Abstract
Several distinct families of endogenous retrovirus-like sequences (HERVs) exist in the genomes of humans and other primates. One of these families, the HERV-K group, contains members that encode functional proteins and that have been implicated in the etiology of insulin-dependent diabetes mellitus (IDDM). Because of potential functional and disease relevance, it is important to determine if there are HERV-K-associated genetic differences between individuals. In this study, we have investigated the divergence and evolutionary age of HERV-K long terminal repeats (LTRs). Thirty-seven LTRs, taken primarily from random human clones in GenBank, were aligned and grouped into nine clusters with decreasing sequence divergence. Cluster 1 sequences are 8.6% divergent, on average, whereas cluster 9 LTRs, represented by the LTRs of the fully sequenced HERV-K10 clone, show an average of only 1.1% divergence from each other. The evolutionary age of 18 LTRs from different clusters was then investigated by genomic PCR to determine presence or absence of the retroviral element in different primate species. LTRs from clusters of higher divergence were detected in monkeys and apes, whereas LTRs in clusters with lower divergence were acquired later in evolution. Notably, LTRs of cluster 9 were found only in humans at all nine loci examined. Genomic Southern analysis with an oligonucleotide probe specific for cluster 9 LTRs suggests that HERV-K elements with this type of LTR expanded independently in the genomes of humans and the great apes. This is the first report of endogenous retroviral integrations that are specific to humans and indicates that some HERVs have amplified much later than previously thought. These elements may still be actively transposing and may therefore represent a source of genetic variation linked to disease development.
Human endogenous retroviral (HERV) elements comprise 1 to 2% of human DNA (26), but their biological relevance is largely unknown. The genomic presence of most HERVs is presumably without a major effect, but it is possible that some could be involved in normal or pathogenic processes (18, 29). Recently, the HERV-K (K denoting a lysine tRNA primer binding site) family of viral sequences has gained attention because a HERV-K env-encoded superantigen was reported to be associated with type I diabetes (7). This autoantigen was detected in insulin-dependent diabetes mellitus (IDDM) patients and raises the possibility that a genetic susceptibility for developing IDDM could be linked to variation in the expression or presence of HERV-K elements in humans. The HERV-K element group (also referred to as HML-2 elements) (21) is present in about 30 full-length copies (18) and approximately 2,000 solitary long terminal repeats (LTRs) (unpublished data). Unlike most HERV families, they have been shown to encode functional enzymatic proteins (14, 22), viral particles (6), and autoimmune antigens (7), indicating that some HERV-K elements have retained retroviral functions. Previous evolutionary analyses suggested that most HERV-K elements found in humans integrated prior to the divergence of hominoids from the Old World monkey lineages (20, 27). Indeed, no retroviral integrations specific to humans have been reported to date. Interestingly, the LTRs flanking the originally sequenced full-length element HERV-K10 are only 0.2% divergent in sequence (24). Integrated LTR sequences will acquire mutations after insertion (19), so this low degree of divergence suggests that HERV-K10 integrated relatively recently, but this possibility has not been investigated. In this study, we have examined different subgroups of HERV-K LTRs to determine their divergence and evolutionary age. Our results indicate that HERV-K elements have spread throughout primate evolution and demonstrate that a subset specifically integrated and amplified in the human lineage.
MATERIALS AND METHODS
Sequence analysis.
GenBank was screened with the HERV-K10 5′ LTR (accession no. M12851) by using BLASTN (1). Identified sequences with structures corresponding to a complete HERV-K LTR were aligned by using CLUSTALW (28). After sequence alignment optimization and sequence divergence calculations, programs NEIGHBOR and DRAWGRAM of PHYLIP (10) were used to calculate the branch length and to draw the neighbor-joining dendrogram, respectively. Statistical significance evaluation of the branching pattern was done with 100 random samplings of the input sequence alignment by using SEQBOOT of PHYLIP. Consensus sequences were derived from clusters 1, 3, 8, and 9, respectively, where a nucleotide had to be present in >60% of the sequences to be considered as a consensus position. CG dinucleotides were excluded in the consensus.
DNA samples and locus-specific typing.
Genomic DNA was prepared by using standard protocols from the same primate cell lines used previously (11). Human DNAs were derived from different healthy individuals. Primer sequences were specified from regions flanking integrated LTRs, and a total of 18 primate loci were investigated for the presence or absence of an LTR, as described previously (11, 25). The primer sequences and annealing temperatures used for the amplification of each locus are available upon request. PCR amplification conditions were as described previously (21). The first primate species having a particular integrated LTR is indicated in Table 1. The absence of an integrated LTR was determined for all 18 loci investigated.
TABLE 1.
Integration pattern of HERV-K elements
| Clustera | Primate speciesb | HERV-K elementc | Mean ± SD of divergence (%)d | Expected divergence (%)e |
|---|---|---|---|---|
| 1 | Old World monkey | AC002049 | 8.6 ± 2.2 | 7.2 |
| 3 | Old World monkey | AB000381 | 4.2 ± 0.7 | 5.0–7.2 |
| Gibbon | AF001550 | |||
| D87002 | ||||
| 5 | Gibbon | Z82901 | 3.0 | 5.0 |
| 8 | Gorilla | HERV-K18 | 2.0 ± 0.4 | 1.8 |
| L35659 | ||||
| AC000401 | ||||
| Chimpanzee | AC003023 | |||
| 9 | Human | HERV-K10 | 1.1 ± 0.2 | <1.8 |
| AC002400 | ||||
| Z84493 | ||||
| AC002508 | ||||
| U47924 | ||||
| AC002350 | ||||
| U73641a | ||||
| L47334 | ||||
| Z80898a |
Refers to the clusters shown in Fig. 1.
Primate DNA where the integrated LTR was first detected.
Only sequences in which the orthologous locus was successfully amplified in both species possessing and lacking the HERV-K element are listed.
Estimated percentage of divergence since integration was calculated as the average divergence between sequences within the cluster.
Percentage sequence difference between η-globin pseudogenes in humans and the species listed (5).
Southern hybridizations.
Primate DNAs were digested with 5 U of EcoRI per μg of DNA, separated on 0.7% Tris-acetate–EDTA gels, and transferred by alkali onto nylon membranes. Hybridization was carried out as previously described (21). A final high-stringency wash at 68°C was done in 1.25× SSPE (5× SSPE is 0.9 M NaCl, 0.05 M NaH2PO4, 5 mM Na2EDTA), 0.1% sodium dodecyl sulfate for 20 min. The oligonucleotide used (5′-CTCAGTAGATGGAGCATACAATCGGGTT-3′) was specified to detect sequences of cluster 9 only (Fig. 1). A complement of this was used in the hybridizations. The washing stringency (described above) was determined by dot hybridizations towards identical and related sequences. Only cluster 9 sequences were detected at this temperature.
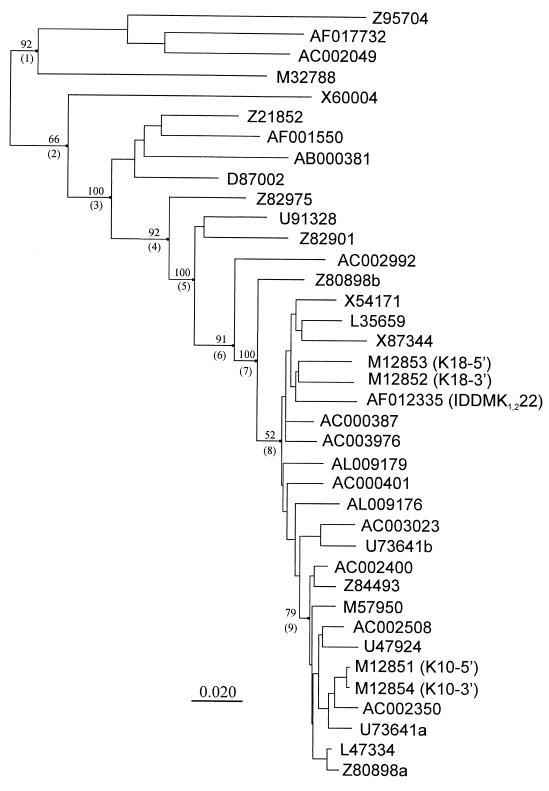
FIG. 1.
Dendrogram derived from 37 HERV-K LTR sequences. The names of each sequence refer to the GenBank accession numbers from which the HERV-K LTRs were identified. The names of previously described HERV-K elements are shown in parentheses, where 5′ and 3′ refer to the LTRs flanking the proviral element. GenBank entries containing more than one LTR sequence are indicated by an a or a b following the accession number. All LTRs used were complete sequences of approximately 970 bp, except for the IDDM-associated element, for which only 809 bp of LTR sequence is available in GenBank (accession no. AF012335). The values at the nodes indicate the percentage of the bootstrapped trees supporting the branching pattern, and the numbers in parentheses indicate the clusters referred to in the text. The length of the bar corresponds to a nucleotide sequence divergence of 2%.
RESULTS
Alignment of HERV-K LTRs.
By searching GenBank with the 968-bp 5′ LTR of the HERV-K10 element, we identified 35 additional LTRs. Two of these came from element K18, from which the LTRs and a short internal sequence have been determined (23). The remaining LTRs represented solitary units (17) and were derived from either random genomic clones or sequences described by others (see legend to Fig. 1). At the time of the search (December 1997), no other full-length HERV-K elements were found in GenBank. Solitary LTRs are the result of homologous recombination between the 5′ and 3′ LTRs of a full-length proviral element (11, 17). Therefore, their sequences and integration patterns can be treated the same way as full-length elements in an evolutionary analysis of this type. A dendrogram derived from sequence alignments of the identified LTRs is shown in Fig. 1. Based on clustering patterns supported by bootstrap analysis, nine subgroups were recognized. The branch lengths within a cluster vary substantially, with cluster 1 sequences being quite divergent and cluster 9 sequences being much more similar to each other. The LTRs of the full-length HERV-K10 element belong to the least-divergent subgroup (cluster 9). The AF012335 GenBank entry from the IDDM-associated element (7) does not correspond to a complete LTR but has typical consensus positions of cluster 8 sequences.
Integration analysis.
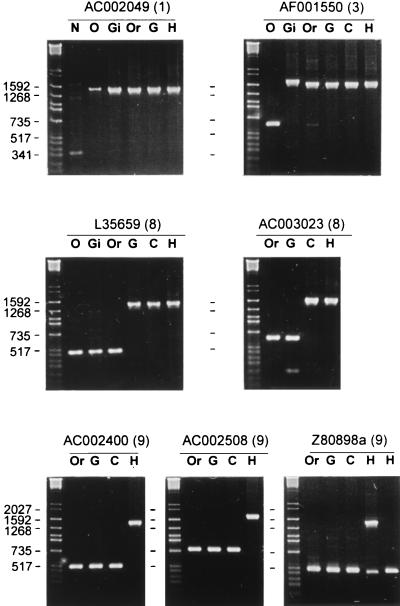
The structure of the HERV-K LTR dendrogram suggests that sequences of the different clusters represent integrations separated in time during primate evolution. To determine whether the clustering pattern and sequence divergence reflect the age of the LTRs, the approximate time of integration of specific LTRs belonging to different clusters was determined by examining selected loci in different primates. Specific primers flanking the sites of the integrated LTRs at 18 loci were used in PCRs of various primate DNAs. Because each primer set flanks the site of an integrated LTR, amplification products from DNA having an LTR at a certain locus will result in a product approximately 970 bp larger than products from DNA lacking the LTR at the corresponding site. Results, shown in Fig. 2 and Table 1, support the branching pattern. In general, LTR sequences of clusters 1 to 5 were first identified in Old World monkey and gibbon DNAs, whereas LTRs of cluster 8 first appeared in DNAs of gorilla and chimpanzee. For example, the AF001550 LTR of cluster 3 is not present in Old World monkeys but is present in gibbon and all higher primates. In contrast, the AC003023 cluster 8 LTR is found only in chimpanzee and human, indicating a more recent integration (Fig. 2). Initial results with primers flanking three of the integrated LTRs of cluster 9 resulted in the expected amplification products in human DNA but not in any of the other primate DNAs (Fig. 2). To demonstrate that sequences of cluster 9 were unique to human DNA, primers flanking the other six identified LTRs of this cluster, including the full-length HERV-K10 element, were used in the amplification of primate DNA. Indeed, all were detected only in human DNA (Table 1), indicating that sequences derived from this cluster integrated after the divergence of the human lineage from the great apes.
FIG. 2.
Examples of PCR amplification results with locus-specific primers flanking the integrated LTR of the sequences indicated. Numbers in parentheses following GenBank accession numbers refer to the sequence clusters in Fig. 1. Numbers to the left correspond to the comigrating DNA size markers of each primate panel. N, New World monkey; O, Old World monkey; Gi, gibbon; Or, orangutan; G, gorilla; C, chimpanzee; H, human. Expected amplification products were always obtained for DNA with integrated LTRs or without an integrated retroviral sequence. A slightly larger product than expected was observed in the gibbon, using primers flanking sequence AF001550.
The loci of the human-specific LTR cluster 9 were investigated by PCR amplification with DNA of 10 different individuals. All individuals were homozygous for the presence of these LTRs except for the Z80898a LTR. This LTR is located upstream of the DQB1 gene of the MHC cluster and is the same as the DQ-LTR reported before (12). Individuals (n = 25) were either heterozygous for the integrated LTR (LTR/locus only) or homozygous for the absence of the LTR in a ratio of about 1:1 (Fig. 2). Presence of the LTR does not appear to be correlated with racial differences. We also found another GenBank entry (U92032), which is about 96% identical in both the DQB1 gene and the flanking region compared to Z80898. This entry did not have the LTR integrated at the corresponding position. DQB1 alleles are highly polymorphic and originated at least 30 million years ago (3), indicating that the polymorphism of this LTR is likely due to its association with the MHC.
To gain insight into the evolutionary history of HERV-K sequences, the divergence values between LTR sequences within a cluster were determined. In a master gene element model, such as that proposed to explain the dispersion of alu sequences (9), the divergence values between sequences within a cluster should reflect the number of nucleotide substitutions that occurred since their dispersion within the primate genomes. Indeed, the observed changes for LTRs of clusters 1, 3, 8, and 9 correspond well to the expected pseudogene divergence rates (5) with respect to their first appearance in the primate lineage (Table 1). Comparison of the divergence rates between the consensus sequences derived from each of these clusters also showed expected divergence rates. Therefore, the results obtained with the sequences used here support an expansion of HERV-K elements according to a master element model (9), with different master progenitors being active at different times in evolution.
Independent amplification of cluster 9 sequences in hominids.
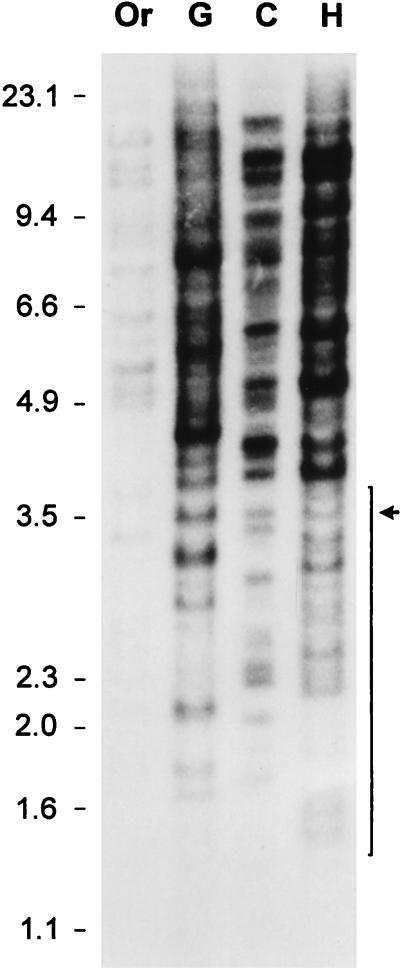
The cluster 9 sequences represent elements that have retrotransposed relatively recently. This cluster also contains one LTR (M57950) that is present in the chimpanzee but not at the corresponding locus in humans (8). Thus, the cluster 9 sequences may represent elements that have expanded independently in the different primate lineages. To investigate this possibility, an oligonucleotide probe specific for the cluster 9 sequences was used in Southern hybridization to a panel of primate DNAs. We estimate that there are at least 300 to 400 of these LTRs in human DNA, assuming that the eight sequences of cluster 9, which were identified in random genomic clones, are a representative number present in the ca. 2% of the human genome sequenced to date. As expected, many hybridizing fragments were seen in the DNAs used, even after stringent hybridization conditions (Fig. 3), making interpretations regarding common and unique hybridizing fragment sizes between the DNAs difficult. The very intense bands of sizes greater than 3.5 kb must represent multiple hybridizing fragments. This pattern suggests that some of these LTRs are part of larger DNA segments that have amplified by other mechanisms. This intense banding pattern precludes examination of bands representing single LTRs in the size range above 3.5 kb. However, several discrete hybridizing fragments in the sizes of 1.5 to 3.5 kb were detected in the DNAs analyzed and likely represent single integrants. Of the fragments in this region, only one was in common for gorilla, chimpanzee, and human DNA, whereas 11 were unique in human DNA, 8 were unique in the chimpanzee, and 6 were unique in the gorilla. These differences are higher than can be explained by restriction enzyme site differences alone, because the expected divergence between human and great ape DNAs is only 2% (5). Even though only a fraction of the total number of cluster 9 LTRs can be analyzed by conventional Southern hybridizations, it is evident that several unique fragments are present in the different primates examined, suggesting an independent amplification of cluster 9 sequences in different primate lineages.
FIG. 3.
Southern hybridization with an oligonucleotide specific for cluster 9 LTRs with a panel of primate DNAs digested with EcoRI (see legend to Fig. 2 for abbreviations). To the left are sizes of the comigrating DNA size markers (in kilobases), and to the right an arrow indicates a hybridizing DNA fragment common to the gorilla, chimpanzee, and human within the 1.5- to 3.5-kb region indicated with a bracket. Fragments larger than 3.5 kb were too numerous to analyze.
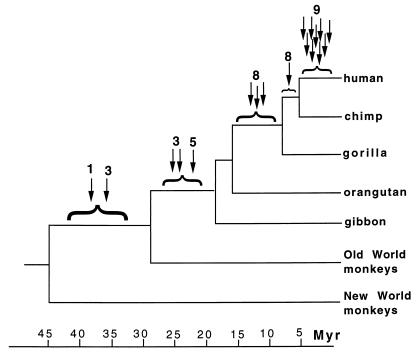
DISCUSSION
The endogenous retroviral sequences found in the human genome are a heterogeneous group of retroelements that were presumably derived from ancient germ line infections of exogenous retroviruses which became fixed in the species. In this study, we have defined distinct subgroups of HERV-K LTRs of different ages. Figure 4 illustrates the approximate time of integration of the particular LTRs investigated by locus-specific PCR analysis. These results, in combination with the cluster analysis, showed a correlation between sequence divergence and the age of the different element subgroups and supports an expansion of HERV-K elements similar to a master gene model (9). That is, the data suggest that different progenitor HERV-K elements amplified in the genome at different stages in primate evolution. The division of HERV-K sequences into subgroups has also been suggested in two recent reports (16, 30) in which other sequence sets were used, derived from either the polymerase gene or from chromosome 19 LTR sequences. However, those previous studies did not investigate the integration time of different subgroups.
FIG. 4.
Approximate integration times of HERV-K elements. Arrows indicate the lineage in which a particular LTR was first detected, and numbers refer to the cluster as identified in Fig. 1. Time estimates for divergence of the different primate lineages were taken from Bailey et al. (5).
During the course of our analysis, we identified a subset of endogenous retroviral elements that has integrated specifically in the human lineage. Interestingly, besides representing a subset with mobile capacity in humans, cluster 9 sequences were shown by Southern analysis to exist in similar numbers in the genomes of humans and the great apes. However, the different banding patterns and the fact that none of the nine specific loci containing cluster 9 elements in humans were occupied in the great apes indicate that most HERV-K cluster 9 elements have amplified independently in chimpanzee and gorilla. This study is the first demonstration of endogenous retroviral integrations specific to humans. Subgroups of another HERV family, HERV-H (formerly RTVL-H), have also been defined based on LTR structure and shown to be of different ages (11). However, the HERV-H subgroups identified to date are older than the HERV-K cluster 9 elements, since their LTRs are more divergent and no human-specific integrations have been detected.
An L1 retrotransposon sequence was earlier shown to have integrated into the factor VIII gene, although the master copy is present at the same chromosomal locus in gorillas, chimpanzees, and humans (13). This finding indicates that retroelements may retain their mobile properties despite a relatively old age. The LTR of the element linked to IDDM falls into cluster 8 (Fig. 1) and is probably of the same age as other sequences of this cluster. This cannot be investigated directly, however, because the corresponding locus has not been cloned. The LTR sequence alignments reveal that regions involved in HERV-K transcription—the hormone-responsive element, the TATA, and polyadenylation motifs (23)—are conserved in sequences belonging to both clusters 8 and 9, suggesting conserved functional properties of these LTRs. Thus, it is possible that some HERV-K elements belonging to cluster 8 have spread after the divergence of humans and the great apes, as was shown for cluster 9 sequences.
Efforts are under way to screen a larger set of human DNA samples to determine whether a copy number variation of cluster 8 or 9 HERV-K elements exists. One polymorphic human cluster 9 LTR locus associated with HLA was identified here. The presence of this LTR was originally described by Kambhu et al. (12) and was also shown to be linked to haplotypes with susceptibility for the development of IDDM (4). The presence of a functionally transposing subset of HERV-K in the human lineage strengthens the possibility that polymorphic HERV-K alleles may be associated with the development of IDDM in some individuals. It is evident that the acquisition of proviruses at novel chromosomal locations may lead to an altered expression pattern of both viral and cellular genes. However, they could also represent alleles fixed in the primate genome that at some stage become transcriptionally activated. Several studies have reported differential expression of HERV-K in leukocytes of different individuals (2, 15). Thus, it is possible that copy number polymorphism or transcriptional activation contributes to variations in gene expression which may lead to the development of diseases having a genetic basis.
ACKNOWLEDGMENTS
We thank Doug Freeman for technical support, Mats Lindeskog and Garvin Hunter for valuable discussions, and Robert Kay for critical reading of the manuscript. We also thank Christine Kelly and Melissa Hudson for help with manuscript preparation.
This work was supported by the Medical Research Council of Canada and the Crafoord Foundation, Lund, Sweden. P.M. received a postdoctoral fellowship from the Cancer Research Society of Canada and an award from the Tage Blücher Foundation, Helsingborg, Sweden.
REFERENCES
- 1.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 2.Andersson M-L, Medstrand P, Yin H, Blomberg J. Differential expression of human endogenous retroviral sequences similar to mouse mammary tumor virus in normal peripheral blood mononuclear cells. AIDS Res Hum Retroviruses. 1996;12:833–840. doi: 10.1089/aid.1996.12.833. [DOI] [PubMed] [Google Scholar]
- 3.Ayala F J, Escalante A A. The evolution of human populations: a molecular perspective. Mol Phylogenet Evol. 1996;5:188–201. doi: 10.1006/mpev.1996.0013. [DOI] [PubMed] [Google Scholar]
- 4.Badenhoop K, Tonjes R R, Rau H, Donner H, Rieker W, Braun J, Herwig J, Mytilineos J, Kurth R, Usadel K H. Endogenous retroviral long terminal repeats of the HLA-DQ region are associated with susceptibility to insulin-dependent diabetes mellitus. Hum Immunol. 1996;50:103–110. doi: 10.1016/0198-8859(96)00146-2. [DOI] [PubMed] [Google Scholar]
- 5.Bailey W J, Fitch D H, Tagle D A, Czelusniak J, Slightom J L, Goodman M. Molecular evolution of the psi-eta globin gene locus: gibbon phylogeny and the human slowdown. Mol Biol Evol. 1991;8:155–184. doi: 10.1093/oxfordjournals.molbev.a040641. [DOI] [PubMed] [Google Scholar]
- 6.Boller K, Konig H, Sauter M, Mueller-Lantzsch N, Lower R, Lower J, Kurth R. Evidence that HERV-K is the endogenous retrovirus sequence that codes for the human teratocarcinoma-derived retrovirus HTDV. Virology. 1993;196:349–353. doi: 10.1006/viro.1993.1487. [DOI] [PubMed] [Google Scholar]
- 7.Conrad B, Weissmahr R N, Boni J, Arcari R, Schupbach J, Mach B. A human endogenous retroviral superantigen as candidate autoimmune gene in type I diabetes. Cell. 1997;90:303–313. doi: 10.1016/s0092-8674(00)80338-4. [DOI] [PubMed] [Google Scholar]
- 8.Craig L C, Pirtle I L, Gracy R W, Pirtle R M. Characterization of the transcription unit and two processed pseudogenes of chimpanzee triosephosphate isomerase (TPI) Gene. 1991;99:217–227. doi: 10.1016/0378-1119(91)90130-4. [DOI] [PubMed] [Google Scholar]
- 9.Deininger P L, Batzer M A, Hutchison C A I, Edgell M H. Master genes in mammalian repetitive DNA amplification. Trends Genet. 1992;8:307–311. doi: 10.1016/0168-9525(92)90262-3. [DOI] [PubMed] [Google Scholar]
- 10.Felsenstein J. PHYLIP version 3.5c. Seattle: Department of Genetics, University of Washington; 1995. [Google Scholar]
- 11.Goodchild N L, Wilkinson D A, Mager D L. Recent evolutionary expansion of a subfamily of RTVL-H human endogenous retrovirus-like elements. Virology. 1993;196:778–788. doi: 10.1006/viro.1993.1535. [DOI] [PubMed] [Google Scholar]
- 12.Kambhu S, Falldorf P, Lee J S. Endogenous retroviral long terminal repeats within the HLA-DQ locus. Proc Natl Acad Sci USA. 1990;87:4927–4931. doi: 10.1073/pnas.87.13.4927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kazazian H H, Jr, Wong C, Youssoufian H, Scott A F, Phillips D G, Antonarakis S E. Hemophilia A resulting from de novo insertion of L1 sequences represents a novel mechanism for mutation in man. Nature. 1988;332:164–166. doi: 10.1038/332164a0. [DOI] [PubMed] [Google Scholar]
- 14.Kitamura Y, Ayukawa T, Ishikawa T, Kanda T, Yoshiike K. Human endogenous retrovirus K10 encodes a functional integrase. J Virol. 1996;70:3302–3306. doi: 10.1128/jvi.70.5.3302-3306.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Krieg A M, Gourley M F, Klinman D M, Perl A, Steinberg A D. Heterogeneous expression and coordinate regulation of endogenous retroviral sequences in human peripheral blood mononuclear cells. AIDS Res Hum Retroviruses. 1992;8:1991–1998. doi: 10.1089/aid.1992.8.1991. [DOI] [PubMed] [Google Scholar]
- 16.Lavrentieva I, Khil P, Vinogradova T, Akhmedov A, Lapuk A, Shakhova O, Lebedev Y, Monastyrskaya G, Sverdlov E D. Subfamilies and nearest-neighbour dendrogram for the LTRs of human endogenous retrovirus HERV-K mapped on human chromosome 19: physical neighbourhood does not correlate with identity level. Hum Genet. 1998;102:107–116. doi: 10.1007/s004390050662. [DOI] [PubMed] [Google Scholar]
- 17.Leib-Mösch C, Haltmeier M, Werner T, Geigl E M, Brack-Werner R, Francke U, Erfle V, Hehlmann R. Genome distribution and transcription of solitary HERV-K LTRs. Genomics. 1993;18:261–269. doi: 10.1006/geno.1993.1464. [DOI] [PubMed] [Google Scholar]
- 18.Löwer R, Löwer J, Kurth R. The viruses in all of us: characteristics and biological significance of human endogenous retrovirus sequences. Proc Natl Acad Sci USA. 1996;93:5177–5184. doi: 10.1073/pnas.93.11.5177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mager D L, Freeman J D. HERV-H endogenous retroviruses: presence in the New World branch but amplification in the Old World primate lineage. Virology. 1995;213:395–404. doi: 10.1006/viro.1995.0012. [DOI] [PubMed] [Google Scholar]
- 20.Mariani-Costantini R, Horn T M, Callahan R. Ancestry of a human endogenous retrovirus family. J Virol. 1989;63:4982–4985. doi: 10.1128/jvi.63.11.4982-4985.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Medstrand P, Blomberg J. Characterization of novel reverse transcriptase encoding human endogenous retroviral sequences similar to type A and type B retroviruses: differential transcription in normal human tissues. J Virol. 1993;67:6778–6787. doi: 10.1128/jvi.67.11.6778-6787.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mueller-Lantzsch N, Sauter M, Weiskircher A, Kramer K, Best B, Buck M, Grasser F. Human endogenous retroviral element K10 (HERV-K10) encodes a full-length gag homologous 73-kDa protein and a functional protease. AIDS Res Hum Retroviruses. 1993;9:343–350. doi: 10.1089/aid.1993.9.343. [DOI] [PubMed] [Google Scholar]
- 23.Ono M. Molecular cloning and long terminal repeat sequences of human endogenous retrovirus genes related to types A and B retrovirus genes. J Virol. 1986;58:937–944. doi: 10.1128/jvi.58.3.937-944.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ono M, Yasunaga T, Miyata T, Ushikubo H. Nucleotide sequence of human endogenous retrovirus genome related to the mouse mammary tumor virus genome. J Virol. 1986;60:589–598. doi: 10.1128/jvi.60.2.589-598.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shih A, Coutavas E E, Rush M G. Evolutionary implications of primate endogenous retroviruses. Virology. 1991;182:495–502. doi: 10.1016/0042-6822(91)90590-8. [DOI] [PubMed] [Google Scholar]
- 26.Smit A F A. The origin of interspersed repeats in the human genome. Curr Opin Genet Dev. 1996;6:743–748. doi: 10.1016/s0959-437x(96)80030-x. [DOI] [PubMed] [Google Scholar]
- 27.Steinhuber S, Brack M, Hunsmann G, Schwelberger H, Dierich M P, Vogetseder W. Distribution of human endogenous retrovirus HERV-K genomes in humans and different primates. Hum Genet. 1995;96:188–192. doi: 10.1007/BF00207377. [DOI] [PubMed] [Google Scholar]
- 28.Thompson J D, Higgins D G, Gibson T J. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, positions-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wilkinson D A, Mager D L, Leong J C. Endogenous human retroviruses. In: Levy J, editor. The Retroviridae. Vol. 3. New York, N.Y: Plenum Press; 1994. pp. 465–535. [Google Scholar]
- 30.Zsiros J, Jebbink M F, Lukashov V V, Voute P A, Berkhout B. Evolutionary relationships within a subgroup of HERV-K-related human endogenous retroviruses. J Gen Virol. 1998;79:61–70. doi: 10.1099/0022-1317-79-1-61. [DOI] [PubMed] [Google Scholar]