 and p34 have been reported to interact physically with T antigen (2, 49, 91), while yeast RPA did not (60), implying that specific T antigen-RPA interactions play a role in viral replication. However, the T-antigen sequences that bind to RPA have not been mapped, nor has the functional relevance of the sites of interaction in the human RPA polypeptides been tested.
In this study, we have sought to define the T-antigen region responsible for physical interaction with RPA and to verify its relevance in the functional interactions between these proteins in the replication of SV40 DNA. We report here a sequence of 85 residues in the DNA binding domain of T antigen (44) that is sufficient for RPA binding. In addition, by screening a panel of T-antigen-specific monoclonal antibodies, we demonstrate that two antibodies whose epitopes map within the DNA binding domain, Pab220 and Pab221, specifically disrupt T antigen’s ability to form complexes with RPA. These monoclonal antibodies have been used to test the physiological importance of this RPA binding site in the early steps of viral DNA replication. Except for weak inhibition of the unwinding of supercoiled SV40 DNA, Pab220 and Pab221 had little effect on other biochemical activities of T antigen that are required for replication. To circumvent possible effects of Pab220 and Pab221 on origin DNA unwinding, we have used a two-step initiation reaction. In the first step, formation of an underwound template was permitted in the absence of primer synthesis. In the second step, primer synthesis was monitored after addition of ribonucleoside triphosphates with or without Pab220 or Pab221, or other monoclonal antibodies. Alternatively, a primed template was formed in the absence of deoxyribonucleotides in the first step, and primer elongation was measured with or without antibodies in the second step. The results show that Pab220 and Pab221 specifically inhibited both primer synthesis and primer elongation, demonstrating that RPA binding to T antigen plays an essential role in both events.
MATERIALS AND METHODS
Protein purification.
SV40 T antigen (25), human RPA (78, 91), and the human Pol/Prim (76, 86) were expressed in Spodoptera frugiperda Sf9 cells infected with recombinant baculoviruses and purified as described elsewhere. T antigen was stored in 20 mM HEPES-KOH (pH 8.5)–50 mM NaCl–0.1 mM EDTA–10% glycerol. Glutathione S-transferase (GST) fusion proteins were expressed and purified on glutathione-agarose as described previously (74). Expression plasmids for GST-T antigen fusion proteins were kindly provided by A. Wildeman, A. Arthur, and I. Moarefi. If the fusion protein, T antigen, or Pol/Prim was to be used for protein-protein interaction studies, the protein was nuclease treated during purification by incubation of the immunoaffinity matrix or the glutathione-agarose beads in a buffer containing benzonase nuclease mixture (0.05 U/μl; Merck) in 30 mM HEPES-KOH (pH 7.8)–5 mM MgCl2–1 mM dithiothreitol (DTT) at room temperature for 30 min prior to elution. After elution from the ssDNA column, RPA was dialyzed against the same buffer and treated with benzonase before MonoQ chromatography (78). Topoisomerase I, purified by the method of Strausfeld and Richter (80) from calf thymus, was kindly provided by I. Moarefi. E. coli SSB was purified from bacterial extracts as described previously (51) and was the kind gift of V. Podust. Monoclonal antibodies from hybridoma culture medium and polyclonal antibodies from serum were purified by ammonium sulfate precipitation and protein A-agarose chromatography as described previously (25) and dialyzed against 20 mM HEPES-KOH (pH 7.8)–50 mM NaCl–0.1 mM EDTA. Isolation and epitope mapping of monoclonal antibodies Pab101 and -108 (35, 36), Pab419, -416, and -414 (37), Pab220 and -221 (62), Pab204 (13), and KT3 (53) against T antigen have been described elsewhere. Monoclonal antibody 70C against the largest RPA subunit was previously characterized (2, 45).
Protein affinity pull-down assay.
A column containing 0.2 ml of glutathione-agarose, to which a GST-T antigen fusion protein had been adsorbed (approximately 1 mg/ml of bed volume), was equilibrated by gravity flow in binding buffer (30 mM HEPES-KOH [pH 7.9]–50 mM KCl–7 mM MgCl2–0.25% inositol–0.25 mM EDTA–0.05% Nonidet P-40 [NP-40]); 20 μg of soluble RPA, diluted to 0.1 mg/ml in binding buffer, was passed three times over the column by gravity flow. The column was washed with 10 column volumes of wash buffer (30 mM HEPES-KOH [pH 7.9], 100 mM KCl, 7 mM MgCl2), and bound RPA was eluted with 5 column volumes of elution buffer (30 mM HEPES-KOH [pH 7.9]–1% sodium dodecyl sulfate [SDS], 300 mM β-mercaptoethanol). The eluted protein was concentrated by adsorption to 20 μl of StrataClean resin (Stratagene), which was then suspended in SDS sample buffer (47). The resin was loaded on a 10% denaturing gel (47), and the proteins were separated by electrophoresis. RPA was detected by immunoblotting (81) using the Amersham enhanced chemiluminescence detection system.
Immunoprecipitation.
Twenty microliters of 50% (vol/vol) protein G-agarose beads and 5 μg of specific monoclonal antibodies were incubated with 2 μg of T antigen for 1 h. After three washes, the beads were resuspended in 100 μl of binding buffer (50 mM HEPES-KOH [pH 7.9], 100 mM KCl, 7 mM MgCl2, 0.25% inositol, 0.25 mM EDTA, 0.05% NP-40, 2% bovine serum albumin [BSA]) and incubated with 1 μg of RPA for 1 h at 4°C. The beads were washed four times with 1 ml of wash buffer (30 mM HEPES-KOH [pH 7.9], 100 mM KCl, 7 mM MgCl2) and boiled in 20 μl of sample buffer. Proteins were electrophoresed on a 10% denaturing gel, transferred to a nitrocellulose filter, and detected by immunoblotting with specific antibody and the enhanced chemiluminescence system. Before being reprobed with a different antibody, the filter was stripped of the first antibody as suggested by the manufacturer.
ELISA.
Enzyme-linked immunosorbent assays (ELISAs) were carried out essentially as described previously (25). Wells of a microtiter ELISA plate were coated with 1 μg of purified protein in 50 μl of phosphate-buffered saline (PBS) for 1 h, washed three times with PBS, blocked with 300 μl of 3% BSA in PBS, and washed again. To screen for the influence of antibodies specific for the solid-phase protein, the wells were incubated with 10 μg of murine monoclonal antibodies in 50 μl of PBS for 1 h. After being washed three times, wells were incubated with 2 μg of a soluble second protein for 2 h at room temperature and then washed again. Binding of the soluble protein was detected by incubation with 20 μg of polyclonal rabbit antibody that had been conjugated with horseradish peroxidase (Zymed, San Francisco, Calif.) according to the supplier’s instructions and a chromogenic substrate and then quantitated spectrophotometrically at 405 nm.
DNA substrates.
pUC-HS DNA (69), containing the complete SV40 origin of DNA replication, was purified by isopycnic centrifugation in CsCl-ethidium bromide gradients and used for unwinding assays with supercoiled template and for in vitro replication assays. pUCmori, containing the minimal SV40 origin, was obtained by insertion of the EcoRI/HindIII fragment of pOR1 (19) into pUC19. For DNA unwinding assays with linear templates, pUCmori was digested with XmnI, NdeI, and HindIII and 5′ end labeled, and the 330-bp origin-containing and 575-bp nonspecific DNA fragments were isolated. For helicase assays, a 5′-end-labeled 30-mer oligodeoxyribonucleotide was hybridized to M13mp18 ssDNA (Pharmacia), and the partial duplex DNA was isolated by agarose gel electrophoresis.
Band shift assays.
The 5′-end-labeled origin-containing 81-bp EcoRI/HindIII fragment of pOR1 was used in band shift experiments. Eight femtomoles of labeled, origin-containing DNA fragment (specific activity, 2,000 cpm/fmol) in 10 μl of 30 mM HEPES-KOH (pH 7.8)–7 mM MgCl2–1 mM DTT–40 mM creatine phosphate–2 μg of creatine kinase–4 mM AMP-PNP–100 pg of pBluescript KSII competitor DNA–1 μg of BSA was incubated with 50 ng of T antigen for 30 min at 37°C (85). Where indicated, 10 μg of monoclonal antibody was present in the reaction. Proteins were cross-linked to DNA by addition of glutaraldehyde to an end concentration of 0.2% and a further 5-min incubation. The reaction was supplemented with 1/5 volume of loading buffer (10 mM HEPES-KOH [pH 7.8], 25% Ficoll 400, 0.2% bromophenol blue, 0.2% xylene cyanol), and protein-DNA complexes were separated by electrophoresis in a 3.5% native polyacrylamide gel in TBE (89 mM Tris-borate, 89 mM boric acid, 0.2 mM EDTA) at 200 V. The gel was dried and autoradiographed. Bound DNA was quantitated by densitometry of the autoradiogram.
ATPase assay.
To measure ATPase activity, 600 ng of T antigen was added to a 20-μl assay mixture containing 50 pmol of ATP and 0.4 μCi of [γ-32P]ATP (3,000 Ci/mmol; ICN) in ATPase buffer (50 mM Tris-HCl [pH 8], 10 mM NaCl, 7 mM MgCl2, 0.05% NP-40, 1 mM DTT). Where stated, 10 μg of the indicated monoclonal antibody was present in the reaction. The ATPase reaction was terminated after 10 min at 37°C by addition of 1 μl of 0.5 M EDTA, 1 μl of the reaction mixture was spotted onto polyethyleneimine-cellulose F thin-layer chromatography plates (Merck), and the plates were developed in 0.75 M NaH2PO4. After drying of the plates, released phosphate (Pi) was quantitated with a PhosphorImager.
Helicase assay.
Helicase assays were performed with 300 ng of T antigen and 10 fmol (corresponding to about 2.5 ng) of oligonucleotide-hybridized M13mp18 DNA (specific activity of 1,000 cpm/ng) in 10 μl of ATPase buffer. Where stated, 10 μg of monoclonal antibody was included in the reaction. After 30 min at 37°C, 2 μl of loading buffer (20 mM HEPES-KOH [pH 7.8], 25% Ficoll 400, 0.01% bromophenol blue, 1% SDS) was added, and the sample was immediately electrophoresed in an 8% polyacrylamide gel in TBE at 80 V until the bromophenol blue marker had migrated 2 cm into the gel. The gel was dried and exposed to X-ray film. Displaced oligonucleotide was quantitated by densitometry of the autoradiogram.
DNA unwinding assays.
Unwinding assays with linear DNA template contained 600 ng of T antigen and 5 fmol each of a 330-bp origin-containing DNA fragment and a 575-bp nonspecific fragment (specific activity, 2,000 cpm/fmol) in 30 μl of ATPase buffer. Where stated, 10 μg of monoclonal antibody was included. After 60 min at 37°C, 10 μl of loading buffer (20 mM HEPES-KOH [pH 7.8], 25% Ficoll 400, 0.01% bromophenol blue, 1% SDS) was added, and the sample was immediately electrophoresed in an 8% polyacrylamide gel in TBE at 80 V until the bromophenol blue marker had reached the bottom of the gel. The gel was dried and exposed to X-ray film, and the unwound DNA was quantitated by densitometry.
Unwinding reactions with supercoiled closed circular DNA (total volume of 20 μl) were performed with 200 ng of pUC-HS DNA, 40 mM HEPES-KOH (pH 7.9), 0.5 mM DTT, 8 mM MgCl2, 4 mM ATP, 40 mM creatine phosphate, 0.5 μg of creatine kinase, 2 μg of BSA, 120 ng of topoisomerase I, and 250 ng of E. coli SSB and were started by adding 800 ng of T antigen. Where indicated, 10 μg of monoclonal antibody or antibody buffer was present. After 1 h at 37°C, the mixture was incubated in 0.2% SDS–400 ng of proteinase K at 37°C for 30 min and then ethanol precipitated. The samples were redissolved in 10 mM EDTA–2% Ficoll–2% sucrose–0.01% bromophenol blue–0.1% SDS and electrophoresed in 1.5% agarose gels. The gel was stained with ethidium bromide and photographed. Unwound DNA fragments were quantitated by densitometry.
SV40 DNA replication.
In vitro replication reactions were carried out essentially as described previously (61), with slight modifications. The reaction mixture (60 μl) contained 30 mM HEPES-KOH (pH 7.8), 7 mM magnesium acetate, 1 mM EGTA, 0.5 mM DTT, 4 mM ATP, 0.2 mM each CTP, GTP, and UTP, 0.1 mM each dGTP and dATP, 0.05 mM each dCTP and dTTP, 5 μCi each of [α-32P]dCTP and [α-32P]dTTP, 40 mM creatine phosphate, 4.8 μg of creatine kinase, 100 ng of pUC-HS DNA, 600 ng of T antigen, and 190 μg of S100 extract prepared from human 293S cells. Where stated, 10 μg of monoclonal antibody was included. After 90 min at 37°C, 5 μl of the reaction mixture was spotted on DE81 paper to quantitate incorporated nucleotides (54). EDTA, SDS, and proteinase K were added to final concentrations of 20 mM, 0.65%, and 1.7 mg/ml, respectively, and incubation was continued for another 30 min. The sample was extracted once with phenol-chloroform, and the DNA was passed over a Sephadex G-50 spin column (Boehringer Mannheim) equilibrated in TE buffer (10 mM Tris-HCl [pH 8], 1 mM EDTA). DNA was ethanol precipitated and dissolved in 20 μl of TE buffer. Then 5-μl aliquots were digested with EcoRI or EcoRI/DpnI, and reaction products were separated by 0.8% agarose gel electrophoresis in TBE. The gel was dried and exposed to X-ray film.
Initiation assays.
Initiation reaction mixtures (69) (40 μl) contained 30 mM HEPES-KOH (pH 7.8), 7 mM magnesium acetate, 1 mM EGTA, 0.5 mM DTT, 4 mM ATP, 0.2 mM each GTP and UTP, 2 μM CTP, 10 μCi of [α-32P]CTP, 40 mM creatine phosphate, 0.4 μg of creatine kinase, 10 μg of BSA, 400 ng of RPA, 600 ng of T antigen, 300 ng of topoisomerase I, 400 ng of Pol/Prim (8 primase units and 15.2 polymerase units [69]), and 100 ng of pUC-HS DNA. Where stated, 10 μg of monoclonal antibody was included. After 60 min at 37°C, 5 μl of the reaction were spotted on DE81 paper to quantitate incorporated nucleotides (54). Reaction products were precipitated in the presence of 0.8 M LiCl–10 mM MgCl2–10 μg of yeast tRNA. The precipitate was dissolved in 35% formamide–8 mM EDTA–0.1% bromophenol blue–0.1% xylene cyanol FF for 30 min at 65°C, heated for 3 min at 95°C, and electrophoresed in 20% denaturing polyacrylamide gels at 600 V until the bromophenol blue had migrated to the bottom of the gel. The gel was exposed wet to an X-ray film.
To uncouple initial unwinding from primer synthesis, a two-step procedure was used. In the first step (unwinding reaction), a 20-μl initiation assay mixture was assembled as described above except that CTP, GTP, UTP and [α-32P]CTP were omitted. After 30 min at 37°C, the reaction mixture was supplemented in the second step (primer synthesis) with the missing nucleotides, adjusting the reaction volume to 40 μl; 10 μg of antibody was added at the beginning of step 1 or step 2, as indicated in the figure legends. After 60 min at 37°C, reaction products were analyzed as described above.
The monopolymerase system.
The monopolymerase system was set up essentially as described elsewhere (66). The standard reaction mixture (40 μl) contained 30 mM HEPES-KOH (pH 7.8), 7 mM magnesium acetate, 1 mM EGTA, 0.5 mM DTT, 4 mM ATP, 0.2 mM each CTP, GTP, and UTP, 0.1 mM each dATP, dGTP, and dTTP, 2 μM dCTP, 10 μCi of [α-32P]dCTP, 40 mM creatine phosphate, 0.4 μg of creatine kinase, 10 μg of BSA, 400 ng of RPA, 600 ng of T antigen, 300 ng of topoisomerase I, 400 ng of Pol/Prim, and 100 ng of pUC-HS DNA. After 60 min at 37°C, 5 μl of the mixture was spotted on DE81 paper to quantitate incorporated nucleotides (54). EDTA, SDS, and proteinase K were added to final concentrations of 20 mM, 0.65%, and 1.7 mg/ml, respectively, and incubation was continued for another 30 min. The sample was extracted once with phenol-chloroform and DNA was passed over a G-50 spin column (Boehringer Mannheim) equilibrated in TE buffer to remove unincorporated nucleotides. DNA was ethanol precipitated in the presence of 10 μg of yeast tRNA, dissolved in 20 μl of alkaline loading buffer (50 mM NaOH, 1 mM EDTA, 5% Ficoll 400, 0.025% bromocresol green), and electrophoresed at 4°C in alkaline 1.5% agarose gels in 50 mM NaOH–1 mM EDTA for 10 h at 150 mA with circulating buffer. The gel was fixed in 10% trichloroacetic acid, dried, and exposed to X-ray film.
To uncouple unwinding/initiation from the elongation reaction, a two-step procedure was used. A 40-μl initiation assay mixture containing 0.2 mM CTP and four times the normal amounts of proteins and DNA, but no labeled CTP, was first assembled. After 30 min at 37°C, unincorporated nucleotides were removed by gel filtration on G-50 spin columns (Boehringer Mannheim). (addition of labeled CTP to the DNA complex recovered after gel filtration and further incubation did not result in any significant incorporation of radioactivity, demonstrating efficient removal of nucleoside triphosphates). In the second step, a 40-μl elongation reaction mixture was assembled as described above except that no nucleoside triphosphates were added and the naked DNA was replaced with one-fourth of the DNA complex recovered after gel filtration. Fresh proteins at the standard concentrations were included, since they increased incorporation rates five- to sevenfold (data not shown).
RESULTS
Physical interaction of RPA with T-antigen sequences within the DNA binding domain.
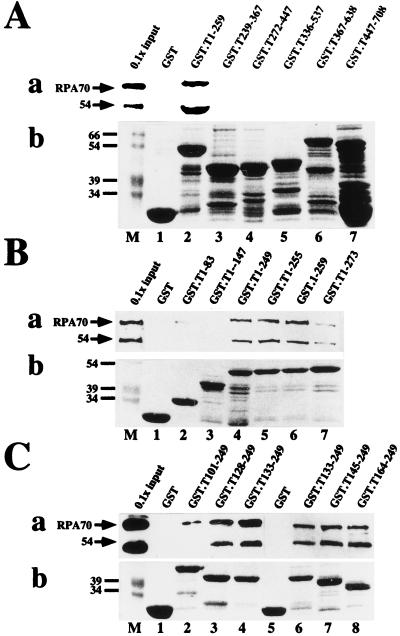
To map the site(s) of interaction of human RPA with T antigen, GST-T antigen fusion proteins bound to glutathione-agarose were tested for the ability to bind to RPA in a protein affinity pull-down assay (Fig. 1). Immunoblotting of the bound material with a monoclonal antibody against RPA70 was used to detect bound RPA. Coarse mapping using large fusion peptides indicated that RPA bound to T-antigen sequences within residues 1 to 259 but not to C-terminal regions of T antigen or to GST used as a negative control (Fig. 1A, panel a). Since the proteins had been treated with nucleases during their purification, this interaction was unlikely to be due to bridging by nucleic acids present in the protein preparations. Furthermore, inclusion of 50 μg of ethidium bromide per ml in the binding reaction (48) did not prevent RPA binding to the fusion proteins (data not shown). Coomassie blue staining of the fusion proteins bound to the beads demonstrated that all of them were present in similar amounts (Fig. 1A, panel b). Fine mapping of the N-terminal 259 residues of T antigen was then performed to define the site of interaction more closely. RPA bound relatively well to T-antigen residues 1 to 249 but poorly to 1 to 83 and 1 to 147 (Fig. 1B, panel a), suggesting that its binding site could be located between residues 147 and 249. Indeed fusion proteins bearing T-antigen residues 128 to 249, 133 to 249, 145 to 249, and 164 to 249 bound well to RPA (Fig. 1C, panel a), demonstrating that a site sufficient for RPA binding resides within the C-terminal portion of the T-antigen DNA binding domain (44).
FIG. 1.
Mapping the RPA binding sequences of SV40 T antigen. The indicated residues of T antigen were expressed as GST fusion proteins and adsorbed to glutathione-agarose. Fusion protein-bound beads were incubated with purified RPA in a pull-down assay. (a) After washing, bound RPA was detected by denaturing gel electrophoresis and immunoblotting with RPA antibody 70C and chemiluminescence (lanes 1 to 7 [A and B] or 1 to 8 [C]). As a marker, 1/10 of the input RPA (lanes M) was analyzed in parallel. Positions of the 70-kDa subunit and a 54-kDa degradation product are indicated by arrows. (b) A 10-μl sample of beads bearing each fusion protein was analyzed by denaturing gel electrophoresis and detected by Coomassie staining (lanes 1 to 7 and 1 to 8). Lanes M show prestained marker proteins. Only the relevant portions of the gels are shown.
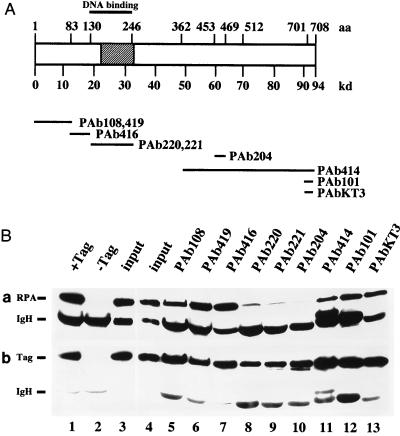
To confirm the location of the T-antigen binding site for RPA, we used a panel of monoclonal antibodies against T antigen whose epitopes had been mapped (Fig. 2A) to immunoprecipitate T antigen and then test for its ability to bind RPA. We reasoned that monoclonal antibodies whose epitopes map outside the region of RPA binding should not interfere with the interaction, while those with epitopes close to or overlapping the RPA binding site might inhibit the interaction. Immunoprecipitation of T antigen was observed with each of the antibodies used (Fig. 2B, panel b). Two antibodies whose epitopes mapped within the T-antigen DNA binding domain, Pab220 and Pab221, precipitated slightly less T antigen than the other antibodies but noticeably diminished the amount of RPA that bound to the T antigen (Fig. 2B, panel a, lanes 8 and 9). This result is consistent with the RPA binding site defined by using T-antigen fusion proteins. However, RPA binding was also inhibited by Pab204, whose epitope was mapped in the C terminus of T antigen well outside the RPA binding site defined by using the fusion proteins (panel a, lane 10). Although this observation was initially surprising, Pab204 was also found to inhibit every other biochemical activity of T antigen that was tested (see Fig. 3 and 4), suggesting that it drastically disrupted the overall structure of the protein.
FIG. 2.
Coimmunoprecipitation of RPA with T antigen. (A) A schematic diagram depicting the amino acid (aa) regions in T antigen (open box) to which the epitopes for the monoclonal antibodies indicated below were mapped. The minimal origin DNA binding domain (44) is indicated by a thick line above the T-antigen diagram. The binding site for RPA determined in Fig. 1 is shown as a hatched box. (B) T antigen was bound to the indicated monoclonal antibody adsorbed to protein G-agarose, and the beads were incubated with RPA. (a) Bound RPA was eluted (lanes 5 to 13), separated by denaturing gel electrophoresis, and detected by immunoblotting with the 70-kDa protein-specific monoclonal antibody 70C. The input T antigen (Tag) and 1/10 of the input RPA were run on the same gel (lane 4). On a separate gel, controls with Pab419 beads loaded with T antigen (lane 1) and without T antigen (lane 2) were analyzed together with a duplicate input control (lane 3). Positions of the 70-kDa subunit (RPA) and the antibody heavy chain (IgH) are indicated. (B) The same blots reprobed with the T-antigen-specific antibody Pab419. Positions of T antigen (Tag) and the heavy chain (IgH) are indicated.
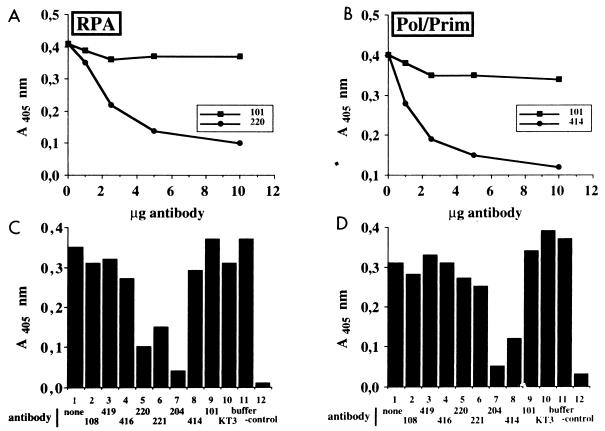
FIG. 3.
Effects of T-antigen-specific monoclonal antibodies on complex formation with cellular initiation proteins. (A and B) T antigen coupled to wells of an ELISA plate was treated with the indicated amounts of monoclonal antibody Pab101 or Pab220 (A) or Pab101 or Pab414 (B). After washing, the wells were incubated with either RPA (A) or Pol/Prim (B). The bound RPA or Pol/Prim was detected by incubation with the corresponding peroxidase-coupled polyclonal antibodies and a chromogenic substrate and then quantitated spectrophotometrically at 405 nm. (C and D) T antigen bound to the wells of the ELISA plate was incubated with T-antigen buffer (column 1), with 10 μg of the indicated monoclonal antibody (columns 2 to 10), or with antibody buffer (column 11). After addition of either RPA (C, columns 1 to 11) or Pol/Prim (D, columns 1 to 11) or neither (column 12), the bound RPA or Pol/Prim was detected as in panels A and B.
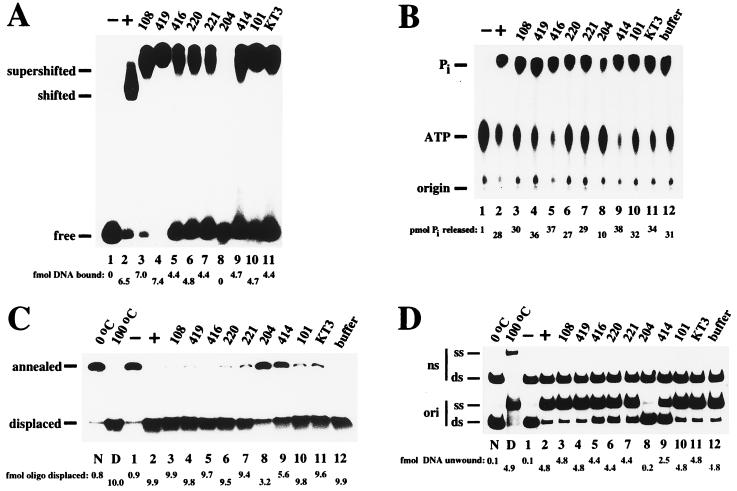
FIG. 4.
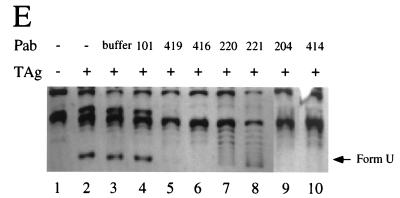
Effects of T-antigen-specific monoclonal antibodies on biochemical activities of T antigen. (A) T-antigen binding to a labeled SV40 origin DNA fragment was tested in a band shift in the presence of the indicated monoclonal antibodies (lanes 3 to 11), without antibody (lane 2), or without T antigen (lane 1). (B) ATPase reactions were carried out without T antigen (lane 1), with T antigen (lane 2), or with T antigen in the presence of the indicated monoclonal antibody (lanes 3 to 11) or buffer (lane 12). The reaction products were separated by ascending thin-layer chromatography. Helicase reactions were performed with M13 DNA annealed to a labeled primer (C), and unwinding reactions were performed with a labeled duplex origin DNA fragment (ori) and a labeled nonspecific DNA fragment (ns) (D), in the presence of the indicated monoclonal antibodies (lanes 3 to 11) or buffer (lanes 12). Negative control reactions were performed without T antigen (lanes 1); positive controls were performed with T antigen and without antibodies (lanes 2) as indicated. (C and D) The substrate DNA in the native conformation (lane N) or after heat denaturation (lane D) was electrophoresed in parallel. Quantitative evaluation of the autoradiograms for each reaction is given below each lane ss and ds, single-stranded and double-stranded DNA, respectively. (E) SV40 DNA unwinding assays contained closed circular supercoiled pUC-HS DNA, T antigen (TAg; lanes 2 to 10), topoisomerase I, and E. coli SSB. Reactions were carried out in the presence of monoclonal antibodies as indicated (lanes 4 to 10) or antibody buffer (lane 3). Reaction products were analyzed by electrophoresis and ethidium bromide staining. Form U, underwound covalently closed circular DNA.
ELISAs were carried out to verify that Pab220 and Pab221 specifically inhibited RPA binding of T antigen. T antigen was immobilized on ELISA plates and incubated with increasing amounts of monoclonal antibody Pab220, Pab414, or Pab101, or buffer as a control (Fig. 3A and B). After washing and incubation with RPA or Pol/Prim, bound protein was detected using peroxidase-conjugated polyclonal rabbit antibodies against either RPA (Fig. 3A) or Pol/Prim (Fig. 3B) and a chromogenic substrate. Maximal inhibition of both RPA binding and Pol/Prim binding to T antigen was observed with 10 μg of monoclonal antibodies Pab220 and Pab414, respectively, while Pab101 displayed little inhibition of either interaction. We then tested the ability of 10 μg of each monoclonal antibody in the panel to inhibit T-antigen interactions with RPA and Pol/Prim (Fig. 3C and D). Pab220 and Pab221 again impaired T-antigen interactions with RPA (Fig. 3C, columns 5 and 6) but had no effect on its interactions with Pol/Prim (Fig. 3D, columns 5 and 6). In agreement with previous reports (14, 25, 31, 69, 72), T-antigen interactions with Pol/Prim were impaired by Pab414 (Fig. 3C and D, columns 8). Pab204 inhibited T-antigen binding to both proteins (columns 7), while the other monoclonal antibodies had little effect on these protein-protein interactions. These results confirm that T-antigen binding to RPA was specifically impaired by Pab220 and Pab221.
Effect of Pab220 and Pab221 on other biochemical activities of T antigen.
The ability of Pab220 and Pab221 to specifically block T-antigen interaction with RPA might provide a way to test the functional relevance of the RPA binding site defined above in viral DNA replication. A clear link between any interference of Pab220 and Pab221 in viral DNA replication with a block in T antigen-RPA binding, however, would require that these antibodies not interfere with other biochemical activities of T antigen. Since Pab220 and Pab221 epitopes map within the DNA binding domain of T antigen, which is involved in multiple functions of the protein (6, 99), specific binding of T antigen to the viral origin of DNA replication and assembly as a double hexamer on the origin seemed the most likely activity with which the antibodies might interfere. An electrophoretic mobility shift assay was used to test binding of T antigen to a labeled origin DNA fragment (Fig. 4A). T antigen-origin DNA complexes migrated more slowly than free DNA (compare lanes 1 and 2). Addition of monoclonal antibodies supershifted the complexes to even lower mobility (lanes 3 to 7 and 9 to 11), except for Pab204, which prevented or disrupted T antigen-origin DNA complex formation (lane 8). The results indicate that Pab220 and Pab221 did not impair origin DNA binding activity of T antigen and that the epitopes were still available for binding in the T antigen-DNA complex.
The ATPase activity of T antigen has been mapped to the C-terminal region of the protein (6, 29) and hence was not expected to be affected by Pab220 or Pab221. In fact, none of the monoclonal antibodies in this panel except Pab204 inhibited the ATPase activity of T antigen (Fig. 4B). Hydrolysis of ATP was reduced to about one-third of the control by Pab204 (compare lanes 2 and 8), in agreement with earlier reports (30, 94). A modest stimulation of ATPase activity was observed with antibodies Pab419, Pab416, and Pab414 (lanes 4, 5, and 9).
The DNA helicase activity of T antigen (6, 29) requires sequences within the origin DNA binding domain (98), suggesting that it might be affected by Pab220 or Pab221. However, in reactions with a partial duplex DNA template, the helicase activity of T antigen was only marginally inhibited by Pab220 or Pab221 (Fig. 4C; compare lanes 6 and 7 with lane 2). Strong inhibition was observed in the presence of Pab204 (lane 8) and Pab414 (lane 9). None of the other antibodies affected helicase activity.
Bidirectional unwinding of SV40 origin DNA requires the coordinated functioning of multiple domains of T antigen: specific binding of T antigen to the origin, assembly as a double hexamer, DNA helicase activity, and probably interactions between the two hexamers (6, 28, 57, 58, 61, 73, 85, 92, 93). The effect of monoclonal antibodies on bidirectional origin DNA unwinding was tested in two different assays, one using linear DNA fragments (Fig. 4D) and one using closed circular supercoiled DNA carrying the origin of replication (Fig. 4E). Since both Pab204 and Pab414 impaired the helicase activity of T antigen, it was not unexpected that they also suppressed origin DNA unwinding in both assays (Fig. 4D, lanes 8 and 9; Fig. 4E, lanes 9 and 10). Four antibodies that had little effect on helicase activity impaired origin DNA unwinding. Pab419 did not inhibit unwinding of linear DNA but did inhibit unwinding of supercoiled DNA (Fig. 4D, lane 4; Fig. 4E, lane 5). Pab416 slightly inhibited unwinding of the linear origin DNA fragment (Fig. 4D, lane 5) and strongly inhibited unwinding of supercoiled DNA (Fig. 4E, lane 6). Pab220 and Pab221 reduced unwinding of the linear origin DNA fragment slightly and also partially inhibited unwinding of supercoiled DNA (Fig. 4D [compare lanes 6 and 7 with lane 2] and 4E [compare lanes 7 and 8 with lane 2]). The other antibodies had no effect on unwinding in either assay.
The effects of this panel of monoclonal antibodies on the biochemical activities of T antigen are summarized in Table 1.
TABLE 1.
Inhibition of biochemical activities of SV40 T antigen by T-antigen-specific monoclonal antibodies
| Antibody | Inhibition ofa:
|
||||||
|---|---|---|---|---|---|---|---|
| SV40 DNA binding | ATPase | Helicase | Unwinding
|
Complex formation
|
|||
| Linear | CCS | RPA | Pol/Prim | ||||
| 108 | − | − | − | − | − | − | − |
| 419 | − | − | − | − | + | − | − |
| 416 | − | − | − | p | + | − | − |
| 220 | − | − | − | p | p | + | − |
| 221 | − | − | − | p | p | + | − |
| 204 | + | + | + | + | + | + | + |
| 414 | − | − | + | + | + | − | + |
| 101 | − | − | − | − | − | − | − |
| KT3 | − | − | − | − | − | − | − |
Purified monoclonal antibodies were tested for the ability to inhibit SV40 origin DNA binding (Fig. 4A); ATPase activity (Fig. 4B); helicase activity using an oligonucleotide-primed M13mp18 ssDNA (Fig. 4C); SV40 origin DNA unwinding activity on linear and closed circular supercoiled (CCS) templates (Fig. 4D and E and data not shown); and complex formation with RPA and Pol/Prim (Fig. 3). +, inhibition; p, partial inhibition; −, lack of inhibition.
Effects of monoclonal antibodies on the early steps in SV40 DNA replication.
The results presented above suggested that with the possible exception of Pab108, Pab101, and KT3, each of the antibodies would be expected to interfere with SV40 DNA replication at one or several of the early steps. Indeed, each of the antibodies in the panel except these three did significantly block SV40 DNA replication in vitro in crude cell extracts (data not shown). However, the functional relevance of the RPA binding site that is blocked by Pab220 and Pab221 cannot be deduced from these experiments, since these antibodies not only inhibited T-antigen interaction with RPA but also partially inhibited origin DNA unwinding, which is known to be independent of a direct physical interaction between T antigen and RPA (3, 14, 45, 60, 97). To distinguish between the effects of Pab220 and Pab221 on origin unwinding and on subsequent steps in replication, we sought to uncouple these events, allowing unwinding to proceed in the absence of antibody and then testing the effect of antibody in subsequent events.
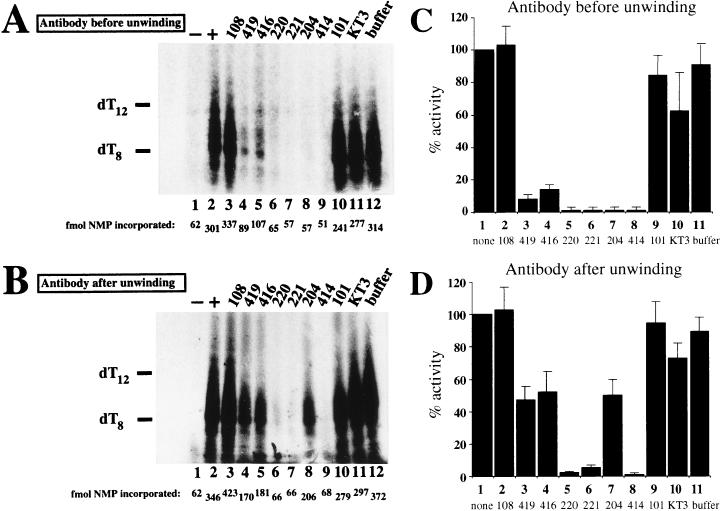
As a foundation for this strategy, conventional coupled initiation reactions containing purified T antigen, RPA, Pol/Prim, and topoisomerase I (56, 84, 89) were first carried out in the presence and absence of each antibody (Fig. 5A). Labeled RNA primers were synthesized in the presence of Pab108, Pab101, and KT3 in amounts similar to those in control reactions (Fig. 5A; compare lanes 3, 10, and 11 with lanes 2 and 12). In contrast, primer synthesis was markedly reduced in the presence of Pab419 and Pab416 (lanes 4 and 5) and nearly absent in the presence of Pab220, Pab221, Pab204, and Pab414 (lanes 6 to 9).
FIG. 5.
Delineation of the step in SV40 initiation at which antibodies interfere. Uncoupled initiation reactions were performed in which antibodies (lanes 3 to 11) or buffer (lanes 12) were added before (A) or after (B) the origin DNA-unwinding step. Negative control reactions contained no T antigen (lanes 1), and positive controls contained no antibodies (lanes 2). NMP, nucleoside monophosphate. (C and D) Mean results from three independent initiation experiments performed as for panels A and B. Brackets indicate the average error of the mean. Incorporation in reactions with antibodies (column 2 to 10) or buffer (column 11) was expressed as a percentage of the value in a reaction without antibody (column 1), defined as 100%.
To determine whether these antibodies interfered with initiation by blocking origin binding and unwinding, or at a later step, the initiation reaction was carried out in two sequential steps (7, 22, 27, 83). In the first step, T antigen, RPA, Pol/Prim, topoisomerase I, and DNA were preincubated with ATP to allow formation of an underwound DNA template, but without the other ribonucleoside triphosphates to prevent primer synthesis. In the second step, ribonucleotides were added in the presence or absence of each monoclonal antibody to assess primer synthesis (Fig. 5B). Primer synthesis was detected at levels resembling the controls in reactions containing Pab108, Pab101, and KT3 (lanes 3 and 10 to 12), and little or no primer synthesis was observed in reactions containing Pab220, Pab221, and Pab414 (lanes 6, 7, and 9). These results were thus largely independent of the time of addition of the antibody to the reaction. Interestingly, however, primer synthesis in the presence of Pab419, Pab416, and Pab204 was clearly less sensitive to inhibition when the antibodies were added after formation of an underwound template DNA (lanes 4, 5, and 8). Quantitative estimates of primer synthesis in three independent experiments with each antibody added to the reaction either prior to origin binding and unwinding, or afterwards, were averaged to give the results depicted in Fig. 5C and D. These results thus separate the antibodies that impaired origin DNA unwinding (Fig. 4D and E) into two classes: those that inhibited DNA replication primarily at the unwinding step and significantly less in primer synthesis (Pab419, Pab416, and Pab204) and those that inhibited both steps (Pab220, Pab221, and Pab414).
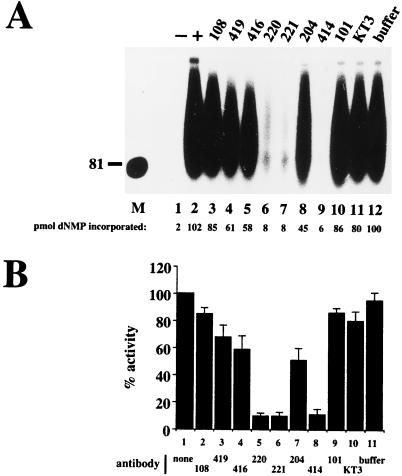
To test whether elongation of RNA primers was also sensitive to inhibition by these monoclonal antibodies, an elongation reaction was carried out in two steps. In the first, origin DNA binding, unwinding, and primer synthesis were permitted in the absence of deoxyribonucleoside triphosphates, and the primed unwound template was isolated by gel filtration to remove unincorporated ribonucleoside triphosphates. Addition of labeled CTP to this isolated template did not support synthesis of labeled products (data not shown), indicating that ribonucleotides had been removed. In the second step, deoxyribonucleoside triphosphates were added to permit primer elongation, either in the presence or in the absence of each monoclonal antibody. Supplementation of the reaction with fresh replication proteins in the second step stimulated incorporation five- to sevenfold, while fresh Pol/Prim alone stimulated incorporation three- to fivefold (data not shown), suggesting that some Pol/Prim, and possibly other proteins, had dissociated from the prereplication complex during gel filtration. The reactions shown here were therefore supplemented with fresh proteins prior to elongation. Primer elongation in the presence of Pab108, Pab101, and KT3 was nearly as efficient in as the control reactions (Fig. 6A; compare lanes 3, 10, and 11 with lanes 2 and 12). Pab419, Pab416, and Pab204 reduced primer elongation to about half of the level observed in the control reactions (lanes 4, 5, and 8). In contrast, primer elongation in the presence of Pab220, Pab221, and Pab414 was almost completely blocked (lanes 6, 7, and 9). Quantitative estimates of primer elongation products formed in three independent experiments in the presence and absence of each antibody were averaged (Fig. 6B) and confirmed this conclusion. These results demonstrate that Pab419, Pab416, and Pab204 inhibited DNA replication primarily at the origin-unwinding step and significantly less in primer synthesis and elongation, while Pab220, Pab221, and Pab414 essentially abolished primer synthesis and elongation even when added after origin unwinding.
FIG. 6.
Influence of antibodies on primer elongation in the absence of primer synthesis. (A) Standard initiation reactions were performed except that T antigen was omitted from the negative control reaction (lane 1). After the reaction, unincorporated ribonucleoside triphosphates were removed by gel filtration, and the primed DNA-protein complex was recovered. Elongation reactions containing the primed DNA-protein complex supplemented with additional proteins and deoxyribonucleoside triphosphates were carried out in the presence of the indicated antibodies (lanes 3 to 11) or buffer (lane 12). A positive control reaction contained no antibody (lane 2). dNMP, deoxynucleoside monophosphate. (B) Quantitation of results of three independent experiments as in panel A. The mean is shown as a percentage of the positive control, which was set at 100%. The brackets indicate the average error of the mean.
DISCUSSION
SV40 T antigen has been previously shown to interact physically and functionally with RPA during initiation of viral DNA replication and lagging-strand DNA synthesis (2, 14, 24, 26, 56, 60, 65, 67). Here we have used fusion peptides of T-antigen and anti-T-antigen monoclonal antibodies whose epitopes have been mapped to localize the sequences in T antigen that interact with RPA and to confirm the functional relevance of this binding site in viral DNA replication.
The region of T antigen (residues 164 to 249) that binds to RPA is localized within the DNA binding domain of T antigen (Fig. 1 and 2). Genetic evidence has implicated the DNA binding domain in multiple functions of T antigen (99). Biochemical studies demonstrate that it not only is essential for sequence-specific binding to the SV40 control region DNA but also participates in multiple interactions with host cell proteins. Among the proteins known to bind within this region of T antigen are the transcription factors TATA binding protein (TBP), TFIIB, several TBP-associated factors, TEF-1, Sp1, RNA polymerase II, and topoisomerase I (1, 16, 34, 39, 43, 71). Finally, functional interactions between T-antigen hexamers during bidirectional origin DNA unwinding appear to require sequences within the DNA binding domain (58, 92). Competition studies indicate that not all of the transcription factors can bind to T antigen at once, suggesting that some of the binding sites for these proteins may overlap (43). However, at least several separate protein interaction sites appear to reside within the DNA binding domain. Preliminary evidence from competition experiments suggests that RPA binds to a region of T antigen that does not overlap with the binding site for TBP or TEF-1 (39). The observation that monoclonal antibodies Pab220 and Pab221 block RPA binding but not DNA binding to T antigen (Fig. 2, 3, and 4A) indicates that the RPA and DNA binding surfaces are unlikely to overlap. However, it remains possible that the topoisomerase I binding site of T antigen may overlap the RPA binding site.
The solution structure of the DNA binding domain of T antigen was recently determined by nuclear magnetic resonance spectroscopy, and on the basis of spectroscopic and genetic data, the origin DNA binding surface has been modeled to include two neighboring loops containing residues 152 to 155 and 203 to 207 (44, 52, 70, 99). A mutation at residue 189 (S189N) impairs T-antigen binding to TEF-1, activation of transcription by TEF-1, stimulation of quiescent cells, and cell transformation by T antigen (1, 21). Residue 189 is located in a loop between β strands B and C that resides on the opposite side of the DNA binding domain from the proposed DNA binding surface (52) and that may comprise part of the TEF-1 binding site. Mutations at residues 173 and 174 (K173A and K174A) were reported to prevent T-antigen interaction with several transcription factors and to block transactivation by T antigen (43). Also, a small in-frame insertion mutation at residue 168 was shown to significantly reduce transactivation activity (16). These three residues are all located in α helix B on one surface of the DNA binding domain (52), which may constitute part of a binding surface for transcription factors that is distinct from that for origin DNA.
Based on the genetic and biochemical evidence above, we suggest that the RPA binding surface is unlikely to overlap with those for either the transcription factors or the viral origin. Functional interactions of RPA with the T-antigen-related proteins, polyomavirus T antigen, and bovine papillomavirus E1 protein, as well as with EBNA-1, have been observed (4, 59, 101) and may reflect similar binding sites in these proteins for RPA. Although there is little homology between SV40 T antigen and EBNA-1, comparison of the RPA binding region of SV40 T antigen with the entire sequences of polyomavirus T antigen and E1 reveals short regions of homology that correspond to SV40 T antigen residues 194 to 199 and 196 to 200, which are located at the junction between β strand C and the second loop postulated for the DNA binding surface (52). Most of the residues between 194 to 200 are not exposed on the surface (52), but it will be interesting to determine whether mutations in this region or neighboring sequences in the three-dimensional structure affect RPA binding activity.
T antigen has recently been reported to associate with the large RPA subunit RPA70 within residues 1 to 326, but not 1 to 168 or 237 to 616 (2), suggesting that the T-antigen binding site probably resides between residues 168 and 237. We previously demonstrated that T antigen associated with native trimeric RPA but not with recombinant RPA70 expressed as an insoluble protein in bacteria (24). However, our more recent studies performed with soluble RPA70 expressed as a fusion protein confirm that RPA70 is sufficient by itself to bind to T antigen (91). Since RPA70 is poorly soluble (32, 38), it seems likely that the concentration of the resolubilized RPA70 used in our early experiments was too low to detect the interaction with T antigen. Consistent with this interpretation, recent evidence from surface plasmon resonance experiments indicates that T-antigen affinity for RPA is about an order of magnitude weaker than its affinity for Pol/Prim (33).
RPA binding to T antigen was specifically inhibited by monoclonal antibodies Pab220 and Pab221 (Fig. 2 and 3). These antibodies recognize native but not denatured sequences within the DNA binding domain of T antigen (62). These antibodies had little effect on other biochemical functions of T antigen that are known to play a role in viral DNA replication (Fig. 4). The partial inhibition of T-antigen-mediated unwinding of closed circular origin DNA detected in the presence of Pab220 and Pab221 may be due to partial interference with the hexamer-hexamer interactions that are implicated in origin DNA unwinding (12, 57, 58, 61, 73, 85, 92, 93). Several other monoclonal antibodies inhibited unwinding of closed circular origin DNA essentially completely (Fig. 4E). Pab419, whose epitope mapped within the J domain at the N terminus of T antigen (Fig. 2) (11, 75), strongly inhibited unwinding, as did Pab416, whose epitope mapped between the J domain and the DNA binding domain. Pab414, whose epitope mapped to the C terminus, inhibited not only unwinding but also DNA helicase activity (Fig. 4C to E) and binding to Pol/Prim (14, 25, 72). Pab204 inhibited virtually every replication-related activity of T antigen (Fig. 2 to 6 and reference 94), suggesting that it probably disrupts the global structure of the protein. All of these antibodies were also potent inhibitors of initiation of SV40 DNA replication (Fig. 5A). Interestingly, when Pab419, -416, or -204 was added to the initiation reaction after origin DNA unwinding, the inhibition was significantly relieved (Fig. 5B), suggesting either that these epitopes were masked in the origin DNA-protein complex or that further unwinding of the template DNA was not required to observe primer synthesis (Fig. 5B) or primer elongation (Fig. 6).
In contrast with these antibodies, Pab414 strongly interfered with primer synthesis and primer elongation even when added to the assays after origin DNA unwinding or after unwinding and primer synthesis (Fig. 5B and 6). This interference may reflect the essential role of T-antigen binding to Pol/Prim in primer synthesis and elongation (14, 15, 23–25, 30, 31, 69, 72, 77), but interference with unwinding during primer synthesis and elongation is difficult to rule out (Fig. 4C to E). Recent evidence from fluorescence spectroscopy indicates that the stoichiometry of Pol/Prim binding to T-antigen monomers in solution is 1:6 (41), consistent with a model in which one Pol/Prim would be located on each lagging-strand template, tethered to a T-antigen double hexamer associated with both replication forks (22, 28, 40, 55, 61, 73, 93). Pab220 and Pab221 interfered with primer synthesis and elongation even when added to the reactions after template unwinding or unwinding and primer synthesis (Fig. 5B and 6), implying that T antigen-RPA interactions are required for both primer synthesis and elongation. These results are consistent with previous data that yeast RPA failed to bind to T antigen and to support primer synthesis on ssDNA (60). The critical role of RPA-T antigen interactions in primer elongation was more unexpected, particularly since much of the Pol/Prim that synthesized primers in the first step of the assay apparently dissociated from the primed unwound template during gel filtration (Fig. 6). T antigen, once assembled as a double hexamer active in unwinding, has been shown to be processive (65), probably due to the toroidal structure of each hexamer encircling the DNA (68). RPA was probably retained on the unwound template DNA during gel filtration through its strong DNA binding affinity (96). Nevertheless, prevention or disruption of RPA binding to T antigen on the primed unwound template appeared to be sufficient to prevent primer elongation by freshly added Pol/Prim (Fig. 6). These observations suggest the existence of a multiprotein complex involving multiple protein-protein interactions in lagging-strand synthesis. This report defining the RPA binding site in T antigen represents one further step toward a clearer understanding of how this complex works.
ACKNOWLEDGMENTS
We thank A. Brunahl for help with antibody purification and ELISAs, I. Moarefi, A. Arthur, and A. Wildeman for sharing plasmids and topoisomerase I, V. Podust for purified SSB, D. von Winkler for antiserum against RPA, H.-P. Nasheuer for antiserum against Pol/Prim, and M. Kenny, J. Hurwitz, D. P. Lane, E. Harlow, E. Gurney, and G. Walter for monoclonal antibodies. We thank T. Melendy and F. Grosse for communication of unpublished data, and we thank V. Podust and U. Herbig for criticism of the manuscript.
The financial support of the NIH (GM52948), Vanderbilt University, and the NSF (Shared Instrumentation grant BIR-9419667) is gratefully acknowledged. These studies were begun with a grant from the German Science Foundation to E.F.
REFERENCES
- 1.Berger L C, Smith D B, Davidson I, Hwang J-J, Fanning E, Wildeman A G. Interaction between T antigen and the TEA domain of the factor derepresses simian virus 40 late promoter in vivo: identification of the T-antigen domains important for transcriptional control. J Virol. 1996;70:1203–1212. doi: 10.1128/jvi.70.2.1203-1212.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Braun K A, Lao Y, He Z, Ingles J, Wold M S. Role of protein-protein interactions in the function of replication protein A (RPA): RPA modulates the activity of DNA polymerase alpha by multiple mechanisms. Biochemistry. 1997;36:8443–8454. doi: 10.1021/bi970473r. [DOI] [PubMed] [Google Scholar]
- 3.Brill S J, Stillman B. Yeast replication factor-A functions in the unwinding of the SV40 origin of DNA replication. Nature. 1989;342:92–95. doi: 10.1038/342092a0. [DOI] [PubMed] [Google Scholar]
- 4.Brinton B, Hassell J A. SV40 and polyoma DNA replication. In: DePamphilis M L, editor. DNA replication in eukaryotic cells. Plainview, N.Y: Cold Spring Harbor Laboratory Press; 1996. pp. 639–677. [Google Scholar]
- 5.Brush G S, Kelly T J. Mechanisms for replicating DNA. In: DePamphilis M L, editor. DNA replication in eukaryotic cells. Plainview, N.Y: Cold Spring Harbor Laboratory Press; 1996. pp. 1–43. [Google Scholar]
- 6.Bullock P A. The initiation of simian virus 40 DNA replication in vitro. Crit Rev Biochem Mol Biol. 1997;32:503–568. doi: 10.3109/10409239709082001. [DOI] [PubMed] [Google Scholar]
- 7.Bullock P A, Seo Y S, Hurwitz J. Initiation of simian virus 40 DNA synthesis in vitro: pulse-chase experiments identify the first labeled species as topologically unwound. Proc Natl Acad Sci USA. 1989;86:3944–3948. doi: 10.1073/pnas.86.11.3944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bullock P A, Seo Y S, Hurwitz J. Initiation of simian virus 40 DNA synthesis in vitro. Mol Cell Biol. 1991;11:2350–2361. doi: 10.1128/mcb.11.5.2350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bullock P A, Tevosian S, Jones C, Denis D. Mapping initiation sites for simian virus 40 lagging-strand DNA synthesis events in vitro. Mol Cell Biol. 1994;14:5043–5055. doi: 10.1128/mcb.14.8.5043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bullock P A, Denis D. DNA synthesis generally initiates outside of the simian virus 40 core origin in vitro. Mol Cell Biol. 1995;15:173–178. doi: 10.1128/mcb.15.1.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Campbell K, Mullane K, Aksoy I, Stubdal H, Pipas J, Silver P, Roberts T, Schaffhausen B, DeCaprio J. DnaJ/hsp40 chaperone domain of SV40 large T antigen promotes efficient viral DNA replication. Genes Dev. 1997;11:1098–1110. doi: 10.1101/gad.11.9.1098. [DOI] [PubMed] [Google Scholar]
- 12.Cegielska A, Moarefi I, Fanning E, Virshup D M. T-antigen kinase inhibits simian virus 40 DNA replication by phosphorylation of intact T antigen on serines 120 and 123. J Virol. 1994;68:269–275. doi: 10.1128/jvi.68.1.269-275.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Clark R, Lane D P, Tjian R. Use of monoclonal antibodies as probes of simian virus 40 T antigen ATPase activity. J Biol Chem. 1981;256:11854–11858. [PubMed] [Google Scholar]
- 14.Collins K L, Kelly T J. The effects of T antigen and replication protein A on the initiation of DNA synthesis by DNA polymerase α-primase. Mol Cell Biol. 1991;11:2108–2115. doi: 10.1128/mcb.11.4.2108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Collins K L, Russo A A R, Tseng B Y, Kelly T J. The role of the 70 kDa subunit of human DNA polymerase α in DNA replication. EMBO J. 1993;12:4555–4566. doi: 10.1002/j.1460-2075.1993.tb06144.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Damania B, Alwine J C. TAF-like function of SV40 large T antigen. Genes Dev. 1996;10:1369–1381. doi: 10.1101/gad.10.11.1369. [DOI] [PubMed] [Google Scholar]
- 17.Dean F B, Bullock P A, Murakami Y, Wobbe C R, Weissbach L, Hurwitz J. Simian virus 40 (SV40) DNA replication: SV40 large T antigen unwinds DNA containing the SV40 origin of replication. Proc Natl Acad Sci USA. 1987;84:16–20. doi: 10.1073/pnas.84.1.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dean F B, Dodson M, Echols H, Hurwitz J. ATP-dependent formation of a specialized nucleoprotein structure by simian virus 40 (SV40) large tumor antigen at the SV40 replication origin. Proc Natl Acad Sci USA. 1987;84:8981–8985. doi: 10.1073/pnas.84.24.8981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.DeLucia A L, Deb S, Partin K, Tegtmeyer P. Functional interactions of the simian virus 40 core origin of replication with flanking regulatory sequences. J Virol. 1986;57:138–144. doi: 10.1128/jvi.57.1.138-144.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Denis D, Bullock P A. Primer-DNA formation during simian virus 40 DNA replication in vitro. Mol Cell Biol. 1993;13:2882–2890. doi: 10.1128/mcb.13.5.2882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dickmanns A, Zeitvogel A, Simmersbach F, Weber R, Arthur A K, Dehde S, Wildeman A G, Fanning E. The kinetics of simian virus 40-induced progression of quiescent cells into S phase depend on four independent functions of large T antigen. J Virol. 1994;68:5496–5508. doi: 10.1128/jvi.68.9.5496-5508.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dodson M, Dean F B, Bullock P, Echols H, Hurwitz J. Unwinding of duplex DNA from the SV40 origin of replication by T antigen. Science. 1987;238:964–968. doi: 10.1126/science.2823389. [DOI] [PubMed] [Google Scholar]
- 23.Dornreiter I, Copeland W C, Wang T S-F. Initiation of simian virus 40 DNA replication requires the interaction of a specific domain of human DNA polymerase α with large T antigen. Mol Cell Biol. 1993;13:809–820. doi: 10.1128/mcb.13.2.809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dornreiter I, Erdile L F, Gilbert I U, von Winkler D, Kelly T J, Fanning E. Interaction of DNA polymerase α-primase with cellular replication protein A and SV40 T antigen. EMBO J. 1992;11:769–776. doi: 10.1002/j.1460-2075.1992.tb05110.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dornreiter I, Höss A, Arthur A K, Fanning E. SV40 T antigen binds directly to the large subunit of DNA polymerase alpha. EMBO J. 1990;9:3329–3336. doi: 10.1002/j.1460-2075.1990.tb07533.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Erdile L F, Heyer W D, Kolodner R, Kelly T J. Characterization of a cDNA encoding the 70-kDa single-stranded DNA-binding subunit of human replication protein A and the role of the protein in DNA replication. J Biol Chem. 1991;266:12090–12096. [PubMed] [Google Scholar]
- 27.Fairman M P, Stillman B. Cellular factors required for multiple stages of SV40 replication in vitro. EMBO J. 1988;7:1211–1218. doi: 10.1002/j.1460-2075.1988.tb02933.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fanning E. Control of SV40 DNA replication by protein phosphorylation: a model for cellular DNA replication? Trends Cell Biol. 1994;4:250–255. doi: 10.1016/0962-8924(94)90123-6. [DOI] [PubMed] [Google Scholar]
- 29.Fanning E, Knippers R. Structure and function of simian virus 40 large tumor antigen. Annu Rev Biochem. 1992;61:55–85. doi: 10.1146/annurev.bi.61.070192.000415. [DOI] [PubMed] [Google Scholar]
- 30.Gannon J V, Lane D P. p53 and DNA polymerase α compete for binding to SV40 T antigen. Nature (London) 1987;329:456–458. doi: 10.1038/329456a0. [DOI] [PubMed] [Google Scholar]
- 31.Gannon J V, Lane D P. Interactions between SV40 T antigen and DNA polymerase α. New Biol. 1990;2:84–92. [PubMed] [Google Scholar]
- 32.Gomes X V, Wold M S. Structural analysis of human replication protein A: mapping functional domains of the 70-kDa subunit. J Biol Chem. 1995;270:4534–4543. doi: 10.1074/jbc.270.9.4534. [DOI] [PubMed] [Google Scholar]
- 33.Grosse, F. Personal communication.
- 34.Gruda M, Zabolotny J, Xiao J, Davidson I, Alwine J. Transcriptional activation by simian virus 40 large T antigen: interactions with multiple components of the transcription complex. Mol Cell Biol. 1993;13:961–969. doi: 10.1128/mcb.13.2.961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gurney E G, Harrison R O, Fenno J. Monoclonal antibodies against simian virus 40 T antigens: evidence for distinct subclasses of large T antigen and for similarities among nonviral T antigens. J Virol. 1980;34:752–763. doi: 10.1128/jvi.34.3.752-763.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gurney E G, Tamowsky D, Deppert W. Antigenic binding sites of monoclonal antibodies specific for simian virus 40 large T. J Virol. 1986;57:1168–1172. doi: 10.1128/jvi.57.3.1168-1172.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Harlow E, Crawford L V, Pim D C, Williamson N M. Monoclonal antibodies specific for simian virus 40 tumor antigen. J Virol. 1981;39:861–869. doi: 10.1128/jvi.39.3.861-869.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Henricksen L A, Umbricht C B, Wold M S. Recombinant replication protein A: expression, complex formation, and functional characterization. J Biol Chem. 1994;269:11121–11132. [PubMed] [Google Scholar]
- 39.Herbig, U., K. Weisshart, P. Taneja, and E. Fanning. Interaction of the transcription factor TFIID with SV40 large T antigen interferes with replication of SV40 DNA in vitro. Submitted for publication. [DOI] [PMC free article] [PubMed]
- 40.Herendeen D, Kelly T J. SV40 DNA replication. In: Blow J J, editor. Eukaryotic DNA replication. New York, N.Y: Oxford University Press; 1996. pp. 29–65. [Google Scholar]
- 41.Huang, S.-G., K. Weisshart, I. Gilbert, and E. Fanning. Stoichiometry and mechanism of assembly of SV40 T antigen complexes with the viral origin of DNA replication and DNA polymerase α-primase. Biochemistry, in press. [DOI] [PubMed]
- 42.Ishimi Y, Claude A, Bullock P, Hurwitz J. Complete enzymatic synthesis of DNA containing the SV40 origin of replication. J Biol Chem. 1988;263:19723–19733. [PubMed] [Google Scholar]
- 43.Johnston S D, Yu X-M, Mertz J E. The major transcriptional transactivation domain of simian virus 40 large T antigen associates nonconcurrently with multiple components of the transcriptional preinitiation complex. J Virol. 1996;70:1191–1202. doi: 10.1128/jvi.70.2.1191-1202.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Joo W X, Luo X, Denis D, Kim H Y, Rainey G J, Jones C, Sreekumar K R, Bullock P A. Purification of the simian virus 40 (SV40) T antigen DNA-binding domain and characterization of its interactions with the SV40 origin. J Virol. 1997;71:3972–3985. doi: 10.1128/jvi.71.5.3972-3985.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kenny M K, Lee S-H, Hurwitz J. Multiple functions of human single-stranded DNA binding protein in simian virus 40 DNA replication: single-strand stabilization and stimulation of DNA polymerase α and δ. Proc Natl Acad Sci USA. 1989;86:9757–9761. doi: 10.1073/pnas.86.24.9757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kenny M K, Schlegel U, Furneaux H, Hurwitz J. The role of human single-stranded DNA binding protein and its individual subunits in simian virus 40 DNA replication. J Biol Chem. 1990;265:7693–7700. [PubMed] [Google Scholar]
- 47.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature (London) 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 48.Lai J, Herr W. Ethidium bromide provides a simple tool for establishing genuine DNA-independent protein associations. Proc Natl Acad Sci USA. 1992;89:6958–6962. doi: 10.1073/pnas.89.15.6958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lee S-H, Kim D K. The role of the 34-kDa subunit of human replication protein in simian virus 40 DNA replication in vitro. J Biol Chem. 1995;270:12801–12807. doi: 10.1074/jbc.270.21.12801. [DOI] [PubMed] [Google Scholar]
- 50.Lee S-H, Eki T, Hurwitz J. Synthesis of DNA containing the simian virus 40 origin of replication by the combined action of DNA polymerase α and δ. Proc Natl Acad Sci USA. 1989;86:7361–7365. doi: 10.1073/pnas.86.19.7361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lohman T M, Green J M, Beyer R S. Large-scale overproduction and rapid purification of the Escherichia coli ssb gene product. Expression of the ssb gene under λ PL control. Biochemistry. 1986;25:21–25. doi: 10.1021/bi00349a004. [DOI] [PubMed] [Google Scholar]
- 52.Luo X, Sanford D G, Bullock P A, Bachovchin W W. Solution structure of the origin specific DNA binding domain from simian virus 40 T-antigen. Nat Struct Biol. 1996;3:1034–1039. doi: 10.1038/nsb1296-1034. [DOI] [PubMed] [Google Scholar]
- 53.MacArthur H, Walter G. Monoclonal antibodies specific for the carboxy terminus of simian virus 40 large T antigen. J Virol. 1984;52:483–491. doi: 10.1128/jvi.52.2.483-491.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Maniatis T, Fritsch E F, Sambrook J. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 55.Mastrangelo I A, Hough P V C, Wilson V G, Wall J S, Hainfeld J F, Tegtmeyer P. ATP-dependent assembly of double hexamers of SV40 T antigen at the viral origin of DNA replication. Nature (London) 1989;338:658–662. doi: 10.1038/338658a0. [DOI] [PubMed] [Google Scholar]
- 56.Matsumoto T, Eki T, Hurwitz J. Studies on the initiation and elongation reactions in the simian virus 40 DNA replication system. Proc Natl Acad Sci USA. 1990;87:9712–9716. doi: 10.1073/pnas.87.24.9712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.McVey D, Ray S, Gluzman Y, Berger L, Wildeman A G, Marshak D R, Tegtmeyer P. cdc2 phosphorylation of threonine 124 activates the origin-unwinding functions of simian virus 40 T antigen. J Virol. 1993;67:5206–5215. doi: 10.1128/jvi.67.9.5206-5215.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.McVey D, Woelker B, Tegtmeyer P. Mechanisms of simian virus 40 T-antigen activation by phosphorylation of threonine 124. J Virol. 1996;70:3887–3893. doi: 10.1128/jvi.70.6.3887-3893.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Melendy, T. Personal communication.
- 60.Melendy T, Stillman B. An interaction between replication protein A and SV40 T antigen appears essential for primosome assembly during SV40 DNA replication. J Biol Chem. 1993;268:3389–3395. [PubMed] [Google Scholar]
- 61.Moarefi I F, Small D, Gilbert I, Hoepfner M, Randall S K, Schneider C, Russo A R R, Ramsperger U, Arthur A K, Stahl H, Kelly T J, Fanning E. Mutation of the cyclin-dependent kinase phosphorylation site in simian virus 40 (SV40) large T antigen specifically blocks SV40 origin DNA binding. J Virol. 1993;67:4992–5002. doi: 10.1128/jvi.67.8.4992-5002.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Mole S E, Gannon J V, Ford M J, Lane D P. Structure and function of large T antigen. Philos Trans R Soc Lond Ser B. 1987;317:455–469. doi: 10.1098/rstb.1987.0072. [DOI] [PubMed] [Google Scholar]
- 63.Murakami Y, Wobbe C R, Weissbach L, Dean F B, Hurwitz J. Role of DNA polymerase α and DNA primase in simian virus 40 DNA replication in vitro. Proc Natl Acad Sci USA. 1986;83:2869–2873. doi: 10.1073/pnas.83.9.2869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Murakami Y, Eki T, Hurwitz J. Studies on the initiation of simian virus 40 replication in vitro: RNA primer synthesis and its elongation. Proc Natl Acad Sci USA. 1992;89:952–956. doi: 10.1073/pnas.89.3.952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Murakami Y, Hurwitz J. Functional interactions between SV40 T antigen and other replication proteins at the replication fork. J Biol Chem. 1993;268:11008–11017. [PubMed] [Google Scholar]
- 66.Murakami Y, Hurwitz J. DNA polymerase α stimulates the ATP-dependent binding of simian virus 40 tumor (T) antigen to the SV40 origin of replication. J Biol Chem. 1993;268:11018–11027. [PubMed] [Google Scholar]
- 67.Nasheuer H-P, von Winkler D, Schneider C, Dornreiter I, Gilbert I, Fanning E. Purification and functional characterization of bovine RP-A in an in vitro SV40 DNA replication system. Chromosoma. 1992;102:S52–S59. doi: 10.1007/BF02451786. [DOI] [PubMed] [Google Scholar]
- 68.San Martin M C, Gruss C, Carazo J M. Six molecules of SV40 large T antigen assemble in a propeller-shaped particle around a channel. J Mol Biol. 1997;268:15–20. doi: 10.1006/jmbi.1997.0952. [DOI] [PubMed] [Google Scholar]
- 69.Schneider C, Weisshart K, Guarino L A, Dornreiter I, Fanning E. Species-specific functional interactions of DNA polymerase α-primase with simian virus 40 (SV40) T antigen require SV40 origin DNA. Mol Cell Biol. 1994;14:3176–3185. doi: 10.1128/mcb.14.5.3176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Simmons D T, Loeber G, Tegtmeyer P. Four major sequence elements of simian virus 40 large T antigen coordinate its specific and nonspecific DNA binding. J Virol. 1990;64:1973–1983. doi: 10.1128/jvi.64.5.1973-1983.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Simmons D T, Melendy T, Usher D, Stillman B. Simian virus 40 large T antigen binds to topoisomerase I. Virology. 1996;222:365–374. doi: 10.1006/viro.1996.0433. [DOI] [PubMed] [Google Scholar]
- 72.Smale S T, Tjian R. T-antigen-DNA polymerase alpha complex implicated in simian virus 40 DNA replication. Mol Cell Biol. 1986;6:4077–4087. doi: 10.1128/mcb.6.11.4077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Smelkova N V, Borowiec J A. Dimerization of simian virus 40 T-antigen hexamers activates T-antigen DNA helicase activity. J Virol. 1997;71:8766–8773. doi: 10.1128/jvi.71.11.8766-8773.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Smith D B, Johnson K S. Single-step purification of polypeptides expressed in Escherichia coli as fusions with glutathione S-transferase. Gene. 1988;67:31–40. doi: 10.1016/0378-1119(88)90005-4. [DOI] [PubMed] [Google Scholar]
- 75.Srinivasan A, McClellan A J, Vartikar J, Marks I, Cantalupo P, Li Y, Whyte P, Rundell K, Brodsky J L, Pipas J M. The amino-terminal transforming region of simian virus 40 large T and small t antigens functions as a J domain. Mol Cell Biol. 1997;17:4761–4773. doi: 10.1128/mcb.17.8.4761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Stadlbauer F, Brückner A, Rehfuess C, Eckershorn C, Lottspeich F, Förster V, Tseng B-Y, Nasheuer H-P. DNA replication in vitro by recombinant DNA-polymerase α–primase. Eur J Biochem. 1994;222:781–793. doi: 10.1111/j.1432-1033.1994.tb18925.x. [DOI] [PubMed] [Google Scholar]
- 77.Stadlbauer F, Voitenleitner C, Brückner A, Fanning E, Nasheuer H-P. Species-specific replication of simian virus 40 DNA in vitro requires the p180 subunit of human DNA polymerase α-primase. Mol Cell Biol. 1996;16:94–1104. doi: 10.1128/mcb.16.1.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Stigger E, Dean F, Hurwitz J, Lee S-H. Reconstitution of functional human single-stranded DNA-binding protein from individual subunits expressed by recombinant baculoviruses. Proc Natl Acad Sci USA. 1994;91:579–583. doi: 10.1073/pnas.91.2.579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Stillman B. Comparison of DNA replication in cells from prokarya and eukarya. In: DePamphilis M L, editor. DNA replication in eukaryotic cells. Plainview, N.Y: Cold Spring Harbor Laboratory Press; 1996. pp. 435–460. [Google Scholar]
- 80.Strausfeld U, Richter A. Simultaneous purification of DNA topoisomerases I and II from eucaryotic cells. Prep Biochem. 1989;19:37–48. doi: 10.1080/10826068908544895. [DOI] [PubMed] [Google Scholar]
- 81.Towbin H, Staehelin T, Gordon J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc Natl Acad Sci USA. 1979;76:4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Tsurimoto T, Stillman B. Multiple replication factors augment DNA synthesis by two eukaryotic DNA polymerases, α and δ. EMBO J. 1989;8:3883–3889. doi: 10.1002/j.1460-2075.1989.tb08567.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Tsurimoto T, Fairman M P, Stillman B. Simian virus 40 DNA replication in vitro: identification of multiple stages of initiation. Mol Cell Biol. 1989;9:3839–3849. doi: 10.1128/mcb.9.9.3839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Tsurimoto T, Melendy T, Stillman B. Sequential initiation of lagging and leading strand synthesis by two different polymerase complexes at the SV40 replication origin. Nature (London) 1990;346:534–539. doi: 10.1038/346534a0. [DOI] [PubMed] [Google Scholar]
- 85.Virshup D M, Russo A A R, Kelly T J. Mechanism of activation of simian virus 40 DNA replication by protein phosphatase 2A. Mol Cell Biol. 1992;12:4883–4895. doi: 10.1128/mcb.12.11.4883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Voitenleitner C, Fanning E, Nasheuer H-P. Phosphorylation of DNA polymerase α-primase by cyclin A-dependent kinases regulates initiation of DNA replication in vitro. Oncogene. 1997;14:1611–1615. doi: 10.1038/sj.onc.1200975. [DOI] [PubMed] [Google Scholar]
- 87.Waga S, Stillman B. Anatomy of a DNA replication fork revealed by reconstitution of SV40 DNA replication in vitro. Nature (London) 1994;369:207–212. doi: 10.1038/369207a0. [DOI] [PubMed] [Google Scholar]
- 88.Waga S, Bauer G, Stillman B. Reconstitution of complete SV40 DNA replication with purified replication factors. J Biol Chem. 1994;269:10923–10934. [PubMed] [Google Scholar]
- 89.Weinberg D H, Collins K L, Simancek P, Russo A, Wold M S, Virshup D M, Kelly T J. Reconstitution of simian virus 40 DNA replication with purified proteins. Proc Natl Acad Sci USA. 1990;87:8692–8696. doi: 10.1073/pnas.87.22.8692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Weisshart K, Bradley M K, Weiner B M, Schneider C, Moarefi I, Fanning E, Arthur A K. An N-terminal deletion mutant of simian virus 40 (SV40) large T antigen oligomerizes incorrectly on SV40 DNA but retains the ability to bind to DNA polymerase α and replicate SV40 DNA in vitro. J Virol. 1996;70:3509–3516. doi: 10.1128/jvi.70.6.3509-3516.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Weisshart, K., P. Taneja, I. Boche, A. Brunahl, and E. Fanning. Unpublished data.
- 92.Weisshart, K., P. Taneja, A. Jenne, U. Herbig, D. T. Simmons, and E. Fanning. Two regions of SV40 T antigen determine cooperativity of double hexamer assembly on the viral origin of DNA replication and promote hexamer interactions during bidirectional origin DNA unwinding. Submitted for publication. [DOI] [PMC free article] [PubMed]
- 93.Wessel R, Schweizer J, Stahl H. Simian virus 40 large T antigen DNA helicase is a hexamer which forms a binary complex during the elongation process of simian virus 40 DNA replication. J Virol. 1992;66:804–815. doi: 10.1128/jvi.66.2.804-815.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Wiekowski M, Droege P, Stahl H. Monoclonal antibodies as probes for a function of large T antigen during the elongation process of simian virus 40 DNA replication. J Virol. 1987;61:411–418. doi: 10.1128/jvi.61.2.411-418.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Wobbe C R, Weissbach L, Borowiec J A, Dean F B, Murakami Y, Bullock P, Hurwitz J. Replication of simian virus 40 origin-containing DNA in vitro with purified proteins. Proc Natl Acad Sci USA. 1987;84:1834–1838. doi: 10.1073/pnas.84.7.1834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Wold M S. Replication protein A: a heterotrimeric, single-stranded DNA-binding protein required for eukaryotic DNA metabolism. Annu Rev Biochem. 1997;66:61–92. doi: 10.1146/annurev.biochem.66.1.61. [DOI] [PubMed] [Google Scholar]
- 97.Wold M S, Kelly T J. Purification and characterization of replication protein A, a cellular protein required for in vitro replication of simian virus 40 DNA. Proc Natl Acad Sci USA. 1988;85:2523–2527. doi: 10.1073/pnas.85.8.2523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Wun-Kim K, Simmons D T. Mapping of helicase and helicase substrate-binding domains on simian virus 40 large T antigen. J Virol. 1990;64:2014–2020. doi: 10.1128/jvi.64.5.2014-2020.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Wun-Kim K, Upson R H, Young W, Simmons D T. The DNA binding domain of simian virus large T antigen has multiple functions. J Virol. 1993;67:7608–7611. doi: 10.1128/jvi.67.12.7608-7611.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Yang L, Wold M S, Li J J, Kelly T J, Liu L F. Roles of DNA topoisomerases in simian virus 40 DNA replication in vitro. Proc Natl Acad Sci USA. 1987;84:950–954. doi: 10.1073/pnas.84.4.950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Zhang D, Frappier L, Gibbs E, Hurwitz J, O’Donnell M. Human RPA (hSSB) interacts with EBNA1, the latent origin binding protein of Epstein-Barr virus. Nucleic Acids Res. 1998;26:631–637. doi: 10.1093/nar/26.2.631. [DOI] [PMC free article] [PubMed] [Google Scholar]